Abstract
Adaptation in the chemosensory pathways of bacteria like Escherichia coli is mediated by the enzyme-catalyzed methylation (and demethylation) of glutamate residues in the signaling domains of methyl-accepting chemotaxis proteins (MCPs). MCPs can be methylated in trans, where the methyltransferase (CheR) molecule catalyzing methyl group transfer is tethered to the C terminus of a neighboring receptor. Here, it was shown that E. coli cells exhibited adaptation to attractant stimuli mediated through either engineered or naturally occurring MCPs that were unable to tether CheR as long as another MCP capable of tethering CheR was also present, e.g., either the full-length aspartate or serine receptor (Tar or Tsr). Methylation of isolated membrane samples in which engineered tethering and substrate receptors were coexpressed demonstrated that the truncated substrate receptors (trTsr) were efficiently methylated in the presence of tethering receptors (Tar with methylation sites blocked) relative to samples in which none of the MCPs had tethering sites. The effects of ligand binding on methylation were investigated, and an increase in rate was produced only with serine (the ligand specific for the substrate receptor trTsr); no significant change in rate was produced by aspartate (the ligand specific for the tethering receptor Tar). Although the overall efficiency of methylation was lower, receptor-specific effects were also observed in trTar- and trTsr-containing samples, where neither Tar nor Tsr possessed the CheR binding site at the C terminus. Altogether, the results are consistent with a ligand-induced conformational change that is limited to the methylated receptor dimer and does not spread to adjacent receptor dimers.
Bacterial chemotaxis signaling pathways are two-component systems in which the sensors are composed of noncovalent ternary complexes of transmembrane receptor proteins, the cytoplasmic adaptor protein CheW and the histidine kinase CheA (9, 35). The transmembrane receptors, which are also known as methyl-accepting chemotaxis proteins (MCPs), are organized as homodimers to bind ligand (28), where each subunit of the dimer consists of a periplasmic ligand-binding domain (27, 49), two α-helical transmembrane segments (31), and a helical cytoplasmic region that contains the highly conserved signaling domain flanked by the methylation helices (16, 20). Receptor dimers of different ligand specificity cluster in the membrane in synergy with CheW and CheA, frequently at the poles of the cell (26). The available evidence suggests that the receptors in these signaling patches possess the trimer-of-dimers organization, first identified in the crystal structure of the cytoplasmic domain (1, 16, 41). The complexity of the organization is further heightened by the possible involvement of interdigitating cytoplasmic domains, which have been observed by an electron microscope study of receptor arrays (46). Evidence consistent with extensive interactions among receptor dimers in the excitation and adaptation phases of signaling has been obtained from both biochemical analyses of reconstituted systems (21-23) and in vivo analyses of chemotactic ability, signaling, and protein-protein interactions (1, 10, 14, 38, 41). Taken together, the results indicate that interdimer interactions are manifested at a number of levels, i.e., from two dimers to possibly very large arrays of dimers.
Adaptation to stimuli is mediated by reversible methylation of the MCPs through a feedback loop that involves a methyltransferase (CheR) (39), which catalyzes methyl group transfer to specific glutamyl residues in the cytoplasmic domain (33, 43), and a methylesterase (CheB), which catalyzes methyl ester hydrolysis (40). It is now appreciated that methylation can occur in trans (21, 23) because CheR tethers to serine and aspartate receptors (Tsr and Tar) through a conserved motif (NWETF) at the C terminus. From this location, CheR can more effectively catalyze methyl group transfer from S-adenosyl-l-methionine (SAM) to the various methylation sites of neighboring dimers (48). In vitro studies of receptor methylation have utilized mixtures of full-length and truncated receptors with the same ligand specificity (21, 23), which demonstrated that efficient methylation of truncated receptors depended on a full-length receptor for a CheR tethering interaction. Methylation may occur by both intradimer and interdimer processes with receptors of the same ligand specificity (21), but interdimer methylation is the only plausible process available for the ribose-galactose and dipeptide receptors in Escherichia coli (Trg and Tap, respectively) because receptor heterodimers do not form (28) and Trg and Tap lack the CheR-docking site at the C terminus (5, 19). These properties of Trg and Tap are consistent with their low methyl-accepting activity and poor ability to adapt to stimuli when they are the only receptors present in the cell (2, 8, 45) and also with the functional rescue of Trg produced by genetically fusing the NWETF binding motif from Tsr to the C terminus of Trg (8).
In spite of the evidence for extensive interactions among receptors, it must be the case, at least to a certain extent, that ligand-specific responses during excitation and adaptation are retained within the heterogeneous array of receptor signaling complexes. Thus, we designed a set of experiments to test the extent to which the activation of transmethylation was ligand specific by using engineered forms of the aspartate and serine receptors (Tar and Tsr), because both of these receptors bind their respective attractants directly and with high specificity (4, 6, 24). The results provided clear evidence that CheR tethering interactions between receptors of different ligand specificities were essential for adaptation and efficient methylation, yet the increases in the methylation rate that resulted from attractant binding were communicated exclusively through receptor dimers accepting the methyl groups and not by the receptor dimers involved in tethering the transferase. The results of these in vitro experiments also provided evidence that attractant binding produced a much larger activation factor than has been observed previously. Taken together, these results contribute to an understanding of the process of adaptation to disparate stimuli experienced by the bacterial cell.
MATERIALS AND METHODS
Cell strains.
The E. coli strains used in this study (followed by their relevant chemotaxis genotypes) were as follows: RP437 was the wild type for chemotaxis (32); HCB433 [Δtsr(7021) Δtrg(100) zbd::Tn5] had deletions in the genes for the serine and ribose-galactose receptors (tsr and trg, respectively) (47); HCB316 [Δtsr(7021) Δtar-tap(5201)] carried deletions in tsr and in the genes for the aspartate and dipeptide receptors (tar and tap, respectively) (47); HCB721 [Δtsr(7021) trg::Tn10 Δ(cheA-cheY)::XhoI(Tn5)] was devoid of tar, tap, tsr, and trg as well as genes required for phosphotransfer activity in the chemotaxis system (cheA and cheW), receptor methylation (cheR), and receptor demethylation-deamidation (cheB) (7).
Expression vectors.
Table 1 lists the plasmids used to express and/or coexpress Tsr and Tar. The pFA plasmids shown in Table 1 were derived from pHSe5 (29) or pBAD (12) to have compatible origins of replication and independently inducible promoters. A combination of standard subcloning (34) and site-directed mutagenesis protocols (Quickchange; Stratagene Corp.) were used to introduce tar (from pAK101R1) (18) into pFA23 and pFA24 and modify it to produce glutamines at the four major methylation sites (295, 302, 309, and 491) (42). The resulting plasmids expressed full-length Tar (TarQQQQ; pFA23) or Tar with 39 amino acids truncated from the C terminus (trTarQQQQ; pFA24). A plasmid that produced the C-terminal truncated form of Tsr (trTsr) with the third methylation site available (trTsrQQEQ; pFA32) was constructed from pJL31 (trTsrQQQQ) by site-directed mutagenesis. The tar and tsr genes in these vectors were verified by sequencing. All plasmids were monitored for protein production and showed bands with the expected molecular masses on sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) gels. For the behavioral studies, pBR322 was used as a control plasmid that did not express MCP.
TABLE 1.
Plasmids used in behavioral experiments and the production of membrane samples
| Plasmid | Receptor expressed | Origin of replication | Inducer | Parent plasmid | Reference or source |
|---|---|---|---|---|---|
| pHSe5.tsrQEQE | TsrQEQE | ColE1 | IPTG | pHSe5a | 33 |
| pJL21 | trTsrQEQE | ColE1 | IPTG | pHSe5.tsrQEQE | 23 |
| pHSe5.tsrQQQQ | TsrQQQQ | ColE1 | IPTG | pHSe5 | 33 |
| pJL31 | trTsrQQQQ | ColE1 | IPTG | pHSe5.tsrQQQQ | 23 |
| pAC03 | TsrQQEQ | ColE1 | IPTG | pHSe5.tsrQQQQ | This study |
| pFA32 | trTsrQQEQ | ColE1 | IPTG | pJL31 | This study |
| pFA23 | TarQQQQ | p15 | Arabinose | pBAD33b | This study |
| pFA24 | trTarQQQQ | p15 | Arabinose | pBAD33 | This study |
Swarm and swimming assays.
Swarm assays were conducted in semisolid (0.3%) agar plates made with M9 medium [0.05 M Na2HPO4 · 7H2O, 0.02 M KH2PO4, 1.0 mM NaCl, 1.0 mM MgSO4, 5.0 mM (NH4)2SO4] supplemented with 0.4% glycerol, thiamine (10 μg/ml), ampicillin (150 μg/ml) and histidine, methionine, leucine, and threonine (each at 50 μg/ml). The attractants aspartate, serine, and ribose were incorporated at 100 μM. Swarm plates were inoculated with log-phase M9 cultures and incubated for 12 to 14 h at 30°C, at which time dark-field observation was used to measure the swarm ring radius every 2 h over a 10-h period. The average swarm rates were determined from nine data sets.
To analyze ligand-stimulated adaptation by free-swimming bacteria, fresh overnight cultures grown in M9 medium were diluted 1:10 into 5 ml of medium and were grown at 30°C until an optical density at 600 nm of ∼0.7 was reached. The culture was prepared for microscope observation by first diluting an aliquot with an equal volume of motility medium (0.67 M NaCl, 0.01 M KH2PO4, 0.01 M Na lactate, 1.0 mM EDTA, 10 μM methionine, 10 μM leucine) and was then mixed with an equal amount of attractant-supplemented motility medium. A 30-μl aliquot of the mixture was immediately applied to a microscope slide within a ring of vacuum grease, which was then sealed beneath a cover glass, at which point the cells were videotaped under the microscope. Videotapes were subsequently analyzed to assess adaptation from the time dependence of tumble frequencies. Tumble events were identified as abrupt cell turnings (∼0.1-s duration) followed by a smooth swimming episode (∼1-s duration). Tumble frequencies (per cell per second) at each time point were determined from an average over a 10-s period for at least 10 different cells. Adaptation times were estimated as the period of time required to achieve 50% adaptation, i.e., halfway between the tumble frequency observed immediately after stimulus application and the fully adapted state. Baseline tumble frequencies were determined in control experiments involving each cell strain-plasmid combination used in which the diluted culture aliquot was mixed with an equal volume of motility medium without attractant. Swarm rates and adaptation times were measured for cell strain-plasmid combinations without added IPTG (isopropyl-β-d-thiogalactopyranoside).
Membrane preparations.
HCB721 was used to isolate membrane samples expressing and/or coexpressing Tsr and Tar, which ensured that the receptors did not undergo the posttranslational modifications catalyzed by CheR and CheB. Receptor-containing inner membrane fractions were isolated on sucrose gradients as previously described (9, 24). The Luria broth cultures of HCB721 containing compatible plasmids were supplemented with ampicillin (150 μg/ml) and/or chloramphenicol (10 μg/ml) and were grown to an optical density at 600 nm of 0.6, at which time expression was induced by the addition of 1 mM IPTG and/or 0.2% arabinose. Estimates of the receptor concentrations in the membrane samples were conducted by comparisons to an affinity-purified trTsr standard on SDS-PAGE gels by using scanning densitometry (GS-700 densitometer with Molecular Analyst version 1.4 software; Bio-Rad).
Methylation assays.
Salmonella enterica serovar Typhimurium CheR was purified according to a published procedure (37). Samples were typically composed of a solution containing 7 μM methylatable receptor, 14 μM [3H-methyl]SAM (15 Ci/mmol; Amersham Biosciences, Piscataway, N.J.), 1 μM CheR, and, as needed, a 1 mM concentration of the attractant in 100 μl of buffer (50 mM sodium phosphate [pH 7.5], 1 mM EDTA, 10% glycerol, 1 mM phenylmethylsulfonyl fluoride). Methylation reactions were initiated by the addition of CheR. At various times, 14-μl aliquots were removed and quenched by the addition of 3× SDS (7 μl) reducing sample buffer followed by 4 min in a boiling water bath. Aliquots of 14 μl were resolved on SDS-PAGE gels (10% [wt/vol] acrylamide), stained for 10 min with Gel Blue Code stain (Pierce Biotechnology, Inc.), and washed with water. Receptor-containing bands were excised and placed in scintillation vials containing 1 ml of 1 M NaOH followed by the addition of 2 ml of scintillation fluid. The extent of tritiated methyl group incorporation was estimated based on the assumption of 100% sample recovery. The dependence of methylation on receptor concentration was measured in samples containing fixed amounts of [3H]SAM and CheR (14 and 1 μM, respectively) but a range of concentrations of methylatable receptor (1.5 to 20 μM).
RESULTS
The CheR-docking segment contributes to efficient cellular adaptation and chemotaxis.
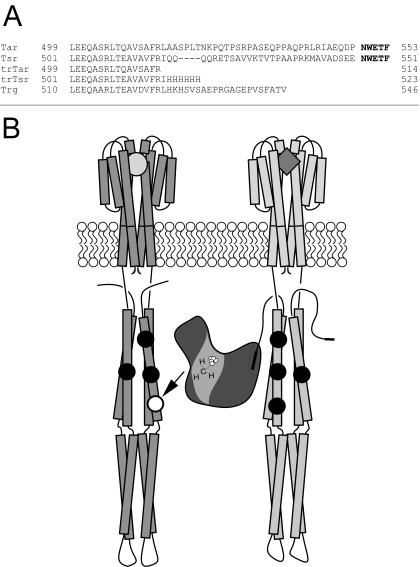
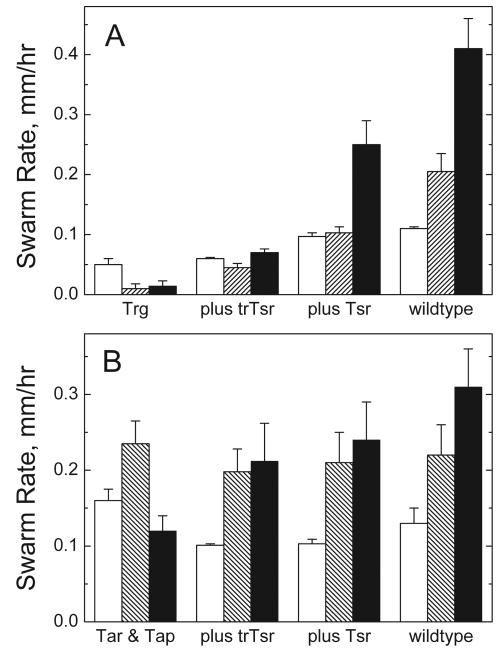
Figure 1A shows the C-terminal sequences of the natural and engineered MCPs used to assess the requirement for the CheR-docking segment (NWETF) in adaptation and to test for the ligand specificity in the activation of methylation. The C-terminal deletions in Tar and Tsr resulted in engineered forms (trTar and trTsr, respectively) that were unable to bind CheR at the docking site (23, 48), analogous to the naturally occurring form of Trg (Fig. 1A). Interdimer methylation was thus the only expected means of efficient methylation (Fig. 1B), since the formation of receptor heterodimers with two subunits of different ligand binding specificity has not been observed (28). Comparison of the swarm rates in Fig. 2 supported the idea that the presence of a receptor with a CheR-docking site was necessary for efficient chemotaxis and could provide assistance to receptors lacking the CheR-docking site since (i) the introduction of a receptor with the CheR-docking site improved the overall chemotactic ability (swarm rate of Trg plus Tsr > Trg plus trTsr ∼ Trg [Fig. 2A]) and (ii) the swarm rate of the truncated form of Tsr (trTsr) was assisted significantly by the presence of full-length Tar. (The swarm rate of Trg plus trTsr in Fig. 2A was significantly less than the swarm rate of Tar & Tap plus trTsr in Fig. 2B.)
FIG. 1.
(A) The aligned C termini of native (Tar, Tsr, and Trg) and engineered (trTar and trTsr) MCPs, with the CheR-docking site (NWETF) shown in boldface type. (B) Cartoon that depicts engineered interdimer methylation between homodimers of trTsr and Tar (left and right, respectively). CheR is bound to the Tar dimer at the docking site (depicted as the C-terminal filled rectangle), which is fully amidated (TarQQQQ) at the sites of methylation (depicted as filled circles). The trTsr dimer has one site available for methylation (site 3, depicted as an open circle) and is unable to bind CheR.
FIG. 2.
Swarm rates of cells expressing either full-length or truncated serine receptor (Tsr or trTsr) in combination with Trg (A) or Tar (B) on semisolid agar plates. Panel A (left to right): swarm rates of plasmid-containing HCB316 expressing Trg (HCB316/pBR322), Trg and trTsr (HCB316/pJL21), Trg and Tsr (HCB316/pHSe5.tsrQEQE), and also RP437/pBR322 (wild type for chemotaxis). Open bars, without attractant; striped bars, 100 μM ribose; filled bars, 100 μM serine. Panel B (left to right): swarm rates of plasmid-containing HCB433 expressing Tar and Tap (HCB433/pBR322); Tar, Tap, and trTsr (HCB433/pJL21); Tar, Tap, and Tsr (HCB433/pHSe5.tsrQEQE); and RP437/pBR322. Open bars, without attractant; striped bars, 100 μM aspartate; filled bars, 100 μM serine. Uncertainties are standard deviations.
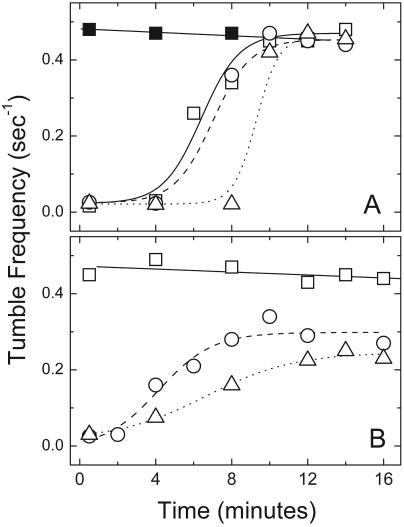
Adaptation to attractants by free-swimming cells expressing various combinations of Trg, trTsr, Tsr, and Tar provided evidence for the universal nature of the assistance by receptors with a CheR-docking segment (Fig. 3 and Table 2). Adaptation times were used as indicators of the relative efficiency of receptor methylation (Fig. 3). The Tar+ HCB433 cells exhibited full adaptation to aspartate, whether the cells were Tsr− or also expressed trTsr or Tsr (Fig. 3A). HCB433 also responded and adapted partially to serine when trTsr (or Tsr) was present (Fig. 3B), which was consistent with functional assistance from Tar. It was noted that the extents of adaptation to aspartate and serine were significantly different; full adaptation to aspartate was observed, while adaptation to serine was partial. Partial adaptation to serine has been observed in a previous study with wild-type chemotaxis strain AW405 (3). We also observed partial adaptation with wild-type strain RP437 (data not shown). Thus, partial adaptation seems to be a normal feature of the serine response and not an artifact of either chromosome-versus-plasmid-based tsr expression or the C-terminal truncation. Consequently, the similar adaptation times in cells expressing either trTsr or Tsr with Tar were interpreted as evidence of functional interaction with Tar in the interdimer methylation of trTsr.
FIG. 3.
Tumble frequencies as a function of time after the introduction of 50 μM aspartate (A) or 50 μM serine (B). HCB433/pBR322, □ (Tap and Tar); HCB433/pHSe5.tsrQEQE, ○ (Tap, Tar, and Tsr); HCB433/pJL21, ▵ (Tap, Tar, and trTsr); HCB433/pBR322 (no attractant control), ▪ (A). The curves drawn through the data are either least-square lines or sigmoid functions to help guide the eyes.
TABLE 2.
Adaptation times of swimming cells
| Sample no. | Strain/plasmid | Receptor(s) with NWETF | Receptor(s) without NWETF | Stimulus (concn [μM]) | Adaptation time (min) |
|---|---|---|---|---|---|
| 1 | HCB433/pBR322 | Tar | Tap | Aspartate (50) | 6 |
| 2 | HCB433/pHSe5.tsr | Tar, Tsr | Tap | Aspartate (50) | 6 |
| 3 | HCB433/pJL21 | Tar | Tap, trTsr | Aspartate (50) | 9 |
| 4 | HCB433/pHSe5.tsr | Tar, Tsr | Tap | Serine (50) | 4 |
| 5 | HCB433/pJL21 | Tar | Tap, trTsr | Serine (50) | 6 |
| 6 | HCB316/pHSe5.tsr | Tsr | Trg | Serine (100) | 6 |
| 7 | HCB316/pJL21 | Trg, trTsr | Serine (50) | >20 | |
| 8 | HCB316/pHSe5.tsr | Tsr | Trg | Ribose (100) | 2 |
| 9 | HCB316/pJL21 | Trg, trTsr | Ribose (100) | >20 | |
| 10 | RP437/pBR322 | Tar, Tsr | Tap, Trg | Aspartate (100) | 8 |
| 11 | RP437/pBR322 | Tar, Tsr | Tap, Trg | Ribose (100) | 2 |
| 12 | RP437/pBR322 | Tar, Tsr | Tap, Trg | Serine (100) | 10 |
Table 2 summarizes the adaptation times of swimming cells to aspartate, ribose, and serine stimuli. The adaptation times were readily divided into two groups, which correlated with the expected efficiencies of methyl group addition to the receptors conveying the attractant stimuli. Receptors were regarded as efficiently methylated when the adaptation times were comparable to those of wild-type cells (Table 2, samples 10 to 12). In contrast, adaptation was not observed with inefficiently methylated receptors. In every situation where adaptation was observed, the receptor conveying the attractant stimulus either had a CheR-docking site or was coexpressed with a receptor with the docking site. Conversely, adaptation was not observed when receptors without the CheR-docking segment were the only receptors present in the cell (Trg and trTsr [Table 2, samples 7 and 9]). Specifically, adaptation to the ribose stimulus sensed through Trg was assisted by the presence of Tsr but not trTsr (Table 2, sample 8 versus sample 9), a result that was in agreement with previous observations of adaptation to ribose stimuli, where the presence of Tar and/or Tsr was required for efficient adaptation (13). The data in Table 2 extended the generality of the result, since adaptation to serine could be conveyed through trTsr when Tar was also present (Table 2, sample 5 versus sample 7). Overall, the observations made in these swarm and adaptation assay experiments were consistent with promiscuous interdimer methylation; i.e., any receptor conveying the signal generated by ligand binding could be assisted by any receptor with a CheR-docking site irrespective of ligand binding specificities of the two receptor dimers involved. The most important factor contributing to efficient adaptation (and thus methylation) was the presence of at least one receptor with a CheR-docking segment.
Ligand specificity of transmethylation.
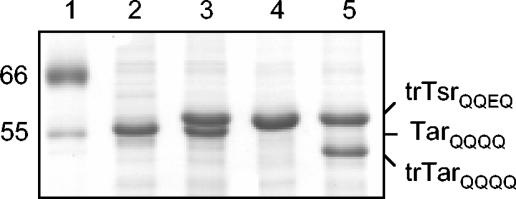
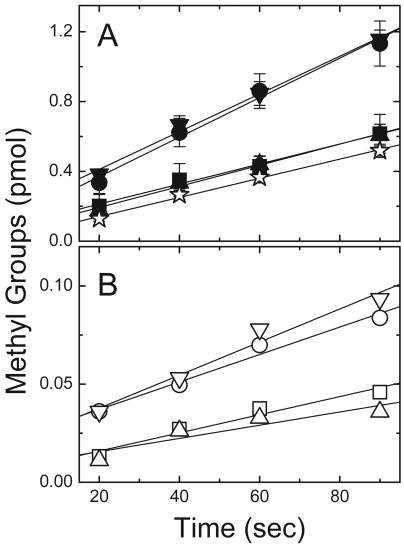
In vitro methylation was used to assess the extent of the interactions among receptor subunits by examining the influence of attractants in inner membrane samples with coexpressed TarQQQQ and trTsrQQEQ (trTsrQQEQ/TarQQQQ). These inner membrane samples, prepared from HCB721, contained MCPs that were in defined levels of covalent modification (Fig. 4) and in comparable amounts when two MCPs were coexpressed (Fig. 4, lanes 3 and 5). Methylation of these samples was expected to proceed by the interdimer process depicted in Fig. 1B, since TarQQQQ provided the transferase-docking site but lacked an available major methylation site and trTsrQQEQ had one available major methylation site (E311) but lacked the transferase-docking site. To test for ligand-specific responses, the methylation of trTsrQQEQ/TarQQQQ-containing membranes was measured in the absence of attractant and in the presence of serine, aspartate, or both serine and aspartate (Table 3). The methylation rate increased significantly when serine was present and to a similar extent when both serine and aspartate were present, but it did not increase significantly when aspartate was the only ligand added (Fig. 5A; Table 3, samples 1 to 4). This increase was evident in ratios of the serine receptor methylation rates with attractant(s) to the rate without attractant (2.0 and 1.9 versus 1.1, respectively, in Table 3). A control sample consisting of coexpressed trTsrQQQQ/TarQQQQ was used to measure incorporation at the minor methylation sites, which are known to contribute to Tsr methylation (33). The structural similarity of Tsr cytoplasmic fragments in the QEQE and QQQQ modification states determined by X-ray crystallography (17) suggests that trTsrQQQQ is well suited as a control for trTsrQQEQ in these experiments, although it is possible that the cytoplasmic domains in the intact receptor molecules adopt different structures. The methylation rate of the trTsrQQQQ/TarQQQQ sample was not affected by either serine or aspartate (data not shown); thus, the rate reported in Table 3 (0.31 pmol/min), which represented an average of several experiments conducted with and without ligand, was subtracted as background. The background-subtracted rates indicated that the contribution to methylation from the minor sites was significant. Thus, in spite of the greater uncertainty in ratios calculated with the background-subtracted data (due to the small value for the background-corrected rate in the absence of serine), it seemed that the rate increase at site 3 produced by serine binding was likely to be larger than the ratios calculated with the uncorrected rates suggested.
FIG. 4.
SDS-PAGE (12.5% gels) of inner membrane samples expressing Tsr and/or Tar. Lanes: 1, molecular weight markers (in thousands); 2, TarQQQQ; 3, coexpressed TarQQQQ and trTsrQQEQ; 4, trTsrQQEQ; 5, coexpressed trTarQQQQ and trTsrQQEQ.
TABLE 3.
Initial rates of serine receptor methylation
| Sample no. | Coexpressed receptors | Liganda |
Methylation (pmol/min) |
Background subtracted (pmol/min)b |
|||
|---|---|---|---|---|---|---|---|
| Serine | Aspartate | Rate ± SD | Ligand effectc | Rate ± SD | Ligand effectc | ||
| 1 | trTsrQQEQ/TarQQQQ | − | − | 0.34 ± 0.03 | 0.03 ± 0.04 | ||
| 2 | trTsrQQEQ/TarQQQQ | + | − | 0.68 ± 0.05 | 2.0 | 0.37 ± 0.06 | 12 |
| 3 | trTsrQQEQ/TarQQQQ | − | + | 0.36 ± 0.05 | 1.1 | 0.05 ± 0.05 | 1.6 |
| 4 | trTsrQQEQ/TarQQQQ | + | + | 0.65 ± 0.04 | 1.9 | 0.33 ± 0.04 | 11 |
| 5 | trTsrQQEQ/trTarQQQQ | − | − | 0.028 | |||
| 6 | trTsrQQEQ/trTarQQQQ | + | − | 0.042 | 1.5 | ||
| 7 | trTsrQQEQ/trTarQQQQ | − | + | 0.020 | 0.7 | ||
| 8 | trTsrQQEQ/trTarQQQQ | + | + | 0.050 | 1.8 | ||
| 9 | trTsrQEQE/TarQQQQ | − | − | 2.64 ± 0.24 | 2.34 ± 0.24 | ||
| 10 | trTsrQEQE/trTarQQQQ | + | − | 3.18 ± 0.42 | 1.2 | 2.88 ± 0.42 | 1.2 |
| 11 | trTsrQEQE/trTarQQQQ | − | + | 3.12 ± 0.18 | 1.2 | 2.82 ± 0.18 | 1.2 |
| 12 | trTsrQEQE/trTarQQQQ | + | + | 3.12 ± 0.06 | 1.2 | 2.82 ± 0.12 | 1.2 |
+ and − indicate the presence and absence, respectively, of serine (1 mM) or aspartate (1 mM).
The rate of methyl group incorporation (0.31 ± 0.02 pmol/min) into samples of coexpressed trTsrQQQQ and TarQQQQ was subtracted.
Rate in the presence of serine, aspartate, or both divided by the rate in the absence of ligand.
FIG. 5.
Methylation of inner membranes coexpressing Tar and Tsr. (A) Coexpressed TarQQQQ and trTsrQQEQ (filled symbols) and a control sample (coexpressed TarQQQQ and trTsrQQQQ [⋆]). (B) Coexpressed trTarQQQQ and trTsrQQEQ (open symbols). ▪ and □, no ligand; • and ○, 1 mM serine; ▴ and ▵, 1 mM aspartate; ▾ and ▿, both serine and aspartate. Reaction conditions included a solution containing 7 μM methylatable receptor (trTsrQQEQ), 1 μM CheR, and 14 μM [3H]SAM.
The third site (E311) was chosen at the outset of these studies to be the only major site available for methylation, based on previous studies of Tar in which site 3 was found to be the most rapidly methylated site by far (44) and on the significant homology between Tar and Tsr in the methylation helices (33, 43). However, rate measurements at the major sites in full-length Tsr have demonstrated that site 4 is methylated over 10-fold more rapidly than either site 2 or site 3 (in TsrQQQE, TsrQEQQ, and TsrQQEQ samples, respectively) (A. Chalah and R. M. Weis, unpublished data). Not only were comparable methylation rates observed for trTsrQQEQ/TarQQQQ (Table 3, sample 1) and TsrQQEQ samples (data not shown) under similar assay conditions, the methylation rates of trTsrQEQE/TarQQQQ and trTsrQQEQ/trTarQQQQ differed by ∼8-fold (Table 3, samples 1 and 9), which was qualitatively consistent with the rate difference in the full-length Tsr molecules. Also, the lack of a significant increase in the methylation rate upon the addition of serine in the trTsrQEQE/TarQQQQ samples (Table 3, samples 9 to 12) mirrored the behavior of (full-length) TsrQQQE, which showed no demonstrable increase in the methylation rate with serine (Chalah and Weis, unpublished). Thus, the methylation rate of trTsrQEQE/TarQQQQ could be plausibly attributed to rapid methylation at site 4, which was unaffected by serine. Fortuitously, serine produced a more pronounced effect on the methylation rate at site 3 (E311) in these transmethylation experiments.
Ligand-specific stimulation does not require CheR tethering.
To test the requirement of the docking segment for efficient methylation and ligand-specific stimulation, methylation assays were performed on coexpressed Tar and Tsr samples in which neither MCP possessed the CheR-docking site (trTsrQQEQ/trTarQQQQ) (Fig. 5B; Table 3, samples 5 to 8). The addition of serine produced the same proportional rate increase in the trTsrQQEQ/trTarQQQQ samples as in trTsrQQEQ/TarQQQQ, although the absolute rate was 10-fold lower with the doubly truncated samples (note the different y axis scales in Fig. 5A and B). Overall, the doubly truncated samples displayed the same ligand-specific response as the trTsrQQEQ/TarQQQQ-containing membranes. The lower methylation rates of the doubly truncated receptor samples (Table 3, samples 5 to 8) compared to the control sample for the minor methylation sites (trTsrQQQQ/TarQQQQ) (Table 3) underscored the requirement of a CheR-docking segment for efficient receptor methylation. In any event, the corresponding background (trTsrQQQQ/trTarQQQQ) sample was not prepared or tested due to the already low initial rate observed with trTsrQQEQ/trTarQQQQ.
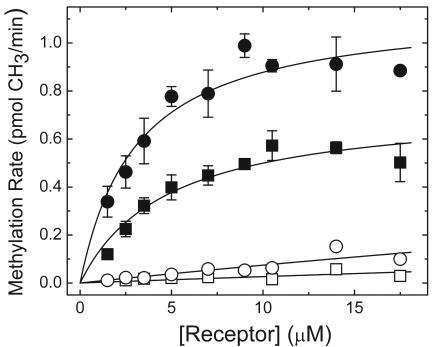
Further evidence for the significant difference in the methylation efficiency of trTsrQQEQ/trTarQQQQ from that of trTsrQQEQ/TarQQQQ was obtained from initial rate measurements conducted as a function of the trTsrQQEQ concentration, which was produced by varying the total amount of membrane (Fig. 6). The trTsrQQEQ/TarQQQQ data resulted in similar Km values in the absence and presence of serine (4 and 3 μM, respectively), and the Vmax increased by ∼2-fold. trTsrQQEQ/trTarQQQQ samples could not be prepared at sufficiently large concentrations to estimate Vmax and Km values independently, yet the low rates of methylation in these samples were plausibly explained by fits in which the Vmax was fixed to the values obtained with coexpressed trTsrQQEQ/TarQQQQ, which led to larger Km values (>100 μM). Altogether, the properties of trTsrQQEQ/TarQQQQ and trTsrQQEQ/trTarQQQQ samples were consistent with a ligand-specific activation of receptor methylation within the receptor dimer accepting the methyl groups. These properties were consistent with the hypothesis that the binding of CheR to the receptor via NWETF served to increase the effective CheR concentration near the sites of methylation.
FIG. 6.
Methylation rates as a function of the trTsrQQEQ concentration in trTsrQQEQ/TarQQQQ samples in the absence of ligand (▪) and in the presence of 1 mM serine (•) and in trTsrQQEQ/trTarQQQQ samples in the absence of ligand (□) and in the presence of 1 mM serine (○) are shown. Samples at the different trTsrQQEQ concentrations also included 1 μM CheR and 14 μM [3H]SAM.
DISCUSSION
The chemoreceptor clusters localized in the inner membrane of E. coli are probably heterogeneous with respect to ligand binding specificity (41). Within this heterogeneous environment, adaptation is achieved by modulating the rates of receptor methylation and demethylation in response to changes in the concentration of attractant. An increase in the attractant concentration speeds methylation (11) and also slows demethylation transiently by inhibiting CheA-mediated phosphorylation (and activation) of CheB until adaptation is reestablished (15, 25). Thus, ligand-specific adaptation may be achieved by ligand-specific increases in methylation, ligand-specific decreases in demethylation, or both. Such ligand-specific effects must be at work in addition to the global influences that have been proposed to contribute to adaptation, e.g., a global activation of methylesterase activity brought about by CheA-mediated phosphorylation of CheB (15).
Here, evidence that defines the features of ligand-specific receptor methylation is presented. The present results, together with those of previous studies (8, 21, 23), indicate that efficient receptor methylation can occur as long as a receptor that possesses the CheR-docking segment (NWETF) (30, 48) is also present. It has been shown previously that methylation rates of truncated receptors, when coexpressed with full-length receptors of the same type, e.g., trTar/Tar and trTsr/Tsr, are similar to the rates measured with their full-length counterparts (21, 23). Moreover, the methylation of Tsr in trTsr/TarQQQQ samples (Table 3) and in full-length Tsr samples (Chalah and Weis, unpublished) occurs at similar rates. The CheR-docking segment is required for efficient methylation and adaptation. When all the MCPs in a membrane sample lack the CheR-docking segment, methylation rates are decreased more than 10-fold compared to samples in which at least one of the MCPs has the docking segment (Table 3 and Fig. 6) (2, 21). Also, cells fail to adapt to an attractant stimulus in behavioral assays whenever all the MCPs lack the CheR-docking segment (Table 2) (13). Altogether, the results of these studies reinforce the notion that interdimer methylation can occur between receptor dimers of any ligand specificity; e.g., Tsr can be assisted by Tar, Trg can be assisted by either Tsr or Tar, etc. The molecular basis for this interaction is interpretable within the framework of interdimer methylation (21, 23), the trimer-of-dimer organization (36) that was first identified in crystals of the Tsr cytoplasmic domain (16), and in vivo evidence which has been obtained through site-directed mutagenesis and site-specific cross-linking (1, 41).
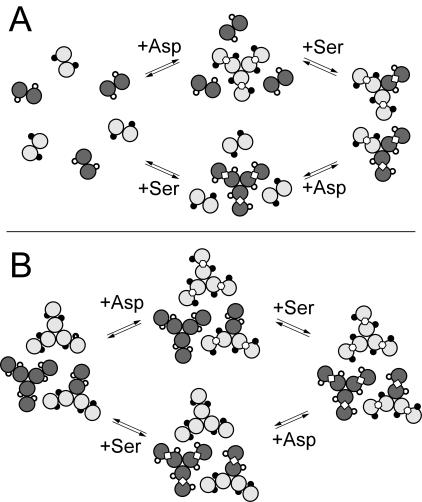
By utilizing an engineered form of Tsr with one available methylation site, but without the CheR-docking segment (trTsrQQEQ), and an engineered form of Tar with the CheR-docking segment, but without a major methylation site available (TarQQQQ), the individual and collective influences of attractant binding to the substrate (trTsrQQEQ) and tethering (TarQQQQ) receptors could be tested. Two possible effects of attractant binding are depicted in Fig. 7, which we imagine could be at work within the context of a cluster of receptors and interdimer methylation. First, a ligand-induced change in proximity could lead to an increase in the methylation rate by reducing distances between receptor dimers, thereby giving tethered CheR molecules access to a larger number of methylation sites (Fig. 7A). Second, a ligand-induced change in receptor conformation could improve the ability of the receptor to function as a substrate (Fig. 7B). The experimental results support a ligand-induced change in conformation, akin to the process depicted in Fig. 7B.
FIG. 7.
Illustrations of ligand-induced changes in methylation based on (A) changes in proximity and (B) changes in receptor dimer conformation. Each pair of circles represents a receptor dimer cytoplasmic domain in cross section (TarQQQQ, light gray; trTsrQQEQ, dark gray), where the sites of methylation are depicted as circles on the surface of the domain (available, open circles; blocked, filled circles). Serine (Ser, ⋄) and aspartate (Asp, ) are shown bound to the dimer interface.
Figure 7A serves to illustrate that ligand-induced changes in receptor proximity might be somewhat ligand specific within the heterogeneous receptor (dimer) environment, where binding affects the proximity in the immediate neighborhood of receptors binding attractant. Yet, according to this line of reasoning, aspartate as well as serine should alter the proximity between TarQQQQ and TsrQQEQ subunits and consequently should also influence the methylation rate. This was not observed, as only serine produced an increase in methylation (Fig. 5 and Table 3). A ligand-specific effect can be more strictly maintained when a propagated conformational change is confined to the dimer that binds ligand, which improves the methylation rate only on that dimer. Figure 7B illustrates one possible way in which ligand binding can influence the receptor dimer as a change in the relative orientation of the methylation helices within the cytoplasmic domain, which positions the methyl-accepting glutamyl residues more favorably for methylation. A mechanism of this kind is consistent with the following experimental observations. (i) Only the attractant that binds to the methyl-accepting dimer (trTsrQQEQ) increases the methylation rate, whereas the attractant that binds to the tethering dimer (TarQQQQ) does not. (ii) The CheR tethering interaction is not required to observe ligand-specific increases in the methylation rate. Although the overall efficiency of methylation is lower when no tethering subunits are present, as in the trTsrQQEQ/trTarQQQQ samples, ligand-specific increases in the methylation rate are still observed.
A consistent mechanism of receptor methylation emerges in which receptor dimers are near enough to mediate efficient interdimer methylation via the CheR tethering interaction but in which ligand-specific effects are confined to the receptor dimer that binds ligand. Thus, the process of receptor methylation represents a ligand-specific process not regulated at a global level; in contrast, the regulation of the kinase CheA seems to be distributed throughout a receptor array, one that is sensitive to both the extent of ligand binding and the level of covalent modification.
Acknowledgments
We thank Anthony Shrout for purified CheR. We also appreciate helpful discussions with Abdalin Asinas, Tatiana Besschetnova, Anas Chalah, David Montefusco, Anthony Shrout, and Li Zhi.
This work was supported by the U.S. Public Health Service NIH grant GM53210 to R.M.W.
REFERENCES
- 1.Ames, P., C. A. Studdert, R. H. Reiser, and J. S. Parkinson. 2002. Collaborative signaling by mixed chemoreceptor teams in Escherichia coli. Proc. Natl. Acad. Sci. USA 99:7060-7065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Barnakov, A. N., L. A. Barnakova, and G. L. Hazelbauer. 1998. Comparison in vitro of a high- and low-abundance chemoreceptor of Escherichia coli: similar kinase activation but different methyl-accepting activities. J. Bacteriol. 180:6713-6718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Berg, H. C., and D. A. Brown. 1972. Chemotaxis in Escherichia coli analysed by three-dimensional tracking. Nature 239:500-504. [DOI] [PubMed] [Google Scholar]
- 4.Biemann, H. P., and D. E. Koshland, Jr. 1994. Aspartate receptors of Escherichia coli and Salmonella typhimurium bind ligand with negative and half-of-the-sites cooperativity. Biochemistry 33:629-634. [DOI] [PubMed] [Google Scholar]
- 5.Bollinger, J., C. Park, S. Harayama, and G. L. Hazelbauer. 1984. Structure of the Trg protein: homologies with and differences from other sensory transducers of Escherichia coli. Proc. Natl. Acad. Sci. USA 81:3287-3291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Clarke, S., and D. E. Koshland, Jr. 1979. Membrane receptors for aspartate and serine in bacterial chemotaxis. J. Bacteriol. 254:9695-9702. [PubMed] [Google Scholar]
- 7.Conley, M. P., A. J. Wolfe, D. F. Blair, and H. C. Berg. 1989. Both CheA and CheW are required for reconstitution of chemotactic signaling in Escherichia coli. J. Bacteriol. 171:5190-5193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Feng, X., A. A. Lilly, and G. L. Hazelbauer. 1999. Enhanced function conferred on low-abundance chemoreceptor Trg by a methyltransferase-docking site. J. Bacteriol. 181:3164-3171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gegner, J. A., D. R. Graham, A. F. Roth, and F. W. Dahlquist. 1992. Assembly of an MCP receptor, CheW and kinase CheA complex in the bacterial chemotaxis signal transduction pathway. Cell 70:975-982. [DOI] [PubMed] [Google Scholar]
- 10.Gestwicki, J. E., and L. L. Kiessling. 2002. Inter-receptor communication through arrays of bacterial chemoreceptors. Nature 415:81-84. [DOI] [PubMed] [Google Scholar]
- 11.Goy, M., M. S. Springer, and J. Adler. 1977. Sensory transduction in Escherichia coli: role of a protein methylation reaction in sensory adaptation. Proc. Natl. Acad. Sci. USA 74:4964-4968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Guzmán, L. M., D. Belin, M. Carson, and J. Beckwith. 1995. Tight regulation, modulation, and high-level expression by vectors containing the arabinose PBAD promoter. J. Bacteriol. 177:4121-4130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hazelbauer, G. L., and P. Engström. 1980. Parallel pathways for transduction of chemotactic signals in Escherichia coli. Nature 283:98-100. [DOI] [PubMed] [Google Scholar]
- 14.Homma, M., D. Shiomi, M. Homma, and I. Kawagishi. 2004. Attractant binding alters arrangement of chemoreceptor dimers within its cluster at a cell pole. Proc. Natl. Acad. Sci. USA 101:3462-3470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kehry, M. R., T. G. Doak, and F. W. Dahlquist. 1984. Stimulus-induced changes in methylesterase activity during chemotaxis in Escherichia coli. J. Biol. Chem. 259:11828-11835. [PubMed] [Google Scholar]
- 16.Kim, K. K., H. Yokota, and S.-H. Kim. 1999. Four-helical-bundle structure of the cytoplasmic domain of the serine chemotaxis receptor. Nature 400:787-792. [DOI] [PubMed] [Google Scholar]
- 17.Kim, S.-K., H. Yokota, and K. K. Kim. 2002. Dynamic and clustering model of bacterial chemotaxis receptors: structural basis for signaling and high sensitivity. Proc. Natl. Acad. Sci. USA 99:11611-11615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Krikos, A., M. P. Conley, A. Boyd, H. C. Berg, and M. I. Simon. 1985. Chimeric chemosensory transducers of Escherichia coli. Proc. Natl. Acad. Sci. USA 82:1326-1330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Krikos, A., N. Mutoh, A. Boyd, and M. I. Simon. 1983. Sensory transducers of E. coli are composed of discrete structural and functional domains. Cell 33:615-622. [DOI] [PubMed] [Google Scholar]
- 20.Le Moual, H., and D. E. Koshland, Jr. 1996. Molecular evolution of the C-terminal cytoplasmic domain of a superfamily of bacterial receptors involved in taxis. J. Mol. Biol. 261:568-585. [DOI] [PubMed] [Google Scholar]
- 21.Le Moual, H., T. Quang, and D. E. Koshland, Jr. 1997. Methylation of the Escherichia coli chemotaxis receptors: intra- and interdimer mechanisms. Biochemistry 36:13441-13448. [DOI] [PubMed] [Google Scholar]
- 22.Li, G., and R. M. Weis. 2000. Covalent modification regulates ligand binding to receptor complexes in the chemosensory system of Escherichia coli. Cell 100:357-365. [DOI] [PubMed] [Google Scholar]
- 23.Li, J., G. Li, and R. M. Weis. 1997. The serine chemoreceptor from Escherichia coli is methylated through an inter-dimer process. Biochemistry 36:11851-11857. [DOI] [PubMed] [Google Scholar]
- 24.Lin, L., J. Li, J. F. Brandts, and R. M. Weis. 1994. The serine receptor of bacterial chemotaxis exhibits half-site saturation for serine binding. Biochemistry 33:6564-6570. [DOI] [PubMed] [Google Scholar]
- 25.Lupas, A., and J. Stock. 1989. Phosphorylation of an N-terminal regulatory domain activates the CheB methylesterase in bacterial chemotaxis. J. Biol. Chem. 264:17337-17342. [PubMed] [Google Scholar]
- 26.Maddock, J. L., and L. Shapiro. 1993. Polar location of the chemoreceptor complex in the Escherichia coli cell. Science 259:1717-1723. [DOI] [PubMed] [Google Scholar]
- 27.Milburn, M. V., G. G. Privé, D. L. Milligan, W. G. Scott, J. Yeh, J. Jancarak, D. E. Koshland, Jr., and S. H. Kim. 1991. Three-dimensional structures of the ligand-binding domain of the bacterial aspartate receptor with and without a ligand. Science 254:1342-1347. [DOI] [PubMed] [Google Scholar]
- 28.Milligan, D. L., and D. E. Koshland, Jr. 1988. Site-directed cross-linking. Establishing the dimeric structure of the aspartate receptor of bacterial chemotaxis. J. Biol. Chem. 263:6268-6275. [PubMed] [Google Scholar]
- 29.Muchmore, D. C., L. P. McIntosh, C. B. Russell, E. Anderson, and F. W. Dahlquist. 1989. Expression and nitrogen-15 labeling of proteins for proton and nitrogen-15 nuclear magnetic resonance. Methods Enzymol. 177:44-73. [DOI] [PubMed] [Google Scholar]
- 30.Okumura, H., S. Nishiyama, A. Sasaki, M. Homma, and I. Kawagishi. 1998. Chemotactic adaptation is altered by changes in the carboxy-terminal sequence conserved among the major methyl-accepting chemoreceptors. J. Bacteriol. 180:1862-1868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Pakula, A. A., and M. I. Simon. 1992. Determination of transmembrane protein structure by disulfide cross-linking: the Escherichia coli Tar receptor. Proc. Natl. Acad. Sci. USA 89:4144-4148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Parkinson, J. S., and S. E. Houts. 1982. Isolation and behavior of Escherichia coli deletion mutants lacking chemotaxis functions. J. Bacteriol. 151:106-113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Rice, M. S., and F. W. Dahlquist. 1991. Sites of deamidation and methylation in Tsr, a bacterial chemotaxis sensory transducer. J. Biol. Chem. 266:9746-9753. [PubMed] [Google Scholar]
- 34.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 35.Schuster, S. C., R. V. Swanson, L. A. Alex, R. B. Bourret, and M. I. Simon. 1993. Assembly and function of a quaternary signal transduction complex monitored by surface plasmon resonance. Nature 365:343-347. [DOI] [PubMed] [Google Scholar]
- 36.Shimizu, T. S., N. Le Nouvere, M. D. Levin, A. J. Beavil, B. J. Sutton, and D. Bray. 2000. Molecular model of a lattice of signalling proteins involved in bacterial chemotaxis. Nat. Cell Biol. 2:792-796. [DOI] [PubMed] [Google Scholar]
- 37.Simms, S. A., A. M. Stock, and J. B. Stock. 1987. Purification and characterization of the S-adenosylmethionine:glutamyl methyltransferase that modifies membrane chemoreceptor proteins in bacteria. J. Biol. Chem. 262:8537-8543. [PubMed] [Google Scholar]
- 38.Sourjik, V., and H. C. Berg. 2004. Functional interactions among receptors in bacterial chemotaxis. Nature 428:437-441. [DOI] [PubMed] [Google Scholar]
- 39.Springer, W., and D. E. Koshland, Jr. 1977. Identification of a protein methyltransferase as the cheR gene product in the bacterial sensing system. Proc. Natl. Acad. Sci. USA 74:533-537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Stock, J. B., and D. E. Koshland, Jr. 1978. A protein methyltransferase involved in bacterial sensing. Proc. Natl. Acad. Sci. USA 75:3659-3663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Studdert, C. A., and J. S. Parkinson. 2004. Crosslinking snapshots of bacterial chemoreceptor squads. Proc. Natl. Acad. Sci. USA 101:2117-2122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Terwilliger, T. C., E. Bogonez, E. Wang, and D. E. Koshland, Jr. 1983. Sites of methyl esterification on the aspartate receptor involved in bacterial chemotaxis. J. Biol. Chem. 258:9608-9611. [PubMed] [Google Scholar]
- 43.Terwilliger, T. C., and D. E. Koshland, Jr. 1984. Sites of methyl esterification and deamination on the aspartate receptor involved in chemotaxis. J. Biol. Chem. 259:7719-7725. [PubMed] [Google Scholar]
- 44.Terwilliger, T. C., J. Y. Wang, and D. E. Koshland, Jr. 1986. Kinetics of receptor modification. The multiply methylated aspartate receptors involved in bacterial chemotaxis. J. Biol. Chem. 261:10814-10820. [PubMed] [Google Scholar]
- 45.Weerasuriya, S., B. M. Schneider, and M. D. Manson. 1998. Chimeric chemoreceptors in Escherichia coli: signaling properties of Tar-Tap and Tap-Tar hybrids. J. Bacteriol. 180:914-920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Weis, R. M., T. Hirai, A. Chalah, M. Kessel, P. J. Peters, and S. Subramaniam. 2003. Electron microscopic analysis of membrane assemblies formed by the bacterial chemotaxis receptor Tsr. J. Bacteriol. 185:3636-3643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wolfe, A. J., M. P. Conley, T. J. Kramer, and H. C. Berg. 1987. Reconstitution of signaling in bacterial chemotaxis. J. Bacteriol. 169:1878-1885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Wu, J., J. Li, G. Li, D. G. Long, and R. M. Weis. 1996. The receptor binding site for the methyltransferase of bacterial chemotaxis is distinct from the sites of methylation. Biochemistry 35:4984-4993. [DOI] [PubMed] [Google Scholar]
- 49.Yeh, J. I., H. P. Biemann, J. Pandit, D. E. Koshland, Jr., and S.-H. Kim. 1993. The three-dimensional structure of the ligand-binding domain of a wild-type bacterial chemotaxis receptor. Structural comparison to the cross-linked mutant forms and conformational changes upon ligand binding. J. Biol. Chem. 268:9787-9792. [PubMed] [Google Scholar]