Abstract
Modern comparative genomics has been established, in part, by the sequencing and annotation of a broad range of microbial species. To gain further insights, new sequencing efforts are now dealing with the variety of strains or isolates that gives a species definition and range; however, this number vastly outstrips our ability to sequence them. Given the availability of a large number of microbial species, new whole genome approaches must be developed to fully leverage this information at the level of strain diversity that maximize discovery. Here, we describe how optical mapping, a single-molecule system, was used to identify and annotate chromosomal alterations between bacterial strains represented by several species. Since whole-genome optical maps are ordered restriction maps, sequenced strains of Shigella flexneri serotype 2a (2457T and 301), Yersinia pestis (CO 92 and KIM), and Escherichia coli were aligned as maps to identify regions of homology and to further characterize them as possible insertions, deletions, inversions, or translocations. Importantly, an unsequenced Shigella flexneri strain (serotype Y strain AMC[328Y]) was optically mapped and aligned with two sequenced ones to reveal one novel locus implicated in serotype conversion and several other loci containing insertion sequence elements or phage-related gene insertions. Our results suggest that genomic rearrangements and chromosomal breakpoints are readily identified and annotated against a prototypic sequenced strain by using the tools of optical mapping.
Microbial genomes are being sequenced at an increasing pace, and we are rapidly building a detailed molecular picture of the microbial world that is yielding new biological paradigms on a weekly basis. With over 140 finished bacterial genomes now publicly available, and a larger number in progress, these data are scientific touchstones for their respective communities, in addition to establishing the molecular basis for microbial diversity studies. Improvements to sequencing technologies have reduced the cost of whole-genome sequencing, bestowing less-well-studied microbes with sequence data sets and the modern analysis approaches they engender. Microbial genomes, however, are characterized by extensive intraspecific variation, in that different strains or types within the same species can vary by as much as 20% in gene content (2, 11, 23, 25). Thus, until radical changes in sequencing technologies become commercial realities, the study of microbial variation represented by countless numbers of strains and isolates will not fully benefit from the advances offered by large-scale sequencing efforts.
To fill this void, a series of genome fingerprinting approaches have been developed that reveal genome alterations but cannot directly link such data with sequence information. This problem is typified by pulsed-field gel electrophoresis (33) analysis or macrorestriction physical mapping, in which gels show novel band patterns between strains that cannot be directly linked to molecular data or annotation. More recently, DNA microarray analysis has solved some of these issues (17), but it generally discerns only the presence, absence, or duplication of open reading frames. Finally, there are also PCR-based approaches used for whole-genome analysis of microbial populations, such as PCR-based subtractive hybridization (15, 35), octamer-based genome scanning (22), and whole-genome PCR scanning, etc. (29), which also allow the detection of genomic variation among different strains or isolates. What all of these approaches share is the inability to achieve a detailed and comprehensive whole-genome view that readily permits the discovery and characterization of novel chromosomal alterations.
We describe here how optical mapping (4-8, 10, 16, 20, 24, 26-28), an emerging single molecule system for whole-genome mapping (Fig. 1), analyzes bacterial strains to identify indels, inversions, and gross rearrangements. Notably, we show analysis based on sequence and optical maps data that identifies breakpoints caused by such genomic events with direct links to sequence information. This is further demonstrated by the discovery of novel indels in the Shigella flexneri serotype Y (AMC[328Y]) genome through the alignment of an optical map against several sequenced S. flexneri strains (2a 2457T and 301) and associated genes contributing to serotype conversion, identified by annotation borrowed from these strains. Such results point to the use of optical mapping to widely characterize genomic variation present in strains against sequenced genomes that have been chosen to represent entire species.
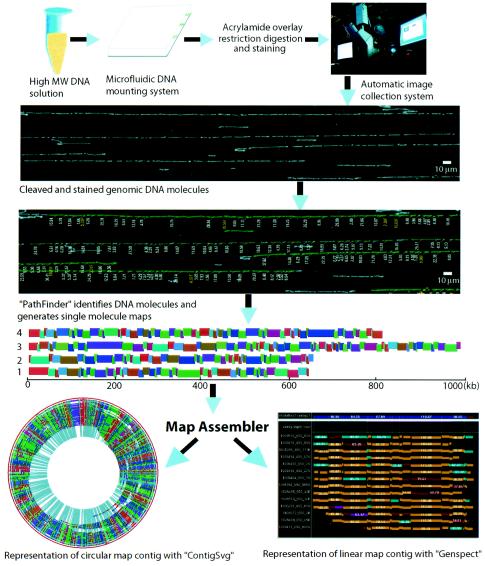
FIG. 1.
Overview of the optical mapping system. The optical mapping system is fully integrated, incorporating microfluidics, surface modalities for molecular interrogation, operator-free image acquisition, machine vision, molecule-to-map analysis, aligning software, database structures for all operations and a myriad of user interfaces for data acquisition and visualization (see Materials and Methods). Analysis starts with the deposition of high-molecular-weight DNA onto optical mapping surfaces by using a microfluidic device (16). Restriction digestion cleaves surface-bound DNA molecules; fluorochrome staining enables fluorescence microscopy to image and size contiguous restriction fragments. This is now automatically performed by Pathfinder machine vision software, whose output is large map files. An interactive image viewer, Omari, enables the user to rapidly browse and interact with large superimages consisting of hundreds of overlapped digital micrographs showing genomic DNA molecules, and the fragment mass measurements are determined by Pathfinder. These map files are overlapped to construct whole-genome maps with the map assembler and viewed by Genspect, which displays aligned maps, linked annotation and presents the user with a variety of editing tools and analysis. A new imaging system (Genome Zephyr) can acquire and process ∼2,000 images/h or 60,000 images in ∼30 h, corresponding to ∼4-fold coverage of the human genome.
MATERIALS AND METHODS
DNA preparation.
S. flexneri serotype Y strain AMC[328Y] (ATCC 9473) genomic DNA gel inserts (33) were prepared from a culture grown in Luria-Bertani medium at 37°C. Prior to use, the DNA gel inserts were washed thoroughly overnight in TE (10 mM Tris, 1 mM EDTA [pH 8.0]) to remove excess EDTA. The washed inserts were melted at 72°C for 7 min, and the melted agarose was digested at 42°C for 2 h in β-agarase solution (New England Biolabs, Boston, Mass.; 100 μl of TE plus 2 μl [1 U] of β-agarase per 20 μl of agarose). Suitable DNA dilutions were made from this sample with TE by using centrifugation (Eppendorf microfuge; 6,000 rpm, 30 min) to ensure uniform dilution and to minimize the presence of supercoiled plasmid DNA on optical mapping surfaces. Lambda DASH II bacteriophage DNA (Stratagene) was added to the genomic DNA solution (10 pg/μl) as an internal standard for fragment sizing. Such DNA samples were mounted onto an optical mapping surface and examined by fluorescence microscopy to check the molecular integrity and concentration.
Surface preparation.
Glass coverslips (22 by 22 mm; Fisher Finest [Fisher Scientific]) were cleaned and derivatized as described previously (42). The surface properties were assayed by digesting lambda DASH II bacteriophage DNA with 40 U of BamHI diluted in 100 μl of digestion buffer with 0.02% Triton X-100 (Sigma) at 37°C to determine optimal digestion times, which ranged from 40 to 120 min.
DNA mounting, overlay, digesting, and staining.
DNA molecules were mounted on derivatized glass surfaces by using a microfluidic device (16). Then, a thin layer of acrylamide (3.3%; 29 parts acrylamide to 1 part N,N′-methylene-bisacrylamide, 0.075% ammonium persulfate, 0.1% tetramethylethylenediamine, and 0.02% Triton X-100 [Sigma]) was applied to the surface, which upon polymerization was washed with 400 μl of TE for 2 min, followed by washing with 200 μl of digestion buffer for another 2 min. To set up the digestion, 200 μl of digestion buffer with enzyme (20 μl of NEB [New England Biolabs] 10× buffer 2, 176 μl of high-purity water, 2 μl of 2% Triton X-100 [Sigma], and 2 μl of NEB-BamHI [20 U/μl]) was added to the surface, followed by incubation in a humidified chamber at 37°C for 40 to 120 min. After digestion, the surface was washed twice by adding 400 μl of TE and aspirated off, 2 to 5 min each time. The surface was mounted onto a glass slide with 12 μl of 0.2 μM YOYO-1 solution (containing 5 parts YOYO-1 {1,1′-[1,3-propanediylbis[(dimethyliminio)-3,1-propanediyl]]bis[4-[(3-methyl-2(3H)-benzoxazolylidene)methyl]]-tetraiodide; Molecular Probes, Eugene, Oreg.} and 95 parts β-mercaptoethanol in 20% [vol/vol] TE). The sample was sealed with nail polish and incubated (4°C, in the dark) for 20 min or overnight for the staining dye to diffuse before checking by fluorescence microscopy.
Image acquisition and processing.
DNA samples were imaged by fluorescence microscopy as previously described with a ×63 objective lens (Zeiss, Oberkochen, Germany) and a high-resolution digital camera (Princeton Instruments) (26, 42). Images were collected by using a fully automated image acquisition system developed by our laboratory (ChannelCollect). Comounted lambda DASH II DNA molecules were used to estimate the digestion rate and to provide internal fluorescence standards for accurately sizing the DNA fragments (6, 27). The image files were processed to create maps by using previously described software (26, 42) and recently developed machine vision software (Pathfinder) (Fig. 1 and unpublished results).
Recent efforts have focused on boosting the throughput of the optical mapping system through enhanced image acquisition (laser illumination and advanced digital cameras) and effective machine vision (Pathfinder) that has enabled operator-free analysis of molecular image data sets. These advances have been incorporated within a fully integrated system featuring linked experimental databases, viewers, and seamless access to large computational clusters. Such developments have made possible the large-scale analysis of bacterial populations.
Optical map assembly.
Individual molecule restriction maps were overlapped by dedicated optical map assembler software (4-8, 10, 20, 24, 26-28). Briefly, the software assembles single molecule restriction maps into a genome-wide map contig by using a computationally efficient algorithm with limited backtracking for finding an almost optimal scoring set of map contigs in order to avoid the high computational complexity that would occur in attempting to find the optimal assembly. Bayesian inference techniques were used to estimate the probability that two distinct single molecule restriction maps could have been derived from the proposed placement while subject to various data errors such as sizing errors, missing restriction sites (missing cuts), and false cut errors. The Bayesian inference approach required the fine tuning of these parameters and a known prior statistical distribution of error sources. Important measures of data quality, such as measurement standard deviations, digestion rate, false cut, and false match probability, can be reestimated from the data by using a limited number of iterations of Bayesian probability density maximization. After these parameters were correctly estimated from the data, the best offset and alignment between a pair of maps was computed by an efficient dynamic programming algorithm.
Map homologies.
Map homologies were scored by first using a sliding window to break a whole-genome restriction map (optical or in silico map) into “segments” consisting of 10 consecutive restriction fragments, at two-fragment intervals. This produced a series of overlapping map segments, which were pairwise aligned and merged (with other alignments) by using a modified version of the map assembler, against a second reference map constructed from a mapped or sequenced genome. Since the map assembler performs global alignments, only highly congruent maps were aligned. Differences stemming from fragment sizing errors and missing or spurious cut sites have been previously modeled and accounted for within the assembly software. However, gross local map differences were not accounted for in this alignment process and were partly compensated for by the alignment of relatively small (10 fragment) maps against the reference. Resulting alignments were merged into single consensus maps for comparison against the reference map. The merging process produces a single consensus map in much the same way single-molecule maps are combined to create a whole-genome map. As such, some regions of homology across a given pair of strains may not have been accounted for. Given these caveats, we estimated the percentage of genome homology by simply summing the fragment sizes of homologous regions, defined as only regions covered by the just-described alignment and merging process.
Coding versus noncoding restriction enzyme cleavage sites were tabulated by comparing the nucleotide coordinates of the given enzyme recognition sites in the genome sequence with the coordinate ranges for the genes in genome sequence annotation. If the coordinates for any given restriction site were within the coordinate range of any given gene, this restriction site was considered within a coding region. All other restriction sites were scored as residing within noncoding regions.
Annotation.
Variant loci detected by map comparisons were characterized by using annotation derived from sequenced S. flexneri strains. Basically, the coordinate ranges for the fragments, which varied among these strains, were guided by the whole-genome in silico maps. Thus, identified, corresponding sequences were aligned at the nucleotide level by using MegAlign (DNAStar, Madison, Wis.) to recognize insertion or deletions between the two sequenced strains and annotation from the National Center for Biotechnology Information (NC_004337 and NC_004741).
RESULTS
Map comparison approach. (i) Strategy.
Bacterial genomes are quite dynamic and frequently exhibit dramatic rearrangement events. Bacterial evolution not only involves DNA sequence divergence among orthologous genes but also the gain of new genes from phages, other clones, or species or the loss of existing genes due to selection or redundancy (11, 25). With this in mind we reasoned that, since large genomic alterations are discernible by ordinary restriction fingerprinting, high-resolution optical maps would provide unique insights into genome dynamics through comparisons of optical maps versus restriction maps constructed in silico from available whole-genome sequence data. Our current map assembler (4-6, 24, 42) was designed to perform global alignments of optical maps; however, we used a sliding window consisting of 10 consecutive restriction fragments over an entire genome to perform a series of alignments to closely approximate local pairwise comparisons (see Materials and Methods). This process produced hundreds of alignments that, after merging, yielded continuous segmental restriction maps; the same map assembler software was used for this purpose. Such comparisons should reveal inversions, insertions, deletions, duplications, and translocations while also providing orientation (Fig. 2). These comparisons would also enable the detailed tabulation of breakpoints within the resolution afforded by the average size of a restriction fragment (6 to 30 kb).
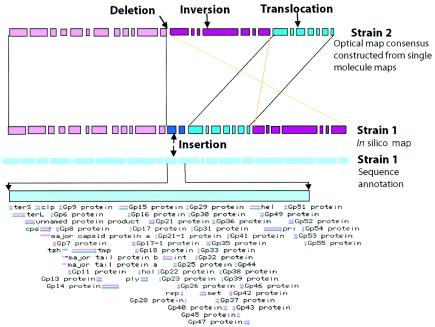
FIG. 2.
Map strategy for strain comparisons. (Bars represent consecutive restriction fragments; spaces show cleavage sites.) Optical maps are high-resolution, whole genome, ordered restriction maps that are compared against restriction maps constructed from whole-genome sequence data. Average restriction fragment sizes depend on the base composition of a strain and enzyme choice. Typically, optical maps show average restriction fragment sizes of from 6 to 30 kb. Map alignments reveal differences at the chromosome level to include: insertions, translocations, inversions, and deletions. These differences are considered at this level of resolution as a collection of breakpoints, indicative of genomic rearrangements, since the probability of picking up point mutations is low by restriction analysis. Breakpoints and their intervening map regions are annotated by direct locus comparisons against the sequenced strain. Confidence in such comparisons can be buttressed by analysis of PCR amplicons from mapped DNA template and primers synthesized from the sequenced strain.
(ii) Consequences of enzyme choice on map comparisons.
Since map comparisons are only meaningful between strains or isolates of a given species, it was useful to parse restriction enzymes into groups that honed into conserved versus variable genomic regions. For example, regions bearing housekeeping genes will be more conserved since operon organization is more likely to be conserved compared to those bearing hypervariable or noncoding regions (31, 40).
We evaluated this concept by comparing the in silico restriction maps of Yersinia pestis CO-92 and KIM strains and S. flexneri 2a 2457T versus Escherichia coli K-12 (9, 14, 30, 37). The map alignments used the procedures described in Materials and Methods, and restriction sites were grouped according to coding versus noncoding loci (Table 1). With Y. pestis CO-92 as the reference genome versus KIM, the extent of map homology ranged from 2.31 to 4.23 Mb or from 49.7 to 90.0% for the listed restriction enzymes (edited for optical mapping use). Enzymes that cleave too frequently produce an excessive number of small (<500 bp) fragments that are not reliably detected by optical mapping, and that is why these enzymes were not included in this analysis. The same analysis for S. flexneri 2a 2457T and E. coli K-12 (the reference strain) showed map homologies ranging from 1.37 to 2.17 Mb or 29.7 to 47.1%.
TABLE 1.
Homologous regions identified by map comparison with different restriction enzymes and distributions of the restriction sites in coding ORF regions
| Strain Comparison | Enzyme (sequence) | Avg fragment size (kb) | Genomes
|
Restriction sites
|
||
|---|---|---|---|---|---|---|
| Size/reference genome size (Mb)a | % | No. of sites in the coding region/total no. of sites | % | |||
| Y. pestis KIM vs. CO-92 | XhoI (CTCGAG) | 17.83 | 2.41/4.65 | 51.8 | 196/265 | 74.0 |
| Y. pestis KIM vs. CO-92 | SacI (GAGCTC) | 22.06 | 3.08/4.65 | 66.2 | 178/212 | 84.0 |
| Y. pestis KIM vs. CO-92 | PvuII (CAGCTG) | 10.34 | 4.23/4.65 | 91.0 | 372/391 | 95.1 |
| Y. pestis KIM vs CO-92 | XbaI (TCTAGA) | 29.67 | 3.36/4.65 | 72.3 | 94/156 | 60.3 |
| Y. pestis KIM vs CO-92 | SwaI (ATTTAAAT) | 27.87 | 2.80/4.65 | 60.2 | 97/165 | 58.8 |
| Y. pestis KIM vs. CO-92 | PacI (TTAATTAA) | 22.54 | 2.31/4.65 | 49.7 | 89/203 | 43.8 |
| S. flexneri 2a 2457T vs 301 | BamHI (GGATCC) | 9.78 | 4.56/4.61 | 98.9 | 444/473 | 93.9 |
| S. flexneri 2a 2457T vs E. coli K-12 | XhoI (CTCGAG) | 28.31 | 1.82/4.61 | 39.5 | 162/177 | 91.5 |
| S. flexneri 2a 2457T vs E. coli K-12 | HindIII (AAGCTT) | 8.27 | 1.85/4.61 | 40.1 | 484/556 | 87.1 |
| S. flexneri 2a 2457T vs E. coli K-12 | BamHI (GGATCC) | 9.62 | 2.17/4.61 | 47.1 | 466/495 | 94.1 |
| S. flexneri 2a 2457T vs E. coli K-12 | PciI (ACATGT) | 10.28 | 1.37/4.61 | 29.7 | 387/477 | 81.1 |
| S. flexneri 2a 2457T vs E. coli K-12 | PspAI/SmaI (CCCGGG) | 8.45 | 2.11/4.61 | 45.8 | 357/400 | 89.3 |
| S. flexneri 2a 2457T vs E. coli K-12 | SalI (GTCGAC) | 9.44 | 2.06/4.61 | 44.7 | 448/526 | 85.2 |
That is, the size of the homologous regions identified versus the reference genome size.
Evaluation of optical mapping errors in comparative analysis.
Since optical maps are accurate but omit map information that is <500 bp in size and show sizing errors of ca. 2 to 5%, it is important to consider how such errors may affect map comparison results across strains. Although we have software (unpublished) that simulates errors common to optical mapping, we chose to use a previously published PvuII optical map of Y. pestis KIM (41) to directly evaluate comparison results against an in silico map derived from sequence data; Y. pestis CO-92 was used as the reference strain for both types of comparisons. The purely in silico comparisons between both Y. pestis strains showed 21 conserved map segments ranging from 15.79 to 639.64 kb, 9 of which were inversions and translocations, whereas 12 were solely translocations. The combined size of the homologous regions was 4.23 Mb, with significant rearrangements apparent (data not shown). There were also six large map segments that did not align since they were composed of ≤3 restriction fragments that cannot be confidently aligned. The optical versus in silico PvuII map comparison (Fig. 3A) produced the same alignments but, as expected, differed slightly in the degree of homology (2.12%) due to optical mapping sizing errors.
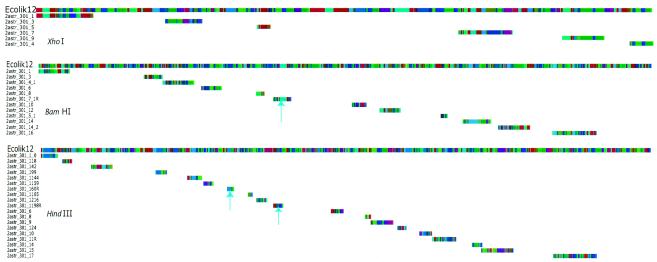
FIG. 3.
Strain comparisons via maps. Map comparisons were performed as described in Materials and Methods. Multicolored tracks represent restriction maps, with each colored bar showing the length of a restriction fragment. Green arrows point to the reference genome. Yellow lines show homologous regions in the same orientation as in the reference genome, and pale blue lines show inverted homologies. (A) Optical versus in silico PvuII map comparisons between Y. pestis CO-92 and KIM strains; extensive rearrangements are apparent. (B) Optical versus in silico BamHI map comparisons of S. flexneri serotype 2a (strains 301and 2457T) and of S. flexneri serotype Y (strain AMC[328Y]). Rearrangements are apparent between 301 and 2457T, whereas AMC[328Y] shows apparent congruency. Fine-scale differences are shown in Fig. 5. (C) Genome sequence comparison between strain 301 and 2457T genomes (37) showing overall homology and inverted regions.
Comparison between S. flexneri serotypes Y and 2a.
Since S. flexneri serotype Y (AMC[328Y]) has not been sequenced, we constructed a BamHI optical map for comparison (see Materials and Methods) to the sequenced strains 301 and 2457T. BamHI was selected to maximize homology based on the analysis of the enzyme choice effect on map comparison. A whole-genome BamHI optical map was generated by aligning 217 of 246 collected single DNA molecular maps (see Materials and Methods) with a total mass of 158.87 Mb, which is ca. 35 times the coverage based on the optically estimated genome size of 4.53 Mb for AMC[328Y] strain (Fig. 4). The comparisons showed a high degree of congruency between the AMC[328Y] optical map and the 301 in silico map from genome sequence (19) (Fig. 3B); however, there were 14 regions that displayed differences either in fragment size, restriction sites, or missing fragments. Some of these differences are shown in Fig. 5. In contrast, the purely in silico comparisons between 2457T and 301 showed more striking differences with seven homologous map segments and two inversions identified between these two strains, totaling 4.52 Mb or 98% of the reference genome size (301 genome). Local differences are presented in Fig. 5.
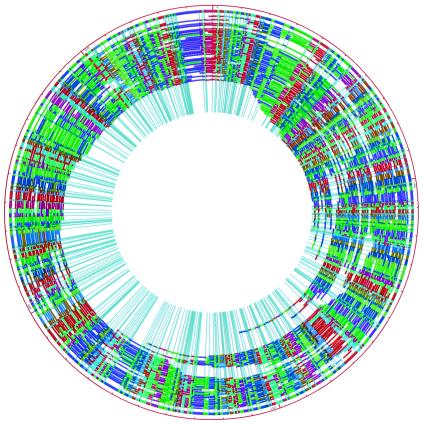
FIG. 4.
Whole-genome BamHI map contig of S. flexneri serotype Y strain AMC[328Y] displayed by using Contigsvg software (written in our laboratory). The outermost color circle is the consensus map generated by the map assembler and is built from the underlying maps represented as arcs. These maps were constructed from individual DNA molecules cleaved with BamHI. Congruent restriction fragments shown in the consensus map are denoted by a common color; the color ordering scheme is random to provide contrast; total genome size is 4.53 Mb.
FIG. 5.
Identification and annotation of fine scale map differences. Genspect views show S. flexneri strain differences at selected map loci. Maps are represented as undulating bars, with restriction fragment sizes noted in kilobases. Pale blue arrows indicate deletions, deep blue arrows indicate insertions, and yellow arrows indicate differences between all strains. (A) Deletion in AMC[328Y] strain. Corresponding fragments in strains 301 and 2457T harbor glucosyltransferase genes. (B) Deletion in 301 strain. The indicated shrinking noted fragment, corresponding to the 2457T fragment, contains phage-related genes. (C) Extra restriction site in strains AMC[328Y] and 2457T caused by an insertion sequence element insertion. (D) 12.48-kb insertion in the 34.65-kb restriction fragment in 2457T. It contains iron uptake genes (sitA, -B, -C, and -D). (E) The 6.68-kb (strain 301) and 9.14-kb (strain 2457T) fragments contain the biosynthesis cofactors, the CobU, CobS, and CobT genes, and an alpha-helix protein gene. The two extra restriction fragments and additional sequence in 9.14-kb (strain 2457T) fragment contain several insertion sequence element insertions.
Annotation (see Materials and Methods) of these local differences showed that a two-fragment deletion (14.29 kb; based on strain 301 genome map) in the serotype Y strain AMC[328Y] (Fig. 5A) corresponded to fragments in strains 301 and 2457T coding for bactoprenol glucosyltransferase (gtrII, gtrAI, and gtrBI) (3). Figure 5D shows an insertion within this locus of the 2457T genome map that was 12.48 kb larger than the corresponding AMC[328Y] fragment and, based on sequence annotation codes for iron uptake genes (SitA, -B, -C, and -D), is commonly associated with intracellular pathogens (32). Other differences are noted and annotated in Fig. 5.
Comparison between S. flexneri 2a 301 and E. coli K-12.
Distantly related bacterial strains are commonly compared at the sequence level, so we wanted to evaluate the efficacy of map comparisons (Fig. 6) for such cases. As an example, we compared the XhoI, BamHI, and HindIII in silico maps of S. flexneri 2a 301 with E. coli K-12. The results varied. XhoI identified six homologous regions (101.46 to 396.06 kb) with a total size of 1.78 Mb, and no inversions. BamHI revealed 12 homologous regions (48.59 to 337.92 kb), with a total size of 2.05 Mb, and 1 of the 12 homologous regions was inverted. HindIII produced 19 homologous map segments (31.62 to 332.09 kb), with a total size of 1.92 Mb, and 2 conserved map segments were inverted; this extra inversion is unique to HindIII. With the E. coli K-12 genome as a reference, the three enzymes largely identified common regions of homology; however, unique differences were also detected by each enzyme (Fig. 6).
FIG. 6.
In silico restriction map comparisons of E. coli K-12 and S. flexneri serotype 2a strain 301 (with XhoI, BamHI, and HindIII). Restriction maps of the respective enzymes are displayed as in Fig. 3 (long multicolored tracks). Pale blue arrows show inversions. Other tracks show maps of S. flexneri homology with E. coli. We used E. coli K-12 as the reference for all shown alignments. These enzymes showed similar, though not congruent, homology patterns.
DISCUSSION
As the reach of modern sequencing technologies extends to cover many of the more well-studied prokaryotic genomes, important insights are being derived from approaches (17) that closely analyze the genomes of strains within a given species, since strains often differ in their ability to adapt to new environments or hosts, to cause disease, to metabolize new substrates, etc. (13, 39). Since sequencing technologies (34) are still not sufficiently advanced to enable the massive analysis of a large number of strains or isolates, high-resolution physical maps can fill this need if such maps can be rapidly constructed and appropriately analyzed. An important difference between optical mapping data and other approaches is that prior hypotheses are not required for analysis, and this virtue enhances discovery, and an informative physical map facilitates the rapid characterization of such findings within the context of whole-genome data. Although optical maps do not approach the resolution of whole-genome sequence data, genomic rearrangements are made obvious and breakpoints are readily indexed against a sequenced prototypic strain. This is difficult to do with common genome fingerprinting approaches, which also reveal genomic alterations but obscure locus information. As such, we predict that optical mapping will supplant pulsed-field gel electrophoresis fingerprinting for the characterization and typing of bacterial strains. Consider that optical map databases can be queried and used in much the same way as Pulsenet (http://www.cdc.gov/pulsenet/) given new alignment tools that might function like BLAST but use map information in lieu of sequence data.
Overall, the results presented here have leveraged publicly available sequence data to construct in silico restriction maps of whole genomes (Y. pestis and S. flexneri strains), for comparison by using the tools of optical mapping. More specifically, we have shown that our previously published optical map alignment software can reveal important genomic structural alterations, including indels, inversions, and gross rearrangements.
The optical versus in silico map comparisons for three strains of S. flexneri also showed the power of optical mapping for comparative genome analysis among closely related strains within a species. We constructed an optical map of an unsequenced strain, S. flexneri serotype Y AMC[328Y], and compared it to the geographically and temporally separated S. flexneri serotype 2a strains 301 and 2457T, which have been sequenced (19, 37) and compared; notably, 2457T showed three inversions versus 301 (Fig. 3C) (37). The largest inversion (900 kb) contained the replication origin, whereas the others were located near the terminus. We used these sequence comparisons to gauge the effectiveness of our in silico versus in silico map comparisons. They were quite similar. 2457T showed seven conserved map segments; two of these were inversions: 876 kb (containing the replication origin) and 72 kb (near the terminus) (Fig. 3C) (37). We were unable to confidently place the small, ∼20-kb inversion identified by sequence analysis.
The map comparison between the S. flexneri serotype Y strain AMC[328Y] and the serotype 2a strain 301 and 2457T showed that the AMC[328Y] genome was highly similar to the strain 301 genome in terms of overall genome structure and organization. However, in some small regions the AMC[328Y] genome was more similar to the 2457T genome than the 301 genome, whereas in other small regions the AMC[328Y] genome differed from both strains 301 and 2457T (Fig. 5). These results were initially surprising; however, serotype conversion due to O-antigen modification mediated by serotype-converting bacteriophages is not uncommon in S. flexneri (1, 3, 12, 18), and further background checks of AMC[328Y] indicated that it was actually a degraded isolate of serotype 2a (38). Consistent with these data, we identified a deletion of the glucosyltransferase genes, that could be linked to the serotype conversion inherent to the mapped, but unsequenced AMC[328Y] strain.
The comparisons of in silico versus in silico maps between the distantly related strains E. coli K-12 and S. flexneri serotype 2a strain 301 differed according to the choice of restriction enzyme. Our study used three restriction enzymes that revealed some differences in the extent and location of chromosome homology. However, most of the homologous regions were identified by all three enzymes (Fig. 6). Although Shigella spp. and E. coli are considered strains within the same species (21), sufficient differences exist that were revealed by the modest degree of map homology (<50%). Comparison of the genome sequences of E. coli K-12 and S. flexneri 2a 301 or 2457T, which suggested that these two strains (301 and 2457T) share ∼80% backbone with K-12, showed that the total homology identified by map comparison is relatively low. These and other results presented here suggest that a battery of enzyme maps be used to fully and confidently discern homologies between mapped strains.
Although we have demonstrated here how optical mapping reveals differences among closely related isolates or strains, the ability of ordered restriction maps to confidently discern genome map homologies naturally diminishes as the evolutionary distance between organisms increase (36), as reflected in part by the variability of genome structural elements, codon usage, and functional motifs represented by vastly different nucleotide compositions. Obviously, analysis of sequence data that enables alignments at the amino acid level represents the most general approach. As such, low-pass shotgun sequencing (one- to threefold coverage), which commonly produces a string of sequence contigs, could be combined with optical maps to reveal large-scale genomic features that would only be apparent after additional sequence coverage and expensive finishing efforts. Essentially, these issues come down to considerations of cost and throughput. Given the recent developments in the optical mapping system (16), the analysis of collections of 100 strains now becomes practical at a cost of about one-tenth that of low-pass sequencing and without the need to construct whole-genome libraries.
In summary, optical mapping was demonstrated to reveal genomic differences that were directly linked to available sequence or map data. This direct linkage to sequence information allows PCR amplicons to be made and sequenced to identify small novel insertions or to confirm results. Such genomic differences might evade detection by common genome fingerprinting approaches, which do not offer simple routes to the full characterization of genomic breakpoints and the identification of novel insertions or elements.
Acknowledgments
This study was supported by a grant from the DOE DE-FC02-01ER63175 to D.C.S.
We thank Thomas Anantharaman for software efforts on map assembly, Matthew Peterson for systems, and Konstantinos Potamousis for workstation engineering.
Footnotes
Presented in part at the 11th International Conference on Microbial Genomes, Durham, N.C., 28 September to 2 October 2003.
REFERENCES
- 1.Adhikari, P., G. Allison, B. Whittle, and N. K. Verma. 1999. Serotype 1a O-antigen modification: molecular characterization of the genes involved and their novel organization in the Shigella flexneri chromosome. J. Bacteriol. 181:4711-4718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Akopyants, N. S., A. Fradkov, L. Diatchenko, J. E. Hill, P. D. Siebert, S. A. Lukyanov, E. D. Sverdlov, and D. E. Berg. 1998. PCR-based subtractive hybridization and differences in gene content among strains of Helicobacter pylori. Proc. Natl. Acad. Sci. USA 95:13108-13113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Allison, G. E., and N. K. Verma. 2000. Serotype-converting bacteriophages and O-antigen modification in Shigella flexneri. Trends Microbiol. 8:17-22. [DOI] [PubMed] [Google Scholar]
- 4.Anantharaman, T. S., B. Mishra, and D. C. Schwartz. 1999. Genomics via optical mapping III: contiging genomic DNA and variations. Int. Conf. Intelligent Systems Mol. Biol. 7:18-27. [PubMed] [Google Scholar]
- 5.Anantharaman, T. S., B. Mishra, and D. C. Schwartz. 1998. Genomics via optical mapping III: contiging genomic DNA and variations. Courant technical report 760. Courant Institute, New York University, New York, N.Y.
- 6.Anantharaman, T. S., B. Mishra, and D. C. Schwartz. 1997. Genomics via optical mapping 2: ordered restriction maps. J. Comput. Biol. 4:91-118. [DOI] [PubMed] [Google Scholar]
- 7.Aston, C., C. Hiort, and D. C. Schwartz. 1999. Optical mapping: an approach for fine mapping. Methods Enzymol. 303:55-73. [DOI] [PubMed] [Google Scholar]
- 8.Aston, C., B. Mishra, and D. C. Schwartz. 1999. Optical mapping and its potential for large-scale sequencing projects. Trends Biotechnol. 17:297-302. [DOI] [PubMed] [Google Scholar]
- 9.Blattner, F. R., G. Plunkett III, C. A. Bloch, N. T. Perna, V. Burland, M. Riley, J. Collado-Vides, J. D. Glasner, C. K. Rode, G. F. Mayhew, J. Gregor, N. W. Davis, H. A. Kirkpatrick, M. A. Goeden, D. J. Rose, B. Mau, and Y. Shao. 1997. The complete genome sequence of Escherichia coli K-12. Science 277:1453-1474. [DOI] [PubMed] [Google Scholar]
- 10.Cai, W., J. Jing, B. Irvin, L. Ohler, E. Rose, H. Shizuya, U. Kim, M. Simon, T. Anantharaman, B. Mishra, and D. C. Schwartz. 1998. High-resolution restriction maps of bacterial artificial chromosomes constructed by optical mapping. Proc. Natl. Acad. Sci. USA 95:3390-3395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Casjens, S. 1998. The diverse and dynamic structure of bacterial genomes. Annu. Rev. Genet. 32:339-377. [DOI] [PubMed] [Google Scholar]
- 12.Chen, J., W. Hsu, C. Chiou, and C. Chen. 2003. Conversion of Shigella flexneri serotype 2a to serotype Y in a shigellosis patient due to a single amino acid substitution in the protein product of the bacterial glucosyltransferase gtrII gene. FEMS Microbiol. Lett. 224:277-283. [DOI] [PubMed] [Google Scholar]
- 13.Claverys, J. P., M. Prudhomme, I. Mortier-Barriere, and B. Martin. 2000. Adaptation to the environment: Streptococcus pneumoniae, a paradigm for recombination-mediated genetic plasticity? Mol. Microbiol. 35:251-259. [DOI] [PubMed] [Google Scholar]
- 14.Deng, W., V. Burland, G. Plunkett III, A. Boutin, G. F. Mayhew, P. Liss, N. T. Perna, D. J. Rose, B. Mau, S. Zhou, D. C. Schwartz, J. D. Fetherston, L. E. Lindler, R. R. Brubaker, G. V. Plano, S. C. Straley, K. A. McDonough, M. L. Nilles, J. S. Matson, F. R. Blattner, and R. D. Perry. 2002. Genome sequence of Yersinia pestis KIM. J. Bacteriol. 184:4601-4611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dietchenko, L., Y. F. C. Lau, A. P. Campell, A. Chenchik, F. Moqadam, B. Huang, S. Lukyanov, K. Lukyanov, N. Gurskaya, E. D. Sverdlov, and P. D. Siebert. 1996. Suppression subtractive hybridization: a method for generating differentially regulated or tissue-specific cDNA probes and libraries. Proc. Natl. Acad. Sci. USA 93:6025-6030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Dimalanta, E. T., R. Runnheim, A. Lim, C. Lamers, C. Churas, D. K. Forrest, M. D. Graham, J. J. de Pablo, S. N. Coppersmith, and D. C. Schwartz. 2004. A microfluidic system for large DNA molecule arrays. Anal. Chem. 76:5293-5301. [DOI] [PubMed] [Google Scholar]
- 17.Hinchliffe, S. J., K. E. Isherwood, R. A. Stabler, M. B. Prentice, A. Rakin, R. A. Nichols, P. C. F. Oyston, J. Hinds, R. W. Titball, and B. W. Wren. 2003. Application of DNA microarrays to study the evolutionary genomics of Yersinia pestis and Yersinia pseudotuberculosis. Genome Res. 13:2018-2029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Huan, P. T., B. L. Whittle, D. A. Bastin, A. A. Lindberg, and N. K. Verma. 1997. Shigella flexneri type-specific antigen V: cloning, sequencing and characterization of the glucosyl transferase gene of temperate bacteriophage SFV. Gene 195:207-216. [DOI] [PubMed] [Google Scholar]
- 19.Jin, Q., Z. Yuan, J. Xu, Y. Wang, Y. Shen, W. Lu, J. Wang, H. Liu, J. Yang, F. Yang, X. Zhang, G. Yang, H. Wu, D. Qu, J. Dong, L. Sun, Y. Xue, A. Zhao, Y. Gao, J. Zhu, B. Kan, K. Ding, S. Chen, H. Cheng, Z. Yao, B. He, R. Chen, D. Ma, B. Qiang, Y. Hou, and J. Yu. 2002. Genome sequence of Shigella flexneri 2a: insights into pathogenicity through comparison with genomes of Escherichia coli K-12 and O157. Nucleic Acids Res. 30:4432-4441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jing, J., Z. Lai, C. Aston, J. Lin, D. J. Carucci, M. J. Gardner, B. Mishra, T. Anantharaman, H. Tettelin, L. M. Cummings, S. L. Hoffman, J. C. Venter, andD. C. Schwartz. 1999. Optical mapping of Plasmodium falciparum chromosome 2. Genome Res. 9:175-181. [PMC free article] [PubMed] [Google Scholar]
- 21.Johnson, J. 2000. Shigella and Escherichia coli at the crossroads: Machiavellian masqueraders or taxonomic treachery? J. Med. Microbiol. 49:583-585. [DOI] [PubMed] [Google Scholar]
- 22.Kim, J., J. Nietfeldt, and A. K. Benson. 1999. Octamer-based genome scanning distinguishes a unique subpopulation of Escherichia coli O157:H7 strains in cattle. Proc. Natl. Acad. Sci. USA 96:13288-13293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kudva, I. T., P. S. Evans, N. T. Perna, T. J. Barret, F. M. Ausubel, F. R. Blattner, and S. B. Calderwood. 2002. Strains of Escherichia coli O157:H7 differ primarily by insertions or deletions, not single-nucleotide polymorphisms. J. Bacteriol. 184:1873-1879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lai, Z., J. Jing, C. Aston, V. Clarke, J. Apodaca, E. T. Dimalanta, D. J. Carucci, M. J. Gardner, B. Mishra, T. S. Anatharaman, S. Paxia, S. L. Hoffman, J. C. Venter, E. J. Huff, and D. C. Schwartz. 1999. A shotgun optical map of the entire Plasmodium falciparum genome. Nat. Genet. 23:309-313. [DOI] [PubMed] [Google Scholar]
- 25.Lan, R., and P. R. Reeves. 2000. Intraspecies variation in bacterial genomes: the need for a species genome concept. Trends Microbiol. 8:396-401. [DOI] [PubMed] [Google Scholar]
- 26.Lim, A., E. T. Dimalanta, K. D. Potamousis, G. Yen, J. Apodoca, C. Tao, J. Lin, R. Qi, J. Skiadas, A. Ramanathan, N. T. Perna, G. Plunkett III, V. Burland, B. Mau, J. Hackett, F. R. Blattner, T. S. Anantharaman, B. Mishra, and D. C. Schwartz. 2001. Shotgun optical maps of the whole Escherichia coli O157:H7 genome. Genome Res. 11:1584-1593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lin, J., R. Qi, C. Aston, J. Jing, T. S. Anatharaman, B. Mishra, O. White, M. J. Daly, K. W. Minton, J. C. Venter, and D. C. Schwartz. 1999. Whole genome shotgun optical mapping of Deinococcus radiodurans. Science 285:1558-1562. [DOI] [PubMed] [Google Scholar]
- 28.Meng, X., K. Benson, K. Chada, J. E. Huff, and D. C. Schwartz. 1995. Optical mapping of lambda bacteriophage clones using restriction endonucleases. Nat. Genet. 9:432-438. [DOI] [PubMed] [Google Scholar]
- 29.Ohnishi, M., J. Terajima, K. Kurokawa, K. Nakayama, T. Murata, K. Tamura, Y. Ogura, H. Watanabe, and T. Hayashi. 2002. Genomic diversity of enteromorrhagic Escherichia coli O157 revealed by whole genome PCR scanning. Proc. Natl. Acad. Sci. USA 99:17043-17048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Parkhill, J., B. W. Wren, N. R. Thomson, R. W. Titball, M. T. Holden, M. B. Prentice, M. Sebaihia, K. D. James, C. Churcher, K. L. Mungall, S. Baker, D. Basham, S. D. Bentley, K. Brooks, A. M. Cerdeno-Tarraga, T. Chillingworth, A. Cronin, R. M. Davies, P. Davis, G. Dougan, T. Feltwell, N. Hamlin, S. Holroyd, K. Jagels, A. V. Karlyshev, S. Leather, S. Moule, P. C. Oyston, M. Quail, K. Rutherford, M. Simmonds, J. Skelton, K. Stevens, S. Whitehead, and B. G. Barrell. 2001. Genome sequence of Yersinia pestis, the causative agent of plague. Nature 413:523-527. [DOI] [PubMed] [Google Scholar]
- 31.Perna, N. T., G. Plunkett III, V. Burland, B. Mau, J. D. Glasner, D. J. Rose, G. F. Mayhew, P. S. Evans, J. Gregor, H. A. Kirpatrick, et al. 2000. Genome sequence of enterohemorrhagic Escherichia coli O157:H7. Nature 409:529-533. [DOI] [PubMed] [Google Scholar]
- 32.Runyen-Janecky, L. J., S. A. Reeves, E. G. Gonzales, and S. M. Payne. 2003. Contribution of the Shigella flexneri Sit, Iuc, and Feo iron acquisition systems to iron acquisition in vitro and in cultured cells. Infect. Immun. 71:1919-1928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Schwartz, D. C., and C. R. Cantor. 1984. Separation of yeast chromosome-sized DNAs by pulsed-field gradient gel electrophoresis. Cell 37:67-75. [DOI] [PubMed] [Google Scholar]
- 34.Sensen, C. W. 1999. Sequencing microbial genomes, p. 1-9. In R. L. Charlebois (ed.), Organization of the prokaryotic genome. ASM Press, Washington, D.C.
- 35.Straus, D., and F. M. Ausubel. 1990. Genomic Subtraction for cloning DNA corresponding to deletion mutations. Proc. Natl. Acad. Sci. USA 87:1889-1893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Waterman, M. S., T. F. Smith, and H. L. Katcher. 1984. Algorithm for restriction map comparison. Nucleic Acids Res. 12:237-242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wei, J., M. B. Goldberg, V. Burland, M. M. Venkatesan, W. Deng, G. Fournier, G. F. Mayhew, G. Plunkett III, D. J. Rose, A. Darling, B. Mau, N. T. Perna, S. M. Payne, L. J. Runyen-Janeky, S. Zhou, D. C. Schwartz, and F. R. Blattner. 2003. Complete genome sequence and comparative genomics of Shigella flexneri serotype 2a strain 2457T. Infect. Immun. 71:2775-2786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wheeler, K. M. 1944. Antigenic relationships of Shigella paradysenteriae. J. Immunol. 48:87-101. [Google Scholar]
- 39.Whittam, T. S., and A. C. Bumbaugh. 2002. Inferences from whole-genome sequences of bacterial pathogens. Curr. Opin. Genet. Dev. 12:719-725. [DOI] [PubMed] [Google Scholar]
- 40.Wolf, Y. I., I. B. Rogozin, A. S. Kondrashov, and E. V. Koonin. 2001. Genome alignment, evolution of prokaryotic genome organization, and prediction of gene function using genomic context. Genome Res. 11:356-372. [DOI] [PubMed] [Google Scholar]
- 41.Zhou, S., W. Deng, T. S. Anatharaman, A. Lim, E. T. Dimalanta, J. Wang, T. Wu, C. Tao, R. Creighton, A. Kile, E. Kvikstad, M. Bechner, G. Yen, A. Garic-Stankovic, J. Severin, D. Forrest, R. Runnheim, C. Churas, C. Lamers, N. T. Perna, V. Burland, F. R. Blattner, B. Mishra, and D. C. Schwartz. 2002. A whole-genome shotgun optical map of Yersinia pestis strain KIM. Appl. Environ. Microbiol. 68:6321-6331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zhou, S., E. Kvikstad, A. Kile, J. Severin, D. Forrest, R. Runnheim, C. Churas, J. W. Hickman, C. Mackenzie, M. Choudhary, T. Donohue, S. Kaplan, and D. C. Schwartz. 2003. Whole-genome shotgun optical mapping of Rhodobacter sphaeroides strain 2.4.1 and its use for whole-genome shotgun sequence assembly. Genome Res. 13:2142-2151. [DOI] [PMC free article] [PubMed]