Abstract
Exposure to blue light of the facultative phototrophic proteobacterium Rhodobacter sphaeroides grown semiaerobically results in repression of the puc and puf operons involved in photosystem formation. To reveal the genome-wide effects of blue light on gene expression and the underlying photosensory mechanisms, transcriptome profiles of R. sphaeroides during blue-light irradiation (for 5 to 135 min) were analyzed. Expression of most photosystem genes was repressed upon irradiation. Downregulation of photosystem development may be used to prevent photooxidative damage occurring when the photosystem, oxygen, and high-intensity light are present simultaneously. The photoreceptor of the BLUF-domain family, AppA, which belongs to the AppA-PpsR antirepressor-repressor system, is essential for maintenance of repression upon prolonged irradiation (S. Braatsch et al., Mol. Microbiol. 45:827-836, 2002). Transcriptome data suggest that the onset of repression is also mediated by the AppA-PpsR system, albeit via an apparently different sensory mechanism. Expression of several genes, whose products may participate in photooxidative damage defense, including deoxypyrimidine photolyase, glutathione peroxidase, and quinol oxidoreductases, was increased. Among the genes upregulated were genes encoding two σ factors: σE and σ38. The consensus promoter sequences for these σ factors were predicted in the upstream sequences of numerous upregulated genes, suggesting that coordinated action of σE and σ38 is responsible for the upregulation. Based on the dynamics of upregulation, the anti-σE factor ChrR or its putative upstream partner is proposed to be the primary sensor. The identified transcriptome responses provided a framework for deciphering blue-light-dependent signal transduction pathways in R. sphaeroides.
Light is an important environmental factor for most organisms. Therefore, organisms must perceive and appropriately respond to light signals. Blue light reaches deepest zones in the water column compared to light of other parts of the spectrum. It is reasonable to envision that aquatic organisms would have receptors tuned to blue light. High-intensity blue light might indicate coexistence of harmful UV light. Therefore, having sensors of blue light might help to protect cells from UV irradiation.
Two types of plant blue-light receptors, i.e., cryptochromes and phototropins, have been known for several years (1, 13). Cryptochromes contain both folate and FAD as cofactors (38), whereas phototropins contain flavins bound by the so-called LOV domains (12). The first putative bacterial blue-light receptor, photoactive yellow protein (PYP) from the α-proteobacterium Ectothiorhodospira halophila was identified in 1985 (41). PYP uses hydroxicinnamoic acid as its chromophore (3, 25). PYP has been proposed to function in the photophobic response in E. halophila (54); however, this has yet to be verified. The Ppr protein from Rhodocista centenaria combines a PYP-like and a phytochrome domain (29) and undergoes a photocycle similar to that for PYP. In a Ppr mutant, expression of the gene for chalcone synthase is no longer blue light dependent, unlike the expression of this gene in the wild type (29). A blue-light-dependent photocycle was revealed for the LOV domain protein YvtA from Bacillus subtilis (36), which serves as an antisigma factor antagonist (2). It is not known what blue-light response is mediated by YvtA. In general, our knowledge of the biological relevance of blue-light responses and blue-light-dependent signal transduction remains poor.
We identified the Rhodobacter sphaeroides AppA protein as a blue-light receptor responsible for repression of selected photosynthesis genes in semiaerobically grown cells irradiated with blue light (8). R. sphaeroides is a metabolically versatile α-proteobacterium. It can perform respiration by using oxygen or, in the absence of oxygen, by using alternative electron acceptors. In the absence of oxygen and in the presence of light, it can use cyclic photosynthetic electron transfer to drive photophosphorylation (28, 40). The two light-harvesting complexes (LHI and LHII) of R. sphaeroides trap light energy and transfer excitation energy to the reaction center (RC), where the photosynthetic electron transport is initiated. The RC, LHI, and LHII complexes comprise the R. sphaeroides photosystem (PS). Most genes required for PS formation are clustered on chromosome I (11, 57). These include bch and crt genes and operons (encoding enzymes involved in bacteriochlorophyll a and carotenoid biosynthesis, respectively), the puf and puhA operons (encoding the structural and assembly proteins of the LHI and RC complexes). The puc1 and puc2 operons (encoding the structural and assembly proteins of the LHII complex) are located separately from the PS gene cluster (66).
Oxygen and light are critical environmental signals that regulate PS formation in R. sphaeroides. Although oxygen-dependent regulation of PS genes has been studied extensively in Rhodobacter species (for reviews, see references 4, 23, 24, 45, and 64), little was known about light-dependent regulation of PS genes until the function of the AppA protein as photoreceptor was unraveled (6, 8).
AppA was originally identified as a protein involved in the regulation of PS genes in response to oxygen (18). Subsequently, Gomelsky and Kaplan (20) provided genetic evidence that AppA functions as an antagonist of the PpsR repressor, which is a key regulator of PS gene expression (19, 22, 47, 48; O. V. Moskvin, L. Gomelsky, and M. Gomelsky, submitted for publication). The N-terminal domain of AppA was found to noncovalently bind FAD; however, it was not involved in oxygen control (21). This domain was later designated BLUF and shown to function as a novel type photoreceptor (8, 17). Thus, AppA is the first identified protein with dual sensing capabilities involving oxygen and light signals. Masuda and Bauer (39) used purified PpsR and AppA proteins to demonstrate that AppA can form an AppA-PpsR2 antirepressor-repressor complex. Blue light promotes dissociation of this complex, thus making PpsR available for DNA binding and PS gene repression. The primary photochemistry of AppA may be based on an unusual mechanism for flavin-based photoreceptors, i.e., reversible light-induced proton transfer within the BLUF domain (32).
To extend our knowledge on blue-light-dependent regulation and to gain insights into additional blue-light sensory mechanisms in R. sphaeroides, we used whole-genome transcriptional profiling of dark to blue-light transition of cells grown semiaerobically, i.e., in the presence of intermediate oxygen concentrations. The experimental setup involved monitoring of transient changes in gene expression, at 0, 5, 45, and 135 min after blue-light irradiation. The 5-min time point was chosen to capture the earliest possible changes in expression, whereas the later time points were chosen based on the blue-light-dependent expression changes observed by us earlier (8).
(Preliminary findings reported here were presented at the XIth International Symposium on Phototrophic Prokaryotes [abstr. P-155] in Tokyo, Japan, in 2003.)
MATERIALS AND METHODS
Strains and growth conditions.
Wild-type R. sphaeroides strain 2.4.1 was grown at 30°C on Sistrom's minimal medium A (14) containing succinate as a carbon source. The cultures were grown in 60-ml glass tubes under continuous sparging with a gas mixture containing 89% N2, 10% O2, and 1% CO2, resulting in a 120 μM dissolved O2 concentration. For light shift experiments, cultures grown in the dark to the early exponential phase were exposed to blue light for 0, 5, 45, or 135 min and harvested at an A600 of 0.20 ± 0.03. Blue light was obtained by passing light from a 500-W halogen lamp through a band-pass filter (BG12; Schott, Mainz, Germany). The maximum transmission of the filter is at 400 nm, with 50% transmission at 370 and 443 nm and no detectable transmission at <317 or >515 nm. The irradiance produced by this light source measured at the culture level was 10 μmol of photons m−2 s−1.
The ΔappA mutant APP11 (18), 2.4.1 and 2.4.1 overexpressing PpsR, and 2.4.1(pPNs) (19) were grown in the dark under continuous sparging with a gas mixture containing 79% N2, 20% O2, and 1% CO2.
RNA extraction and quantitative RT-PCR.
RNA extraction and quantitative reverse transcription-PCR (RT-PCR) was performed as described earlier (16, 46). In brief, rifampin was added to R. sphaeroides cultures at the final concentration of 200 μg ml−1 to halt transcription initiation. Cells were collected into centrifugation bottles containing shaved ice and pelleted by brief centrifugation; the cell pellets were then disrupted with a Mini-BeadBeater (Biospec Products). RNA was extracted by use of the RNEasy midikit (Qiagen) and tested for the lack of genomic DNA contamination by quantitative real-time PCR by using the SYBR Green method and the iCycler iQ Real-Time PCR detection system (Bio-Rad). The quality and yield of RNA and cDNA were tested by using capillary electrophoresis (BioAnalyzer, Agilent Technologies).
DNA microarray experiments and data analysis.
A high-density oligonucleotide R. sphaeroides microarray, GeneChip, corresponding to the whole genome was used for transcriptome profiling (46). cDNA synthesis, fragmentation, labeling, and genechip hybridization were performed according to the protocol for Pseudomonas aeruginosa genechips recommended by Affymetrix (http://www.affymetrix.com/support/technical/manuals.affx). Robust Multi-Array Analysis (RMA) with quantile normalization (27; http://www.stat.berkeley.edu/users/bolstad/RMAExpress/RMAExpress.html) and GeneSpring 4.2 software package (Silicon Genetics) were used for genechip data analysis and presentation. We recorded and analyzed transcriptome profiles from two to three independent cultures grown under semiaerobic growth conditions after blue-light irradiation for 0 (dark), 5, 45, or 135 min. The experimental reproducibility (r values) between replicates from the same growth conditions were in the range of 0.96 to 0.99.
Unreliably measured and unchanged genes were filtered out by using two criteria. Criterion 1 was used to filter out unreliable changes. We retained only genes, whose expression values from each replicate at one time point during irradiation (ai) differed by at least 15% from values from each replicate at 0 min (dark) of irradiation, (aj), i.e., ai ≥ 1.15 aj or ai ≤ 0.85 aj. Average values from all replicates were derived from the replicates that passed the first criterion; fold changes were calculated based on these average values and compared to 0 min (dark). The average expression values are presented in the figures and tables. Criterion 2 was used to filter out potentially physiologically meaningless changes. We defined “meaningful changes” as those where a fold change between an average value at least at one time point during irradiation (Âi) was ≥1.5-fold higher or lower than the average value at 0 min (Âj), i.e., Âi ≥ 1.50 Âj or Âi ≤ 0.67 Âj.
To increase the number of potentially meaningful changes that did not pass these criteria, we retained for analysis those genes that belonged to clearly identifiable operons, if several genes from the operon passed only the first criterion. This is justified because (i) according to our own and others' observations, the magnitude of expression changes measured by genechips are often lower than that measured by alternative techniques, e.g., RT-PCR (26, 33, 46, 63); (ii) the primary statistical analysis tool applied here, RMA, has been shown to produce a low percentage of false-positive results but a relatively high percentage of false-negative changes compared to other methods (27).
The expression data obtained here were deposited in the Gene Expression Omnibus (GEO) database of the National Center for Biotechnology Information (www.ncbi.nih.gov/geo), platform GPL162, under the “blue-light” series.
RESULTS AND DISCUSSION
Overview of the R. sphaeroides transcriptome changes in response to blue light under semiaerobic conditions.
We recently showed that major known metabolic and physiological responses of R. sphaeroides are mediated via modification of gene expression patterns (46), which probably holds true for most prokaryotes. Therefore, we anticipated that transcriptome analysis of blue-light-exposed cultures under semiaerobic conditions would reveal most metabolic and physiological trends in R. sphaeroides and provide insights into the sensory mechanisms involved.
The total number of genes whose expression changed in response to exposure to blue light was relatively limited. Expression of none of the general stress-related genes was changed. The dark-grown and irradiated cultures had identical growth rates (not shown). These observations suggest that blue-light irradiation did not result in significant stress in R. sphaeroides and that response to blue light was specific. A total of 110 ORFs and 12 intergenic regions were up- or downregulated at least at one time point during irradiation after application of the two cutoff criteria described in Materials and Methods. Of these, mRNA levels of approximately one-half was upregulated and one-half was downregulated. Based on the dynamics of expression, several clusters of genes were evident.
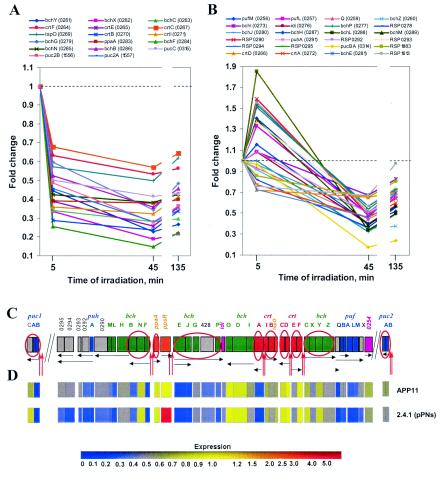
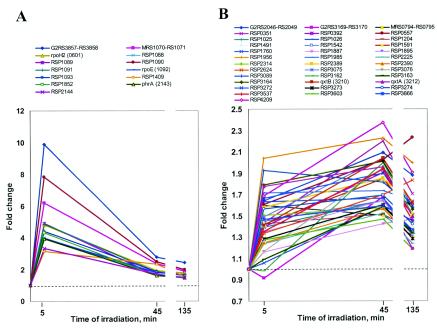
Genes from cluster 1 were downregulated at 5 min of blue-light irradiation and remained downregulated for the duration of the experiment (Fig. 1A). Genes belonging to cluster 2 at 5 min were unchanged, downregulated insignificantly, or slightly upregulated but were significantly downregulated later, at 45 and 135 min of irradiation (Fig. 1B). Genes from cluster 3 were severalfold upregulated at 5 min; however, their expression was attenuated to lower levels later in the time course (Fig. 2A). Genes from cluster 4 were upregulated at 5 and/or 45 min to a much lesser extent, usually ≤2-fold, and remained upregulated at similar levels for the duration of irradiation (Fig. 2B). We found that for most genes expression levels observed at 45 min remained changed very little at 135 min, suggesting that the long-term effects of blue-light irradiation were already evident at 45 min. This observation simplified our analysis to comparisons between the early (5 min) and late (45 and 135 min) time points. The genes from clusters 1 to 4 were analyzed with regard to (i) their function and (ii) the presence of common regulatory motifs in their upstream regions.
FIG. 1.
Major expression patterns of the downregulated genes in R. sphaeroides in response to blue light. Clusters were derived by using GeneSpring software. (A) Cluster 1 represents genes whose expression was downregulated at 5 min of irradiation and remained low during irradiation. (B) Cluster 2 represents genes whose expression was significantly downregulated at 45 min but not at 5 min of irradiation. (C) PS gene cluster and puc operons. PpsR-binding sites are shown as red vertical arrows. Putative transcripts are shown as black horizontal arrows. Colors: green, bacteriochlorophyll biosynthesis genes; red, carotenoid biosynthesis genes; blue, genes encoding structural polypeptides of photocomplexes; gray, genes encoding assembly factors or proteins of unknown function; orange, genes encoding regulatory factors; pink, genes encoding enzymes common to bacteriochlorophyll and ubiquinone biosynthesis. Circled genes were directly repressed by PpsR (Moskvin et al., submitted). (D) Relative expression of the PS genes in the ΔappA mutant APP11 and the wild type expressing higher levels of PpsR, 2.4.1(pPNs), compared to the wild-type strain. The expression of every gene in the wild type (not shown) was assigned to 1. The expression levels in APP11 and 2.4.1(pPNs) are according to the color scheme shown.
FIG. 2.
Major expression patterns of the upregulated genes in R. sphaeroides in response to blue light. (A) Cluster 3 represents genes whose expression was severalfold upregulated at 5 min of irradiation and remained upregulated at the attenuated levels upon prolonged irradiation. (B) Cluster 4 represents genes whose expression was upregulated ≤2.5-fold at 45 min of irradiation and did not change drastically at other time points.
Functional analysis of downregulated genes (clusters 1 and 2). (i) PS genes.
The majority of downregulated genes from clusters 1 and 2 were PS genes (Fig. 1C). This result is consistent with the earlier observations that puc and puf mRNA levels were decreased in semiaerobically grown cells exposed to blue light (8, 53). Coordinated downregulation of PS development may reflect an adaptation process to photooxidative stress conditions. Photooxidative stress is presumed to result from interactions between photoexcited bacteriochlorophyll or its precursors and molecular oxygen that results in formation of highly reactive singlet oxygen. Decreased expression of bacteriochlorophyll biosynthetic genes and genes encoding bacteriochlorophyll-binding proteins presents an obvious way to limit photooxidative damage. Interestingly, along with the downregulatioin of bacteriochlorophyll biosynthesis genes, we also observed downregulation of carotenoid biosynthesis genes, although carotenoids are known as major scavengers of singlet oxygen. This somewhat counterintuitive response may be explained by the common mechanisms that regulate expression of the photopigment genes (see below).
To test for the presence of singlet oxygen, we applied the fluorescence dye DanePy. The fluorescence of this dye is decreased by singlet oxygen-mediated oxidation (30). We were unable to detect increased production of singlet oxygen by using this method (data not shown). Further, growth of cultures was not impaired during blue-light irradiation, indicating that no or limited stress actually occurred. It is therefore conceivable that blue light in the presence of oxygen elicits an adaptive response, which prepares cells for photooxidative stress that may arise if light intensity increases or if UV light appears. Such adaptive responses to low levels of the stressor have been described for the oxidative stress response in bacteria (55, 56).
The phenomenon of adaptation to an anticipated stress resembles preparation of the Rhodobacter species to photosynthetic growth upon a decrease in oxygen tension. Photosynthesis is the preferred energy generation mode under anoxic conditions; however, it obviously requires the presence of light. Expression of the PS genes in Rhodobacter is induced in response to low oxygen independently of the presence or absence of light (reviewed in references 4, 23, 24, 45, 65, and 64).
(ii) Non-PS genes.
A cluster of genes, RSP2661 to RSP2669, predicted to encode iron-containing alcohol dehydrogenase, glycerol, or carbohydrate kinase and sugar transport proteins were downregulated during blue-light irradiation. For unknown reasons, expression of this gene cluster was found to be highly variable between replicates, both in this experiment and in other, unrelated, genechip experiments (data not shown). Three additional genes encoding a putative transmembrane protein (RSP2879) and proteins involved in transport (RSP1883 and RSP3287) were downregulated. The biological consequences of downregulation of any of these genes by blue light are currently unknown.
Regulatory mechanisms involved in blue-light-dependent downregulation of gene expression.
Based on the dynamics of responses to blue light, PS genes are clearly divided into two clusters. Genes from cluster 1 were downregulated as early as 5 min of irradiation, whereas genes from cluster 2 were downregulated only at the later time points (Fig. 1A and B). No functional distinction could be found between these clusters. However, analysis of the upstream regions of operons, to which genes in cluster 1 belong, revealed that all of them contain PpsR-binding sites (bchCXY, crtC, crtIB tspO, crtEF, ppaA, bchFBN, pucC, and puc2BA) and are directly controlled by the PpsR repressor (18, 66; Moskvin et al., submitted) (Fig. 1C). Apparently, the AppA/PpsR pathway is responsible for downregulation of these PS genes. Several genes known to be under direct PpsR control did not fall into cluster 1. Some of these belong to the operons listed above (bchZ, crtD, and puc1BA), whereas others (crtA) do not. Expression of bchZ, crtD, and crtA was decreased but did not pass the second cutoff criterion. We speculate that transcription initiation of all of these genes decrease in response to blue light. Because the extent of the decrease in mRNA levels depends on the rate of mRNA synthesis, the rate of degradation, and the original mRNA abundance, the more stable and abundant transcripts or transcript segments (e.g., puc1BA) are expected to show less variation in mRNA levels after a short (5-min) period of irradiation than other transcripts. This could explain why some genes directly controlled by PpsR did not fall into cluster 1.
Earlier, we presented a model, according to which the BLUF-domain flavoprotein AppA acts as a direct blue-light sensor and showed that its activity as a PpsR antirepressor is diminished upon blue-light irradiation (8). This was later confirmed by Masuda and Bauer (39), who showed that affinity of the AppA protein to PpsR in vitro is decreased upon blue-light irradiation. Although this model is consistent with the role of AppA as a sensor of blue light responsible for downregulation of PS genes, it may not account for downregulation of PS genes immediately after irradiation is applied. A mutation in AppA that destroys FAD binding, or a deletion of the FAD-binding BLUF domain do not prevent the immediate decrease in PS gene expression upon blue-light irradiation (8). However, both of these mutations result in an inability to maintain repression during prolonged irradiation. This suggests that an additional blue-light sensory mechanism is involved in the onset of PS gene repression. Given that the genes downregulated at 5 min of irradiation clearly belong to the AppA/PpsR pathway, it is likely that this pathway is responsible for the onset of repression. One possibility is that, under semiaerobic conditions, AppA senses blue light by two different mechanisms: (i) via FAD bound to the BLUF domain (repression maintenance) and (ii) via a different protein domain. However, we cannot exclude the existence of an additional protein involved in the AppA/PpsR pathway. These possibilities are currently under investigation.
The expression of PS genes belonging to cluster 2 was either unchanged, insignificantly downregulated or slightly upregulated at 5 min of irradiation but decreased at 45 min and remained low at 135 min (Fig. 1B). All PS genes, whether they do or do not contain PpsR-binding sites, were recently shown to be regulated via the AppA/PpsR pathway (Moskvin et al., submitted). It was suggested that the AppA/PpsR system controls activity or expression of an additional regulator of PS gene expression that is responsible for downregulation of those genes, whose upstream regions do not contain PpsR-binding sites. The separation of PS genes into two clusters based on the dynamics of blue-light responses observed here lends further support to this hypothesis. One candidate for such regulator is PpaA (photopigment and puc activator protein [16]). The expression of the ppaA gene is repressed by PpsR by ∼2.5-fold at 5 min of blue-light irradiation (Fig. 1A). Whether or not a lower level of PpaA during prolonged irradiation is responsible for the downregulation of cluster 2 genes remains to be tested.
Does AppA affect gene expression independently of PpsR?
Since the expression of many non-PS genes was upregulated by blue light and AppA was the only known factor in R. sphaeroides capable of blue-light sensing, we investigated the possibility that AppA affects gene expression via a PpsR-independent pathway. To this end, we compared transcriptome profiles of the ΔappA mutant, APP11 (18) with that of the wild type (strain 2.4.1) and the wild type overexpressing PpsR, strain 2.4.1(pPNs) (19). The latter strain expresses the ppsR gene at the ∼12-fold-higher level than the wild type (Fig. 1D). If an AppA-dependent but PpsR-independent pathway existed, the ΔappA mutant would show different expression patterns than strain 2.4.1(pPNs). However, if an AppA-dependent but PpsR-independent pathway did not exist, the transcriptome profiles of both strains would be very similar.
We analyzed the transcriptome profiles of the APP11 and 2.4.1(pPNs) grown under aerobic conditions. Compared to the wild type, the ΔappA mutation resulted in significant changes in the expression of PS genes, as well as of other, non-PS genes, suggesting that AppA is functional under these conditions. Significant expression changes were observed in 2.4.1(pPNs), compared to 2.4.1, suggesting that PpsR present in excess is also functional. Detailed analysis of the expression changes in these strains will be described elsewhere (Moskvin et al., submitted). Here, it is important to note that the transcriptome profiles of these two strains were qualitatively very similar. Compared to the wild type, the directions of expression changes of all genes were identical in these two strains. The expression of none of the genes upregulated by blue light in the wild type was lower in the APP11 strain but not in strain 2.4.1(pPNs). The observed quantitative differences in expression levels between these two strains can be readily explained by different levels of PpsR. The similarity in expression patterns in strains APP11 and 2.4.1(pPNs) is illustrated in Fig. 1D, where the expression profiles of PS genes in these strains are compared to those of the wild type.
These data argue against existence of an AppA-dependent but PpsR-independent regulatory pathway. We therefore conclude that upregulation of non-PS genes after blue-light irradiation most likely results from an AppA/PpsR-independent regulatory pathway(s). Such pathways are described below.
Functional analysis of upregulated genes (clusters 3 and 4).
Among upregulated genes there were several genes putatively involved in photooxidative stress defense.
(i) Deoxypyrimidine photolyase.
The expression of RSP2143 encoding a protein from the deoxypyrimidine photolyase/cryptochrome family (50) was upregulated as early as 5 min of irradiation (Fig. 2A). These proteins bind flavin and folate ligands, which are involved in blue-light sensing and/or DNA repair. Photolyases repair cyclobutane rings formed upon UV irradiation of the two adjacent thymine residues. The protein architecture of both cryptochromes and photolyases is similar. However, cryptochromes are not involved in DNA repair. Instead, they function as blue-light receptors in eukaryotes and bacteria (9, 31, 50, 59). The R. sphaeroides 2.4.1 genome sequence contains two ORFs, RSP2143 and RSP1981, that belong to this family. Since R. sphaeroides uses sunlight as an energy source and is constantly exposed to UV light in natural environments, it is likely to possess at least one functional photolyase. This assumption is consistent with our recent identification and characterization of a photolyase from a related organism, Rhodobacter capsulatus (7).
The RSP2143 gene product exhibits 56% sequence identity to the R. capsulatus photolyase and lacks residues believed to be unique to cryptochromes (9). The induction of RSP2143 expression by blue light is more consistent with its role in protection against photooxidative stress as opposed to the role in photoreception. Blue light has been shown to induce the transcription of photolyase genes in the goldfish Carasius auratus (42, 62) and in the fungus Trichoderma harzianum (5). In these organisms, light induction can be mimicked by treatment with H2O2 and suppressed by radical scavengers, indicating the involvement of reactive oxygen species in signaling (reference 50 and references therein). R. sphaeroides RSP2143 showed a fast fourfold induction after treatment of cultures with 1 mM H2O2 but did not respond to other chemicals causing oxidative stress, such as the superoxide anion-generating paraquat or diamide that causes glutathione depletion (Table 1 and unpublished data). Based on sequence analysis and its expression pattern, we postulate that RSP2143 is a photolyase and not a cryptochrome and so designate it phrA. Further experiments will be aimed at elucidation of the physiological function of phrA.
TABLE 1.
Comparative gene expression in R. sphaeroides in response to H2O2 or blue-light irradiation under semiaerobic conditions
| Gene | Gene description | Fold change in expressiona after exposure to:
|
|||
|---|---|---|---|---|---|
| H2O2 for 7 min | Blue light for:
|
||||
| 5 min | 45 min | 135 min | |||
| RSP0601 | σ38 factor, rpoH2 | 12.7 | 4.8 | 1.9 | 1.8 |
| RSP1092 | σE factor, rpoE | 10.8 | 4.0 | 1.8 | 1.7 |
| RSP1981 | Putative DNA photolyase/cryptochrome | (1.3) | (0.9) | (0.9) | (0.9) |
| RSP2143 | DNA photolyase, phrA | 4.4 | 3.9 | 1.6 | 1.5 |
| RSP2146 | Putative NTP pyrophosphohydrolase | (1.0) | (1.4) | (1.1) | (1.1) |
| RSP2388 | Putative NTP pyrophosphohydrolase | 2.1 | (1.4) | (1.4) | (1.3) |
| RSP2389 | Glutathione peroxidase | 3.5 | 1.6 | 1.8 | 1.6 |
| RSP2390 | Putative deacetylase | (1.1) | (1.2) | 1.6 | (1.3) |
| RSP2987 | Putative acetyltransferases | 2.2 | 1.5 | (1.1) | (1.1) |
| RSP3162 | Putative heme-binding protein | 2.8 | (1.0) | 2.0 | 1.5 |
| RSP3163 | Aerobic-type carbon monoxide dehydrogenase, large subunit | 4.2 | (1.1) | 1.6 | (1.3) |
| RSP3164 | Aerobic-type carbon monoxide dehydrogenase, small subunit | 7.4 | 1.8 | 2.0 | 1.6 |
| RSP3210 | Quinol oxidoreductase subunit 2, qxtB | 1.8 | (1.4) | 2.0 | 1.6 |
| RSP3212 | Quinol oxidoreductase subunit 1, qxtA | (1.3) | 1.8 | 2.0 | 1.5 |
| RSP4209 | Putative acetyltransfcrase | 4.1 | 1.7 | 2.4 | 1.9 |
That is, the fold change compared to a no-treatment control. Values in parentheses correspond to insignificant changes according to the chosen criteria.
RSP2146 encoding a nucleoside triphosphate (NTP) pyrophosphohydrolase implicated in oxidative damage repair was slightly upregulated, and so was the unlinked gene, RSP2388, with the same predicted function. However, these genes did not pass the second cutoff parameter.
In contrast to phrA, RSP1981 is expressed independently of blue light at a low level. The RSP1981 gene product exhibits only 26% sequence identity with the R. capsulatus photolyase and lacks the three conserved tryptophanes, which are supposed to be involved in photolyase intraprotein electron transfer to the catalytic cofactor FAD (10). There was no response of RSP1981 to H2O2, paraquat, or diamide (Table 1). The function of the RSP1981 gene product remains to be investigated.
(ii) Glutathione peroxidase.
Another gene upregulated by blue light is RSP2389, which encodes a putative glutathione peroxidase (Fig. 2B). Its expression was increased at 5 min of irradiation and remained upregulated by 1.6- to 1.8-fold. Glutathione peroxidase is believed to contribute to the oxidative stress defense (34). We observed a threefold induction of RSP2389 by H2O2 (Table 1). Interestingly, the gene downstream of the putative glutathione peroxidase, RSP2390, encoding a putative deacetylase was upregulated by blue light in a fashion similar to that observed for glutathione peroxidase. RSP2987 and RSP4209, other genes encoding putative acetyltransferases, were also upregulated. The significance of the apparent activation of acetyl group transfers during blue-light irradiation is currently unknown.
(iii) Quinol oxidoreductases.
Expression of genes encoding putative aerobic carbon monoxide dehydrogenase or quinol oxidoreductase (RSP3162 to RSP3164) and cytochrome bd-type quinol oxidase (qxtB, RSP3210; RSP3212) was increased. Blue-light irradiation is expected to cause formation of semiquinol radicals. We hypothesize that these enzymatic complexes, which are involved in quinol oxidation, may help scavenging semiquinol radicals and therefore may be important for photooxidative stress defense. The validity of this hypothesis is yet to be tested. The expression of RSP3162 to RSP3164 was also increased by H2O2 treatment, whereas the expression of RSP3210 and RSP3212 was only weakly affected or unaffected by H2O2 (Table 1).
Many other upregulated genes encode hypothetical proteins. We were unable to predict any physiological role of these proteins in responses of R. sphaeroides to blue light under the semiaerobic conditions.
Analyses of the transcriptome responses to different stresses revealed that many expression changes, considered in the past to be specific to a certain stress, in fact overlap, e.g., responses to heat shock, oxidative stress, and stationary phase (15, 35, 67). The changes in the R. sphaeroides transcriptome in response to blue-light irradiation are different from the changes observed in response to oxidative stress agents; however, certain overlaps clearly exist (Table 1 and unpublished data). It will be important to unravel the regulatory pathways underlying the blue-light responses in order to understand its place among other stress responses.
Regulatory mechanisms involved in blue-light-dependent upregulation of gene expression.
Genes encoding two different σ-factor proteins, rpoE (RSP1092) and rpoH2 (RSP0601), were significantly upregulated—4.0- and 4.8-fold, respectively—at 5 min of irradiation by blue light and remained upregulated until 135 min, albeit at attenuated levels (Fig. 2A). rpoE encodes a σE-factor, which belongs to the extracytoplasmic σ70-factor family, i.e., the ECF family (49). rpoH2 encodes a σ70-type factor, which belongs to the heat shock family of σ-factors (58).
The products of rpoH2, σ38 (37), and rpoE, σE (44), have been shown to drive transcription of the cytochrome c2 gene, cycA (RSP0296). However, under the conditions used here, cycA expression was unchanged. This does not necessarily contradict the published data because transcription of cycA is driven by at least three promoters, yet the overall range of variation in cycA expression is small (44).
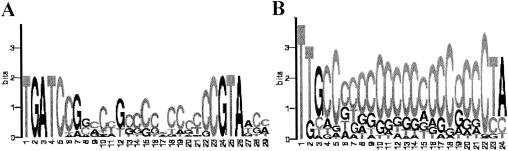
We reasoned that, if rpoE and rpoH2 expression is increased, so should be expression of genes transcribed by σE and σ38. We analyzed the upstream regions of all upregulated genes by using GeneSpring software with the aim of finding possible consensus sequences. Two such sequences having low false discovery rates (FDRs) were found upstream of numerous upregulated genes. One consensus sequence, TGATC-N17-CGTA with no mismatches (FDR 1.37e−6), was present in the upstream regions of six upregulated genes. This sequence is reminiscent of a promoter, i.e., it has a putative “-35” and “-10” areas separated by 17 nucleotides. The following promoter sequence recognizable by σE was deduced earlier: TGATCC-N18-TAAGA (44). The perfect match to the consensus derived by Newman et al. (44) is found upstream of the rpoE and phrA genes overlapping with the consensus identified here. When mismatches are allowed, both putative consensus sequences converge. This suggests that they represent one and the same promoter sequence. Given that our consensus is found upstream of the larger number of coregulated genes than that derived by Newman et al. (44), we conclude that the sequence found here best describes consensus for the σE promoter (Fig. 3A).
FIG. 3.
Deduced consensus sites presented as sequence logos (52). (A) Putative σE promoter; (B) putative σ38 promoter.
Interestingly, the expression pattern of all genes containing the deduced σE promoter, as well as genes apparently cotranscribed with them, formed a cluster, which is clearly different from the expression pattern of the remaining upregulated genes (Fig. 2A, cluster 3, and Table 2). The expression of genes in cluster 3 increased the most at 5 min of irradiation and declined with time to the levels, which were still higher than those in the dark.
TABLE 2.
Consensus sequences upstream of the genes upregulated by blue light
| Cluster and gene | Description | Coordinatea |
|---|---|---|
| Cluster 3 | TGATC(N17)CGTAb | |
| G2RS3857-RS3858 | Intergenic region | −110 |
| RSP0601 | σ38, rpoH2 | −66 |
| RSP1091 (RSP1087 to RSP1090)c | Flavin containing amine oxidoreductase | −54 |
| RSP1092 (RSP1093) | σE, rpoE | −130 |
| RSP1852d | Putative UV-endonuclease | −103 |
| RSP2143 (RSP2144) | DNA photolyase, phrA | −49 |
| Cluster 4 | TTG(N18)CTAe | |
| MRS0794-RS0795 | Intergenic region | −34 |
| RSP0351 | Putative oxidoreductase | −133 and −53 |
| RSP0392 | Putative lactoylglutathione lyase | −56 |
| RSP0557 | Hypothetical protein | −86 |
| RSP1025 (RSP1026) | Conserved hypothetical protein | −63 |
| RSP1204 | ABC-type transport system, ATPase component | −52 |
| RSP1760 | Conserved hypothetical protein | −149 |
| RSP1887 | Ribosomal protein L33 | −85 and −40 |
| RSP1895 | Putative transport protein | −35 |
| RSP1985 | Hypothetical protein | −56 |
| RSP2314 | Oxidoreductase of aldo/keto reductase family | −57 |
| RSP2389 (RSP2390) | Putative glutathione peroxidase | −102 |
| RSP2624 | Hypothetical protein | −40 |
| RSP3075 (RSP3076) | Conserved hypothetical protein | −54 |
| RSP3164 (RSP3162 to RSP3163) | Putative oxidoreductase/aldehyde dehydrogenase subunit | −54 |
| RSP3272 | Gamma-glutamyltranspeptidase | −47 |
| RSP3274 (RSP3273) | ABC multidrug/carbohydrate transporter, ATPase subunit | −91 |
| RSP3537 | Alcohol dehydrogenase, zinc containing | −52 |
| RSP3866 | Hypothetical protein | −111 |
| RSP4209 | Putative acyltransferase | −107 |
That is, the coordinate(s) of the 5′-end nucleotide(s) of the consensus sequence upstream of the first translated nucleotide (+1).
There were no mismatches in the conserved nucleotides.
Genes in parentheses are proposed (based on gene arrangement) to be cotranscribed with the gene containing the consensus sequence.
This gene also has the putative consensus of σ38 promoter located at −63.
No more than one mismatch in the conserved nucleotides.
The rpoE gene is transcribed by its own product, σE. The fast increase in rpoE gene expression suggests that its product, σE, directly or indirectly senses blue light or a product(s) formed as a result of blue-light irradiation in the presence of oxygen. The fact that a photolyase gene phrA is apparently transcribed by σE further strengthens this suggestion.
The activity of σE is known to be regulated by the anti-σ-factor ChrR. The latter protein has been proposed to respond to a signal from the porphyrin biosynthetic pathway (44, 51). Porphyrins absorb in the blue area of light spectrum (Sorret band) and may be susceptible to blue-light irradiation. It is also possible that singlet oxygen produced under these conditions at low levels interacts with a porphyrin intermediate(s) and through such intermediate(s) affects ChrR. The accumulation of porphyrin intermediates in the Escherichia coli heme biosynthetic mutants sensitizes cells to blue light (60, 61). Alternatively, ChrR may interact with an as-yet-unidentified upstream protein sensor of blue light. Newman et al. showed that activity of ChrR in vitro is regulated by zinc (43). At present, it is unclear whether or how zinc binding and porphyrin intermediates are related.
Until the present study, nothing has been known about the environmental factors involved in activation of rpoH2 expression and about genes, other than cycA (37), transcribed by σ38. Apparently, blue-light irradiation in the presence of oxygen is one of the conditions activating rpoH2 gene expression and the σ38 regulon. It is not known whether such activation works via activation of the σ38 protein or its upstream partner, if any, or via upregulation of rpoH2 gene expression by σE. However, because rpoH2 belongs to cluster 3 of the σE-dependent genes and a predicted σE-dependent promoter is present upstream of rpoH2, we favor the latter possibility (Fig. 2A and Table 2).
What genes are transcribed by σ38? The second consensus sequence (FDR 1.19e−3) identified in the upstream regions of the upregulated genes, TTG-N18-CTA, may hold the answer to this question (Fig. 3B). This sequence resembles a truncated promoter sequence. Further, it shares significant similarity with the sequence of the σ37-dependent P1 promoter of cycA. σ37 is a member of the heat shock family of σ-factors in R. sphaeroides, to which σ38 belongs. The P1 promoter is recognizable by both σ37 and σ38 (37). Twenty-one genes from cluster 4 contain the putative σ38-promoter. These are likely to constitute the σ38 regulon. Because most of these genes are hypothetical, it is difficult to predict functional significance of their upregulation. An exception is the glutathione peroxidase gene, RSP2389, anticipated to be involved in photooxidative damage defense.
Concluding remarks.
The described R. sphaeroides transcriptome analysis showed that blue light causes downregulation and upregulation of expression of a limited number of genes. We predicted regulatory mechanisms that are likely to control changes in expression of >81% of all genes. PS genes comprise the largest cluster of downregulated genes. Their downregulation most likely represents an adaptation process, which prepares cells for photooxidative stress. This process is under the control of the AppA/PpsR antirepressor-repressor system. AppA has been shown to function as the primary photoreceptor for maintenance of PS gene repression upon long-term blue-light irradiation (8). Our transcriptome analysis suggests that AppA is also responsible for the onset of repression by an as-yet-unknown sensory mechanism. However, involvement of an additional factor cannot be excluded. Consistent with our interpretation of the significance of downregulation of PS development, several genes involved in photooxidative damage defense are upregulated by blue light. Physiological roles of other upregulated genes, many of which are annotated as encoding hypothetical proteins, is unclear. A combination of downregulation of PS development and upregulation of photooxidative stress defense mechanisms apparently prepares R. sphaeroides for photooxidative stress that could occur if the light intensity rises to higher levels. We revealed that expression of the vast majority of upregulated genes is controlled by the σE and σ38 pathways. We deduced promoter sequences for these σ-factors. We predicted that the anti-σE-factor ChrR or its upstream partner is involved in sensing of blue light or a product of blue-light irradiation in the presence of oxygen, possibly singlet oxygen. This sensory event is likely to result in the release of inhibition of the σE protein, which positively regulates expression of its own gene and thus promotes expression of the σE regulon. Initiation of expression of one of the σE-dependent genes, σ38, promotes expression of the σ38 regulon. The present study has provided the framework and generated a number of testable hypotheses for the in-depth analysis of the blue-light-dependent signal transduction pathways in R. sphaeroides.
Acknowledgments
This study was supported by grants from NIH NCRR (COBRE) P20 RR15640 (M.G.) and DFG Kl563/15-1/15-2 (G.K.), as well as by startup funds from the University of Wyoming (M.G.).
We thank the staff of the University of Colorado (Denver) Cancer Center Microarray Core Facility where R. sphaeroides genechips were processed. Real-time PCR was performed at the Macromolecular Core Facility of the University of Wyoming.
REFERENCES
- 1.Ahmad, M., and A. R. Cashmore. 1993. HY4 gene of A. thaliana encodes a protein with characteristics of a blue-light photoreceptor. Nature 366:162-166. [DOI] [PubMed] [Google Scholar]
- 2.Akbar, S., T. A. Gaidenko, C. M. Kang, M. O'Reilly, K. M. Devine, and C. W. Price. 2001. New family of regulators in the environmental signaling pathway which activates the general stress transcription factor σB of Bacillus subtilis. J. Bacteriol. 183:1329-1338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Baca, M., G. E. O. Borgstahl, M. Boissinot, P. M. Burke, D. R. Williams, K. A. Slater, and E. D. Getzoff. 1994. Complete chemical structure of photoactive yellow protein: novel thioester-linked 4-hydroxy-cinnamyl chromophore and photocycle chemistry. Biochemistry 33:14369-14377. [DOI] [PubMed] [Google Scholar]
- 4.Bauer, C. E., S. Elsen, and T. H. Bird. 1999. Mechanisms for redox control of gene expression. Annu. Rev. Microbiol. 53:495-523. [DOI] [PubMed] [Google Scholar]
- 5.Berrocal-Tito, G., L. Sametz-Baron, K. Eichenberg, B. A. Horwitz, and A. Herrera-Estrella. 1999. Rapid blue light regulation of a Trichoderma harzianum photolyase gene. J. Biol. Chem. 274:14288-14294. [DOI] [PubMed] [Google Scholar]
- 6.Braatsch, S., and G. Klug. 2004. Blue light perception in bacteria. Photosynth. Res. 79:45-57. [DOI] [PubMed] [Google Scholar]
- 7.Braatsch, S., and G. Klug. 2004. ORF90, a gene required for photoreactivation in Rhodobacter capsulatus SB1003 encodes a cyclobutane pyrimidine dimer photolyase. Photosynth. Res. 79:167-177. [DOI] [PubMed] [Google Scholar]
- 8.Braatsch, S., M. Gomelsky, S. Kuphal, and G. Klug. 2002. A single flavoprotein, AppA, integrates both redox and light signals in Rhodobacter sphaeroides. Mol. Microbiol. 45:827-836. [DOI] [PubMed] [Google Scholar]
- 9.Brudler, R., K. Hitomi, H. Daiyasu, H. Toh, K. I. Kucho, M. Ishiura, M. Kanehisa, V. A. Roberts, T. Todo, J. A. Tainer, and E. D. Getzoff. 2003. Identification of a new cryptochrome class: structure, function, and evolution. Mol. Cell 11:59-67. [DOI] [PubMed] [Google Scholar]
- 10.Byrdin, M., A. P. Eker, M. H. Vos, and K. Brettel. 2003. Dissection of the triple tryptophan electron transfer chain in Escherichia coli DNA photolyase: Trp382 is the primary donor in photoactivation. Proc. Natl. Acad. Sci. USA 100:8676-8681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Choudhary, M., and S. Kaplan. 2000. DNA sequence analysis of the photosynthesis region of Rhodobacter sphaeroides 2.4.1. Nucleic Acids Res. 28:862-867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Christie, J. M., M. Salomon, K. Nozue, M. Wada, and W. R. Briggs. 1999. LOV (light, oxygen, or voltage) domains of the blue-light photoreceptor phototropin (nph1): binding sites for the chromophore flavin mononucleotide. Proc. Natl. Acad. Sci. USA 96:8779-8783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Christie, J. M., P. Reymond, G. K. Powell, P. Bernasconi, A. A. Raibekas, E. Liscum, and W. R. Briggs. 1998. Arabidopsis NPH1: a flavoprotein with the properties of a photoreceptor for phototropism. Science 282:1698-1701. [DOI] [PubMed] [Google Scholar]
- 14.Cohen-Bazire, G., W. R. Sistrom, and R. V. Stanier. 1957. Kinetic studies of pigment synthesis by non-sulfur bacteria. J. Cell. Comp. Physiol. 49:25-68. [DOI] [PubMed] [Google Scholar]
- 15.Desikan, R., S. A.-H.-Mackerness, J. T. Hancock, and S. J. Neill. 2001. Regulation of the Arabidopsis transcriptome by oxidative stress. Plant Physiol. 127:159-172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gomelsky, L., J. Sram, O. V. Moskvin, I. M. Horne, H. N. Dodd, J. M. Pemperton, A. G. McEwan, S. Kaplan, and M. Gomelsky. 2003. Identification and in vivo characterization of PpaA, a regulator of photosystem formation in Rhodobacter sphaeroides. Microbiology 149:377-388. [DOI] [PubMed] [Google Scholar]
- 17.Gomelsky, M., and G. Klug. 2002. BLUF: a novel FAD-binding domain involved in sensory transduction in microorganisms. Trends Biochem. Sci. 27:497-500. [DOI] [PubMed] [Google Scholar]
- 18.Gomelsky, M., and S. Kaplan. 1995. appA, a novel gene encoding a trans-acting factor involved in the regulation of photosynthesis gene expression in Rhodobacter sphaeroides 2.4.1. J. Bacteriol. 177:4609-4618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gomelsky, M., and S. Kaplan. 1995. Genetic evidence that PpsR from Rhodobacter sphaeroides 2.4.1 functions as a repressor of puc and bchF expression. J. Bacteriol. 177:1634-1637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gomelsky, M., and S. Kaplan. 1997. Molecular genetic analysis suggesting interactions between AppA and PpsR in regulation of photosynthesis gene expression in Rhodobacter sphaeroides 2.4.1. J. Bacteriol. 179:128-134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gomelsky, M., and S. Kaplan. 1998. AppA, a redox regulator of photosystem formation in Rhodobacter sphaeroides 2.4.1, is a flavoprotein. Identification of a novel FAD binding domain. J. Biol. Chem. 273:35319-35325. [DOI] [PubMed] [Google Scholar]
- 22.Gomelsky, M., I. M. Horne, H. J. Lee, J. M. Pemberton, A. G. McEwan, and S. Kaplan. 2000. Domain structure, oligomeric state, and mutational analysis of PpsR, the Rhodobacter sphaeroides repressor of photosystem gene expression. J. Bacteriol. 182:2253-2261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gregor, J., and G. Klug. 1999. Regulation of bacterial photosynthesis genes by oxygen and light. FEMS Microbiol. Lett. 179:1-9. [DOI] [PubMed] [Google Scholar]
- 24.Gregor, J., and G. Klug. 2002. Oxygen-regulated expression of genes for pigment binding proteins in Rhodobacter capsulatus. J. Mol. Microbiol. Biotechnol. 4:249-253. [PubMed] [Google Scholar]
- 25.Hoff, W. D., I. H. Stokkum, M. E. van Brederode, A. M. Brouwer, J. C. Fitch, T. E. Meyer, R. van Grondelle, and K. J. Hellingwerf. 1994. Measurement and global analysis of the absorbance changes in the photocycle of the photoactive yellow protein from Ectothiorhodospira halophila. Biophys. J. 67:1691-1705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Holland, M. J. 2002. Transcript abundance in yeast varies over six orders of magnitude. J. Biol. Chem. 277:14363-14366. [DOI] [PubMed] [Google Scholar]
- 27.Irizarry, R. A., B. M. Bolstad, F. Collin, L. M. Cope, B. Hobbs, and T. P. Speed. 2003. Summaries of Affymetrix GeneChip probe level data. Nucleic Acids Res. 31:e15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Jackson, J. B. 1988. Bacterial photosynthesis, p. 317-376. In C. Anthony (ed.), Bacterial energy transduction. Academic Press, Ltd., London, United Kingdom.
- 29.Jiang, Z. Y., L. R. Swem, B. G. Rushing, S. Devanathan, G. Tollin, and C. E. Bauer. 1999. Bacterial photoreceptor with similarity to photoactive yellow protein and plant phytochromes. Science 285:406-409. [DOI] [PubMed] [Google Scholar]
- 30.Kalai, T., E. Hideg, I. Vass, and K. Hideg. 1998. Double (fluorescent and spin) sensors for detection of reactive oxygen species in the thylakoid membrane. Free Radic. Biol. Med. 24:649-652. [DOI] [PubMed] [Google Scholar]
- 31.Kanai, S., R. Kikuno, H. Toh, H. Ryo, and T. Todo. 1997. Molecular evolution of the photolyase-blue-light photoreceptor family. J. Mol. Evol. 45:535-548. [DOI] [PubMed] [Google Scholar]
- 32.Laan, W., M. A. van der Horst, I. H. van Stokkum, and K. J. Hellingwerf. 2003. Initial characterization of the primary photochemistry of AppA, a blue-light-using flavin adenine dinucleotide-domain containing transcriptional antirepressor protein from Rhodobacter sphaeroides: a key role for reversible intramolecular proton transfer from the flavin adenine dinucleotide chromophore to a conserved tyrosine? Photochem. Photobiol. 78:290-297. [DOI] [PubMed] [Google Scholar]
- 33.Lee, J. M., S. Zhang, S. Saha, S. Santa Anna, C. Jiang, and J. Perkins. 2001. RNA expression analysis using an antisense Bacillus subtilis genome array. J. Bacteriol. 183:7371-7380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lei, X. G. 2001. Glutathione peroxidase-1 gene knockout on body antioxidant defense in mice. Biofactors 14:93-99. [DOI] [PubMed] [Google Scholar]
- 35.Leichert, L., C. Scharf, and M. Hecker. 2003. Global characterization of disulfide stress in Bacillus subtilis. J. Bacteriol. 185:1967-1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Losi, A., E. Polverini, B. Quest, and W. Gartner. 2002. First evidence for phototropin-related blue-light receptors in prokaryotes. Biophys. J. 82:2627-2634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.MacGregor, B. J., R. K. Karls, and T. J. Donohue. 1998. Transcription of the Rhodobacter sphaeroides cycA P1 promoter by alternative RNA polymerase holoenzymes. J. Bacteriol. 180:1-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Malhotra, K., S. T. Kim, A. Batschauer, L. Dawut, and A. Sancar. 1995. Putative blue-light photoreceptors from Arabidopsis thaliana and Sinapis alba with a high degree of sequence homology to DNA photolyase contain the two photolyase cofactors but lack DNA repair activity. Biochemistry 34:6892-6899. [DOI] [PubMed] [Google Scholar]
- 39.Masuda, S., and C. E. Bauer. 2002. AppA is a blue light photoreceptor that antirepresses photosynthesis gene expression in Rhodobacter sphaeroides. Cell 110:613-623. [DOI] [PubMed] [Google Scholar]
- 40.McEwan, A. G. 1994. Photosynthetic electron transport and anaerobic metabolism in purple non-sulfur bacteria. Antonie Leeuwenhoek 66:151-164. [DOI] [PubMed] [Google Scholar]
- 41.Meyer, T. E. 1985. Isolation and characterization of soluble cytochromes, ferredoxins, and other chromophoric proteins from the halophilic phototrophic bacterium Ectothiorhodospira halophila. Biochim. Biophys. Acta 806:175-183. [DOI] [PubMed] [Google Scholar]
- 42.Mitani, H., N. Uchida, and A. Shima. 1996. Induction of cyclobutane pyrimidine dimer photolyase in cultured fish cells by UVA and blue light. Photochem. Photobiol. 64:943-948. [DOI] [PubMed] [Google Scholar]
- 43.Newman, J. D., L. C. Anthony, and T. J. Donohue. 2001. The importance of zinc-binding to the function of Rhodobacter sphaeroides ChrR as an anti-sigma factor. J. Mol. Biol. 313:495-499. [DOI] [PubMed] [Google Scholar]
- 44.Newman, J. D., M. J. Falkowski, B. A. Schilke, L. C. Anthony, and T. J. Donohue. 1999. The Rhodobacter sphaeroides ECF sigma factor, σE, and the target promoters cycA P3 and rpoE P1. J. Mol. Biol. 294:307-320. [DOI] [PubMed] [Google Scholar]
- 45.Oh, J. I., and S. Kaplan. 2000. Redox signaling: globalization of gene expression. EMBO J. 19:4237-4247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Pappas, C. T., J. Sram, O. V. Moskvin, P. S. Ivanov, R. C. Mackenzie, M. Choudhary, M. L. Land, F. W. Larimer, S. Kaplan, and M. Gomelsky. 2004. Construction and validation of the Rhodobacter sphaeroides 2.4.1 DNA microarray: transcriptome flexibility at diverse growth modes. J. Bacteriol. 186: in press. [DOI] [PMC free article] [PubMed]
- 47.Penfold, R. J., and J. M. Pemberton. 1991. A gene from the photosynthetic cluster of Rhodobacter sphaeroides induces trans suppression of bacteriochlorophyll and carotenoid levels in R. sphaeroides and R. capsulatus. Curr. Microbiol. 23:259-263. [Google Scholar]
- 48.Penfold, R. J., and J. M. Pemberton. 1994. Sequencing, chromosomal inactivation, and functional expression in Escherichia coli of ppsR, a gene which represses carotenoid and bacteriochlorophyll synthesis in Rhodobacter sphaeroides. J. Bacteriol. 176:2869-2876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Raivio, T. L., and T. J. Silhavy. 2001. Periplasmic stress and ECF sigma factors. Annu. Rev. Microbiol. 55:591-624. [DOI] [PubMed] [Google Scholar]
- 50.Sancar, A. 2003. Structure and function of DNA photolyase and cryptochrome blue-light photoreceptors. Chem. Rev. 103:2203-2237. [DOI] [PubMed] [Google Scholar]
- 51.Schilke, B. A., and T. J. Donohue. 1995. ChrR positively regulates transcription of the Rhodobacter sphaeroides cytochrome c2 gene. J. Bacteriol. 177:1929-1937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Schneider, T. D., and R. M. Stephens. 1990. Sequence logos: a new way to display consensus sequences. Nucleic Acids Res. 18:6097-6100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Shimada, H., K. Iba, and K. Takamiya. 1992. Blue-light irradiation reduces the expression of puf and puc operons of Rhodobacter sphaeroides under semi-aerobic conditions. Plant Cell Physiol. 33:471-475. [Google Scholar]
- 54.Sprenger, W. W., W. D. Hoff, J. P. Armitage, and K. J. Hellingwerf. 1993. The eubacterium Ectothiorhodospira halophila is negatively phototactic, with a wavelength dependence that fits the absorption spectrum of the photoactive yellow protein. J. Bacteriol. 175:3096-3104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Storz, G., and J. A. Imlay. 1999. Oxidative stress. Curr. Opin. Microbiol. 2:188-194. [DOI] [PubMed] [Google Scholar]
- 56.Storz, G., and M. Zheng. 2000. Oxidative stress, p. 47-59. In G. Storz and R. Hengge-Aronis (ed.), Bacterial stress responses. ASM Press, Washington, D.C.
- 57.Suwanto, A., and S. Kaplan. 1989. Physical and genetic mapping of the Rhodobacter sphaeroides 2.4.1 genome: genome size, fragment identification, and gene localization. J. Bacteriol. 171:5850-5859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Wosten, M. M. 1998. Eubacterial sigma-factors. FEMS Microbiol. Rev. 22:127-150. [DOI] [PubMed] [Google Scholar]
- 59.Worthington, E. N., I. H. Kavakli, G. Berrocal-Tito, B. E. Bondo, and A. Sancar. 2003. Purification and characterization of three members of the photolyase/cryptochrome family blue-light photoreceptors from Vibrio cholerae. J. Biol. Chem. 278:39143-39154. [DOI] [PubMed] [Google Scholar]
- 60.Yang, H., A. Sasarman, H. Inokuchi, and J. Adler. 1996. Non-iron porphyrins cause tumbling to blue light by an Escherichia coli mutant defective in hemG. Proc. Natl. Acad. Sci. USA 93:2459-2463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Yang, H., H. Inokuchi, and J. Adler. 1995. Phototaxis away from blue light by an Escherichia coli mutant accumulating protoporphyrin IX. Proc. Natl. Acad. Sci. USA 92:7332-7336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Yasuhira, S., H. Mitani, and A. Shima. 1991. Enhancement of photorepair of ultraviolet-damage by preillumination with fluorescent light in cultured fish cells. Photochem. Photobiol. 53:211-215. [DOI] [PubMed] [Google Scholar]
- 63.Yuen, T., E. Wurmbach, R. L. Pfeffer, B. J. Ebersole, and S. C. Sealfon. 2002. Accuracy and calibration of commercial oligonucleotide and custom cDNA microarrays. Nucleic Acids Res. 30:E48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Zeilstra-Ryalls, J. H., and S. Kaplan. 2004. Oxygen intervention in the regulation of gene expression: the photosynthetic bacterial paradigm. Cell Mol. Life Sci. 61:417-436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Zeilstra-Ryalls, J. H., M. Gomelsky, A. A. Yeliseev, J. M. Eraso, and S. Kaplan. 1998. Transcriptional regulation of photosynthesis operons in Rhodobacter sphaeroides 2.4.1. Methods Enzymol. 297:151-166. [DOI] [PubMed] [Google Scholar]
- 66.Zeng, X., M. Choudhary, and S. Kaplan. 2003. A second and unusual pucBA operon of Rhodobacter sphaeroides 2.4.1: genetics and function of the encoded polypeptides. J. Bacteriol. 185:6171-6184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Zheng, M., X. Wang, L. J. Templeton, D. R. Smulski, R. A. LaRossa, and G. Storz. 2001. DNA microarray-mediated transcriptional profiling of the Escherichia coli response to hydrogen peroxide. J. Bacteriol. 183:4562-4570. [DOI] [PMC free article] [PubMed] [Google Scholar]