Abstract
Bathynellacea (Crustacea, Syncarida, Parabathynellidae) are subterranean aquatic crustaceans that typically inhabit freshwater interstitial spaces (e.g., groundwater) and are occasionally found in caves and even hot springs. In this study, we sequenced the whole transcriptome of Allobathynella bangokensis using RNA-seq. De novo sequence assembly produced 74,866 contigs including 28,934 BLAST hits. Overall, the gene sequences were most similar to those of the waterflea Daphnia pulex. In the A. bangokensis transcriptome, no opsin or related sequences were identified, and no contig aligned to the crustacean visual opsins and non-visual opsins (i.e. arthropsins, peropsins, and melaopsins), suggesting potential regressive adaptation to the dark environment. However, A. bangokensis expressed conserved gene family sets, such as heat shock proteins and those related to key innate immunity pathways and antioxidant defense systems, at the transcriptional level, suggesting that this species has evolved adaptations involving molecular mechanisms of homeostasis. The transcriptomic information of A. bangokensis will be useful for investigating molecular adaptations and response mechanisms to subterranean environmental conditions.
Introduction
Subterranean fauna form below the surface of the earth. Hyporheic/groundwater environments are harsh for animals due to limited space, permanent darkness, low dissolved oxygen concentrations, and limited energy/food inputs. The environment includes two major ecosystems, namely stygofauna (aquatic and living in groundwater) and troglofauna (air-breathing and living in caves and voids) [1]. Particularly, subterranean fauna exhibit several ecological and physiological characteristics that are evolutionary adaptations to the extreme environmental conditions. These adaptations include a high tolerance to hypoxia, low metabolic rates, longevity, delayed maturity, smaller clutch size, and simple food webs with few trophic links [2, 3]. Also, many hyporheic/groundwater organisms show phenotypical or morphological convergence such as reduced pigment, poorly functioning eyes or eye loss, development of non-optic sensory organs, and/or relative lengthening of appendages [4]. Although species richness is relatively restricted in the subterranean fauna, groundwater habitats develop unique biodiversity [5]. The groundwater ecosystem is composed mainly of tiny crustaceans, oligochaetes, nematodes, acari, and molluscs that have small body sizes of < 1mm to several centimeters [6]. Due to limited distribution, poor competitive ability, and low reproduction, the hyporheic/groundwater ecosystem is particularly vulnerable to environmental stressors and anthropogenic contamination.
Crustaceans such as amphipods, isopods, copepods, and bathynellaceans are most dominant animal groups found in most groundwater habitats [7]. Almost all major taxonomic groups of crustaceans in groundwater also occur in surface water. Bathynellaceans, however, have been known to have exclusively occurred only in groundwater since the Palaeozoic. Bathynellacea (Crustacea, Syncarida, Parabathynellidae), are widely distributed in most parts of the world except Antarctica, but its species have been poorly studied due to the interstitial aquatic environments they inhabit [8, 9]. The genus Allobathynella Morimoto and Miura, 1957 has been characterized mostly in Eastern Asia (e.g., Japan and South Korea) and includes many species that were formerly assigned to Parabathynella [10]. Recently, 14 new species of Allobathynella were identified and characterized in South Korea [11]. Although molecular tools, such as allozyme and mitochondrial DNA analyses, have been successfully applied to investigate endemism, cryptic species, and the distribution patterns of subterranean syncarids [12], little is known regarding gene sequences, expression, and molecular evolution.
Here, we present the first report on the transcriptome of the subterranean crustacean Allobathynella bangokensis Park and Cho, 2016 including analyses of basal-level mRNA expression. We analyzed distinct or conserved gene families in A. bangokensis in comparison with the transcriptional profiles of several crustacean species. This information provides a whole transcriptomic dataset that will help our understanding of the molecular characteristics of subterranean syncarids.
Materials and Methods
Allobathynella bangokensis
Allobathynella bangokensis was collected from the subterranean region of Hongcheon-Gun, Gangwon-Do, South-Korea (37° 41‘N, 127° 40‘E). Species identification was confirmed by assessing its morphological characteristics using stereomicroscopy and mitochondrial cytochrome oxidase subunit 1 (CO1) sequence analysis. Collected A. bangokensis specimens were stored immediately at −80°C for subsequent RNA extraction.
Ethics statement
No specific permits were required for the described field studies: a) no specific permissions were required for these locations/activities; b) location are not privately-owned or protected; c) the field studies did not involve endangered or protected species.
RNA extraction and library construction
Total RNA was extracted using the the RNeasy® Micro Kit (Qiagen) and the RNase-Free DNase I Kit (Qiagen, Valencia, CA, USA) according to manufacturers’ instructions. Whole bodies of 15 adult A. bangokensis were pooled and homogenized in RLT buffer (Qiagen). Extracted RNA was stored in RNA stable® (Biometrica, San Diego, CA, USA) to prevent RNA degradation during long-term storage. Extracted RNA quality and concentration were determined using the 2100 Bioanalyzer (Agilent Technologies, Santa Clara, CA, USA). A next-generation sequencing (NGS) library was constructed from 2 μg total RNA using NuGEN Encore® Complete RNA-Seq Library Systems (NuGEN, San Carlos, CA, USA). Final transcriptomic library lengths and concentrations were determined using the 2100 Bioanalyzer. Transcriptomic libraries were sequenced by the MiSeq® System (Illumina) platform using sequenced runs of 300×2 paired-end reads. Index and adaptor sequences were trimmed using Trimmomatic [13] and low quality reads were removed using the FASTX tool kit [14].
De novo assembly and annotation
We performed de novo assembly using software packages designed for short read sequence assembly, including Abyss [15], Velvet [16], CLC Genomics Workbench 7.5 environment (CLC Bio Aarhus, Denmark), and Oases (D.R. Zerbino, European Bioinformatics Institute). We assembled each sample using the same assembly parameters (K-mer length = 27, coverage cutoff = 10, minimum contig length = 200 bp). Consideration of the assembly statistics (N50, longest contig, number of contigs, proportion of reads assembled) led us to finally choose Oases, which generated the longest assembled ESTs.
Data deposition
The raw sequencing reads of A. bangokensis were deposited in the Sequence Read Archive in GenBank (SAMN05712658).
Annotation and gene ontology (GO) analysis
GO and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway analyses of the contigs were performed using the Blast2GO sequence annotation tool [17]. The three main categories biological processes, cellular components, and molecular functions were obtained after aligning contigs using default parameters. The assembled data were arranged including read length, gene annotation, GenBank number, E-value, species, and the species accession number. The assembled data, including GO terms, were deposited as supplementary material (S3–S5 Tables). In each section, the specific GO term composition was calculated and presented as a percentage.
Gene expression analysis
The gene expression level of the A. bangokensis transcriptome was calculated using the reads per kilobase transcriptome per million mapped reads (RPKM) method [18]. Heat map analysis was conducted to represent the transcriptomic profiles using MeV software (ver. 7.4; Dana-Farber Cancer Institute, Boston, MA, USA).
Results and Discussion
Transcriptome assembly and gene annotation
To establish the transcriptomic database of Bathynellacea, we performed RNA-seq using the subterranean crustacean Allobathynella bangokensis. After trimming and assembly, a total of 63 Mb including 74,866 contigs of A. bangokensis was obtained by Illumina sequencing (Table 1). The lengths of the A. bangokensis contigs ranged from 200 to 26,238 bp, with an average length of 843 bp, and the N50 values of those contigs were 1,302 bp. To our knowledge, this is the first whole transcriptome study conducted in Bathynellacea.
Table 1. Summary of the statistics from the transcriptomic analysis of the subterranean crustacean Allobathynella bangokensis.
| Raw data | |
| Reads no. | 16,865,850 |
| Reads length (bp) | 4,010,699,130 |
| Raw sequence after QC | |
| Reads no. | 15,954,043 |
| Reads length (bp) | 3,703,731,082 |
| De novo assembly | |
| Contigs no. | 74,866 |
| Contig length (bp) | 63,112,330 |
| Length distribution (bp) | 200 to 26,238 |
| Average length (bp) | 843 |
| N50 (bp) | 1,302 |
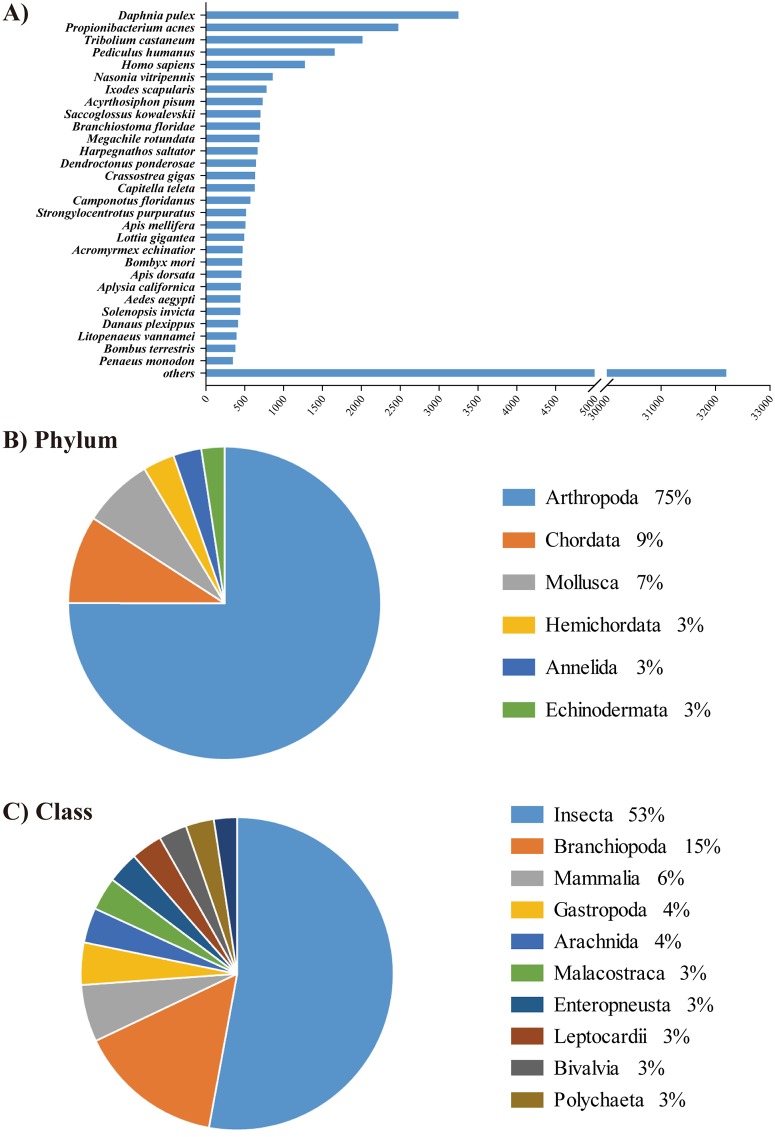
Gene annotation of the transcripts was performed by BLASTx analysis using the NCBI non-redundant (NR) protein database. The results showed that 28,934 contigs (38.6%) had at least one positive BLAST hit (E-value < 1e-04) representing 11,751 annotated genes (S1 Table). Distribution analysis showed that 18 species had more than 500 BLAST transcript hits, and the waterflea Daphnia pulex (Crustacea, Branchiopoda) showed the highest similarity with 3,250 reads (S2 Table; Fig 1A). Among the top hit species, 75% of contigs matched sequences of the phylum Arthropoda, while the other 25% was comprised of Chordata, Mollusca, Hemichordata, Annelida, and Echinodermata (Fig 1B). In addition, 53% and 15% of contigs showed homologies to insects and crustaceans (Branchiopoda), respectively, at the class level (Fig 1C). In the NCBI NR database, a relatively a huge amount of gene information of insects was appended compared to those of crustaceans. Thus, although Bathynellacean is an order of crustaceans, the overall gene annotation results represent high quality of assembled transcripts of A. bangokensis [19].
Fig 1. BLAST top hit species distribution.
Number of top BLAST hit species identified for the Allobathynella bangokensis transcript contigs (A) and pie charts presenting the annotated genes and their corresponding phylum (B) and class (C). Detailed information is appended in the supplementary materials (S2 Table).
Functional annotation
InterProScan protein sequence analysis and classification can be used to effectively classify protein functions by predicting domains and important sites [20]. The most abundant InterPro domains are presented in Table 2 (S1 Fig). The InterPro domains with the highest numbers of hits were immunoglobulin (IG)-like domain (IPR007110; 2433 hits), followed by P-loop containing nucleoside triphosphate hydrolase (IPR027417; 2342 hits) and fibronectin type III (IPR003961; 1761 hits). The immunoglobulin superfamily (IgSF) is a large group of soluble cell surface proteins that are mostly involved in adaptive immune defenses (e.g., recognition, binding, or adhesion processes), which are believed to be restricted to vertebrates [21]. All members of IgSF possess at least one IG-like domain or IG fold. The basic molecular mechanisms of the IG-like domain have rarely been investigated in invertebrates, but several genes possessing IG domains such as the C-type lectin domain, Down syndrome cell adhesion molecules, fibrinogen-related proteins, and hemolin have been suggested to be involved in host defense mechanisms of arthropods including crustaceans and mollusks [22–28]. Based on the relationship between IgSF and invertebrate immunity, it is possible that A. bangokensis has a robust innate immune defense system involving IG-like domain-containing proteins.
Table 2. The most abundant InterPro domain classifications of Allobathynella bangokensis transcript contigs.
| InterPro ID | Domain description | Number of matched contigs |
|---|---|---|
| IPR007110 | Immunoglobulin-like domain | 2,433 |
| IPR027417 | P-loop containing nucleoside triphosphate hydrolase | 2,342 |
| IPR003961 | Fibronectin type III | 1,761 |
| IPR013783 | Immunoglobulin-like fold | 1,583 |
| IPR007087 | Zinc finger, C2H2 | 1,491 |
| IPR002172 | Low-density lipoprotein (LDL) receptor class A repeat | 1,327 |
| IPR001478 | PDZ domain | 1,085 |
| IPR000719 | Protein kinase domain | 1,040 |
| IPR002110 | Ankyrin repeat | 997 |
| IPR020683 | Ankyrin repeat-containing domain | 986 |
| IPR001680 | WD40 repeat | 864 |
| IPR001611 | Leucine-rich repeat | 820 |
| IPR013087 | Zinc finger C2H2-type/integrase DNA-binding domain | 809 |
| IPR000504 | RNA recognition motif domain | 773 |
| IPR012677 | Nucleotide-binding alpha-beta plait domain | 742 |
| IPR001452 | SH3 domain | 687 |
| IPR001781 | Zinc finger, LIM-type | 685 |
| IPR000742 | EGF-like domain | 650 |
| IPR002126 | Cadherin | 647 |
| IPR000859 | CUB domain | 641 |
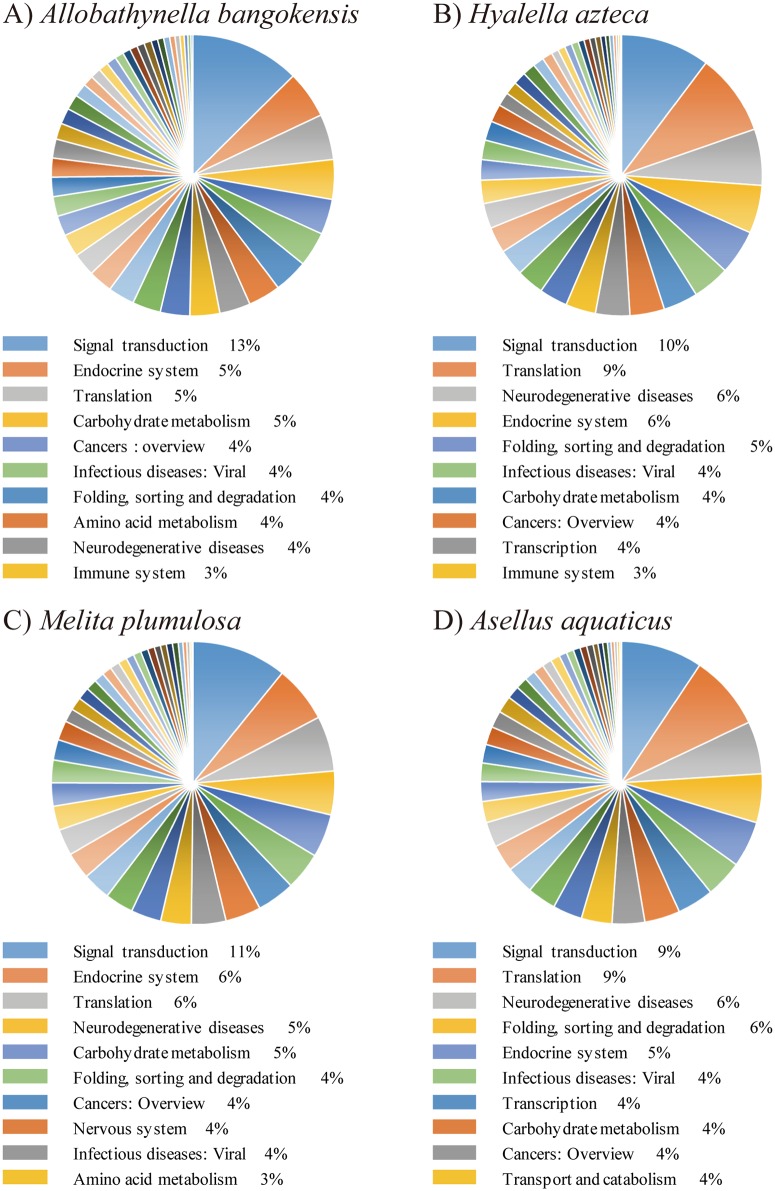
GO terms related to the top domains were described at the second level (S2 Fig). Detailed GO distributions in three GO categories (biological process, cellular component, and molecular function) are shown in the supplementary material (S3–S5 Tables). In the biological process category, many genes are classified as metabolic processes (28%), cellular processes (25%), and single-organism processes (19%). In the molecular function category, the vast majority of genes are involved in catalytic activities (43%) and binding functions (40%). In the cellular component class, most genes are related to the cell (36%) and cell membrane (24%). The overall proportions of the major GO categories in A. bangokensis sequences were very similar to those of the transcriptomes of several other crustaceans [29–32], and we were not able to identify significant differences in the proportions of the three GO term categories. KEGG analysis of the A. bangokensis transcriptome revealed that the vast majority of KEGG pathways are involved in signal transduction (13%), followed by the endocrine system (5%), translation (5%), and carbohydrate metabolism (5%) (Fig 2A; S6 Table). The composition rate and percentage rankings of the top 10 A. bangokensis KEGG pathways were similar to those of amphipods [30, 33] (Fig 2B; Hyalella azteca and Fig 2C; Melita plumulosa) and an isopod [34] (Fig 2D; Asellus aquaticus). Thus, these results suggest the intactness of the A. bangokensis transcriptome, since the information does not lack major functional GO categories or KEGG pathways compared with the transcriptomes of arthropods.
Fig 2. The composition of Malacostraca KEGG analysis.
The compositions and percentage rankings of the A) Allobathynella bangokensis, B) amphipod Hyalella azteca, C) amphipod Melita plumulosa, and D) isopod Asellus aquaticus KEGG pathways. Detailed information is appended in the supplementary materials (S6 Table).
Opsin repertoire
Opsins have been classified into four major monophyletic groups: 1) ciliary photoreceptors (‘c-opsins’), 2) rhabdomeric photoreceptors (‘r-opsins’), 3) cnidarian opsins (‘Cnidops’), and 4) a mixed group consisting of ‘retinal G-protein coupled receptors’, peropsins and neuropsins [35]. Interestingly, in the A. bangokensis transcriptome, we were not able to identify an opsin contig. Of the 28,934 Blast hit results, several candidates identified as putative opsin or relevant genes were searched, but our additional analysis using an in-depth annotation platform (e.g., domain analysis, phylogenetic analysis) revealed that the sequences were named incorrectly, because the contigs were matched to previous incorrectly registered opsin genes of other species in the NR database. In addition, short partial contigs of A. bangokensis that showed low similarity in various regions of the amino acid sequence could not be annotated to an opsin gene. To investigate putative opsin transcripts in A. bangokensis, a total of 50 opsin amino acid sequences from arthropods and onychophorans (numbered by 1–50 in S7 Table; i.e., long wavelength sensitive; middle wavelength sensitive; short wavelength sensitive groups, Blue and UV; onychopsin) annotated in a previous report [36], 7 amphipod opsins (numbered by 51–57 in S7 Table), and 21 arthropsins (numbered by 58–78 in S7 Table) and 25 peropsins (numbered by 79–103 in S7 Table) of arthropods, and additional 8 invertebrate melanopsins (numbered by 104–111 in S7 Table) registered in GenBank were mapped directly to 74,866 contigs of A. bangokensis using an internal local BLASTx platform. However, no contig of A. bangokensis was aligned to the gene pool comprised of opsin, arthropsin, peropsin, and eye pigmentation genes. In addition, we attempted to identify several eye-related genes previously annotated in the crustacean lineage [37]. Although these genes are involved in diverse functions beyond eye development or vision, many were transcriptionally not detected in the A. bangokensis (Table 3). Expression of the Dachshund (Dac) gene is important for controlling cell fate determination in eye, limb, brain, and muscle development [38]. Drosophila eyes absent (Eya) plays an essential role in retinal cell survival and differentiation [39], while multiple functions of Eya homologues (Eya1, Eya2, Eya3, and Eya4) have been identified continuously in mammals [40]. These two genes, Dac and Eya, act synergistically to induce ectopic retinal development and positively regulate each other’s expression through conserved domains in Drosophila [41]. Eyegone, which is a member of the Pax transcription factor family, was discovered for its essential requirement in retinal primordium growth in Drosophila [42, 43]. We were unable to determine the transcriptional roles of these genes in A. bangokensis, but overall transcriptional patterns that are not detected in the transcriptome would affect phenotypical or morphological adaptations in A. bangokensis. Further identification in embryonic and/or early developmental stage should be expanded in future study, as we analyzed the transcriptome of adult A. bangokensis.
Table 3. List of eye-related genes identified in the Allobathynella bangokensis transcriptome, presented with RPKM values and matching species information.
The genes and reference protein IDs of Daphnia pulex were adopted from a previous study [37].
| Gene family | Reference protein ID (Daphnia pulex) | Matched sequence ID of A. bangokensis | RPKM | Matched species | E-value | Accession No. |
|---|---|---|---|---|---|---|
| Visual system specification gene family | ||||||
| Decapentaplegic | Dappu-347232 | - | - | - | - | - |
| Engrailed (En) | Dappu-290630 | Allo_10518 | 8.43 | Pholcus phalangioides | 1.00E-43 | CYF18415 |
| Dappu-290638 | - | - | - | - | - | |
| Hedgehog (Hh) | Dappu-347555 | Allo_11417 | 10.35 | Daphnia pulex* | 3.00E-07 | EFX65836 |
| Wnt1 | Dappu-44743 | - | - | - | - | |
| Retinal determination network gene family | ||||||
| Dachshund (Dac) | Dappu- 310049 | - | - | - | - | - |
| Eyes-absent (Eya) | Dappu- 204955 | - | - | - | - | - |
| Eyegone (Eyg/Toe) | Dappu- 253988 | - | - | - | - | - |
| Pax-6 | Dappu- 249978 | Allo_67774 | 4.98 | Parasteatoda tepidariorum | 1.00E-79 | XP_015917132 |
| Dappu-249991 | - | - | - | - | - | |
| Six 1/2 | Dappu-65962 | Allo_28121 | 2.71 | Pediculus humanus corporis** | 2.00E-89 | XP_002431193 |
| Photoreceptor differentiation gene family | ||||||
| Epidermal Growth Factor Receptor (EGFR) | Dappu-324147 | Allo_53476 | 1.90 | Halyomorpha halys** | 0 | XP_014284376 |
| Dappu- 321139 | Allo_40048 | 27.72 | Daphnia magna* | 0 | KZS10602 | |
| Kruppel (Kr) | Dappu-290527 | Allo_38368 | 7.75 | Strigamia maritima** | 2.00E-30 | AAY45764 |
| Glass (Gl) | Dappu-234903 | - | - | - | - | - |
| Notch | Dappu-328760 | Allo_53041 | 27.09 | Parhyale hawaiensis* | 0 | ABK56706 |
| Spitz (Spi) | Dappu-271304 | - | - | - | - | - |
| CVC Homeobox (Vsx) | Dappu-323346 | - | - | - | - | - |
| Phototransduction gene family | ||||||
| Arrestin (Arr) | Dappu-216585 | Allo_38980 | 10.80 | Marsupenaeus japonicas* | 0 | AME17864 |
| Dappu-207575 | - | - | - | - | - | |
| Gq-alpha | Dappu-211929 | Allo_36916 | 0.44 | Marsupenaeus japonicas* | 0 | BAH98115 |
| Dappu-188187 | - | - | - | - | - | |
| Phospholipase-C (PLC) | Dappu- 226357 | Allo_48820 | 9.30 | Homarus americanus* | 1.00E-99 | AAD32609 |
| Dappu- 304714 | - | - | - | - | - | |
| Transient Receptor Potential Channel (TRPC) | Dappu-54362 | Allo_59564 | 0.37 | Daphnia magna* | 9.00E-13 | EFX75016 |
| Dappu- 309057 | Allo_30981 | 10.46 | Tribolium castaneum** | 3.00E-52 | KYB28200 | |
* Crustacea
** Insecta
Since stygofauna have reduced or absent eyes and have enhanced non-optic sense organs without pigmentation [44, 45], the absence of pigments and eyes is observed in A. bangokensis. A total of 25 and 26 eye pigmentation genes that were retrieved from Drosophila melanogaster and Tribolium castaneum respectively [46], were also aligned to the entire transcripts of A. bangokensis, but there was no matched transcripts in A. bangokensis transcriptome. Although mRNA expression of putative opsin genes was not observed in the A. bangokensis transcriptome, it will be interesting to investigate the presence or absence of the opsin repertoire in the genome and the evolutionary development of alternative sensory organs in future studies. Several previous examples showed the potential correlation between reduced or absent eyes and transcriptional expression of the opsin repertoire. No loss of gene function in opsin gene paralogs with a reduced level of gene expression was reported in the cave-adapted amphipod Gammarus minus [36]. The authors suggested that loss of expression of opsin genes without loss of gene function is explained by the pleiotropic roles of opsin genes [36]. Extensive transcriptomic analysis revealed that three independently evolved subterranean diving beetles lack transcripts of nearly all opsin photoreceptor genes, whereas the two surface beetles showed evidence of transcriptional expression of a full suite of insect visual and non-visual opsin genes [47]. Thus, research on the absence or presence of putative opsin genes from the A. bangokensis genome will be useful to understand the regressive evolution of eye reduction in the Bathynellacea lineage.
Heat shock protein superfamily
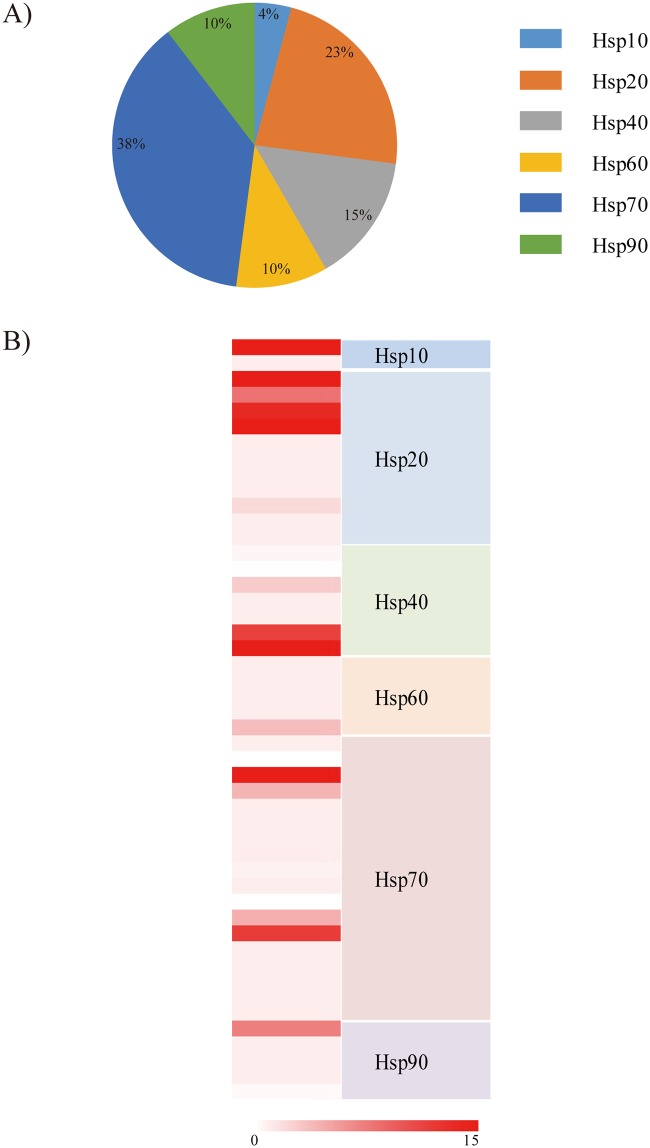
Environmental changes or stress factors induce molecular and systemic metabolism to maintain cellular and physiological homeostasis. The heat shock protein (Hsp) superfamily is the most conserved protein present in both prokaryotes and eukaryotes. In general, their expression can be induced by a wide variety of physiological and environmental stimuli [48]. Proteins of the Hsp superfamily function as molecular chaperones and are the key components responsible for assisting the correct folding of nascent or stress-accumulated misfolded proteins and for preventing aggregation [49]. In the A. bangokensis transcriptome, 48 Hsp genes distributed among five subfamilies (i.e., Hsp10, Hsp20, Hsp40, Hsp60, Hsp70, and Hsp90) were annotated (Fig 3A; S8 Table). The diversity of the A. bangokensis Hsp family was unpredictable, since a drastic reduction in the ranges of daily and annual temperatures is a distinguishing characteristic of the subterranean zone [50]. Overall, the RPKM values for most Hsp genes were relatively low; 31 Hsp genes (65%) showed RPKM values < 1 (Fig 3B; S8 Table). Some Hsp genes are expressed constitutively at minimal or basal levels, while other Hsp genes can be induced rapidly in response to environmental stimuli [51]. Thus, we expect that A. bangokensis employs an effective holding and folding defense system using a combination of differentially expressed Hsp genes at low levels. This defense system diminished cellular stress, which can be triggered by even small changes in subterranean environmental conditions.
Fig 3. Analysis of heat shock protein superfamily of the Allobathynella bangokensis.
The compositions and percentage rankings of the Allobathynella bangokensis heat shock protein (Hsp) superfamily (A) and their basal transcript levels represented by RPKM values (B). Detailed information is appended in the supplementary materials (S8 Table).
Regardless of their chaperonin function, a previous report highlighted a distinct gain of function of the hsp90α gene in the cavefish Astyanax mexicanus [52]. hsp90α is expressed specifically in the cavefish lens starting just prior to apoptosis, while the expression was not observed in the lens of eyed surface-dwelling A. mexicanus (surface fish), suggesting that activation of the hsp90α gene may be required for eye degeneration and apoptosis in the lens in cavefish [52]. Therefore, further study of Hsp function in A. bangokensis is needed to understand its phenotypic adaptation. Also, it has been widely suggested that Hsp genes are good biomarkers for numerous environmental changes [48, 49]. Sequence information of the A. bangokensis Hsp family can be applied to subterranean environmental monitoring by analyzing mRNA and protein expression profiles after induction by environmental stressors.
Innate immune system
In general, metazoans have an innate immune system, which consists of cellular and humoral metabolism, as the first line of defense against pathogenic bacteria, fungi, viruses, and metazoan parasites using recognition, regulation, and response [53, 54]. Many key pathways employing genes involved in innate immunity are conserved between mammals and arthropods including crustaceans [53, 55]. In the A. bangokensis transcriptome, we found conservation of most immunity-related gene families previously analyzed in arthropods [55–58]. As shown in Table 4, a variety of innate immunity-related sequences corresponding to adhesive proteins, antimicrobial proteins, apoptosis- and cell cycle-related proteins, cellular defense effectors, immune regulators, pattern recognition proteins, proteases, protease inhibitors, reduction/oxidation-related proteins, signal transduction-related proteins, and stress proteins were observed. These results suggest that complex immune-relevant gene sets are actively expressed to maintain cellular homeostasis even in subterranean animals. Immune-relevant gene sets will help extend our knowledge on the immune systems of syncarids in comparative aspects and ecological genetics within Arthropoda.
Table 4. Immune-relevant genes annotated in the Allobathynella bangokensis transcriptome database.
| Seq ID | Gene name | Matched species | E-value | Accession No. | RPKM |
|---|---|---|---|---|---|
| Adhesive protein | |||||
| Allo_01147 | Cadherin 87 | Cephus cinctus** | 8.00E-07 | XP_015588457 | 9.55 |
| Allo_18151 | Galectin | Litopenaeus vannamei* | 3.00E-29 | AGV04659 | 4.61 |
| Allo_40297 | multiple PDZ domain protein isoform X2 | Acyrthosiphon pisum** | 1.00E-147 | XP_008181772 | 20.81 |
| Allo_74308 | Tetraspanin-5-like isoform 1 | Athalia rosae** | 7.00E-152 | XP_012258733 | 14.26 |
| Allo_63960 | Transglutaminase | Pacifastacus leniusculus* | 7.00E-47 | AAK69205 | 4.53 |
| Allo_31604 | Histone H2A-like | Diuraphis noxia** | 2.00E-68 | XP_015363720 | 26.19 |
| Allo_69601 | Histone H3.3 | Oncorhynchus mykiss | 1.00E-41 | ACO08041 | 8.80 |
| Allo_29964 | integral membrane, | Ixodes scapularis | 1.00E-68 | XP_002412964 | 0.79 |
| Apoptosis and cell cycle | |||||
| Allo_61561 | inhibitor of apoptosis protein | Penaeus monodon* | 3.00E-166 | ABO38431 | 0.00 |
| Allo_68354 | β-catenin-like protein 1 | Limulus polyphemus | 1.00E-167 | XP_013791309 | 0.70 |
| Allo_12816 | Catenin delta-2 | Lasius niger** | 2.00E-56 | KMQ88737 | 0.74 |
| Allo_14751 | Programmed cell death protein 4 | Zootermopsis nevadensis** | 2.00E-146 | KDR08650 | 38.58 |
| Allo_56958 | Rab-1 | Macrobrachium rosenbergii* | 7.00E-104 | AJC97112 | 0.77 |
| Allo_55831 | ras-related Rab-32B-like | Aplysia californica | 3.00E-60 | XP_005097933 | 0.32 |
| Cellular defense effecter | |||||
| Allo_70113 | Dual oxidase | Marsupenaeus japonicas* | 0.0 | BAM76968 | 12.50 |
| Allo_25381 | fibrinogen-like protein | Fenneropenaeus merguiensis* | 9.00E-58 | AKR15662 | 0.00 |
| Allo_27844 | Thioredoxin reductase 3 | Crassostrea gigas | 0.0 | XP_011419860 | 15.24 |
| Allo_56333 | c-X-C motif chemokine 15-like | Bos taurus | 1.00E-10 | XP_003582386 | 2.95 |
| Immune regulator | |||||
| Allo_24131 | signal sequence receptor beta-like protein | Plectreurys tristis | 8.00E-59 | AJD25284 | 16.15 |
| Allo_72274 | Translocon-associated protein subunit α | Zootermopsis nevadensis** | 2.00E-73 | KDR17543 | 36.74 |
| Allo_00305 | Carboxypeptidase B2 | Daphnia magna* | 5.00E-134 | KZS09622 | 82.73 |
| Protease and protease inhibitor | |||||
| Allo_47130 | 26S protease regulatory subunit 10B X2 | Myotis lucifugus | 5.00E-135 | XP_014318322 | 2.01 |
| Allo_23607 | α-2-macroglobulin 2 isoform 3 | Pacifastacus leniusculus* | 0.0 | AEC50083 | 0.64 |
| Allo_40857 | trypsin | Litopenaeus vannamei* | 6.00E-112 | CAA75311 | 0.00 |
| Allo_30909 | Cystatin 2 | Cherax quadricarinatus* | 7.00E-141 | ALC79585 | 0.00 |
| Allo_37572 | Cathepsin A | Eriocheir sinensis* | 0.0 | ADO65982 | 14.52 |
| Allo_54726 | Cathepsin B | Fenneropenaeus chinensis* | 1.00E-175 | AHA83423 | 1.27 |
| Allo_56730 | Cathepsin C | Fenneropenaeus chinensis* | 3.00E-122 | ACG60902 | 0.00 |
| Allo_64644 | Cathepsin D | Palaemon carinicauda* | 0.0 | AGJ03549 | 33.05 |
| Allo_54848 | Cathepsin L protein | Cherax quadricarinatus* | 0.0 | AJS13771 | 0.00 |
| Allo_54759 | inter-α-trypsin inhibitor heavy chain H6 | Monodelphis domestica | 1.00E-14 | XP_016282619 | 6.49 |
| Allo_54846 | matrix metalloproteinase-14 | Papilio Xuthus** | 1.00E-173 | XP_013166430 | 5.18 |
| Allo_29726 | protein phosphatase | Fenneropenaeus chinensis* | 0.0 | AHE40944 | 34.04 |
| Allo_00427 | serine carboxypeptidase CPVL-like | Neolamprologus brichardi | 1.00E-159 | XP_006801474 | 26.79 |
| Redox | |||||
| Allo_39210 | hypothetical ferritin light-chain subunit | Rimicaris exoculata* | 5.00E-41 | ACR43472 | 0.44 |
| Allo_37530 | Peroxiredoxin | Penaeus monodon* | 1.00E-129 | ABZ80828 | 52.99 |
| Allo_23907 | selenium-binding protein 1-A-like | Corvus brachyrhynchos | 1.00E-126 | XP_008627838 | 14.72 |
| Signal transduction | |||||
| Allo_28397 | calmodulin-A-like isoform X1 | Microplitis demolitor** | 9.00E-68 | XP_008550049 | 6.82 |
| Allo_50919 | cAMP-dependent kinase | Nasonia vitripennis** | 4.00E-16 | ACH99585 | 1.20 |
| Allo_69397 | Casein kinase II subunit beta | Cerapachys biroi** | 4.00E-133 | XP_011340131 | 8.24 |
| Allo_55642 | COP9 signalosome complex subunit 2 | Athalia rosae** | 0.0 | XP_012264747 | 20.29 |
| Allo_14758 | mitogen-activated protein kinase | Fenneropenaeus chinensis* | 2.00E-131 | AHA83424 | 12.99 |
| Allo_20185 | tyrosine- kinase Src64B-like isoform 1 | Zootermopsis nevadensis** | 0.0 | KDR17588 | 0.00 |
| Allo_04375 | Protein vav | Neodiprion lecontei** | 2.00E-34 | XP_015521070 | 0.34 |
| Stress protein | |||||
| Allo_50356 | ATP synthase F0 subunit 6 | Aradacanthia heissi | 8.00E-30 | ADQ64032 | 12.38 |
| Allo_22874 | heat shock factor binding 1-like | Trachymyrmex zeteki** | 3.00E-22 | KYQ58790 | 2.21 |
| Allo_40206 | melanoma-associated antigen G1 | Myotis davidii | 2.00E-08 | XP_006771818 | 3.54 |
| Allo_33230 | CD63 antigen | Lepeophtheirus salmonis* | 3.00E-61 | ACO12374 | 10.02 |
| Allo_21482 | B-cell lymphoma leukemia 11B-like | Zootermopsis nevadensis** | 2.00E-05 | KDR23016 | 0.69 |
| Allo_23197 | Transferrin | Zootermopsis nevadensis** | 4.00E-144 | KDR19744 | 0.00 |
| Allo_21081 | ubiquitin specific peptidase 25 | Astyanax mexicanus | 6.00E-17 | XP_007244811 | 6.83 |
| Allo_26367 | LITAF | Litopenaeus vannamei* | 2.00E-25 | AEK86526 | 4.32 |
| Acute phase response/inflammation | |||||
| Allo_05913 | heme oxygenase 1-like isoform X2 | Apis dorsata** | 1.00E-06 | XP_006620087 | 5.49 |
* Crustacea
** Insecta
In arthropods, pathogens are selectively recognized by major signaling pathways, such as Toll, immune deficiency (IMD), and Janus kinase (JAK)-signal transducer of activators of transcription (STAT) for activation of immune effectors [57]. Since there is no information on the intactness of the major pathways in subterranean crustaceans, we evaluated the absence or presence of pathways using in silico approaches (e.g., domain analysis, phylogenetic analysis) based on KEGG pathway analysis of the A. bangokensis transcriptome. Detailed protein interactions and their functions for all members involved in each pathway are mostly omitted in this manuscript, as several valuable publications have extensively reviewed their roles in arthropods [56–60]. The identified genes and information pertaining to the absence or presence of expression, sequence IDs, matching species, E-values, and GenBank accession numbers are presented in Supplementary material (S9 Table).
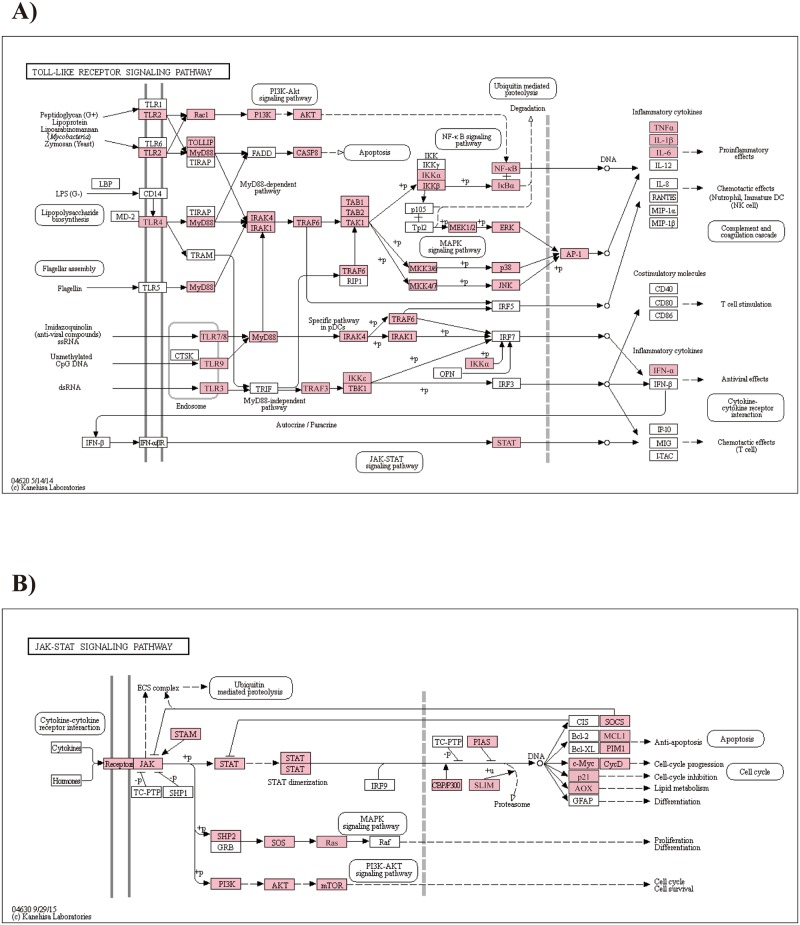
Of the pattern recognition receptors, the Toll-like receptors (TLRs) represent an evolutionarily conserved host defense mechanism in both invertebrates and vertebrates [61]. Based on the type of adaptor molecule, individual TLR signaling pathways differentially activate downstream components in the signaling cascade, causing activation of the transcription factor NF-κB and interferon (IFN) regulatory factors [62]. Although there are no reports regarding MyD88-independent pathways in invertebrates, crustacean TLRs and MyD88 are transcriptionally responsive to immune stimulation [63–67]. We found all putative homologs of TLR signaling pathway members to be highly conserved across arthropods, including Spätzle cytokine (Allo_71790), MyD88 (Allo_03624), Pelle (interleukin-1 receptor-associated kinase 1, IRAK1, Allo_70004), Tube (IRAK4, Allo_32335), Pelle-interacting protein Pellino (Allo_43963), Toll-interacting protein (TOLLIP, Allo_08036), sterile alpha- and armadillo-motif-containing protein (SARM, Allo_67175), evolutionarily conserved signaling intermediate in the Toll pathway (ECSIT, Allo_69618), tumor necrosis factor receptor-associated factor (TRAF, Allo_61987), Cactin (Allo_58700), NF-κB inhibitor Cactus (IκB, Allo_32054), and Dorsal (Rel/NF-κB, Allo_30610) (Fig 4A; S9 Table). Thus, the Toll pathway appears to be intact in A. bangokensis.
Fig 4. Toll-like receptor and JAK-STAT signaling pathway.
Allobathynella bangokensis transcripts coding for the corresponding enzymes of the A) Toll-like receptor and B) JAK-STAT signaling pathways from the KEGG database. The annotated enzymes are shown in pink boxes.
The JAK-STAT signaling pathway is evolutionarily conserved and mediates the response to chemical messenger molecules such as diverse cytokines, IFNs, growth factors, and related molecules [68]. In the A. bangokensis transcriptome, putative homologs of the transmembrane cytokine receptor Domeless (Allo_26652), JAK (Allo_51891), STAT (Allo_07305), signal transducing adaptor molecule (Allo_52924), suppressor of cytokine signaling (Allo_16562), and protein inhibitors of activated STAT (PIAS, Allo_52571) were all identified by KEGG analysis as commonly observed key components in crustaceans (Fig 4B; S9 Table) [60]. The transcriptional presence of key modulators of the JAK-STAT pathway, as well as downstream components of the pathway, suggests that the JAK-STAT pathway is strongly involved in the regulation of A. bangokensis immunity.
Similar to the JAK-STAT pathway, many downstream members of the IMD signaling pathway are also conserved in the A. bangokensis transcriptome. The IMD pathway is activated mainly by Gram-negative bacteria and is separated into the NF-κB/Relish and Jun N-terminal kinase (JNK) branches [69]. Recently, Rosa et al [60] summarized 23 key component proteins of the insect IMD pathway, 17 of which are commonly identified in crustaceans. Since the KEGG database does not include the IMD pathway, manual identification was employed in the A. bangokensis transcriptome, which showed that most of the components of the IMD pathway have a homologue in the A. bangokensis transcriptome. Among the 17 crucial proteins involved in the crustacean IMD pathway, 16 were annotated in the A. bangokensis transcriptome (S9 Table), and only defense repressor 1 was not identified at the transcriptional level. These 16 proteins are IMD (Allo_52952), enzymes involved in ubiquitination (UEV1a, Allo_70733; Effete/Ubc13, Allo_20821; Bendless/Ubc5, Allo_28290), the negative regulators Caspar (Fas-associated factor 1, Allo_72340) and POSH (E3 ligase Plenty of SH3, Allo_27226), inhibitor of apoptosis IAP2 (Allo_73002), transforming growth factor-β-activated kinase 1 (TAK1, Allo_01961), TAK1-binding protein 2 (TAB2, Allo_72573), IκB kinase IKK-α (Allo_18196), a Relish-like Rel/NF-κB transcription factor (Allo_40693), Caudal homeobox protein (Allo_52669), mitogen-activated protein kinase kinase Hemipterous (Allo_31075), the JNK homolog Basket (Allo_17845), the negative regulator Puckered (Allo_58869), Activator protein 1, and the transcription factor Jun-related antigen (Allo_32187). Overall, the TAK1/TAB2 complex-activated JNK pathway was apparently conserved in A. bangokensis, but several members involved in the NF-kB/Relish branch were missing from the transcriptome database.
In crustaceans, the absence or presence of several components of the IMD pathway (i.e., peptidoglycan recognition proteins, Fas associated protein with death domain (FADD), death related ced-3/Nedd2-like (DREDD), poor IMD response upon knock-in (Pirk), IκB kinase Kenny/NEMO, Fos-related antigen/Kayak (Fra)) is controversial [60], and A. bangokensis also did not express FADD, DREDD, or Pirk at the transcriptional level. Thus, this result supports that A. bangokensis lacks several crucial members of a well-conserved IMD pathway as shown by the missing sequences in Insecta (e.g., Hemiptera), Crustacea, and Chelicerata [60]. Further experiments are needed to clarify the absence/presence of these genes at the genome level. However, Kenny/NEMO (Allo_64535) and Fra (Allo_55867) contigs were observed in the database. Taken together, we have confirmed that major immune responsive pathways, such as Toll, JAK-STAT, and the JNK branch of IMD pathway, involved in the innate immune system of A. bangokensis are evolutionarily conserved across crustaceans at the transcriptional level.
Antioxidant defense system
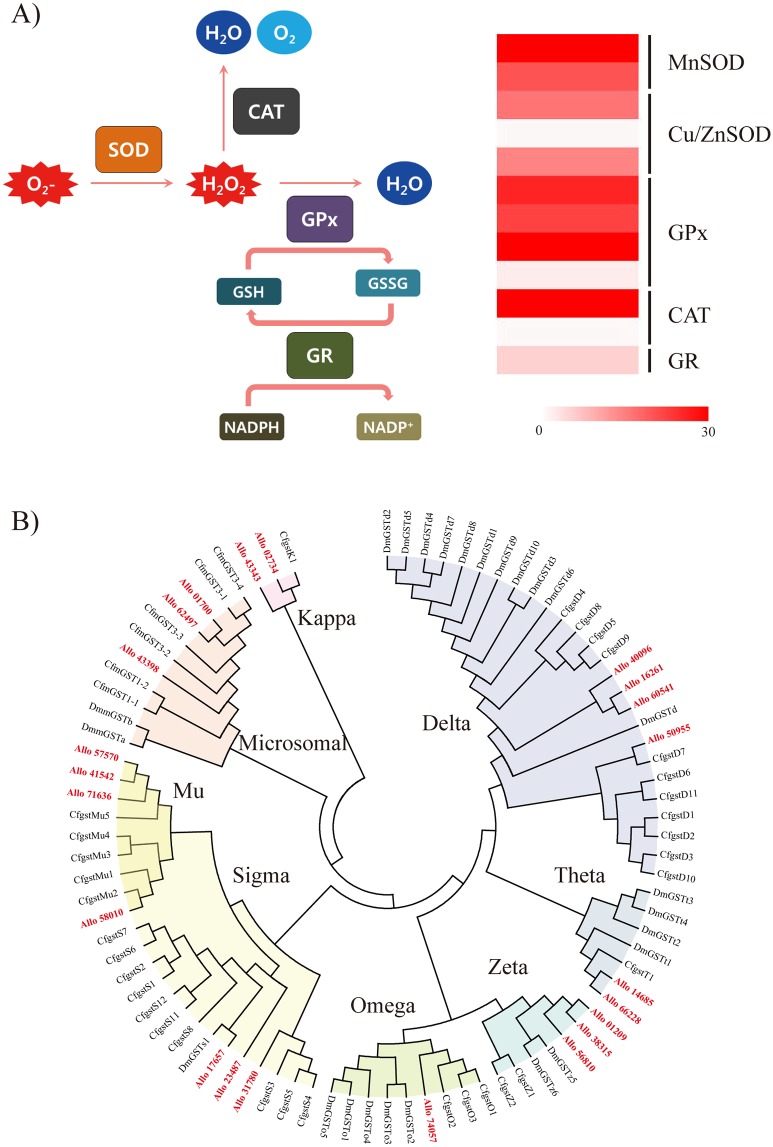
Reactive oxygen species (ROS) such as superoxide anion, hydrogen peroxide, and hydroxyl radical are generated during mitochondrial oxidative phosphorylation and during the cellular response to xenobiotics, cytokines, and bacterial invasion [70]. Oxidative stress results in direct or indirect ROS-mediated damage to nucleic acids, proteins, and lipids. To maintain cellular homeostasis, antioxidant defense systems composed of catalase, glutathione depletion, glutathione reductase, glutathione peroxidase, and superoxide dismutase (SOD) can be induced to eliminate excess ROS levels. Based on the annotation results, we found that A. bangokensis has an antioxidant defense system similar to those of other arthropods (Fig 5A). In addition, the basal RPKM values of all genes indicated relatively high transcription levels, and thus it remains to be determined how A. bangokensis protects its cells and bodies from exogenous oxidative stress.
Fig 5. Antioxidant defense system in Allobathynella bangokensis.
A) Schematic diagram representing a proposed cascade for the antioxidant defense system, along with RPKM values of the Allobathynella bangokensis transcripts coding for the corresponding enzymes in this system. B) Phylogenetic tree of glutathione S-transferase (GST) proteins from A. bangokensis and from arthropods constructed by the Bayesian method.
Glutathione S-transferases (GSTs) and their isoforms play important roles in the cellular antioxidant defense system. In addition to their activities under oxidative stress conditions, GST family members play crucial roles in drug metabolism and the detoxification pathway. Phase I, phase II, and phase III detoxification systems are an efficient means of protecting against the potential impacts via both metabolic homeostasis and elimination of exogenous molecules (e.g., xenobiotics) in animals [71]. Of these detoxification systems, phase II is characterized by reductive or conjugative modification reactions of phase I metabolites to endogenous compounds through GST enzymatic activity. Thus, GSTs and their isoforms have been commonly used as strong biomarkers of both oxidative stress and cellular toxicity. In the A. bangokensis transcriptome, 22 GST genes were annotated and grouped into eight well-characterized GST subfamilies (Delta, Kappa, Mu, Omega, Sigma, Theta, Zeta, and microsomal GST) of arthropods (Fig 5B). Taken together, we provide evidence for a robust antioxidant defense system in A. bangokensis and maintenance of cellular homeostasis in subterranean crustaceans. Elucidating the mechanisms underlying activation of the antioxidant defense system by environmental changes is crucial for the development of effective monitoring strategies for subterranean ecosystems.
Conclusions
The ability of A. bangokensis to survive in such extreme subterranean environments suggests that they have evolved adaptations by employing molecular homeostatic mechanisms. The relationships between animals and their hyporheic/groundwater environments are being investigated continuously, and transcriptomic sequencing of A. bangokensis, as a sentinel species, renders it an optimal model for studying molecular adaptation and response mechanisms to harsh environmental conditions. Although the transcriptional responses induced by environmental changes or the unique adaptive metabolism of A. bangokensis are not discussed in this study due to the limited samples from subterranean regions, the transcriptomic database and gene composition provide the basis for clarifying the adaptive and responsive metabolic pathways. Thus, further identification and confirmation of the functions of conserved genes or pathways as well as the dissection of the genetic architecture of response genes will be useful for the study of adaptive mechanisms in subterranean ecosystems.
Supporting Information
(TIF)
(TIF)
Detailed information is appended in S3–S5 Tables.
(TIF)
(XLSX)
(XLSX)
(XLSX)
(XLSX)
(XLSX)
(XLSX)
(XLSX)
(XLSX)
(XLSX)
Data Availability
All relevant data are within the paper and its Supporting Information files.
Funding Statement
This work was supported by a grant, entitled “Polar Genomics 101 Project (PE17080)”, funded by the Korea Polar Research Institute.
References
- 1.Schmidt SI, Hahn HJ. What is groundwater and what does this mean to fauna?–An opinion. Limnologica-Ecology and Management of Inland Waters. 2012;42(1):1–6. [Google Scholar]
- 2.Danielopol D, des Châtelliers MC, Mösslacher F, Pospisil P, Popa R. Adaptation of Crustacea to interstitial habitats: a practical agenda for ecological studies. Groundwater Ecology Academic Press, San Diego: 1994:217–43. [Google Scholar]
- 3.Malard F, Hervant F. Oxygen supply and the adaptations of animals in groundwater. Freshwater Biology. 1999;41(1):1–30. [Google Scholar]
- 4.Porter ML, Crandall KA. Lost along the way: the significance of evolution in reverse. Trends in Ecology & Evolution. 2003;18(10):541–7. [Google Scholar]
- 5.Lefébure T, Douady C, Malard F, Gibert J. Testing dispersal and cryptic diversity in a widely distributed groundwater amphipod (Niphargus rhenorhodanensis). Molecular phylogenetics and evolution. 2007;42(3):676–86. 10.1016/j.ympev.2006.08.020 [DOI] [PubMed] [Google Scholar]
- 6.Gibert J, Culver DC, DOLE-OLIVIER MJ, Malard F, Christman MC, Deharveng L. Assessing and conserving groundwater biodiversity: synthesis and perspectives. Freshwater Biology. 2009;54(4):930–41. [Google Scholar]
- 7.Humphreys WF. Aquifers: the ultimate groundwater-dependent ecosystems. Australian Journal of Botany. 2006;54(2):115–32. [Google Scholar]
- 8.Vandel A. Biospeleology: the biology of cavernicolous animals: Elsevier; 2013. [Google Scholar]
- 9.Schminke HK, Noodt W. Groundwater crustacea of the order Bathynellacea (Malacostraca) from North America. Journal of crustacean biology. 1988:290–9. [Google Scholar]
- 10.Morimoto Y. Allobathynella japonica gen. et sp. nov., a new bathynellid from Japan. Proceedings of the Japan Academy. 1957;33(3):145–8. [Google Scholar]
- 11.Park J-G, Cho J-L. Fourteen new species of Allobathynella Morimoto and Miura, 1957 from South Korea: with a redescription of A. coreana Morimoto, 1970 (Crustacea, Bathynellacea, Parabathynellidae). Journal of Species Research. 2016;5(1):49–156. [Google Scholar]
- 12.Asmyhr MG, Hose G, Graham P, Stow AJ. Fine-scale genetics of subterranean syncarids. Freshwater Biology. 2014;59(1):1–11. [Google Scholar]
- 13.Bolger AM, Lohse M, Usadel B. Trimmomatic: A flexible trimmer for Illumina Sequence Data. Bioinformatics. 2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gordon A, Hannon G. Fastx-toolkit. FASTQ/A short-reads pre-processing tools. http://hannonlab.cshl.edu/fastx_toolkit. 2010.
- 15.Simpson JT, Wong K, Jackman SD, Schein JE, Jones SJ, Birol I. ABySS: a parallel assembler for short read sequence data. Genome research. 2009;19(6):1117–23. 10.1101/gr.089532.108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zerbino DR, Birney E. Velvet: Algorithms for de novo short read assembly using de Bruijn graphs. Genome Research. 2008;18(5):821–9. 10.1101/gr.074492.107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Conesa A, Götz S, García-Gómez JM, Terol J, Talón M, Robles M. Blast2GO: a universal tool for annotation, visualization and analysis in functional genomics research. Bioinformatics. 2005;21(18):3674–6. 10.1093/bioinformatics/bti610 [DOI] [PubMed] [Google Scholar]
- 18.Mortazavi A, Williams BA, McCue K, Schaeffer L, Wold B. Mapping and quantifying mammalian transcriptomes by RNA-Seq. Nature methods. 2008;5(7):621–8. 10.1038/nmeth.1226 [DOI] [PubMed] [Google Scholar]
- 19.Jenner RA, Dhubhghaill CN, Ferla MP, Wills MA. Eumalacostracan phylogeny and total evidence: limitations of the usual suspects. BMC Evolutionary Biology. 2009;9(1):1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zdobnov EM, Apweiler R. InterProScan—an integration platform for the signature-recognition methods in InterPro. Bioinformatics. 2001;17(9):847–8. [DOI] [PubMed] [Google Scholar]
- 21.Williams AF, Barclay AN. The immunoglobulin superfamily-domains for cell surface recognition. Annual review of immunology. 1988;6(1):381–405. [DOI] [PubMed] [Google Scholar]
- 22.Bettencourt R, Lanz-Mendoza H, Lindquist KR, Faye I. Cell Adhesion Properties of Hemolin, an Insect Immune Protein in the Ig Superfamily. European Journal of Biochemistry. 1997;250(3):630–7. [DOI] [PubMed] [Google Scholar]
- 23.Léonard PM, Adema CM, Zhang S-M, Loker ES. Structure of two FREP genes that combine IgSF and fibrinogen domains, with comments on diversity of the FREP gene family in the snail Biomphalariaglabrata. Gene. 2001;269(1):155–65. [DOI] [PubMed] [Google Scholar]
- 24.Watson FL, Püttmann-Holgado R, Thomas F, Lamar DL, Hughes M, Kondo M, et al. Extensive diversity of Ig-superfamily proteins in the immune system of insects. Science. 2005;309(5742):1874–8. 10.1126/science.1116887 [DOI] [PubMed] [Google Scholar]
- 25.Brites D, McTaggart S, Morris K, Anderson J, Thomas K, Colson I, et al. The Dscam Homologue of the Crustacean Daphnia Is Diversified by Alternative Splicing Like in Insects. Molecular Biology and Evolution. 2008;25(7):1429–39. 10.1093/molbev/msn087 [DOI] [PubMed] [Google Scholar]
- 26.Chou P-H, Chang H-S, Chen IT, Lin H-Y, Chen Y-M, Yang H-L, et al. The putative invertebrate adaptive immune protein Litopenaeus vannamei Dscam (LvDscam) is the first reported Dscam to lack a transmembrane domain and cytoplasmic tail. Developmental & Comparative Immunology. 2009;33(12):1258–67. [DOI] [PubMed] [Google Scholar]
- 27.Watthanasurorot A, Jiravanichpaisal P, Liu H, Söderhäll I, Söderhäll K. Bacteria-induced Dscam isoforms of the crustacean, Pacifastacus leniusculus. PLoS pathog. 2011;7(6):e1002062 10.1371/journal.ppat.1002062 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 28.Wang X-W, Wang J-X. Diversity and multiple functions of lectins in shrimp immunity. Developmental & Comparative Immunology. 2013;39(1):27–38. [DOI] [PubMed] [Google Scholar]
- 29.Zeng V, Villanueva KE, Ewen-Campen BS, Alwes F, Browne WE, Extavour CG. De novo assembly and characterization of a maternal and developmental transcriptome for the emerging model crustacean Parhyale hawaiensis. BMC genomics. 2011;12(1):1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hook SE, Twine NA, Simpson SL, Spadaro DA, Moncuquet P, Wilkins MR. 454 pyrosequencing-based analysis of gene expression profiles in the amphipod Melita plumulosa: transcriptome assembly and toxicant induced changes. Aquatic Toxicology. 2014;153:73–88. 10.1016/j.aquatox.2013.11.022 [DOI] [PubMed] [Google Scholar]
- 31.Kang S, Kim S, Park H. Transcriptome of the Antarctic amphipod Gondogeneia antarctica and its response to pollutant exposure. Marine genomics. 2015;24:253–4. 10.1016/j.margen.2015.07.012 [DOI] [PubMed] [Google Scholar]
- 32.Yednock BK, Sullivan TJ, Neigel JE. De novo assembly of a transcriptome from juvenile blue crabs (Callinectes sapidus) following exposure to surrogate Macondo crude oil. BMC genomics. 2015;16(1):1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Weston DP, Poynton HC, Wellborn GA, Lydy MJ, Blalock BJ, Sepulveda MS, et al. Multiple origins of pyrethroid insecticide resistance across the species complex of a nontarget aquatic crustacean, Hyalella azteca. Proceedings of the National Academy of Sciences. 2013;110(41):16532–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Stahl BA, Gross JB, Speiser DI, Oakley TH, Patel NH, Gould DB, et al. A Transcriptomic Analysis of Cave, Surface, and Hybrid Isopod Crustaceans of the Species Asellus aquaticus. PLoS ONE. 2015;10(10):e0140484 10.1371/journal.pone.0140484 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Porter ML, Blasic JR, Bok MJ, Cameron EG, Pringle T, Cronin TW, et al. editors. Shedding new light on opsin evolution. Proc R Soc B; 2012: The Royal Society. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Carlini DB, Satish S, Fong DW. Parallel reduction in expression, but no loss of functional constraint, in two opsin paralogs within cave populations of Gammarus minus (Crustacea: Amphipoda). BMC Evolutionary Biology. 2013;13(1):1–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Rivera AS, Pankey MS, Plachetzki DC, Villacorta C, Syme AE, Serb JM, et al. Gene duplication and the origins of morphological complexity in pancrustacean eyes, a genomic approach. BMC evolutionary biology. 2010;10(1):1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kim S-S, Zhang R-g, Braunstein SE, Joachimiak A, Cvekl A, Hegde RS. Structure of the retinal determination protein Dachshund reveals a DNA binding motif. Structure. 2002;10(6):787–95. [DOI] [PubMed] [Google Scholar]
- 39.Bonini NM, Leiserson WM, Benzer S. The eyes absent gene: Genetic control of cell survival and differentiation in the developing Drosophila eye. Cell. 1993;72(3):379–95. 10.1016/0092-8674(93)90115-7. [DOI] [PubMed] [Google Scholar]
- 40.Rebay I. Multiple functions of the Eya phosphotyrosine phosphatase. Molecular and cellular biology. 2016;36(5):668–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Chen R, Amoui M, Zhang Z, Mardon G. Dachshund and Eyes Absent Proteins Form a Complex and Function Synergistically to Induce Ectopic Eye Development in Drosophila. Cell. 1997;91(7):893–903. 10.1016/S0092-8674(00)80481-X. [DOI] [PubMed] [Google Scholar]
- 42.Chao JL, Tsai YC, Chiu SJ, Sun YH. Localized Notch signal acts through eyg and upd to promote global growth in Drosophila eye. Development. 2004;131(16):3839–47. Epub 2004/07/16. 10.1242/dev.01258 [DOI] [PubMed] [Google Scholar]
- 43.Dominguez M, Ferres-Marco D, Gutierrez-Aviño FJ, Speicher SA, Beneyto M. Growth and specification of the eye are controlled independently by Eyegone and Eyeless in Drosophila melanogaster. Nature genetics. 2004;36(1):31–9. 10.1038/ng1281 [DOI] [PubMed] [Google Scholar]
- 44.Coineau N. Adaptations to interstitial groundwater life. Ecosystems of the World. 2000:189–210. [Google Scholar]
- 45.Hancock PJ, Boulton AJ, Humphreys WF. Aquifers and hyporheic zones: towards an ecological understanding of groundwater. Hydrogeology Journal. 2005;13(1):98–111. [Google Scholar]
- 46.Friedrich M, Chen R, Daines B, Bao R, Caravas J, Rai PK, et al. Phototransduction and clock gene expression in the troglobiont beetle Ptomaphagus hirtus of Mammoth cave. The Journal of experimental biology. 2011;214(21):3532–41. [DOI] [PubMed] [Google Scholar]
- 47.Tierney SM, Cooper SJ, Saint KM, Bertozzi T, Hyde J, Humphreys WF, et al. Opsin transcripts of predatory diving beetles: a comparison of surface and subterranean photic niches. Royal Society open science. 2015;2(1):140386 10.1098/rsos.140386 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Lindquist S, Craig EA. The Heat-Shock Proteins. Annual Review of Genetics. 1988;22(1):631–77. [DOI] [PubMed] [Google Scholar]
- 49.Feder ME, Hofmann GE. HEAT-SHOCK PROTEINS, MOLECULAR CHAPERONES, AND THE STRESS RESPONSE: Evolutionary and Ecological Physiology. Annual Review of Physiology. 1999;61(1):243–82. [DOI] [PubMed] [Google Scholar]
- 50.Gibert J, Deharveng L. Subterranean Ecosystems: A Truncated Functional Biodiversity This article emphasizes the truncated nature of subterranean biodiversity at both the bottom (no primary producers) and the top (very few strict predators) of food webs and discusses the implications of this truncation both from functional and evolutionary perspectives. BioScience. 2002;52(6):473–81. [Google Scholar]
- 51.Morimoto RI, Kline MP, Bimston DN, Cotto JJ. The heat-shock response: regulation and function of heat-shock proteins and molecular chaperones. Essays in biochemistry. 1997;32:17–29. Epub 1997/01/01. [PubMed] [Google Scholar]
- 52.Hooven TA, Yamamoto Y, Jeffery WR. Blind cavefish and heat shock protein chaperones: a novel role for hsp90alpha in lens apoptosis. International Journal of Developmental Biology. 2004;48(8–9):731–8. 10.1387/ijdb.041874th [DOI] [PubMed] [Google Scholar]
- 53.Hoffmann JA, Kafatos FC, Janeway CA, Ezekowitz R. Phylogenetic perspectives in innate immunity. Science. 1999;284(5418):1313–8. [DOI] [PubMed] [Google Scholar]
- 54.Iwanaga S, Lee B-L. Recent advances in the innate immunity of invertebrate animals. BMB Reports. 2005;38(2):128–50. [DOI] [PubMed] [Google Scholar]
- 55.McTaggart SJ, Conlon C, Colbourne JK, Blaxter ML, Little TJ. The components of the Daphnia pulex immune system as revealed by complete genome sequencing. Bmc Genomics. 2009;10(1):1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Gerardo NM, Altincicek B, Anselme C, Atamian H, Barribeau SM, De Vos M, et al. Immunity and other defenses in pea aphids, Acyrthosiphon pisum. Genome biology. 2010;11(2):1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Palmer WJ, Jiggins FM. Comparative genomics reveals the origins and diversity of arthropod immune systems. Molecular biology and evolution. 2015;32(8):2111–29. 10.1093/molbev/msv093 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Verbruggen B, Bickley LK, Santos EM, Tyler CR, Stentiford GD, Bateman KS, et al. De novo assembly of the Carcinus maenas transcriptome and characterization of innate immune system pathways. BMC genomics. 2015;16(1):1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Smith AA, Pal U. Immunity-related genes in Ixodes scapularis—perspectives from genome information. Lyme Disease: Recent Advances and Perspectives. 2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Rosa RD, Capelli-Peixoto J, Mesquita RD, Kalil SP, Pohl PC, Braz GR, et al. Exploring the immune signalling pathway-related genes of the cattle tick Rhipicephalus microplus: From molecular characterization to transcriptional profile upon microbial challenge. Developmental & Comparative Immunology. 2016;59:1–14. [DOI] [PubMed] [Google Scholar]
- 61.Aderem A, Ulevitch RJ. Toll-like receptors in the induction of the innate immune response. Nature. 2000;406(6797):782–7. 10.1038/35021228 [DOI] [PubMed] [Google Scholar]
- 62.Takeda K, Akira S, editors. TLR signaling pathways Seminars in immunology; 2004: Elsevier. [DOI] [PubMed] [Google Scholar]
- 63.Coscia M, Giacomelli S, Oreste U. Toll-like receptors: an overview from invertebrates to vertebrates. ISJ. 2011;8:210–26. [Google Scholar]
- 64.Li X-C, Zhu L, Li L-G, Ren Q, Huang Y-Q, Lu J-X, et al. A novel myeloid differentiation factor 88 homolog, SpMyD88, exhibiting SpToll-binding activity in the mud crab Scylla paramamosain. Developmental & Comparative Immunology. 2013;39(4):313–22. [DOI] [PubMed] [Google Scholar]
- 65.Yu A-Q, Jin X-K, Guo X-N, Li S, Wu M-H, Li W-W, et al. Two novel Toll genes (EsToll1 and EsToll2) from Eriocheir sinensis are differentially induced by lipopolysaccharide, peptidoglycan and zymosan. Fish & shellfish immunology. 2013;35(4):1282–92. [DOI] [PubMed] [Google Scholar]
- 66.Huang Y, Chen Y-H, Wang Z, Wang W, Ren Q. Novel myeloid differentiation factor 88, EsMyD88, exhibits EsTube-binding activity in Chinese mitten crab Eriocheir sinensis. Developmental & Comparative Immunology. 2014;47(2):298–308. [DOI] [PubMed] [Google Scholar]
- 67.Valenzuela-Munoz V, Gallardo-Escarate C. TLR and IMD signaling pathways from Caligus rogercresseyi (Crustacea: Copepoda): in silico gene expression and SNPs discovery. Fish & shellfish immunology. 2014;36(2):428–34. [DOI] [PubMed] [Google Scholar]
- 68.Rawlings JS, Rosler KM, Harrison DA. The JAK/STAT signaling pathway. Journal of cell science. 2004;117(8):1281–3. [DOI] [PubMed] [Google Scholar]
- 69.Waterhouse RM, Kriventseva EV, Meister S, Xi Z, Alvarez KS, Bartholomay LC, et al. Evolutionary dynamics of immune-related genes and pathways in disease-vector mosquitoes. science. 2007;316(5832):1738–43. 10.1126/science.1139862 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Apel K, Hirt H. REACTIVE OXYGEN SPECIES: Metabolism, Oxidative Stress, and Signal Transduction. Annual Review of Plant Biology. 2004;55(1):373–99. [DOI] [PubMed] [Google Scholar]
- 71.Xu C, Li CY-T, Kong A-NT. Induction of phase I, II and III drug metabolism/transport by xenobiotics. Archives of Pharmacal Research. 2005;28(3):249–68. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(TIF)
(TIF)
Detailed information is appended in S3–S5 Tables.
(TIF)
(XLSX)
(XLSX)
(XLSX)
(XLSX)
(XLSX)
(XLSX)
(XLSX)
(XLSX)
(XLSX)
Data Availability Statement
All relevant data are within the paper and its Supporting Information files.