Abstract
Plasmid p42a from Rhizobium etli CFN42 is self-transmissible and indispensable for conjugative transfer of the symbiotic plasmid (pSym). Most pSym transconjugants also inherit p42a. pSym transconjugants that lack p42a always contain recombinant pSyms, which we designated RpSyms*. RpSyms* do not contain some pSym segments and instead have p42a sequences, including the replication and transfer regions. These novel recombinant plasmids are compatible with wild-type pSym, incompatible with p42a, and self-transmissible. The symbiotic features of derivatives simultaneously containing a wild-type pSym and an RpSym* were analyzed. Structural analysis of 10 RpSyms* showed that 7 shared one of the two pSym-p42a junctions. Sequencing of this common junction revealed a 53-bp region that was 90% identical in pSym and p42a, including a 5-bp central region flanked by 9- to 11-bp inverted repeats reminiscent of bacterial and phage attachment sites. A gene encoding an integrase-like protein (intA) was localized downstream of the attachment site on p42a. Mutation or the absence of intA abolished pSym transfer from a recA mutant donor. Complementation with the wild-type intA gene restored transfer of pSym. We propose that pSym-p42a cointegration is required for pSym transfer; cointegration may be achieved either through homologous recombination among large reiterated sequences or through IntA-mediated site-specific recombination between the attachment sites. Cointegrates formed through the site-specific system but resolved through RecA-dependent recombination or vice versa generate RpSyms*. A site-specific recombination system for plasmid cointegration is a novel feature of these large plasmids and implies that there is unique regulation which affects the distribution of pSym in nature due to the role of the cointegrate in conjugative transfer.
Bacteria belonging to the genus Rhizobium are able to establish a symbiotic relationship with the roots of leguminous plants, in which the bacteria provide fixed nitrogen to the plants in exchange for a carbon source and a secure environment. Usually, the bacterial genetic information required for establishment of this symbiotic relationship is localized on symbiotic plasmids (pSyms) (16, 39).
Self-transmissible pSyms have been described in Rhizobium leguminosarum (4, 24). A recent report indicated that transfer of one of these plasmids (pRL1JI) is regulated by a quorum-sensing mechanism (29, 51). It has been shown that transfer genes localized on the pSym of Rhizobium sp. strain NGR234 are induced by quorum-sensing regulators, although the conjugal efficiency of this plasmid is extremely low (21). Also, self-transmissible cryptic plasmids have been described in Sinorhizobium meliloti (31). However, data regarding the transfer mechanism of these plasmids are scarce. Sequence analysis of various rhizobial symbiotic regions has revealed the presence of transfer-related genes and sequences homologous to the luxI-luxR type of quorum-sensing regulators (2, 15, 19, 30). Nevertheless, the conditions under which they are functional remain to be elucidated. In Mesorhizobium loti, the transfer of a chromosomally integrated symbiotic island has been documented (46); this element integrates into a phenylalanine tRNA gene in a process mediated by a P4-type integrase encoded in the element (47).
Rhizobium etli is the symbiont of Phaseolus vulgaris. R. etli type strain CFN42, contains six plasmids, designated p42a to p42f, whose sizes range from 185 to 643 kb. Plasmid p42d has been identified as the symbiotic plasmid (pSym), because it carries most of the information required for nodulation and nitrogen fixation (38). The physical map of this plasmid has been determined (18), and complete sequencing has recently been concluded (19).
Analyzing the plasmids of R. etli CFN42 for transfer functions, we found that only p42a is self-transmissible at a high frequency (∼10−2) and that the transfer is regulated by quorum sensing. The transfer region (oriT, tra, and trb genes) of p42a is highly similar to the transfer region of the Agrobacterium tumefaciens Ti plasmid (48). On the other hand, the sequence of the pSym has shown that this plasmid carries traA and traCDG genes (19) (National Center for Biotechnology Information [NCBI] accession number NC_004041). The products of these genes could participate in DNA processing during conjugation. In recent work Pérez-Mendoza et al. (36) identified a functional mob region in the pSym of R. etli CFN42. Furthermore, they showed that this pSym is able to perform self-transfer under special conditions that include artificial overexpression of a possible conjugation activator. However, in a normal laboratory environment, we have always found that conjugative transfer of the pSym is fully dependent on the presence of p42a and that the plasmid transfers at a frequency on the order of 10−6 (7). Interestingly, if a recA mutant strain is used as the donor, the pSym transfer frequency is diminished (to ∼10−7). Analysis of pSym transconjugants has shown that they usually contain p42a in addition to the pSym; alternatively, a plasmid corresponding to a cointegrate of these two plasmids has also been visualized. A small fraction (about 10%) of the transconjugants contain only a plasmid whose size and overall composition resemble the pSym (7). These data led us to propose that pSym transfer is achieved through cointegration with p42a and thus is an example of conduction. Cointegrates may be formed either through homologous recombination or through a mechanism independent of RecA. Reiteration of DNA sequences is common among the rhizobia (16, 39). The presence of large regions (>500 bp) shared by the pSym and p42a has been described previously (18), and these regions may be substrates for RecA-dependent cointegration. To gain further insight into the mechanisms that allow pSym conjugative transfer, we performed a thorough analysis of pSym transconjugants. This analysis led us to determine that some transconjugants carry recombinant pSyms, which we designated RpSyms*. The characteristics of the RpSyms* allow them to coexist with the wild-type pSym. In this paper we describe a sequence analysis of the pSym-p42a junction of a recombinant plasmid. Through this analysis we identified a site-specific recombination system that is responsible for the RecA-independent cointegration event. A model explaining the mechanism of pSym conjugative transfer and the generation of new RpSyms* is discussed. The symbiotic features of a laboratory-constructed chimera that simultaneously contains a wild-type pSym and an RpSym* are also presented below.
MATERIALS AND METHODS
Bacterial strains and growth conditions.
The strains used in this work are listed in Table 1. Rhizobium strains were grown at 30°C in PY medium (35). Escherichia coli and A. tumefaciens strains were grown on Luria-Bertani medium (34). When required, antibiotics were added at the following concentrations: nalidixic acid, 20 μg ml−1; neomycin, 60 μg ml−1; spectinomycin, 75 μg ml−1; rifampin, 100 μg ml−1, ampicillin, 100 μg ml−1; erythromycin, 100 μg ml−1; and tetracycline, 10 μg ml−1 for E. coli or 5 μg ml−1 for Rhizobium.
TABLE 1.
Bacterial strains and plasmids
| Strain or plasmid | Relevant features | Reference or source |
|---|---|---|
| R. etli strains | ||
| CFN42 | Wild type (p42a, p42b, p42c, p42d, p42e, p42f) | 38 |
| CFNX5 | CFN42 derivative (nifHc::GDYN-1) | 40 |
| CFN2001 | CFN42 derivative (p42a− p42d−) | 28 |
| CFNX192 | CFNX89/p42d::Tn5mob | 6 |
| CFN42recA | recA derivative of CFN42 | J. Martínez, CIFN |
| CFNX247 | CFN42 derivative (nifA::ΩSp) | 17 |
| CFNX643 | CFN2001 with Rp42d* (nifA::ΩSp) from CFN247 | This study |
| CFNX644 | CFN2001 with Rp42d* (nifA::ΩSp) from CFN247 | This study |
| CFNX645 | CFN2001 with p42d (nifA::ΩSp) and p42a from CFN247 | This study |
| CFNX646 | CFN2001 transconjugant with Rp42d* from CFNX5 | This study |
| CFNX647 | CFN2001 transconjugant with Rp42d* from CFNX5 | This study |
| CFNX648 | CFN2001 transconjugant with Rp42d* from CFNX5 | This study |
| CFNX649 | CFN2001 transconjugant with Rp42d* from CFNX5 | This study |
| CFNX650 | CFN2001 transconjugant with Rp42d* from CFNX5 | This study |
| CFNX651 | CFN2001 transconjugant with Rp42d* from CFNX5 | This study |
| CFNX652 | CFN2001 transconjugant with Rp42d* from CFNX5 | This study |
| CFNX653 | CFN42recA Rifr with p42d::Tn5mob from CFNX192 | This study |
| CFNX654 | CFNX653 with Rp42d* from CFNX646 | This study |
| CFNX655 | CFNX653 with Rp42d* from CFNX648 | This study |
| CFNX656 | CFNX653 with Rp42d* from CFNX648 | This study |
| CFNX657 | CFNX653 with Rp42d* from CFNX647 | This study |
| CFNX658 | CFNX653 with Rp42d* from CFNX649 | This study |
| CFNX659 | CFNX653 with Rp42d* from CFNX650 | This study |
| CFNX660 | CFNX653 with Rp42d* from CFNX652 | This study |
| CFNX661 | CFN42 intA::pJQ200mp18, Gmr | This study |
| CFNX662 | CFNX661 complemented with pAGS4 | This study |
| CFNX663 | CFN42 intA::ΩSpr Smr | This study |
| CFNX664 | CFNX663 complemented with pAGS4 | This study |
| A. tumefaciens strains | ||
| GMI9023 | C58 cured of its native plasmids | 42 |
| UIA143 | recA pTi− derivative of C58 | 12 |
| GMI9023/p42a | GMI9023 containing p42a::Tn5mob | 6 |
| GMI9023/p42d | GMI9023 containing p42d::Tn5mob | 6 |
| GMI9023/p643 | GMI9023 containing RpSym* from CFNX643 | This study |
| GMI9023/p644 | GMI9023 containing RpSym* from CFNX644 | This study |
| GMI9023/p645 | GMI9023 containing pSym from CFNX645 | This study |
| GMI9023/p646 | GMI9023 containing RpSym* from CFNX646 | This study |
| GMI9023/p648-1 | GMI9023 containing RpSym* from CFNX648 | This study |
| GMI9023/p648-2 | GMI9023 containing RpSym* from CFNX648 | This study |
| GMI9023/p649 | GMI9023 containing RpSym* from CFNX649 | This study |
| GMI9023/p651-1 | GMI9023 containing RpSym* from CFNX651 | This study |
| GMI9023/p651-2 | GMI9023 containing RpSym* from CFNX651 | This study |
| GMI9023/p652-1 | GMI9023 containing RpSym* from CFNX652 | This study |
| GMI9023/p652-2 | GMI9023 containing RpSym* from CFNX652 | This study |
| E. coli strains | ||
| HB101/pRK2013 | Conjugation helper | 14 |
| DH5α | Receptor for transformation | 43 |
| S17-1 | C600::RP-4-2 (Tc::Mu)(Km::Tn7) | 44 |
| Plasmids | ||
| pCR2.1 | TA cloning vector for PCR products, Kmr Apr | Invitrogen |
| pMOSblue | Blunt-ended cloning vector, Apr | Amersham Pharmacia Biotech |
| pBluescript II SK | Sequencing vector, Apr | Stratagene |
| pJQ200mp18 | Suicide vector for gene replacement, GmrsacB Mob | 37 |
| pJQ200SK | Suicide vector for gene replacement, GmrsacB Mob | 37 |
| pBBRMCS3 | Broad-host-range cloning vector, Tcr | 28 |
| pAGS1 | 543-bp PCR fragment from the 3′ end of intA, cloned in pMOSblue | This study |
| pAGS2 | 570-bp BamHI-SalI fragment from pAGS1 cloned in pJQ200mp18, Gmr | This study |
| pAGS3 | 2,064-bp XbaI-KpnI PCR fragment containing the entire intA gene cloned in pCR2.1 | This study |
| pAGS4 | 2,064-bp XbaI-KpnI PCR fragment containing the entire intA gene cloned in pBBRMCS3 | This study |
| pAGS10 | 1.6-kb XhoI-PstI fragment with ΩSpr Smr insertion in a unique EcoRI site within the intA gene, cloned in pJQ200SK | This study |
Genetic manipulations.
Conjugation experiments were performed on PY plates at 30°C by using overnight cultures grown to the stationary phase. Donors and recipients were mixed at a 1:2 ratio, and suitable markers were used for transconjugant selection. Standard molecular cloning techniques were performed as described previously (43). A 760-bp PCR product covering the left junction of the pSym-p42a border of an RpSym* was obtained by using DNA of transconjugant CFNX643 as the template with one primer from the pSym (primer Up 78-1 [5′-CCCATCCTCAGCAGCATCC-3′]) and another primer from p42a (primer Lw 4-20EcoR1 [5′-ATAGAATTCAGGGCAGGGGAGGCGAAGG-3′; the underlined sequence, containing an EcoRI restriction site, was added to facilitate the cloning procedure]). This PCR product was cloned in the pSK+ phagemid vector (Stratagene, La Jolla, Calif.). A 2,529-bp PCR fragment from p42a containing DNA adjacent to the site where recombination with the pSym occurred was obtained by using DNA of the wild-type strain as the template and primers Up4r-3EcoR1 (5′-TATGAATTCAAGCCGAAGTCACCGAACG-3′) and Lw 4-20EcoR1 (5′-ATAGAATTCAGGGCAGGGGAGGCGAAGG-3′). This PCR product was also cloned in pSK+.
All the oligonucleotide primers were purchased from Unidad de Síntesis Química IBT-UNAM. PCR amplification was performed in a Mastercycler 5330 (Eppendorf, Hamburg, Germany). PCR products were generated with a Gene-Amp XL PCR kit (Perkin-Elmer, Branchburg, N.J.) by using an initial denaturation step consisting of 94°C for 2 min. The cycling regimen consisted of 16 cycles of 94°C for 30 s and 68°C for 10 min, followed by 12 cycles of 94°C for 30 s and 68°C for 11 min. PCR fragments cloned in the pBluescript II SK(+) vector were sequenced with a combination of custom-made and universal oligonucleotide primers by using a Taq DyeDeoxy terminator cycle sequencing kit and an automatic 373A DNA sequencing system (Applied Biosystems, Foster City, Calif.). DNA was sequenced at least twice on both strands. Plasmids from Agrobacterium transconjugants were purified as described by Hirsch et al. (22).
Insertional mutagenesis.
In order to construct mutant CFNX661 (intA::pJQ200mp18), a 543-bp internal fragment of the intA gene was amplified by PCR with primers 5′-AAGGCGGTCACCGGCGACGTA-3′ and 5′-TTGCAACGACCGATGTCGCGA-3′ and cloned by the T-A annealing method into the pMOSblue vector (Amersham Pharmacia Biotech, Little Chalfont, Buckinghamshire, England). The resulting plasmid, designated pAGS1, was restricted with SalI and BamHI, and the 570-bp fragment obtained was cloned into the suicide vector pJQ200mp18 (37). The plasmid obtained (pAGS2) was integrated into the intA gene of R. etli CFNX192 by single recombination, producing two incomplete copies of the gene. One of these copies lacked 63 bp of the 3′ end, where the potential catalytic Tyr-324 is encoded. The second copy lacked 640 bp at the 5′ end. The disruption of intA was confirmed by Southern blot analysis (data not shown).
A 2,064-bp KpnI-XbaI fragment containing the entire intA gene was amplified by PCR with primers 5′-GGGTACCCTGGCCGCAGCAAGGTAAG-3′ and 5′-CTCTAGAGCACCCGGCAGAGACGCATTTC-3′and cloned into the pCR2.1 vector (Invitrogen, Carlsbad, Calif.); the resulting plasmid was designated pAGS3. The insert from pAGS3 was then subcloned into the broad-host-range vector pBBRMCS3 (27). The plasmid obtained, designated pAGS4, was used to complement the intA mutants.
Mutant CFNX663 (intA::ΩSp) was constructed by cloning a 2.1-kb SpeI-PstI fragment from pAGS3 in pBluescript II SK(+). An ΩSp cassette (13) was inserted into this plasmid in the unique BamHI site located 130 bp downstream of the first initiation codon. A 1.6-kb XhoI-PstI fragment containing intA::ΩSp was subcloned into pJQ200SK (37). The resulting plasmid, designated pAGS10, was used for gene replacement by double homologous recombination. Southern blotting (data not shown) was used to verify the replacement of the wild-type gene with the mutagenized gene.
Filter blot hybridization and plasmid visualization.
For Southern-type hybridizations (45), genomic DNA was digested with appropriate restriction enzymes, electrophoresed in 1% (wt/vol) agarose gels, blotted onto nylon membranes, and hybridized under stringent conditions, as previously reported (18), by using Rapid-hyb buffer. Probes were linearized by digesting them with appropriate restriction enzymes and were labeled with [α-32P]dCTP by using a Rediprime DNA labeling system. Plasmid profiles were visualized by the Eckhardt technique (10), as modified by Hynes and McGregor (23), and hybridized similarly. All restriction endonucleases, [α-32P]dCTP, hybridization buffer, and labeling systems were purchased from Amersham Pharmacia Biotech.
Sequence analysis.
Nucleotides and amino acid sequences were analyzed by using the following programs. From the Wisconsin Package, version 10.0 (Genetics Computer Group), we used PILEUP to create a multiple-sequence alignment, PRETTY to calculate a consensus sequence, and BESTFIT to find the segment with the best similarity between two sequences. The NCBI ORF FINDER program was used to find possible open reading frames. Amino acid sequence comparisons and conserved domain searches were done with the NCBI BLASTP program (1). Conserved domain comparisons were done against the Pfam database of protein domains and HMMs (hidden Markov models) at Washington University, St. Louis, Mo. (http://pfam.wustl.edu/index.html).
Plant assays.
P. vulgaris cv. Negro Jamapa plants were inoculated with the desired strain and grown in a greenhouse. After 32 days, acetylene reduction assays for nitrogenase activity were carried out as previously described (41).
Nucleotide sequence accession numbers.
The nucleotide sequences reported in this paper have been deposited in the GenBank database under accession numbers AF538364 and AF538365.
RESULTS
Features of pSym transconjugants and identification of RpSyms*.
To analyze the characteristics of pSym transconjugants, we performed crosses with R. etli CFN2001 by using two strains with different genetic labels on the pSym as donors. One of these strains was strain CFNX5, which carries an insertion of the GDYN-1 (Km/Gm Sp/Str) element in nifHc, one of the three reiterations of this gene (40), while the other, strain CFNX247, carries a ΩSp cassette inserted into nifA (17). Transconjugants were selected based on inheritance of these pSym localized markers. The plasmid patterns of the transconjugants were analyzed on Eckhardt-type gels. For both crosses the results showed (Table 2) that most of the transconjugants (∼80%) inherited p42a in addition to the pSym, while a smaller fraction (∼20%) apparently inherited only the pSym. Some of the transconjugants were analyzed further in order to determine the transfer frequency of the pSym, the compatibility with the wild-type pSym, and hybridization to a cosmid (C-13) which contains the tra and rep regions of p42a (47). All of the transconjugants analyzed which inherited only the pSym (seven transconjugants from conjugation 1 and two transconjugants from cross 2 [Table 2]) showed an increase (from 10−6 to ∼10−3) in the transfer frequency of the pSym to another recipient (A. tumefaciens strain GMI9023). Additionally, a strong hybridization signal of the pSym with cosmid C-13 was detected. Incompatibility properties were also affected, since the pSyms were able to coexist with a wild-type pSym (labeled with Tn5mob [44]) and were unable to coexist with a differently labeled wild-type p42a. Symbiotic plasmids with these characteristics were designated RpSyms*. On the other hand, analysis of the pSyms of transconjugants containing both the pSym and p42a (seven transconjugants from cross 1 and one transconjugant from cross 2) indicated that they were transferred at the same frequency as previously detected (in the range of 10−6), were unable to coexist with another, differently labeled pSym, and showed no difference in hybridization with p42a (data not shown).
TABLE 2.
Features of pSym transconjugants
| Donor | Recipient | No. of pSym transconjugants containing pSym and p42a/total no. (%) | No. of pSym transconjugants containing RpSym*/total no. (%)a |
|---|---|---|---|
| CFNX5 (nifHc::GDYN) | CFN2001 (p42a− p42d−) | 82/93 (88) | 11/93 (12) |
| CFNX247 (nifA::ΩSp) | CFN2001 (p42a− p42d−) | 64/85 (76) | 21/85 (24) |
RpSym* was defined by its high transfer frequency, compatibility with wild-type pSym, and hybridization to p42a tra genes.
These results indicate that the RpSyms* had integrated segments of p42a, which included the genes that participate in conjugative transfer and the origin of replication, and probably did not some contain segments of the pSym, as they were usually the size of a wild-type pSym.
Mapping of RpSyms*.
To determine which segments of the pSym were conserved in the RpSyms*, we transferred two RpSyms* (from strains CFNX643 and CFNX644) and the wild-type pSym from strain CFNX645 to the plasmidless A. tumefaciens strain GMI9023. The three plasmids were purified from this background by employing the method described by Hirsch et al. (22). Each of the purified plasmids was digested with BamHI, blotted onto nylon membranes, and hybridized with the ordered set of cosmids covering the whole pSym, whose physical map contains 85 BamHI bands (18). These experiments allowed us to determine which segments of the RpSyms* differed from the wild-type pSym. Subsequently, Agrobacterium transconjugants containing the RpSyms* from eight other strains were constructed (Table 1). DNA from all these strains were hybridized with individual BamHI bands from the cosmids that showed differences compared to the wild type, which allowed us to further delimit the pSym borders. A similar experiment was done to find out which segments of p42a were present in the RpSyms*. Unfortunately, the set of cosmids from p42a did not cover the whole plasmid, and only some cosmids were ordered. Nevertheless, we could identify the band that joins p42a to the pSym in most of them. The results are shown in Fig. 1.
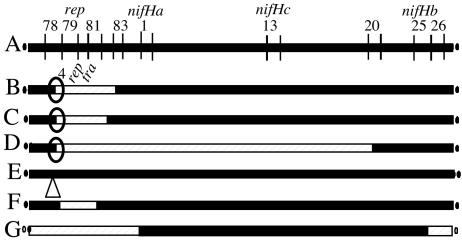
FIG. 1.
Structural analysis of RpSyms*. The pSym is represented by solid lines, and p42a is represented by cross-hatched lines. (A) Map of the wild-type pSym. The numbers represent BamHI fragments; the localization of the three nifH reiterations and the localization of the replication region are indicated. (B to G) Structure of 10 different RpSyms*, indicating BamHI bands deleted in the pSym and substituted by p42a. (B) RpSyms* lacking sequences from the end of band 78 to band 83 of the pSym. The relative localization of the rep and tra regions is indicated. The strains examined were GMI9023/p643, GMI9023/p646, GMI9023/p648-1 (this strain additionally had a duplication of the nifHa-nifHb region), GMI9023/p648-2, and GMI9023/p649. (C) RpSym* lacking sequences from the end of band 78 to the middle of band 82. The strain examined was GMI9023/p651-1. (D) RpSym* lacking sequences from the end of band 78 to band 20. The strain examined was GMI9023/p644. (E) RpSym* containing the whole pSym, with p42a integrated in band 78. The strain examined was GMI9023/p651-2. (F) RpSym* lacking bands 79 to 81. The strain examined was GMI9023/p652-1. (G) RpSym* lacking bands 26 to 1. The strain was GMI9023/p652-2. Similar endpoints for the pSym-p42a border are indicated by ellipses; the number 4 in panel B refers to EcoRI band 4 mentioned in the text. The maps are not drawn to scale.
The results show that the (arbitrarily designed) left borders of most RpSyms* (7 of 10 plasmids) had the same endpoint for splicing of pSym and p42a, which is localized in pSym BamHI band 78 and p42a EcoRI band 4 (Fig. 1B, C, and D). The relative localization of the p42a rep and tra genes is indicated in Fig. 1B. The RpSym* p648-1 additionally had a duplication of the 120-kb region bordered by nifHa and nifHb reiterations. This type of amplification has been described previously (39). The RpSym* shown in Fig. 1E (p651-2) also had the pSym joined to p42a in pSym band 78, but we were not able to identify the p42a border; this plasmid still contained the whole pSym sequence. This is the only RpSym* that remained incompatible with a wild-type pSym, in agreement with the fact that it retained the plasmid's replication zone, localized in BamHI band 79. The RpSym* of strain GMI9023/p652-1 (Fig. 1F) contained a complete band 78 but lacked band 79.
The right border of the p42a-pSym junction varied more in the different RpSyms*, although five of these plasmids (Fig. 1A) also had a similar endpoint in the pSym. The RpSym* of strain GMI9023/p652-2 (Fig. 1G) was very different, as only the region of the pSym bordered by the nifHa and nifHb reiterations was conserved in this plasmid.
An example of how we obtained the map of the RpSym* presented in Fig. 1B is shown in Fig. 2. Figures 2A and B show the hybridization of a wild-type pSym and the RpSym* of GMI9023/p643 to cosmids 47 and 7, which were two of the overlapping cosmids covering the pSym (18). Cosmid 47 contained BamHI bands 73 to 80, and cosmid 7 contained bands 80 to 85, 1, and 2. The results show that the RpSym* differed from the wild-type plasmid. While bands 73, 74, 75, 76, 77 (cosmid 47) and bands 84, 85, 1, and 2 (cosmid 7) were present, band 78 appeared to be smaller (indicated by 78*). Band 79 (cosmid 47) and bands 81 and 82 (cosmid 7) were absent, while the presence of bands 80 and 83 was not easily discerned. In order to determine if the actual pSym bands were present or were masked by hybridization to reiterations localized elsewhere, we performed hybridization experiments using the purified bands as probes. Figures 2C, D, E, and F show the hybridization to bands 79, 80, 83, and 84, respectively. The results show that bands 79, 80, and 83 were absent in the RpSym*, although reiterations were still present. Band 84 was similar to the wild-type band. Figure 2H shows the hybridization to band 78 of the pSym. The results confirm that band 78 was truncated in the RpSym* analyzed (designated band 78*). This truncated band 78 was also found to hybridize with EcoRI band 4 of p42a (Fig. 2G). The size of band 4 in the RpSym* (designated band 4*) was greater than the size of the wild-type band localized in p42a. The controls analyzed allowed us to determine the presence of reiterations of p42a EcoRI band 4 localized elsewhere in the R. etli genome, as well as in the A. tumefaciens genome. The data show that the RpSym* plasmids of strains of the type represented by CFNX643 lost a segment of pSym band 78 and the region from band 79 to band 83 and retained a wild-type band 84. Additionally, the truncated band 78* was shown to be spliced to p42a, as it was able to hybridize to a probe from this plasmid.
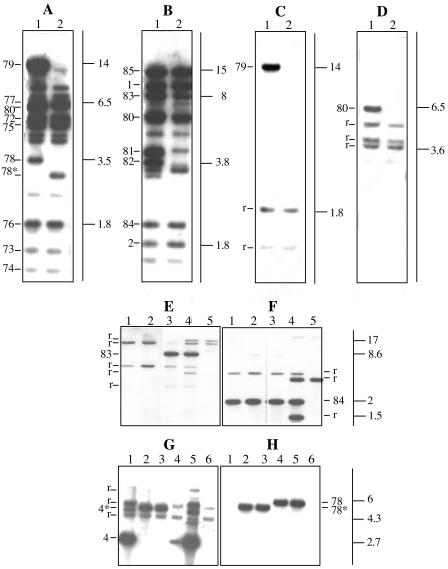
FIG. 2.
Southern hybridization of restricted DNA from different strains with whole cosmids and individual bands from the pSym and p42a. The probes used were pSym cosmid 47 (A), pSym cosmid 7 (B), pSym BamHI band 79 (C), pSym BamHI band 80 (D), pSym BamHI band 83 (E), pSym BamHI band 84 (F), p42a EcoRI band 4 (G), and pSym BamHI band 78 (H). (A to D) BamHI-digested DNA from wild-type pSym from GMI9023/p645 (lane 1) and RpSym* from GMI9023/p643 (lane 2). (E and F) BamHI-digested DNA from RpSym* from GMI9023/p646 (lane 1), RpSym* from GMI9023/p643 (lane 2), wild-type pSym from GMI9023/p42d (lane 3), wild-type CFN42 (lane 4), and p42a from GMI9023/p42a (lane 5). (G and H) EcoRI-digested DNA from wild-type p42a from GMI9023/p42a (lane 1), RpSym* from GMI9023/p646 (lane 2), RpSym* from GMI9023/p643 (lane 3), wild-type pSym from GMI9023/p42d (lane 4), wild-type CFN42 (lane 5), and GMI9023 (lane 6), r, reiteration. The sizes of markers (in kilobases) are indicated on the right in all of the panels.
Cloning and sequence analysis of the left pSym-p42a border of the most frequently found RpSym*.
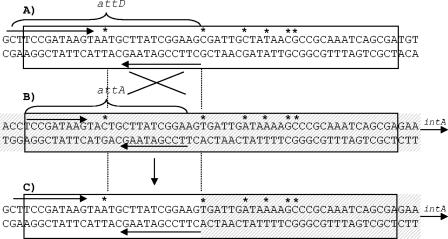
In order to clone the border region of the pSym and p42a plasmids found in most of the RpSyms*, we obtained a PCR product using one oligonucleotide corresponding to band 78 of the pSym and one oligonucleotide from band 4 of p42a. To design the oligonucleotide for the pSym, we used the sequence of this plasmid (accession number NC_004041). Band 4 of p42a was cloned in pSK and was sequenced with universal oligonucleotide primers. The sequence obtained was used to design custom-made oligonucleotide primers. A 760-bp PCR product was obtained by using the primers mentioned above and DNA of strain CFNX643 as the template. This PCR product hybridized with band 78 of the wild-type pSym, band 4 of the wild-type p42a, truncated band 78*, and the larger band 4* in the RpSym* of CFNX643 (data not shown). A similar product was obtained by using DNA from strains GMI9023/p646, GMI9023/p648-1, GMI9023/p648-2, GMI9023/p649, GMI9023/p651-1, and GMI9023/p644, confirming that the left pSym-p42a borders were similar in the RpSyms* of all these strains (data not shown). The PCR product of strain CFNX643 was cloned as described in Materials and Methods. Its complete sequence was obtained (accession number AF538365) and compared to the sequences of pSym band 78 (accession number NC_004041) and the sequence obtained from p42a band 4 (accession number AF538364). The results (Fig. 3) showed the presence of a 53-bp region that was similar in the pSym and p42a; this region overlapped a 27-bp sequence from the pSym, designated attD, and a 23-bp sequence from p42a, designated attA. attD consisted of a 5-bp central region flanked by an 11-bp inverted repeat, while attA contained a 5-bp central region flanked by a 9-bp inverted repeat. Six of the 53 bases were different in the two plasmids, and one of them was in the central region bound by the inverted repeat. The structural organization of the attD and attA sequences was highly reminiscent of bacterial and phage attachment sites (20). The sequence of the RpSym* was identical to the pSym sequence in the 27-bp attD region and to the p42a sequence in the rest of the homologous region. These results indicate that the RpSym* was probably generated by recombination between the pSym attD and p42a attA sites in the interval between the 12th base of the homologous region (A in pSym and RpSym* and C in p42a) and base 26 (C in the pSym and T in p42a and RpSym*).
FIG. 3.
Sequence of the left pSym-p42a border in most RpSyms* and the attachment-like sequences shared by the pSym and p42a. (A) pSym; (B) p42a; (C) RpSym* from CFNX643. The sequence of p42a is indicated by cross-hatching, the homologous region between the pSym and p42a is enclosed in a box, and the inverted repeats are underlined with arrows; asterisks indicate the bases that are different in the two plasmids. The possible recombination region is indicated by two crossed lines framed by dotted lines. The locations of the attD and attA sites are indicated. The relative location of the open reading frame encoding a putative integrase (intA) is also indicated.
Sequence analysis of 1,322 bp from R. etli plasmid p42a containing the intA gene.
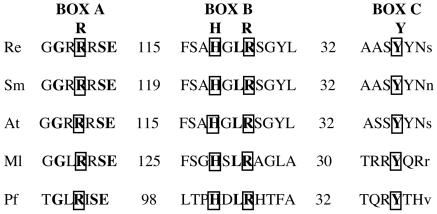
Computer analysis predicted an open reading frame (ORF1) that was 1,251 bp long downstream of the attA site of p42a (accession number AF538364). Two potential start codons preceded by a putative ribosome binding site were identified at positions 108 (UUG) and 165 (UUG). The deduced amino acid sequence encoded by ORF1 (416 amino acids) showed 80% identity over the entire length of the protein to a putative phage-related integrase of A. tumefaciens encoded in plasmid pATC58 (52) (NCBI accession number AAL45708). The S. meliloti integrase-like protein encoded in plasmid pSymA had the second-highest level of homology (65% identity) (2) (NCBI accession number AAK65874). The levels of identity to the putative integrases encoded in plasmids of M. loti (26) (NCBI accession number BAB54967), Coxiella burnetii (S. Lautenschlaeger et al., unpublished data) (NCBI accession number CAA75853), and Pseudomonas sp. (B. M. Martínez et al., unpublished data) (NCBI accession number AAK50335) were low (30%). Through a conserved domains search (NCBI), we found that the C-terminal region of the protein encoded by ORF1 aligned very well with the catalytic domains (box A, box B, and box C) of a representative consensus sequence for 38 members of the phage integrase family (Pfam00589). The members of this family include integrases like Cre (phage P1), XerC, XerD, and FimB (E. coli). Based on these similarities, we designated this ORF1 intA. The alignment shown in Fig. 4 revealed that R. etli IntA and the integrases found in other members of the Rhizobiaceae family contain the invariant catalytic site residues (R-H-R-Y) shared by all the tyrosine recombinases (11). This alignment also showed that the spacing between the two conserved arginine residues varies from 98 amino acids for the Pfam consensus sequence to 125 amino acids for the M. loti integrase. Similar variability was found in the alignment of 81 integrases reported by Esposito and Scocca (11). In contrast, the distance between the second conserved arginine and the invariant tyrosine varies from 30 to 32 amino acids, a range consistently observed in the majority of prokaryotic integrases (11). The results suggest that the specific recombination between the pSym and p42a may be mediated by the putative integrase IntA.
FIG. 4.
Alignment of the conserved regions (boxes A, B, and C) of tyrosine recombinases from R. etli (Re) (accession number AF538364), S. meliloti (Sm) (accession number AAK65874), A. tumefaciens (At) (accession number AAL45708), and M. loti (Ml) (accession number BAB54967) and a representative consensus sequence for 38 members of the phage integrase family Pfam00589 (Pf) (accession number PF00589). The invariant catalytic site residues (R-H-R-Y) are enclosed in boxes, and the length of the space between these residues is indicated by numbers. Residues that are identical in all five sequences are indicated by boldface type, and residues that are identical in >50% of the sequences are indicated by normal uppercase letters. Lowercase letters indicate residues that are identical in <50% of the sequences.
Transfer of the pSym requires cointegration with p42a.
Data from this and previous work (7) suggest that cointegration of the pSym with p42a is required for conjugative transfer of the pSym. We have shown that a pSym labeled with Tn5 is able to perform conjugative transfer in the presence of a self-transmissible cosmid (pC-13) derived from p42a. This cosmid carries the whole transfer region but lacks the attA and intA sequences. All the transconjugants analyzed contained cosmid pC-13, which either formed a cointegrate with the pSym or was an independent replicon (48). We repeated this experiment using a recA strain as the receptor (Table 3). The transconjugants were selected for the presence of the pSym localized marker; nevertheless, we found that all of them contained a stable pC-13-pSym cointegrate. This finding was supported by the following evidence: (i) all transconjugants were Tcr (this marker was localized on pC-13 in 100 colonies analyzed), and (ii) Southern blots of Eckhardt-type gels of the transconjugants (20 colonies analyzed) showed that the same plasmid hybridized with a probe of the pSym and a probe of pC-13 (data not shown). Finally, we tested whether pC-13 could promote transfer of a Tn5-labeled pSym when the donor was recA. In this case, we could not detect transfer of the pSym (Table 3). Our interpretation of the experiments described above is as follows. pC-13 and the pSym share the traA, traCDG, and repABC genes. RecA-dependent homologous recombination among these sequences allows cointegration of the pSym with pC-13, and the cointegrate is able to perform conjugative transfer. This mechanism requires homologous recombination and consequently does not operate in the absence of RecA.
TABLE 3.
Transfer frequency of R. etli plasmids pSym and p42a from different donorsa
| Donor | Relevant genotype | Transfer frequencyb
|
|
|---|---|---|---|
| pSym | p42a | ||
| CFNX192/pC-13 | pSym::Tn5, p42a tra region | 3 × 10−6 | —c |
| CFNX653/pC-13 | recA derivative of CFNX192, p42a tra region | NDd | — |
| CFNX653 | recA derivative of CFNX192 | 6.2 × 10−6 | — |
| CFNX661 | intA::pJQ200mp18 derivative of CFNX653 | 1 × 10−9 | 3 × 10−1 |
| CFNX662 | CFNX661 complemented with pAGS4 carrying wild-type intA | 1.7 × 10−6 | 3 × 10−1 |
| CFNX663 | intA::ΩSpr derivative of CFNX653 | ND | 3 × 10−1 |
| CFNX664 | CFNX663 complemented with pAGS4 carrying wild-type intA | 2.9 × 10−7 | 2.7 × 10−1 |
| CFNX661/pBBR-Tc | CFNX661 with vector pBBRMCS3 | ND | 5.3 × 10−2 |
| CFNX663/pBBR-Tc | CFNX663 with vector pBBRMCS3 | ND | 6 × 10−1 |
All crosses were repeated at least three times. Strain UIA143 was always used as the receptor.
Expressed as the number of transconjugants per donor.
—, not determined.
ND, not detected (transfer frequency, <1 × 10−9).
IntA is required for RecA-independent cointegration of the pSym with p42a.
We constructed two derivatives of CFN42 containing a p42a which carried an interrupted intA gene and a Tn5-labeled pSym (see Materials and Methods). The mutations disrupting intA drastically decreased conjugative transfer of the labeled pSym in the absence of RecA (Table 3). Complementation of the intA mutant with the wild-type gene restored the phenotype, while introduction of the vector without the intA gene did not restore the phenotype. The transfer frequency of p42a was not affected by the mutation in intA (Table 3). Utilization of a recA strain as the receptor in the crosses in which the complemented strains were used as donors allowed us to confirm that the transconjugants contained a stable cointegrate of the pSym with p42a (data not shown). These data strongly support the hypothesis that the RecA-independent cointegration mechanism involves site-specific recombination at the att sites, mediated by the product of intA. The fact that a mutation in intA prevents transfer of the pSym in the recA donor confirms its participation in the cointegration event.
Construction and analysis of derivatives simultaneously containing a wild-type plasmid and an RpSym*.
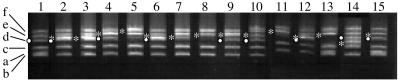
The RpSyms* from six strains, including CFNX646, CFNX647, CFNX648, CFNX649, CFNX650, and CFNX652, were introduced by conjugation into strain CFNX653 (recA pSym::Tn5mob). Figure 5 shows the plasmid patterns of the parental strains containing either a wild-type pSym from strain CFNX653 (lanes 1 and 15) or an RpSym* from strain CFNX646, CFNX647, CFNX649, CFNX648, CFNX650, or CFNX652 (lanes 3, 5, 7, 8, 11, and 13) and the plasmid patterns of the derivatives containing both pSyms from transconjugants CFNX654, CFNX657, CFNX658, CFNX655, CFNX656, CFNX659, and CFNX660 (lanes 2, 4, 6, 9, 10, 12, and 14). All of the derivatives contained the wild-type pSym from strain CFNX653; additionally, strain CFNX654 contained the RpSym* from CFNX646, strain CFNX657 contained the RpSym* from CFNX647, strain CFNX658 contained the RpSym* from CFNX649, strains CFNX655 and CFNX656 contained the RpSym* from CFNX648, strain CFNX659 contained the RpSym* from CFNX650, and strain CFNX660 contained the RpSym* from CFNX652. Variations in RpSym* size can be appreciated. Also, the RpSym* bands tended to be more intense than the wild-type pSym bands; this may have reflected a higher copy number of these plasmids. Most of the strains containing either an RpSym* or the two pSyms lacked p42a; the only exception was CFNX652 (lane 13), which still contained the replication machinery of the pSym. New variations in RpSym* size were generated in the crosses used for construction of the strains containing the two pSyms; lane 10 shows a larger RpSym* migrating above p42f, and a smaller RpSym* migrating below the wild-type pSym is shown in lane 14. These new variations could be due to amplifications or deletions of different regions of the plasmid, generated by a mechanism described previously (40), in the intraplasmid reiterations present in each of the replicons or in the repeated sequences shared by the pSym and p42a.
FIG. 5.
Plasmid patterns of CFN42 derivatives with an RpSym* and constructions simultaneously containing a wild-type pSym and an RpSym*. a to f indicate the positions of p42a (195 kb), p42b (185 kb), p42c (270 kb), p42d (pSym, 371 kb), p42e (500 kb), and p42f (600 kb), respectively. The position of wild-type pSym is indicated by white dots, and the position of RpSym* is indicated by asterisks. Lane 1, CFNX653; lane 2, CFNX654; lane 3, CFNX646; lane 4, CFNX657; lane 5, CFNX647; lane 6, CFNX658; lane 7, CFNX649; lane 8, CFNX648; lane 9, CFNX655; lane 10, CFNX656; lane 11, CFNX650; lane 12, CFNX659; lane 13, CFNX652; lane 14, CFNX660; lane 15, CFNX653.
The symbiotic features of three of the strains (Fig. 5, lanes 2, 9, and 10) were analyzed and compared to the features of their parental strains (lanes 1, 3, and 8), as shown in Table 4. The results show that in the derivatives containing only an RpSym* there was an increase in specific nitrogenase activity; however, this increase was not reflected in the plant shoot dry weight. These results could have been due to an increased copy number of the RpSym* (see above), or alternatively, the increased nitrogen fixation activity may have reflected the loss of a repressor activity in the RpSyms*. In support of the latter interpretation, in derivatives that simultaneously contained a wild-type plasmid and an RpSym* (CFNX654 and CFNX655) there was a decrease in nitrogenase activity. Interestingly, although transconjugant CFNX656 harbored a wild-type plasmid and an RpSym*, there was a severe reduction in nitrogenase activity in this strain. This strain also had an uncharacterized rearrangement that increased the size of the RpSym*. It is not known if the reduction in nitrogen fixation was due solely to the increase in the size of the RpSym*.
TABLE 4.
Nitrogen fixation of derivatives with a wild-type plasmid and an RpSym*a
| Strain | Relevant genotype | Total nitrogenase activity (μmol of ethylene/plant/h)b | Specific nitrogenase activity (μmol of ethylene/g of nodules/h)b | Shoot dry wt weight (g/plant)b |
|---|---|---|---|---|
| CFN42 | Wild-type pSym | 2.09 ± 0.67 | 25.8 ± 10.2 | 0.41 ± 0.08 |
| CFNX646 | RpSym* | 3.28 ± 0.53 | 44.9 ± 6.2 | 0.46 ± 0.05 |
| CFNX648 | RpSym* | 2.60 ± 0.63 | 45.1 ± 15.8 | 0.46 ± 0.05 |
| CFNX654 | Wild-type pSym + RpSym* | 1.17 ± 0.82 | 20.7 ± 14.4 | 0.46 ± 0.09 |
| CFNX655 | Wild-type pSym + RpSym* | 1.63 ± 0.82 | 34.6 ± 15.8 | 0.42 ± 0.08 |
| CFNX656 | Wild-type pSym + RpSym* | 0.06 ± 0.03 | 2.6 ± 1.4 | 0.37 ± 0.06 |
| CFNX653 | Wild-type pSym | 0.87 ± 0.30 | 14.7 ± 7.5 | 0.43 ± 0.13 |
Preparations were analyzed 32 days after inoculation.
The values are means ± standard deviations.
DISCUSSION
Cointegration among plasmids from gram-negative bacteria is usually carried out through RecA-dependent recombination among homologous sequences or through specific cointegration involving transposon or insertion sequences (32). Many examples have been found in E. coli plasmids (32, 33). Phage-like systems are characteristic of conjugative transposons (32). The symbiotic island of M. loti may be considered a large (500-kb) conjugative transposon (47). Some small plasmids from gram-positive bacteria, such as pSAM2 (11 kb), pSE101 (11.3 kb), and pMEA300 (13.3 kb), are also able to cointegrate by using a phage-like system (3, 8, 50). A recent review proposed that pSAM2 should not be considered a genuine plasmid but is a site-specific integrative and conjugative element, because although it is able to replicate, the replication of the circular form is not involved in the maintenance of the element but is necessary for its transfer (9) Plasmid cointegration among large plasmids from the Rhizobiaceae (A. tumefaciens, Agrobacterium rhizogenes, and R. leguminosarum) has been proposed to occur through the same mechanisms described for E. coli plasmids: homologous RecA-dependent recombination or specific cointegration with transposon or insertion sequences (25, 49, 53).
In this paper, we describe a phage-like system, localized in two large plasmids (194 kb for p42a and 371 kb for the pSym), in the gram-negative bacterium R. etli.
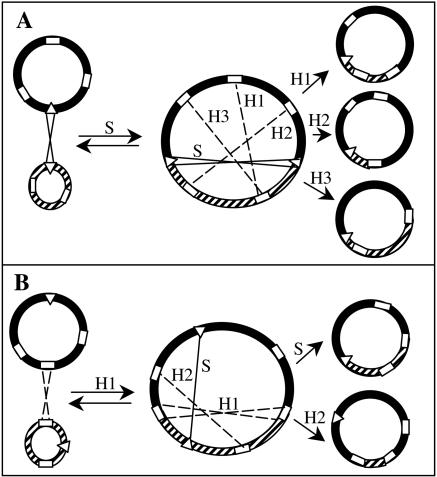
The data presented here show that conjugative transfer of the pSym from R. etli CFN42 depends on its cointegration with p42a, and the results allowed us to propose a model for the mechanism responsible for this cointegration event and for the generation of RpSyms*. A diagram of this model is shown in Fig. 6. A specific recombination event between the unique attachment-like sites present on the pSym and p42a leads to cointegration of both plasmids, which is mediated by the product of the intA gene localized downstream of the p42a attachment site. The pSym-p42a cointegrate is able to perform conjugative transfer. In most cases, resolution of this cointegrate is mediated through specific recombination at the same site, regenerating the wild-type symbiotic and p42a plasmids. In a few cases, the cointegrate may be resolved through recombination among some of the other sequences, which are reiterated in the pSym and p42a (18), thus generating the RpSyms* (Fig. 6A). These RpSyms* all share one endpoint of the pSym-p42a junction. Alternatively, cointegration of p42a and p42d may occur through recombination at large homologous sequences in a RecA-dependent mechanism (Fig. 6B). When the cointegrate is resolved through recombination at the same sequences that generated it, the wild-type plasmids are recovered, but if recombination occurs through the site-specific mechanism or through other homologous sequences, RpSyms* are also generated. The pSym map contains 85 BamHI bands, 19 of which are reiterated on the p42a map (18). In support of our proposition, we found that the right endpoints of the different RpSyms* are always located in bands containing some of these reiterations. The sequence of the whole pSym (19) indicates that most of the reiterations correspond to insertion sequences. In this work we found that transfer of pSym can occur only in the presence of a functional recombination system (either RecA or IntA dependent).
FIG. 6.
Model for the mechanism of pSym conjugative transfer and generation of RpSyms*. The pSym is indicated by solid lines, and p42a is indicated by cross-hatched lines. Triangles represent the attA and attD sequences, and open rectangles represent large sequences shared by both plasmids. Solid lines and arrows labeled with the letter S indicate site-specific recombination among attachment-like sequences. Dashed lines and arrows labeled with the letter H followed by a number indicate homologous recombination among large repeated sequences. (A) When cointegration is the result of site-specific recombination, resolution through the same system regenerates the wild-type plasmids (S), but if the cointegrate is resolved through homologous recombination among other sequences (H1, H2, or H3), RpSyms* are generated. (B) Cointegrates formed and resolved through homologous recombination between two repeated sequences regenerate the wild-type plasmids (H1) but give rise to RpSyms* when they are resolved through homologous recombination among different repeats (H2) or through site-specific recombination (S).
The model proposed above suggests a very interesting mechanism for the conjugative transfer of a symbiotic plasmid, which involves a specific recombination event. Thus, pSym transfer would be subject to very tight regulation, with at least two levels of control: factors affecting cointegration between pSym and p42a and quorum-sensing regulation of p42a tra genes (48). Interestingly, in 1982 Brewin et al. (5) reported transfer of nod and nif genes from a nontransmissible symbiotic plasmid from R. leguminosarum through cointegration with a transmissible plasmid (5). These findings could have been the result of a mechanism similar to the one proposed here for R. etli. Also, integrase-like open reading frames have been described in various rhizobial strains (2, 26, 52). Interestingly, the orthologous genes found in different members of the Rhizobiaceae family, in Pseudomonas sp., and in C. burnetii are also plasmid borne. Studies regarding the cointegration of these plasmids have not been reported, and the putative integrases remain uncharacterized.
Finally, this model has direct evolutionary implications, as the resolution of cointegrates through sites different from those leading to their formation results in the generation of symbiotic plasmids containing novel sequence combinations.
Acknowledgments
We are grateful to Laura Cervantes and Javier Rivera for excellent technical assistance, to Rosa Isela Santamaría and Mariana Canedo for DNA sequencing, and to Paul Gaytán and Eugenio López for synthesis of oligonucleotides.
This work was supported in part by grants IN202599 and IN226802 from DGAPA, UNAM.
REFERENCES
- 1.Altschul, S. F., T. L. Madden, A. A. Schäffer, J. Zhang, Z. Zhang, W. Miller, and D. J. Lipman. 1997. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25:3389-3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Barnett, M. J., R. F. Fisher, T. Jones, C. Komp, A. P. Abola, F. Barloy-Hubler, L. Bowser, D. Capela, F. Galibert, J. Gouzy, M. Gurjal, A. Hong, L. Huizar, R. Palm, M. C. Peck, R. Surzycki, D. H. Wells, K. C. Yeh, R. W. Davis, N. A. Federspiel, and S. R. Long. 2001. Nucleotide sequence and predicted functions of the entire Sinorhizobium meliloti pSymA megaplasmid. Proc. Natl. Acad. Sci. USA 98:9883-9888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Boccard, F., T. Smokvina, J. L. Pernodet, A. Friedmann, and M. Guérineau. 1989. Structural analysis of loci involved in pSAM2 site-specific integration in Streptomyces. Plasmid 21:59-70. [DOI] [PubMed] [Google Scholar]
- 4.Brewin, N. J., J. E. Beringer, A. V. Buchanan-Wollaston, A. W. B. Johnston, and P. R. Hirsch. 1980. Transfer of symbiotic genes with bacteriocinogenic plasmids in Rhizobium leguminosarum. J. Gen. Microbiol. 116:261-270. [Google Scholar]
- 5.Brewin, N. J., E. A. Wood, A. W. B. Johnston, N. J. Dibb, and G. Hombrecher. 1982. Recombinant nodulation plasmids in Rhizobium leguminosarum. J. Gen. Microbiol. 128:1817-1827. [Google Scholar]
- 6.Brom, S., A. García de los Santos, T. Stepkowsky, M. Flores, G. Dávila, D. Romero, and R. Palacios. 1992. Different plasmids of Rhizobium leguminosarum bv. phaseoli are required for optimal symbiotic performance. J. Bacteriol. 174:5183-5189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Brom, S., A. García de los Santos, L. Cervantes, R. Palacios, and D. Romero. 2000. In Rhizobium etli symbiotic plasmid transfer, nodulation competivity and cellular growth require interaction among different replicons. Plasmid 44:34-43. [DOI] [PubMed] [Google Scholar]
- 8.Brown, D. P., K. B. Idler, D. M. Backer, S. Donadio, and L. Katz. 1994. Characterization of the genes and attachment sites for site-specific integration of plasmid pSE101 in Saccharopolyspora erythrea and Streptomyces lividans. Mol. Gen. Genet. 242:185-193. [DOI] [PubMed] [Google Scholar]
- 9.Burrus, V., G. Pavlovic, B. Decaris, and G. Guédon. 2002. Conjugative transposons: the tip of the iceberg. Mol. Microbiol. 46:601-610. [DOI] [PubMed] [Google Scholar]
- 10.Eckhardt, T. 1978. A rapid method for the identification of plasmid deoxyribonucleic acid in bacteria. Plasmid 1:584-588. [DOI] [PubMed] [Google Scholar]
- 11.Esposito, D., and J. J. Scocca. 1997. The integrase family of tyrosine recombinases: evolution of a conserved active site domain. Nucleic Acids Res. 25:3605-3614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Farrand, S. K., S. P. O'Morchoe, and J. McCutchan. 1989. Construction of an Agrobacterium tumefaciens C58 recA mutant. J. Bacteriol. 171:5314-5321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fellay, R., J. Frey, and H. Krisch. 1987. Interposon mutagenesis of soil and water bacteria: a family of DNA fragments designed for in vitro insertional mutagenesis of Gram-negative bacteria. Gene 52:147-154. [DOI] [PubMed] [Google Scholar]
- 14.Figurski, D. H., and D. R. Helinski. 1979. Replication of an origin containing derivative of plasmid RK2 dependent on a plasmid function provided in trans. Proc. Natl. Acad. Sci. USA 76:1648-1652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Freiberg, C., R. Fellay, A. Bairoch, W. J. Broughton, A. Rosenthal, and X. Perret. 1997. Molecular basis of symbiosis between Rhizobium and legumes. Nature 387:394-401. [DOI] [PubMed] [Google Scholar]
- 16.García de los Santos, A., S. Brom, and D. Romero. 1996. Rhizobium plasmids in bacteria-legume interactions. World J. Microbiol. Biotechnol. 12:119-125. [DOI] [PubMed] [Google Scholar]
- 17.Girard, L., B. Valderrama, R. Palacios, D. Romero, and G. Dávila. 1996. Transcriptional activity of the symbiotic plasmid of Rhizobium etli is affected by different environmental conditions. Microbiology 142:2847-2856. [Google Scholar]
- 18.Girard, M. L., M. Flores, S. Brom, D. Romero, R. Palacios, and G. Dávila. 1991. Structural complexity of the symbiotic plasmid of Rhizobium leguminosarum bv. phaseoli. J. Bacteriol. 173:2411-2419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.González, V., P. Bustos, M. A. Ramírez-Romero, A. Medrano-Soto, H. Salgado, I. Hernández-González, J. C. Hernández-Celis, V. Quintero, G. Moreno-Hagelsieb, L. Girard, O. Rodríguez, M. Flores, M. A. Cevallos, J. Collado-Vides, D. Romero, and G. Dávila. 2003. The mosaic structure of the symbiotic plasmid of Rhizobium etli CFN42 and its relation to other symbiotic genome compartments. Genome Biol. 4:R36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gottfried, P., E. Yagil, and M. Kolot. 2000. Core-binding specificity of bacteriophage integrases. Mol. Gen. Genet. 263:619-624. [DOI] [PubMed] [Google Scholar]
- 21.He, X., W. Chang, D. L. Pierce, L. O. Seib, J. Wagner, and C. Fuqua. 2003. Quorum sensing in Rhizobium sp. strain NGR234 regulates conjugal transfer (tra) gene expression and influences growth rate. J. Bacteriol. 185:809-822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hirsch, P. R., M. Van Montagu, A. W. B. Johnston, N. J. Brewin, and J. Schell. 1980. Physical identification of bacteriocinogenic, nodulation and other plasmids in strains of R. leguminosarum. J. Gen. Microbiol. 120:403-412. [Google Scholar]
- 23.Hynes, M. F., and N. F. McGregor. 1990. Two plasmids other than the nodulation plasmid are necessary for formation of nitrogen-fixing nodules by Rhizobium leguminosarum. Mol. Microbiol. 4:567-574. [DOI] [PubMed] [Google Scholar]
- 24.Johnston, A. W. B., G. Hombrecher, N. J. Brewin, and M. C. Cooper. 1982. Two transmissible plasmids in Rhizobium leguminosarum strain 300. J. Gen. Microbiol. 128:85-93. [Google Scholar]
- 25.Jouanin, L., J. Tourneur, C. Tourneur, and F. Casse-Delbart. 1986. Restriction maps and homologies of the three plasmids of Agrobacterium rhizogenes strain A4. Plasmid 16:124-134. [DOI] [PubMed] [Google Scholar]
- 26.Kaneko, T., Y. Nakamura, S. Sato, E. Asamizu, T. Kato, S. Sasamoto, A. Watanabe, K. Idesawa, A. Ishikawa, K. Kawashima, T. Kimura, Y. Kishida, C. Kiyokawa. M. Kohara, M. Matsumoto, A. Matsuno, Y. Mochizuki, S. Nakayama, N. Nakazaki, S. Shimpo, M. Sugimoto, C. Takeuchi, M. Yamada, and S. Tabata. 2000. Complete genome structure of the nitrogen-fixing symbiotic bacterium Mesorhizobium loti. DNA Res. 7:331-338. [DOI] [PubMed] [Google Scholar]
- 27.Kovach, M. E., P. H. Elzer, D. S. Hill, G. T. Robertson, M. A. Farris, R. M. Roop, and K. M. Peterson. 1995. Four new derivatives of the broad-host-range cloning vector pBBRMCS, carrying different antibiotic-resistance cassettes. Gene 166:175-176. [DOI] [PubMed] [Google Scholar]
- 28.Leemans, J., G. Soberón, M. A. Cevallos, L. Fernández, M. A. Pardo, H. de la Vega, M. Flores, C. Quinto, and R. Palacios. 1984. General organization in R. phaseoli nif plasmids, p. 710. In C. Veeger and W. E. Newton (ed.), Advances in nitrogen fixation research. Nijhoff, Junk and Pudoc, The Hague, The Netherlands.
- 29.Lithgow, J. K., A. Wilkinson, A. Hardman, B. Rodelas, F. Wisniewski-Dyé, P. Williams, and J. A. Downie. 2000. The regulatory locus cinR in Rhizobium leguminosarum controls a network of quorum-sensing loci. Mol. Microbiol. 37:81-97. [DOI] [PubMed] [Google Scholar]
- 30.Marketon, M. M., and J. E. González. 2002. Identification of two quorum-sensing systems in Sinorhizobium meliloti. J. Bacteriol. 184:3466-3475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Mercado-Blanco, J., and J. Olivares. 1993. Stability and transmissibility of the cryptic plasmids of Rhizobium meliloti GR4. Arch. Microbiol. 160:477-485. [Google Scholar]
- 32.Merlin, C., J. Mahillon, J. Nesvera, and A. Toussaint. 2000. Gene recruiters and transporters: the modular structure of bacterial mobile elements, p. 363-409. In C. M. Thomas (ed.), The horizontal gene pool. Harwood Academic Publishers, Amsterdam, The Netherlands.
- 33.Miller, C. A., and S. N. Cohen. 1980. F plasmid provides a function that promotes recA-independent site-specific fusions of pSC101 replicon. Nature (London) 285:577-579. [DOI] [PubMed] [Google Scholar]
- 34.Miller, J. H. 1972. Experiments in molecular genetics. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 35.Noel, K. D., A. Sánchez, L. Fernández, J. Leemans, and M. A. Cevallos. 1984. Rhizobium phaseoli symbiotic mutants with transposon Tn5 insertions. J. Bacteriol. 158:148-155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Pérez-Mendoza, D., A. Domínguez-Ferrer, S. Muñoz, M. J. Soto, J. Olivares, S. Brom, L. Girard, J. A. Herrera-Cervera, and J. Sanjuan. 2004. Identification of functional mob regions in Rhizobium etli: evidence for self-transmissibility of the symbiotic plasmid pRetCFN42d. J. Bacteriol 186:5753-5761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Quandt, J., and M. F. Hynes. 1993.Versatile suicide vectors which allow direct selection for gene replacement in Gram-negative bacteria. Gene 127:15-21. [DOI] [PubMed] [Google Scholar]
- 38.Quinto, C., H. de la Vega, M. Flores, L. Fernández, T. Ballado, G. Soberón, and R. Palacios. 1982. Reiteration of nitrogen fixation gene sequences in Rhizobium phaseoli. Nature (London) 299:724-728. [Google Scholar]
- 39.Romero, D., and S. Brom. 2004. The symbiotic plasmids of the Rhizobiaceae, p. 271-290. In B. E. Funell and G. E. Phillips (ed.), Plasmid biology. American Society for Microbiology, Washington, D.C.
- 40.Romero. D., S. Brom, J. Martínez-Salazar, L. Girard, R. Palacios, and G. Dávila. 1991. Amplification and deletion of a nod-nif region in the symbiotic plasmid of Rhizobium phaseoli. J. Bacteriol. 173:2435-2441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Romero, D., P. W. Singleton, L. Segovia, E. Morett, B. B. Bohlool, R. Palacios, and G. Dávila. 1988. Effect of naturally occurring nif reiterations on symbiotic effectiveness in Rhizobium phaseoli. Appl. Environ. Microbiol. 54:848-850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Rosenberg, C., and T. Hughet. 1984. The pATC58 plasmid of Agrobacterium tumefaciens is not essential for tumor induction. Mol. Gen. Genet. 196:533-536. [Google Scholar]
- 43.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 44.Simon, R. 1984. High frequency mobilization of gram-negative bacterial replicons by the in vitro constructed Tn5-Mob transposon. Mol. Gen. Genet. 196:413-420. [DOI] [PubMed] [Google Scholar]
- 45.Southern, E. M. 1975. Detection of sequences among DNA fragments separated by gel electrophoresis. J. Mol. Biol. 98:503-517. [DOI] [PubMed] [Google Scholar]
- 46.Sullivan, J. T., and C. W. Ronson. 1998. Evolution of rhizobia by acquisition of a 500 kb symbiosis island that integrates into a phe-tRNA gene. Proc. Natl. Acad. Sci. USA 95:5145-5149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Sullivan, J. T., J. R. Trzebiatowski, R. W. Cruickshank, J. Gouzy, S. D. Brown, R. M. Elliot, D. J. Fleetwood, N. G. McCallum, U. Rossbach, G. S. Stuart, J. E. Weaver, R. J. Webby, F. J. de Bruijn, and C. W. Ronson. 2002. Comparative sequence analysis of the symbiosis island of Mesorhizobium loti strain R7A. J. Bacteriol. 184:3086-3095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Tun-Garrido, C., P. Bustos, V. González, and S. Brom. 2003. The conjugative transfer of p42a from Rhizobium etli CFN42, which is required for mobilization of the symbiotic plasmid, is regulated by quorum sensing. J. Bacteriol. 185:1681-1692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Vaudequin-Dransart, V., A. Petit, W. S. Chilton, and Y. Dessaux. 1998. The cryptic plasmid of Agrobacterium tumefaciens cointegrates with the Ti plasmid and cooperates for opine degradation. Mol. Plant-Microbe Interact. 11:583-591. [Google Scholar]
- 50.Vrijbloed, J. W., J. Madon, and L. Dijkhizen. 1994. A plasmid from the methylotrophic actinomycete Amycolatopsis methanolica capable of site-specific integration. J. Bacteriol. 176:7087-7090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Wilkinson, A., V. Danino, F. Wisniewski-Dyé, J. K. Lithgow, and J. A. Downie. 2002. N-Acyl-homoserine lactone inhibition of rhizobial growth is mediated by two quorum-sensing genes that regulate plasmid transfer. J. Bacteriol. 184:4510-4519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Wood, D. W., J. C. Setubal, R. Kaul, D. Monks, J. P. Kitajima, V. K. Okura, Y. Zhou, L. Chen, G. E. Wood, N. F. Almeida, Jr., L. Woo, Y. Chen, I. T. Paulsen, J. A. Eisen, P. D. Karp, D. Bovee, Sr., P. Chapman, J. Clendenning, G. Deatherage, W. Gillet, C. Grant, T. Kutyavin, R. Levy, M. J. Li, E. McClelland, A. Palmieri, C. Raymond, G. Rouse, C. Saenphimmachak, Z. Wu, P. Romero, D. Gordon, S. Zhang, H. Yoo, Y. Tao, P. Biddle, M. Jung, W. Krespan, M. Perry, B. Gordon-Kamm, L. Liao, S. Kim, C. Hendrick, Z. Y. Zhao, M. Dolan, F. Chumley, S. V. Tingey, J. F. Tomb, M. P. Gordon, M. V. Olson, and E. W. Nester. 2001. The genome of the natural genetic engineer Agrobacterium tumefaciens C58. Science 294:2317-2323. [DOI] [PubMed] [Google Scholar]
- 53.Zhang, X.-X., B. Kosier, and U. B. Priefer. 2001. Symbiotic plasmid rearrangement in Rhizobium leguminosarum bv. viciae VF39SM. J. Bacteriol. 183:2141-2144. [DOI] [PMC free article] [PubMed] [Google Scholar]