Abstract
Most flagellar proteins are exported via a type III export apparatus which, in part, consists of the membrane proteins FlhA, FlhB, FliO, FliP, FliQ, and FliR and is housed within the membrane-supramembrane ring formed by FliF subunits. Salmonella FlhA is a 692-residue integral membrane protein with eight predicted transmembrane spans. Its function is not understood, but it is necessary for flagellar export. We have created mutants in which potentially important sequences were deleted. FlhA lacking the amino-terminal sequence prior to the first transmembrane span failed to complement and was dominant negative, suggesting that the sequence is required for function. Similar effects were seen in a variant lacking a highly conserved domain (FHIPEP) within a putative cytoplasmic loop. Scanning deletion analysis of the cytoplasmic domain (FlhAc) demonstrated that substantially all of FlhAc is required for efficient function. Affinity blotting showed that FlhA interacts with several other export apparatus membrane proteins. The implications of these findings are discussed, and a model of FlhA within the export apparatus is presented.
The bacterial flagellum is a complex molecular machine. It is self-assembling, containing its own type III export apparatus (17, 28). Almost all flagellar proteins that reside beyond the cell membrane are exported via the flagellum-specific pathway. The flagellar components (basal body, hook, and filament) all contain a continuous interior channel of about 2 to 3 nm (29, 42) through which exported proteins travel to their final destination in a cell-proximal to cell-distal fashion.
The machinery which effects export consists of both soluble and membrane proteins. A patch of membrane containing the membrane-integrated components is presumed to lie within the basal-body MS ring, forming a selective pore through which exported substrates pass. This substructure of the export machinery is assumed to consist of integral membrane proteins FlhA, FlhB, FliO, FliP, FliQ, and FliR. While specific functions have yet to be determined for all but FlhB (8, 25, 40), they are all required for export (24). Intergenic suppression data suggest that FlhA is in contact with FliF, which forms the MS ring (14). FliP and FliR localize to the basal body (7). Analysis of a FliR-FlhB fusion protein demonstrated close contact between FliR and FlhB as well as basal-body localization of FlhB (36).
FlhA is a 692-residue protein that is organized into two large domains. Its N-terminal membrane domain is highly conserved among homologs and variously predicted to contain six to eight transmembrane spans (see Discussion). It also has a large, less well-conserved, approximately 365-residue C-terminal cytoplasmic domain (FlhAc) that interacts with both soluble export apparatus components and export substrates (27), although the nature and consequences of these interactions are unknown.
FlhA contains a particularly well-conserved hydrophilic domain of unknown function in what we predict to be a cytoplasmic loop between TMs 4 and 5. This domain, FHIPEP (for “flagellum/hypersensitive response/invasion protein export pore”), is found in FlhA homologs from various species, including HrpI from Erwinia amylovora and Pseudomonas syringae, as well as InvA from Salmonella (2, 11, 39).
On the cytoplasmic side of the MS ring are several soluble proteins that play key roles: FliI, an ATPase which couples ATP hydrolysis with export; FliH, which negatively regulates FliI; and FliJ, a general flagellar chaperone. Two FliH and one FliI form a heterotrimer (10) for which there is evidence of association with the soluble domains of the membrane export apparatus (22, 43). FliJ is necessary for efficient export of many substrates (21). It associates with FliH and FliI. Purified FliI forms a homohexamer in the presence of ATP (3). It may be that a multimeric FliH-FliI-FliJ complex delivers export substrates to the membrane components and that FliI-catalyzed ATP hydrolysis provides the energy necessary to push the substrate across the membrane. FliH and FliI partition with the cellular membrane (1). Recently, Minamino et al. (22) showed that overexpression of FliI could compensate for the absence of FliH in restoring export. Mutations resulting in altered residues within the likely cytoplasmic amino-terminal region of FlhA (at I21 and L22) enhanced the bypass effect.
To better understand the role of FlhA in flagellar export, we undertook a study to systematically analyze the importance of the cytoplasmic domains: the amino-terminal juxtamembrane sequence, the FHIPEP domain, and FlhAc with a series of deletion mutants. We also examined the interactions of purified native FlhA with other export components. What has emerged is a picture of FlhA as a central element of the export machinery. A model of the export apparatus incorporating these findings is presented.
MATERIALS AND METHODS
Abbreviations used.
BSA, bovine serum albumin; CHAPS, 3-[(3-cholamidopropyl)dimethylammonio]-l-propanesulfonate; GST, glutathione S-transferase; IPTG, isopropyl-β-d-thiogalactopyranoside; MS, membrane-supramembrane; NTA, nitrilotriacetic acid; Sarkosyl, N-lauroylsarcosine; TM, transmembrane segment.
Strains and media.
The bacterial strains and plasmids used in this study are listed in Table 1. Luria broth and soft tryptone agar plates were prepared as described previously (27). T7 medium contains, per liter, 20 g of tryptone, 10 g of yeast extract, 5 g of NaCl, and 8.7 g of K2HPO4 (pH 7.2). Ampicillin (50 μg ml−1), tetracycline (12.5 μg ml−1), and chloramphenicol (25 μg ml−1) were added where appropriate.
TABLE 1.
Constructs used in this study
| Strain or plasmid | Relevant characteristics | Source or reference |
|---|---|---|
| E. coli strains | ||
| BL21(DE3)pLysS | For overproduction of proteins | Novagen |
| NovaBlue | Recipient for cloning experiments | Novagen |
| Salmonella strains | ||
| JR501 | For converting E. coli plasmids to Salmonella compatibility | 33 |
| SJW1103 | Wild type for motility | 41 |
| SJW1364 | flhA mutant (Δ917-929, frameshift) | 15 |
| Plasmids | ||
| pET19b | N-terminal His tag T7 expression vector | Novagen |
| pGEX-2T | GST fusion expression vector | Amersham |
| pTrc99A | Expression vector | Amersham |
| pTrc99A-FF4 | Modified pTrc expression vector | 30 |
| pFFF1300 | pET19b/His-FliF | 7 |
| pGFBN269A | pET19b/His-FlhBN269A | 8 |
| pGS20 | pET19b/His-FliO | G. Schoenhals, unpublished |
| pJM200. . . pJM218 | pTrc99A-FF4/FlhAΔ1 (residues 328-357). . .pTrc99A-FF4/FlhA Δ19 (residues 688-692) | This study |
| pJM219 | pTrc99A-FF4/FlhAΔFHIPEP | This study |
| pJM272 | pTrc99A/His-FlhAΔ18QWQIL22 | This study |
| pJM273 | pTrc99A/His-FlhAΔ1-22 | This study |
| pJMA1 | pET19b/His-FlhA | This study |
| pJMP1 | pGEX-2T/GST-FliP (residues 22-245)-His | This study |
| pJMQ1 | pGEX-2T/GST-FliQ-His | This study |
| pJMR1 | pGEX-2T/GST-FliR-His | This study |
| pMM101 | pTrc99A-FF4/His-FLAG-FlhAc | 27 |
| pMM108 | pTrc99A/His-FLAG-FlhA | T. Minamino, unpublished |
| pMM130 | pTrc99A-FF4/FlhA | 14 |
| pMM309 | pTrc99A-FF4/FliH | 26 |
| pMM310 | pET19b/His-FliH | 27 |
| pMM404 | pTrc99A-FF4/FliJ | 21 |
| pMM405 | pET19b/His-FliJ | 27 |
| pMM1701 | pET19b/His-FliI | 27 |
| pMM1719 | pTrc99A-FF4/FliI | 26 |
Construction of plasmids.
pJMA1 was produced by ligation of the NdeI-BamHI fragment from pMM130, which contains the entire flhA sequence, into pET19b (Novagen, Madison, Wis.). The resulting sequence produces an otherwise wild-type FlhA sequence with a N-terminal His encoded 5′ to the NdeI site.
Deletion mutations were made by a method adapted from that of Toker et al. (34). Primers complementary to the sequence flanking each side of the segment to be deleted were used with outside primers to generate fragments of FlhA from pJMA1 using Pfu Turbo DNA polymerase (Stratagene, La Jolla, Calif.). Products were gel purified (Qiagen, Valencia, Calif.) and used as templates for a second round of amplification that produced the otherwise full-length gene lacking the deleted codons. These products were restricted with NdeI and BamHI and cloned into pTrc99A-FF4 for pJM200-pJM219. pJM272 and pJM273 were digested with NcoI and BamHI and cloned into pTrc99A, resulting in the retention of the pET19b-based His tag sequence 5′ to the flhA sequence. The entire coding region of each plasmid was sequenced prior to use.
pJMP1, pJMQ1, and pJMR1 were produced by amplification of the gene sequences of FliP (without its signal sequence), FliQ, and FliR with primers that contained BamHI and EcoRI sites as well as codons for a His6 sequence. The products were restricted and ligated into pGEX-2T, resulting in plasmids coding for N-terminal GST fusion proteins with C-terminal His tags.
Motility analysis.
For complementation and dominance studies, SJW1364 or SJW1103 cells were transformed with plasmids of interest, inoculated onto soft tryptone agar plates containing ampicillin, and incubated at 30°C. To test for dominant negative effects, 0.1 mM IPTG was included in the plates.
Cell fractionation, NaOH and detergent extraction, and protease protection analysis.
To assay the membrane assembly of variant FlhAs, cell fractionation, NaOH extraction and protease protection analysis were carried out essentially as described previously (19), and fragments were detected by Western blotting using anti-FlhAc antisera (a gift of K. Kutsukake) (23) and enhanced chemiluminescence immunodetection as specified by the manufacturer (Amersham, Piscataway, N.J.). Briefly, protease protection analysis was performed by treating spheroplasts made from Escherichia coli BL21(DE3)pLysS overexpressing the protein of interest. Three aliquots were taken: one was a no-treatment control, another was treated with 25 μg of proteinase K (Roche, Mannheim, Germany) per ml, and the last was treated with the same concentration of proteinase K in the presence of 0.2% CHAPS (Anatrace, Maumee, Ohio) to solubilize the membranes and allow the protease access to cytoplasmic components. Samples were precipitated with 10% trichloroacetic acid rinsed with acetone, resuspended in sample buffer, subjected to sodium dodecyl sulfate-polyacrylamide gel electrophoresis, transferred to nitrocellulose membranes, and analyzed by Western blotting.
Expression and purification of FlhA and affinity blotting target proteins.
For nondenaturing purification of His-tagged FlhA, an overnight culture of E. coli BL21(DE3)pLysS carrying pJMA1 was subcultured 1:100 into 6 liters of T7. The culture was grown with vigorous shaking at 30°C until it reached an optical density at 600 nm of 0.4. IPTG was then added to a final concentration of 0.2 mM, and growth continued for 4 h. Cells were harvested by centrifugation and stored at −85°C until used.
Cell pellets were thawed on ice, resuspended in 120 ml of lysis buffer (50 mM sodium phosphate [pH 8.0], 500 mM NaCl, 20% glycerol, 10 mM β-mercaptoethanol, and 0.1 mM phenylmethylsulfonyl fluoride), sonicated, and centrifuged at 10,000 × g to pellet unbroken cells and inclusion bodies. The supernatant was then centrifuged at 100,000 × g for 45 min. Pellets (crude membrane fractions) were retained and resuspended by homogenization in 60 ml of solubilization buffer (lysis buffer with 1% [vol/vol] Triton X-100 and 10 mM imidazole). The suspension was stirred at 4°C for 1 h and then subjected to centrifugation at 100,000 × g for 45 min. This solubilized membrane fraction was retained as the supernatant.
The supernatant was then loaded by gravity flow onto a 5-ml Ni-NTA (Qiagen) column, washed with 20 column volumes of wash buffer (solubilization buffer with 20 mM imidazole), and eluted in the same buffer with 250 mM imidazole. Near homogeneity was achieved in this one-column process. Protein-containing fractions were pooled and stored at −85°C until used.
FlhB(N269A), a noncleavable variant of FlhB (8), was expressed from pGFBN269A. FliF was expressed from pFFF1300. FliP, FliQ, and FliR were expressed as N-terminal GST fusion proteins with C-terminal His tags from pJMP1, pJMQ1, and pJMR1, respectively. All target membrane proteins except FliO were purified as above on a smaller scale using Ni-NTA chromatography. Ni-NTA purification of His-FliO expressed from pGS20 required 6 M urea. Other proteins, including FlhAc, were purified as described previously (27).
Affinity blotting.
Samples to be probed were subjected to sodium dodecyl sulfate-polyacrylamide gel electrophoresis (15% polyacrylamide) and transferred onto nitrocellulose membranes with a Hoefer transblotting apparatus. The gels were loaded such that substantially equal amounts of each target protein (as determined by the Bradford assay) were in each lane. Efficient transfer was verified by Ponceau S staining. Membranes were blocked in Tris-buffered saline with 0.1% Tween 20 plus 5% nonfat milk overnight. Tris-buffered saline with 0.1% Tween 20 plus 5% nonfat milk was used in all steps and for the extensive washing performed between each step. Purified probe protein was added to 10 μg ml−1 and allowed to bind for 2 h. Membrane-bound probe was immunodetected using anti-FlhAc antisera and enhanced chemiluminescence. Antibodies demonstrated no cross-reactivity to membrane-bound target proteins in affinity blot analyses performed without probe protein.
RESULTS
The amino-terminal soluble domain is critical for function.
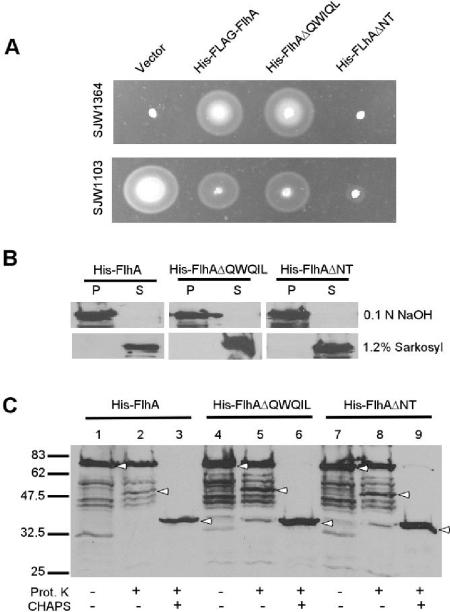
Minamino et al. (22) reported that mutations resulting in amino acid substitutions within the amino-terminal juxtamembrane sequence at I21 and L22 were pseudorevertants of a fliH null strain. To investigate the importance of this region, plasmids containing two deletion mutations, pJM272 (encoding FlhAΔQWQIL [residues 18 to 22 deleted]) and pJM273 (FlhAΔNT [residues 1 to 22 deleted]) were constructed. Both, although pTrc based, contained the pET19b-derived N-terminal His tag containing sequences of 23 residues to minimize the likelihood that the deletions would affect membrane assembly due to loss of soluble sequence prior to the first TM. pJM272 complemented flhA mutant strain SJW1364 as well as pMM108 (His-FLAG-FlhA) (Fig. 1A). (SJW1364 has a 13-bp deletion from bp 917 to 929, resulting in a frameshift that reaches a stop codon at bp 1022.) pJM273 failed to complement. Both variants had dominant negative effects when expressed in wild-type-motility strain SJW1103; the effect of FlhAΔQWQIL was similar to that of FlhA, whereas FlhAΔNT had a much more severe effect. Addition of IPTG inhibited complementation and increased dominant negative effects (data not shown).
FIG. 1.
Deletion of 18QWQIL22 and amino-terminal sequence. (A) Motility assay of SJW1364 and SJW1103 transformed with pMM108 (encoding His-FLAG-FlhA), pJM272 (His-FlhAΔQWQIL), or pJM273 (His-FlhAΔNT). Transformants were incubated at 30°C for 5 h on semisolid tryptone agar. (B) Extraction of His-FlhA, His-FlhAΔQWQIL, and His-FlhAΔNT in NaOH and Sarkosyl. P, pellet fraction; S, supernatant fraction. (C) Protease protection assay of His-FlhA, His-FlhAΔQWQIL, and His-FlhAΔNT. Addition of proteinase K (Prot. K) and CHAPS is indicated by +. Bands of interest are indicated by arrowheads, and positions of molecular weight standards are shown on the left in thousands.
FlhAΔQWQIL and FlhAΔNT are stably expressed and partition with the membrane fraction. Both are resistant to NaOH extraction and are solubilized by 1.2% Sarkosyl (Fig. 1B). Further, protease protection analysis demonstrates that both are able to properly assemble in the membrane. In spheroplasts made from cells overexpressing FlhAΔQWQIL and FlhAΔNT, both variants had protease protection profiles identical to that of FlhA expressed from pJMA1 (Fig. 1C). While the majority of the expressed protein remained full length in the proteinase K-treated samples (Fig. 1C, lanes 2, 5, and 8), a band consistent with protection of a fragment corresponding to FlhAc with a transmembrane span was also seen, indicating that at least a fraction of the population is assembled. The presence of full-length FlhA in these lanes is probably a result of inclusion body formation or a failure to assemble of much of the population of FlhA on overexpression. Alternatively, the protein could be properly assembled but incompletely digested due to partial inaccessibility. Nevertheless, the variants were able to assemble at least as well as the wild type. Addition of 0.2% CHAPS to solubilize the membranes resulted in retention only of a band similar to but smaller than the 38.5-kDa V8 and trypsin-resistant FlhAc fragments previously observed by Saijo-Hamano et al. (33a), confirming the proper cytoplasmic localization of FlhAc and thus assembly.
The FHIPEP motif is critical for export function.
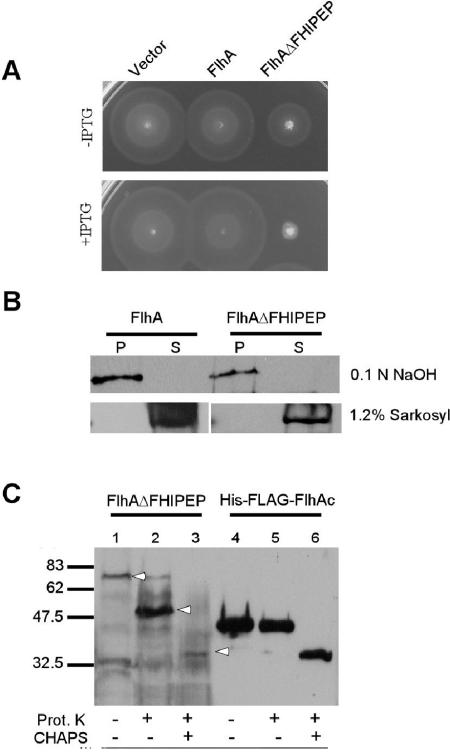
A PROSITE (6) search of the primary sequence of FlhA identifies a motif, FHIPEP (PS00994), that is conserved among FlhA homologs. This sequence, which in Salmonella FlhA is RIAEVGARFVLDGMPGKQMAIDAD (residues 147 to 170), is within what is probably its second cytoplasmic loop (see Discussion). pJM219, a plasmid encoding FlhA without the FHIPEP sequence (FlhAΔFHIPEP), failed to complement SJW1364. SJW1364 transformed with pJM219 failed to export both FlgD and FliC, indicating an inability to export either rod/hook or filament-type substrates (data not shown). However, after an extended incubation time (24 h), SJW1364 transformed with pJM219 was motile in the absence but not the presence of IPTG, suggesting that there is minor FlhA function at low levels of production. FlhAΔFHIPEP exerted a substantial dominant negative effect on motility when expressed in SJW1103 (Fig. 2A). The addition of IPTG reduced motility to a greater extent. After 24 h of incubation, SJW1103 with pJM219 exhibited limited motility in the absence but not the presence of IPTG (data not shown).
FIG. 2.
Deletion of the FHIPEP motif. (A) Motility assay of SJW1103 transformed with pMM130 (encoding wild-type FlhA) and pJM219 (FlhAΔFHIPEP) with and without the addition of 0.1 M IPTG. Transformants were incubated at 30°C for 5 h on semisolid tryptone agar. (B) Extraction of FlhA and FlhAΔFHIPEP in NaOH and Sarkosyl. P, pellet fraction; S, supernatant fraction. (C) Protease protection assay of FlhAΔFHIPEP and HisFLAG-FlhAc. Addition of proteinase K (Prot. K) and CHAPS is indicated by +. Bands of interest are indicated by arrowheads, and positions of molecular weight standards are shown on the left in thousands.
FlhAΔFHIPEP is also stably expressed, partitions with the membrane fraction, is resistant to NaOH extraction, and is solubilized by 1.2% (wt/vol) Sarkosyl (Fig. 2B). Furthermore, protease protection analysis demonstrates that with FlhAΔFHIPEP, the FlhAc domain is fully protected from proteinase K (Fig. 2C), confirming its correct cytoplasmic location and indicating that deletion of the motif does not affect proper membrane assembly. The protected fragment is somewhat larger than His-FLAG-FlhAc produced from pMM101 (Fig. 2C, lane 5), consistent with protection of the cytoplasmic domain and a transmembrane span. Addition of CHAPS in both cases reduced the protected fragment to the proteinase K-resistant band as seen with FlhA, FlhAΔQWQIL, and FlhAΔNT (Fig. 1C and 2C). Interestingly, the intensity of the protected band in the FlhAΔFHIPEP sample is much greater than that of wild-type FlhA, which is almost fully protected in the proteinase K-added lane (compare lanes 2 of Fig. 1C and 2C). FlhAΔFHIPEP may assemble more efficiently on overexpression, perhaps due to the loss of a net charge of −1, as would be expected in obedience to the positive-inside rule (38), or because relatively less goes to inclusion bodies.
Complementation and dominance properties of FlhAc deletion mutants.
A set of 19 pTrc-based plasmids (pJM200 to pJM218) containing flhA with deletion mutations of 20 codons each (except the last, in which a stop codon was created at position 2062, functionally deleting the last 5 codons) that together comprise systematic partial deletion of the entire FlhAc domain were analyzed; e.g., pJM200 encodes FlhAΔ1, which lacks residues 328 to 347, whereas pJM201 encodes FlhAΔ2, which lacks residues 347 to 368. In SJW1364, only Δ19 resulted in motility after 5 h of incubation at 30°C (data not shown). However, after extended incubation (24 h), all deletion mutants but Δ18 were motile to some extent. Vector-only transformed SJW1364 was not motile, even after 24 h. In all cases, failure to complement coincided with failure to export both rod/hook and filament-type substrates since FlgD and FliC were not detected in culture supernatants (data not shown). When expressed in SJW1103, Δ1, Δ6, and Δ17 exhibited substantial inhibition of motility in the presence of 0.1 mM IPTG, suggesting that the deleted sequences are important for function. All FlhA variants are detectable by anti-FlhAc in Western blotting and migrate at the correct position, indicating that they are all stably expressed (data not shown).
Purification of FlhA.
Previously, FlhA had been purified under denaturing conditions (43). However, a nondenaturing purification scheme was desirable since interactions of the native protein were of interest and an assay for proper refolding is not currently available. Therefore, a method was developed using a mild detergent (Triton X-100) and glycerol to solubilize the protein from crude membrane preparations, seeking to avoid misfolded protein in inclusion bodies, include properly membrane-assembled protein, and not subject it to harsh, denaturing conditions. Full-length His-FlhA was purified under nondenaturing, solubilizing conditions to near homogeneity in a one-column purification. A typical yield was about 8 mg per liter of culture medium.
Affinity blotting demonstrates interactions between FlhA and other membrane export components.
To test for interactions between FlhA and other membrane export components, affinity blotting of the membrane components using purified, solubilized FlhA against targets of purified components was performed. It was necessary to express FliP, FliQ, and FliR as GST fusion proteins in order to achieve detectable expression and sufficient yield. A plasmid encoding GST-FliO caused cell lysis on induction. Therefore, denatured His-tagged FliO expressed from pGS20 was used instead.
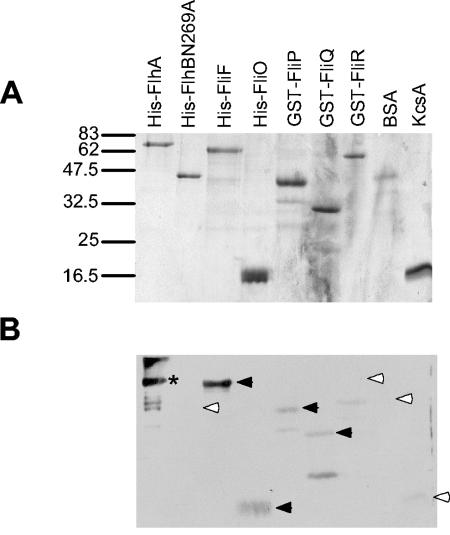
A clear positive signal was seen with FliF (Fig. 3). Weaker but probably significant signals were also present for FliO, GST-FliP, and GST-FliQ. No band was detected at the FlhB(N269A) or GST-FliR positions. Negative controls BSA and KcsA, an E. coli integral membrane protein (16), failed to produce significant signal, suggesting that the interactions seen are specific. Crude membrane fractions of cells overexpressing target proteins were also probed, with the same results (data not shown). The band seen for FlhA is assuredly direct antibody binding to the target, but this does not rule out a FlhA-FlhA interaction. Likewise, a paucity of signal for any target protein does not rule out an interaction. Affinity blotting of the same set of target proteins with purified FlhAc gave no signal (data not shown), suggesting that the interactions observed are between transmembrane domains.
FIG. 3.
Affinity blotting of membrane components of the export apparatus versus purified His-FlhA. (A) Coomassie blue-stained gel of purified membrane export apparatus components. Positions of molecular weight standards are shown on the left in thousands. (B) Affinity blot of the same samples with 10 μg of His-FlhA and anti-FlhAc antibody per ml. Significant signals are indicated by black arrowheads; absence of signal and weak or insignificant signals are indicated by white arrowheads. The asterisk denotes an anti-FlhA-reactive band that represents direct antibody binding to FlhA.
Affinity blotting demonstrates interactions between FlhA and soluble export components FliH, FliI, and FliJ.
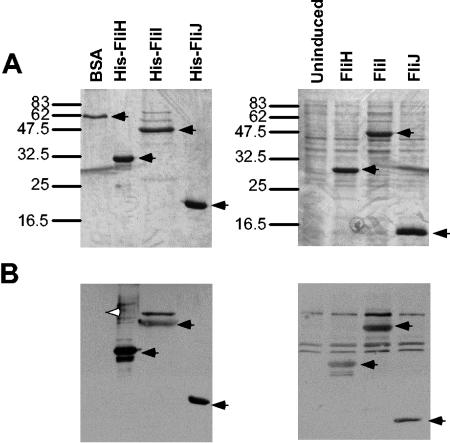
Full-length FlhA, like FlhAc only (27), bound purified FliH, FliI, and FliJ, but not the negative-control protein BSA (Fig. 4). Clear positive signals were also obtained with FliH, FliI, and FliJ in overexpressed cell lysates, suggesting specificity.
FIG. 4.
Affinity blotting of soluble export proteins versus purified His-FlhA. (A) Coomassie blue-stained gel of purified BSA and His-tagged FliH, FliI, and FliJ (left) and cell lysates of uninduced cells and untagged overexpressed FliH, FliI, and FliJ (right). Positions of molecular weight standards are shown on the left in thousands. (B) Affinity blotting of the same samples with 10 μg of His-FlhA and anti-FlhAc antibody per ml. Significant signals are indicated by black arrowheads, while lack of signal of the control protein, BSA, is indicated by a white arrowhead.
DISCUSSION
Topology of FlhA.
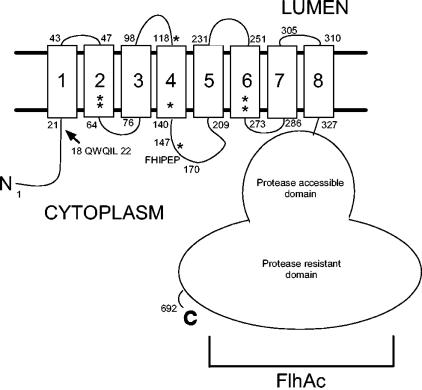
FlhA has been previously predicted to have eight transmembrane spans (14). Several topological prediction algorithms (shown in Table 2) generate predictions between six and eight spans. The majority, five of eight, predict seven spans (4, 5, 9, 12, 13, 32, 35, 38). TopPred2 similarly predicts seven spans for homologs Pseudomonas solanacearum HrpO, Bacillus subtilis FlhA, and Salmonella InvA. However, the positive-charge balance of the N terminus (38) and extensive experimental evidence for cytoplasmic localization of both the N and C termini, demonstrated previously (22, 27) and herein by protease protection of the FlhAc domain, led us to predict either six or eight spans, since an odd number would place the termini on opposite sides of the membrane. Indeed, TMHMM predicts eight spans when the N and C termini are fixed to the cytoplasmic side of the membrane (E. Granseth, personal communication), a tactic proven to increase the accuracy of TM prediction (20, 31). We further speculate that FlhA has eight transmembrane spans because experimental data suggest that its Salmonella virulence factor secretion apparatus homolog, InvA, has eight spans (J. E. Galan, personal communication), reiterating our earlier prediction, albeit with differing delineations of transmembrane span boundaries. While the precise topology of FlhA remains unknown, a schematic of likely FlhA topography incorporating the transmembrane boundaries as predicted by the fixed-termini version of TMHMM is presented in Fig. 5. The fact that our candidate for the likely membrane organization differs from the results of the majority of prediction programs underscores the still speculative nature of topological prediction and the need for experimental determination of the topology of FlhA.
TABLE 2.
Topology predictions for FlhA by topological prediction algorithms
| TM | Residues in FlhA primary sequence predicted to constitute the TM
|
|||||||
|---|---|---|---|---|---|---|---|---|
| SwissProt | DAS | HMMTOP | PHDtm | SOSUI | TMHMM | TMpred | TopPred2 | |
| TM1 | 24-44 | 23-63 | 22-41 | 26-50 | 19-41 | 21-43 | 21-44 | 24-44 |
| TM2 | 46-66 | 46-66 | 46-64 | 44-66 | 47-64 | 46-65 | ||
| TM3 | 71-91 | 75-86 | 73-97 | 72-89 | 72-94 | 76-98 | 68-93 | 68-88 |
| TM4 | 95-115 | |||||||
| TM5 | 121-141 | 123-140 | 118-142 | 120-139 | 122-144 | 118-140 | 123-140 | 122-142 |
| TM6 | 209-229 | 211-228 | 210-229 | 210-227 | 208-230 | 213-235 | 209-229 | 209-229 |
| TM7 | 247-267 | 258-264 | 250-269 | 250-267 | 248-270 | 250-272 | 244-266 | 250-270 |
| TM8 | 296-316 | 293-304 | 292-316 | 296-320 | 298-320 | 293-315 | 310-327 | 307-327 |
FIG. 5.
Schematic of the predicted topology of FlhA. Transmembrane segment boundaries as predicted by terminus-fixed TMHMM (see Discussion) are shown with terminal residue numbers shown at either end of each TM. The positions of the QWQIL and FHIPEP sequences are indicated. Asterisks denote the positions of residues encoded by intergenic suppressors of fliF mutants previously reported (14).
The amino terminus and FHIPEP motif are critical for function.
Within the transmembrane domain, defined as the first 327 residues, all soluble loops are short except the one containing the FHIPEP motif between predicted TM4 and TM5, which is about 66 residues in length. Deletion of the 24 residues constituting the FHIPEP motif results in failure to complement in the flhA mutant and a dominant negative phenotype in the wild-type strain. It is unlikely that the deletion causes a large perturbation in the protein structure for several reasons: there is still ample loop length to allow proper transmembrane domain assembly; the protein is still able to integrate into the membrane properly, and perhaps better than the wild type, as determined by protease protection and base and detergent extraction; and its dominant negative effect on the wild-type strain strongly suggests that it competes with FlhA produced from the chromosome for assembly into the membrane export complex.
The nature of the specific function of the FHIPEP motif remains a matter for speculation. Its likely cytoplasmic location suggests interactions with the soluble export apparatus component FliH, FliI, or FliJ, export substrates, or soluble domains of other membrane export components, of which FlhB is a candidate since it possesses a large cytoplasmic domain. Attempts to copurify FHIPEP expressed as a polypeptide fused to GST with FliH, FliI, and FliJ were unsuccessful, suggesting that any such interaction is dynamic or that FHIPEP itself is necessary but insufficient for stable binding. Since it is possible that FHIPEP is within the basal body lumen, we cannot rule out a very different role.
The amino-terminal sequence prior to TM1 is required for export function. Juxtamembrane residues I21 and L22 are sites for pseudoreversion variants, indicating their importance perhaps as a docking site for FliI (22). However, deletion of 18QWQIL22 does not noticeably affect motility, although deletion of residues 1 to 22 does. The QWQIL sequence is not highly conserved, although polarity at positions 18 and 20 and an aliphatic residue at position 22 seem to be important. I21 and L22 are thus probably constituent, but not absolutely required, elements of an important binding site for FliI, other soluble export components, or substrates.
Deletion analysis of FlhAc.
It would seem from the observation that all of the plasmids encoding FlhAc deletion variants in full-length flhA failed to complement and that substantially all of the C-terminal domain is necessary for efficient function. However, all of the deletions except Δ18 resulted in complementation at extended incubation times, suggesting that there is some residual activity in each. The dominant negative effects exhibited by some, including Δ1, Δ6, and Δ17, suggest a particular importance of those deleted sequences. Of note in this regard is Δ1, which is very similar to His-FLAG-FlhAΔ328-351 described by Saijo-Hamano et al. (33a). Thus, the N-terminal sequence of the FlhAc domain may play an important role, perhaps as a flexible hinge between the transmembrane and FlhAc domains.
The C-terminal portion of FlhAc is probably in a tightly folded conformation. Our observation of a proteinase K-resistant fragment of FlhAc that is smaller than the 39- and 38-kDa trypsin-resistant and V8-resistant fragments observed by Saijo-Hamano et al. (33a) suggests that the majority of FlhAc is in a very tightly folded structure. It may be that deletions of 20 residues disrupt this tight folding and hence disrupts the function of FlhA.
Interactions between full-length FlhA and other export components.
Kihara et al. (14) found intergenic suppressors of FliF mutations within the sequence coding for the membrane domain of FlhA, suggesting a FliF-FlhA interaction. Our results support this contention and provide direct biochemical evidence of FlhA-FliF binding. Further, the improved topological prediction places all but one of the fliF suppressors within TMs or at the terminus of a presumed TM (L118Q) (Fig. 5), more strongly suggesting that the FlhA-FliF interaction is between transmembrane domains. The other suppressor results in an I148S substitution at the N-terminal side of the FHIPEP motif, although it, too, is near a TM. FlhA also probably interacts with FliO, FliP, and FliQ. Given the unknown roles of these proteins and the fact that FliO is not completely conserved among export systems (18), the implications of these interactions are unclear. FlhAc and the cytoplasmic domain of FlhB (FlhBc) have previously been shown to interact (27). No signal was seen between full-length FlhA and full-length FlhB(N269A) or between FlhAc and full-length FlhB(N269A). The noncleavable FlhB(N269A) variant was used in this study to avoid loss of the FlhBcc domain (25) during solubilizing purification. It may be that cleavage of FlhBc is required for interaction with FlhAc.
Previously, FlhAc had been demonstrated to interact with FliH, FliI, and FliJ (27, 43). These results, for FlhA, suggest that the presence of the membrane domain does not prohibit soluble interactions. The cytoplasmic face of FlhA, including FlhAc, may act as a docking platform for the substrate-bound HIJ complex.
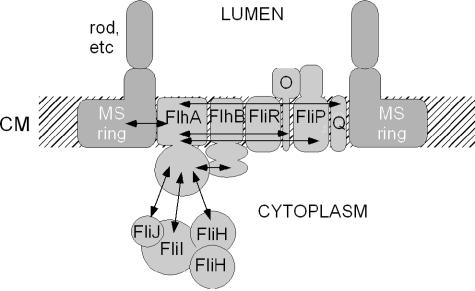
A cartoon model of the export apparatus proteins incorporating interactions involving FlhA discussed in this study and others previously found is shown in Fig. 6. Interactions among membrane export proteins shown to date are FliF-FlhA, FliR-FlhB, and FlhAc-FlhBc (14, 27, 36). Other interactions suggested by affinity blotting, i.e., FlhA-FliO, FlhA-FliP, and FlhA-FliQ, are also included. Affinity blotting also suggests that FliO but not FliF, FliP, FliQ, or FliR also interacts with the membrane domain of FlhB (unpublished data). No suggestions of interactions between FliR and FliO or FliP and FliQ are intended; no conclusions should be drawn in this respect from the way they are depicted. A presumed interaction between FliO and FliP due to the existence of a gene fusion in Buchnera aphidicola (37) is also incorporated.
FIG. 6.
Cartoon model of FlhA interactions with export apparatus proteins. Interactions of FlhA with other proteins are indicated by double-headed arrows. CM, cytoplasmic membrane.
Acknowledgments
We thank Lise Heginbotham for the gift of purified KcsA, Gary Schoenhals for plasmid pGS20, Tohru Minamino for helpful criticism and for sharing data prior to publication, Gunnar von Heijne and Erik Granseth for assistance with the revised topology prediction and use of the “fixing” version of TMHMM, and Hedda Ferris for helpful discussion and critical reading of the manuscript.
This work was supported by Public Health Service grants AI12202 (to R.M.M.) and GM070333 (to J.L.M.).
REFERENCES
- 1.Auvray, F., A. J. Ozin, L. Claret, and C. Hughes. 2002. Intrinsic membrane targeting of the flagellar export ATPase FliI: interaction with acidic phospholipids and FliH. J. Mol. Biol. 318:941-950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Carpenter, P. B., and G. W. Ordal. 1993. Bacillus subtilis FlhA: a flagellar protein related to a new family of signal-transducing receptors. Mol. Microbiol. 7:735-743. [DOI] [PubMed] [Google Scholar]
- 3.Claret, L., S. R. Calder, M. Higgins, and C. Hughes. 2003. Oligomerization and activation of the FliI ATPase central to bacterial flagellum assembly. Mol. Microbiol. 48:1349-1355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Claros, M., and G. von Heijne. 1994. TopPred II: an improved software for membrane protein structure predictions. Comput. Appl. Biosci. 10:685-686. [DOI] [PubMed] [Google Scholar]
- 5.Cserzö, M., E. Wallin, I. Simon, G. von Heijne, and A. Elofsson. 1997. Prediction of transmembrane α-helices in procaryotic membrane proteins: the dense alignment surface method. Protein Eng. 10:673-676. [DOI] [PubMed] [Google Scholar]
- 6.Falquet, L., M. Pagni, P. Bucher, N. Hulo, C. J. A. Sigrist, K. Hofmann, and A. Bairoch. 2002. The PROSITE database, its status in 2002. Nucleic Acids Res. 30:235-238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Fan, F., K. Ohnishi, N. R. Francis, and R. M. Macnab. 1997. The FliP and FliR proteins of Salmonella typhimurium, putative components of the type III flagellar export apparatus, are located in the flagellar basal body. Mol. Microbiol. 26:1035-1046. [DOI] [PubMed] [Google Scholar]
- 8.Fraser, G. M., T. Hirano, H. U. Ferris, L. L. Devgan, M. Kihara, and R. M. Macnab. 2003. Substrate specificity of type III flagellar protein export in Salmonella is controlled by subdomain interactions in FlhB. Mol. Microbiol. 48:1043-1057. [DOI] [PubMed] [Google Scholar]
- 9.Gasteiger, E., E. Jung, and A. Bairoch. 2001. SWISS-PROT: connecting biological knowledge via a protein database. Curr. Issues Mol. Biol. 3:47-55. [PubMed] [Google Scholar]
- 10.González-Pedrajo, B., G. M. Fraser, T. Minamino, and R. M. Macnab. 2002. Molecular dissection of Salmonella FliH, a regulator of the ATPase FliI and the type III flagellar protein export pathway. Mol. Microbiol. 45:967-982. [DOI] [PubMed] [Google Scholar]
- 11.Gough, C. L., S. Genin, V. Lopes, and C. A. Boucher. 1993. Homology between the HrpO protein of Pseudomonas solanacearum and bacterial proteins implicated in a signal peptide-independent secretion mechanism. Mol. Gen. Genet. 239:378-392. [DOI] [PubMed] [Google Scholar]
- 12.Hirokawa, T., S. Boon-Chieng, and S. Mitaku. 1998. SOSUI: classification and secondary structure prediction system for membrane proteins. Bioinformatics 14:378-379. [DOI] [PubMed] [Google Scholar]
- 13.Hofmann, K., and W. Stoffel. 1993. TMbase—a database of membrane spanning protein segments. Biol. Chem. Hoppe-Seyler 347:166. [Google Scholar]
- 14.Kihara, M., T. Minamino, S. Yamaguchi, and R. M. Macnab. 2001. Intergenic suppression between the flagellar MS ring protein FliF of Salmonella and FlhA, a membrane component of its export apparatus. J. Bacteriol. 183:1655-1662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kutsukake, K., and T. Iino. 1985. Refined genetic analysis of the region II che mutants in Salmonella typhimurium. Mol. Gen. Genet. 199:406-409. [DOI] [PubMed] [Google Scholar]
- 16.LeMasurier, M., L. Heginbotham, and C. Miller. 2001. KcsA: it's a potassium channel. J. Gen. Physiol. 118:303-314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Macnab, R. M. 2003. How bacteria assemble flagella. Annu. Rev. Microbiol. 57:77-100. [DOI] [PubMed] [Google Scholar]
- 18.Macnab, R. M. Type III flagellar protein export and flagellar assembly. BBA Mol. Cell Res., in press. [DOI] [PubMed]
- 19.McMurry, J. L., and D. A. Kendall. 1999. An artificial transmembrane segment directs SecA, SecB, and electrochemical potential-dependent translocation of a long amino-terminal tail. J. Biol. Chem. 274:6776-6782. [DOI] [PubMed] [Google Scholar]
- 20.Melén, K., A. Krogh, and G. von Heijne. 2003. Reliability measures for membrane protein topology prediction algorithms. J. Mol. Biol. 327:735-744. [DOI] [PubMed] [Google Scholar]
- 21.Minamino, T., R. Chu, S. Yamaguchi, and R. M. Macnab. 2000. Role of FliJ in flagellar protein export in Salmonella. J. Bacteriol. 182:4207-4215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Minamino, T., B. González-Pedrajo, M. Kihara, K. Namba, and R. M. Macnab. 2003. The ATPase FliI can interact with the type III flagellar protein export apparatus in the absence of its regulator, FliH. J. Bacteriol. 185:3983-3988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Minamino, T., T. Iino, and K. Kutsukake. 1994. Molecular characterization of the Salmonella typhimurium flhB operon and its protein products. J. Bacteriol. 176:7630-7637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Minamino, T., and R. M. Macnab. 1999. Components of the Salmonella flagellar export apparatus and classification of export substrates. J. Bacteriol. 181:1388-1394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Minamino, T., and R. M. Macnab. 2000. Domain structure of Salmonella FlhB, a flagellar export component responsible for substrate-specificity switching. J. Bacteriol. 182:4906-4914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Minamino, T., and R. M. Macnab. 2000. FliH, a soluble component of the type III flagellar export apparatus of Salmonella, forms a complex with FliI and inhibits its ATPase activity. Mol. Microbiol. 37:1494-1503. [DOI] [PubMed] [Google Scholar]
- 27.Minamino, T., and R. M. Macnab. 2000. Interactions among components of the Salmonella flagellar export apparatus and its substrates. Mol. Microbiol. 35:1052-1064. [DOI] [PubMed] [Google Scholar]
- 28.Minamino, T., and K. Namba. 2004. Self-assembly and type III protein export of the bacterial flagellum. J. Mol. Microbiol. Biotechnol. 7:5-17. [DOI] [PubMed] [Google Scholar]
- 29.Morgan, D. G., C. Owen, L. A. Melanson, and D. J. DeRosier. 1995. Structure of bacterial flagellar filaments at 11 Å resolution: packing of the α-helices. J. Mol. Biol. 249:88-110. [DOI] [PubMed] [Google Scholar]
- 30.Ohnishi, K., F. Fan, G. J. Schoenhals, M. Kihara, and R. M. Macnab. 1997. The FliO, FliP, FliQ, and FliR proteins of Salmonella typhimurium: putative components for flagellar assembly. J. Bacteriol. 179:6092-6099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Rapp, M., D. Drew, D. O. Daley, J. Nilsson, T. Carvalho, K. Melén, J.-W. de Gier, and G. von Heijne. 2004. Experimentally based topology models for E. coli inner membrane proteins. Protein Sci. 13:937-945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Rost, B., R. Casadio, P. Fariselli, and C. Sander. 1995. Transmembrane helices predicted at 95% accuracy. Protein Sci. 4:521-533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ryu, J., and R. J. Hartin. 1990. Quick transformation in Salmonella typhimurium LT2. BioTechniques 8:43-44. [PubMed] [Google Scholar]
- 33a.Saijo-Hamano, Y., T. Minamino, R. M. Macnab, and K. Namba. 2004. Structural and functional analysis of the C-terminal cytoplasmic domain of FlhA, an integral membrane component of the type III flagellar protein export apparatus in Salmonella. J. Mol. Biol. 343:457-466. [DOI] [PubMed] [Google Scholar]
- 34.Toker, A. S., M. Kihara, and R. M. Macnab. 1996. Deletion analysis of the FliM flagellar switch protein of Salmonella typhimurium. J. Bacteriol. 178:7069-7079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Tusnády, G. E., and I. Simon. 2001. The HMMTOP transmembrane topology prediction server. Bioinformatics 17:849-850. [DOI] [PubMed] [Google Scholar]
- 36.Van Arnam, J. S., J. L. McMurry, M. Kihara, and R. M. Macnab. 2004. Analysis of an engineered Salmonella flagellar fusion protein, FliR-FlhB. J. Bacteriol. 186:2495-2498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.van Ham, R. C. H. J., J. Kamerbeek, C. Palacios, C. Rausell, F. Abascal, U. Bastolla, J. M. Fernández, L. Jiménez, M. Postigo, F. J. Silva, J. Tamames, E. Viguera, A. Latorre, A. Valencia, F. Morán, and A. Moya. 2003. Reductive genome evolution in Buchnera aphidicola. Proc. Natl. Acad. Sci. USA 100:581-586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.von Heijne, G. 1992. Membrane protein structure prediction. Hydrophobicity analysis and the positive-inside rule. J. Mol. Biol. 225:487-494. [DOI] [PubMed] [Google Scholar]
- 39.Wei, Z.-M., and S. V. Beer. 1993. HrpI of Erwinia amylovora functions in secretion of harpin and is a member of a new protein family. J. Bacteriol. 175:7958-7967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Williams, A. W., S. Yamaguchi, F. Togashi, S.-I. Aizawa, I. Kawagishi, and R. M. Macnab. 1996. Mutations in fliK and flhB affecting flagellar hook and filament assembly in Salmonella typhimurium. J. Bacteriol. 178:2960-2970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Yamaguchi, S., H. Fujita, K. Sugata, T. Taira, and T. Iino. 1984. Genetic analysis of H2, the structural gene for phase-2 flagellin in Salmonella. J. Gen. Microbiol. 130:255-265. [DOI] [PubMed] [Google Scholar]
- 42.Yonekura, K., S. Maki-Yonekura, and K. Namba. 2003. Complete atomic model of the bacterial flagellar filament by electron cryomicroscopy. Nature (London) 424:643-650. [DOI] [PubMed] [Google Scholar]
- 43.Zhu, K., B. González-Pedrajo, and R. M. Macnab. 2002. Interactions among membrane and soluble components of the flagellar export apparatus of Salmonella. Biochemistry 41:9516-9524. [DOI] [PubMed] [Google Scholar]