Abstract
The molecular identification of the Corynebacterium glutamicum urea uptake system is described. This ABC-type transporter is encoded by the urtABCDE operon, which is transcribed in response to nitrogen limitation. Expression of the urt genes is regulated by the global nitrogen regulator AmtR, and an amtR deletion strain showed constitutive expression of the urtABCDE genes. The AmtR repressor protein also controls transcription of the urease-encoding ureABCEFGD genes in C. glutamicum. The ure gene cluster forms an operon which is mainly transcribed in response to nitrogen starvation. To confirm the increased synthesis of urease subunits under nitrogen limitation, proteome analyses of cytoplasmic protein extracts from cells grown under nitrogen surplus and nitrogen limitation were carried out, and five of the seven urease subunits were identified.
Urea is a readily available nitrogen source, since it is excreted by a variety of organisms into the environment. In bacteria able to metabolize this solute, urea is hydrolyzed by the cytoplasmic urease enzyme complex, leading to one CO2 and two ammonium molecules (for review, see reference 20). Although urea is small and uncharged and can therefore easily pass the bacterial membrane, many prokaryotes, including Corynebacterium glutamicum, synthesize energy-dependent transport systems for its uptake. When present in high concentrations, sufficient urea crosses the C. glutamicum cytoplasmic membrane by passive diffusion, and only under conditions of nitrogen starvation is an energy-dependent urea uptake system synthesized (27). With a Km of 8 μM for urea, the affinity of this uptake system is much higher than the affinity of urease for its substrate (Km of approximately 55 mM). The maximum urea uptake rate depends on the level of expression and is relatively low at 2.0 to 3.5 nmol mg (dry weight)−1 min−1 (27). Genes coding for the C. glutamicum urea uptake system have not been previously identified.
The genes encoding the urease enzyme complex were isolated and sequenced (21, 24). While ureA, ureB, and ureC encode the urease structural subunits, the ureE, ureF, ureG, and ureD genes code for accessory proteins. As shown by mutant analyses, at least the ureC and the ureD products are essential for a functional urease (21). Depending on the growth conditions, urease activity observed in the wild type varies between 0.9 and 2.2 U mg of protein−1 for cells grown in ammonium-rich minimal medium (21, 24), between 1.0 and 1.6 U mg of protein−1 when glutamine is used as nitrogen source (24), and between 2.0 and 6.1 U mg of protein−1 when different concentrations of urea were added to the medium as sole nitrogen source (21, 24). These medium-dependent changes in urease activity indicate that the enzyme is moderately regulated, either on the level of activity or on the level of gene expression.
Here, we describe the identification of genes encoding the C. glutamicum urea uptake system and their transcriptional organization and regulation, as well as the organization and control of the genes coding for the urease enzyme complex.
MATERIALS AND METHODS
Bacterial strains and growth.
C. glutamicum and Escherichia coli strains and plasmids used in this study are listed in Table 1. E. coli cells were routinely grown in Luria-Bertani medium (25) at 37°C, and C. glutamicum strains were grown in brain heart infusion (Difco, Sparks, Md.) at 30°C. To study the effects of nitrogen starvation under comparable conditions, a standard inoculation scheme was applied. A fresh C. glutamicum culture grown in brain heart infusion medium was used to inoculate minimal medium [per liter, 42 g of morpholinepropanesulfonic acid, 20 g of (NH4)2SO4, 5 g of urea, 0.5 g of K2HPO4 · 3H2O, 0.5 g of KH2PO4, 0.25 g of MgSO4 · 7H2O, 0.01 g of CaCl2, 50 g of glucose, 0.2 mg of biotin, 10 mg of FeSO4, 10 mg of MnSO4, 1 mg of ZnSO4, 0.2 mg of CuSO4, 0.02 mg of NiCl2 · 6H2O, 0.09 mg of H3BO3, 0.06 mg of CoCl2 · 6H2O, 0.009 mg of NaMoO4 · 2H2O; pH adjusted to pH 7.0 using NaOH (16)] for overnight growth. This culture, with an overnight optical density at 600 nm (OD600) of approximately 25 to 30, was used to inoculate fresh minimal medium to an OD600 of approximately 1, and cells were grown until the exponential growth phase was reached (OD600 of approximately 4 to 5). To induce nitrogen starvation, cells were harvested by centrifugation and the pellet was suspended in and transferred to prewarmed minimal medium without a nitrogen source. Media were supplemented with antibiotics (50 μg of carbenicillin/ml, 25 μg of chloramphenicol/ml, 25 μg of kanamycin/ml [final concentrations]) if appropriate.
TABLE 1.
Strains and plasmids used in this study
| Strain or plasmid | Relevant genotype or descriptiona | Reference or source |
|---|---|---|
| C. glutamicum strains | ||
| ATCC 13032 | Type strain | 1 |
| MJ6-18 | ATCC 13032 ΔamtR | 14 |
| RES167 | Restriction-deficient mutant of type strain ATCC 13032; Δ(cgIIM-cgIIR-cgIIIR) | 28 |
| ATCC::pDRIVE-urtC | urtC insertion in wild-type strain | This study |
| RES167::pDRIVE-urtA | urtA insertion in restriction-deficient strain RES167 | This study |
| RES167::pDRIVE-urtE | urtE insertion in restriction-deficient strain RES167 | This study |
| RES167::pDRIVE-ureR | ureR insertion in restriction-deficient strain RES167 | This study |
| E. coli strain | ||
| DH5αmcr | supE44 hsdR17 recA1 endA1 gyrA96 thi1 relA mcrA Δ(mrr-hsdRMS-mcrBC) | 7 |
| Plasmids | ||
| pDRIVE | Apr Kmr; A-T cloning vector | Qiagen |
| pDRIVE-urtA | pDRIVE carrying an an internal 1.1-kb urtA fragment | This study |
| pDRIVE-urtC | pDRIVE carrying an an internal 0.8-kb urtC fragment | This study |
| pDRIVE-urtE | pDRIVE carrying an an internal 0.6-kb urtE fragment | This study |
| pDRIVE-ureR | pDRIVE carrying an internal 0.5-kb ureR fragment | This study |
| pGEM-3z | E. coli plasmid for in vitro transcription, Apr | Promega |
| pGEM-urtA | 0.5-kb internal urtA fragment in pGEM-3z | This study |
| pGEM-ureA | 0.3-kb internal ureA fragment in pGEM3z | This study |
| pUC11-1.8 | AmtR delivery plasmid for gel retardation experiments | 14 |
| pUC18 | plac, Apr | 32 |
Abbreviations: Ap, ampicillin; Km, kanamycin.
Determination of urea and amino acid uptake.
Since urea is immediately decomposed within the cell by the action of urease and, as a consequence, it is not possible to follow its accumulation within the cell, the decrease of the urea concentration in the medium was determined as described previously (21, 27). For this purpose, log-phase cells grown in minimal medium were harvested, washed, and suspended in nitrogen-free minimal medium to an OD600 of 0.5. After 3 h of nitrogen starvation at 30°C, [14C]urea (specific activity, 55 mCi mmol−1) was added (4.0 μM final concentration). Samples were taken at intervals and directly added to 2 ml of scintillation cocktail (Roth, Karlsruhe, Germany). The [14C]urea concentration in the medium was determined using a Beckman LS6500 multipurpose scintillation counter (Beckman, Munich, Germany), and uptake rates were calculated from the decrease in the label.
For measurement of amino acid uptake rates, cells were incubated as described for the urea transport measurements. The uptake determination was started by the addition of 14C-labeled l-amino acids (specific activity, 254 mCi mmol−1; 1.5 μM final concentration). In this case, radioactivity transported into the cell was measured directly by a filtration assay as described earlier (18).
Determination of urease activity.
Urease activity was measured by the indophenol method (13). Cells were harvested and washed once with 50 mM potassium phosphate buffer at pH 7.0, resuspended to an OD600 of 200, and disintegrated by ultrasonic treatment with a Branson sonifier 250 at an output control of 2.5 and a duty cycle of 25% for 4 min. After centrifugation at 4°C for 30 min at 18,000 × g, urease activity was determined immediately in the cell-free supernatant by measuring the amount of ammonia produced from urea in 15 min at 30°C. A calibration curve was obtained from appropriate dilutions of a freshly prepared NH4Cl solution. One unit of enzyme activity was defined as 1 micromole of ammonia released per minute.
Measurement of the intracellular ATP pool.
Intracellular ATP concentrations were determined as described previously by using a modified firefly luciferin-luciferase assay (17).
General molecular biology techniques.
Standard techniques for plasmid isolation, transformation, and cloning were used (2, 25).
Construction of mutant strains.
For gene disruption mutagenesis, internal gene fragments of the genes ureR, urtA, urtC, and urtE were amplified by PCR and ligated to pDRIVE vector DNA, resulting in the plasmids pDRIVE-ureR, pDRIVE-urtA, pDRIVE-urtC, and pDRIVE-urtE. Plasmids pDRIVE-ureR, pDRIVE-urtA, and pDRIVE-urtE were used to create gene disruption mutants with the restriction-deficient C. glutamicum strain RES167 as parental strain; for plasmid pDRIVE-urtC, the type strain ATCC 13032 was used as parental strain. For the construction of the different gene disruption plasmids, internal DNA fragments of the corresponding genes were amplified via PCR and ligated to DNA of the pDRIVE AT cloning vector as recommended by the supplier (QIAGEN, Hilden, Germany). The following primer combinations were used for PCR: 5′-AGCAACACCGCAGCTTCC-3′ and 5′-GCAGCGGTGGTCTTGGC-3′ (urtA); 5′-TGCGGTATTGCTGTGCGC-3′ and 5′-CCACCCAGCCCAGCACC-3′ (urtC); 5′-ATTTGTGTGCAGGTTATGGC-3′ and 5′-GCGCCTTGTCCCGATTCC-3′ (urtE); 5′-TTACGAACCTTGGATGCGCG-3′ and 5′-CGACAGGCCGACTCCCATT-3′ (ureR). Mutant strains generated were controlled by PCR experiments, which verified correct gene disruption and absence of wild-type alleles.
Gel retardation experiments.
To investigate AmtR binding, gel shift assays were carried out. For the upstream region of the urtA and ureA genes, PCR-generated DNA fragments spanning the corresponding putative AmtR binding sites were labeled with digoxigenin using the DIG oligonucleotide 3′-end labeling kit, second generation (Roche, Mannheim, Germany). The labeled DNA fragments were incubated for 20 min on ice with different amounts of AmtR-containing E. coli cell extract (DH5αmcr pUC11-1.8) or control extract lacking AmtR (DH5αmcr pUC18). The formation of unspecific DNA-protein complexes was minimized by adding sheared salmon sperm DNA (Eppendorf, Hamburg, Germany) and bovine serum albumin (AppliChem, Darmstadt, Germany). DNA-protein complexes were separated from free DNA on a nondenaturing 6% polyacrylamide-Tris-borate-EDTA gel by electrophoresis at 20 mA. The DNA was blotted onto a positively charged nylon membrane (Roche), and the digoxigenin-labeled DNA was detected with a Fuji luminescent image analyzer LAS1000 (Raytest, Straubenhardt, Germany).
RNA preparation and hybridization.
Total RNA was prepared after disruption of the C. glutamicum cells with glass beads by using the NucleoSpin RNA II kit as recommended by the supplier (Macherey-Nagel, Düren, Germany). The RNA was blotted onto positively charged nylon membranes (BioBond Sigma, Taufkirchen, Germany) using a Minifold I dot blotter (Schleicher & Schuell, Dassel, Germany). Hybridization of digoxigenin-labeled RNA probes was detected with a Fuji luminescent image analyzer LAS1000 (Raytest) or Kodak X-Omat X-ray films (Sigma-Aldrich, Darmstadt, Germany) using alkaline phosphatase-conjugated antidigoxigenin Fab fragments and CSPD as light-emitting substrate as recommended by the supplier (Roche Diagnostics, Mannheim, Germany). In order to quantify hybridization signals, the total amount of RNA applied was determined using the RiboGreen RNA quantitation kit (Molecular Probes, Eugene, Oreg.).
For the generation of antisense probes, internal DNA fragments of the corresponding genes were amplified by PCR and cloned into plasmid pGEM plasmids (Promega, Mannheim, Germany) using restriction sites added to the primer sequence. For cloning of the ureA gene fragment EcoRI and HindIII sites were introduced (primer sequences, 5′-GCGCGCGAATTCCGTCGCCGTAAAGATCG-3′ and 5′-GCGCGCAAGCTTATCAAACGTTGCTTCAACC-3′), and for the urtA probe EcoRI and HindIII restriction sites were added (5′-GCGCGCGAATTCGGCTTGGACTTTGAAGACG-3′ and 5-GCGCGCAAGCTTCACCGTGATCAACAAATGG-3′) (restriction sites are shown in bold) to insert the PCR products into plasmid pGEM-3z. The hybridization probes were produced by in vitro transcription with the SP6 polymerase.
RT-PCR.
For reverse transcriptase (RT)-PCR the OneStep RT-PCR kit was used as recommended by the supplier (QIAGEN) together with the following primers: 5′-AGCAACACCGCAGCTTCC-3′ (urtA); 5′-GCGCCTTGTCCCGATTCC-3′ (urtE); 5′-GCGATCTTTACGGCGACGTGCAA-3′ (ureA); 5′-AGCAACCGCACTGAGGAGTTGAT-3′ (ureD).
Real-time PCR.
For real-time PCR, a TaqMan device (Applied Biosystems), the QuantiTect SYBR Green RT-PCR kit (QIAGEN), 100 ng of template RNA, and the following primer combinations were used: 5′-TCGCCCTCATCACGTATGAA-3′ and 5′-TCAAAATGGTGCTTCCCCAG-3′ (urtA); 5′-CGATGCCTCCGACTTCTTCTT-3′ and 5′-TTCTTCCGCCAGTACAACAGC-3′ (ureA). Reverse transcription was carried out at 50°C for 30 min, the RT was inactivated and the polymerase was activated by 15 min of incubation at 94°C, and PCR was carried out for 40 cycles of the following program: DNA denaturation for 15 s at 94°C, primer annealing for 30 s at 60°C, and DNA polymerization for 30 s at 72°C. All experiments were carried out at least in triplicate with templates from independent cultures. For an exact quantification of real-time PCR results, dilution series were tested for each pair of primers to determine the PCR efficiency, and the total amount of RNA applied in the real-time PCR experiments was determined using the RiboGreen RNA quantitation kit (Molecular Probes). This method allows normalization of the results without additional control reactions with housekeeping genes or 16S rRNA (8).
Two-dimensional polyacrylamide gel electrophoresis and matrix-assisted laser desorption ionization-time of flight mass spectrometry (MALDI-TOF MS).
C. glutamicum cells were disrupted using glass beads and a Q-BIOgene FastPrep FP120 instrument (Q-BIOgene, Heidelberg, Germany) by lysing the cells four times for 30 s and 6.5 m/s in the presence of Complete proteinase inhibitor as recommended by the supplier (Roche, Basel, Switzerland). Proteins were separated in the cytoplasmic and membrane-associated protein fractions by ultracentrifugation (10-12). In this study, only the cytoplasmic proteins were further analyzed. Protein concentrations were determined using the Roti-Nanoquant assay (Roth).
For isoelectric focusing, 24-cm precast IPG strips of pI 4 to 7 and an IPGphor isoelectric focusing unit (Amersham Biosciences, Freiburg, Germany) were used as described elsewhere (11). A 120-μg aliquot of protein was focused for 68,000 V · h in a sample buffer containing 6 M urea, 2 M thiourea, 4% 3-[(3-cholamidopropyl)-dimethylammonio]-1-propanesulfonate, 0.5% Pharmalyte (3-10), and 0.4% dithiothreitol. For the second dimension, electrophoresis was carried out using precast 12-to-14% polyacrylamide linear gradient gels (ExcelGel Gradient XL 12-14; Amersham Biosciences, Freiburg, Germany) in a Multiphor II apparatus as described previously (11, 12). After electrophoresis, two-dimensional gels were stained with colloidal Coomassie brilliant blue. The Coomassie-stained gels were aligned using the Delta2D software, version 3.1 (Decodon, Greifswald, Germany).
MALDI-TOF-MS for peptide mass fingerprint analyses was carried out by the bioanalytics service unit at the Center for Molecular Medicine Cologne.
RESULTS
Identification of the C. glutamicum urea uptake system.
While urea transport has been characterized biochemically in a wide variety of microorganisms, including C. glutamicum, only a few urea uptake systems have been characterized at the genetic level. These transporters can be differentiated into those facilitating diffusion (9, 26, 30) and active, energy-dependent uptake systems (19, 29). Since we were interested in identifying the C. glutamicum urea uptake system, we used the known genes as query sequences for searches in the recently published C. glutamicum genome (15). Genes with high similarity to ureI from Helicobacter pylori (30) or yut from Yersinia pseudotuberculosis (26) were not observed, but a gene cluster similar to the fmd genes of Methylophilus methyltrophus (19) and the urt gene cluster from Anabaena (29) was found. The amino acid sequence deduced from the NCgl0893 gene exhibited 43.3% identical amino acids with the fmdD gene product and 49.2% identity with the UrtA protein. The identity of the putative NCgl0894 protein was 40.8% to FmdE and 49.3% to UrtB, while the NCgl0895 gene product showed an identity of 35.2% with FmdF and 40.4% with UrtC. The NCgl0893, NCgl0894, and NCgl0895 genes encode components of an ABC-type transport system, namely, a lipid-anchored substrate binding protein and two membrane-integral permease subunits. Downstream, two open reading frames, NCgl0986 and NCgl0987, code for two putative ATP binding proteins, completing the proteins necessary for a typical ABC-type transporter. The proteins encoded by the latter two genes showed an identity of 34.0 and 38.7% with the corresponding Urt subunits. Besides the Fmd and Urt system, the transporter encoded by the NCgl0893-NCgl0897 cluster showed similarity (between 21.6 and 37.0% identical amino acids depending on the different subunits) to the LIV transporter of E. coli for the uptake of l-leucine, l-isoleucine, and l-valine. Based on the highest homologies observed, the NCgl0893-NCgl0897 genes are designated as urtABCDE in the following text.
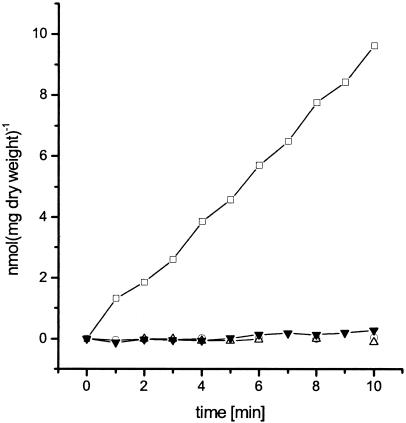
To demonstrate that the urtABCDE gene cluster encodes the C. glutamicum urea uptake system, the urtA, urtC, and urtE genes encoding the substrate binding protein, one permease subunit, and one ATP binding protein were inactivated by insertion mutagenesis. Each single gene disruption resulted in loss of urea transport activity (0.01 to 0.04 nmol min−1 mg [dry weight]−1 compared to 0.8 ± 0.2 nmol min−1 mg [dry weight]−1 determined for the wild type) (Fig. 1), indicating a crucial role of the ABC transporter for urea uptake. Polar effects of the mutations on downstream genes cannot be excluded; however, these are unlikely due to the chromosomal organization of the gene cluster.
FIG. 1.
Urea consumption in C. glutamicum wild type (open squares) and urtA, urtC, and urtE mutant strains (open triangles, open circles, and inverted closed triangles, respectively) grown in minimal medium with urea as single nitrogen source. Under the experimental conditions used (urea concentration, 4 μM), diffusion of urea across the cytoplasmic membrane was negligible.
To exclude that the loss of urea transport is the result of a secondary effect, i.e., that the ABC transporter takes up Ni2+ ions, which are essential for urease function, urease activities in the different mutant strains were determined. For the wild type a urease activity of 0.32 ± 0.13 U min−1 was measured; the mutant strains showed activities of 0.24 ± 0.10 (urtA), 0.48 ± 0.12 (urtC), and 0.25 ± 0.07 (urtE) U min−1. Hence, urease activity is not greatly influenced by urt mutations.
Because the C. glutamicum urea transporter showed similarity to the Fmd and LIV systems, competition of urea uptake was tested with Fmd and LIV substrates as well as other nitrogen sources. While addition of a 10-fold excess of unlabeled urea reduced the apparent uptake rate as expected by about 90%, 10-fold concentrations of ammonium, formamide, γ-amino butyric acid, l-glutamate, l-glutamine, l-histidine, l-isoleucine, l-alanine, l-threonine, and l-valine had no significant impact on urea uptake (data not shown). Furthermore, when uptake of labeled l-arginine (50 or 500 μM final concentration), l-serine (0.5 to 500 μM final concentration), or l-leucine (50 to 500 μM final concentration) was tested in the wild type and the urtA and urtC mutant strains, no differences in their uptake rates were observed (data not shown).
Since the urt genes encode an ABC-type urea uptake system, an ATP-dependent transport mechanism was expected. However, an influence of the protonophore CCCP (carbonyl cyanide m-chlorophenylhydrazone) on urea transport was reported previously (27). Addition of this uncoupler leads to immediate dissipation of the membrane potential, while in C. glutamicum the intracellular ATP pool is only marginally affected by this substance for several minutes (27). As a consequence, ATP-driven transport systems should not be affected by CCCP addition. However, as described before (27), here also an influence of CCCP on urea transport was observed. Urea uptake rates decreased from 0.9 ± 0.3 nmol min−1 mg (dry weight)−1 to 0.5 ± 0.1 nmol min−1 mg (dry weight)−1 when 5 μM CCCP was added to the transport assay mixture 30 s prior to the start of the measurement. To characterize whether this CCCP effect was specific for the urea transporter, l-glutamate uptake via the GluABCD ABC transporter (18) was determined. These measurements showed that the uptake of l-glutamate via the GluABCD transport system was significantly less affected by the addition of the uncoupler than was urea uptake by the UrtABCDE transporter. An l-glutamate uptake rate of 1.3 nmol min−1 mg (dry weight)−1 was obtained without the addition of CCCP, and addition of 5 μM CCCP hardly decreased this rate (1.2 nmol min−1 mg [dry weight]−1).
To investigate the CCCP sensitivity of the urea transporter in more detail, the intracellular ATP pool of the cells was determined before CCCP addition and after 6 min of incubation with the uncoupler. Without the addition of CCCP, the internal ATP concentrations decreased from 4.8 to 4.1 mM after 6 min. Addition of 5 μM CCCP had no effect on the ATP pool compared to the control (4.6 mM before addition, 4.1 mM after 6 min of CCCP treatment). Consequently, a decrease of the ATP concentration can be excluded as a reason for the CCCP effect on urea transport.
Regulation of urtABCDE transcription.
C. glutamicum exhibits active urea transport only under conditions of nitrogen starvation, and synthesis of this transporter can be suppressed by the addition of chloramphenicol, indicating a regulation on the level of gene expression (27). Therefore, we investigated transcription of the urtABCDE genes.
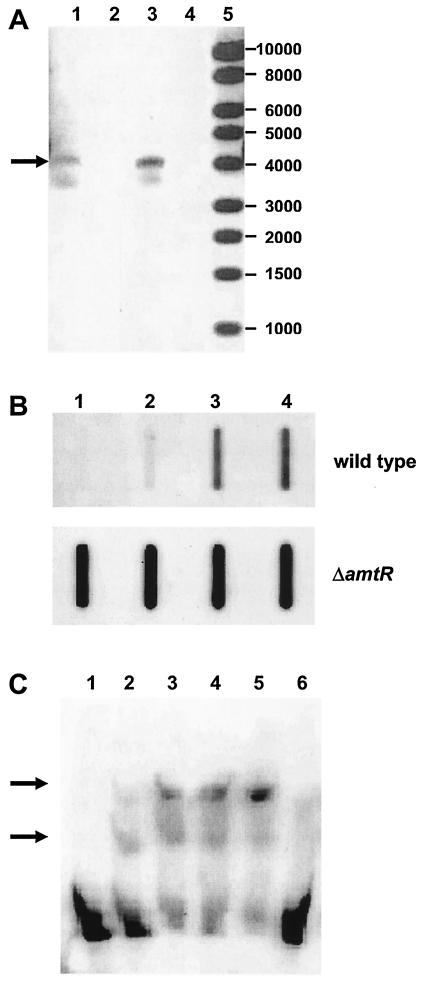
In our first approach, RT-PCR experiments were carried out using RNA extracted from nitrogen-starved cells as template to investigate the transcriptional coupling suggested by the genetic organization of the urt genes. Control reactions carried out without RT gave no PCR product, showing that the RNA preparation was DNA free. Primers annealing to urtA and urtE sequences led to a DNA fragment of 4.1 kb in an RT-PCR (Fig. 2A), indicating the presence of a joint transcript spanning the complete urtABCDE gene cluster. The result was confirmed by Northern hybridization experiments which showed an approximately 4,000-base-long mRNA which hybridized with a urtA probe (data not shown).
FIG. 2.
Transcription of the genes encoding the C. glutamicum urea uptake system. (A) RT-PCR using primers annealing to urtA and urtE. Total RNA isolated from wild-type strain ATCC 13032 after 30 min of nitrogen starvation (lane 1) and from amtR deletion strain MJ6-18 grown under nitrogen surplus (lane 3) was used as template. The RT-PCR products (indicated by the arrow) had an expected size of 4.0 kb. Control reactions without the addition of RT (lanes 2 and 4) gave no PCR product, validating that the RNA preparations used were DNA free (lane 5). Lane M, marker DNA (1-kb DNA ladder; New England Biolabs). (B) RNA hybridization experiments with an urtA probe and total RNA (1 μg per slot) prepared from the wild type and from the amtR deletion strain MJ6-18 grown in nitrogen-rich minimal medium and after 5, 15, and 30 min of nitrogen limitation (lanes 1 to 4). (C) Gel retardation experiment. Increasing amounts of AmtR-containing E. coli cell extracts (lanes 1 to 5: 0, 5, 10, 20, and 30 μg of protein, respectively) were added to a digoxigenin-labeled 122-bp DNA fragment from the urtA promoter region spanning the two identified putative AmtR binding sites. As a control, cell extract lacking AmtR was used (lane 6; 30 μg). DNA fragments with shifted mobilities depending on AmtR are indicated by arrows.
RNA slot blot hybridizations were carried out to investigate the regulation of urtABCDE expression. An antisense urtA probe as a representative of the complete operon was hybridized to total RNA isolated from cells grown in nitrogen-rich medium and then incubated for different time periods in nitrogen-free minimal medium. As shown, urtA expression was enhanced within 5 min of nitrogen starvation and stayed at a high level for at least 30 min (Fig. 2B). To determine the factor by which transcription of urtA was increased more accurately, real-time PCR experiments were carried out in triplicate with independent RNA preparations. These revealed that urtA expression in the wild type was upregulated after 30 min of N starvation by a factor of 344 ± 2.
In C. glutamicum, a repressor protein designated AmtR controls the nitrogen starvation-dependent expression of several genes (3, 14, 22; for review, see references 4 to 6). We tested if AmtR were involved in the regulation of the urtABCDE operon. Two AmtR binding sites with the sequences CTATAG and CTAT-N4-CTATAGA were observed at positions 63 to 58 and 37 to 23 upstream of the urtA gene. To prove AmtR-dependent regulation, RNA hybridization experiments were carried out with a urtA probe and total RNA isolated from amtR deletion strain MJ6-18. In the amtR deletion strain, high and unregulated expression of urtA was observed (Fig. 2B). In contrast to the wild-type ATCC 13032, transcription of urtA in the amtR deletion strain MJ6-18 was found even in nitrogen-rich growth medium, and transcription stayed constant when strain MJ6-18 was incubated in nitrogen-free minimal medium. These results were quantified in real-time PCR experiments. Expression of urtA under nitrogen excess was increased by a factor of 1,504 ± 2 compared to the wild-type transcription level, and after 30 min of nitrogen deprivation a factor of 1,882 ± 2 was observed, verifying the RNA hybridization experiments. The higher factors obtained for upregulation of urtA transcription in strain MJ6-18 compared to the wild type might be the result of another level of transcriptional regulation independent from AmtR.
The binding of AmtR to the urt operon promoter DNA was verified by gel retardation experiments. A 122-bp fragment of the urtA upstream region was changed in its electrophoretic mobility depending on the AmtR protein. In the presence of the repressor, two additional bands with lower mobility were detectable, indicating that the two putative AmtR binding sites upstream of urtA in fact bind the repressor protein (Fig. 2C).
Transcription of urease-encoding genes.
Although the expression of the urtABCDE operon, which codes for the urea uptake system, is nitrogen starvation dependent, the regulation of the urease-encoding ureABCEFGD cluster was less clear. In a previous study, regulation of expression depending on the urea or ammonium concentration in the growth medium was not observed (21). However, urease activity was found to increase during nitrogen starvation (21, 27). Therefore, the transcriptional control of the ureABCEFGD cluster was revisited in this study.
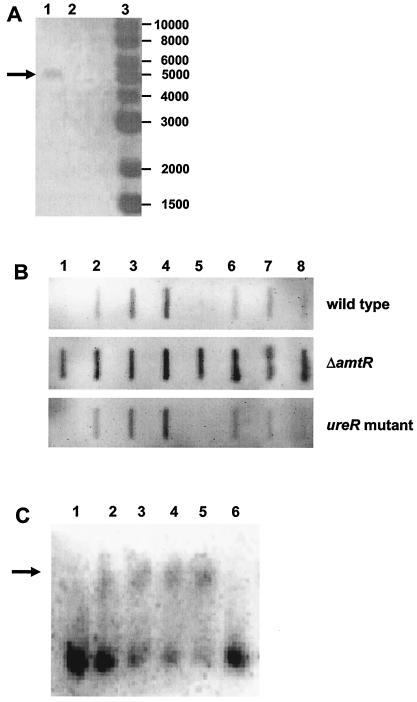
RT-PCR experiments were carried out using total RNA extracted from nitrogen-starved cells as template to reveal the organization of the ure genes. Control reactions carried out without RT gave no PCR product and thus showed that the RNA preparation used was DNA free. RT-PCR with primers annealing to ureA and ureD sequences gave a DNA fragment of 5.1 kb (Fig. 3A), which proved that a common mRNA of the complete ureABCEFGD gene cluster is transcribed. This result was validated by Northern hybridization experiments. Using a ureA probe, an mRNA of approximately 5,000 bases was observed, spanning the complete gene cluster, in addition to at least two smaller transcripts which corresponded to 3′-truncated forms (data not shown).
FIG. 3.
Transcription of the urease-encoding genes in C. glutamicum. (A) RT-PCR with primers annealing to ureA and ureD cluster. As template, total RNA isolated from amtR deletion strain MJ6-18 was used. The RT-PCR product (lane 1; indicated by an arrow) had an expected size of 5.1 kb. A PCR control without addition of RT (lane 2) showed no signal, indicating DNA-free RNA preparation; lane 3, marker DNA (1-kb DNA ladder; New England Biolabs). (B) RNA hybridization experiments with a ureA probe and total RNA (1 μg per slot) prepared from the wild type, from amtR deletion strain MJ6-18, and from the ureR gene disruption mutant Res167::pDRIVE-ureR under nitrogen surplus (lanes 1 and 5); after 5, 15, and 30 min of incubation with no nitrogen source starvation (lanes 2 to 4); and after 5, 15, and 30 min of incubation in the presence of 15 mM urea (lanes 6 to 8). (C) Gel retardation experiment using increasing amounts of AmtR-containing E. coli cell extracts (lanes 1 to 5: 0, 5, 10, 20, and 30 μg of protein, respectively) and a digoxigenin-labeled 216-bp DNA fragment from the ure promoter region spanning the identified putative AmtR binding site. As a control, cell extract lacking AmtR was used (lane 6; 30 μg). The DNA fragment that shifted in its electrophoretic mobility due to the presence of AmtR is indicated by an arrow.
RNA hybridization experiments were carried out using the ureA gene as a probe and total RNA isolated from cells grown in nitrogen-rich medium and incubated for different time periods in nitrogen-free minimal medium (Fig. 3B). Transcription of ureA increased within 5 min of nitrogen starvation and stayed constant for at least 30 min. As in the case of the urtA gene, real-time PCR experiments were carried out to quantify the data obtained by RNA hybridization. Again, this approach showed a clear upregulation of ureA transcription by a factor of 14 ± 2 when cells were incubated for 30 min without nitrogen source compared to the basal level during nitrogen-rich growth.
For the regulation of the ure cluster, two proteins might function as expression regulators, the putative ureR gene product and the global regulator AmtR. The ureR gene lies adjacent to ureA but is transcribed in the opposite direction. Due to its location, UreR might function in the regulation of ureABCEFGD expression. To investigate its importance, a ureR mutant was constructed and tested for ureA transcription. In comparison to the wild type, no significant changes were observed with the ureR mutant when RNA preparations from cells grown in nitrogen-rich standard minimal medium and those from nitrogen-starved cells were studied (Fig. 3B). Additionally, no influence of UreR on transcription of the ureABCEFGD operon was seen when cells were grown in the presence of different urea concentrations.
Since mutagenesis of ureR had no influence on the nitrogen starvation response of ureABCEFGD expression, the amtR deletion strain MJ6-18 was investigated. In this strain, constitutive transcription of ureA was observed, demonstrating an important function of AmtR in expression control of the ureABCEFGD operon. Compared to the wild type, expression of ureA was high during nitrogen-rich growth, and no further increase during incubation in nitrogen-free medium for 5 to 30 min was observed (Fig. 3B). Real-time experiments confirmed these results. Compared to the basal level of transcription observed in the wild type during growth under nitrogen surplus, expression of ureA in strain MJ6-18 was increased by a factor of 20 ± 3 under nitrogen excess and by a factor of 12 ± 4 after 30 min of nitrogen shortage.
The binding of AmtR to the ure operon promoter region was verified by gel retardation assays. A 216-bp fragment of the ureA upstream region was retarded in its electrophoretic mobility in the presence of the AmtR protein (Fig. 3C). In accordance with the presence of only one putative AmtR binding site, only one band with lower mobility was observed in the presence of AmtR.
Two-dimensional gel electrophoresis studies.
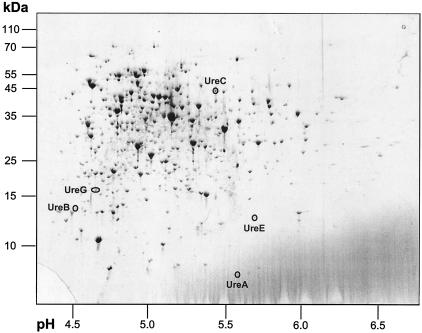
In this study, transcriptional regulation of the ureABCEFGD genes has been shown which should, in principle, be detected at the protein level as well. To investigate putative changes in the amount of urease subunits, two-dimensional gel electrophoresis experiments and MALDI-TOF MS fingerprint analyses were carried out with protein extracts from cells grown under nitrogen surplus and incubated in nitrogen-free minimal medium. In fact, protein spots with molecular masses and pI values in the range expected for different urease subunits were detected in cytoplasmic protein fractions from nitrogen-starved wild-type cells (Fig. 4), while these spots were not detectable in protein extracts from ATCC 13032 cells grown in nitrogen-rich standard minimal medium (data not shown). The corresponding spots were excised from the gel, and proteins were subjected to tryptic in-gel digest followed by MALDI-TOF MS and peptide mass fingerprint analyses. Using this approach, five out of seven urease subunits were identified, namely, UreA, UreB, UreC, UreE, and UreG; the UreD and UreF proteins were not detected. These results on the protein level validate the data obtained for transcription control.
FIG. 4.
Identification of urease subunits in the protein extract of nitrogen-starved cells. Cytoplasmic proteins (120 μg) of C. glutamicum ATCC 13032 grown in standard minimal medium and incubated for 3 h in nitrogen-free minimal medium were separated by two-dimensional gel electrophoresis and stained with colloidal Coomassie brilliant blue. Marked protein spots, which were not detectable in extracts from cells grown in nitrogen-rich medium (data not shown), were identified by MALDI-TOF MS and peptide mass fingerprint analysis. Experiments were carried out independently in duplicate; molecular mass markers and pH values of the gel section are indicated.
DISCUSSION
In this communication, we describe the molecular identification of the C. glutamicum urea uptake system. This ABC-type transporter is specific for urea; additionally, it has a low affinity to thiourea and hydroxyurea (27). The protein complex is encoded by the urtABCDE operon, which is transcribed in response to nitrogen limitation. Expression of this operon is controlled by the global nitrogen regulator AmtR, and an amtR deletion strain shows a constitutive expression of the urtABCDE genes.
In a previous communication (27), urea uptake in C. glutamicum was biochemically described as most likely linked to the symport of protons, since addition of CCCP, an uncoupler of the membrane potential, significantly reduced the transport rate while the internal ATP pool was only marginally affected. Changes in the membrane potential caused by the addition of valinomycin and potassium ions impaired urea uptake, indicating a secondary active transport system (27). However, in this communication, we have demonstrated that an ATP-driven ABC-type transporter is responsible for urea uptake. The reason for the decrease in urea uptake upon CCCP addition is unclear and might be due to secondary effects of the uncoupler on the internal pH or an unknown regulatory mechanisms.
Once inside the cell, urea is hydrolyzed by the urease enzyme complex, which is encoded by the ureABCEFGD gene cluster (21, 24). While coupling of transcription was previously shown for the ureA, ureB, and ureC genes (21), the operon structure of ureABCEFGD was demonstrated here. Moreover, while regulation of the urease-encoding gene cluster ureABCEFGD on the level of transcription had not been observed in RNA hybridization experiments or RT-PCR in a previous study, most likely due to problems with the preparation of total RNA (21), we showed upregulation of transcription levels in response to nitrogen starvation. The latter data are in accordance with urease activity measurements. These data show a low activity of 0.9 U mg of protein−1 when cells are grown in nitrogen-rich complex medium and a high activity of 7.8 U mg of protein−1 in extracts from nitrogen-starved cells (21). The results of the transcription analysis were validated on the protein level for five of the seven urease subunits.
Mutant analyses showed that AmtR regulates transcription of the ureABCEFGD operon, and no influence of ureR was observed. This gene, which encodes a putative transcriptional regulator, lies adjacent to ureA but is transcribed in the opposite direction. Our experiments indicate that AmtR alone is sufficient to regulate expression of the urease-encoding genes under the conditions tested. However, a function of UreR for a fine tuning of transcription or responding to different environmental conditions cannot be excluded, especially in the light of the fact that multiple regulators of urease-encoding operons have been described in other bacteria, namely, Bacillus subtilis (31) and Proteus mirabilis (23).
Acknowledgments
We thank U. Hildebrandt and H. Bothe for help with the real-time PCR experiments.
This work was supported by the Bundesministerium für Forschung und Technologie (GenoMik program) and the Deutsche Forschungsgemeinschaft (BU894/1-3).
REFERENCES
- 1.Abe, S., K. Takayama, and S. Kinoshita. 1967. Taxonomical studies on glutamic acid-producing bacteria. J. Gen. Microbiol. 13:279-301. [Google Scholar]
- 2.Ausubel, F. M., R. Brent, R. E. Kingston, D. D. Moore, J. G. Seidman, J. A. Smith, and K. Struhl. 1987. Current protocols in molecular biology. John Wiley and Sons, New York, N.Y.
- 3.Beckers, G., L. Nolden, and A. Burkovski. 2001. Glutamate synthase of Corynebacterium glutamicum is not essential for glutamate synthesis and is regulated by the nitrogen status. Microbiology 147:2961-2970. [DOI] [PubMed] [Google Scholar]
- 4.Burkovski, A. 2003. I do it my way: regulation of ammonium uptake and ammonium assimilation in Corynebacterium glutamicum. Arch. Microbiol. 179:83-88. [DOI] [PubMed] [Google Scholar]
- 5.Burkovski, A. 2003. Ammonium assimilation and nitrogen control in Corynebacterium glutamicum and its relatives: an example for new regulatory mechanisms in actinomycetes. FEMS Microbiol. Rev. 27:617-628. [DOI] [PubMed] [Google Scholar]
- 6.Burkovski, A. Nitrogen metabolism and its regulation. In M. Bott, and L. Eggeling (ed.), Handbook of Corynebacterium glutamicum, in press. CRC Press LLC, Boca Raton, Fla.
- 7.Grant, S. N. G., J. Jessee, F. R. Bloom, and D. Hanahan. 1990. Differential plasmid rescue from transgenic mouse DNAs into Escherichia coli methylation-restriction mutants. Proc. Natl. Acad. Sci. USA 87:4645-4649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hashimoto, J. G., A. S. Beadles-Bohling, and K. M. Wiren. 2004. Comparison of RiboGreen and 18S rRNA quantitation for normalizing real-time RT-PCR expression analysis. BioTechniques 36:54-60. [DOI] [PubMed] [Google Scholar]
- 9.Heller, K. B., E. C. Lin, and T. H. Wilson. 1980. Substrate specificity and transport properties of the glycerol facilitator of Escherichia coli. J. Bacteriol. 144:274-278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hermann, T., G. Wersch, E.-M. Uhlemann, R. Schmid, and A. Burkovski. 1998. Mapping and identification of Corynebacterium glutamicum proteins by two-dimensional gel electrophoresis and microsequencing. Electrophoresis 19:3217-3221. [DOI] [PubMed] [Google Scholar]
- 11.Hermann, T., M. Finkemeier, W. Pfefferle, G. Wersch, R. Krämer, and A. Burkovski. 2000. Two-dimensional electrophoretic analysis of Corynebacterium glutamicum membrane fraction and surface proteins. Electrophoresis 21:654-659. [DOI] [PubMed] [Google Scholar]
- 12.Hermann, T., W. Pfefferle, C. Baumann, E. Busker, S. Schaffer, M. Bott, H. Sahm, N. Dusch, J. Kalinowski, A. Pühler, A. K. Bendt, R. Krämer, and A. Burkovski. 2001. Proteome analysis of Corynebacterium glutamicum. Electrophoresis 22:1712-1723. [DOI] [PubMed] [Google Scholar]
- 13.Jahns, T., A. Zobel, D. Kleiner, and H. Kaltwasser. 1988. Evidence for carrier-mediated, energy-dependent uptake of urea in some bacteria. Arch. Microbiol. 149:377-383. [Google Scholar]
- 14.Jakoby, M., L. Nolden, J. Meier-Wagner, R. Krämer, and A. Burkovski. 2000. AmtR, a global repressor in the nitrogen regulation system of Corynebacterium glutamicum. Mol. Microbiol. 37:964-977. [DOI] [PubMed] [Google Scholar]
- 15.Kalinowski, J., B. Bathe, N. Bischoff, M. Bott, A. Burkovski, N. Dusch, L. Eggeling, B. J. Eikmanns, L. Gaigalat, A. Goesmann, M. Hartmann, K. Huthmacher, R. Krämer, B. Linke, A. C. McHardy, F. Meyer, B. Möckel, W. Pfefferle, A. Pühler, D. Rey, C. Rückert, H. Sahm, V. F. Wendisch, I. Wiegräbe, and A. Tauch. 2003. The complete Corynebacterium glutamicum ATCC 13032 genome sequence and its impact on the production of l-aspartate-derived amino acids and vitamins. J. Biotechnol. 104:5-25. [DOI] [PubMed] [Google Scholar]
- 16.Keilhauer, C., L. Eggeling, and H. Sahm. 1993. Isoleucine synthesis in Corynebacterium glutamicum: molecular analysis of the ilvB-ilvN-ilvC operon. J. Bacteriol. 175:5595-5603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Krämer, R., and C. Lambert. 1990. Uptake of glutamate in Corynebacterium glutamicum. 2. Evidence for a primary active transport system. Eur. J. Biochem. 194:937-944. [DOI] [PubMed] [Google Scholar]
- 18.Kronemeyer, W., N. Peekhaus, R. Krämer, L. Eggeling, and H. Sahm. 1995. Structure of the gluABCD cluster encoding the glutamate uptake system of Corynebacterium glutamicum. J. Bacteriol. 177:1152-1158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mills, J., N. R. Wyborn, J. A. Greenwood, S. G. Williams, and C. W. Jones. 1998. Characterisation of a binding-protein-dependent, active transport system for short-chain amides and urea in the methylotrophic Methylophilus methylotrophus. Eur. J. Biochem. 251:45-53. [DOI] [PubMed] [Google Scholar]
- 20.Mobley, H. L., M. D. Island, and R. P. Hausinger. 1995. Molecular biology of microbial ureases. Microbiol. Rev. 59:451-480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Nolden, L., G. Beckers, B. Möckel, W. Pfefferle, K. M. Nampoothiri, R. Krämer, and A. Burkovski. 2000. Urease of Corynebacterium glutamicum: organization of corresponding genes and investigation of activity. FEMS Microbiol. Lett. 189:305-310. [DOI] [PubMed] [Google Scholar]
- 22.Nolden, L., M. Farwick, R. Krämer, and A. Burkovski. 2001. Glutamine synthetases in Corynebacterium glutamicum: transcriptional control and regulation of activity. FEMS Microbiol. Lett. 201:91-98. [DOI] [PubMed] [Google Scholar]
- 23.Poore, C. A., and H. L. T. Mobley. 2003. Differential regulation of the Proteus mirabilis urease gene cluster by UreR and H-NS. Microbiology 149:3383-3394. [DOI] [PubMed] [Google Scholar]
- 24.Puskás, L. G., M. Inui, and H. Yukawa. 2000. Structure of the urease operon of Corynebacterium glutamicum. DNA Seq. 11:383-394. [DOI] [PubMed] [Google Scholar]
- 25.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 26.Sebbane, F., S. Bury-Moné, K. Cailliau, E. Browaeys-Poly, H. De Reuse, and M. Simonet. 2002. The Yersinia pseudotuberculosis Yut protein, a new type of urea transporter homologous to eukaryotic channels and functionally interchangeable in vitro with the Helicobacter pylori UreI protein. Mol. Microbiol. 45:1165-1174. [DOI] [PubMed] [Google Scholar]
- 27.Siewe, R. M., B. Weil, A. Burkovski, L. Eggeling, R. Krämer, and T. Jahns. 1998. Urea uptake and urease activity in Corynebacterium glutamicum. Arch. Microbiol. 169:411-416. [DOI] [PubMed] [Google Scholar]
- 28.Tauch, A., O. Kirchner, B. Löffler, S. Götker, A. Pühler, and J. Kalinowski. 2002. Efficient electrotransformation of Corynebacterium diphtheriae with a mini-replicon derived from the Corynebacterium glutamicum plasmid pGA1. Curr. Microbiol. 45:362-367. [DOI] [PubMed] [Google Scholar]
- 29.Valladares, A., M. L. Montesinos, A. Herrero, and E. Flores. 2002. An ABC-type, high-affinity urea permease identified in cyanobacteria. Mol. Microbiol. 43:703-715. [DOI] [PubMed] [Google Scholar]
- 30.Weeks, D. L., S. Eskandari, D. R. Scott, and G. Sachs. 2000. A H+-gated urea channel: the link between Helicobacter pylori urease and gastric colonization. Science 287:482-485. [DOI] [PubMed] [Google Scholar]
- 31.Wray, L. V., Jr., A. E. Ferson, and S. H. Fisher. 1997. Expression of the Bacillus subtilis ureABC operon is controlled by multiple regulatory factors including CodY, GlnR, TnrA, and Spo0H. J. Bacteriol. 179:5494-5501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Yanisch-Perron, C., L. Vieira, and J. Messing. 1985. Improved M13 phage cloning vectors and host strains: nucleotide sequences of M13mp18 and pUC19 vectors. Gene 33:103-119. [DOI] [PubMed] [Google Scholar]