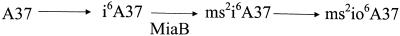Abstract
As components involved in Fe-S cluster metabolism are described, the challenge becomes defining the integrated process that occurs in vivo based on the individual functions characterized in vitro. Strains lacking yggX have been used here to mimic chronic oxidative stress and uncover subtle defects in Fe-S cluster metabolism. We describe the in vivo similarities and differences between isc mutants, which have a known function in cluster assembly, and mutants disrupted in four additional loci, gshA, apbC, apbE, and rseC. The latter mutants share similarities with isc mutants: (i) a sensitivity to oxidative stress, (ii) a thiamine auxotrophy in the absence of the YggX protein, and (iii) decreased activities of Fe-S proteins, including aconitase, succinate dehydrogenase, and MiaB. However, they differ from isc mutants by displaying a phenotypic dependence on metals and a distinct defect in the SoxRS response to superoxides. Results presented herein support the proposed role of YggX in iron trafficking and protection against oxidative stress, describe additional phenotypes of isc mutants, and suggest a working model in which the ApbC, ApbE, and RseC proteins and glutathione participate in Fe-S cluster repair.
Studies in bacteria have uncovered a number of loci involved in iron-sulfur (Fe-S) cluster metabolism in addition to the well-characterized isc and suf operons. The suf operon is proposed to be a system for Fe-S cluster assembly that functions under conditions of iron limitation and oxidative stress (22, 28, 45). In Salmonella enterica, mutations in gshA (14), apbC, apbE (41), rseC (E. Skovran, unpublished data), and yggX (13) result in impaired Fe-S cluster metabolism. While most of the Isc proteins and several of the Suf proteins have clearly defined roles in Fe-S cluster assembly (10), the functions of the remaining loci that affect Fe-S cluster metabolism are not well understood.
The gshA gene encodes γ-l-glutamyl-l-cysteine synthetase, which functions in the first step of glutathione synthesis (1) and has been implicated in the assembly and repair of Fe-S clusters (11). While the specific biochemical functions of ApbC, ApbE, and RseC are unknown, several reports have suggested these proteins and/or their homologs are involved in metal cluster metabolism (16, 35, 37, 38). ApbC, also called Mrp in Escherichia coli, is a member of the MinD subfamily of proteins and has ATPase activity anticipated by sequence analysis (41). A homolog of apbC in Saccharomyces cerevisiae, cfd1, was identified by a mutant phenotype that implicated CFD1 in cytosolic Fe-S cluster assembly (36). In plants, mutants lacking the ApbC homolog (FSC) displayed phenotypes that were interpreted to mean this protein was involved specifically in synthesis of 4Fe-4S clusters (24). ApbE is a periplasmic lipoprotein (3) that is similar to a gene in the rnf operon involved in nitrogen fixation in Rhodobacter capsulatus (38). In S. enterica, rseC is the fourth gene in the rpoE operon, although no connection between rseC and rpoE regulation has been found. In E. coli, mutations in this gene were isolated because they caused constitutive expression of the SoxRS regulon (18), leading to the hypothesis that RseC is involved in the reduction of the Fe-S clusters in SoxR after recovery from oxidative stress (18).
Along with mutations in the isc operon, mutations in the four loci described above cause a conditional thiamine auxotrophy in S. enterica attributed to impaired assembly or repair of the Fe-S cluster in the ThiH enzyme (14, 25, 42). In addition to this thiamine requirement, one of the defining features of this class of mutants was the observation that the expression of yggX suppressed the thiamine auxotrophy of each (13). YggX is a 91-amino-acid protein that provides resistance to oxidative stress and is proposed to have a role in iron trafficking (13, 15). It was recently shown that yggX is a part of the soxRS regulon, members of which are up-regulated in response to superoxide stress (34). In E. coli K-12 and the S. enterica serovar Typhimurium LT series of strains (LT1 to -22), YggX accumulates to a high level (∼10,000 copies/cell) (15). The single exception seems to be the LT2 strain, which has been propagated as a laboratory wild-type strain and fails to express detectable YggX, though the intact coding sequence is present (13).
This study was initiated to gain a better understanding of the multiple components participating in Fe-S cluster metabolism in vivo, specifically taking advantage of yggX expression to manipulate the state of oxidative stress in the cell. The results presented further define this mutant class and identify a number of differences between isc mutants and the other members. Taken together, the phenotypic similarities and differences are consistent with a working model in which the status of ApbC, ApbE, RseC, and glutathione in the cell impacts the repair of Fe-S clusters, while the Isc proteins function in their synthesis.
MATERIALS AND METHODS
Bacterial strains, media, and chemicals.
All strains used in this study were derived from S. enterica LT2 (lab stock DM1) and are listed with their respective genotypes in Table 1. The NCE medium of Berkowitz et al. (4) supplemented with 1 mM MgSO4 was used as a minimal medium with 11 mM glucose or 10 mM gluconate added as a sole carbon source. Trace minerals (2) (including or lacking iron) were added to minimal media as indicated in the text. When present in the culture medium, these compounds were used at the following final concentrations: nicotinic acid, 20 μM; thiamine, 100 nM; CoCl2, 10 μM; FeCl3, 20 μM or as indicated in the text. Luria broth and Difco nutrient broth (NB) (8 g/liter) with NaCl (5 g/liter) were used as rich media with Difco BiTek agar added to a final concentration of 1.5% for solid media. The final concentrations of antibiotics were as follows: gentamicin (Gm), 6 μg/ml; tetracycline (Tc), 20 μg/ml; kanamycin (Kn), 50 μg/ml; ampicillin (Ap), 30 μg/ml; chloramphenicol (Cm), 20 μg/ml. All chemicals were purchased from Sigma-Aldrich (St. Louis, Mo.).
TABLE 1.
Strains
| Strain | Genotype |
|---|---|
| DM1 | Wild-type LT2 strain |
| PP103 | sin3::MudJa,c |
| DM5015 | zxx::8077 Tn10db (Cm) gshA102::MudJ |
| DM5647 | yggX::Gm |
| DM5986 | yggX::Gm apbC55::Tn10d(Tc) |
| DM5988 | yggX::Gm apbE64::MudJ |
| DM5990 | yggX::Gm gshA102::MudJ |
| DM5992 | yggX::Gm isc1::MudJ |
| DM5994 | yggX::Gm iscA2::MudJ |
| DM6692 | yggX::Gm apbE42::Tn10d (Tc) |
| DM7149 | yggX::Gm rseC::Kn |
| DM7616 | yggX::Gm isc5::Cm (replacement of iscS through fdx with Cm cassette) |
| DM7218 | zxx::8077 Tn10d (Cm) yggX::Gm isc1::MudJ |
| DM7220 | zxx::8077 Tn10d (Cm) isc1::MudJ |
| DM7221 | zxx::8077 Tn10d (Cm) yggX::Gm iscA2::MudJ |
| DM7223 | zxx::8077 Tn10d (Cm) iscA2::MudJ |
| DM7225 | zxx::8077 Tn10d (Cm) yggX::Gm |
| DM7226 | zxx::8077 Tn10d (Cm) |
| DM7306 | zxx::8077 Tn10d (Cm) yggX::Gm apbC55::Tn10d (Tc) |
| DM7307 | zxx::8077 Tn10d (Cm) apbC55::Tn10d (Tc) |
| DM7308 | zxx::8077 Tn10d (Cm) yggX::Gm apbE64::MudJ |
| DM7309 | zxx::8077 Tn10d (Cm) apbE64::MudJ |
| DM7310 | zxx::8077 Tn10d (Cm) yggX::Gm rseC::Kn |
| DM7311 | zxx::8077 Tn10d (Cm) rseC::Kn |
| DM7312 | zxx::8077 Tn10d (Cm) yggX::Gm gshA102::MudJ |
| DM7313 | zxx::8077 Tn10d (Cm) gshA102::MudJ |
| DM7433 | yggX::Gm rseC3Δ |
| DM7618 | yggX::Gm isc5Δ (deletion of iscS through fdx) |
| DM7622 | zxx::8077 Tn10d (Cm) yggX::Gm sin3::MudJ |
| DM7623 | zxx::8077 Tn10d (Cm) sin3::MudJ |
| DM7624 | yggX::Gm apbE42::Tn10d (Tc) sin3::MudJ |
| DM7626 | zxx::8077 Tn10d (Cm) yggX::Gm apbE42::Tn10d (Tc) sin3::MudJ |
| DM7627 | zxx::8077 Tn10d (Cm) apbE42::Tn10d (Tc) sin3::MudJ |
| DM7628 | yggX::Gm gshA101::Tn10d (Tc) sin3::MudJ |
| DM7630 | yggX::Gm apbC55::Tn10d (Tc) sin3::MudJ |
| DM7632 | yggX::Gm rseC3 sin3::MudJ |
| DM7633 | zxx::8077 Tn10d (Cm) yggX::Gm rseC3Δ sin3::MudJ |
| DM7634 | zxx::8077 Tn10d (Cm) rseC3Δ sin3::MudJ |
| DM7635 | yggX::Gm apbC55::Tn10d (Tc) sin3::MudJ pET20b (Ap) |
| DM7636 | yggX::Gm apbC55::Tn10d (Tc) sin3::MudJ pCTH-ApbCd |
| DM7814 | zxx::8077 Tn10d (Cm) yggX::Gm isc5Δ sin3::MudJ |
| DM7815 | zxx::8077 Tn10d (Cm) isc5Δ sin3::MudJ |
| DM7819 | zxx::8077 Tn10d (Cm) yggX::Gm apbC55::Tn10d (Tc) sin3::MudJ |
| DM7820 | zxx::8077 Tn10d (Cm) apbC55::Tn10d (Tc) sin3::MudJ |
| DM7822 | zxx::8077 Tn10d (Cm) yggX::Gm gshA101::Tn10d (Tc) sin3::MudJ |
| DM7823 | zxx::8077 Tn10d (Cm) gshA101::Tn10d (Tc) sin3::MudJ |
MudJ is used throughout the text to refer to the MudI1734 transposon (6).
Tn10d refers to the transposition-defective mini-Tn10 (Tn10Δ-16 Δ-17) (44), with the relevant drug resistance designated in parentheses.
sin3 (sox inducible) designates a MudJ insertion in the fpr gene and was provided by P.J. Pomposiello (32).
Plasmid pCTH-ApbC and the parent vector pET20b are described in reference 41.
Strain construction.
For clarity, the yggX* nomenclature of past publications has been dropped (13, 15). Instead, we use the standard genetic nomenclature, where yggX indicates no detectable gene product and the wild-type state indicates expression of the gene. To generate one set of strains, a yggX::Gm cassette was introduced into the wild type (DM1) followed by transduction of insertions in isc, apbC, apbE, rseC, or gshA. Two previously characterized insertions in isc were used (42). The isc1 insertion is located between the iscR and iscS genes and is polar on the downstream genes in the operon. The iscA2 insertion is located in the iscA gene and is polar onto the downstream genes. The resulting strains were used as recipients in transductions with DM5015 [zxx-8077::Tn10d(Cm) yggX+] as a donor. Cmr transductants were screened for Gms, and an isogenic pair (yggX::Gm yggX+) was saved in each case. The presence of the yggX+ allele was confirmed by immunoblot analysis (15).
A similar strategy was used to construct mutant strains with the fpr::MudJ insertion. A Tn10d insertion conferring tetracycline resistance in the apbC, apbE, and gshA genes was transduced into a starting strain that contained the yggX::Gm cassette. For the rseC and isc strains, deletion mutants were constructed by the method of Datsenko and Wanner (8), through a Kn cassette intermediate replacing the rseC gene and a Cm cassette replacing the entire isc operon. Subsequently, the cassettes were deleted using FLP recombinase carried on plasmid pCP20 as described elsewhere (7). Finally, the fpr::MudJ insertion was transduced into the above five strains and the wild-type strain. The yggX::Gm/yggX+ isogenic pairs were constructed as described above.
Phenotypic analysis. (i) Growth in rich media.
For assessment of growth in rich media, overnight cultures grown at 37°C in NB were subcultured, with 150 μl added into 5 ml of NB, resulting in an initial optical density at 650 nm (OD650) of 0.03 to 0.06. Culture tubes were placed in an air shaker at 37°C, and growth was monitored by the OD650 using a Bausch & Lomb Spectronic 20. Doubling times were determined as previously described (30).
(ii) PQ sensitivity.
For paraquat (PQ) sensitivity analysis, cultures were prepared and growth was monitored as described above. Strains were subcultured into 5 ml of NB containing 0 to 100 μM PQ as indicated below. The final OD650 reported reflected the density reached after 24 h, since the absorbance for all strains had reached a plateau by that time.
(iii) Nutritional requirements and metal sensitivity.
Nutritional requirements were assessed by growth in liquid or on solid media. To test for growth using solid media, strains were patched onto nutrient agar plates and incubated at 37°C for 4 h. Plates were then replica printed to NCE agar plates containing carbon sources and nutritional supplements as indicated. For analysis of nutritional requirements in liquid media, cells were grown overnight at 37°C in NB diluted 1:1 with double-distilled H2O in new disposable culture tubes (14-961-32; Fisher) to minimize iron content. Cells were harvested and resuspended in an equal volume of NCE medium and subsequently inoculated (200 μl of resuspended culture) into 5 ml of the appropriate medium. Culture tubes were placed in an air shaker at 37°C, and growth was monitored as described above. Doubling times were determined as previously described (30). In growth curves, the starting OD650 was routinely between 0.02 and 0.06. When final cell density is reported, it reflects the OD650 after 24 h of growth. MgSO4 (1 mM), iron, and cobalt were added as indicated.
Enzyme assays. (i) Aconitase assays.
Aconitase activity was determined as previously described (42) with the following exceptions. For each experiment, cell extracts from six independent cultures were prepared from cells grown in minimal gluconate-thiamine nicotinic acid medium that had been passed over Chelex 100 resin. MgSO4 (1 mM) and trace minerals (2) lacking iron were subsequently added. Cells were sonicated for a total of 5 s, with 0.5-s pauses between bursts.
(ii) Succinate dehydrogenase assays.
For succinate dehydrogenase assays, cells were grown, resuspended, and sonicated as described above for aconitase assays. Assays were performed as described previously (42).
(iii) MiaB-catalyzed ms2 modification levels in vivo.
Cells from duplicate 100-ml cultures of each mutant strain were pelleted and resuspended in 2 ml of 10 mM Tris (pH 7.5) and 5 mM MgCl2. An equal volume of buffered phenol was added, the mixture was vortexed for 1 min, and the layers were separated by centrifugation at 10,000 × g for 30 min. The top aqueous layer was transferred to a new tube, and nucleic acids were precipitated by the addition of 0.1 volume of 3 M NH4O-acetate (NH4OAc), pH 5.3, and 2.5 volumes of cold ethanol. The crude tRNA pellet was dissolved in 0.5 ml of water and 0.5 ml of 8 M urea, 0.05% bromophenol blue was added, and the tRNA was purified by polyacrylamide gel electrophoresis on an 8% denaturing gel. The band corresponding to tRNA was excised, and tRNA was eluted in 0.5 M NaCl and then precipitated and dissolved in water and stored at −20°C. RNA concentration was measured by absorbance at 260 nm (1 OD unit = 40 μg/ml). For high-performance liquid chromatography analysis, 50 μg of tRNA was first digested for 16 h at 37°C with 10 μg of nuclease P1 in a 100-μl reaction mixture containing 30 mM NaOAc (pH 5.3) and 0.2 mM Zn(OAc)2. To this reaction mixture was added 10 μl of 1 M Tris-HCl, pH 8, and 2 U of bacterial alkaline phosphatase (Sigma). After >3 h at 37°C, 50 μl of this mixture was loaded onto a Supelco LC-18 column and eluted with a 30-min linear gradient (1.5 ml/min) of 10 to 35% acetonitrile in 5 mM ammonium phosphate, pH 5.3. Levels of i6A, ms2i6A, and ms2io6A were measured by peak area at 260 nm and reported as fractions of the total to compensate for minor variations in the amount of tRNA loaded onto the column. Internal comparisons of the total area of these three peaks with those of other modified bases (t6A, m2A) showed no significant variation in the sum of i6A-derived modification levels for the different strains.
(iv) β-Galactosidase assays.
Overnight cultures grown at 37°C in NB were subcultured, 150 μl into 5 ml of NB. Culture tubes were placed in an air shaker at 37°C, and growth was monitored by the OD650 with a Bausch & Lomb Spectronic 20. At an OD650 of ∼0.3, the cultures were divided and induced (or not) with 250 μM PQ for 1 h with shaking. Cells were harvested and resuspended in NaCl and assayed for β-galactosidase activity according to the method of Miller (26).
RESULTS
Lack of yggX increases sensitivity to oxidative stress in mutants compromised for Fe-S cluster metabolism.
A set of isogenic strains was constructed to address the effect of YggX expression on various phenotypes. Fourteen strains were generated; each pair contained a strain lacking yggX and one expressing yggX in a background that was either wild type or mutant for isc, apbC, apbE, gshA, or rseC (Table 1).
When grown aerobically in liquid rich medium, the two isc mutants had a significant growth defect in the absence of yggX, as judged by the ∼2.5-fold increase in doubling time (Table 2). The presence of yggX (wild type) significantly decreased the doubling time of both isc mutants but failed to completely restore a wild-type rate.
TABLE 2.
Suppression of growth defects by yggX expression
| Relevant genotype | Doubling time (h) in NB | Final OD650 in NB medium with paraquat at:
|
|||
|---|---|---|---|---|---|
| 0 μM | 10 μM | 20 μM | 100 μM | ||
| yggX::Gm | 0.98 ± 0.02 | 1.16 ± 0.01 | 1.14 ± 0.01 | 0.57 ± 0.01 | 0.52 ± 0.01 |
| WTc | 0.96 ± 0.02 | 1.15 ± 0.01 | 1.14 ± 0.01 | 1.14 ± 0.01 | 0.60 ± 0.03 |
| yggX::Gm isc1::MudJa | 2.71 ± 0.04 | 1.00 ± 0.01 | 0.52 ± 0.01 | 0.39 ± 0.02 | 0.33 ± 0.01 |
| isc1::MudJ | 1.58 ± 0.02 | 1.16 ± 0.01 | 1.11 ± 0.01 | 1.03 ± 0.00 | 0.48 ± 0.01 |
| yggX::Gm iscA2::MudJb | 2.31 ± 0.10 | 1.01 ± 0.01 | 0.46 ± 0.01 | 0.40 ± 0.04 | 0.36 ± 0.01 |
| iscA2::MudJ | 1.44 ± 0.05 | 1.10 ± 0.01 | 1.02 ± 0.01 | 1.03 ± 0.02 | 0.45 ± 0.01 |
| yggX::Gm gshA102::MudJ | 1.10 ± 0.06 | 1.18 ± 0.01 | 0.49 ± 0.03 | 0.51 ± 0.00 | 0.50 ± 0.02 |
| gshA102::MudJ | 0.95 ± 0.03 | 1.17 ± 0.01 | 1.13 ± 0.01 | 1.14 ± 0.01 | 0.70 ± 0.06 |
| yggX::Gm apbC55::Tn10d | 1.03 ± 0.03 | 1.16 ± 0.01 | 0.54 ± 0.02 | 0.52 ± 0.03 | 0.51 ± 0.02 |
| apbC55::Tn10d | 0.99 ± 0.00 | 1.15 ± 0.01 | 1.15 ± 0.01 | 0.85 ± 0.01 | 0.53 ± 0.01 |
| yggX::Gm apbE64::MudJ | 1.07 ± 0.03 | 1.17 ± 0.01 | 0.66 ± 0.03 | 0.56 ± 0.01 | 0.52 ± 0.02 |
| apbE64::MudJ | 1.00 ± 0.01 | 1.15 ± 0.01 | 1.15 ± 0.01 | 1.15 ± 0.01 | 0.56 ± 0.01 |
| yggX::Gm rseC::Kn | 0.98 ± 0.01 | 1.17 ± 0.02 | 0.62 ± 0.08 | 0.53 ± 0.01 | 0.54 ± 0.01 |
| rseC::Kn | 0.98 ± 0.01 | 1.14 ± 0.01 | 1.13 ± 0.01 | 1.13 ± 0.01 | 0.55 ± 0.01 |
isc1 contains a polar insertion between iscR and iscS in the isc operon.
iscA2 contains a polar insertion in iscA.
WT indicates the allele of yggX that results in accumulation of ∼10,000 copies of YggX/cell (15).
Lack of yggX increased the sensitivity of all strains to oxidative stress, as judged by their growth in the presence of the hydroxyl radical-generating compound PQ. In these experiments final cell yield (OD650) was measured, and the data are shown in Table 2. In an otherwise-wild-type genetic background, a yggX mutant showed reduced growth in 20 μM but not 10 μM PQ; however, the strain expressing yggX was resistant to the higher concentration (Table 2). In the absence of yggX, null mutations in the isc, gshA, apbC, apbE, or rseC genes prevented strains from growing to full density in 10 μM PQ. The expression of yggX allowed full growth in media containing either 10 or 20 μM PQ. Only the apbC mutant maintained a slight sensitivity to 20 μM in the presence of YggX (OD650, 0.85 versus 1.15). Growth of all 14 strains was compromised in the presence of 100 μM PQ, and at this concentration expression of yggX had little effect.
Expression of a Sox reporter is altered in strains defective in Fe-S cluster metabolism.
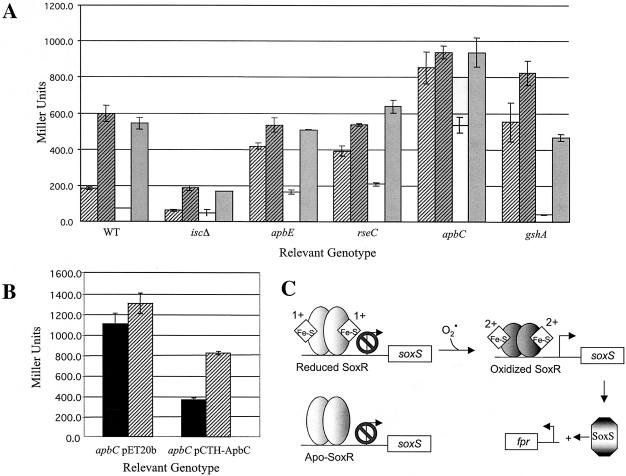
Because these mutant strains were sensitive to PQ, their ability to mount a SoxRS defense response was investigated. The SoxR-mediated response to oxidative stress involves oxidation of a resident 2Fe-2S cluster, allowing activation of gene expression, schematically represented in Fig. 1C (for a review, see reference 33). A MudJ::lac fusion in fpr [ferredoxin (flavodoxin)-NADP(H) reductase] was used as a reporter of SoxR activity (32). Strains mutant for isc, gshA, apbC, apbE, or rseC and containing the Sox reporter were constructed lacking or expressing yggX as described in Materials and Methods. Following the outgrowth of each strain with or without exposure to PQ (inducing level, 250 μM), β-galactosidase activity was assayed. The results are shown in Fig. 1A. The strains fell into three general classes based on their responses to both PQ and the expression of yggX. In the presence of yggX, PQ induced expression from fpr as expected (32). In the yggX mutant strain, expression of fpr was increased twofold in the absence of PQ, confirming that yggX mutants continuously perceive oxidative stress. Consistent with impaired Fe-S cluster assembly, the isc mutant strains (with or without yggX) maintained a regulatory response but failed to reach a significant level of fpr transcription upon induction with PQ. The response in the remaining strains, while differing quantitatively, had similar features. When lacking yggX, mutants of apbC, apbE, rseC, or gshA had high levels of fpr expression, even in the absence of applied stress (i.e., PQ). Expression of yggX restored regulation (i.e., induction) in response to PQ, but only in the gshA mutant was the noninduced level of expression completely repressed. Consistent with these data, an rseC mutant of E. coli has been reported to be constitutive for expression of the Sox regulon (18). With the exception of isc, the full induction of fpr expression in response to PQ suggests that the 2Fe-2S clusters in SoxR are present at wild-type levels and that the sensitivity to PQ seen in these mutants is not due to an inability to mount an effective SoxRS response.
FIG. 1.
SoxRS response as determined by expression of the fpr lacZ fusion. (A) Strains lacking the indicated locus were grown aerobically in NB to an OD650 of 0.3, divided, and induced (gray) or not (white) with 250 μM PQ. Cultures were shaken aerobically for 1 h, and β-galactosidase activity was measured. Data for strains expressing (solid bars) and lacking (striped bars) yggX are shown. (B) Strains were grown in the presence of ampicillin and assayed as described above. Strains lacked yggX and contained either the vector pET20b or apbC cloned into pET20b, as indicated. Solid bars depict expression without induction, and striped bars depict expression with induction by 250 μM PQ. (C) Schematic of the elements required for fpr expression. Neither apo-SoxR nor SoxR with a reduced cluster allows transcription of the fpr gene.
Since mutants lacking apbC displayed the largest deviation from the wild-type response, a plasmid expressing apbC was introduced into the yggX fpr apbC mutant strain and expression of fpr was monitored (Fig. 1B). When apbC was expressed, fpr expression was repressed in the absence of PQ, indicating that the apbC lesion was responsible for the constitutive expression in the double mutant. Although it has been shown that lack of fpr does not affect the Sox-dependent response (19), it remains formally possible that the regulatory changes observed are due to the combination of an fpr mutation and the additional lesion under study.
Lack of yggX can alter the biochemical properties of strains compromised in Fe-S cluster metabolism.
In the absence of yggX, mutants lacking the isc, apbC, apbE, or gshA loci have reduced activities of Fe-S cluster-containing enzymes, specifically succinate dehydrogenase and aconitase (41, 42). Mutants lacking rseC showed a similar defect in both succinate dehydrogenase and aconitase activity (71.0 and 68.2% of wild-type specific activities, respectively). Expression of yggX increased succinate dehydrogenase activity (from 32.2% of wild-type specific activity to 77.8%) and restored wild-type levels of aconitase activity to a gshA mutant (13), but it did not reproducibly alter activity in other strains (data not shown).
MiaB activity.
Modified bases in the tRNA were measured and used to quantify the relative activity of the Fe-S cluster-containing enzyme MiaB (31). The miaB gene is required to convert i6A to the 6-N-dimethylallyl-2-methylthioadenosine modification (ms2i6A37) of certain tRNAs (9) and encodes a member of the radical SAM family of proteins (43). Mutants lacking MiaB (9) or isc (21, 23) produce (and accumulate) i6A instead of the fully modified base. In S. enterica, an additional oxidation occurs, generating ms2io6A as the end product of this pathway (29). Figure 2 schematically represents the relevant pathway, and Table 3 shows the MiaB activities in each of the 14 strains described above as determined by the relative ratio of i6A to the sum of the i6A-derived species (i6A + ms2i6A37 + ms2i6oA37). Several points were noted. As previously reported, isc mutants had low levels of MiaB activity. Second, a yggX mutant had less MiaB activity (i.e., higher relative amount of i6A) than the isogenic yggX-expressing strain, consistent with more damage occurring to the labile cluster. Although strains lacking apbC were no different than the parental strain when assayed under these conditions, the remaining three mutant strains lacking yggX had severely reduced MiaB activities. The expression of yggX increased activity in all strains except the isc mutants, though full activity was restored only in the gshA mutant.
FIG. 2.
Schematic representation of the position of MiaB in the biosynthetic pathway of 6-N-dimethylallyl-2-methyl-thioadenosine.
TABLE 3.
2-Methylthio (ms2) modification levels in tRNA isolated from mutant strainsa
| Mutant locus |
yggX deficient
|
yggX+
|
||
|---|---|---|---|---|
| i6A | ms2i6A + ms2io6A | i6A | ms2i6A + ms2io6A | |
| WT | 0.14 ± 0.03 | 0.86 ± 0.04 | <0.05 | >0.95 |
| isc1 | >0.95 | <0.05 | >0.95 | <0.05 |
| iscA | >0.95 | <0.05 | >0.95 | <0.05 |
| apbC | 0.17 ± 0.03 | 0.83 ± 0.03 | <0.05 | >0.95 |
| apbE | 0.69 ± 0.06 | 0.31 ± 0.06 | 0.45 ± 0.03 | 0.55 ± 0.03 |
| rseC | 0.59 ± 0.01 | 0.41 ± 0.01 | 0.33 ± 0.04 | 0.67 ± 0.04 |
| gshA | 0.69 ± 0.02 | 0.31 ± 0.02 | <0.05 | >0.95 |
Values were derived from high-performance liquid chromatography peak areas and are represented as the fraction of total i6 A-derived species (i6A + ms2i6A + ms2io6A). Errors represent deviations for duplicate experiments. The position of MiaB in the biosynthetic pathway is shown in Fig. 2.
Nutritional requirements are altered by lack of yggX.
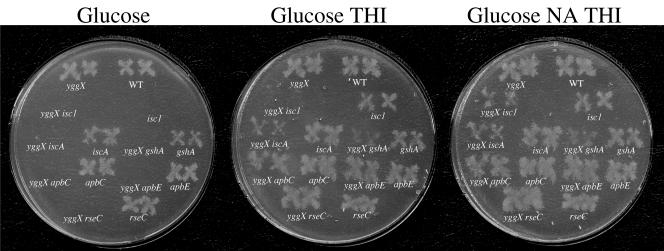
Mutants defective in Fe-S cluster metabolism have a reported thiamine requirement likely due to impaired synthesis or repair of the Fe-S cluster in ThiH (20, 39, 41, 42). Under defined conditions, in a strain background lacking yggX, each of the six mutations described above caused a requirement for thiamine on solid medium. The isc1 insertion is polar onto iscS, generating a mutant that also requires nicotinic acid (42) due to impaired synthesis of the oxygen-labile Fe-S cluster in NadA (12). The 14 isogenic strains were plated on solid medium with various supplements, and results are shown in Fig. 3. Expression of yggX restored prototrophic growth to all strains except the isc1 mutant, where it eliminated the nicotinic acid but not the thiamine requirement. The inability of yggX expression to restore thiamine prototrophy was expected, since isc1 is polar onto iscS, which is required for sulfane-sulfur transfer in thiamine synthesis in addition to its role in Fe-S cluster assembly (17, 20). Growth of all strains was restored by the presence of nicotinic acid and thiamine.
FIG. 3.
YggX accumulation restores thiamine prototrophy to the mutant class. Duplicates of each strain isogenic for yggX were patched onto NB agar plates, incubated for 4 h at 37°C, and replica printed to NCE glucose agar plates containing 20 μM nicotinic acid (NA) and/or 100 nM thiamine (THI), as indicated. Relevant genotypes are depicted below each patch. YggX is present unless indicated by genotypic designation (yggX).
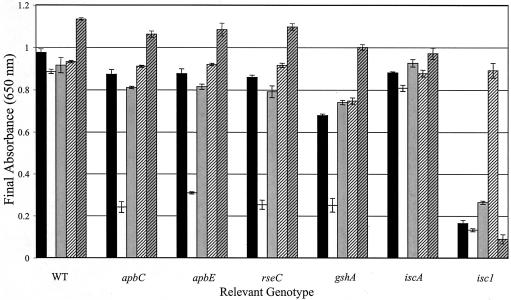
Expression of yggX alters sensitivity of thiamine synthesis to metals.
A change in the water supply source to the laboratory resulted in loss of the well-documented thiamine requirement and aconitase defect of apbC mutants grown in liquid media (30, 41). The addition of trace minerals restored the auxotrophic phenotype to an apbC mutant (data not shown), which led to a meticulous dissection of the effect trace minerals had on the growth of the class of mutants relevant here. While several sensitivities that were specific to the mutant locus (apbC, Zn sensitive; apbE, Zn and Se sensitive; apbE, growth stimulation by Cu) were noted, the most striking result involved inhibition by cobalt. Results from the growth experiments dissecting this effect are shown in Fig. 4. With no additions (except nicotinic acid to the isc1 mutant), only the strain containing the polar isc1 insertion was unable to grow. When yggX was absent and cobalt (10 μM) was provided, growth of the gshA, apbC, apbE, and rseC strains was inhibited while the yggX and iscA mutant strains were unaffected. The addition of thiamine restored full growth to each strain and indicated that the most sensitive target of the cobalt effect was thiamine synthesis. The thiamine requirement in the presence of cobalt was also eliminated by expression of yggX or by addition of iron. The latter result suggested that a competition between Fe and Co might be occurring in vivo. As anticipated by the dual role of IscS in thiamine synthesis (sulfur insertion into the thiazole moiety of thiamine and assembly of the Fe-S cluster in ThiH), thiamine prototrophy of strains carrying the isc1 insertion could not be restored by yggX expression or iron.
FIG. 4.
Inhibition of thiamine synthesis by cobalt is alleviated by exogenous iron or by expression of yggX. Strains lacking yggX and the indicated locus were grown in NCE glucose medium with no addition (black bars), 10 μM CoCl2 (white bars), 100 nM thiamine and 10 μM CoCl2 (cross-hatched white bars), or 20 μM FeCl3 and 10 μM CoCl2 (cross-hatched gray bars). Solid gray bars represent the growth of strains expressing yggX in NCE glucose medium containing 10 μM CoCl2. Growth was assessed after 24 h, and the final OD650 is reported.
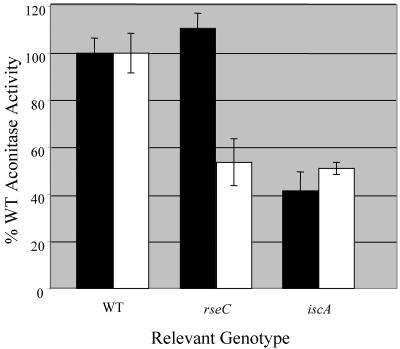
The water supply change that affected growth phenotypes similarly altered the level of aconitase activity seen in the apbC, apbE, and rseC mutants, but not in strains lacking isc or gshA (all strains analyzed were in the yggX background). Representative data with rseC and iscA mutants are shown in Fig. 5. As with the nutritional requirements, addition of trace minerals (lacking iron) to the lab medium decreased the level of aconitase detected in the rseC mutant. Based on the analysis described above, it was considered that this effect was likely due to the Co present in the trace minerals solution.
FIG. 5.
Trace minerals affect the activity of aconitase in an rseC mutant. Strains were grown at 37°C until mid-log phase in NCE gluconate medium containing 20 μM nicotinic acid and 100 nM thiamine and assayed for aconitase activity. Strains were grown in NCE gluconate medium with (open bars) or without (solid bars) trace minerals (2) lacking iron. Activity is reported as the percentage of the activity obtained with the wild-type strain under the same growth conditions.
DISCUSSION
Cellular components involved in the Fe-S cluster synthesis and repair processes in vivo are not yet completely defined. While studies in a variety of organisms continue to identify relevant loci, the challenge is to determine how multiple components work together to ensure that the complex homeostasis of Fe-S cluster metabolism is maintained in vivo. Work in our laboratory defined a class of mutants with lesions in one of five loci (gshA, apbC, apbE, rseC, and isc) that had in common (i) a thiamine requirement reflecting an impaired ThiH enzyme, (ii) decreased activities of Fe-S cluster enzymes, and (iii) nutritional phenotypes suppressed by expression of yggX.
The work presented here was initiated to extend the correlation between mutants of this class and to identify differences between them that might provide insight into individual and collective roles the gene products have in the cell. Results herein emphasized the central role that YggX has in preventing oxidative damage in the cell. Cells lacking this protein are under constant oxidative stress, as indicated by the increase in Sox-dependent expression under aerobic growth, increased sensitivity to PQ, and the reduction in activity of the labile Fe-S cluster-containing protein MiaB. Lack of YggX compromised metabolism such that otherwise-subtle defects in Fe-S cluster metabolism were detected by significant growth phenotypes. This conclusion was emphasized by the finding that the majority of phenotypes assessed herein were partly or completely suppressed by the expression of yggX. For instance, mutants defective in Fe-S cluster metabolism were more sensitive to oxidative stress when lacking yggX. In yeast, mutants compromised in Fe-S cluster metabolism are sensitive to oxidative stress (27, 40), but we are unaware of a report directly linking impaired Fe-S cluster metabolism with a sensitivity to oxidative stress in bacteria. Lack of such reports could reflect the presence of YggX in most bacteria, which obscures the sensitivity. Thus, a yggX mutant background has proven to be a valuable tool for identification and characterization of a distinct set of functions involved in Fe-S cluster metabolism and oxidative damage repair.
Lesions in the five loci discussed herein cause a diverse set of phenotypes that can be explained by a defect in Fe-S cluster metabolism. By extending the phenotypic and biochemical characterization, properties unique to some members became apparent. For instance, the severely reduced activity of Fe-S cluster enzymes (41, 42), their general growth defect (Table 2), and lack of suppression by yggX expression separated isc mutants from those with lesions in the other loci. Additionally, isc mutants were distinct in their SoxRS response, as monitored by the pattern of fpr expression. Unlike the remaining mutants, isc mutants were unable to mount a full SoxRS response to PQ. A simple interpretation of these results is that the Isc proteins, but not ApbCE and RseC, are required for the synthesis of the 2Fe-2S clusters contained in SoxR. The constitutive level of fpr expression in the gshA, apbC, apbE, and rseC mutants was consistent with these strains perceiving high levels of oxidative stress in the absence of PQ and/or a role for these gene products in the reduction of the cluster in SoxR (reduction of the SoxR Fe-S clusters is required to cease activation of soxS). It is likely that the different responses reflect a combination of effects, both direct and indirect, caused by lack of the relevant gene products.
Extensive growth analyses determined that the nutritional requirements of this class of mutant strains (lacking yggX) are medium dependent, affected by the level of certain minerals and amount of aeration. The apbC, apbE, rseC, and gshA mutants were distinguished from strains lacking isc by a cobalt-induced thiamine requirement. The finding that cobalt sensitivity could be relieved by thiamine or exogenous iron suggested that a competition between the transition metals was occurring in the absence of yggX. One interpretation of these data is that the ApbCE and RseC proteins, in addition to glutathione (11), function in Fe-S cluster repair. We would suggest that, when yggX and one of these gene products are missing, Co if present in excess can be incorporated into damaged Fe-S clusters, resulting in loss of function of the enzyme. Consistent with this hypothesis, Camba and Armstrong have shown that after 4Fe-4S clusters are damaged, metals such as zinc will trap the 3Fe-4S intermediate as a 3Fe-4S zinc adduct (5). The oxygen-labile 4Fe-4S cluster in ThiH would provide an obvious and sensitive target for this effect. The ability of yggX expression to suppress the cobalt-induced thiamine requirement is consistent with a function of YggX in iron trafficking and suggests a further role for YggX as a “specificity factor,” making iron a better competitor in the presence of cobalt. The suppression by YggX may also be facilitated by the fact that less oxidative damage occurs in the presence of YggX (15), decreasing the need for cluster repair.
While the above model is consistent with the data presented herein and is the one we favor, it is not the only model that could explain several of the defects seen in these mutant strains. For instance, it is formally possible that mutations in apbC/E and rseC increase the level of oxidative stress in the cell, thereby perpetuating damage to Fe-S clusters. While oxygen-reactive species have not been directly measured in these mutants, they do not display the increased mutation frequency associated with mutagenesis mediated by the Fenton reaction (data not shown). Another possibility that was previously considered is that ApbC/E and/or RseC work with the Suf proteins to facilitate Fe-S cluster assembly (41). Such a hypothesis does not easily explain the cobalt sensitivity of these mutants or why the apbC/E and rseC mutants have phenotypic defects that suf mutants do not (data not shown). These models are not mutually exclusive and, considering the complexity of the in vivo system, it is likely that some combination of these models will prove to be most correct.
Data from many labs have shown that the metabolism of Fe-S clusters is complex and involves multiple loci. Several of the proteins involved in Fe-S cluster assembly have been elucidated and functionally characterized in vitro. However, it remains unclear how clusters are repaired and which proteins traffic the iron for cluster assembly and/or repair in vivo. By starting with a strain under low levels of constant oxidative stress, we have identified additional proteins that can affect Fe-S cluster metabolism in vivo. While providing evidence for a number of functional distinctions between gene products, the data do not yet support a specific mechanistic model for the gene products in question. Instead, this work provides a basis for additional studies to address the broad working model that has been generated as a result of these studies. The results described herein are consistent with a working model in which YggX participates in iron trafficking and the ApbC, GshA, ApbE, and RseC proteins are distinguished from the isc gene products by their participation in the repair of Fe-S clusters.
Acknowledgments
We thank P. Pomposiello for providing the fpr reporter strain. We thank Jenny Gross and Jeff Gralnick for technical assistance in constructing the rseC and isc deletion strains, respectively.
This work was supported by competitive grants GM47296 (D.M.D.) and GM57002 (C.L.) from the National Institutes of Health and MCB0096513 from NSF (D.M.D.). Funds were also provided from a 21st Century Scientists Scholars Award from the J. M. McDonnell fund to D.M.D. Elizabeth Skovran was supported by the William H. Peterson predoctoral fellowship from the Department of Bacteriology.
REFERENCES
- 1.Apontoweil, P., and W. Berends. 1975. Mapping of gshA, a gene for the biosynthesis of glutathione in Eschericha coli K12. Mol. Gen. Genet. 141:91-95. [DOI] [PubMed] [Google Scholar]
- 2.Balch, W. E., G. E. Fox, L. J. Magrum, C. R. Woese, and R. S. Wolfe. 1979. Methanogens: reevaluation of a unique biological group. Microbiol. Rev. 43:260-296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Beck, B. J., and D. M. Downs. 1999. A periplasmic location is essential for the role of the ApbE lipoprotein in thiamine synthesis in Salmonella typhimurium. J. Bacteriol. 181:7285-7290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Berkowitz, D., J. M. Hushon, H. J. Whitfield, J. Roth, and B. N. Ames. 1968. Procedure for identifying nonsense mutations. J. Bacteriol. 96:215-220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Camba, R., and F. A. Armstrong. 2000. Investigations of the oxidative disassembly of Fe-S clusters in Clostridium pasteurianum 8Fe ferredoxin using pulsed-protein-film voltammetry. Biochemistry 39:10587-10598. [DOI] [PubMed] [Google Scholar]
- 6.Castilho, B. A., P. Olfson, and M. J. Casadaban. 1984. Plasmid insertion mutagenesis and lac gene fusion with mini Mu bacteriophage transposons. J. Bacteriol. 158:488-495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cherepanov, P. P., and W. Wackernagel. 1995. Gene disruption in Escherichia coli: TcR and KmR cassettes with the option of Flp-catalyzed excision of the antibiotic-resistance determinant. Gene 158:9-14. [DOI] [PubMed] [Google Scholar]
- 8.Datsenko, K. A., and B. L. Wanner. 2000. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc. Natl. Acad. Sci. USA 97:6640-6645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Esberg, B., H. C. Leung, H. C. Tsui, G. R. Bjork, and M. E. Winkler. 1999. Identification of the miaB gene, involved in methylthiolation of isopentenylated A37 derivatives in the tRNA of Salmonella typhimurium and Escherichia coli. J. Bacteriol. 181:7256-7265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Frazzon, J., J. R. Fick, and D. R. Dean. 2002. Biosynthesis of iron-sulphur clusters is a complex and highly conserved process. Biochem. Soc. Trans. 30:680-685. [DOI] [PubMed] [Google Scholar]
- 11.Gardner, P. R., and I. Fridovich. 1993. Effect of glutathione on aconitase in Escherichia coli. Arch. Biochem. Biophys. 301:98-102. [DOI] [PubMed] [Google Scholar]
- 12.Gardner, P. R., and I. Fridovich. 1991. Quinolinate synthetase: the oxygen-sensitive site of de novo NAD(P)+ biosynthesis. Arch. Biochem. Biophys. 284:106-111. [DOI] [PubMed] [Google Scholar]
- 13.Gralnick, J., and D. Downs. 2001. Protection from superoxide damage associated with an increased level of the YggX protein in Salmonella enterica. Proc. Natl. Acad. Sci. USA 98:8030-8035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gralnick, J., E. Webb, B. Beck, and D. Downs. 2000. Lesions in gshA (encoding gamma-l-glutamyl-l-cysteine synthetase) prevent aerobic synthesis of thiamine in Salmonella enterica serovar Typhimurium LT2. J. Bacteriol. 182:5180-5187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gralnick, J. A., and D. M. Downs. 2003. The YggX protein of Salmonella enterica is involved in Fe(II) trafficking and minimizes the DNA damage caused by hydroxyl radicals: residue CYS-7 is essential for YggX function. J. Biol. Chem. 278:20708-20715. [DOI] [PubMed] [Google Scholar]
- 16.Jeon, W., J. Cheng, and P. Ludden. 2001. Purification and characterization of membrane-associated CooC protein and its functional role in the insertion of nickel into carbon monoxide. J. Biol. Chem. 276:38602-38609. [DOI] [PubMed] [Google Scholar]
- 17.Kambampati, R., and C. T. Lauhon. 2000. Evidence for the transfer of sulfane sulfur from IscS to ThiI during the in vitro biosynthesis of 4-thiouridine in Escherichia coli tRNA. J. Biol. Chem. 275:10727-10730. [DOI] [PubMed] [Google Scholar]
- 18.Koo, M. S., J. H. Lee, S. Y. Rah, W. S. Yeo, J. W. Lee, K. L. Lee, Y. S. Koh, S. O. Kang, and J. H. Roe. 2003. A reducing system of the superoxide sensor SoxR in Escherichia coli. EMBO J. 22:2614-2622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Krapp, A. R., R. E. Rodriguez, H. O. Poli, D. H. Paladini, J. F. Palatnik, and N. Carrillo. 2002. The flavoenzyme ferredoxin (flavodoxin)-NADP(H) reductase modulates NADP(H) homeostasis during the SoxRS response of Escherichia coli. J. Bacteriol. 184:1474-1480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lauhon, C. T., and R. Kambampati. 2000. The iscS gene in Escherichia coli is required for the biosynthesis of 4-thiouridine, thiamin, and NAD. J. Biol. Chem. 275:20096-20103. [DOI] [PubMed] [Google Scholar]
- 21.Lauhon, C. T., E. Skovran, H. D. Urbina, D. M. Downs, and L. E. Vickery. 2004. Substitutions in an active site loop of Escherichia coli IscS result in specific defects in Fe-S cluster and thionucleoside biosynthesis in vivo. J. Biol. Chem. 279:19551-19558. [DOI] [PubMed] [Google Scholar]
- 22.Lee, J. H., W. S. Yeo, and J. H. Roe. 2004. Induction of the sufA operon encoding Fe-S assembly proteins by superoxide generators and hydrogen peroxide: involvement of OxyR, IHF and an unidentified oxidant-responsive factor. Mol. Microbiol. 51:1745-1755. [DOI] [PubMed] [Google Scholar]
- 23.Leipuviene, R., Q. Qian, and G. R. Bjork. 2004. Formation of thiolated nucleosides present in tRNA from Salmonella enterica serovar Typhimurium occurs in two principally distinct pathways. J. Bacteriol. 186:758-766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lezhneva, L., K. Amann, and J. Meurer. 2004. The universally conserved HCF101 protein is involved in assembly of [4Fe-4S]-cluster-containing complexes in Arabidopsis thaliana chlorplasts. Plant J. 37:174-185. [DOI] [PubMed] [Google Scholar]
- 25.Martinez-Gomez, N. C., M. Robers, and D. M. Downs. Mutational analysis of ThiH, a member of the radical S-adenosylmethionine (SAM) protein superfamily. J. Biol. Chem., in press. [DOI] [PubMed]
- 26.Miller, J. H. 1972. Experiments in molecular genetics. Cold Spring Harbor Laboratory, Cold Spring Harbor, N.Y.
- 27.Muhlenhoff, U., N. Richhardt, M. Ristow, G. Kispal, and R. Lill. 2002. The yeast frataxin homolog Yfh1p plays a specific role in the maturation of cellular Fe/S proteins. Hum. Mol. Genet. 11:2025-2036. [DOI] [PubMed] [Google Scholar]
- 28.Outten, F. W., O. Djaman, and G. Storz. 2004. A suf operon requirement for Fe-S cluster assembly during iron starvation in Escherichia coli. Mol. Microbiol. 52:861-872. [DOI] [PubMed] [Google Scholar]
- 29.Persson, B. C., and G. R. Bjork. 1993. Isolation of the gene (miaE) encoding the hydroxylase involved in the synthesis of 2-methylthio-cis-ribozeatin in tRNA of Salmonella typhimurium and characterization of mutants. J. Bacteriol. 175:7776-7785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Petersen, L. A., and D. M. Downs. 1997. Identification and characterization of an operon in Salmonella typhimurium involved in thiamine biosynthesis. J. Bacteriol. 179:4894-4900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Pierrel, F., G. R. Bjork, M. Fontecave, and M. Atta. 2002. Enzymatic modification of tRNAs: MiaB is an iron-sulfur protein. J. Biol. Chem. 277:13367-13370. [DOI] [PubMed] [Google Scholar]
- 32.Pomposiello, P. J., and B. Demple. 2000. Identification of SoxS-regulated genes in Salmonella enterica serovar Typhimurium. J. Bacteriol. 182:23-29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Pomposiello, P. J., and B. Demple. 2001. Redox-operated genetic switches: the SoxR and OxyR transcription factors. Trends Biotechnol. 19:109-114. [DOI] [PubMed] [Google Scholar]
- 34.Pomposiello, P. J., A. Koutsolioutsou, D. Carrasco, and B. Demple. 2003. SoxRS-regulated expression and genetic analysis of the yggX gene of Escherichia coli. J. Bacteriol. 185:6624-6632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Rangaraj, P., P. Shah, and P. Ludden. 1997. ApoNifH functions in iron-molybdenum cofactor synthesis and apodinitrogenase maturation. Proc. Natl. Acad. Sci. USA 94:11250-11255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Roy, A., N. Solodovnikova, T. Nicholson, W. Antholine, and W. E. Walden. 2003. A novel eukaryotic factor for cytosolic Fe-S cluster assembly. EMBO J. 22:4826-4835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Saunders, N. F. W., J. J. Hornberg, W. N. M. Reijnders, H. V. Westerhoff, S. De Vries, and R. J. M. Van Spanning. 2000. The NosX and NirX proteins of Paracoccus denitrificans are functional homologues: their role in maturation of nitrous oxide reductase. J. Bacteriol. 182:5211-5217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Schmehl, M., A. Jahn, A. Meyer zu Vilsendorf, S. Hennecke, B. Masepohl, M. Schuppler, M. Marxer, J. Oelze, and W. Klipp. 1993. Identification of a new class of nitrogen fixation genes in Rhodobacter capsulatus: a putative membrane complex involved in electron transport to nitrogenase. Mol. Gen. Genet. 241:602-615. [DOI] [PubMed] [Google Scholar]
- 39.Schwartz, C. J., O. Djaman, J. A. Imlay, and P. J. Kiley. 2000. The cysteine desulfurase, IscS, has a major role in in vivo Fe-S cluster formation in Escherichia coli. Proc. Natl. Acad. Sci. USA 97:9009-9014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sipos, K., H. Lange, Z. Fekete, P. Ullmann, R. Lill, and G. Kispal. 2002. Maturation of cytosolic iron-sulfur proteins requires glutathione. J. Biol. Chem. 277:26944-26949. [DOI] [PubMed] [Google Scholar]
- 41.Skovran, E., and D. M. Downs. 2003. Lack of the ApbC or ApbE protein results in a defect in Fe-S cluster metabolism in Salmonella enterica serovar Typhimurium. J. Bacteriol. 185:98-106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Skovran, E., and D. M. Downs. 2000. Metabolic defects caused by mutations in the isc gene cluster in Salmonella enterica serovar Typhimurium: implications for thiamine synthesis. J. Bacteriol. 182:3896-3903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Sofia, H. J., G. Chen, B. G. Hetzler, J. F. Reyes-Spindola, and N. E. Miller. 2001. Radical SAM, a novel protein superfamily linking unresolved steps in familiar biosynthetic pathways with radical mechanisms: functional characterization using new analysis and information visualization methods. Nucleic Acids Res. 29:1097-1106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Way, J. C., M. A. Davis, D. Morisato, D. E. Roberts, and N. Kleckner. 1984. New Tn10 derivatives for transposon mutagenesis and for construction of lacZ operon fusions by transposition. Gene 32:369-379. [DOI] [PubMed] [Google Scholar]
- 45.Zheng, M., X. Wang, L. J. Templeton, D. R. Smulski, R. A. LaRossa, and G. Storz. 2001. DNA microarray-mediated transcriptional profiling of the Escherichia coli response to hydrogen peroxide. J. Bacteriol. 183:4562-4570. [DOI] [PMC free article] [PubMed] [Google Scholar]