Abstract
We have found CueO from Escherichia coli to have a robust cuprous oxidase activity, severalfold higher than any homologue. These data suggest that a functional role for CueO in protecting against copper toxicity in vivo includes the removal of Cu(I).
Copper is an essential micronutrient, serving as a cofactor for numerous enzymes. However, being a redox-active transition metal, it is also cytotoxic through generation of reactive oxygen species in aerobic cells. Organisms in all kingdoms of life have therefore evolved elaborate copper homeostatic mechanisms (1, 17). Disturbances in copper homeostasis have been implicated in several diseases, including Menkes and Wilson disease (12), Parkinson's disease (25), and Alzheimer's disease and related aging disorders (19).
Escherichia coli, an enteric bacterium living in the digestive tract of warm-blooded animals, has a two-component copper regulatory system, enabling it to survive high concentrations of copper that may be generated through proteolytic and acidic degradation of food in the animal gut (18). One system activates expression of CopA and CueO by the MerR homologue CueR, which is activated upon Cu(I) binding (15). CopA was shown to be a Cu(I)-translocating P-type ATPase that transports cytosolic Cu(I) into the periplasm, and CueO was shown to be a copper-regulated multicopper oxidase located in the periplasm (4, 5). The other system is a two-component signal transduction system, with the cusRS genes forming a sensor-regulator pair that activates the divergently transcribed cusCFBA genes encoding a four-component copper efflux pump (2, 13). Both systems sense and respond to copper primarily in the cuprous oxidation state, the state in which it can diffuse through the cytoplasmic membrane and is most toxic (18).
CueO is an essential component of the copper regulatory mechanism in E. coli under aerobic conditions (4). Multicopper oxidases such as CueO couple four one-electron substrate oxidation steps to the reduction of dioxygen to water, generally through a T1-type copper at the substrate-binding site and a trinuclear copper center at the oxygen-binding site (23). Two aspects of CueO differ from other multicopper oxidases. First, CueO requires a fifth copper adjacent to the T1 site for activity (10, 21). Second, CueO displays a methionine-rich helix that lies over the T1 site and may function in sensing copper (20).
CueO has a broad substrate specificity and will oxidize numerous compounds in vitro, including ferrous iron, catechols, and iron-chelating siderophores (4, 10), and these characteristics have made the role of CueO in copper homeostasis difficult to discern. Most suggestions have centered on either a direct link to copper homeostasis through oxidation of Cu(I) (4, 6, 14) or on an indirect link via iron homeostasis, through oxidation of Fe(II) or the iron chelator enterobactin (4, 6, 7, 10). Attempts to measure cuprous activity in CueO have been reported to fail (10) but are difficult to measure due to the propensity for oxidation by molecular oxygen. Recently, however, this difficulty was overcome through use of a caged Cu(I) compound, allowing cuprous oxidase activity to be measured in the Saccharomyces cerevisiae (Fet3) and human (ceruloplasmin) homologues of CueO (24), leading to the suggestion that these proteins play an essential role in both iron and copper homeostasis (22). Here, we report that CueO has an excellent cuprous oxidase activity that is the highest among all the functional homologues, suggesting that this activity is central to the mechanism by which CueO protects E. coli against copper toxicity.
Two proteins, recombinant wild-type CueO and C500S mutant CueO (designed to incapacitate binding of the T1 copper), were used to establish the cuprous oxidase activity of CueO. The C500S mutant was constructed using a PCR-based approach as previously described (21); DNA sequencing of both strands confirmed the site-specific mutation. As expected, the mutant protein would not support resistance to copper toxicity when placed in a cell line missing a functional CueO gene (data not shown) in a procedure previously described for other CueO mutants (21). Both wild-type and C500S proteins were affinity purified from an overexpressing E. coli strain as previously described (4). An additional gel filtration purification step, using a Sephacryl column, was added to remove an apparently dimeric, iron-containing contaminant that ran identically to CueO on sodium dodecyl sulfate-polyacrylamide gel electrophoresis gels. The contaminant, which bound to a Strep-tag column and displayed an electronic absorption band at 410 nm, was completely removed by this final step, leaving highly pure CueO protein. CueO protein concentrations were determined spectroscopically (Varian Cary 300) using an ɛ280 of 63,036 M−1 cm−1.
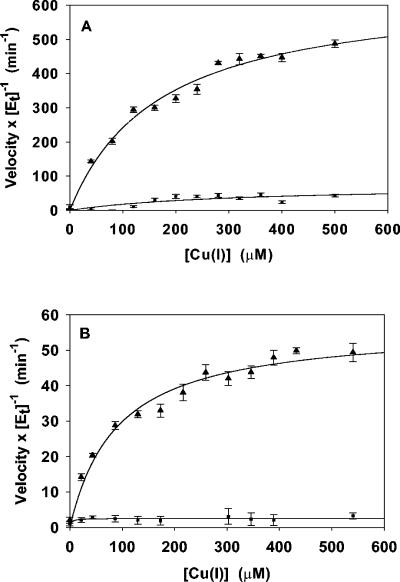
Cuprous oxidase activities at two different pH values were measured in terms of rates of oxygen consumption by using an oxygraph (Hansatech) as previously described (21). Initial rates were calculated manually from the recorded curves of oxygen concentration versus time. The substrate, Cu(I), was added as a complex [Cu(I)(MeCN)4]PF6 (Sigma-Aldrich), which releases free Cu(I) in solution. Stock solutions of Cu(I) were freshly prepared in degassed and argon-purged acetonitrile and subsequently diluted anaerobically by using gas-tight syringes. Reactions were initiated by addition of the substrate to an air-saturated mixture containing the enzyme, 100 mM buffer, 5% acetonitrile, and 1 mM Cu(II), added as CuSO4, for stimulating activity through binding at the fifth copper site (21). Buffers were chosen that provided the best stability for the Cu(I) substrate. Measurements at pH 5.0 were in 100 mM Tris-acetate, pH 7.0, in 100 mM morpholineethanesulfonic acid. The means of three activity measurements for each substrate concentration at each of the two pH values were plotted using SigmaPlot 7.0 (SSI, Richmond, Calif.) (Fig. 1), and kinetic constants Km and kcat were evaluated by using a least-squares fit of a Michaelis-Menten curve to a plot of the enzyme activities, after subtracting the baseline enzyme-independent oxidation of Cu(I) (Table 1).
FIG. 1.
Steady-state cuprous oxidase activity catalyzed by wild-type CueO (triangles) and C500S mutant CueO (squares) at pH 5.0 (A) or 7.0 (B). Aerial oxidation of Cu(I) in the absence of enzyme was subtracted from the wild-type and C500S mutant CueO activities to obtain fitted values of Km and kcat. Plotted are the means and standard errors (error bars) for three measurements at each substrate concentration, as the initial velocity of oxygen consumption normalized by the enzyme concentration, along with fitted curves. The average of three measurements of enzyme-independent Cu(I) oxidation was subtracted from the enzyme-dependent value, and the standard errors were obtained through combination of unbiased standard deviations for the two averages.
TABLE 1.
Multicopper oxidase activities
| Protein | Substratea | Km (μM) | kcat (min−1) | kcat/Km (μM−1 min−1) |
|---|---|---|---|---|
| CueO | Cu(I) | 165 ± 24b | 914 ± 55 | 5.5 |
| CueO + Cu(II)c | Cu(I) | 169 ± 24 | 651 ± 37 | 3.9 |
| CueO + Cu(II) | Cu(I) (pH 7.0) | 90 ± 14 | 57 ± 2 | 0.63 |
| CueO | Fe(II) | NMd | NM | |
| CueO + Cu(II) | Fe(II) | 129 ± 15 | 215 ± 9 | 1.7 |
| CueO | Mn(II) | NM | NM | |
| CueO + Cu(II) | Mn(II) | NM | NM | |
| Yeast Fet3e | Cu(I) | 38 ± 3 | 79 ± 3 | 2.1 |
| Human Cpe | Cu(I) | 37 ± 5 | 22.5 ± 1 | 0.61 |
Substrates were added as Cu(I)(MeCN)4, Fe(NH4)2(SO4)2, and MnCl2. Measurements were at pH 5.0 except where noted.
Value ± estimated error from nonlinear fit.
Cu(II) was included at 1 mM.
NM, no measurable activity.
Fet3 and ceruloplasmin data were taken from reference 24.
The kinetic analyses established wild-type CueO as an excellent cuprous oxidase, with a kcat 8- to 30-fold higher and a Km ∼4-fold higher than for yeast Fet3 and human ceruloplasmin (Table 1). The measured Km should be viewed as an upper limit, since it does not directly measure the free concentration of Cu(I) but rather the concentration of Cu(I) released from the acetonitrile complex. The higher kcat observed at pH 5.0 than at pH 7.0 in CueO was consistent with the pH dependence of phenoloxidase activity previously described (21). Surprisingly, however, the cuprous oxidase activity was independent of Cu(II) addition (Table 1), unlike with other substrates. To ensure the oxidase activity was due to CueO catalysis, since the Cu(I)-acetonitrile complex is labile, we repeated the measurements with C500S CueO, which is completely inactive. As expected, oxygen consumption in the presence of the mutant protein was the same as background (Fig. 1). We also examined Mn(II), which was inactive as a substrate, and Fe(II), which was an excellent substrate in the presence of Cu(II) (Table 1) but inactive in the absence of Cu(II), consistent with previous measurements (10, 21).
Thus, CueO is an excellent cuprous oxidase, displaying kcat/Km values two- to fourfold greater than for Fet3 and human ceruloplasmin (24) and kcat/Km values two- to threefold greater than for ferroxidase activity (Table 1). Based on these findings, it seems likely that CueO functions in vivo to convert extremely toxic Cu(I) to Cu(II), which is less toxic. During copper stress, CueO is coinduced with CopA, which pumps Cu(I) from the cytoplasm into the periplasm, where CueO resides, providing a linked pathway for detoxification. Cu(I) is the only CueO substrate so far identified that is active in the absence of Cu(II). Preliminary crystallographic data (S. K. Singh and W. R. Montfort, unpublished data) suggest that Cu(I) binds in the same labile site previously identified as the regulatory Cu(II) binding site (21), where it could directly reduce the T1 copper.
The identification of cuprous oxidase activity may also serve to resolve another puzzling aspect of CueO, that of the copper-stimulated catecholate oxidase activity. Such compounds will reduce Cu(II) to Cu(I) (6, 9, 11), which is then a substrate for CueO. Thus, some of the Cu(II)-stimulated catecholate oxidase activity is, in actuality, cuprous oxidase activity. In this scenario, oxidation of catecholate may be from reduction of Cu(II) in addition to any direct oxidation by CueO. This possibility has implications for human health: environmental exposure to copper, or lower ceruloplasmin levels as in Wilson's disease or aceruloplasminemia, is correlated with an increased incidence of Parkinson's disease (3, 8, 16). If, in fact, copper toxicity due to the loss of ceruloplasmin activity leads to neuronal loss, administration of the current drug of choice, l-DOPA, could lead to an unintended increase in copper toxicity through the reduction of Cu(II) to Cu(I).
Acknowledgments
This work was supported in part by National Institute of Environmental Health Sciences grant ESO4940 with funds from the U.S. Environmental Protection Agency to C.R. and by National Institutes of Health grant HL62969 to W.R.M.
We thank Michael Wells for use of his oxygraph.
REFERENCES
- 1.Finney, L. A., and T. V. O'Halloran. 2003. Transition metal speciation in the cell: insights from the chemistry of metal ion receptors. Science 300:931-936. [DOI] [PubMed] [Google Scholar]
- 2.Franke, S., G. Grass, and D. H. Nies. 2001. The product of the ybdE gene of the Escherichia coli chromosome is involved in detoxification of silver ions. Microbiology 147:965-972. [DOI] [PubMed] [Google Scholar]
- 3.Gorell, J., E. Peterson, B. Rybicki, and C. Johnson. 2004. Multiple risk factors for Parkinson's disease. J. Neurol. Sci. 217:169-174. [DOI] [PubMed] [Google Scholar]
- 4.Grass, G., and C. Rensing. 2001. CueO is a multi-copper oxidase that confers copper tolerance in Escherichia coli. Biochem. Biophys. Res. Commun. 286:902-908. [DOI] [PubMed] [Google Scholar]
- 5.Grass, G., and C. Rensing. 2001. Genes involved in copper homeostasis in Escherichia coli. J. Bacteriol. 183:2145-2147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Grass, G., K. Thakali, P. E. Klebba, D. Thieme, A. Müller, G. F. Wildner, and C. Rensing. 2004. Linkage between catecholate siderophores and the multicopper oxidase CueO in Escherichia coli. J. Bacteriol. 186:5826-5833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Huston, W., M. Jennings, and A. McEwan. 2002. The multicopper oxidase of Pseudomonas aeruginosa is a ferroxidase with a central role in iron acquisition. Mol. Microbiol. 45:1741-1750. [DOI] [PubMed] [Google Scholar]
- 8.Johnson, S. 2001. Is Parkinson's disease the heterozygote form of Wilson's disease: PD = 1/2 WD? Med. Hypotheses 56:171-173. [DOI] [PubMed] [Google Scholar]
- 9.Kamau, P., and R. Jordan. 2002. Kinetic study of the oxidation of catechol by aqueous copper(II). Inorg. Chem. 41:3076-3083. [DOI] [PubMed] [Google Scholar]
- 10.Kim, C., W. W. Lorenz, J. T. Hoopes, and J. F. D. Dean. 2001. Oxidation of phenolate siderophores by the multicopper oxidase encoded by the Escherichia coli yacK gene. J. Bacteriol. 183:4866-4875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Li, Y., M. Trush, and J. Yager. 1994. DNA damage caused by reactive oxygen species originating from a copper-dependent oxidation of the 2-hydroxy catechol of estradiol. Carcinogenesis 15:1421-1427. [DOI] [PubMed] [Google Scholar]
- 12.Mercer, J. F., and R. M. Llanos. 2003. Molecular and cellular aspects of copper transport in developing mammals. J. Nutr. 133:1481S-1484S. [DOI] [PubMed] [Google Scholar]
- 13.Munson, G. P., D. L. Lam, F. W. Outten, and T. V. O'Halloran. 2000. Identification of a copper-responsive two-component system on the chromosome of Escherichia coli K-12. J. Bacteriol. 182:5864-5871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Outten, F. W., D. L. Huffman, J. A. Hale, and T. V. O'Halloran. 2001. The independent cue and cus systems confer copper tolerance during aerobic and anaerobic growth in Escherichia coli. J. Biol. Chem. 276:30670-30677. [DOI] [PubMed] [Google Scholar]
- 15.Outten, F. W., C. E. Outten, J. Hale, and T. V. O'Halloran. 2000. Transcriptional activation of an Escherichia coli copper efflux regulon by the chromosomal MerR homologue, CueR. J. Biol. Chem. 40:31024-31029. [DOI] [PubMed] [Google Scholar]
- 16.Paris, I., A. Dagnino-Subiabre, K. Marcelain, L. Bennett, P. Caviedes, R. Caviedes, C. Azar, and J. Segura-Aguilar. 2001. Copper neurotoxicity is dependent on dopamine-mediated copper uptake and one-electron reduction of aminochrome in a rat substantia nigra neuronal cell line. J. Neurochem. 77:519-529. [DOI] [PubMed] [Google Scholar]
- 17.Peña, M. M. O., J. Lee, and D. J. Thiele. 1999. A delicate balance: homeostatic control of copper uptake and distribution. J. Nutr. 129:1251-1260. [DOI] [PubMed] [Google Scholar]
- 18.Rensing, C., and G. Grass. 2003. Escherichia coli mechanisms of copper homeostasis in a changing environment. FEMS Microbiol. Rev. 27:197-213. [DOI] [PubMed] [Google Scholar]
- 19.Requena, J. R., D. Groth, G. Legname, E. R. Stadtman, S. B. Prusiner, and R. L. Levine. 2001. Copper-catalyzed oxidation of the recombinant SHa(29-231) prion protein. Proc. Natl. Acad. Sci. USA 98:7170-7175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Roberts, S. A., A. Weichsel, G. Grass, K. Thakali, J. T. Hazzard, G. Tollin, C. Rensing, and W. R. Montfort. 2002. Crystal structure and electron transfer kinetics of CueO, a multicopper oxidase required for copper homeostasis in Escherichia coli. Proc. Natl. Acad. Sci. USA 99:2766-2771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Roberts, S. A., G. F. Wildner, G. Grass, A. Weichsel, A. Ambrus, C. Rensing, and W. R. Montfort. 2003. A labile regulatory copper ion lies near the T1 copper site in the multicopper oxidase CueO. J. Biol. Chem. 278:31958-31963. [DOI] [PubMed] [Google Scholar]
- 22.Shi, X., C. Stoj, A. Romeo, D. J. Kosman, and Z. Zhu. 2003. Fre1p Cu2+ reduction and Fet3p Cu1+ oxidation modulate copper toxicity in Saccharomyces cerevisiae. J. Biol. Chem. 278:50309-50315. [DOI] [PubMed] [Google Scholar]
- 23.Solomon, E. I., U. M. Sundaram, and T. E. Machonkin. 1996. Multicopper oxidases and oxygenases. Chem. Rev. 96:2563-2605. [DOI] [PubMed] [Google Scholar]
- 24.Stoj, C., and D. Kosman. 2003. Cuprous oxidase activity of yeast Fet3p and human ceruloplasmin: implication for function. FEBS Lett. 554:422-426. [DOI] [PubMed] [Google Scholar]
- 25.Torsdottir, J., S. Kristinsson, J. Sveinbjornsdottir, J. Snaedal, and T. Johannesson. 1999. Copper, ceruloplasmin, superoxide dismutase and iron parameters in Parkinson's disease. Pharmacol. Toxicol. 85:239-243. [DOI] [PubMed] [Google Scholar]