Abstract
Here we describe ISHp609 of Helicobacter pylori, a new member of the IS605 mobile element family that is novel and contains two genes whose functions are unknown, jhp960 and jhp961, in addition to homologs of two other H. pylori insertion sequence (IS) element genes, orfA, which encodes a putative serine recombinase-transposase, and orfB, whose homologs in other species are also often annotated as genes that encode transposases. The complete four-gene element was found in 10 to 40% of strains obtained from Africa, India, Europe, and the Americas but in only 1% of East Asian strains. Sequence comparison of 10 representative ISHp609 elements revealed higher levels of DNA sequence matches (99%) than those seen in normal chromosomal genes (88 to 98%) or in other IS elements (95 to 97% for IS605, IS606, and IS607) from the same H. pylori populations. Sequence analysis suggested that ISHp609 can insert at many genomic sites with its left end preferentially next to TAT, with no target specificity for its right end, and without duplicating or deleting target sequences. A deleted form of ISHp609, containing just jhp960 and jhp961 and 37 bp of orfA, found in reference strain J99, was at the same chromosomal site in 15 to 40% of the strains from many geographic regions but again in only 1% of the East Asian strains. The abundance and sequence homogeneity of ISHp609 and of this nonmobile remnant suggested a recent bottleneck and then rapid spread in H. pylori populations, possibly selected by the contributions of the elements to bacterial fitness.
Insertion sequence (IS) elements are a diverse group of specialized DNA segments that move to new sites in prokaryotic and eukaryotic genomes by mechanisms that do not require extensive DNA sequence homology (for reviews see references 5, 7, and 19). They cause insertion mutations and genome rearrangements, affect nearby gene expression, and help mediate the spread of resistance and virulence determinants within and among species. Many IS elements specify just a single transposase protein that acts in concert with host proteins at each end of the element to mediate insertion. Other, more complex elements, such as Tn7, specify two or more proteins that act together as the transposase, plus additional proteins that help select insertion sites or affect the efficiency of transposition. Host proteins can also affect the frequency or specificity of transposition of certain elements. Many, but not all, species of elements terminate in short inverted repeats (size range, 9 to 40 bp) and generate short direct repeats of target sequences (typically 2 to 9 bp) when they transpose.
Each of the four known species of IS elements in H. pylori (IS605, IS606, IS607, and ISHp608) belongs to the distinctive IS605 mobile element family, and each seems to be chimeric, containing two transposition-related genes, orfA and orfB, that may have different phylogenetic origins (12, 14, 16). The IS605 element family is divisible into two subfamilies based on orfA homologies; in one subfamily, represented by IS607, orfA encodes a putative serine recombinase that helps IS607 transpose to multiple sites (in contrast, most serine recombinases mediate site-specific recombination), and in the other subfamily, represented by IS605, IS606, and ISHp608 (28 to 36% amino acid identity for sequences encoded by orfA genes), orfA encodes a transposase related to that encoded by IS200, a single-gene element that is widespread in enteric species (5, 19, 25) whose product is distinct from serine recombinase proteins. The proteins encoded by orfB genes of IS605 family members exhibit 25 to 35% amino acid identity to one another; their homologs in other species are often annotated in sequence databases as putative transposases, and they are also homologs of the protein encoded by gipA, a Salmonella prophage gene that enhances bacterial growth in Peyer's patches (22).
IS605, IS607, and ISHp608 each have been found to transpose in Escherichia coli (12, 14, 16). With each of the two elements tested (IS607 and ISHp608), transposition depended on orfA and not on orfB. Hence, the constant presence of orfB in IS605 family members suggested either involvement in transposition in certain species or a contribution to bacterial fitness (11a). Inspection of sequences at sites of insertion in H. pylori and E. coli indicated that (i) IS605, IS607, and ISHp608 insert with their left (orfA) ends immediately downstream of specific AT-rich sequences (5′-TTTAA or 5′-TTTAAC for IS605, 5′-TTTAT for IS606, and 5′-TTAC for ISHp608), and their right (orfB) ends seem to join to target DNAs nonspecifically; (ii) in contrast, IS607 whose orfA transposase gene is unrelated to the other genes, inserted preferentially between adjacent GG nucleotides in target DNA; (iii) none of these elements duplicated or deleted sequences at sites of insertion; and (iv) none contained terminal inverted repeats (12, 14, 16). The H. pylori ISHp609 element described here is nonrandomly distributed geographically and is unique among elements of the IS605 family in containing four open reading frames instead of the usual two.
MATERIALS AND METHODS
General methods.
Standard procedures were used for H. pylori growth on brain heart infusion agar (Difco) containing 10% horse blood in a microaerobic atmosphere (27). High-molecular-weight genomic DNA was isolated by a hexadecyltrimethylammonium method (3).
Specific PCR were carried out in 20-μl mixtures containing 5 to 10 ng of DNA, 0.1 U of Taq polymerase (Biolase; Midwest Scientific, St. Louis, Mo.) or the Expand High Fidelity Taq-Pwo polymerase mixture (Boehringer-Mannheim, Indianapolis, Ind.), 2.5 pmol of each primer, and each deoxynucleoside triphosphate at a concentration of 0.2 mM in a standard buffer for 30 cycles with the following cycling parameters: denaturation at 94°C for 30 s, annealing at a temperature appropriate for the primer sequence (generally 50°C) for 30 s, and DNA synthesis at 72°C for an appropriate time (1 min per kb). PCR primers are listed in Table 1.
TABLE 1.
Primersa
| Primer | Sequence | Location |
|---|---|---|
| ISHp609 primers | ||
| 609LE | CATTAAACTTTCAATTTAATTTTG | At the left end, forward |
| 609R-t1 | CTAAAAACTCTTTAATCTTATTAAAAGTA | At the right end, reverse |
| 609.F1 | CACAACAGGTATTAATGCTT | 1,083 bp from the right end, in orfB, forward |
| 609.R1 | CTTTAGCTCTTGTTTCCAGC | 1,985 bp from the left end, in orfB, reverse |
| 609t2-1b | TTACCTGCAAGCCATTAAGG | 1,037 bp from the right end, in orfB, forward |
| 609t2-2b | TTATGCATAAAGTTATTCAGC | 1,535 bp from the left end, in orfB, reverse |
| FlankL | CTTTATTTGGGAGATTTAGAAGC | 2,266 bp from the right end, in orf1, forward |
| 609.R6 | CCATTCTGCAATAATAGCTTCTC | 346 bp from the left end, in orf2, reverse |
| 609R3t1 | TAAATAGCTTTTCTACTAACTCATA | 1,001 bp from the left end, in orfA, reverse |
| 609.R5 | ATTAGCGTAGTTGTAAAGGTT | 1,199 bp from the left end, in orfB, reverse |
| Other primers | ||
| jhp959 | GTAGCACAACTCTTATGCGATG | 177 bp from the 3′ end of jhp959, forward |
| jhp962 | TGTATGTCATGCTGAGCGAAAAC | 371 bp from the 3′ end of jhp962, reverse |
| CR2-LF | CATACTAGACTTAAAGGACAGCA | Left of ISHp609 in CR2 clonec |
| CR2-RF | ATTTTGATTTTGTCTGTTACTTCGCAC | Right of ISHp609 in CR2 clonec |
| B43-LF | GCTATGGCAGAAAACATCTAT | Left of ISHp609 in HUP-B43 |
| B43-RF | GAGTTTCTAAATAATTCTCAT | Right of ISHp609 in HUP-B43 |
| B79-LF | CAGATTTACCCAAACTCACT | Left of ISHp609 in HUP-B79 |
| B79-RF | AAGGTTTTATCAAAGCCTATGC | Right of ISHp609 in HUP-B79 |
| LitA7-LF | GAGGTAATTTTTGAAATTTTAACAC | Left of ISHp609 in LitA7 |
| LitA7-RF | ATCAGGCTTTAGGGTATTCTTT | Right of ISHp609 in LitA7 |
| LitA38-LF | TTTATCCTTTCTTTATTATTAAACTTTC | Left of ISHp609 in LitA38 |
| LitA38-RF | ATTTTATGCGATTGCATTGAAAG | Right of ISHp609 in LitA38 |
Sequences of additional primers used for sequencing are available on request.
ISHp609var specific (Chennai4 strain).
See reference 6.
DNA sequencing was carried out by using a Big Dye Terminator DNA sequencing kit (Perkin-Elmer) and an ABI automated sequencer. Direct sequencing on chromosomal DNA was done with 5 μl of chromosomal DNA (1 to 2 μg), 1 μl of primer (10 pmol per μl), and 6 μl of Big Dye under the following conditions: 96°C for 5 min and then 90 cycles of denaturation at 96°C for 10 s, annealing at a temperature appropriate for the primer for 5 s, and extension at 60°C for 4 min under oil-free conditions (Perkin-Elmer 2400).
DNA sequence editing and analysis were performed with programs in the GCG package (Genetics Computer Group, Madison, Wis.), programs and data in the H. pylori genome sequence databases (2, 24), and BLAST and Pfam (version 14.0) homology search programs (http://www.ncbi.nlm.nih.gov/BLAST/BLAST.cgi; http://pfam.wustl.edu/hmmsearch.shtml).
Phylogenetic analysis.
H. pylori OrfA and OrfB IS element protein sequences were aligned with CLUSTALX by using a PAM250 amino acid substitution matrix. OrfA and OrfB neighbor-joining phylogenies were constructed by using the Jones-Taylor-Thornton distance matrix and variable rates among sites (modeled with a gamma distribution shape parameter, α = 0.5). Gaps in the protein alignments were deleted in pairwise comparisons. Phylogenetic analysis and DNA diversity calculation were done with Mega2.1 (www.megasoftware.net).
Bacterial strains and plasmids.
Most H. pylori strains used were obtained from the Berg laboratory collection and have been described in detail elsewhere (4, 13, 15, 16, 20). The H. pylori strains used in this study included 71 Peruvian strains, 24 Spanish strains, 47 Lithuanian strains from Vilnius, 28 African strains (16 strains from Soweto in South Africa and 12 strains from Gambia), 69 Indian strains (48 strains from Calcutta, 15 strains from Chennai, and 6 strains from the Santal tribe), 71 Japanese strains (24 strains from Ube and 47 strains from Fukui), 46 strains from Alaskan natives, 47 Chinese strains (40 strains from YunNan and 7 strains from Changle), and 47 Korean strains.
Plasmids carrying an intact ISHp609 element marked with chloramphenicol resistance downstream of the orfB stop codon were constructed, and transposition assays in E. coli were carried out as described previously for ISHp608 (16).
GenBank accession numbers of DNA sequences.
The sequence of ISHp609 from strain HUP-B43 corresponds to nucleotides 14337 through 16733 of the 33,671-nucleotide sequence of the plasticity zone in GenBank accession number AY487825. The other three full-length ISHp609 elements were accessioned in GenBank as follows: from strain HUP-B79, nucleotides 287 through 2684 of the 3,084 nucleotides in accession number AY639112; from strain LitA7, nucleotides 381 through 2794 of the 2,875 nucleotides in accession number AY639110; and from strain LitA38, nucleotides 20 through 2417 of the 2,603 nucleotides in accession number AY639111. The full length of the rare type of ISHp609 element (ISHp609var) from strain Chen4 corresponds to nucleotides 313 through 2346 of the 2,496 nucleotides in accession number AY639119. The GenBank accession numbers for partial ISHp609 sequences (all but 24 nucleotides on the left and 34 nucleotides on the right due to the positions of PCR binding sites) were as follows: strain HUP-B80, AY639113; Alaska64, AY639115; Alaska97, AY639116; AfricaR48, AY639114; I-86, AY639117; and SJM27, AY639118.
RESULTS
ISHp609 discovery and sequence.
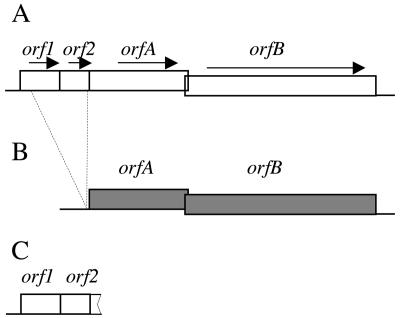
The 2.4-kb ISHp609 element was discovered while the plasticity zone of strain HUP-B43 (accession number AY487825) was being sequenced as an insertion in gene jhp928 (the gene designations used match those of reference strains J99 [jhp] and 26695 [hp] when homologies are high [2, 24]). It contained four open reading frames designated orf1, orf2, orfA, and orfB (Fig. 1A).
FIG. 1.
Structures of ISHp609 and related elements. (A) Full-length predominant ISHp609 type with four characteristic open reading frames and 99% DNA identity, independent of geographic origin. Sequence analysis of 10 full-length elements from Spanish strains (HUP-B43, HUP-B79, and HUP-B80; accession numbers AY487825, AY639112, and AY639113), Lithuanian strains (Lit-7 and Lit38; accession numbers AY639110 and AY639111), Alaskan strains (Al64 and Al97; accession numbers AY639115 and AY639116), a Peruvian strain (SJM27; accession number AY639118), an Indian strain (I-86; accession number AY639117), and an African strain (R48; accession number AY639114) showed limited internal divergence. In strain Lit7 orfA contained a frameshift due to a 14-bp duplication; in strain HUP-B79 orfA contained an in-frame stop codon due to a G-to-T substitution; and in strain I-86 orf1 contained an in-frame stop codon due to a C-to-T substitution. (B) ISHp609var. This rare variant element was found in one Indian strain (Chennai4; accession number AY639119) with 81% DNA identity to the predominant type. ISHp609var has full-length orfA and orfB genes, although orfB is probably inactive due to a frameshift. It lacks orf2 and most of orf1, and it has the first 79 bp of orf1, but there is no start codon at its left end. (C) orf1-2 remnant. This DNA segment consists of orf1 (jhp960 in strain J99), orf2 (jhp961 in strain J99), and the first 37 bp of orfA and is located between homologs of jhp959 and jhp962 in most or all strains (as determined by PCR with primers jhp959 and jhp962, primers jhp959 and 609.R6, and primers FlankL and jhp962).
The ISHp609 orfA sequence (210 codons) is homologous to the orfA sequence of IS607 (32% protein identity and 58% protein similarity) (Fig. 2A), and its 5′ end specifies a helix-turn-helix domain (conserved domain cd01104, HTH_MlrA; E value, 3e-04) that might mediate DNA binding. Extrapolation from IS607 studies (14) suggested that orfA might encode a transposase. The ISHp609 orfA product also belongs to a widespread protein family whose members are generally designated serine recombinases or site-specific integrase-resolvases. One of the strongest homologies is with a putative serine recombinase gene of Thermoanaerobacter tengcongensis MB4T (tte0714; 47% protein identity and 68% protein similarity).
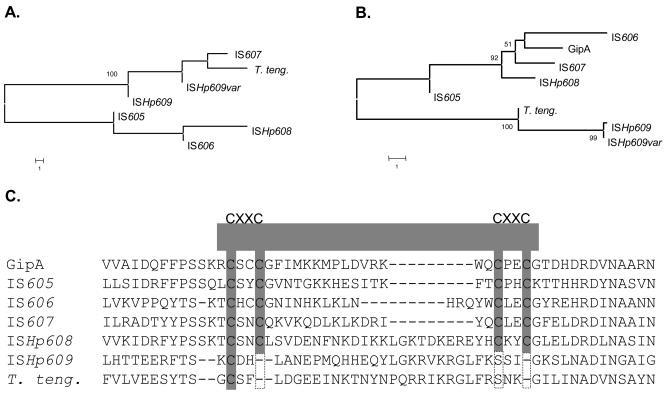
FIG. 2.
Phylogenetic relationships among H. pylori IS elements. (A) OrfA and homologs. Two subfamilies were identified, one represented by OrfAs of IS605 (accession number NP_208326), IS606 (accession number AAD11513), and ISHp608 (accession number AAL06576), which are not considered to encode serine recombinases based on amino acid homologies, and the other represented by IS607 (accession number AAF05600), ISHp609 (accession number AAR83266.1), ISHp609var (accession number AY639119), and the closest homolog in T. tengcongensis (T. teng.) (tte0714; accession number AAM23976), which are thought to encode serine recombinases. Branches with significant bootstrap support (≥50) are indicated. Bar = 1 amino acid substitution per site. (B) OrfB and homologs. OrfBs of H. pylori IS605 (accession number NP_208324), IS606 (accession number AAD11514), IS607 (accession number AAF05601), ISHp608 (accession number AAL06577), and Salmonella's GipA protein (accession number NP_752781) form an OrfB subfamily different from that of ISHp609 (accession number AAR83267.1), ISHp609var (accession number AY639119), and the closest homolog in T. tengcongensis (tte0715; accession number AAM23977). (C) Partial C-terminal sequence alignment of H. pylori IS element OrfBs, GipA (22), and the corresponding sequence in T. tengcongensis. A single C-terminal Zn(II) binding tetracysteine motif, CX(2)CX(15)CX(2)C (C4-type zinc finger), is well conserved among IS605, IS606, IS607, and ISHp608 OrfBs and GipA and might potentially facilitate DNA or RNA binding or protein-protein interaction. Notably, this motif is not present in ISHp609 OrfB (both predominant and variant).
The ISHp609 orfB sequence (409 codons) exhibits weak homology to the IS605 orfB sequence (22% protein identity and 42% protein similarity) (Fig. 2B) and more generally to members of a widespread gene family that are annotated as genes that encode transposases but that also include the gipA virulence gene of Salmonella. Again, one of the strongest homologies is with an open reading frame in T. tengcongensis MB4T (tte0715; 42% protein identity and 58% protein similarity), which is adjacent to the orfA homolog, tte0714. The C-terminal tetracysteine Zn(II) binding motif CX(2)CX(15)CX(2)C (C4-type zinc finger) found in GipA (22) and the OrfB proteins encoded in IS605, IS606, IS607, and ISHp608 was absent from the ISHp609 OrfB protein and also from its close homolog in T. tencongensis (Fig. 2C).
Two short coding sequences in ISHp609, orf1 (85 codons) and orf2 (61 codons), closely matched two genes of reference strain J99 whose functions are unknown, jhp960 (99% DNA identity and 100% protein identity) and jhp961 (98% DNA identity, 96% protein identity, and 98% protein similarity). These open reading frames are located between jhp959 and jhp962 in strain J99, not within jhp928, but they are absent from the other fully sequenced genome (strain 26695) (24). Although annotated as a gene whose function is unknown, orf1 belongs to a gene family whose protein products generally contain a characteristic ligand binding domain next to a helix-turn-helix DNA binding domain (Pfam domain 03681, UPF0150; 14 to 68 bp; E value, 1e−10), and the protein encoded by orf2 exhibits a conserved predicted periplasmic or secreted lipoprotein motif throughout its length (61 amino acids; cluster of orthologous groups, COG1724; E value, 6e−13). Small genes related to orf1 and orf2 are found in many bacterial species, in some cases together (for example, ssr1765 and ssr1766 in Synechocystis sp. strain PCC 6803 [GenBank accession numbers BAA16930 and BAA16931], with 46% amino acid identity and 70% amino acid similarity to orf1 and 35% amino acid identity and 38% amino acid similarity to orf2, respectively). However, most homologs of either orf1 or orf2 occur singly and are not linked to a homolog of the other gene in other bacterial species. No orfA or orfB homologs were found next to ssr1765 and ssr1766 in Synechocystis, nor were orf1 or orf2 homologs found next to the orfA and orfB homologs (tte0714 and tte0715) in T. tengcongensis.
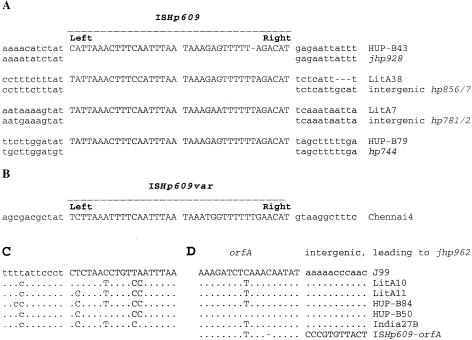
ISHp609 elements and adjacent DNAs from three additional strains (strains HUP-B79, LitA7, and LitA38) were sequenced by primer walking on genomic DNAs in order to better understand the structure of the element and its insertion specificity. All three elements were 2.4 kb long, contained four open reading frames (orf1, orf2, orfA, and orfB) matching the open reading frames found in strain HUP-B43, and were 99% identical in DNA sequence to one another. A comparison of sequences flanking ISHp609 with corresponding empty sites in the 26695 and/or J99 genome showed that the element was located at a different genetic locus in each strain (Fig. 3A). These results, coupled with PCR data (see below), indicate that ISHp609 is transposable and that it can insert into many genomic sites. Sequence comparisons also identified the probable termini of the element and its insertion specificity. Based on data summarized in Fig. 3A, the ends of ISHp609 seemed to be 5′-TAT or CAT on the left and 5′-CAT on the right. The left end was inserted preferentially next to 5′-TAT, whereas there seemed to be no target specificity for the right end, and there was no evidence of target sequence duplication or deletion during insertion.
FIG. 3.
Terminal sequences of ISHp609, ISHp609var, and the orf1-2 remnant and their sites of insertion in H. pylori. ISHp609, ISHp609var, and orf1-2 remnant termini are in uppercase type. Flanking DNA and empty sites in reference strains 26695 and J99 (gene designations beginning with hp and jhp, respectively) are in lowercase type. (A) ISHp609 predominant type. (B) ISHp609var type. Terminal sequences were extrapolated from a comparison with sequences of the predominant ISHp609 type, because its flanking sequences did not have homology with known H. pylori sequences and therefore the site of insertion (empty site) was not known. (C) Predicted left end of the orf1-2 remnant and its flanking sequences. Sites of insertion could not be determined precisely due to local sequence heterogeneity in strains lacking this element (intergenic region between jhp959 and jhp962) (see Fig. S2 in the supplemental material). (D) Predicted right end of the orf1-2 remnant compared to the corresponding region of orfA in the ISHp609 sequence.
Further IS element sequencing was done by using nearly full-length ISHp609 PCR products from six additional strains from five geographic regions (Spain, Africa, Alaska, India, and Peru) and with primers located just within the ISHp609 ends (primers 609LE and 609R-t1) (GenBank accession numbers AY636113 to AY636118). Each element had the same number and organization of open reading frames, and each was ≥99% identical in DNA sequence to other elements, regardless of the geographic origin. The ratio of synonymous to nonsynonymous changes in the four open reading frames ranged from 3.2 to 7.0 (average, 5.0) (Table 2), values that are typical of H. pylori housekeeping genes (1, 9).
TABLE 2.
DNA divergence in ISHp609
| Gene | Length (bp) | πJCa | πSJCb | πNSJCc | Ks/Kad |
|---|---|---|---|---|---|
| orf1 | 255 | 0.009 | 0.028 | 0.004 | 7 |
| orf2 | 183 | 0.02 | 0.045 | 0.014 | 3.2 |
| orfA | 633 | 0.008 | 0.018 | 0.005 | 3.6 |
| orfB | 1,227 | 0.01 | 0.03 | 0.005 | 6 |
| Overall | 2,298 | 0.01175 | 0.03025 | 0.007 | 4.95 |
Nucleotide diversity per site. Nucleotide diversity calculations were corrected for multiple hits by using the Jukes-Cantor (JC) correction.
Nucleotide diversity per synonymous site.
Nucleotide diversity per nonsynonymous site.
Ratio of synonymous to nonsynonymous changes.
In addition to this predominant ISHp609 type, we also found one variant element (strain Chennai4) based on weaker-than-normal PCR amplification with orfB-specific primers 609.F1 and 609.R1. This element, ISHp609var, exhibited only 81% DNA homology to other ISHp609 elements and consisted of the 5′ end of orf1 (79 bp, 79% DNA homology) without an obvious start codon, linked directly to full-length orfA and orfB (although orfB contains a frameshift due to a 5-bp internal deletion) (Fig. 1B). ISHp609var was inserted into an unknown chromosomal site with its inferred left end next to a TAT sequence, which suggested insertion specificity matching that of the predominant ISHp609 type (Fig. 3B).
In further experiments, a plasmid clone of ISHp609 from strain HUP-B43, marked with a chloramphenicol resistance determinant, was used to select for transposition to the F factor pOX38 in E. coli in a standard mating-out assay, essentially as done previously with IS607 and ISHp608 (14, 16). However, no transposition (<10−9) of this marked ISHp609 element was detected in any of four repetitions of this assay. In contrast, IS607 and ISHp608 transposed at frequencies of about 10−7 in equivalent assays (14, 16).
ISHp609 geographic distribution and structural analysis.
The frequency of ISHp609 carriage in various H. pylori populations was estimated by PCR with 479 strains by using two orfB-specific primers (primers 609.F1 and 609.R1). ISHp609 was found in 35 to 40% of the strains from Europe (Spain and Lithuania), in 20 to 40% of the strains from the Americas (Peru, Guatemala, and Alaska), and in 10 to 15% of the strains from India and Africa but in only 1% of the strains from East Asia (Table 3). All 479 strains, including 68 additional strains from India, were tested for the variant type (ISHp609var) that had been found in one Indian strain by using specific primers (primers 609t2-1 and 609t2-2). No additional strains harboring this variant were found. This seemed to be reminiscent of the single case of a rare type 3 ISHp608 element found in one Indian strain but not in 116 other Indian strains or in 606 strains from other regions (16).
TABLE 3.
Distribution of ISHp609 in different geographic regions
| Strain origin | No. of strains | No. (%) ISHp609 positivea | No. (%) with orf1-2 remnant onlyb |
|---|---|---|---|
| Europe | |||
| Lithuania | 47 | 17 (36) | 11 (23) |
| Spain | 24 | 10 (42) | 3 (13) |
| The Americas | |||
| Alaska | 46 | 20 (43) | 12 (26) |
| Peru | 71 | 12 (17) | 13 (18) |
| Guatemala | 29 | 8 (28) | 6 (21) |
| Africa | 28 | 3 (11) | 11 (39) |
| South Asia: India | 69 | 11 (16) | 14 (20) |
| East Asia | |||
| Japan | 71 | 0 | 1 (1) |
| China | 47 | 0 | 1 (2) |
| Korea | 47 | 2 (4) | 0 |
ISHp609-positive strains were identified by PCR with four sets of primers (primers 609.F1 and 609.R1, primers FlankL and 609R3t1, primers FlankL and 609.R5, and primers FlankL and 609.R1 [Table 1]). Thirty-three of 83 ISHp609 elements were truncated, mostly from the right end. PCR tests indicated that 16 of the 83 ISHp609-positive strains also contained the orf1-2 remnant (see below), in each case at the location occupied in strain J99, between jhp959 and jhp962.
The orf1-2 remnant contains orf1, orf2, and the 5′ end of orfA, as in strain J99 (see text). PCR (with primers jhp959 and jhp962, primers jhp959 and 609.R6, and primers FlankL and jhp962) located it between jhp959 and jhp962 in 64 of 72 cases, as in strain J99 (exceptions might have been due to sequence divergence in primer binding sites).
Strain J99 contains close homologs of orf1 and orf2 (jhp960 and jhp961) and the first 37 bp of orfA, as noted above. This suggested that this strain might contain a remnant (deleted) version of ISHp609 (Fig. 1C). Related segments of the same size were found by PCR (with primers FlankL and 609.R6) in 15 to 40% of the strains from various geographic regions, some of which also contained full-length ISHp609 elements in other genome locations. As observed with full-length ISHp609, however, this remnant was also rare in East Asian strains (1%) (Table 3). Further PCR tests with jhp959- and jhp962-specific primers located the remnant between jhp959 and jhp962, as in strain J99, in at least 64 of the 72 cases tested. This inference was confirmed by sequencing PCR products generated with jhp959 and jhp962 primers from five strains (LitA10, LitA11, HUP-B50, HUP-B84, and India27); in each case orf1 (jhp960), orf2 (jhp961), and the 5′ end of orfA were inserted between jhp959 and jhp962, exactly as they are in J99 (predicted left and right ends of the remnant element and insertion site are shown in Fig. 3C and D, respectively). The conservation of the chromosomal location and of orf1, orf2, and the 5′ end of orfA sequences suggests that this block of DNA (called the orf1-2 remnant) resulted from a deletion within a full-length ISHp609 element and loss of transposition ability and that it spread through H. pylori populations by interstrain recombination. High sequence homology (99%) was also found throughout this segment, except for the 3′ end of the jhp959 sequence, which is highly variable (70 to 90% DNA identity) (see Fig. S1 in the supplemental material).
The connection between jhp959 and jhp962 was highly conserved among all H. pylori strains, independent of the geographic origin; PCR with primers jhp959 and jhp962 showed that jhp959 and jhp962, which flanked the orf1-2 remnant whenever it was present, were next to one another in at least 96% of the orf1-2 remnant-free strains. This included reference strain 26695, in which the hp422 gene, a close homolog of jhp962 (100% protein identity), is adjacent to hp423, a distant homolog of jhp959 (49% protein identity and 64% protein similarity).
To try to define a precise insertion site of the orf1-2 remnant, we sequenced the putative empty site region in 15 strains that lacked this remnant. Here too jhp959 sequences and the intergenic region leading to jhp962 were highly divergent, both in length and in base substitution differences (25 to 30% DNA divergence) (see Fig. S2 in the supplemental material). Consequently, no unique empty site sequence could be discerned. The reason for this extreme diversity is not known. One possible explanation involves slipped strand mispairing during normal DNA replication; an alternative explanation involves deletion of an element and error-prone gap repair.
Other deletions in ISHp609 were also common; up to one-half of ISHp609 elements were truncated, mostly from the right end, terminating at different points in orfB or orfA sequences (Table 2). A distinct 0.9-kb internal deletion due to recombination between 11-bp direct repeats (CCTT[T/G]CTAAAA) located in orf1 and the 3′ end orfA was found in ISHp609 in three of nine Peruvian strains and two of nine Spanish strains (this study). This sequence matched a separately reported sequence from a Costa Rican strain (clone CR2 in reference 6; accession number AF326626). This 0.9-kb deletion was not found in any ISHp609 element from 15 Lithuanian, 10 Alaskan, 8 Guatemalan, 4 Indian, 3 African, and 2 Korean strains. In all six cases the ISHp609 element with the 0.9-kb internal deletion was inserted at the same chromosomal site that was defined by PCR (with primers CR2-LF and CR2-RF). Because the 0.9-kb deletion is less widely disseminated than the orf1-2 remnant, it may have arisen more recently, but it also may have been spread by interstrain recombination. Full-length ISHp609 sequences from two other strains (one Lithuanian strain and one Guatemalan strain) were found in the same location as the elements containing the 0.9-kb internal deletion.
PCR was used to test for ISHp609 at each of the other four chromosomal sites identified by sequencing (Fig. 3A) (locations in strains HUP-B43, HUP-B79, LitA7, and LitA38) and also the jhp959-jhp962 location of the orf1-2 remnant (with primers in Table 2) in 60 ISHp609-positive strains from Spain, Peru, Guatemala, Lithuania, Alaska, Africa, India, and Korea. ISHp609 was found at the HUP-B43 site (in the jhp928 homolog) in two of four ISHp609-positive Indian strains but not in the other 56 ISHp609-positive strains from various countries. No other strain was found to carry ISHp609 at the specific sites containing this element in strains LitA7, LitA38, and HUP-B79. Also, no full-length ISHp609 element was found between jhp959 and jhp962, the site normally occupied by the orf1-2 remnant.
DISCUSSION
The ISHp609 element described here is the fifth member of the IS605 mobile element family found in H. pylori and is novel since it contains four open reading frames instead of the usual two. In addition to orfA and orfB, homologs of which are present in other H. pylori IS elements, it also has two other open reading frames, orf1 and orf2, which are members of short, conserved gene families whose functions are unknown that are widespread among eubacterial species. A divergent variant of ISHp609 was also found, but in only 1 of the 479 strains tested.
ISHp609 was more abundant in strains from Europe, the Americas, and South Asia (10 to 40%) than in strains from East Asia (1%) (Table 3). This is reminiscent of the ISHp608 distribution (16). The difference between East Asian and other strains in terms of the relative abundance of the ISHp608 and ISHp609 elements is in accord with the major geographic differences in the H. pylori gene pool, as determined by PCR or multilocus sequence typing of virulence-associated and ordinary housekeeping genes (1, 9, 15, 26, 28), and also with the geographic differences in the sequences of orfA and orfB from IS605 and IS607 elements (11a). This geographic partitioning can be ascribed to H. pylori's preferential transmission within families and local communities rather than in sweeping epidemics (10, 21).
The sequences of ISHp609 elements from various geographic regions are more closely related to one another (99%) than normal chromosomal gene sequences from the same populations are. This is reminiscent of patterns seen with IS elements of natural isolates of E. coli, where the sequences within each IS element type are also more highly conserved than the sequences of chromosomal genes are (11). This, plus the frequent occurrence of elements on bacterial plasmids, suggested that E. coli IS elements were spread easily among bacterial lineages by conjugation and transposition (18). Such easy spread would have allowed little time to accumulate neutral variation and would have resulted in the observed sequence homogeneity. Most strains of H. pylori are readily transformable, and extensive interstrain recombination is evident in the gene pool (1, 8, 13, 23), although it is restricted geographically because transmission is highly localized. The distribution and sequence uniformity of ISHp609 suggest that this element entered the H. pylori gene pool recently on the evolutionary time scale (perhaps after separation of East Asian strains from other H. pylori strains) and spread rapidly, leading to its present abundance in some regions (up to 40% of the strains). This suggests that there was selection for ISHp609 carriage, stemming from a significant contribution to H. pylori host fitness and/or strong molecular drive (selfish DNA). The similar sequence uniformity and distribution of the orf1-2 remnant (orf1, orf2, and 5′ end of orfA) also favor a selection model, but one in which orf1 and/or orf2 contributes to fitness. The abundance of truncated (inactive) ISHp609 elements also suggests such dynamics, but with selection for mutations that rendered many elements inactive. This is reminiscent of the dynamics of P elements in Drosophila (17).
The ISHp609 gene(s) that mediates transposition has not been identified experimentally because we could not, in several tries, detect this element's movement in E. coli. The many genomic locations of ISHp609 in H. pylori populations imply that there is easy transposition in this host and thus perhaps involvement of a host factor that E. coli K-12 lacks. The constancy of the genomic positions of two ISHp609 deletion variants (orf1-2 remnant and 0.9-kb internal deletion) suggests that neither orfB (intact in the 0.9-kb deletion element) nor orf1 and orf2 (intact in the orf1-2 remnant) are sufficient for ISHp609 transposition. Rather, orfA of ISHp609 may be needed for movement of this element, as is the case with orfA homologs of IS607 and ISHp608 (14, 16).
Other questions concerning transposition mechanisms emerged from the analysis of ISHp609 sequences. The OrfA phylogenies (Fig. 2A) illustrate two distinct subfamilies, one represented by the orfA genes of IS607 and ISHp609, which appear to encode serine recombinases, and the other represented by the orfA genes of IS605, IS606, and ISHp608, which encode another type of transposition-associated function. Despite ISHp609 orfA's homology to IS607's putative transposase gene, its insertion specificity (downstream of 5′-TAT) seems to be more closely related to that of the IS605-IS606-ISHp608 subfamily (downstream of 5′-TTTAA or 5′-TTTAAC, 5′-TTTAT, and 5′-TTAC) than to that of IS607 (between GG). Further studies are needed to determine if this stems from functional differences in IS element-encoded proteins or in host factors with which they interact. In terms of orfB phylogenies (Fig. 2B), the orfB gene of ISHp609 is more distant from the gipA Salmonella virulence gene than the genes of other IS605 family members are. Therefore, together with its homolog in T. tengcongensis, this gene could be considered a member of a distinct subfamily. Most striking in this regard is the C-terminal tetracysteine Zn(II) binding motif CX(2)CX(15)CX(2)C (C4-type zinc finger) that could potentially facilitate DNA or RNA binding or protein-protein interaction. This motif is found in GipA and the orfB products of IS605, IS606, IS607, and ISHp608, but it is absent from the ISHp609 orfB product (both predominant and rare variant types) and is also absent from the close homolog in T. tencongensis (Fig. 2C). Although the functions of IS element orfB genes are not known, the zinc finger motif difference between OrfB of ISHp609 and the other elements hints at possible differences in the interactions with nucleic acids or other cellular constituents, which in turn might affect transposition or control of host gene expression.
Supplementary Material
Acknowledgments
We thank Giedrius Dailide and Shumin Tan for valuable discussions and help.
This research was supported by grants RO1 AI38166, RO3 AI49161, RO1 DK53727, RO1 DK63041, and P30 DK52574 from the National Institutes of Health.
Footnotes
Supplemental material for this article may be found at http://jb.asm.org.
REFERENCES
- 1.Achtman, M., T. Azuma, D. E. Berg, Y. Ito, G. Morelli, Z. J. Pan, S. Suerbaum, S. A. Thompson, A. van der Ende, and L. J. van Doorn. 1999. Recombination and clonal groupings within Helicobacter pylori from different geographical regions. Mol. Microbiol. 32:459-470. [DOI] [PubMed] [Google Scholar]
- 2.Alm, R. A., L. S. Ling, D. T. Moir, B. L. King, E. D. Brown, P. C. Doig, D. R. Smith, B. Noonan, B. C. Guild, B. L. deJonge, G. Carmel, P. J. Tummino, A. Caruso, M. Uria-Nickelsen, D. M. Mills, C. Ives, R. Gibson, D. Merberg, S. D. Mills, Q. Jiang, D. E. Taylor, G. F. Vovis, and T. J. Trust. 1999. Genomic-sequence comparison of two unrelated isolates of the human gastric pathogen Helicobacter pylori. Nature 397:176-180. [DOI] [PubMed] [Google Scholar]
- 3.Ausubel, F. M., R. Brent, R. E. Kingston, D. D. Moore, J. G. Seidman, J. A., Smith, and K. A. Struhl. 1994. Current protocols in molecular biology, supplement 27 CPMB, page 2.4.1. Greene Publishing and Wiley Interscience, New York, N.Y.
- 4.Berg, D. E., R. H. Gilman, J. Lelwala-Guruge, K. Srivastava, Y. Valdez, J. Watanabe, J. Miyagi, N. S. Akopyants, A. Ramirez-Ramos, T. H. Yoshiwara, S. Recavarren, and R. Leon-Barua. 1997. Helicobacter pylori populations in individual Peruvian patients. Clin. Infect. Dis. 25:996-1002. [DOI] [PubMed] [Google Scholar]
- 5.Berg, D. E., and M. M. Howe (ed.). 1989. Mobile DNA. American Society for Microbiology, Washington, D.C.
- 6.Chanto, G., A. Occhialini, N. Gras, R. A. Alm, F. Megraud, and A Marais. 2002. Identification of strain-specific genes located outside the plasticity zone in nine clinical isolates of Helicobacter pylori. Microbiology 148:3671-3680. [DOI] [PubMed] [Google Scholar]
- 7.Craig, N. C., R. Craigie, M. Gellert, and A. M. Lambowitz (ed.). 2002. Mobile DNA II. ASM Press, Washington, D.C.
- 8.Falush, D., C. Kraft, N. S. Taylor, P. Correa, J. G. Fox, M. Achtman, and S. Suerbaum. 2001. Recombination and mutation during long-term gastric colonization by Helicobacter pylori: estimates of clock rates, recombination size, and minimal age. Proc. Natl. Acad. Sci. USA 98:15056-15061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Falush, D., T. Wirth, B. Linz, J. K. Pritchard, M. Stephens, M. Kidd, M. J. Blaser, D. Y. Graham, S. Vacher, G. I. Perez-Perez, Y. Yamaoka, F. Megraud, K. Otto, U. Reichard, E. Katzowitsch, X. Wang, M. Achtman, and S. Suerbaum. 2003. Traces of human migrations in Helicobacter pylori populations. Science 299:1582-1585. [DOI] [PubMed] [Google Scholar]
- 10.Han, S. R., H. C. Zschausch, H. G. Meyer, T. Schneider, M. Loos, S. Bhakdi, and M. J. Maeurer. 2000. Helicobacter pylori: clonal population structure and restricted transmission within families revealed by molecular typing. J. Clin. Microbiol. 38:3646-3651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hartl, D. L., and S. A. Sawyer. 1988. Why do unrelated insertion sequences occur together in the genome of Escherichia coli? Genetics 118:537-541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11a.Kalia, A., A. K. Mukhopadhyay, G. Dailide, Y. Ito, T. Azuma, B. C. Y. Wong, and D. E. Berg. 2004. Evolutionary dynamics of insertion sequences in H. pylori. J. Bacteriol. 186:7508-7520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kersulyte, D., N. S. Akopyants, S. W. Clifton, B. A. Roe, and D. E. Berg. 1998. Novel sequence organization and insertion specificity of IS605 and IS606: chimaeric transposable elements of Helicobacter pylori. Gene 223:175-186. [DOI] [PubMed] [Google Scholar]
- 13.Kersulyte, D., H. Chalkauskas, and D. E. Berg. 1999. Emergence of recombinant strains of Helicobacter pylori during human infection. Mol. Microbiol. 31:31-43. [DOI] [PubMed] [Google Scholar]
- 14.Kersulyte, D., A. K. Mukhopadhyay, M. Shirai, T. Nakazawa, and D. E. Berg. 2000. Functional organization and insertion specificity of IS607, a chimeric element of Helicobacter pylori. J. Bacteriol. 182:5300-5308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kersulyte, D., A. K. Mukhopadhyay, B. Velapatino, W. W. Su, Z. J. Pan, C. Garcia, V. Hernandez, Y. Valdez, R. S. Mistry, R. H. Gilman, Y. Yuan, H. Gao, T. Alarcon, M. Lopez-Brea, G. Balakrish Nair, A. Chowdhury, S. Datta, M. Shirai, T. Nakazawa, R. Ally, I. Segal, B. C. Wong, S. K. Lam, F. O. Olfat, T. Boren, L. Engstrand, O. Torres, R. Schneider, J. E. Thomas, S. Czinn, and D. E. Berg. 2000. Differences in genotypes of Helicobacter pylori from different human populations. J. Bacteriol. 182:3210-3218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kersulyte, D., B. Velapatino, G. Dailide, A. K. Mukhopadhyay, Y. Ito, L. Cahuayme, A. J. Parkinson, R. H. Gilman, and D. E. Berg. 2002. Transposable element ISHp608 of Helicobacter pylori: nonrandom geographic distribution, functional organization, and insertion specificity. J. Bacteriol. 184:992-1002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kidwell, M. G., and D. R. Lisch. 2001. Perspective: transposable elements, parasitic DNA, and genome evolution. Evol. Int. J. Org. Evol. 55:1-24. [DOI] [PubMed] [Google Scholar]
- 18.Lawrence, J. G., H. Ochman, and D. L. Hartl. 1992. The evolution of insertion sequences within enteric bacteria. Genetics 131:9-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mahillon, J., and M. Chandler. 1998. Insertion sequences. Microbiol. Mol. Biol. Rev. 62:725-774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mukhopadhyay, A. K., D. Kersulyte, J. Y. Jeong, S. Datta, Y. Ito, A. Chowdhury, S. Chowdhury, A. Santra, S. K. Bhattacharya, T. Azuma, G. B. Nair, and D. E. Berg. 2000. Distinctiveness of genotypes of Helicobacter pylori in Calcutta, India. J. Bacteriol. 182:3219-3227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Owen, R. J., and J. Xerry. 2003. Tracing clonality of Helicobacter pylori infecting family members from analysis of DNA sequences of three housekeeping genes (ureI, atpA and ahpC), deduced amino acid sequences, and pathogenicity-associated markers (cagA and vacA). J. Med. Microbiol. 52:515-524. [DOI] [PubMed] [Google Scholar]
- 22.Stanley, T. L., C. D. Ellermeier, and J. M. Slauch. 2000. Tissue-specific gene expression identifies a gene in the lysogenic phage Gifsy-1 that affects Salmonella enterica serovar Typhimurium survival in Peyer's patches. J. Bacteriol. 182:4406-4413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Suerbaum, S., J. M. Smith, K. Bapumia, G. Morelli, N. H. Smith, E. Kunstmann, I. Dyrek, and M. Achtman. 1998. Free recombination within Helicobacter pylori. Proc. Natl. Acad. Sci. USA. 95:12619-12624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Tomb, J. F., O. White, A. R. Kerlavage, R. A. Clayton, G. G. Sutton, R. D. Fleischmann, K. A. Ketchum, H. P. Klenk, S. Gill, B. A. Dougherty, K. Nelson, J. Quackenbush, L. Zhou, E. F. Kirkness, S. Peterson, B. Loftus, D. Richardson, R. Dodson, H. G. Khalak, A. Glodek, K. McKenney, L. M. Fitzegerald, N. Lee, M. D. Adams, E. K. Hickey, D. E. Berg, J. D. Gocayne, T. R. Utterback, J. D. Peterson, J. M. Kelley, M. D. Cotton, J. M. Weidman, C. Fujii, C. Bowman, L. Watthey, E. Wallin, W. S. Hayes, M. Borodovsky, P. D. Karp, H. O. Smith, C. M. Fraser, and J. C. Venter. 1997. The complete genome sequence of the gastric pathogen Helicobacter pylori. Nature 388:539-547. [DOI] [PubMed] [Google Scholar]
- 25.Turlan, C., and M. Chandler. 2000. Playing second fiddle: second-strand processing and liberation of transposable elements from donor DNA. Trends Microbiol. 8:268-274. [DOI] [PubMed] [Google Scholar]
- 26.van der Ende, A., Z. J. Pan, A. Bart, R. W. van der Hulst, M. Feller, S. D. Xiao, G. N. Tytgat, and J. Dankert. 1998. cagA-positive Helicobacter pylori populations in China and The Netherlands are distinct. Infect. Immun. 66:1822-1826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Westblom, T. U. 1991. Laboratory diagnosis and handling of Helicobacter pylori, p 81-91. In B. J. Marshall, R. W. McCallum, and R. L. Guerrant (ed.), Helicobacter pylori in peptic ulceration and gastritis. Blackwell Scientific Publications, Boston, Mass.
- 28.Yamaoka, Y., M. S. Osato, A. R. Sepulveda, O. Gutierrez, N. Figura, J. G. Kim, T. Kodama, K. Kashima, and D. Y. Graham. 2000. Molecular epidemiology of Helicobacter pylori: separation of H. pylori from East Asian and non-Asian countries. Epidemiol. Infect. 124:91-96. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.