Abstract
We show that for two well-characterized regulatory circuits in Escherichia coli, Tn10 tetracycline resistance and porin osmoregulation, the transcriptional outputs in individual cells are graded functions of the applied stimuli. These systems are therefore examples of naturally occurring regulatory circuits that exhibit continuous control of transcription. Surprisingly, however, we find that porin osmoregulation is open loop; i.e., the porin expression level does not feed back into the regulatory circuit. This mode of control is particularly interesting for an organism such as E. coli, which proliferates in diverse environments, and raises important questions regarding the biologically relevant inputs and outputs for this system.
Cell signaling circuits, like electrical circuits, can be based on continuous or discrete control. With continuous control the system functions like a rheostat, i.e., the output is a graded function of the input or applied stimulus, whereas with discrete control the system is more akin to an on-off switch, with an output that is a steep or discontinuous function of the input. For systems under discrete control, an intermediate level of stimulus will give rise to a mixed population of cells; in some cells the regulatory circuit will be on and in others it will be off. For systems under continuous control, on the other hand, an intermediate level of stimulus will give rise to a uniform population in which all of the cells have essentially the same response. In both cases, however, the population-averaged response will have an intermediate value. For this reason, to distinguish between discrete and continuous control, one must measure or infer the stimulus-response behavior of individual cells. In both prokaryotes and eukaryotes, a number of wild-type regulatory circuits that have been analyzed at the single-cell level have been shown to exhibit discrete control (5, 9, 18, 25, 28, 34, 43, 48). In addition, modified or synthetic circuits with either discrete or continuous control have been constructed (7, 21, 27, 40). Wild-type circuits that exhibit continuous control of transcription in single cells, on the other hand, are less well studied, although recently two examples in yeast have been described (9, 37). Here we describe two different regulatory circuits in Escherichia coli that are based on continuous control, Tn10 tetracycline resistance and porin osmoregulation.
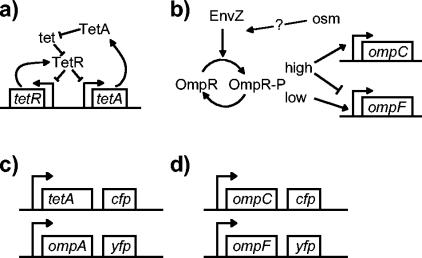
The tetracycline resistance determinant derived from the transposon Tn10 is one of the best-characterized regulatory circuits (Fig. 1a) (23). The primary protein components of this system are the efflux pump TetA and the repressor TetR. The corresponding genes tetA and tetR are transcribed from divergent promoters, both of which are repressed when TetR binds operator sites on the DNA. The antibiotic tetracycline binds TetR with high affinity, which releases the repressor from the DNA and enables transcription of tetA and tetR. This in turn results in production of TetA, which pumps tetracycline out of the cell.
FIG. 1.
(a) The Tn10 tetracycline resistance circuit. (b) The porin osmoregulatory circuit. (c, d) To measure transcription in single cells, strains were constructed in which operon fusions of tetA with cfp and ompA with yfp (c) or ompC with cfp and ompF with yfp (d) were integrated into the chromosome.
Porin osmoregulation is another well-studied regulatory circuit (Fig. 1b), although our understanding of this system is less complete than that of the tetracycline resistance circuit. The porins OmpF and OmpC are homologous proteins that form pores in the outer membrane of E. coli. OmpF generally has higher permeability than OmpC, depending on the properties of the solute, such as size, charge, and hydrophobicity (33). The best-studied environmental condition that affects porin expression is osmolarity (38). With increasing osmolarity of the extracellular medium, OmpF levels decrease and OmpC levels increase (45). Thus, porin osmoregulation controls the ratio of OmpC expression to OmpF expression. The key part of the network controlling this differential regulation of ompF and ompC transcription is the two-component signaling system consisting of the histidine kinase EnvZ and the response regulator OmpR (Fig. 1b) (16, 26, 45). Although EnvZ is often referred to as an osmosensor, the signal that stimulates this histidine kinase has not been determined, and there is evidence that a variety of environmental factors contribute to porin regulation (20, 29, 38).
Discrete control tends to render circuits insensitive to environmental perturbations so long as they operate far from the threshold for switching (32, 46). In contrast, systems based on continuous control generally employ negative feedback to ensure that the appropriate output level is attained (11, 14, 41, 42). Such systems are referred to as closed loop, whereas systems that lack feedback from the output are referred to as open loop. It is well known in both engineering and physiology that open-loop circuits tend to be susceptible to variations in the environment and have difficulty maintaining homeostatic control (14, 42). For the tetracycline resistance system we show explicitly that the circuit is closed loop. In addition, an open-loop version provides an example of the loss of homeostatic control described above. For the porin osmoregulatory system, on the other hand, we find the surprising result that the wild-type circuit is open loop.
MATERIALS AND METHODS
Cell growth.
Cells were grown aerobically at 37°C in minimal A medium (31) with 0.2% (vol/vol) glycerol, or in Luria-Bertani (LB) broth (31), as noted. Additional supplements, sucrose, tetracycline, ampicillin, and isopropyl-β-d-thiogalactoside (IPTG), were added when appropriate as noted below.
Plasmid and strain construction.
The strains and plasmids used in this study are listed in Table 1. MDG147 is a fluorescent reporter strain derived from MG1655 (4) that contains chromosomal operon fusions of cfp with ompC and yfp with ompF and was constructed in a manner similar to that for MDG131 (6). To create an in-frame deletion in ompC, the plasmid pMG35 (6) was digested with PshAI and religated. This removes 537 bp from the middle of ompC. The resulting plasmid was then used to construct EPB16 [MC4100 Φ(ΔompC-cfp+)Φ(ompF+-yfp+)] by homologous recombination as described previously (6). To construct an in-frame deletion in ompF, the primers 5′-TGAGGGTAATAAATAATGATGAAGCGCAATATTCTGGTGTAGGCTGGAGCTGCTTC-3′and 5′-CTGGTAAACGATACCCACAGCAACGGTGTCGTCTGACATATGAATATCCTCCTTAG-3′ were used to amplify the chloramphenicol resistance cassette from pKD3 (12). This PCR product was introduced into MDG147 by following the protocols described in reference 12 and then introduced into a clean MDG147 background by P1 transduction. The chloramphenicol resistance cassette, which is flanked by FRT sites, was removed by using the FLP-recombinase-expressing plasmid pCP20 (followed by plasmid curing by growth at 42°C) as described previously (12). This resulted in EPB24 [MG1655 Φ(ompC+-cfp+)Φ(ΔompF-yfp+)].
TABLE 1.
Strains and plasmids used in this study
| Strain or plasmid | Relevant genotype | Source or reference |
|---|---|---|
| Strains | ||
| MC4100 | F−araD139 Δ(argF-lac)169 λ−flhD5301 fruA25 relA1 rpsL150(Strr) rbsR22 deoC1 | 10 |
| MG1655 | λ−rph-1 | E. coli Genetic Stock Center, CGSC no. 7740 |
| MDG131 | MC4100 Φ(ompF+-yfp+) Φ(ompC+-cfp+) | 6 |
| MDG147 | MG1655 Φ(ompF+-yfp+) Φ(ompC+-cfp+) | This study |
| EPB16 | MC4100 Φ(ompF+-yfp+) Φ(ΔompC-cfp+) | This study |
| EPB2 | MC4100 Φ(ompA+-yfp+) | This study |
| EPB24 | MG1655 Φ(ΔompF-yfp+) Φ(ompC+-cfp+) | This study |
| MDG149 | MC4100 Φ(ompA+-yfp+) attλ::pMG53 [tetR+ Φ(tetA+-cfp+)] | This study |
| MDG150 | MC4100 Φ(ompA+-yfp+) attλ::pMG56 [tetR+ Φ(ΔtetA-cfp+)] | This study |
| XL1-Blue | [F′ Tn10] | Stratagene |
| Plasmids | ||
| pTrc99a | lacIqPtrc-MCS bla | 3 |
| pEB19 | pTrc99a Ptrc-ompC | This study |
| pCAH63 | oriRλ cat attPλPsynl-uidAf | 22 |
| pTM5 | pCAH63 Δ(Psynl-uidAf) cfp | Goulian lab stock |
| pMG53 | pTM5 tetR+ Φ(tetA+-cfp+) | This study |
| pMG56 | pTM5 tetR+ Φ(ΔtetA-cfp+) | This study |
To construct the OmpC expression plasmid pEB19, the ompC gene was isolated by PCR from MC4100 genomic DNA using the primers 5′-AAAGTTAAAGTACTGTCCCTCC-3′ and 5′-CGGGATCCATCGAGATTAGAACTGGTAA-3′. The underlined bases introduce a BamHI site. This fragment was cloned into pTrc99a (3), which had been digested with NcoI, polished with T4 DNA polymerase, and then digested with BamHI.
A fluorescent reporter of ompA transcription was constructed by assembling a cassette containing the last ∼1 kb of ompA, followed by a promoterless yfp with a ribosome binding site, followed by ∼1 kb of the DNA downstream from ompA. The site of insertion of yfp is between the ompA stop codon and the ompA transcription terminator. This cassette was cloned into pCVD442 (15), and the resulting plasmid was introduced into MC4100 by electroporation. Cells were selected for ampicillin resistance followed by sucrose resistance as described in reference 6. The resulting strain, EPB2 [MC4100 Φ(ompA+-yfp+)], contains a chromosomal operon fusion of yfp to ompA at the wild-type ompA locus in the genome.
To construct an operon fusion between tetA and cfp, a segment containing tetR and tetA was isolated by PCR from XL1-Blue [F′ Tn10] genomic DNA (Stratagene, La Jolla, Calif.) with primers 5′-CGTTGGATCCGCATTATTTTCGC-3′ and 5′-GAGGGTACCTATATTTCGCGGAATAAC-3′. The underlined bases introduce BamHI and KpnI sites, respectively. This fragment was cloned into pTM5 (Goulian lab stock), which is derived from the vector pCAH63 (22) and contains a promoterless cfp (and ribosome binding site) in place of the synthetic promoter and uidAf gene in pCAH63. The resulting plasmid, pMG53, was integrated into the chromosome of EPB2 at the phage lambda attachment site attλ and verified to be in single copy by following the protocols in reference 22. This resulted in MDG149, which has the genotype MC4100 Φ(ompA+-yfp+) attλ::pMG53 [tetR+ Φ(tetA+-cfp+)]. To construct a strain with an in-frame deletion in tetA, pMG53 was digested with AgeI and XmnI. The large fragment was polished with T4 DNA polymerase and ligated to give pMG56. This removes 645 bp from the middle of tetA. The plasmid pMG56 was then integrated into EPB2 in the same manner as was done for pMG53, resulting in MDG150, which has the genotype MC4100 Φ(ompA+-yfp+) attλ::pMG56 [tetR+ Φ(ΔtetA-cfp+)].
Analysis of single cells.
For analysis of porin regulation, colonies were inoculated into 2 ml of minimal A medium supplemented with glycerol and various sucrose concentrations and grown to an optical density at 600 nm (OD600) of ∼0.1. The cultures were then diluted 1:500 into fresh prewarmed medium. When the cultures reached an OD600 of ∼0.2, 100 μg of chloramphenicol/ml was added, and the cultures were rapidly cooled in an ice-water slurry.
Similarly, for single-cell analysis of MDG149 and MDG150, single colonies were inoculated into 2 ml of minimal A glucose medium with 0.1% (wt/vol) Vitamin Assay Casamino Acids (Difco) or LB that was supplemented with various concentrations of tetracycline. Cultures were grown overnight to saturation and then diluted 1:1,000 into fresh prewarmed medium and grown to an OD600 of ∼0.5. After 30 μg of streptomycin/ml was added, the cultures were chilled on ice. To eliminate autofluorescence from the growth medium, the cultures were centrifuged at 5,000 × g for 2 min, and the pellets were resuspended in phosphate-buffered saline (PBS) containing 30 μg of streptomycin/ml. For analysis of growth on solid media, cells were streaked on LB or minimal A glycerol plates containing 1.5% agar and 50 ng or 12 μg of tetracycline/ml and grown overnight. The next day, single colonies were picked from the plates, resuspended in 100 μl of PBS containing 3 μg of streptomycin, and cooled on ice.
Fluorescence measurements.
For microscopy, cells were immobilized on glass number 1.5 coverslips with agarose pads. Approximately 50 μl of molten 1% agarose in PBS was deposited on a microscope slide, and a coverslip was immediately applied. When the agarose had hardened, the coverslip was carefully removed, leaving a thin pad of agarose on the slide. Ten microliters of a bacterial culture was then deposited on a fresh coverslip, and the microscope slide and pad were placed on top with the agarose facing down so that the culture was spread between the pad and coverslip. Fluorescence microscopy was performed on a Zeiss Standard microscope with a 2FL fluorescence adaptor, a 100 W mercury lamp, and a Nikon 60X PlanApo NA 1.4 objective lens. Fluorescence filter sets were D436/20 excitation, 455dclp beam splitter, and D480/40 emission for cyan fluorescent protein (CFP) and HQ500/20 excitation, Q515lp beam splitter, and HQ535/30 emission for yellow fluorescent protein (YFP) (Chroma, Brattleboro, Vt.). Images were acquired with a Hamamatsu (Bridgewater, N.J.) C4742-95 cooled charged coupled device camera and analyzed using our own software, which was written in the G programming language (National Instruments, Austin, Tex.) using IMAQ Vision libraries (National Instruments).
To identify bacteria and quantify cellular fluorescence we used an image erosion method. Briefly, the images taken with the CFP and YFP filters were added together. The summed image was then converted to a binary image by setting all pixel values above a threshold level to 1 and setting the remaining pixels to zero. This was repeated for the full range of possible thresholds, and the number of particles (connected regions with nonzero pixels) was determined as a function of the threshold value. A plateau, i.e., a range of thresholds for which the number of particles does not change, indicates a level of erosion such that the identified particles in the image correspond to cells. The binary image constructed from the lowest threshold value in this interval was then used as a mask to identify cells. Using this mask, integrated CFP and YFP intensities for each cell were extracted from the original (unthresholded) images. The inverse mask was used to determine the average background levels for the CFP and YFP images, which were used for background subtraction.
Analysis of porin deletion strains.
Two milliliters of minimal A glycerol medium supplemented with 0, 5, or 15% sucrose was inoculated and grown aerobically at 37°C overnight. For fluorescence measurements, the saturated cultures were then diluted 1:200 into 2 ml of fresh prewarmed medium. When the cultures reached an OD600 of ∼0.2, chloramphenicol was added to 100 μg/ml, and the cultures were rapidly cooled as described above. CFP and YFP fluorescence levels were measured with a fluorometer as described in reference 6. To measure OmpC and OmpF protein levels, the saturated cultures were diluted 1:200 into 7 ml of fresh prewarmed medium and grown to an OD of ∼0.2. Pelleted cultures were separated by urea-sodium dodecyl sulfate-polyacrylamide gel electrophoresis as described in reference 6 and analyzed by Western blotting using antibodies that cross-react with OmpC, OmpF, and OmpA. Blots were visualized with alkaline phosphatase-conjugated secondary antibodies as described in reference 6 and digitized on a flatbed scanner with a number 12 Wratten filter (Kodak, Rochester, N.Y.) placed between the blot and the scanner.
Analysis of OmpC overexpression.
Two milliliters of minimal A glycerol medium supplemented with 5% sucrose, 50 μg of ampicillin/ml, and the appropriate level of IPTG was inoculated with MDG131/pEB19 or MDG131/pTrc99a and grown aerobically at 37°C to saturation. Cultures were then diluted 1:200 into 7 ml of fresh prewarmed medium and grown to an OD of ∼0.2. At this time, 100 μg of chloramphenicol/ml was added, and the cultures were rapidly cooled as described above. Two milliliters of the cultures was used to measure CFP and YFP fluorescence with a fluorometer as described in reference 6. The remainders of the cultures were used to quantify OmpC protein levels from cell envelopes by urea-sodium dodecyl sulfate-polyacrylamide gel electrophoresis followed by Coomassie staining as described in reference 6.
RESULTS
Continuous transcriptional control in the porin osmoregulation and tetracycline resistance circuits.
To measure the transcriptional output of the porin osmoregulation and tetracycline resistance systems in single cells, we have constructed two-color fluorescence reporter strains in which the genes for cyan fluorescent protein (cfp) and yellow fluorescent protein (yfp) were integrated into the chromosome as operon fusions (Fig. 1c and d). For the porin osmoregulatory system, cfp and yfp were integrated downstream of ompC and ompF, respectively, at the wild-type loci (6). Similarly, for the tetracycline resistance system cfp was integrated into the chromosome downstream of tetA (in a strain containing a chromosomal copy of tetR and tetA), and yfp was integrated downstream of ompA, which codes for an outer membrane structural protein. The YFP fluorescence from ompA transcription provides a convenient normalization when quantifying CFP from the tetA-cfp fusion.
These operon fusions allow rapid and precise measurements of transcriptional activity in whole cultures by use of a fluorometer or in single cells by use of a fluorescence microscope. Since CFP and YFP are expressed in the same cell, the ratio of the corresponding fluorescence signals provides a sensitive measure of the relative transcription of ompC to ompF or of tetA to ompA. This ratio is insensitive to factors affecting the total protein content within the cell and also to various sources of variability in fluorescence measurements. The precision of the fluorescence measurements is evident from the small error bars (many of which are smaller than the data symbols) in the figures.
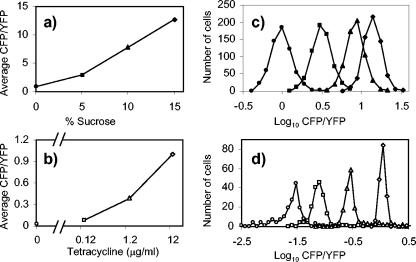
Using the above strains we measured the CFP and YFP fluorescence levels of individual cells for cultures at several different levels of stimulus (osmolarity or tetracycline concentration). Transcription of ompA was unaffected by levels of tetracycline that did not inhibit cell growth (data not shown). As expected, in both cases the population-averaged response was a graded function of the applied stimulus (Fig. 2a and b). However, we also found that this behavior was reflected in the responses of individual cells. For each growth condition, the distribution of CFP/YFP fluorescence for the population of cells has an approximate Gaussian profile, indicating that, within some deviation about the mean, all of the cells in the culture exhibit essentially the same response (Fig. 2c and d). The standard deviation of the distribution presumably reflects a combination of fluctuations in gene expression (8, 17, 36) and errors in the measurements of cellular fluorescence. We conclude that for both tetracycline resistance and porin osmoregulation, cells make use of continuous control; as the stimulus is varied, cells do not exhibit a discontinuous switch-like change in transcription but instead show a continuous or graded response.
FIG. 2.
Histograms of cellular CFP/YFP fluorescence of cultures with varying osmolarity (a, c) or varying tetracycline concentration (b, d). The values in panels a and b are the averages of the corresponding values from the histograms in panels c and d. E. coli cells were MDG131 (a, c) and MDG149 (b, d). Cultures were grown in minimal glycerol medium supplemented with 0 (•), 5 (▪), 10, (▴), or 15% (⧫) sucrose (a, c) or 0 (○), 0.12 (□), 1.2 (▵), or 12.0 μg (◊) of tetracycline/ml (b, d). For 0 μg of tetracycline/ml, the CFP fluorescence was so low that the signal was dominated by cellular autofluorescence (data not shown). The scale for fluorescence measurements in the CFP and YFP channels is arbitrary.
Closed-loop control in the tetracycline resistance circuit.
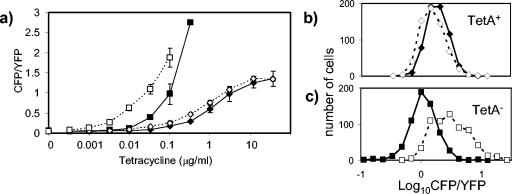
As discussed above, regulatory circuits based on continuous control generally make use of negative feedback to attain the appropriate output level and maintain homeostasis. It is thus natural to ask whether the above systems are closed or open loop—that is, whether or not there is negative feedback from the outputs back into the regulatory circuits. To test this for the tetracycline resistance circuit, we constructed a strain in which tetA was disrupted with an in-frame deletion that is not polar on the downstream cfp gene. We then compared CFP/YFP fluorescence as a function of tetracycline concentration in the growth medium for TetA− and TetA+ (wild-type) cells. Tetracycline concentrations up to 100 ng/ml did not affect the growth rates of TetA− cells (data not shown). We found that under inducing conditions, the CFP fluorescence was greater in TetA− cells than in TetA+ cells (Fig. 3a). These results indicate that there is negative feedback from TetA when tetracycline is present, i.e., the circuit is closed loop, as would be expected from the model shown in Fig. 1a. An increase in production of the efflux pump TetA will result in a drop in intracellular levels of tetracycline, which in turn leads to a drop in induction of the tetA promoter and hence a drop in production of TetA. In the TetA− cells the feedback loop has been disrupted (i.e., the circuit is open loop), resulting in increased intracellular levels of tetracycline and hence increased induction of the tetA promoter.
FIG. 3.
(a) Transcription from the tetA promoter in MDG150 (TetA−) and MDG149 (TetA+) cells in response to tetracycline in rich and minimal media; MDG150 in LB (▪), MDG150 in minimal A glucose medium plus Casamino Acids (□), MDG149 in LB (⧫), and MDG149 in minimal A glucose medium plus Casamino Acids (◊). The growth rates were identical for all samples grown in minimal medium. The growth rates for cells grown in LB were identical except for MDG150 in the presence of 333 μg of tetracycline/ml (the highest concentration of tetracycline shown for MDG150), which exhibited a decreased growth rate. Each measurement was the average fluorescence ratio of at least 100 cells. The CFP/YFP values shown are the averages of results from at least three independent experiments, and the error bars are the corresponding standard deviations (the error bars are smaller than the data symbols in some cases). (b) The distributions of cellular CFP/YFP fluorescence are similar for MDG149 (TetA+) colonies growing on minimal glycerol agar with 12 μg of tetracycline/ml (◊) and on LB agar with 12 μg of tetracycline/ml (⧫). (c) The distribution of cellular CFP/YFP fluorescence for MDG150 (TetA−) colonies growing on minimal glycerol agar with 50 ng of tetracycline/ml (□) is broader than for growth on LB agar with 50 ng of tetracycline/ml (▪). Higher levels of tetracycline were used for MDG149 in order to obtain average levels of fluorescence that were comparable to the levels seen for MDG150.
We can also see the effect of loss of negative feedback on homeostatic control. For a given concentration of tetracycline, tetA transcription in the TetA− strain was higher in minimal A glucose medium than in LB medium (Fig. 3a). In contrast, tetA transcription in the TetA+ strain was comparable in the two media (Fig. 3a). We also examined the distribution of cellular fluorescence for colonies growing on solid media, i.e., minimal A glycerol agar plates or LB agar plates. While the strain with the closed-loop (TetA+) circuit showed similar distributions for growth on the two types of agar (Fig. 3b), the strain with the open-loop (TetA−) circuit showed greater cell-to-cell variability in tetA transcription for growth on minimal A glycerol agar than for growth on LB agar (Fig. 3c). Presumably this variability arises because the open-loop circuit is sensitive to conditions that vary within the microenvironment of bacterial colonies on minimal agar plates. Regardless of the underlying physiological mechanisms that cause the variability in tetA transcription described above for the open-loop (TetA−) circuit, it is clear that the closed-loop (TetA+) circuit is able to maintain homeostatic control in these differing environments.
Open-loop control in the porin osmoregulatory circuit.
From the results of previous studies, there was reason to suspect that porin osmoregulation was open loop. Promoter swap (30) and OmpF overexpression (39) experiments were consistent, at least qualitatively, with the absence of feedback. In addition, transcriptional reporter strains in which either ompC or ompF was disrupted with an insertion of lacZ showed osmoregulation of beta-galactosidase activity (44). However, the conclusions from other studies of ompF deletions were less clear (35, 39). We therefore wanted to check more quantitatively whether there was negative feedback from the output of this circuit.
The porin osmoregulatory circuit controls the ratio of OmpC expression to OmpF expression. To determine whether there is feedback, we looked at the effect of perturbing this ratio by perturbing the OmpC and OmpF expression levels. The porins are under complex control, and such perturbations may well affect many aspects of the network controlling porin expression. However, we are specifically looking at the question of feedback into the circuit controlling porin osmoregulation. For this reason, we are only interested in responses to the above perturbations that affect the relative expression of the two porins. Disruption of ompC will lower the ratio of OmpC expression to OmpF expression (i.e., the ratio will be set to zero). If there is negative feedback, the osmoregulatory circuit will respond by attempting to increase the ratio, either by increasing ompC expression, decreasing ompF expression, or both. This response should be stronger under conditions where the wild-type system (with a functional copy of ompC) has a higher ratio of OmpC expression to OmpF expression, i.e., at high osmolarity. Disruption of OmpF, on the other hand, will cause the circuit to try to lower the ratio, i.e., by trying to decrease ompC expression, increase ompF expression, or both, and this response should be stronger at low osmolarity, where OmpF is the more abundant porin in the wild-type system. Similarly, overexpression of OmpC (from a separately controllable promoter) should affect the osmoregulatory circuit in a manner that is qualitatively similar to disruption of ompF, since both perturbations increase the ratio of OmpC expression to OmpF expression.
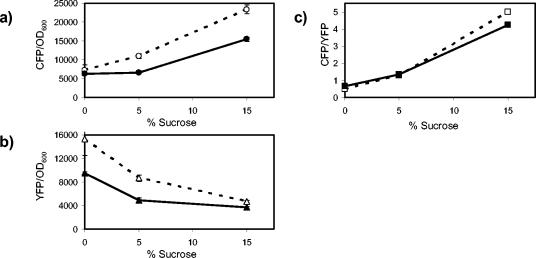
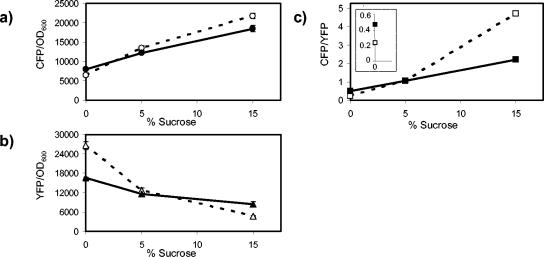
To disrupt porin expression, we constructed two-color fluorescent reporter strains with in-frame deletions in either ompC or ompF that are not polar on cfp or yfp, respectively. The OmpC− strain, compared with the OmpC+ strain, showed a drop in both ompC and ompF transcription (Fig. 4a and b). However, the ratio of transcription of ompC to transcription of ompF showed no difference between the two strains for growth at low and intermediate osmolarities and showed only a small decrease at high osmolarity (Fig. 4c). Thus, loss of OmpC expression does not feed back into the porin osmoregulatory circuit.
FIG. 4.
Effect of in-frame deletions in ompC on ompC and ompF transcription. Open symbols, MDG131 (OmpC+); filled symbols, EPB16 (OmpC−). (a) CFP fluorescence (ompC transcription) normalized by culture OD600. (b) YFP fluorescence (ompF transcription) normalized by OD600. (c) The ratio of CFP fluorescence to YFP fluorescence. All points represent the averages of results from at least three independent experiments. The error bars, which denote the corresponding standard deviations, are smaller than the data symbols in most cases.
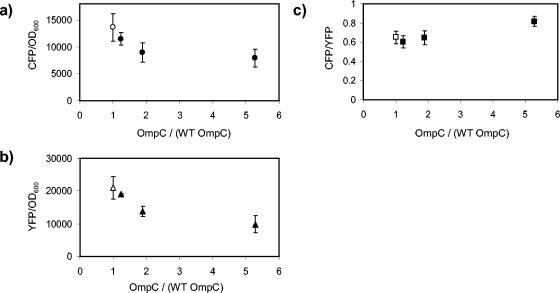
For the OmpF− strain, ompC transcription decreased slightly at high osmolarity compared with that for the OmpF+ strain (Fig. 5a), while ompF transcription decreased at low osmolarity and increased at high osmolarity (Fig. 5b). The ratio of ompC transcription to ompF transcription at high osmolarity was approximately twofold lower for the OmpF− strain than for the OmpF+ strain (Fig. 5c). However, at low osmolarity the ompC-to-ompF transcription ratio was twofold higher for the OmpF− strain (Fig. 5c, see inset for an expanded scale on the vertical axis), which is the opposite response from what would be expected with negative feedback. As discussed above, if there were negative feedback, then the strongest response to an ompF deletion should be at low osmolarity. Thus, while the absence of OmpF has a small effect on transcription of the porin promoters, it is not due to feedback through the porin osmoregulatory circuit.
FIG. 5.
Effect of in-frame deletions in ompF on ompC and ompF transcription. Open symbols, MDG147 (OmpF+); filled symbols, EPB24 (OmpF−). (a) CFP fluorescence (ompC transcription) normalized by culture OD600. (b) YFP fluorescence (ompF transcription) normalized by culture optical density. (c) The ratio of CFP fluorescence to YFP fluorescence. The inset in panel c displays the 0% sucrose points for MDG147 and EPB24 with an expanded scale on the y axis; see the text for a discussion. All points represent the averages of results from at least three independent experiments. The error bars, which denote the corresponding standard deviations, are smaller than the data symbols in most cases.
We also increased the expression of OmpC above the wild-type level for cultures grown in intermediate osmolarity (5% sucrose [Fig. 6]). Increasing OmpC expression levels led to a decrease in transcription of both ompC (Fig. 6a) and ompF (Fig. 6b). However, there was no significant variation in the ratio of transcription of the ompC promoter to transcription of the ompF promoter except for a small increase at the highest levels of OmpC (Fig. 6c). We observed similar results for cultures grown at low and high osmolarities (0 and 15% sucrose [data not shown]). Therefore, OmpC overexpression does not feed back into the porin osmoregulatory circuit.
FIG. 6.
Effect of OmpC overexpression on ompC and ompF transcription. Fluorescence measurements of CFP and YFP normalized by culture OD600 and the corresponding fluorescence ratios for MDG131/pEB19 with various levels of IPTG induction (filled symbols). The OmpC protein level was normalized by the wild-type (WT) value, which was taken to be the OmpC level of MDG131/pTrc99a (open symbols). Cultures were grown in an intermediate-osmolarity medium (5% sucrose). Similar results were obtained for high- and low-osmolarity cultures (15 and 0% sucrose [data not shown]). The points represent the averages of results from at least three independent experiments, and the error bars denote the corresponding standard deviations.
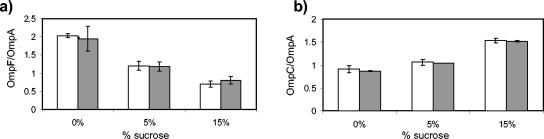
Measurements with transcriptional reporters cannot rule out the possibility that there is feedback at the posttranscriptional level, e.g., via control of translation or of mRNA or protein stability. We therefore also examined porin protein levels with Western blots using antibodies that cross-react with OmpF, OmpC, and OmpA. The expression level of OmpA is only weakly dependent on osmolarity and provides a convenient normalization for band quantification from multiple blots (2). We found that OmpF protein levels were unaffected by the presence or absence of OmpC (Fig. 7a), and similarly, OmpC levels were unaffected by the presence or absence of OmpF (Fig. 7b). Quantification of individual blots without normalizing by OmpA similarly showed no effect on absolute levels of OmpF or OmpC (data not shown). To check that the above results are not specific to laboratory (K-12-derived) strains of E. coli, we also examined the K-1 strain RS218 (1). We again found that deletion of ompF did not affect OmpC protein levels and deletion of ompC did not affect OmpF protein levels (data not shown). We thus conclude that the absence of OmpC or OmpF does not feed back into the porin osmoregulatory circuit at the posttranscriptional level.
FIG. 7.
Effects of an ompC deletion on OmpF protein levels and an ompF deletion on OmpC protein levels. (a) White bars, MDG131 (OmpC+); grey bars, EPB16 (OmpC−). (b) White bars, MDG147 (OmpF+); grey bars, EPB24 (OmpF−). Cultures were grown in minimal glycerol medium supplemented with the indicated concentrations of sucrose. Western blots were performed with antibodies that cross-react with both porins and with the structural protein OmpA. The data in panels a and b represent the averages of results from three and two independent measurements, respectively, and the error bars denote the corresponding standard deviations.
Taken together, the above results imply that the expression of OmpC relative to that of OmpF does not feed back into the porin osmoregulatory circuit, and they lead to the conclusion that porin osmoregulation is under open-loop, continuous control. Unfortunately, without a closed-loop version of the circuit, we cannot explore to what extent the open-loop nature of the circuit results in environmental sensitivity for this system. However, it is worth noting that the expression of OmpC relative to that of OmpF is affected by many environmental factors in addition to osmolarity (20, 29, 38). An example can be seen in the difference in ompC and ompF transcription in strains containing ampicillin-resistant plasmids (and grown in medium containing ampicillin) compared to that in strains without plasmids (which were grown without ampicillin): the CFP/YFP ratio for MDG131 at 5% sucrose was 1.2 (Fig. 4c), whereas the corresponding ratio for MDG131/pTrc99a was 0.65 (Fig. 6c).
DISCUSSION
To our knowledge, our results for the tetracycline resistance and porin osmoregulation circuits are the first demonstration of continuous transcriptional control of wild-type regulatory circuits in bacteria. The fact that cells contain circuits with this form of control suggests that there is a need to adjust or tune the outputs to levels that depend on the strengths of the applied stimuli. In physiology it is generally expected that in such cases the circuits will be closed loops, i.e., there will be negative feedback from the outputs in order to ensure homeostatic control (42).
For the case of tetracycline resistance, we demonstrated explicitly that the circuit is closed loop. One can rationalize the organization of this circuit by the fact that expression of the efflux pump TetA is quite costly for cells (23). It would make sense to express the minimal amount of TetA that is necessary to lower intracellular tetracycline to tolerable levels. The tetracycline resistance circuit possesses two negative feedback loops: one mediated by TetR alone and the second mediated by TetA, tetracycline, and TetR (Fig. 1a). It has been previously demonstrated in a synthetic circuit that negative autoregulation decreases the variability in protein expression among cells (8). Since TetR represses both the tetR and tetA promoters, TetR auto-regulation presumably will decrease cell-to-cell variation not only of tetR transcription but also of tetA transcription. However, if we view the expression level of TetA as the circuit output, then TetR autorepression is an internal feedback, which does not by itself render the circuit closed loop. The TetA-mediated feedback loop, on the other hand, does make the circuit closed loop. As expected, disruption of tetA, which results in an open-loop version of the circuit, showed an increased susceptibility to variations in the environment, even in the presence of the internal feedback (Fig. 3).
For the case of porin osmoregulation we have shown that the circuit is open loop. Thus, there does not appear to be any mechanism for monitoring the relative expression of OmpC and OmpF and for ensuring that the appropriate ratio is achieved. It is possible that there is feedback into the porin osmoregulatory network from cellular components other than OmpC and OmpF, e.g., from other proteins that are controlled by EnvZ and OmpR or from regulatory RNAs such as MicF (13). However, from the point of view of porin osmoregulation this would be an internal feedback loop, and the control system would still be open loop.
We used defined media and mid-log growth for our experiments so that we could more readily distinguish feedback into the porin osmoregulatory circuit from feedback into some other part of the porin regulatory network. These growth conditions show a smaller modulation of porin expression (e.g., Fig. 7 and reference 2) compared with, for example, the rich media and late-log growth that have been used in some studies. However, as is evident from the error bars in the figures, our measurements were clearly sensitive enough to detect feedback into the porin osmoregulatory system, had it been present.
The fact that porin osmoregulation is open loop suggests that cells will have difficulty regulating the relative levels of OmpC and OmpF in diverse environments. It is well known that the OmpC-to-OmpF ratio is affected by many different environmental conditions in addition to osmolarity, including pH, temperature, toxins, culture medium, and growth phase (20, 29, 38; also see the comment above regarding ampicillin). It may be that the porin regulatory network evolved to specifically respond to all of these types of stimulus. However, it is also possible that some of these responses provide no significant survival advantage or disadvantage for the bacterium and simply reflect the limited range of homeostatic control for the regulatory system.
Open-loop control of porin osmoregulation seems surprising and somewhat puzzling. How are E. coli cells able to set the appropriate ratio of OmpC to OmpF expression if the control circuit has no way of monitoring the ratio? It is possible that the sloppy control provided by the open-loop circuit is not sufficiently severe to be a problem for cell survival, or perhaps it even provides a survival advantage. However, this seems unlikely for an organism such as E. coli, which proliferates in diverse environments. Instead, it is more likely that there is a defect in our understanding of the system. In the absence of any known function of the output (the ratio of OmpC expression to OmpF expression) other than to modulate outer membrane permeability, the defect is most likely in our understanding of the circuit input.
Although there has been a substantial amount of work on the regulation of porin expression by the EnvZ/OmpR two-component system, the role of osmolarity remains confusing. In fact, osmolarity has never seemed to be a particularly good signal for controlling the EnvZ/OmpR circuit. As discussed above, the changes in porin expression as a function of osmolarity in some growth conditions are relatively small (e.g., two- to threefold changes). Furthermore, strains with a deletion of envZ or with a nonphosphorylatable allele of ompR still show some level of osmoregulation (19, 24, 47). It is thus possible that osmolarity is not the biologically relevant input for the circuit and, in the presence of the relevant signal, the system exhibits discrete (switch-like) control or continuous control with negative feedback from the OmpC/OmpF expression level. This could occur, for example, if the true stimulus for EnvZ were a small molecule that has different permeability through OmpC and OmpF. However, a proof will require, at the very least, determining the chemical or physical stimulus to which EnvZ responds.
Acknowledgments
We thank A. N. Binns, W. Hillen, E. Sontag, and M. van der Woude for helpful discussions.
This work was supported by NSF grant MCB0212925 (to M.G.), NIGMS grant GM65216 (to T.J.S.), and an NSF graduate fellowship (to E.B.).
REFERENCES
- 1.Achtman, M., A. Mercer, B. Kusecek, A. Pohl, M. Heuzenroeder, W. Aaronson, A. Sutton, and R. P. Silver. 1983. Six widespread bacterial clones among Escherichia coli K1 isolates. Infect. Immun. 39:315-335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Alphen, W. V., and B. Lugtenberg. 1977. Influence of osmolarity of the growth medium on the outer membrane protein pattern of Escherichia coli. J. Bacteriol. 131:623-630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Amann, E., B. Ochs, and K. J. Abel. 1988. Tightly regulated tac promoter vectors useful for the expression of unfused and fused proteins in Escherichia coli. Gene 69:301-315. [DOI] [PubMed] [Google Scholar]
- 4.Bachman, B. J. 1996. Derivations and genotypes of some mutant derivatives of Escherichia coli K-12, p. 2460-2488. In F. C. Neidhardt, R. Curtis, J. L. Ingraham, E. C. C. Lin, K. B. Low, B. Magasanik, W. S. Reznikoff, M. Riley, M. Schaecter, and H. E. Umbarger (ed.), Escherichia coli and Salmonella: cellular and molecular biology, 2nd ed., vol. 2. ASM Press, Washington, D.C. [Google Scholar]
- 5.Bagowski, C. P., and J. E. Ferrell, Jr. 2001. Bistability in the JNK cascade. Curr. Biol. 11:1176-1182. [DOI] [PubMed] [Google Scholar]
- 6.Batchelor, E., and M. Goulian. 2003. Robustness and the cycle of phosphorylation and dephosphorylation in a two-component regulatory system. Proc. Natl. Acad. Sci. USA 100:691-696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Becskei, A., B. Seraphin, and L. Serrano. 2001. Positive feedback in eukaryotic gene networks: cell differentiation by graded to binary response conversion. EMBO J. 20:2528-2535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Becskei, A., and L. Serrano. 2000. Engineering stability in gene networks by autoregulation. Nature 405:590-593. [DOI] [PubMed] [Google Scholar]
- 9.Biggar, S. R., and G. R. Crabtree. 2001. Cell signaling can direct either binary or graded transcriptional responses. EMBO J. 20:3167-3176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Casadaban, M. J. 1976. Transposition and fusion of the lac genes to selected promoters in Escherichia coli using bacteriophage lambda and Mu. J. Mol. Biol. 104:541-555. [DOI] [PubMed] [Google Scholar]
- 11.Csete, M. E., and J. C. Doyle. 2002. Reverse engineering of biological complexity. Science 295:1664-1669. [DOI] [PubMed] [Google Scholar]
- 12.Datsenko, K. A., and B. L. Wanner. 2000. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc. Natl. Acad. Sci. USA 97:6640-6645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Delihas, N., and S. Forst. 2001. MicF: an antisense RNA gene involved in response of Escherichia coli to global stress factors. J. Mol. Biol. 313:1-12. [DOI] [PubMed] [Google Scholar]
- 14.Doebelin, E. O. 1985. Control system principles and design. John Wiley and Sons, New York, N.Y.
- 15.Donnenberg, M. S., and J. B. Kaper. 1991. Construction of an eae deletion mutant of enteropathogenic Escherichia coli by using a positive-selection suicide vector. Infect. Immun. 59:4310-4317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Egger, L. A., H. Park, and M. Inouye. 1997. Signal transduction via the histidyl-aspartyl phosphorelay. Genes Cells 2:167-184. [DOI] [PubMed] [Google Scholar]
- 17.Elowitz, M. B., A. J. Levine, E. D. Siggia, and P. S. Swain. 2002. Stochastic gene expression in a single cell. Science 297:1183-1186. [DOI] [PubMed] [Google Scholar]
- 18.Ferrell, J. E., Jr., and E. M. Machleder. 1998. The biochemical basis of an all-or-none cell fate switch in Xenopus oocytes. Science 280:895-898. [DOI] [PubMed] [Google Scholar]
- 19.Forst, S., J. Delgado, G. Ramakrishnan, and M. Inouye. 1988. Regulation of ompC and ompF expression in Escherichia coli in the absence of envZ. J. Bacteriol. 170:5080-5085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Forst, S., and M. Inouye. 1988. Environmentally regulated gene expression for membrane proteins in Escherichia coli. Annu. Rev. Cell Biol. 4:21-42. [DOI] [PubMed] [Google Scholar]
- 21.Gardner, T. S., C. R. Cantor, and J. J. Collins. 2000. Construction of a genetic toggle switch in Escherichia coli. Nature 403:339-342. [DOI] [PubMed] [Google Scholar]
- 22.Haldimann, A., and B. L. Wanner. 2001. Conditional-replication, integration, excision, and retrieval plasmid-host systems for gene structure-function studies of bacteria. J. Bacteriol. 183:6384-6393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hillen, W., and C. Berens. 1994. Mechanisms underlying expression of Tn10 encoded tetracycline resistance. Annu. Rev. Microbiol. 48:345-369. [DOI] [PubMed] [Google Scholar]
- 24.Kanamaru, K., and T. Mizuno. 1992. Signal transduction and osmoregulation in Escherichia coli: a novel mutant of the positive regulator, OmpR, that functions in a phosphorylation-independent manner. J. Biochem. (Tokyo) 111:425-430. [DOI] [PubMed] [Google Scholar]
- 25.Karttunen, J., and N. Shastri. 1991. Measurement of ligand-induced activation in single viable T cells using the lacZ reporter gene. Proc. Natl. Acad. Sci. USA 88:3972-3976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kenney, L. J. 2002. Structure/function relationships in OmpR and other winged-helix transcription factors. Curr. Opin. Microbiol. 5:135-141. [DOI] [PubMed] [Google Scholar]
- 27.Khlebnikov, A., O. Risa, T. Skaug, T. A. Carrier, and J. D. Keasling. 2000. Regulatable arabinose-inducible gene expression system with consistent control in all cells of a culture. J. Bacteriol. 182:7029-7034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ko, M. S., H. Nakauchi, and N. Takahashi. 1990. The dose dependence of glucocorticoid-inducible gene expression results from changes in the number of transcriptionally active templates. EMBO J. 9:2835-2842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Liu, X., and T. Ferenci. 2001. An analysis of multifactorial influences on the transcriptional control of ompF and ompC porin expression under nutrient limitation. Microbiology 147:2981-2989. [DOI] [PubMed] [Google Scholar]
- 30.Matsuyama, S.-I., K. Inokuchi, and S. Mizushima. 1984. Promoter exchange between ompF and ompC, genes for osmoregulated major outer membrane proteins of Escherichia coli K-12. J. Bacteriol. 158:1041-1047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Miller, J. H. 1992. A short course in bacterial genetics: a laboratory manual and handbook for Escherichia coli and related bacteria. Cold Spring Harbor Laboratory Press, Plainview, N.Y.
- 32.Morohashi, M., A. E. Winn, M. T. Borisuk, H. Bolouri, J. Doyle, and H. Kitano. 2002. Robustness as a measure of plausibility in models of biochemical networks. J. Theor. Biol. 216:19-30. [DOI] [PubMed] [Google Scholar]
- 33.Nikaido, H., and M. Vaara. 1985. Molecular basis of bacterial outer membrane permeability. Microbiol. Rev. 49:1-32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Novick, A., and M. Weiner. 1957. Enzyme induction as an all-or-none phenomenon. Proc. Natl. Acad. Sci. USA 43:553-556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ozawa, Y., and S. Mizushima. 1983. Regulation of outer membrane porin protein synthesis in Escherichia coli K-12: ompF regulates the expression of ompC. J. Bacteriol. 154:669-675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ozbudak, E. M., M. Thattai, I. Kurtser, A. D. Grossman, and A. van Oudenaarden. 2002. Regulation of noise in the expression of a single gene. Nat. Genet. 31:69-73. [DOI] [PubMed] [Google Scholar]
- 37.Poritz, M. A., S. Malmstrom, M. K. Kim, P. J. Rossmeissl, and A. Kamb. 2001. Graded mode of transcriptional induction in yeast pheromone signalling revealed by single-cell analysis. Yeast 18:1331-1338. [DOI] [PubMed] [Google Scholar]
- 38.Pratt, L. A., W. Hsing, K. E. Gibson, and T. J. Silhavy. 1996. From acids to osmZ: multiple factors influence synthesis of the OmpF and OmpC porins in Escherichia coli. Mol. Microbiol. 20:911-917. [DOI] [PubMed] [Google Scholar]
- 39.Ramakrishnan, G., K. Ikenaka, and M. Inouye. 1985. Uncoupling of osmoregulation of the Escherichia coli K-12 ompF gene from ompB-dependent transcription. J. Bacteriol. 163:82-87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Rossi, F. M., A. M. Kringstein, A. Spicher, O. M. Guicherit, and H. M. Blau. 2000. Transcriptional control: rheostat converted to on/off switch. Mol. Cell 6:723-728. [DOI] [PubMed] [Google Scholar]
- 41.Savageau, M. A. 1976. Biochemical systems analysis. Addison-Wesley, Reading, United Kingdom.
- 42.Selkurt, E. E. 1984. Physiology, 5th ed. Little, Brown and Company, Boston, Mass.
- 43.Siegele, D. A., and J. C. Hu. 1997. Gene expression from plasmids containing the araBAD promoter at subsaturating inducer concentrations represents mixed populations. Proc. Natl. Acad. Sci. USA 94:8168-8172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Slauch, J. M., F. D. Russo, and T. J. Silhavy. 1991. Suppressor mutations in rpoA suggest that OmpR controls transcription by direct interaction with the α subunit of RNA polymerase. J. Bacteriol. 173:7501-7510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Slauch, J. M., and T. J. Silhavy. 1996. The porin regulon: a paradigm for the two-component regulatory systems, p. 383-417. In E. C. C. Lin and A. S. Lynch (ed.), Regulation of gene expression in Escherichia coli. Chapman & Hall, New York, N.Y.
- 46.Thattai, M., and A. van Oudenaarden. 2002. Attenuation of noise in ultrasensitive signaling cascades. Biophys. J. 82:2943-2950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Villarejo, M., and C. C. Case. 1984. envZ mediates transcriptional control by local anesthetics but is not required for osmoregulation in Escherichia coli. J. Bacteriol. 159:883-887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Walters, M. C., W. Magis, S. Fiering, J. Eidemiller, D. Scalzo, M. Groudine, and D. I. Martin. 1996. Transcriptional enhancers act in cis to suppress position-effect variegation. Genes Dev. 10:185-195. [DOI] [PubMed] [Google Scholar]