Abstract
Adenosylcobalamin (Ado-B12) is both the cofactor and inducer of ethanolamine ammonia lyase (EA-lyase), a catabolic enzyme for ethanolamine. De novo synthesis of Ado-B12 by Salmonella enterica occurs only under anaerobic conditions. Therefore, aerobic growth on ethanolamine requires import of Ado-B12 or a precursor (CN-B12 or OH-B12) that can be adenosylated internally. Several known enzymes adenosylate corrinoids. The CobA enzyme transfers adenosine from ATP to a biosynthetic intermediate in de novo B12 synthesis and to imported CN-B12, OH-B12, or Cbi (a B12 precursor). The PduO adenosyl transferase is encoded in an operon (pdu) for cobalamin-dependent propanediol degradation and is induced by propanediol. Evidence is presented here that a third transferase (EutT) is encoded within the operon for ethanolamine utilization (eut). Surprisingly, these three transferases share no apparent sequence similarity. CobA produces sufficient Ado-B12 to initiate eut operon induction and to serve as a cofactor for EA-lyase when B12 levels are high. Once the eut operon is induced, the EutT transferase supplies more Ado-B12 during the period of high demand. Another protein encoded in the operon (EutA) protects EA-lyase from inhibition by CN-B12 but does so without adenosylation of this corrinoid.
The eut operon of Salmonella enterica encodes enzymes for use of ethanolamine as a carbon and nitrogen source (28, 29). Only 4 of its 17 genes (19, 33), eutD, eutE, eutB, and eutC, have been correlated directly with an enzymatic activity known to be required for ethanolamine utilization (9, 29). The operon and metabolic pathway are diagrammed in Fig. 1. A fifth gene, eutR, encodes a positive regulatory protein that activates transcription of the eut operon in response to the simultaneous presence of ethanolamine plus adenosylcobalamin (Ado-B12) (27, 32). The eutT and eutA genes described here have not been associated with any specific enzymatic function.
FIG. 1.
Pathway and operon for ethanolamine degradation. Ado-B12 serves as a coinducer of the operon and cofactor for EA-lyase. Degradation of ethanolamine provides both a carbon source and nitrogen source (30). Cbi (cobinamide) is a corrinoid lacking upper and lower ligands that can be adenosylated by CobA to produce Ado-Cbi, a biosynthetic precursor of B12. All eut genes are indicated above the map with the two normal promoters: the major regulated promoter (PI) and the minor constitutive eutR promoter (PII) (19, 27-29). Transposon-associated introduced promoters are below the map.
Use of ethanolamine as a carbon or nitrogen source requires Ado-B12, which is the cofactor of ethanolamine ammonia lyase (EA-lyase; EC 4.3.1.7) (2, 6, 8, 12, 28). Ado-B12 also serves as a coinducer (with ethanolamine) of eut operon transcription (18, 27, 32). A rationale for using Ado-B12 as a coinducer is the fact that operon expression is futile without Ado-B12 to support EA-lyase and this cofactor (unlike most others) is not always present. This is the case because S. enterica synthesizes Ado-B12 de novo only under anaerobic conditions (1, 15, 20, 30). Therefore under aerobic conditions, cells can grow on ethanolamine only if the environment can provide either Ado-B12 or a suitable precursor. Since Ado-B12 is subject to photolysis (4), exogenous precursors are likely to lack the upper (adenosyl) ligand and require internal adenosylation.
It is clear that S. enterica can add the upper adenosyl ligand to cobalamins since commercial CN-B12, which lacks this ligand, allows aerobic growth on ethanolamine (28). The CobA enzyme, ATP:cob(I)alamine adenosyl transferase, adenosylates assimilated CN-B12 and also contributes to de novo B12 synthesis (anaerobic) by adenosylating a biosynthetic intermediate (11, 34). CobA also adenosylates the assimilated precursor cobinamide (Cbi) and converts it to the biosynthetic intermediate Ado-Cbi (11). Another cobalamin transferase (PduO) is encoded within the pdu operon and is induced only during growth on propanediol; this transferase is not involved in any of the metabolism described here (17).
Existence of a third adenosyl transferase was suggested by the fact that CN-B12 allows a cobA mutant to use ethanolamine as a nitrogen source when glycerol is the carbon source (see Table 2), but not with glucose (11). This implied existence of another transferase, perhaps subject to catabolic repression. Evidence is presented that EutT is this transferase and contributes to ability of cells to grow on ethanolamine, especially with low levels of exogenous CN-B12. Another transferase candidate (EutA) helps cells resist inhibitory effects of CN-B12 on EA-lyase, but does so without converting CN-B12 to the normal cofactor Ado-B12.
TABLE 2.
Aerobic growth phenotypes of eut point and deletion mutants
| Strain | Relevant genotype
|
Eut(N) growth phenotypea
|
No. of mutations examinedb | |||
|---|---|---|---|---|---|---|
| eut | cobA | None | +CN−B12 (0.08 μM) | +Ado−B12 (0.08 μM) | ||
| TT20042 | eut+ | − | − | + | + | |
| TT10674 | + | − | + | + | ||
| TT20078 | eutT86(Am) | − | − | − | + | 9c |
| TT20079 | + | − | + | + | ||
| TT20026 | eutA111 | − | − | − | + | 2e |
| TT20025 | + | − | (−)d | + | ||
| TT20138 | eutPQTD333(Δ)f | − | − | − | + | 1 |
| TT20139 | + | − | + | + | ||
| TT20136 | eutDM302(Δ)f | − | − | + | + | 1 |
| TT20137 | + | − | + | + | ||
| TT20064 | eutD101(Am) | − | − | + | + | 3g |
| TT17319 | + | − | + | + | ||
| TT20024 | eutE56 | − | − | + | + | 3h |
| TT17310 | + | − | + | + | ||
Growth tests were done in aerobic liquid medium containing glycerol as a carbon source and ethanolamine as a nitrogen source with indicated additions; growth was scored after 2 days.
Several different mutations in each gene were tested; all mutations in the same gene exhibited the same phenotype. Full genotypes are in Table 1.
All known eutT mutants are nonsense mutations (19). The alleles tested were eutT67 [gln229(Am)], eutT74 [gln529(Am)], eutT75 [gln379(Oc)]eutT77[gln181(Am)], eutT78 [gln402(Op)], eutT86 [pro227(Leu)] [gln229(Am)], eutT88[gln184(Am)], eutT90[gln184(Am)], and eutT104[trp374(Op)].
This strain has a Eut(N+) phenotype at 0.02 μM CN-B12 but fails to grow at 0.08 μM CN-B12; this inhibition phenotype will be described later.
The alleles tested were eutA111 and eutA114.
In-frame nonpolar deletion mutations (19).
The alleles tested were eutD53[gln103(Oc)], eutD101[gln4656(Am)] and eutD124[arg518(Op)].
The alleles tested were eutE56, eutE65, and eutE79.
MATERIALS AND METHODS
Bacterial strains and transposons.
Strains were derived from S. enterica (serovar Typhimurium) LT2 (Table 1). Normal and introduced promoters are diagrammed in Fig. 1. The cobA366::Tn10d(Cm) mutation maps far from the eut operon and eliminates the general B12 adenosyl transferase (11, 34). An inserted element referred to as eutA::Pint throughout this paper is the eutA208::Tn10d(Tc) insertion (28), carrying an outward-directed constitutive promoter, Pint (32), created by a point mutation within the Tn10d(Tc) element (37). This strong promoter expresses eut genes downstream of eutA, including EutBC (EA-lyase) and EutR (32). Another Tn10 derivative (T-POP) (25) inserted in the eutJ gene (or in eutP) allowed tetracycline-inducible expression of genes downstream of its insertion site. Nonpolar deletion mutants eutPQTD333 and eutDM302 were isolated as tetracycline-sensitive Eut+ revertants of T-POP insertions in the eutQ or eutD genes (19) and a set of constructed single-gene, in-frame deletions will be described (23). The eut-38::MudA insertion lies just within the distal end of the eut transcript but outside of all reading frames; it reports operon transcription without impairing any eut function (19, 27). Other eut mutations have been described previously (19, 27).
TABLE 1.
Bacterial strains used in this study
| Strain | Genotype |
|---|---|
| BE119 | E. coli ompT hsdS (rB− mB−) dcm+ Tetrgal λ(DES) endA DE3[argU ileY leuW Camr]/pTA925 (T7 expression vector without insert) |
| BE205 | E. coli ompT hsdS (rB− mB−) dcm+ Tetrgal λ(DES) endA DE3[argU ileY leuW Camr]/pTA1019 (T7 expression vector with eutT insert) |
| TT10000 | LT2 wild type |
| TT10674 | eut-38::MudA |
| TT17300 | eutA208::Tn10d(Tc)Pint eutR156::MudJ |
| TT17306 | eutA208::Tn10d(Tc)Pint |
| TT17310 | eutE56 eut-38::MudA |
| TT17312 | eutA114 eut-38::MudA |
| TT17319 | eutD101 eut-38::MudA |
| TT17320 | eutQ18::Mu dJ eutA208::Tn10d(Tc) Pint |
| TT20024 | eutE56 eut-38::MudA cobA366::Tn10d(Cm) |
| TT20025 | eutA111 eut-38::MudA |
| TT20026 | eutA111 eut-38::MudA cobA366::Tn10d(Cm) |
| TT20033 | eutQ18::MudJ eutA208::Tn10d(Tc)Pint cobA366::Tn10d(Cm) |
| TT20039 | eutE163::MudJ eutA208::Tn10d(Tc)Pint |
| TT20041 | eutE163::MudJ eutA208::Tn10d(Tc)Pint cobA366::Tn10d(Cm) |
| TT20042 | eut-38::MudA cobA366::Tn10d(Cm) |
| TT20043 | eutT11::MudA eutA208::Tn10d(Tc)Pint |
| TT20044 | eutT11::MudA eutA208::Tn10d(Tc)Pint cobA366::Tn10d(Cm) |
| TT20045 | eutP171::MudJ eutA208::Tn10d(Tc)Pint cobA366::Tn10d(Cm) |
| TT20046 | eutP171::MudJ eutA208::Tn10d(Tc)Pint |
| TT20047 | eutG3::MudA eutA208::Tn10d(Tc)Pint |
| TT20054 | eutE10::Mud eutA208::Tn10d(Tc)Pint cobA366::Tn10d(Cm) |
| TT20069 | eutE6::MudA eutA208::Tn10d(Tc)Pint |
| TT20071 | eutE10::MudA eutA208::Tn10d(Tc)Pint |
| TT20078 | eutT86 eut-38::MudA cobA366::Tn10d(Cm) |
| TT20079 | eutT86 eut-38::MudA |
| TT20114 | cobA366::Tn10d(Cm) cobR4 cob24::MudJ metE205 ara-9 zeb-1845::Tn10 |
| TT20124 | eutQ18::MudJ eutJ269::Tn10(Tc)T-POP |
| TT20125 | eutQ18::MudJ eutJ269::Tn10(Tc)T-POP cobA366::Tn10d(Cm) |
| TT20126 | eutT11::MudA eutJ269::Tn10(Tc)T-POP |
| TT20127 | eutT11::MudA eutJ269::Tn10(Tc)T-POP cobA366::Tn10d(Cm) |
| TT20128 | eutE10::MudA eutJ269::Tn10(Tc)T-POP |
| TT20129 | eutE10::MudA eutJ269::Tn10(Tc)T-POP cobA366::Tn10d(Cm) |
| TT20136 | eutDM302 eut-38::MudA |
| TT20137 | eutDM302 eut-38::MudA cobA366::Tn10d(Cm) |
| TT20138 | eutPQTD333 eut-38::MudA |
| TT20139 | eutPQTD333 eut-38::MudA cobA366::Tn10d(Cm) |
| TT24803 | eutT371::FRT(Δ) |
| TT25127 | cobA366::Tn10d(Cm) |
| TT25128 | cobA366::Tn10d(Cm)eutT371::FRT |
Media.
Rich medium (NB) was nutrient broth (0.8%; Difco Laboratories) supplemented with NaCl (5 g/liter). Minimal media were variants of E medium, which contains citrate. NCE medium lacks citrate (26), and NCN medium lacks both citrate and a nitrogen source (5). Ethanolamine hydrochloride (20.5 mM; Aldrich) was used as a carbon source in NCE medium, as a nitrogen source in NCN medium with glycerol (20 mM), or as a carbon and nitrogen source in NCN medium. CN-B12 (80 nM; Sigma Chemical Co.) was the usual exogenous vitamin B12 source. Amino acids and purines were added to minimal medium at the concentrations previously recommended (10). NB agar media contained antibiotics at the following concentrations: ampicillin, 30 μg/ml; chloramphenicol, 20 μg/ml; kanamycin, 50 μg/ml; and tetracycline, 20 μg/ml. Solid media were prepared by adding agar (1.5%, Difco) to E, NCN, NCE, or NB medium.
Assay of β-galactosidase.
Strains were grown in NCE or NCN medium containing glycerol (0.2%) as a carbon source and ethanolamine (20.5 mM) plus CN-B12 (80 nM) as inducers. Following growth to mid-log phase at 37°C on a New Brunswick Shaker, model 50, cells were held at 4°C until assayed. Enzyme activity was determined in chloroform-sodium dodecyl sulfate-permeabilized cells (21). Activity was expressed as nanomoles of nitrophenol produced per minute per optical density at 650 nm (OD650) of cell culture turbidity. All presented values are the average of at least four determinations on two independent cultures.
Cloning the eutT gene for expression.
The eutT coding sequence was amplified by PCR using forward primer GCCGCCAGATCTGATGAACGATTTCATCACCGAAACGTGG and reverse primer GCCGCCAAGCTTTCATGGCTTCTCTCCCAACCGTTG (17). Template DNA from S. enterica was prepared with a Bio-Rad Aquapure genomic DNA isolation kit. Polymerase for PCR was a mixture of Pfu and Taq polymerases (7:1 [unit/unit]). The PCR products were cloned into the T7 expression plasmid pTA925 with BglIII and HinDIII (17). The DNA sequences of selected clones were verified. Plasmids pTA925 (no eutT insert) and pTA1019(eutT insert) were introduced into expression strain Escherichia coli BL21 DE3RIL (Stratagene, La Jolla, Calif.), to form strains BE119 and BE205, which were used for EutT production. Adenosyl transferase activity was assayed as described previously (17).
Overexpression of EutT and preparation of cell extracts.
EutT expression strains were grown at 30oC with shaking in 1,000-ml baffled Erlenmeyer flasks containing 500 ml of Luria broth supplemented with 25-μg/ml kanamycin. Cells were grown to an OD600 of 0.6 to 0.8. At that density, protein expression was induced by the addition of isopropyl-β-d-thiogalactopyranoside (IPTG) to a final concentration of 1 mM. Cultures were shaken for an additional 3 h at 30°C, placed on ice for 5 min, and collected by centrifugation for 5 min at 4,650 × g (maximum) using a Beckman J2-HS centrifuge and a Beckman JLA10.500 rotor. The cell pellet was frozen at −80°C until used. Cell pellets were resuspended in 50 mM potassium phosphate buffer at pH 7, containing 50 mM NaCl, and cells were disrupted in a 5-ml French pressure cell (SLM Aminco, Urbana, Ill.). Cell debris and unbroken cells were pelleted by centrifugation at 31,000 × g (maximum) with a Beckman JA20 rotor. The supernatant served as the soluble fraction for enzyme assays and polyacrylamide gel electrophoresis. The pellet obtained by centrifugation was extracted with Bacterial Protein Extraction Reagent II (B-PERII; Pierce, Rockford, Ill.) according to the manufacturer's directions, but with the following modifications. The B-PERII solution used for the first extraction was supplemented with the protease inhibitor phenylmethylsulfonyl fluoride at a concentration of 100 μg/ml. The B-PERII solution used for the second extraction was supplemented with DNase at a concentration of 20 μg/ml. Inclusion bodies were washed twice with 10 ml of a 1/20 dilution of B-PERII and then resuspended in 50 mM potassium phosphate pH 7.0 containing 50 mM NaCl; this suspension was used for assay of inclusion bodies (17).
Growth inhibition by CN-B12.
A eutA mutant fails to grow on ethanolamine as a carbon source, regardless of the cobalamin form provided. It also fails to use ethanolamine as a nitrogen source when CN-B12 is provided at a high level (80 nM) but does grow if Ado-B12 is given. The Ado-B12-stimulated growth is inhibited by high CN-B12 levels (29). We show here that growth on ethanolamine as a nitrogen source is allowed (inhibition is avoided) if the level of CN-B12 is reduced to 20 nM. To measure this inhibition, overnight nutrient broth cultures were diluted 1/100 into 200 μl of NCN medium containing glycerol (0.2%), ethanolamine (20.5 mM), and CN-B12 at 0, 20, 80, or 140 nM. Cultures were incubated, with aeration, at 37°C in a Bio-Tec automated plate reader. Three parallel cultures were grown at each concentration of CN-B12, and OD650 turbidity was measured every hour.
Effect of cobA and eutT mutations on growth rates.
Three independent NB cultures of each strain were inoculated from single colonies grown on NB plates. After overnight growth, cells were pelleted, washed in minimal medium (pH 7.0), and used (35 μl of final suspension) to inoculate 6-ml cultures in minimal medium. Liquid minimal medium was a mixture of 5 mM KH2PO4, 5 mM NaNH4HPO4, and 1 mM MgSO4 buffered at pH 7.0 by 50 mM MOPS (morpholinepropanesulfonic acid). The carbon source was 41 mM ethanolamine hydrochloride (0.04%). Cyanocobalamin concentrations are indicated later. Minimal medium also contained biotin; Ca-d-pantothenic acid, nicotinamide, and pyridoxine HCl at 4 × 10−4% (wt/vol); and thiamine and riboflavin at 2 × 10−5% (wt/vol) and trace metals as previously described (24). Cultures were aerated slightly by shaking at 240 rpm in tubes standing upright. Growth was monitored by observing OD650 on a Spectronic 20D+ spectrophotometer.
RESULTS
Evidence for a CobA-independent route of cobalamin adenosylation.
Mutants of S. enterica lacking the CobA adenosyl transferase cannot use ethanolamine as a nitrogen source when provided with CN-B12 and glucose as a carbon source (11). This failure is due to inability to convert CN-B12 to the required EA-lyase cofactor, Ado-B12; growth is restored if Ado-B12 replaces CN-B12. We noted that CN-B12 allowed cobA mutants to use ethanolamine as a nitrogen source when glycerol rather than glucose provided carbon (Table 2, top row) or when ethanolamine provided both nitrogen and carbon. This implied that an adenosyl transferase (other than CobA) was expressed during growth on poor carbon sources. Conditions under which this additional activity was inferred were ones that induce the eut operon (27, 32), suggesting that a gene in the eut operon might encode the inferred transferase.
Preliminary evidence for two eut genes that might provide Ado-B12.
Several eut mutations were combined with a cobA mutation and tested for their effect on the inferred ability to adenosylate cobalamin. When CN-B12 was provided aerobically, only eutT and eutA point mutants failed to use ethanolamine as a nitrogen source on glycerol, Eut(N−). Below we characterize these eutT mutations and then return to eutA.
In an otherwise wild-type strain, available eutT mutations (all nonsense types) eliminated growth on ethanolamine as the sole carbon source even when Ado-B12 is provided (19). This is due to their polar effect on expression of the downstream genes for EA-lyase (eutB and eutC), which is essential for use of ethanolamine. However, these simple eutT mutants can use ethanolamine as a nitrogen source, demonstrating that their level of EA-lyase is sufficient for the less-demanding task of supplying nitrogen (Table 2, rows 1 to 4). In strains lacking cobA, eutT mutations eliminate use of ethanolamine as a nitrogen source with provided CN-B12, a Eut(N−) phenotype. This defect is corrected by providing Ado-B12 (Table 2). The Eut(N−) phenotype on CN-B12 was caused by nonpolar deletion mutations that remove eutT (eutPQTD333) but was not seen in eutT+ strains carrying a nonpolar deletion (eutDM302) or in strains with a polar point mutation downstream of the eutT gene (e.g., eutD101). Since the Eut(N−) phenotype of eutT mutants on CN-B12 was corrected by providing either Ado-B12 or a functional CobA transferase activity, EutT was a prime candidate for the eut-specific adenosyl transferase.
Genetic evidence that EutT contributes to cobalamin adenosylation in vivo.
The cofactor Ado-B12 is required to induce the eut operon and to serve as cofactor for EA-lyase. Thus, if eutT mutants fail to make Ado-B12, both induction and enzyme activity should be impaired. These processes were tested independently in strains with insertion eutA::Pint (32), which constitutively expresses the EA-lyase genes (eutBC). Fusions of lac to the eut operon upstream of eutA reported operon induction (Table 3). Operon induction was tested on glycerol-ammonia medium, where no EA-lyase activity is required. All cobA+ strains showed induction. However, in cobA mutants, no induction was seen for Mud insertions that eliminated EutT activity either directly or by polarity (eutP, eutQ, or eutT; Table 3, rows 3 to 6). Inducibility was normal in strains with a eutE::Mud fusion, distal to the eutT gene (Table 3, rows 7 and 8). The induction defect in strains lacking EutT (and CobA) was corrected by Ado-B12 (Table 3, right column). Thus, EutT allows CN-B12 to induce the operon without CobA and is therefore inferred to contribute to conversion of CN-B12 to the inducer Ado-B12.
TABLE 3.
Two tests of EutT-mediated Ado-B12 production, induction and activity
| Strain | Genotypea
|
Eut(N) growth phenotypeb
|
eut operon induction (β-galactosidase activity)c
|
||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| eut | cobA | None | +CN-B12 | +Ado-B12 | None | +EA | +EA +CN-B12 | Fold increase by CN-B12d | Effect of CobA+e | +EA +Ado-B12 | |
| TT20045 | eutP171::lac (EutT−) | − | − | − | + | 2.7 | 3.9 | 6.2 | 1.6 | 21 | 130 |
| TT20046 | + | − | + | + | 1.6 | 4.6 | 130 | 28 | 110 | ||
| TT20033 | eutQ18::lac (EutT−) | − | − | − | + | 9.8 | 32 | 46. | 1.4 | 28 | 1350 |
| TT17320 | + | − | + | + | 21 | 56 | 1300 | 23 | 960 | ||
| TT20044 | eutT11::lac (EutT−)f | − | − | − | + | 5.5 | 18 | 27. | 1.5 | 34 | 590 |
| TT20043 | + | − | + | + | 6.0 | 20 | 910 | 46 | 590 | ||
| TT20041 | eutE163::lac (EutT+)g | − | − | + | + | 5.8 | 29 | 960 | 33 | 0.9 | 740 |
| TT20039 | + | − | + | + | 5.1 | 30 | 850 | 29 | 760 | ||
All strains contain the eutA::Pint mutation that eliminates EutA protein and highly expresses the EutBC (lyase) and EutR (transcription activator) proteins (Fig. 2A). Complete genotypes are in Table 1.
Growth phenotypes were scored + or − by replica-printing cell patches from an NB plate to an NCN plate with glycerol as a carbon source and ethanolamine as a nitrogen source.
Cells were grown on NCE medium containing glycerol as a carbon source and ammonia with additions as shown. β-Galactosidase activity was determined as described in Materials and Methods.
Induction caused by CN-B12.
This is the ratio (cobA+/cobA mutant) of operon induction in strains isogenic except for the cobA mutation.
Essentially the same results were seen with eutT17::Mud-lac.
Essentially the same results were seen with all other Mud-lac insertions located promoter distal to eutT (Fig. 2A). MutA insertion alleles tested were eut-6 (between eutN and eutE), eutE12, eutE10, eutE1, eutJ26, and eutH25. The MudJ alleles tested were eut-168 (between eutD and eutM) eutE163, and eutE181.
Further evidence for a role of EutT in Ado-B12 production was its effect on EA-lyase-dependent growth. In the strains used above, EA-lyase (EutBC) was expressed constitutively from a strong promoter (Pint) within the inserted eutA::Pint element. Since EA-lyase is the only enzyme required to produce ammonia from ethanolamine, these strains can use ethanolamine with or without operon induction but do so only if cofactor Ado-B12 is available (32). Mud insertions that eliminate EutT expression (those in the eutP, eutQ, or eutT genes) caused a Eut(N−) phenotype in the absence of CobA (Table 3, first six rows), whereas a eutE::Mud insertion (distal to eutT) is Eut(N+) (Table 3, last two rows). The eutT growth defect on CN-B12 was corrected by Ado-B12 or by restoring a cobA+ allele (Table 3). Similar results were seen for point mutations in genes eutT, eutD, and eutE (data not shown). Thus, EutT served to provide Ado-B12 by the criterion of ethanolamine growth, as well as operon induction. (Effects of a simple in-frame eutT deletion on growth will be shown later.)
Assays of EutT adenosyl transferase activity.
The genetic evidence above suggested that EutT was a B12 adenosyl transferase. This conclusion was confirmed by transferase assays. Since assay of the CobA and PduO adenosyl transferases requires overproduction of the protein (34), the EutT protein was produced in a T7 expression system (17). A strain (BE205) with eutT inserted in the expression plasmid produced relatively large amounts of a 30.2-kDa protein in both the soluble and inclusion body fractions; this protein was not found in a strain (BE119) whose expression plasmid lacked eutT. Extracts of strains expressing eutT (BE205) showed 50- to 100-fold more transferase activity than the control strain (BE119) (Table 4). These assays were done exactly as described previously and used OH-B12 (reduced to the CobI form) as an adenosyl-accepting substrate (17). It should be noted that a substantial fraction of adenosyl transferase activity sedimented with cell debris and may be present as inclusion bodies or associated with membranes. These results demonstrate that EutT protein has ATP:cob(I)alamin adenosyl transferase activity; the phenotypes above show that this activity is relevant in vivo.
TABLE 4.
ATP:cob(I)alamin adenosyl transferase activity
| Straina | Plasmidb | Gene expressed | Fractionc | Sp act (nmol/ min/mg)d |
|---|---|---|---|---|
| BE119 | pTA925 | Vector only | Soluble | 0.3 |
| BE119 | pTA925 | Vector only | Inclusion bodies | 0.4 |
| BE205 | pTA1019 | EutT | Soluble | 33.2 |
| BE205 | pTA1019 | EutT | Inclusion bodies | 21.1 |
Complete genotypes are in Table 1.
Plasmid construction is described in Materials and Methods.
Preparation of soluble and inclusion body fractions for enzyme assay is described in Materials and Methods.
ATP:cob(I)alamin specific activities were determined as described in Materials and Methods. The values shown are the averages of three trials.
Phenotypes of eutA mutations.
Like eutT mutants, eutA mutants failed to use ethanolamine as either a carbon or nitrogen source when standard levels of CN-B12 (80 nM) were provided. It was shown earlier that growth on ethanolamine as a nitrogen source was restored when Ado-B12 replaces CN-B12, but not when Ado-B12 and high CN-B12 were supplied together (29), suggesting an inhibitory effect of CN-B12. A lower CN-B12 concentration (20 nM) allowed eutA mutants to use ethanolamine as a nitrogen source. The inhibition of growth by high levels of CN-B12 agrees with the previous in vitro demonstration that purified EA-lyase (EutBC) is inhibited by CN-B12 (3, 16, 31). Thus EutA seems to protect against inhibition by CN-B12, reminiscent of reactivating factors that remove inappropriate cofactors from other B12-dependent enzymes (22, 35, 36). Since EutA might protect EA-lyase by adenosylating excess CN-B12, it was tested further as a candidate for an adenosyl transferase.
EutA protein does not provide an adenosylation function.
The above tests of EutT function were done in strains that carried the eutA::Pint insertion and thus lacked EutA function, showing that EutA protein is not required for EutT transferase activity. To test whether EutA can provide adenosylation independent of EutT, the eutA gene was expressed from a tetracycline-inducible promoter (T-POP) inserted in the eutJ gene; this promoter also expressed EA-lyase (eutBC) genes needed for growth on ethanolamine. In this strain, ability to convert CN-B12 to Ado-B12 could be assessed as ability to grow on ethanolamine as a nitrogen source (which is independent of operon induction). Alternatively, Ado-B12 production can be scored by operon induction (which does not require EA-lyase activity). Promoter proximal insertions of Mud-lac in the eutQ, eutT, or eutE genes were added to the eutA::Pint strain in order to block expression of upstream genes and as reporters of operon induction. The eutQ and eutT insertions eliminate EutT expression, while the eutE::MudJ insertion leaves eutT expression intact. As seen in Table 5, high-level expression of the eutA gene (from the eutJ::T-POP promoter) does not provide Ado-B12 in strains that lack EutT and CobA. That is, EutA could not substitute for the EutT or CobA adenosyltransferases in supporting either induction or EA-lyase-dependent growth. We conclude that EutA does not provide an adenosyl transferase activity and must provide resistance to the inhibitory effect of high CN-B12 in some other way. Additional tests of this inhibition are below.
TABLE 5.
The EutA protein cannot provide adenosyl transferase activity
| Strain | Genotypea
|
Eut(N) growth phenotypeb
|
Operon induction (β-galactosidase activity)c
|
||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| eut insertion | cobA | None | +CN-B12 | +Ado-B12 | None | +CN-B12 | Fold increase by CN-B12d | Effect of CobAe | +Ado-B12 | Fold increase by Ado-B12f | |
| TT20125 | eutQ18::lac (EutT−A+) | − | − | − | + | 5.7 | 5.8 | 1.0 | 240 | 1,200 | 220 |
| TT20124 | + | − | + | + | 5.6 | 1,400 | 250 | 1,300 | 230 | ||
| TT20127 | eutT11::lac (EutT− A+) | − | − | − | + | 4.2 | 3.8 | 0.9 | 190 | 640 | 150 |
| TT20126 | + | − | + | + | 5.3 | 710 | 135 | 670 | 130 | ||
| TT20129 | eutE163::lac (EutT+ A+) | − | − | + | + | 4.2 | 680 | 160 | 1.2 | 870 | 210 |
| TT20128 | + | − | + | + | 6.3 | 900 | 140 | 920 | 150 | ||
All strains carry (distal to the lac fusion) a eutJ::T-POP insertion, which expresses the eutGHABCR genes in the presence of tetracycline. Complete genotypes are in Table 1.
All strains were tested for growth on NCN glycerol medium with 20.5 mM ethanolamine, 0.08 μM CN-B12 or Ado-B12, and 2-μg/ml tetracycline.
All strains were grown on NCE glycerol NH3 medium with 20.5 mM ethanolamine, 0.08 μM CN-B12 or Ado-B12, and 2-μg/ml tetracycline. β-Galactosidase activity was determined as described in Materials and Methods.
Induction caused by CN-B12; large induction is due to high expression of EutR from T-POP.
This is the ratio (cobA+/cobA mutant) of operon induction in strains isogenic except for the cobA mutation.
Induction caused by Ado-B12.
Inhibition of eutA mutants by CN-B12.
The inhibitory effect of CN-B12 (80 nM) on eutA mutants noted above was relieved at lower concentrations of CN-B12 (20 nM), which allowed these mutants to grow on glycerol using ethanolamine as a nitrogen source. The growth inhibition by high CN-B12 is not due to a failure to express the operon, since normal induction was seen in eutA mutant strains by 80 nM CN-B12 (in combination with ethanolamine). We presume that CN-B12 inhibits growth by inhibiting EA-lyase directly as seen previously for the pure enzyme (2). However this sensitivity seemed to require other genes in the eut operon.
A role of other genes in sensitivity to CN-B12 was demonstrated with the eutA::Pint insertion (described above), which disrupts eutA and expresses EA-lyase (EutBC) from a constitutive promoter. A strain with this insertion grew on ethanolamine as a nitrogen source (with 20 nM CN-B12) but was inhibited by a higher concentration (80 nM) of CN-B12. The inhibition by high CN-B12 was abolished by an insertion in the promoter proximal eutQ gene, but not by an insertion in the eutG gene. This suggested that one or more genes upstream of eutG (eutS, -P, -Q, -D, -T, -M, -N, -E, or -J) must be needed for sensitivity. Consistent with this, a eutR mutation, which prevents expression of these genes, eliminated sensitivity to high CN-B12. (As described above, the strains used have no EutA function and express EA-lyase [EutBC] constitutively from the Pint promoter.)
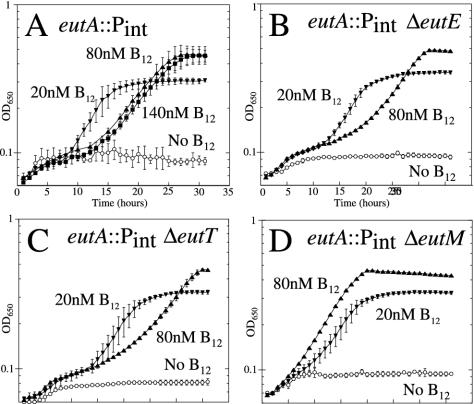
To identify the gene or genes necessary for inhibition by CN-B12, a series of in-frame deletions of individual eut genes were added to the eutA::Pint mutant. The sensitivity of the parent strain to high CN-B12 (Fig. 2A) appears as delayed growth of liquid cultures. This sensitivity was not relieved by deletion of eutT (adenosyl transferase) or the eutD, eutE, or eutJ genes (encoding metabolic enzymes acting after acetaldehyde). Growth of the eutT and eutE mutants is shown in Fig. 2B and C. Inhibition by high CN-B12 was eliminated by removal of any one of the eutM, -N, -P, -Q, or -S genes. Growth of the eutM deletion mutant was typical and is shown in Fig. 2D. The EutM, -N, and -S proteins are homologues of carboxysome shell proteins; the functions of EutP and -Q are unknown. These effects are discussed below. It should be noted that delayed growth caused by high CN-B12 occurs in tests reported in Table 3, but growth is scored after 48 h in these tests so the inhibition does not interfere.
FIG. 2.
Genes required for inhibition of eutA mutants by CN-B12. Mutants lacking EutA function can grow aerobically on ethanolamine as a nitrogen source when supplied with a low level of CN-B12 but are inhibited by higher concentrations of CN-B12 (A). This sensitivity requires expression of some eut genes upstream of the eutA gene (eutS, -P, -Q, -M, and -N) but not others (eutT, -D, -E, -J, -G, and -H). All strains carry the eutA::Pint insertion and therefore express EA-lyase constitutively. Added mutations are constructed nonpolar deletion mutations. (B) ΔeutE. (C) ΔeutT. (D) ΔeutM. The responses shown are typical of mutations in the two groups. Growth rates were determined with a Biotec automated plate reader.
Testing autogenous induction of the eut operon by EutT and EutR.
Evidence is described above that EutT and CobA (but not EutA) contribute to formation of Ado-B12, the inducer and required cofactor for ethanolamine degradative enzymes. This raises the question of the relative roles of EutT and CobA during normal growth on ethanolamine. The positive regulatory protein EutR is encoded within the eut operon (Fig. 1) and thus serves to activate its own production (autogeneous regulation) (27, 32). This autoinduction circuit is required for maximal operon expression (27, 32). The EutT protein also contributes to its own expression by producing Ado-B12, a coinducer of the operon. Thus a strain with a cobA mutation and an uninduced eutT in the operon should not convert CN-B12 to Ado-B12 and therefore should show no induction of its eut operon. However, this situation might be unstable if a momentary stochastic expression of the operon provided enough EutT adenosyl transferase to initiate an autocatalytic induction (i.e., if a tiny bit of EutT produces enough Ado-B12 to supply more EutT and launch a full induction).
To test this, cobA strains with three different preinduced levels of EutT were examined for operon induction by CN-B12 as the Ado-B12 precursor (Table 6). The first two rows describe a normal eut operon with lac genes that were fused at a point within the operon distal to all structural genes (eut-38::lac), leaving EutT expression dependent on induction. With CobA (row 2), normal induction is seen, but without CobA, uninduced EutT allows very little induction (1.8-fold) by CN-B12 ethanolamine on NH3-glycerol-ethanolamine medium. A slightly greater induction by CN-B12 (fivefold) was seen when ethanolamine must provide the nitrogen source (Table 6, row 1, numbers in parentheses). Previous work has revealed no effect of nitrogen limitation on eut operon transcription (27, 32). We suggest that when ethanolamine must provide a nitrogen source, the only cells that grow are those that have stochastically achieved a slightly higher operon expression level, which can be maintained by the produced EutT enzyme.
TABLE 6.
Initiating operon induction with EutT as the sole Ado-B12 source
| Strain | Genotypea
|
eut operon induction (β-galactosidase activity)b
|
|||||||
|---|---|---|---|---|---|---|---|---|---|
| eut | cobA | None | +EA | Fold increase by EAc | +EA + CN-B12 | Fold increase by CN-B12d | +EA + Ado-B12 | Fold increase by Ado-B12e | |
| TT20042 | eut-38:Mudlac (Eut+)f | − | NA | 6.0 | 11 (34) | 1.8 (5.6) | 110 (74) | 19 (12) | |
| TT10674 | + | NA | 5.8 | 240 (164) | 42 (19) | 150 (110) | 25 (19) | ||
| TT20129 | eutE10:Mudlac eutJ269::T-POP | − | NA | 4.2 | 680 | 160 | 870 | 210 | |
| TT20128 | + | NA | 6.3 | 900 | 140 | 920 | 150 | ||
| TT20127 | eutT11::Mudlac eutJ269::T-POP | − | NA | 4.2 | 3.8 | 0.9 | 640 | 150 | |
| TT20126 | + | NA | 5.3 | 710 | 130 | 670 | 130 | ||
| TT20054 | eutE10:Mudlac eutA::Pint | − | 3.6 | 36 | 10.0 | 1,300 | 37 | 690 | 19 |
| TT20071 | + | 3.5 | 27 | 7.7 | 920 | 34 | 590 | 22 | |
| TT20044 | eutT11::Mudlac eutA::Pint | − | 5.5 | 18 | 3.3 | 27 | 1.5 | 590 | 33 |
| TT20043 | + | 6.0 | 20 | 3.3 | 910 | 46 | 590 | 30 | |
Complete genotypes are in Table 1.
Strains were grown on NCE glycerol medium; 20.5 mM ethanolamine, and either 80 nM CN-B12 or 80nM Ado-B12. Strains with a T-POP insertion were grown with 2-μg/ml tetracycline. NA, not assayed. Numbers in parentheses are for strains grown without NH3; ethanolamine must serve as a nitrogen source.
Induction by ethanolamine.
Induction by CN-B12 plus ethanolamine over that seen for ethanolamine alone.
Induction by Ado-B12 plus ethanolamine over that seen for ethanolamine alone.
This strain has a functional eut operon with a lac operon fusion to the transcribed region distal to all genes.
To test the effect of a higher basal level of EutT on operon inducibility, a eutJ::T-POP insertion was added to strains carrying various eut-lac reporter fusions; the T-POP insertion allows tetracycline-inducible expression of downstream genes including eutR. Increasing the level of EutR, is known to allow modest operon expression without regulatory effectors (27, 32). In eutT+ strains expressing eutR from the eutJ::T-POP (Table 6, rows 3 and 4), ethanolamine alone did not induce operon induction with or without CobA, but ethanolamine plus CN-B12 caused full operon induction independent of CobA (compare rows 3 and 4). The full induction depended on the eutT+ gene in strains lacking cobA: that is, when the reporter disrupted eutT, no induction was seen without CobA (rows 5 and 6). Thus a very slight increase in basal operon expression provided enough EutT (and adenosylated CN-B12) for full operon induction by CN-B12 independent of CobA.
A still higher basal (uninduced) level of operon expression is seen in strains that express eutR from the stronger Pint promoter (Table 6, rows 7 to 10). When such strains had a functional eutT gene (rows 7 and 8), ethanolamine alone caused a slight induction but ethanolamine plus CN-B12 caused full induction independent of CobA (compare rows 7 and 8). This CobA-independent induction depended on EutT (compare rows 9 and 10 to 7 and 8). In all of the strains above, Ado-B12 induced the operon even when both CobA and EutT are limiting (see right column).
Physiological roles of EutT and CobA enzymes in ethanolamine metabolism.
The above conclusions raise the question of the relative importance of EutT and CobA to growth on ethanolamine. To test this, an in-frame (nonpolar) deletion of the eutT gene was constructed and placed in strains with and without a cobA mutation. These strains were compared for their ability to grow on ethanolamine (Fig. 3). Lack of EutT caused little impairment of growth on high levels of exogenous B12, but reduced growth rate at low B12 levels. Lack of CobA alone delayed growth initiation regardless of B12 level, presumably by making it difficult to induce the operon, as described above.
FIG. 3.
Effects of cobA and eutT mutations on use of ethanolamine as a carbon source. Growth and media are described in Materials and Methods. In panel A, CN-B12 was added at 10 nM. In panel B, CN-B12 was added at 100 nM. All cultures were grown under low aerobic conditions at 30°C. The isogenic strains used were TT10000 (wild type; squares), TT25127 (cobA; upward triangles), TT24803 (eutT; downward triangles), and TT25128 (cobA eutT; circles).
Evidence that EutT adenosyl transferase does not contribute to de novo B12 synthesis or assimilation of cobinamide.
The CobA adenosyl transferase acts on several substrates, including CN-B12 or the incomplete corrinoid cobinamide (Cbi; the corrinoid ring lacking both the upper and lower ligands); CobA also acts on some biosynthetic intermediate (perhaps cobyric acid) leading to production of Ado-Cbi, the precursor of Ado-B12 (11). The experiments above demonstrated that EutT adenosylates assimilated CN-B12 but do not test its ability to use Cbi or a biosynthetic intermediate.
EutT, unlike CobA, does not adenosylate either Cbi or the biosynthetic intermediate. Strains for these tests expressed eutT from the tetracycline-inducible promoter in a eutP::TPOP insertion (19). Production of B12 in vivo was monitored by using a metE mutant, whose synthesis of methionine depends on the alternative B12-dependent enzyme MetH (reviewed in reference 30). Results are outlined below.
First, a metE mutant can grow aerobically without methionine if Cbi is provided as a B12 precursor. This growth occurs because CobA enzyme adenosylates Cbi to Ado-Cbi, which can be converted to B12 (11). A metE cobA double mutant lacks the CobA adenosyl transferase and cannot produce B12 from Cbi; therefore, it cannot grow on minimal medium without added methionine (or B12). This defect was not corrected by inducing EutT, suggesting that (unlike CobA) EutT cannot adenosylate assimilated Cbi.
Second, a metE mutant can grow anaerobically on minimal medium because B12 is synthesized de novo under these conditions. The de novo B12 pathway depends on adenosylation of an intermediate by CobA (11). A metE cobA double mutant cannot synthesize B12 de novo and fails to grow anaerobically on minimal medium. This defect was not corrected by expressing EutT, suggesting the EutT cannot adenosylate the intermediate in the normal biosynthetic pathway.
DISCUSSION
Ethanolamine metabolism requires Ado-B12 as a coinducer (with ethanolamine) of operon transcription and as a cofactor for the first enzyme (EA-lyase). Under natural conditions, Ado-B12 is not always available because it is synthesized only anaerobically and adenosylated corrinoids may not be present in all aerobic environments (30). By requiring Ado-B12 (plus ethanolamine) for induction of the eut operon, cells ensure that this gene system (17 proteins) is expressed only if both its substrate and required enzyme cofactor are present.
Induction of eut operon transcription is autocatalytic in the sense that the positive regulatory protein EutR is encoded within the operon that it controls (27). This arrangement is thought to be needed because the EutBC (EA-lyase) and EutR (regulator) proteins compete for binding Ado-B12 (32). By producing these proteins at a constant ratio (from genes in the same operon), the cell can respond to a wide range of Ado-B12 levels and remain sensitive to induction at all levels of operon expression; this can be achieved with very little investment in EutR protein prior to induction. The EutT adenosyl transferase adds a second autocatalytic element to this regulatory circuit by contributing to the level of a coinducer of the operon (Ado-B12). Because of this circuit, cells need maintain only a minimal pool of Ado-B12 (made by CobA)—just sufficient to initiate eut operon induction. As the operon (and its eutT gene) is induced, EutT may supplement the Ado-B12 level during high-demand growth on ethanolamine.
The three cobalamin adenosyl transferases, CobA, PduO, and EutT, play distinct biological roles. The CobA enzyme may be the basal housekeeping activity. It supports de novo B12 synthesis (anaerobically) and can adenosylate the assimilated B12 precursor cobinamide, allowing its conversion to Ado-B12. This basal Ado-B12 level initiates eut operon induction. The EutT enzyme adds Ado-B12 to support full induction and allows full growth ability during the period of high demand. The PduO enzyme is thought to serve an analogous function during growth on propanediol, in which CobA seems to play a very minor aerobic role. The pdu operon is induced by propanediol alone (with no need for cofactor), and mutants lacking CobA or PduO singly show only slightly impaired growth on propanediol using CN-B12, while the cobA, pduO double mutant fails to grow (17). It is not known whether PduO can substitute for CobA in de novo B12 synthesis or in assimilation of cobinamide.
It is surprising that the three cobalamin adenosyl transferases of S. enterica, EutT, PduO, and CobA, show no obvious amino acid sequence similarity (17). This could either reflect independent evolutionary origins or such extensive divergence from a common ancestor that homology is not apparent. If the three adenosyl transferases are derived from a common ancestor, their divergence may have been driven by the need (for PduO and EutT) to form close interactions with distinct groups of proteins contributing to the two pathways. The function of both these pathways is associated with formation of carboxysomes (7, 13, 14, 23).
The EutA protein appears to protect EA-lyase from inhibition by excessive CN-B12 (29). Evidence is provided that this protection does not involve conversion of inhibitory CN-B12 to Ado-B12. It seems likely that EutA serves as a reactivating factor that removes damaged or inappropriate cofactor from the enzyme, as has been shown for several B12-dependent enzymes (22, 35, 36). It is curious that while purified EA-lyase is strongly inhibited by CN-B12 (2) in vitro, this inhibition depends in vivo on the EutM, -N, -P, -Q, and -S proteins. While the functions of EutP and -Q are not known, the EutM, -N, and -S proteins resemble structural proteins or the carboxysome, an organelle thought to contain enzymes of the ethanolamine pathway. Thus sensitivity of eutA mutants to CN-B12 may require an intact carboxysome. We suggest that adenosylation (by both CobA and EutT) may occur primarily outside this compartment, and CN-B12 that enters the compartment may escape adenosylation and inhibit EA-lyase. Disruption of the carboxysome may make it impossible for excessive CN-B12 to escape adenosylation. Alternatively, these proteins may form complexes with EA-lyase that increase its sensitivity to CN-B12.
ADDENDUM IN PROOF
Evidence that EutA removes CN-B12 from lyase was published recently (K. Mori, R. Bando, N. Hieda, and T. Turaya, J. Bacteriol. 186:6845-6854, 2004).
Acknowledgments
This work was supported by NIH grant GM34804 to J.R.R.
We thank Tom Fazzio, Chad Rappleye, Marian Price-Carter, Chris Mace, and Peter Anderson for helpful suggestions and conversations.
REFERENCES
- 1.Andersson, D. L., and J. R. Roth. 1989. Mutations affecting regulation of cobinamide biosynthesis in Salmonella typhimurium. J. Bacteriol. 171:6726-6733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Babior, B. M. 1969. The mechanism of action of ethanolamine deaminase. I. Inhibition by coenzyme B12 analogues. J. Biol. Chem. 244:2917-2926. [PubMed] [Google Scholar]
- 3.Babior, B. M., T. J. Carty, and R. H. Abeles. 1974. The mechanism of action of ethanolamine ammonia-lyase, a B12-dependent enzyme. J. Biol. Chem. 249:1689-1695. [PubMed] [Google Scholar]
- 4.Babior, B. M., H. Kon, and H. Lecar. 1969. The mechanism of action of ethanolamine. V. The photolysis of enzyme-bound alkylcobalamins. Biochemistry 8:2662-2889. [DOI] [PubMed] [Google Scholar]
- 5.Berkowitz, D., J. M. Hushon, H. J. Whitfield, Jr., J. Roth, and B. N. Ames. 1968. Procedure for identifying nonsense mutations. J. Bacteriol. 96:215-220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Blackwell, C. M., and J. M. Turner. 1978. Microbial metabolism of amino alcohols: purification and properties of coenzyme B12-dependent ethanolamine ammonia-lyase of Escherichia coli. Biochem. J. 175:555-563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bobik, T. A., G. D. Havemann, R. J. Busch, D. S. Williams, and H. C. Aldrich. 1999. The propanediol utilization (pdu) operon of Salmonella enterica serovar Typhimurium LT2 includes genes necessary for formation of polyhedral organelles involved in coenzyme B12-dependent 1,2-propanediol degradation. J. Bacteriol. 181:5967-5975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bradbeer, C. 1965. The clostridial fermentations of choline and ethanolamine. II. Requirement for a cobamide coenzyme by an ethanolamine deaminase. J. Biol. Chem. 240:4675-4681. [PubMed] [Google Scholar]
- 9.Brinsmade, S. R., and J. C. Escalante-Semerena. 2004. The eutD gene of Salmonella enterica encodes a protein with phosphotransacetylase enzyme activity. J. Bacteriol. 186:1890-1892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Davis, R. W., D. Botstein, and J. R. Roth. 1980. A manual for genetic engineering advanced bacterial genetics. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 11.Escalante-Semerena, J. C., S.-J. Suh, and J. R. Roth. 1990. cobA function is required for both de novo cobalamin biosynthesis and assimilation of exogenous corrinoids in Salmonella typhimurium. J. Bacteriol. 172:273-280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Faust, L. R., and B. M. Babior. 1992. Over expression, purification, and some properties of the Ado Cbl-dependent ethanolamine ammonia-lyase from Salmonella typhimurium. Arch. Biochem. Biophys. 294:50-54. [DOI] [PubMed] [Google Scholar]
- 13.Havemann, G. D., and T. A. Bobik. 2003. Protein content of polyhedral organelles involved in coenzyme B12-dependent degradation of 1,2-propanediol in Salmonella enterica serovar Typhimurium LT2. J. Bacteriol. 185:5086-5095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Havemann, G. D., E. M. Sampson, and T. A. Bobik. 2002. PduA is a shell protein of polyhedral organelles involved in coenzyme B12-dependent degradation of 1,2-propanediol in Salmonella enterica serovar Typhimurium LT2. J. Bacteriol. 184:1253-1261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Jeter, R., B. M. Olivera, and J. R. Roth. 1984. Salmonella typhimurium synthesizes cobalamin (vitamin B12) de novo under anaerobic growth conditions. J. Bacteriol. 159:206-213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Joblin, K., A. Johnson, M. Lappert, and O. Wallis. 1976. Coenzyme B12 dependent reactions. Part IV. Observations on the purification of ethanolamine ammonia-lyase. Biochim. Biophys. Acta 452:262-270. [DOI] [PubMed] [Google Scholar]
- 17.Johnson, C. L. V. J., E. Pechonick, S. D. Park, G. D. Havemann, N. A. Leal, and T. A. Bobik. 2001. Functional genomic, biochemical, and genetic characterization of the Salmonella pduO gene, an ATP:cob(I)alamin adenosyltransferase gene. J. Bacteriol. 183:1577-1584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Jones, P. W., and J. M. Turner. 1984. A model for common control of enzymes of ethanolamine metabolism in Escherichia coli. J. Gen. Microbiol. 130:849-860. [DOI] [PubMed] [Google Scholar]
- 19.Kofoid, E., C. Rappleye, I. Stojiljkovic, and J. Roth. 1999. The 17-gene ethanolamine (eut) operon of Salmonella typhimurium encodes five homologues of carboxysome shell proteins. J. Bacteriol. 181:5317-5329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lawrence, J. G., and J. R. Roth. 1995. The cobalamin (coenzyme B12) biosynthetic genes of Escherichia coli. J. Bacteriol. 177:6371-6380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Miller, J. H. 1972. Experiments in molecular genetics. Cold Spring Harbor Laboratory, Cold Spring Harbor, N.Y.
- 22.Mori, K., T. Tobimatsu, T. Hara, and T. Toraya. 1997. Characterization, sequencing, and expression of the genes encoding a reactivating factor for glycerol-inactivated adenosylcobalamin-dependent diol dehydratase. J. Biol. Chem. 272:32034-32041. [DOI] [PubMed] [Google Scholar]
- 23.Penrod, J., K. Czymmek, D. Sheppard, E. Kofoid, and J. Roth. Unpublished results.
- 24.Price-Carter, M., J. Tingey, T. A. Bobik, and J. R. Roth. 2001. The alternative electron acceptor tetrathionate supports B12-dependent anaerobic growth of Salmonella enterica serovar Typhimurium on ethanolamine or 1,2-propanediol. J. Bacteriol. 183:2463-2475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Rappleye, C. A., and J. R. Roth. 1997. A Tn10 derivative (T-POP) for isolation of insertions with conditional (tetracycline-dependent) phenotypes. J. Bacteriol. 179:5827-5834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ratzkin, B., and J. Roth. 1978. Cluster of genes controlling proline degradation in Salmonella typhimurium. J. Bacteriol. 133:744-754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Roof, D. M., and J. R. Roth. 1992. Autogenous regulation of ethanolamine utilization by a transcriptional activator of the eut operon in Salmonella typhimurium. J. Bacteriol. 174:6634-6643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Roof, D. M., and J. R. Roth. 1988. Ethanolamine utilization in Salmonella typhimurium. J. Bacteriol. 170:3855-3863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Roof, D. M., and J. R. Roth. 1989. Functions required for vitamin B12-dependent ethanolamine utilization in Salmonella typhimurium. J. Bacteriol. 171:3316-3323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Roth, J. R., J. G. Lawrence, and T. A. Bobik. 1996. Cobalamin (coenzyme B12): synthesis and biological significance. Annu. Rev. Microbiol. 50:137-181. [DOI] [PubMed] [Google Scholar]
- 31.Scarlett, F. A., and J. M. Turner. 1976. Microbial metabolism of amino alcohols. Ethanolamine catabolism mediated by coenzyme B12-dependent ethanolamine ammonia-lyase in Escherichia coli and Klebsiella aerogenes. J. Gen. Microbiol. 95:173-176. [DOI] [PubMed] [Google Scholar]
- 32.Sheppard, D. E., and J. R. Roth. 1994. A rationale for autoinduction of a transcriptional activator: ethanolamine ammonia-lyase (EutBC) and the operon activator (EutR) compete for adenosyl-cobalamin in Salmonella typhimurium. J. Bacteriol. 176:1287-1296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Stojiljkovic, I., A. J. Bäumler, and F. Heffron. 1995. Ethanolamine utilization in Salmonella typhimurium: nucleotide sequence, protein expression, and mutational analysis of the cchA cchB eutJ eutG eutH gene cluster. J. Bacteriol. 177:1357-1366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Suh, S.-J., and J. C. Escalante-Semerena. 1995. Purification and initial characterization of the ATP:corrinoid adenosyltransferase encoded by the cobA gene of Salmonella typhimurium. J. Bacteriol. 177:921-925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Tobimatsu, T., H. Kajiura, M. Yunoki, M. Azuma, and T. Toraya. 1999. Identification and expression of the genes encoding a reactivating factor for adenosylcobalamin-dependent glycerol dehydratase. J. Bacteriol. 181:4110-4113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Toraya, T., and K. Mori. 1999. A reactivating factor for coenzyme B12-dependent diol dehydratase. J. Biol. Chem. 274:3372-3377. [DOI] [PubMed] [Google Scholar]
- 37.Way, J. C., M. A. Davis, D. Morisato, D. E. Roberts, and N. Kleckner. 1984. New Tn10 derivatives for transposon mutagenesis and for construction of lacZ operon fusions by transposition. Gene 32:369-379. [DOI] [PubMed] [Google Scholar]