Abstract
Rhodococcus sp. strain RHA1, a potent polychlorinated-biphenyl (PCB)-degrading strain, contains three linear plasmids ranging in size from 330 to 1,100 kb. As part of a genome sequencing project, we report here the complete sequence and characterization of the smallest and least-well-characterized of the RHA1 plasmids, pRHL3. The plasmid is an actinomycete invertron, containing large terminal inverted repeats with a tightly associated protein and a predicted open reading frame (ORF) that is similar to that of a mycobacterial rep gene. The pRHL3 plasmid has 300 putative genes, almost 21% of which are predicted to have a catabolic function. Most of these are organized into three clusters. One of the catabolic clusters was predicted to include limonene degradation genes. Consistent with this prediction, RHA1 grew on limonene, carveol, or carvone as the sole carbon source. The plasmid carries three cytochrome P450-encoding (CYP) genes, a finding consistent with the high number of CYP genes found in other actinomycetes. Two of the CYP genes appear to belong to novel families; the third belongs to CYP family 116 but appears to belong to a novel class based on the predicted domain structure of its reductase. Analyses indicate that pRHL3 also contains four putative “genomic islands” (likely to have been acquired by horizontal transfer), insertion sequence elements, 19 transposase genes, and a duplication that spans two ORFs. One of the genomic islands appears to encode resistance to heavy metals. The plasmid does not appear to contain any housekeeping genes. However, each of the three catabolic clusters contains related genes that appear to be involved in glucose metabolism.
Rhodococcus is a widely occurring genus of aerobic, nonmotile soil bacteria that are closely related to three other genera of GC-rich actinomycetes: Gordonia, Nocardia, and Mycobacterium. Rhodococci degrade an extraordinarily wide variety of organic substrates and thus play an important role in the global C cycle. The unusual armamentarium of enzymatic activities involved in these processes has been exploited in applications ranging from commodity chemical production to the desulfurization of fossil fuels (6). Consequently, the metabolic capabilities of rhodococci are of interest to the pharmaceutical, environmental, chemical, and energy sectors.
Rhodococcus sp. strain RHA1 is characterized by its exceptional ability to transform polychlorinated biphenyls (PCBs) (53), a particularly widespread and persistent class of environmental pollutants. It is generally thought that in aerobic bacteria, PCBs are cometabolized by the bph pathway, which is responsible for the aerobic degradation of biphenyl (23). The upper bph pathway consists of four enzymatic activities that together transform biphenyl to benzoate and 2-hydroxypenta-2,4-dienoate. For each of these four steps, RHA1 appears to possess multiple isozymes, which may help explain the strain's superior PCB-transforming capabilities. Thus, the strain contains at least three bph-type ring-hydroxylating dioxygenases (33) and at least seven different bph-type ring cleavage enzymes (51). It is unclear which of these isozymes is involved in the catabolism of biphenyl or closely related compounds and how these different activities are regulated.
The genome of Rhodococcus sp. strain RHA1 is organized into a chromosome of unknown topology and three large linear plasmids: pRHL1 (1,100 kb), pRHL2 (450 kb), and pRHL3 (330 kb). Most of the genes of the upper biphenyl catabolic pathway are located on the two largest linear plasmids (56). However, genes encoding related isozymes are distributed throughout the genome, as are the genes involved in the degradation of benzoate and 2-hydroxypenta-2,4-dienoate. Analysis of the telomeres of pRHL2 revealed the presence of terminal inverted repeats with covalently associated proteins (56). This structure is characteristic of invertrons, a class of linear elements found in a variety of bacteria, bacteriophages, and viruses (50). A second class of linear elements, found thus far in Borrelia spp. and prophage, has covalently closed hairpin loops at the termini. A probe derived from the right end of pRHL2 cross-hybridized to the pRHL1 and pRHL3 termini, suggesting that these plasmids may also be invertrons (56).
Actinomycete invertrons include plasmids and chromosomes. Although the latter have only been definitively reported to occur in streptomycetes (63), linear plasmids have been characterized in most genera of actinomycetes, including rhodococci (60), streptomycetes (28, 59, 65), planobisporetes (47), and mycobacteria (38, 50). It has been proposed that linear plasmids evolved from bacteriophages (27) and that linear chromosomes arose from the recombination of linear plasmids with circular chromosomes (12). The cores of large linear plasmids replicate bidirectionally from a unique internal origin, similar to replication of circular plasmids (9), and some linear replicons can replicate in circular forms when their telomeres are deleted (55). Regardless of their precise origins, it is clear that actinomycete invertrons are dynamic genetic elements. For example, plasmids can exchange ends with the host chromosome, mobilizing large regions of the chromosomal ends (44), and large regions of linear chromosomes can be duplicated.
As part of an effort to characterize metabolism and its genetic regulation in Rhodococcus sp. strain RHA1, we are determining the sequence of this organism's genome. Within the context of this project, we report here the complete sequence of pRHL3, the smallest of the strain's three plasmids. Sequence analysis of pRHL3 revealed the presence of several interesting plasmid-borne genes, including clusters of catabolic genes, and enabled the identification of regions that may have been acquired by horizontal transfer.
MATERIALS AND METHODS
Chemicals.
The following compounds of the indicated purity were purchased from Sigma-Aldrich: (S)-(−)-limonene (96%), (R)-(+)-limonene (97%), (−)-carveol (mixture of isotopes, 97%), and (S)-(+)-carvone (96%). All other chemicals were of analytical grade and used without further purification.
Strains, media, and growth.
Rhodococcus sp. strain RHA1 was grown at 30°C on Luria-Bertani (LB) broth or W medium supplemented with an appropriate carbon source (53). Liquid cultures of 25 ml were incubated in 125-ml Erlenmeyer flasks shaken at 200 rpm. Limonene, carvone, and carveol were provided in the vapor form to cultures on W medium. For solid medium, 5 μl of the specified compound was placed in a sterile Eppendorf tube placed in a 50-ml tube (Sardstedt) attached to the lid of a petri plate. Several holes in the lid of the petri plate permitted vapors to pass from the tube into the petri plate. The petri plates were sealed with parafilm and incubated lid down. For liquid medium, an Eppendorf tube containing substrate was suspended in the headspace of a flask. Plasmid and fosmid libraries were propagated in Escherichia coli strains DH10V and EPI10, respectively. Genomic libraries were plated on 2xYT supplemented with appropriate antibiotics and, in the case of plasmid libraries, X-Gal (5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside) and IPTG (isopropyl-β-d-thiogalactopyranoside). For cloning the telomeres and the origin of replication, E. coli JM109 was used for DNA propagation and was cultured on LB medium. All E. coli strains were grown at 37°C on medium containing the appropriate antibiotics.
Preparation of RHA1 DNA.
Linear plasmid DNA was prepared, detected, digested with restriction enzymes, and subjected to pulsed-field gel electrophoresis (PFGE) and Southern hybridization analysis as described previously (56). Genomic DNA was prepared for library construction essentially as described by Marmur (39).
Cloning of the telomeres.
Plasmid DNA was extracted by electroelution from pulsed field gels, digested with PstI, and ligated into pBluescript II SK(+) that had been linearized with PstI and EcoRV. The ligation mixture was transformed into E. coli JM109, and transformants were selected on LB agar plates containing 50 mg of ampicillin/liter, 2 mM IPTG, and 0.04% X-Gal. Plasmids pTPE1R and pTPE1L, containing 1.2- and 5.0-kb inserts, respectively, were recovered from the transformants and corresponded to the right and left telomeres of pRHL1. Plasmids pTPE3R and pTPE3L, containing 1.8- and 3.5-kb inserts, respectively, were recovered and corresponded to the right and left telomeres of pRHL3. The telomeres were subcloned into pUC18 and pUC19 and were sequenced by using the dideoxy termination method (52) and a CEQ2000XL sequencer (Beckman Coulter, Inc., Fullerton, Calif.).
Cloning of the replication region.
RHA1 genomic DNA was partially digested with MboI, and the resultant fragments were separated by agarose gel electrophoresis. Fragments of 9 to 23 kb were extracted and ligated into BamHI-digested Charomid 9-28::tsr, constructed by inserting the thiostrepton resistance gene (tsr) into the SmaI site of Charomid 9-28 (32). The resulting DNA was introduced into E. coli DH5α via in vitro packaging. Plasmid DNA was isolated from the transformants and transformed into Rhodococcus sp. strain RHA1 by electrotransformation (66). RHA1 transformants were selected on LB agar plates containing 10 μg of thiostrepton/ml. The plasmid DNA was recovered from each transformant and subjected to Southern hybridization analysis with a tsr-derived probe to detect plasmids after separation of DNA by agarose gel electrophoresis.
Fragments of the replication region were generated by digestion with restriction enzymes and were subcloned into pIJ702 (32) or pBSSK::tsr, two thiostrepton resistance vectors that are unable to replicate in RHA1. Constructs were transformed into RHA1 and selected on LB agar plates containing thiostrepton as described above. In determining the incompatibility of replication region-containing plasmids with pRHL3, the plasmid content was examined by PFGE in at least 10 independently selected transformants. For these analyses, transformants were grown in 10 ml of threefold-diluted LB medium at 30°C.
Genomic libraries.
Plasmid libraries were constructed by using one of two methods. In one method, Rhodococcus sp. strain RHA1 genomic DNA was manually sheared by using a syringe and a 25-gauge needle. Fragments of 2 to 3 kb were double gel purified, end repaired with T4 DNA polymerase plus Klenow fragment, and phosphorylated by using the T4 polynucleotide kinase. Blunt-end fragments were cloned into HincII-linearized, dephosphorylated pUC19. Colonies were analyzed for insertions by PCR (M13F-21/M13R) and restriction double digests (HindIII/XbaI).
In the second method, RHA1 genomic DNA was sheared by sonication and end repaired by limited Bal31 nuclease digestion. End-repaired DNA was run on a 1% low-melting-point agarose gel, and 2- to 3-kb fragments were excised and recovered by β-agarase digestion, phenol extraction, and ethanol precipitation. Size-selected fragments were ligated by using an ∼1,000-fold excess of BstXI adapters (Invitrogen). Excess adapter was removed by three rounds of agarose gel purification. Purified, BstXI-adapted fragments were inserted into BstXI-linearized pBR194c plasmid.
A fosmid library containing 40-kb inserts of manually sheared genomic DNA was constructed by using an EpiFOS fosmid library production kit (catalog no. FOS0901; Epicentre) according to the manufacturer's instructions.
Fingerprint map.
Fingerprints were generated by digesting RHA1 fosmid clones with BamHI and separating the resulting fragments on 1.2% agarose gels (40, 52a). Gel images were processed and captured with IMAGE (http://www.sanger.ac.uk/Software/Image), and the fragments were called using BANDLEADER software (22). A total of 4,973 fingerprints were obtained from 4992 fosmid clones that were analyzed. Fingerprints were automatically assembled into contigs by using FPC (42, 57, 58; see also http://www.genome.clemson.edu/fpc/) based on the restriction fragment overlap determined by the probability of coincidence score. A probability of 1e−10 and the default parameters were used for this map, yielding 417 contigs and 1,260 singletons. After the automated fingerprint binning, each contig was manually edited by using FPC tools. This involved refining order and overlaps based on the fingerprint similarities. Each contig was then extended in both directions from comparing the fingerprints of each contig with all other fingerprints within the FPC database at a less stringent cutoff and permitting the joins that did not contradict the high-stringency data. Some singletons were also used to bridge contigs. The final physical map of the RHA1 genome contained 25 contigs and 366 singletons.
Sequencing.
As part of the project to sequence the genome of RHA1, a total of 76,416 plasmid clones and 9,984 fosmid clones were grown on 2xYT agar containing 100 μg of ampicillin/ml with IPTG and X-Gal (pUC19 vector) or 25 μg of chloramphenicol/ml (pEpiFos5 vector). Sequencing was accomplished by using a combination of universal primers that include M13-Reverse, M13-40-Forward, pEpiFos5-Forward, and pEpiFos5-Reverse. The sequence reactions were performed in a total reaction volume of 5 μl containing 0.54 μl of Applied Biosystems BigDye v.3.1 cycle sequencing reaction mix (Applied Biosystems) and 3 μl of alkaline lysis purified plasmid DNA.
Sequence gaps caused by hard stops were closed by using an alternate 10-μl total volume sequencing reaction mix. The chemistry contained 2 μl of dGTP reaction mix (Applied Biosystems), 5% dimethyl sulfoxide, and 2 μl of alkaline lysis purified plasmid DNA. Reactions were primed by custom oligonucleotides designed to anneal to sequence flanking each hard stop region.
ABI Prism 3100, 3700, and 3730XL DNA analyzer sequencing instruments were used. Base calls of the trace data were performed by the program PHRED (19, 20) with default parameters, and the sequence was trimmed for quality and vector.
Sequence assembly and finishing.
Sequence reads were concurrently assembled by using Arachne 1.0 (4) and Phrap (25), and the assembly progress was monitored by a sequence assembly manager (R. Warren et al., unpublished results). In the latest stage of genome assembly, reads were binned into supercontigs (a higher arrangements of contigs based on read pairs information) by using Arachne and reassembled with Phrap to allow low-quality read bases to be included in the assembly (R. S. Fulton, unpublished data). The clone tiling path deduced from the RHA1 fingerprint map was used to align and orient supercontigs into ultracontigs based on the exact position on the fosmid clone end reads in our sequence assembly. This information, along with self-sequence alignment of the supercontig bins of Phrap contigs were used to rebin the reads for every RHA1 genetic element (chromosome and three plasmids). Consed (24) and Autofinish (25) were used to select primers and clones to finish low-quality regions, telomeres, and gaps in the pRHL3 sequence. Consed was also used to inspect sequence quality and integrity, as well as to edit the final assembly.
Annotation.
Putative genes were identified and annotated by using integrated automated and manual approaches. In the automated step, open reading frames (ORFs) were independently predicted by Glimmer2.10 (14) trained on a set of 500 known rhodococcal genes and by GeneMark-prokaryote (8) trained by using the supplied Mycobacterium tuberculosis model. RBSfinder (The Institute for Genomic Research), a ribosome-binding site prediction program, was used to help determine the start codons of our gene set.
Hand curation was facilitated by using Acedb (17) and an in-house interface to our pRHL3 annotation MySQL database (http://www.mysql.com). Each ORF predicted by the Hidden Markov Model (HMM) was inspected in the context of the plasmid sequence. ORF function and position were confirmed with BLASTP alignments (1) and BLASTX alignments of pRHL3 sequences to nr-SPtrEMBL, NCBI-nr, and Rhodococcus sp. strain I24 (www.integratedgenomics.com) protein databases. Interproscan (2) was used with the PROSITE, PRINTS, Pfam, SMART, TIGRFAMs, PIR SuperFamily, SUPERFAMILY, and ProDom databases to search for conserved domains and motifs and to validate predicted gene function. Finally, BLASTX alignments were used to identify genes that were not predicted by the gene finders used in the present study.
Sequence analyses (alignments, phylogenetic analyses, and genomic islands).
Sequences were aligned by using CLUSTAL W (61) with all parameters set to their default values. For phylogenetic analyses, CLUSTAL W alignments were used as input for the algorithm of the PHYLIP (version 3.6) package (21). Phylogenetic analyses were performed on 24 sequences: six RHA1 telomere sequences corresponding to the three RHA1 plasmids. The first 800 nucleotides of each were used for the analysis due to high base conservation between pRHL1 and pRHL3. The SEQBOOT program of the PHYLIP package was used to generate 100 data sets that were used in conjunction with the DNAPARS (DNA parsimony) program of PHYLIP, forcing 10 permutations per data set. The best tree was obtained by using CONSENSE (PHYLIP) and plotted by using TREEVIEW (43).
Clusters of genes that were potentially acquired through horizontal transfer (genomic islands) were identified by using IslandPath (29). The G+C content variation and dinucleotide bias were calculated with reference to the plasmid average (as opposed to the genome average). Dinucleotide bias was calculated by using both the “ORF clusters” method previously reported for IslandPath and the “whole plasmid sequence” method, with a sliding window size of 3 kb shifted every 0.5 kb. Putative insertion sequence (IS) elements were identified by BLASTN search against the IS Finder database (http://www-is.biotoul.fr/). Repeats were detected by using Reputer's REPFIND program (35) and MUMmers v.3.10 (36). Large repeats were investigated for possible gene duplication by using NCBI-bl2seq and MIROPEATS (45). To identify putative integrons (49), BLASTP and regular expressions were used to search for integron-associated integrases (IntI) and the core attachment site (attI) consensus sequence, respectively.
RESULTS AND DISCUSSION
Characterization of RHA1 linear plasmid termini.
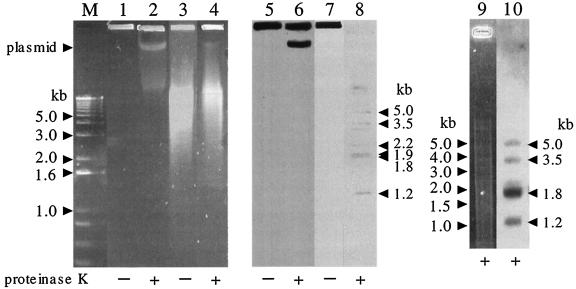
The ends of pRHL1 and pRHL3 were cloned based on the assumption that the termini of the RHA1 linear plasmids are blunt, which is similar to the strategy used to clone the ends of pRHL2 (56). The terminal fragments were confirmed by Southern hybridization analysis of total RHA1 genomic DNA. As seen in Fig. 1, for samples that were prepared without proteinase K, the terminal restriction fragments remained at the origin of electrophoresis. This is probably because the termini are covalently bound by a specific protein(s) (56). In samples digested with proteinase K and PstI, the probe (derived from the right telomere of pRHL3) hybridized to fragments derived from the telomeres of pRHL1 (1.2, 2.2, and 5.0 kb), pRHL2 (1.9 kb), and pRHL3 (1.8 and 3.5 kb). Under conditions of higher stringency, only four of these were observed: the 1.2-, 1.8-, 3.5-, and 5.0-kb fragments. Consistent with the origin of the probe, the strongest signal resulted from hybridization to the 1.8-kb fragment, which is derived from the right terminus of pRHL3. The identity of the 2.2-kb band derived from pRHL1 is unclear. It was also observed in a hybridization analysis of PstI-digested, purified pRHL1 DNA by using a probe derived from the right terminus of pRHL1. However, the 2.2-kb fragment was not recovered in our attempts to clone the plasmid telomeres. Moreover, this fragment was not observed in the hybridization analysis of PstI-digested total DNA prepared without proteinase K. It is possible that the fragment originated from an internal fragment of pRHL1 having an affinity with a terminal protein(s).
FIG. 1.
Analysis of RHA1 telomere fragments. RHA1 cells were lysed in an agarose plug with (+) or without (−) proteinase K treatment. Agarose plugs containing RHA1 DNA were subjected to PFGE directly (lanes 1, 2, 5, and 6) or after PstI digestion (lanes 3, 4, 7, and 8). Electrophoresis was conducted for 6 h with a voltage of 6 V/cm and a pulse time that was increased from 2 to 10 s as the electrophoresis progressed. Lanes M to 4 and 9 were stained with ethidium bromide. Lanes 5 to 10 represent Southern blots with a probe derived from the right telomere of pRHL3. The experiment shown in lanes 9 and 10 was performed by using conditions of higher stringency. Lane M, 1-kb plus DNA ladder size marker (Invitrogen, Carlsbad, Calif.). The position of intact linear plasmid DNA containing pRHL3 is indicated on the left. The estimated sizes of the fragments detected by hybridization are indicated on the right.
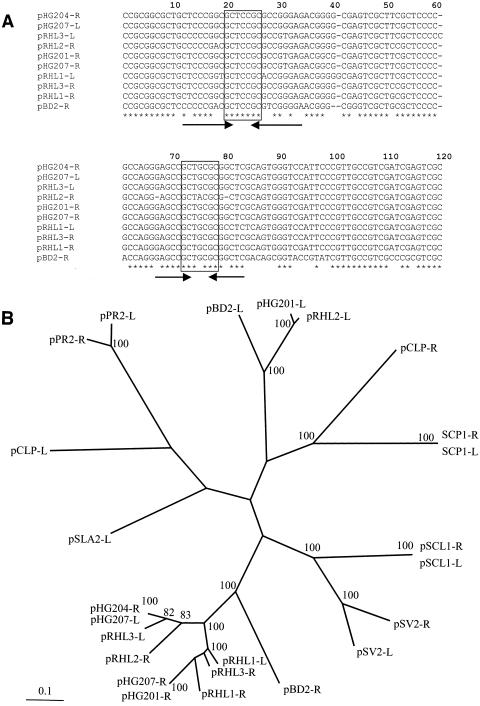
Nucleotide sequences of greater than 600 bp were determined for the cloned pRHL1 and pRHL3 termini and match perfectly (results not shown) the corresponding sequences in the current RHA1 genome assembly (http://www.bcgsc.bc.ca/cgi-bin/rhodococcus/blast_rha1.pl). These sequences are very similar to each other and, as shown in Fig. 2A, to telomeric regions of pHG207 of Rhodococcus sp. strain MR2253 (30) and to the right telomeres of pRHL2 (56), pBD2 of R. erythropolis (60), pHG201 of R. opacus MR11, and pHG204 of R. opacus MR22 (31). Each of these telomere sequences contains two sets of inverted repeats flanking the GCTXCGC central motif (Fig. 2A) originally identified in pHG201 of R. opacus MR11 (31).
FIG. 2.
Sequence analysis of actinomycete invertron telomeres. (A) Alignment of rhodococcal invertron telomere nucleotide sequences. The nucleotide sequences are derived from each of the three RHA1 invertrons (except for pRHL2-L), as well as pHG201 of R. opacus MR11, pHG204 of R. opacus MR22 (31), pBD2 of R. erythropolis (60), and pHG207 of R. sp. strain MR2253 (30). Strictly conserved nucleotides are indicated with hodococcus asterisks. The two sets of inverted repeats are indicated with arrows. The GCTXCGC central motif is boxed. (B) Radial view of best maximum-parsimony tree obtained by PHYLIP analyses of actinomycete telomeres. The first 800 nucleotides of each telomere were aligned. Sequences were taken from each of the plasmids in 2a, as well as the following invertrons: S. clavuligerus pSCL1 (65), S. coelicolor A3 SCP1, S. violaceoruber pSV2 (59), S. rochei 7434AN4 pSLA2-L (28), Planobispora rosea pPR1 and pPR2 (47), and M. celatum pCLP (46).
The long terminal inverted repeats (TIR) in the telomeres, together with evidence of covalently bound protein at their ends, indicate that pRHL1 and pRHL3 are typical actinomycete invertrons. Such telomeres appear to share a common mechanism for replication and/or maintenance. The inverted repeats with the central motif GCTXCGC may play a role in such a mechanism.
Phylogenetic analyses of the TIRs of actinomycete invertrons reveal the presence of at least four distinct groups of telomeres (Fig. 2B). The group formed by the telomeres of pSV2 and pSCL1 have a single set of the inverted repeats with the GCTXCGC motif found in the pRHL1 and pRHL3 telomeres. Interestingly, the telomeres do not group according to species or plasmid. Thus, the high divergence between the left and right ends of pRHL2 and pBD2, respectively, clearly indicate that functional invertrons do not require perfectly matching TIRs.
In streptomycetes, linear plasmids can exchange ends with the host chromosome, mobilizing large regions of the chromosomal ends (44). It seems equally likely that linear plasmids could also exchange ends with each other. This would facilitate recombination and exchange and may partly explain the apparent duplications that occur in pRHL3 (see below) and in the other RHA1 plasmids, as exemplified by the duplications of aromatic hydroxylation dioxygenase genes (30; W. Kitagawa, unpublished results). In the current assembly of the RHA1 genome, several regions of pRHL3 had identity to regions of the chromosome or one of the other two plasmids. Regions of 100% sequence identity were as long as 1.5 kb, and most of these included at least part of a gene putatively involved in recombination. A more complete analysis awaits completion of the genome sequence.
Replication machinery.
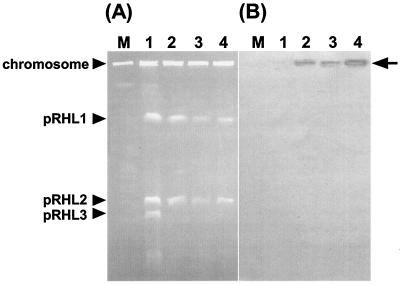
When an RHA1 DNA library constructed by using Charomid 9-28::tsr (which is unable to replicate in rhodococci) was introduced into RHA1, 17 transformants were obtained. Two of these transformants yielded plasmid DNA which, when analyzed by Southern hybridization, carried the tsr gene. The other transformants may have originated from the integration of the tsr sequence into the RHA1 genome. These plasmids contained the same 18-kb insert and restriction fragments, and one of them was designated pCHB79. Southern hybridization analysis after PFGE revealed that the 4-kb HindIII fragment of pCHB79 hybridized specifically to pRHL3. All of the RHA1 transformants containing pCHB79 lost pRHL3, suggesting incompatibility of pCHB79 with pRHL3 (Fig. 3) and that pCBH79 contains the replication origin of pRHL3.
FIG. 3.
Analysis of RHA1 transformants containing origin of replication of pRHL3. Transformants were analyzed by PFGE (A) and Southern hybridization with a probe derived from the tsr gene (B). Lanes were loaded with the following: M, a chromosome size marker derived from Saccharomyces cerevisiae; 1, wild-type RHA1; and 2 to 4, independent transformants of RHA1 containing pCHB79. The positions of the RHA1 chromosome and each plasmid are indicated on the left. An arrow on the right indicates the deduced position of pCHB79, which corresponds to the origin of electrophoresis.
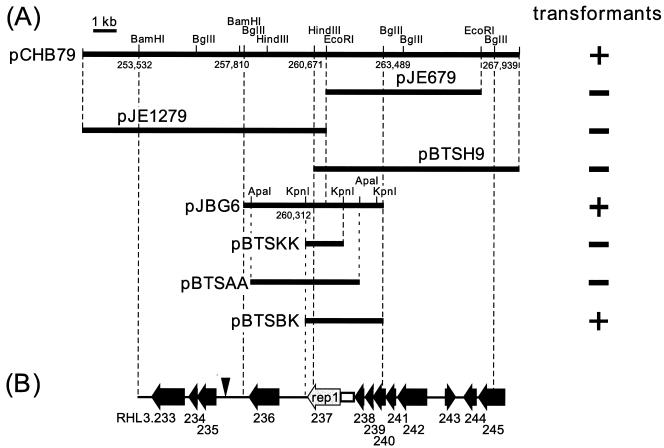
Subcloning of the pCHB79 insert into pIJ702 yielded a single construct that propagated in RHA1. This plasmid, designated pJBG6, contains a 5.7-kb BglII insert (Fig. 4). All independently isolated transformants of RHA1 carrying pJBG6 had lost pRHL3, suggesting incompatibility of pJBG6 with pRHL3 (data not shown). Further subcloning of the 5.7-kb insert into pBSSK::tsr yielded pBTSBK, which replicated in RHA1. Comparison with the other subclones, pBTSKK, pBTSAA, and pBTSH9, which did not propagate in RHA1, suggested that the insert of pBTSBK possesses the minimal requirement for plasmid replication and/or maintenance in RHA1.
FIG. 4.
Replication origin region of pRHL3. (A) Subcloning of the origin of replication. Subclone plasmids are labeled, and their inserts are represented by horizontal thick bars. The results of transformation experiments are presented on the right: +, clones that yielded transformants of RHA1; −, clones that yielded no transformants. The six-digit numbers indicate the nucleotide positions of restriction sites on pRHL3. (B) Annotated ORFs in the replication origin region. ORFs are numbered according to Fig. 5 and are represented by horizontal arrows; rep1 is presented by a shaded arrow. A closed vertical arrowhead and an open box indicate the respective locations of the direct repeats and AT-rich region described in the text.
Nucleotide sequence of the pCHB79 insert revealed two ORFs (RHL3.237 and RHL3.235) and several features that are similar to what is found in pCLP, a mycobacterial plasmid (46). RHL3.237, designated rep1, encodes a 444-residue protein that shares up to 25% amino acid sequence identity with rep genes of mycobacterial and rhodococcal plasmids (3, 5, 16, 46) (Table 1). RHL3.235 encodes a protein that shares 34% sequence identity to ParA protein from R. erythropolis (15). Both pJBG6 and pBTSBK contain rep1. However, neither plasmid contains RHL3.235. A parA homolog located elsewhere on the RHA1 genome may compensate for the lack of RHL3.235 in these subclones. The closest direct repeats (TCGC[GA]CTCATAGCTCTG; one mismatch) occur downstream of rep1, beginning at position 257072, but do not occur on either pJBG6 or pBTSBK. A 600-bp region immediately upstream of rep1 is slightly AT rich (G+C content = 59.8%). No homologs to known parB and parS genes were found on pRHL3.
TABLE 1.
Selected ORFs predicted on pRHL3 of Rhodococcus sp. strain RHA1
| RHL3 ORF | Start coordi- nate (bp) | Stop coordi- nate (bp) | Strand type | Product size (no. of amino acids) | % Identitya | Gene product descriptionb | Closest organism(s)a |
|---|---|---|---|---|---|---|---|
| RHL3.14 | 12172 | 11930 | − | 81 | Hypothetical protein/some similarity with two- component histidine kinase | ||
| RHL3.15 | 13116 | 12187 | − | 310 | Conserved hypothetical protein/putative endonuclease | ||
| RHL3.16a | 13528 | 13815 | + | 96 | Hypothetical protein/methyltransferase/possible pseudogene | ||
| RHL3.16b | 13814 | 14131 | + | 106 | 20 | Hypothetical protein/possible pseudogene | C. equii (R. equi) |
| RHL3.24 | 22119 | 21229 | − | 297 | 43 | Putative transposase | S. violaceoruber |
| RHL3.25 | 22466 | 22119 | − | 116 | 82 | Conserved transposase | Rhodococcus sp. strain I24 |
| RHL3.36 | 34389 | 33994 | − | 132 | Putative recombinase/integrase | ||
| RHL3.37 | 34679 | 35092 | + | 138 | 27 | Hypothetical protein/putative transposase | Micrococcus sp. strain 28 |
| RHL3.41 | 40363 | 39539 | − | 275 | 50 | Conserved carveol dehydrogenase | R. erythropolis |
| RHL3.42 | 41024 | 42184 | + | 387 | 44 | Putative limonene monooxygenase | R. erythropolis |
| RHL3.46 | 44886 | 44254 | − | 211 | 20 | Hypothetical protein/putative transposase | R. erythropolis |
| RHL3.55 | 58568 | 59461 | + | 298 | 67 | Conserved GND protein/6-phosphogluconate dehydrogenase | M. smegmatis |
| RHL3.56 | 59461 | 60462 | + | 334 | 85 | Conserved F420-dependent glucose 6-phosphate dehydrogenase | M. phlei |
| RHL3.57 | 60462 | 62114 | + | 551 | 67 | Conserved glucose 6-phosphate isomerase | M. tuberculosis and M. bovis |
| RHL3.59a | 63480 | 63989 | + | 170 | 31 | Putative transposase/possible pseudogene | S. coelicolor |
| RHL3.59b | 63956 | 64327 | + | 134 | 30 | Putative transposase/possible pseudogene | S. avermitilis |
| RHL3.60 | 65785 | 64430 | − | 452 | 43 | Conserved reductase | P. putida |
| RHL3.61 | 66164 | 65754 | − | 137 | 43 | Putative ferredoxin | B. japonicum |
| RHL3.62 | 67411 | 66167 | − | 415 | 27 | Putative cytochrome P450 | S. avermitilis |
| RHL3.65 | 70574 | 70368 | − | 69 | − | Hypothetical protein | |
| RHL3.66 | 71454 | 70582 | − | 291 | Conserved hypothetical protein/possible endonuclease | ||
| RHL3.85 | 91424 | 92161 | + | 246 | Putative transposase | ||
| RHL3.88 | 95128 | 97482 | + | 785 | 44 | Conserved putative ABC transporter ATP-binding protein | S. avermitilis |
| RHL3.91 | 102553 | 100637 | − | 639 | 25 | Probable serine/threonine-protein kinase | M. tuberculosis and M. bovis |
| RHL3.112 | 119957 | 121129 | + | 391 | 53 | Conserved transposase | S. coelicolor |
| RHL3.135 | 144075 | 143851 | − | 75 | Hypothetical protein | ||
| RHL3.136 | 144427 | 146160 | + | 578 | 52 | Putative methylase | C. crescentus |
| RHL3.137 | 146217 | 147332 | + | 372 | 25 | Putative type I restriction modification system enzyme | M. acetivorans |
| RHL3.138 | 147322 | 150444 | + | 1,041 | 41 | Putative DNA helicase | C. efficiens |
| RHL3.139 | 150662 | 152125 | + | 488 | 42 | Putative ABC transporter ATP-binding protein | Synechococcus sp. strain PCC 7942) (A. nidulans R2) |
| RHL3.141a | 152739 | 153395 | + | 219 | 26 | Putative transposase/possible pseudogene | M. abscessus |
| RHL3.141b | 153148 | 153597 | + | 150 | 30 | Putative transposase/possible pseudogene | M. abscessus |
| RHL3.150 | 161508 | 162518 | + | 337 | 84 | Conserved F420-dependent glucose 6-phosphate dehydrogenase | M. phlei |
| RHL3.151 | 162550 | 164199 | + | 550 | 69 | Conserved glucose 6-phosphate isomerase | M. tuberculosis and M. bovis |
| RHL3.156 | 172382 | 171159 | − | 408 | 31 | Putative permease transporter | S. coelicolor |
| RHL3.157 | 174191 | 173097 | − | 365 | 21 | Putative transporter bacterial inner membrane translocator RbsC | P. multocida |
| RHL3.158 | 175723 | 174203 | − | 507 | 38 | Putative ABC transporter ATP-binding protein | S. coelicolor |
| RHL3.159 | 176971 | 175808 | − | 388 | Putative ABC-type sugar transport system peri- plasmic component | ||
| RHL3.183 | 202742 | 203497 | + | 252 | 53 | Putative sensory transduction protein/two-component regulatory system | C. efficiens |
| RHL3.184 | 203497 | 204651 | + | 385 | 36 | Putative two-component system/sensory transduction histidine kinase | C. glutamicum (B. flavum) |
| RHL3.235 | 256695 | 255916 | − | 260 | 34 | Putative plasmid partioning protein (ParA) | R. erythropolis |
| RHL3.237 | 261733 | 260402 | − | 444 | 25 | Rep-like protein | M. celatum |
| RHL3.246 | 269639 | 268476 | − | 388 | 31 | Putative cytochrome P450 | S. avermitilis |
| RHL3.276 | 307112 | 306021 | − | 364 | 46 | Conserved alcohol dehydrogenase class IV | C. acidovorans (P. acidovorans) |
| RHL3.277 | 308001 | 307105 | − | 299 | 43 | Conserved dioxygenase | A. tumefaciens strain C58 (ATCC 33970) |
| RHL3.279 | 308851 | 308336 | − | 172 | 36 | Putative oxidoreductase | S. coelicolor |
| RHL3.280 | 310077 | 308857 | − | 407 | 30 | Putative indole dioxygenase | S. avermitilis |
| RHL3.281 | 310249 | 311304 | + | 352 | 30 | Probable the operon regulatory protein/AraC family | R. erythropolis |
| RHL3.284 | 313807 | 314808 | + | 334 | 33 | Probable the operon regulatory protein/AraC family | R. erythropolis |
| RHL3.286 | 315759 | 316727 | + | 323 | 20 | Putative ferredoxin/reductase | Rhodococcus sp. strain NCIMB 9784 |
| RHL3.287 | 316809 | 318125 | + | 439 | 26 | Conserved cytochrome P450 | Rhodococcus sp. strain NCIMB 9784 |
| RHL3.294 | 324660 | 322984 | − | 559 | 70 | Conserved G6PI | M. tuberculosis and M. bovis |
| RHL3.295 | 325563 | 324667 | − | 299 | 63 | Conserved GND protein/6-phosphogluconate dehydrogenase | M. smegmatis |
| RHL3.299 | 329871 | 328591 | − | 427 | 90 | Putative G6PDH | R. opacus |
Based on BLASTP alignment with members of the nonredundant SPtrEMBL protein database only. B. japonicum, Bradyrhizobium japonicum; M. acetivorans, Methanosarcina acetivorans; A. nidulans, Anacystis nidulans; B. flavum, Brevibacterium flavum; P. multocida, Pasteurella multocida; C. acidovorans, Comamonas acidovorans; C. crescentus, Caulobacter crescentus; C. efficiens, Corynebacterium efficiens.
Gene product description was inferred by BLAST sequence similarity of predicted genes with annotated proteins of the nr-SPtrEMBL, NCBI-nr, and Rhodococcus sp. strain I24 protein databases and identification of protein motifs from a collection of protein domain databases as described in Materials and Methods.
Plasmid sequence and ORF analysis.
The 332,361 bases that comprise pRHL3 were sequenced by using the whole-genome shotgun sequencing (WGS) approach (11, 18, 34) to the quality of the Bermuda standard. The G+C content of pRHL3 is 64.9%. This is similar to that of pRHL1 (64.9%) and pRHL2 (64.0%) in the current assembly but is lower than that of the chromosome (67.5%). This may reflect the shorter residence of the plasmids than of the chromosome in RHA1.
The plasmid was predicted to contain 300 genes (Table 1 and Fig. 5), including three possible pseudogenes, each of which contained a single frameshift (RHL3.16, RHL3.59, and RHL3.141). Two of these pseudogenes encode putative transposases. The coding region covered 79% of the plasmid. This is lower than what has been reported for other actinomycete invertrons, whose coding regions cover >85% (7, 60). There is a slight bias for genes on the lower strand at 60.4%. Interestingly, the largest gene clusters are arranged in operon-like structures and are all located on the lower strand.
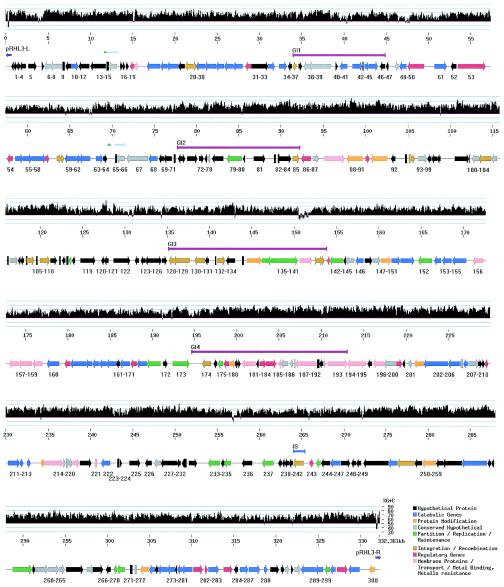
FIG. 5.
Physical map and G+C content of pRHL3. The G+C content is depicted in a histogram in which each vertical bar indicates the G+C composition calculated over a 100-bp interval by using a sliding window of 10 bp. The bottom bar depicts predicted ORFs grouped into eight functional categories (a color code is provided in the lower right corner of the figure). The orientation of each ORF is indicated by an arrowhead. The symbols between the upper and lower bars indicate the respective positions of genomic islands (GI1 to -4; fuchsia-colored bars), IS elements (light blue bar), TIRs (dark blue arrows), and possible duplications (light blue and green arrows).
The functional classes of the predicted genes of pRHL3 are summarized in Table 2. Over half of the predicted genes (153 [51%]) have no known function, including 39.7% that had no match in the searched protein databases. The largest functional class (20.7% of the predicted genes) comprise those that are most similar to reported catabolic genes. Approximately half of these encode dehydrogenases, and six are predicted to code for oxygenases. Regulatory genes constitute the second largest class of functional genes, followed by those involved in transport processes and DNA recombination.
TABLE 2.
Functional classification of pRHL3 ORFs
| Functional classification | No. of genes | % |
|---|---|---|
| Catabolism | 62 | 20.67 |
| Membrane proteins/transport/metal binding | 18 | 6.00 |
| Partition/replication/maintenance | 17 | 5.67 |
| Integration/recombination | 19 | 6.33 |
| Regulation (transcription) | 21 | 7.00 |
| Protein modification | 10 | 3.33 |
| Conserved genes | 34 | 11.33 |
| Hypothetical | 119 | 39.67 |
| Total | 300 | 100 |
G+C composition.
The G+C composition of pRHL3 was calculated for each window of 100 bases, with a sliding window of 10 bp (Fig. 5, graph). Although the overall G+C content of pRHL3 is 64.9%, the analysis reveals several regions of 100 to 500 bp in size that are more than 75% G+C, as well as stretches of 100 to 1,300 bp that are less than 50% G+C. Most of the latter appear to be intergenic and are typically 5′ to coding regions. As noted below, one of the regions of high G+C content is associated with a putative genomic island, RHL3-GI4 (Fig. 5). This island contains many genes coding for transporters and metal-binding proteins, as well as a two-component signal transduction system. There are several 100-bp stretches in this island that have more than 75% G+C.
The telomeres have a distinctive G+C content: the first 100 nucleotides have a very high G+C content (79 to 80%), followed by a considerable decrease in G+C content in the region between positions 300 and 400 bp (36 and 39% G+C for pRHL3-L and pRHL3-R, respectively). A similar pattern is evident for the related telomeres shown in Fig. 2A.
Catabolic gene clusters.
Many of the predicted catabolic genes of pRHL3 appear to be arranged in one of three clusters. Genes within these clusters have the structural characteristic of functional operons, including overlapping stop and start codons, unidirectional transcription, and nearby genes encoding transcriptional regulators. Each of the three catabolic clusters contains up to three genes that encode enzymes with high similarity to 6-phosphogluconate dehydrogenase (gnd gene), glucose 6-phosphate dehydrogenase (G6PDH), and glucose 6-phosphate isomerase (G6PI), respectively. These correspond to RHL3.55, RHL3.56, and RHL3.57 in the first cluster and to RHL.295, RHL3.299, and RHL3.294 in the third cluster. The second catabolic cluster only contains genes encoding G6PDH and G6PI: RHL3.150 and RHL3.151. The three putative isomerases share ca. 66% amino acid sequence identity. Similarly, the two putative 6-phosphogluconate dehydrogenases, encoded by RHL3.55 and RHL3.295, share 67% amino acid sequence identity. In contrast, the putative G6PDHs are not all identical: RHL3.56 and RHL3.150 appear to encode F420-dependent G6PDHs and share 86% amino acid sequence identity, whereas RHL3.299 is similar to NADP-dependent G6PDHs. These findings suggest that at least some of the genes utilized by RHA1 to metabolize glucose may originate from pRHL3.
The first catabolic cluster (RHL3.20 to RHL3.63) spans a region of 54 kb and includes 13 dehydrogenase genes, all of whose products share >23% sequence identity. The substrates of most of these enzymes have yet to be identified. However, RHL3.41 and RHL3.42 of this region encode enzymes that share high sequence identity with carveol dehydrogenase and limonene monooxygenase, respectively, from R. erythropolis DCL14 (62). Rhodococcus sp. strain RHA1 grew on carveol or limonene as the sole organic substrate.
The second catabolic cluster, spanning kilobase positions 158 to 190 of the plasmid, is characterized by the presence of genes encoding metal and permease transporters, inner membrane translocators, and members of the ABC transport system for glucose. The transporter genes are located just downstream of the genes encoding the possible G6PDH (RHL3.150) and G6PI (RHL3.151). Located 10 kb downstream of the second catabolic gene cluster is a 40-kb region containing at least four genes predicted to be involved in heavy metal transport (metal-associated proteins and ATPases), as well as membrane proteins, permeases, and members of the ABC transport system.
The last 30 kb of pRHL3 harbors the third and shortest catabolic gene cluster. This region appears to contain at least two operons. The first of these contains genes predicted to code for a dioxygenase (RHL3.280), a flavoreductase (RHL3.279), an intradiol dioxygenase (RHL3.277), and an iron-containing dehydrogenase (RHL3.276) that appear to constitute an operon involved in the degradation of an aromatic compound. The RHL3.277-encoded protein shares 43% sequence identity with an intradiol dioxygenase from Agrobacterium tumefaciens C58. The protein encoded by RHL3.280 has some similarity to poorly characterized indole dioxygenases from R. opacus and Streptomyces avermitilis. It does not appear to be either a flavin-type oxygenase or a ring-hydroxylating dioxygenase, since the appropriate sequence motifs were not found. This putative operon appears to be regulated by an AraC-type transcriptional regulator (RHL3.281), whose best hit (30% sequence identity) is ThcR from R. erythropolis (41). The second operon contains a cytochrome P450 gene (RHL3.287), as described in the next section.
Cytochrome P450s.
Genes encoding three cytochrome P450s (CYPs) were found on pRHL3 based on sequence alignments to known CYPs and the presence of a cysteine-containing heme-binding motif (Table 3). Of the three putative P450s, only that encoded by RHL3.287 may be assigned to an existing CYP family based on sequence identity, and this one appears to belong to a new class. RHL3.287 lies in the middle of the third cluster of catabolic genes on pRHL3, ∼5 kb downstream of a putative operon that appears to specify the catabolism of an aromatic compound. RHL3.287 encodes a family 116 CYP, showing highest identities to the N-terminal portion of P450RhF from Rhodococcus sp. strain NCIMB9784 (47%) and a thcB-encoded P450 from Rhodococcus sp. strain NI86/21 (48%). The latter is involved in the degradation of two herbicides: EPTC (S-ethyl dipropylthiocarbamate) and atrazine (41). The substrate of P450RhF is unknown. However, P450RhF has an intriguing multidomain structure: a C-terminal domain is similar to the reductase of some dioxygenases, harboring a 2Fe-2S cluster and an FMN (48). Accordingly, P450RhF was proposed to belong to a newly identified class of CYPs (class IV). The RHL3.287-encoded P450 is not a multidomain protein: it corresponds to the N-terminal heme-binding domain of P450RhF. However, RHL3.286, which apparently forms an operon with RHL3.287, encodes a protein whose sequence is 50% identical to the C-terminal reductase domain of P450RhF and is also predicted to harbor a 2Fe-2S cluster and an FMN. Thus, RHL3.286 and RHL3.287 potentially encode a class V system, predicted by Roberts et al. (48).
TABLE 3.
CYP genes of pRHL3
| ORF | Closest CYPa | % Identityb | Speciesc | Family | Heme-binding motifd |
|---|---|---|---|---|---|
| RHL3.287 | CYP116A1 | 48 | R. erythropolis | Actinomycetes | FGFGRHLCLG |
| RHL3.62 | CYP225A1 | 27 | N. aromaticivorans | α-Proteobacteria | FGAGAHRCIG |
| CYP125A2 | 25 | S. avermitilis | Actinomycetes | FGAGAHRCIG | |
| RHL3.246 | CYP107X1 | 37 | S. avermitilis | Actinomycetes | FGRGPHYCLG |
CYP names and sequences as annotated at the following web address: drnelson.utem.edu/CytochromeP450.html.
The percent identity represents the number of exact matches over the closest CYP length.
N. aromaticivorans, Novosphingobium aromaticivorans.
Residues conserved in the heme-binding motif are indicated in boldface.
The putative operon to which RHL3.286 and RHL3.287 belong appears to be regulated by an AraC-type transcriptional regulator (RHL3.284) whose best hits are CYP gene regulators: ThcR and EthR (33.1 and 33.4% sequence identity, respectively). ThcR is proposed to regulate the transcription of a CYP system that degrades the thiocarbamate herbicide EPTC. EthR regulates the transcription of a CYP system that degrades ethyl tert-butyl ether (ETBE) by R. ruber (10). Indeed, the sequence and genetic organization similarities between the eth genes of R. ruber and this putative operon on pRHL3 are remarkable.
The other two P450s, encoded by RHL3.62 and RHL3.246, cannot be assigned to existing families. The sequence of the RHL3.62-encoded P450 is most similar to family 125 and 225 P450s. The genes coding for the putative cognate ferredoxin (RHL3.61) and reductase (RHL3.60) are located immediately downstream of RHL3.62: the three genes appear to be arranged in a transcriptional unit. The physiological role of this system is unclear, and no substrate of a CYP125 or CYP225 has been identified to date. However, the function of RHL3.62 may be linked to the catabolic genes with which it clusters on pRHL3. The sequence of the RHL3.246-encoded P450 is most similar to family 107 CYPs. Many family 107 CYPs have been linked to macrolide biosynthesis. Interestingly, the genes surrounding RHL3.246 encode proteins of no known function, and genes encoding a ferredoxin or a reductase do not appear to be in its vicinity.
The relatively high number of CYP-encoding genes on pRHL3 reflects the total number of CYP genes in the RHA1 genome, currently estimated to be 12. More generally, the number of CYP genes in RHA1 seems to be a hallmark of actinomycete biology. For example, the genome sequences of Streptomyces coelicolor A3 (2), S. avermitilis, and Mycobacterium tuberculosis have 18, 33, and 20 CYP genes, respectively (7, 13, 37). Many of the streptomycete P450s are predicted to be involved in secondary metabolite biosynthesis. In the rhodococci, catabolic function may dominate.
Horizontal gene transfer and recombination.
Clusters of horizontally acquired genes, or “genomic islands,” are frequently associated with a particular adaptation of the recipient microorganism, such as increased virulence, a particular catabolic capability, or resistance to an antimicrobial or heavy metal (26). Moreover, identification of horizontally acquired regions can help elucidate the evolutionary history of a genome, providing insights into recent adaptations. Analyses included IslandPath analysis (29) and searches for mobility genes, repeats, IS elements, and integrons.
Plasmid pRHL3 carries at least 19 putative “mobility” genes that encode proteins likely to be involved in DNA recombination, including 15 possible transposases and 2 possible integrases. Two of the transposase-encoding genes, RHL3.59 and RHL3.141, are pseudogenes; each contains a single frameshift mutation. Six copies of a tandem repeat of GGCGGTC lie immediately upstream of pseudogene RHL3.59. Five integration/recombination genes are found within the first catabolic gene cluster. There appears to be a “hot spot” for transposase genes in the 105- to 154-kb region: 11 transposases are located in this region, many of which are clustered side by side (Fig. 5). The plasmid also contains an intact IS element at position 265 kb that is highly similar to IS1164 (85% nucleotide identity) identified in R. rhodochrous J1. There are several IS-like elements and IS remnants. Thus, RHL3.112 shows 58% identity at the amino acid level to the IS110 transposase, and RHL3.46 has some similarity to the IS1676 transposase but is shorter and possibly truncated. Finally, RHL3.24 and RHL3.25 are most similar to two genes of the transposase operon found in IS1206, and RHL3.85 is highly similar to a transposase found in IS1295 (82% nucleotide identity). However, RHL3.24 and RHL3.25 lack detectable inverted repeats characteristic of IS elements, and RHL3.85 has a 3′ deletion compared to IS1295. Consequently, these three ORFs in pRHL3 may represent IS remnants. No obvious integron-like element was detected on the plasmid.
IslandPath (29), which combines sequence analysis features (%G+C and dinucleotide bias) and annotation features (mobility genes and tRNAs), was used to improve the prediction of genomic islands on pRHL3. Dinucleotide bias was initially calculated by using ORF clusters, as in previous IslandPath analyses. However, due to the smaller size of this plasmid versus the size of the genomic sequences previously analyzed by IslandPath, dinucleotide bias was also calculated over the entire plasmid sequence by using a window of 3 kb, shifted every 0.5 kb. Three regions showed dinucleotide bias by using both approaches. These three regions, together with a fourth that has an unusual G+C content, are described below as putative genomic islands. Each region is proximal to probable mobility genes. This is the first time that IslandPath has been adapted for plasmid analysis.
The first putative island of pRHL3, GI1 in Fig. 5, contains no apparent dinucleotide bias. However, several other considerations indicate that this region may be a genomic island. First, four of eight genes in this region have a G+C value >1 standard deviation below the mean ORF G+C content of the plasmid. Second, these genes are adjacent to a gene predicted to encode a recombinase/integrase (RHL3.36) and two genes predicted to encode transposases (RHL3.37 and RHL3.46). Finally, a pair of 8-bp direct repeats flanks the region. Although repeats of this size often occur by chance, they may represent a duplicated insertion point. GI1 contains the genes predicted to encode limonene monooxygenase, carveol dehydrogenase, and an oxidoreductase.
The second putative island of pRHL3, RHL3-GI2 (Fig. 5), spans positions 78 to 92 kb, includes RHL3.72 to RHL3.85, and is flanked by a pair of 9-bp direct repeats. The majority of these predicted genes encode proteins of no known function (hypothetical or conserved hypothetical). The only annotated gene product corresponds to histone protein H1. Several of the genes in this region have moderate similarity to chromosomally located genes from other actinomycetes. The G+C content of ORFs in this region averages 67%, which is close to the chromosomal G+C content.
The third putative genomic island, RHL3-GI3 (Fig. 5), spans positions 135 to 153 kb and includes RHL3.128 to RHL3.141. The region appears to be an insertion hot spot since there are six transposases flanking this region. At least three pairs of 7- to 8-bp direct repeats also flank this region, suggesting a complex recombination history. As noted above, the significance of such short repeats is unclear. Interestingly, three of the predicted ORFs (RHL3.136, RHL3.137, and RHL3.138) in this island together encode a putative type I restriction modification (R-M) system that has moderate similarity to a Caulobacter system. It has been suggested that horizontal exchange of type I R-M systems may contribute to sequence divergence within R-M families (54). There are also several genes with unknown functions in this second island. The G+C content of each ORF in this region varies. However, the average G+C content, 63%, is slightly below the mean for all ORFs in the plasmid (65%).
The fourth putative island, RHL3-GI4 (Fig. 5), spans positions 194 to 213 kb, includes ORFs RHL3.174 to RHL3.193, and is flanked by a pair of 9-bp direct repeats. The G+C of the region, at 68.1%, is almost 1 standard deviation above the mean for all plasmid ORFs. This island is notable for the presence of several genes encoding probable metal transporters. No other metal transporter genes were found elsewhere on this plasmid. The island also contains two genes, RHL3.183 and RHL3.184, which are predicted to encode the sensor kinase and regulator, respectively, of a two-component signal transduction system.
Finally, pRHL3 contains evidence for a duplication event that involves two ORFs of unknown function (Fig. 5). The respective regions spanning 11.8 to 13.3 kb (RHL3.14 and RHL3.15) and 69.7 to 71.7 kb (RHL3.65, RHL3.66) share an overall nucleotide identity of ca. 92%, which rises to 98% over the first 160 bp. The second region is slightly larger due to a possible insertion between nucleotides 69880 and 70396. The insertion effectively truncates RHL3.65 with respect to RHL3.14. As more sequences from related bacterial species become available, comparative genomic analysis will be helpful in further ascertaining the evolution history of pRHL3.
Regulatory genes.
A significant proportion (7%) of the pRHL3 genes are predicted to be transcriptional regulators. This preponderance of regulatory processes correlates well with the proportion of catabolic genes found on the plasmid and their tightly regulated expression. Indeed, more than half of the putative regulatory genes are distributed within the three main catabolic gene clusters described above. Members of the following subfamilies of transcriptional regulators containing a helix-turn-helix motif are predicted to be encoded by pRHL3 genes: TetR/AcrR, LuxR/UhpA, GntR, MarR/EmrR, DeoR, ArsR, and AraC/XylS. Particular examples of such regulatory proteins are discussed above with respect to catabolic gene clusters. Other putative regulatory genes include those encoding a two-component signal transduction system that are part of the putative genomic island RHL3-GI4 (see above) and RHL3.91 which is predicted to encode a serine/threonine kinase.
Transport.
As noted above, genes involved in transport constitute a significant proportion of the pRHL3 genes. Except for genes encoding putative ATP-binding proteins at 95.1 kb (RHL3.88) and at 150.7 kb (RHL3.139), all of the ORFs predicted to encode transporters, metal-carrying proteins, membrane proteins, and transport-related proteins occur within or near the second catabolic gene cluster in a 70-kb region. Within this region, there are three distinct clusters of transport-related genes located at 171, 202, and 234 kb, respectively. The first of these comprises four genes that appear to encode a functional ABC transport system including a permease (RHL3.156), an RbsC-like inner membrane translocator (RHL3.157), an ATP-binding protein (RHL3.158), and a periplasmic or lipoprotein component (RHL3.159). In gram-positive bacteria, these systems are also known as binding-lipoprotein-dependent transport systems. The substrate for this ABC system is unknown. However, two lines of evidence suggest that it is a sugar. First, sequence analyses indicate that the translocator is most similar to RbsC, a ribose translocator, and the lipoprotein component is most similar to a putative periplasmic component of a sugar transport system. Second, the putative transporter genes are close to genes that are apparently involved in glucose catabolism. The second “transport cluster” is located in the putative genomic island RHL3-GI4 and is characterized by genes whose products show significant sequence similarity with actinomycete clusters involved in heavy metal transport.
Concluding remarks.
The analyses of telomeres and replication components indicate that pRHL3 is an actinomycete invertron typical of those thus far characterized. The analysis of the genes suggests that the principal function of pRHL3 is to increase the catabolic capabilities of RHA1. However, the overall organization of the plasmid is very different from that of the classic catabolic plasmids of pseudomonads, such as pWWO, NAH, and CAM (64). In the latter plasmids, a significant proportion of the genes are responsible for the transformation of a specific compound to tricarboxylic acid cycle intermediates. Moreover, these genes occur in well-organized operons. The high number of mobility genes detected in pRHL3, together with the presence in RHA1 of other invertrons with which it can exchange ends, may help to explain the mosaic nature of this plasmid.
Due to the high number of unknown genes in catabolic gene clusters and the fact that the RHA1 genome is not yet completely known, it is difficult to assess the full catabolic role of pRHL3 in RHA1. However, the presence of several oxygenase- and numerous dehydrogenase-coding genes, known to play central roles in the degradation of aromatic compounds, along with the structural organization of these genes, and the putative nature of some of their regulatory genes strongly suggest that it plays a role in the assimilation of such compounds. Further studies are necessary to shed more light on the true catabolic nature of this plasmid and how it interacts with the RHA1 genome.
ADDENDUM IN PROOF
For the most current annotation of pRHL3 and the rest of the RHA1 genome, see http://www.rhodococcus.ca.
Supplementary Material
Acknowledgments
Work at the University of British Columbia and the Genome Sciences Center was supported by grants from Genome Canada and Genome BC. Work in Japan was supported in part by the Program for Promotion of Basic Research Activities for Innovative Biosciences (PROBRAIN) and the Ministry of Education, Culture, Sports, Science, and Technology of Japan. W.W.L.H. was supported by a Michael Smith Foundation for Health Research (MSFHR) scholarship and the Genome Prairie Pathogenomics Project. F.S.L.B. is an MSFHR scholar.
Footnotes
Supplemental material for this article may be found at http://jb.asm.org/.
Presented in part at the 11th International Conference on Microbial Genomes, Durham, N.C., 28 September to 2 October 2003.
REFERENCES
- 1.Altschul, S. F., W. Gish, W. Miller, E. W. Myers, and D. J. Lipman. 1990. Basic local alignment search tool. J. Mol. Biol. 215:403-410. [DOI] [PubMed] [Google Scholar]
- 2.Apweiler, R., T. K. Attwood, A. Bairoch, A. Bateman, E. Birney, M. Biswas, P. Bucher, L. Cerutti, F. Corpet, M. D. R. Croning, R. Durbin, L. Falquet, W. Fleischmann, J. Gouzy, H. Hermjakob, N. Hulo, I. Jonassen, D. Kahn, A. Kanapin, Y. Karavidopoulou, R. Lopez, B. Marx, N. J. Mulder, T. M. Oinn, M. Pagni, F. Servant, C. J. A. Sigrist, and E. M. Zdobnov. 2001. The InterPro database, an integrated documentation resource for protein families, domains, and functional sites. Nucleic Acids Res. 29:37-40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bachrach, G., M. J. Colston, H. Bercovier, D. Bar-Nir, C. Anderson, and K. G. Papavinasasundaram. 2000. A new single-copy mycobacterial plasmid, pMF1, from Mycobacterium fortuitum which is compatible with the pAL5000 replicon. Microbiology 146:297-303. [DOI] [PubMed] [Google Scholar]
- 4.Batzoglou, S. B. B. Jaffe, K. Stanley, J. Butler, S. Gnerre, E. Mauceli, B. Berger, J. P. Mesirov, and E. S. Lander. 2002. ARACHNE: a whole-genome shotgun assembler. Genome Res. 12:177-189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Beggs, M. L., J. T. Crawford, and K. D. Eisenach. 1995. Isolation and sequencing of the replication region of Mycobacterium avium plasmid pLR7. J. Bacteriol. 177:4836-4840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bell, K. S., J. C. Philip, D. W. J. Aw, and N. Christofi. 1998. The genus Rhodococcus. J. Appl. Microbiol. 85:195-210. [DOI] [PubMed] [Google Scholar]
- 7.Bentley, S. D., K. F. Chater, A.-M. Cerdeno-Tarraga, G. L. Challis, N. R. Thomson, K. D. James, D. E. Harris, M. A. Quail, H. Kieser, D. Harper, A. Bateman, S. Brown, G. Chandra, C. W. Chen, M. Collins, A. Cronin, A. Fraser, A. Goble, J. Hidalgo, T. Hornsby, S. Howarth, C.-H. Huang, T. Kieser, L. Larke, L. Murphy, K. Oliver, E. Rabbinowitsch, M.-A. Rajandream, K. Rutherford, S. Rutter, K. Seeger, D. Saunders, S. Sharp, R. Squares, S. Squares, K. Taylor, T. Warren, A. Wietzorrek, Z. Woodward, B. G. Barrell, J. Parkhill, and D. A. Hopwood. 2002. Complete genome sequence of the model actinomycete Streptomyces coelicolor A3(2). Nature 417:141-147. [DOI] [PubMed] [Google Scholar]
- 8.Borodovsky, M., and J. McIninch. 1993. GeneMark: parallel gene recognition for both DNA strands. Comput. Chem. 17:123-133. [Google Scholar]
- 9.Chang, P. C., and S. N. Cohen. 1994. Bidirectional replication from an internal origin in a linear Streptomyces plasmid. Science 265:952-954. [DOI] [PubMed] [Google Scholar]
- 10.Chauvaux, S., F. Chevalier, C. Le Dantec, F. Fayolle, I. Miras, F. Kunst, and P. Beguin. 2001. Cloning of a genetically unstable cytochrome P-450 gene cluster involved in the degradation of the polluant ethyl tert-butyl ether by Rhodococcus ruber. J. Bacteriol. 183:6551-6557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chen, E., D. Schlessinger, and J. Kere. 1993. Ordered shotgun sequencing, a strategy for integrated mapping and sequencing YAC clones. Genomics 17:651-656. [DOI] [PubMed] [Google Scholar]
- 12.Chen, C. W., C. H. Huang, H. H. Lee, H. H. Tsai, and R. Kirby. 2002. Once the circle has been broken: dynamics and evolution of Streptomyces chromosomes. Trends Genet. 18:522-529. [DOI] [PubMed] [Google Scholar]
- 13.Cole, S. T., R. Brosch, J. Parkhill, T. Garnier, C. Churcher, D. Harris, S. V. Gordon, K. Eiglmeier, S. Gas, C. E. Barry III, F. Tekaia, K. Badcock, D. Basham, D. Brown, T. Chillingworth, R. Connor, R. Davies, K. Devlin, T. Feltwell, S. Gentles, N. Hamlin, S. Holroyd, T. Hornsby, K. Jagels, B. G. Barrell, et al. 1998. Deciphering the biology of Mycobacterium tuberculosis from the complete genome sequence. Nature 393:537-544. [DOI] [PubMed] [Google Scholar]
- 14.Delcher, A. L., D. Harmon, S. Kasif, O. White, and S. L. Salzberg. 1999. Improved microbial gene identification with Glimmer. Nucleic Acids Res. 27:4636-4641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.De Mot, R., I. Nagy, A. De Schrijver, P. Pattanapipitpaisal, G. Schoofs, and J. Vanderleyden. 1997. Structural analysis of the 6-kb cryptic plasmid pFAJ2600 from Rhodococcus erythropolis NI86/21 and construction of Escherichia coli-Rhodococcus shuttle vectors. Microbiology 143:3137-3147. [DOI] [PubMed] [Google Scholar]
- 16.Denis-Larose, C., H. Bergeron, D. Labbe, C. W. Greer, J. Hawari, M. J. Grossman, B. M. Sankey, and P. C. Lau. 1998. Characterization of the basic replicon of Rhodococcus plasmid pSOX and development of a Rhodococcus-Escherichia coli shuttle vector. Appl. Environ. Microbiol. 64:4363-4367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Durbin, R., and J. T. Mieg. 1991. Caenorhabditis elegans database. http://lirmm.lirmm.fr, http://cele.mrc-lmb.cam.ac.uk, and http://ncbi.nlm.nih.gov.
- 18.Edwards, A., and C. Caskey. 1990. Closure stategies for random DNA sequencing. Methods Companion Methods Enzymol. 3:41-47. [Google Scholar]
- 19.Ewing, B., L. Hillier, M. C. Wendl, and P. Green. 1998. Bases-calling of automated sequencer traces using Phred. I. Accuracy assessment. Genome Res. 8:175-185. [DOI] [PubMed] [Google Scholar]
- 20.Ewing, B., and P. Green. 1998. Bases-calling of automated sequencer traces using Phred. II. Error probabilities. Genome Res. 8:186-194. [PubMed] [Google Scholar]
- 21.Felsenstein, J. 1993. PHYLIP (Phylogeny Inference Package), version 3.6. Department of Genetics, University of Washington, Seattle.
- 22.Fuhrmann, D. R., M. I. Krzywinski, R. Chiu, P. Saeedi, J. E. Schein, I. E. Bosdet, A. Chinwalla, L. W. Hillier, R. H. Waterston, J. D. McPherson, S. J. Jones, and M. A. Marra. 2003. Software for automated analysis of DNA fingerprinting gels. Genome Res. 13:940-953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Furukawa, K. 2000. Biochemical and genetic bases of microbial degradation of polychlorinated biphenyls (PCBs). J. Gen. Appl. Microbiol. 46:283-296. [DOI] [PubMed] [Google Scholar]
- 24.Gordon, D., C. Abajian, and P. Green. 1998. Consed: a graphical tool for sequence finishing. Genome Res. 8:195-202. [DOI] [PubMed] [Google Scholar]
- 25.Green, P. 1994. Phrap documentation. http://www.phrap.org/.
- 26.Hentschel, U., and J. Hacker. 2001. Pathogenicity islands: the tip of the iceberg. Microbes Infect. 3:545-548. [DOI] [PubMed] [Google Scholar]
- 27.Hinnebusch, J., and K. Tilly. 1993. Linear plasmids and chromosomes in bacteria. Mol. Microbiol. 10:917-922. [DOI] [PubMed] [Google Scholar]
- 28.Hiratsu, K., S. Mochizuki, and H. Kinashi. 2000. Cloning and analysis of the replication origin and the telomeres of the large linear plasmid pSLA2-L in Streptomyces rochei. Mol. Gen. Genet. 263:1015-1021. [DOI] [PubMed] [Google Scholar]
- 29.Hsiao, W., I. Wan, S. J. Jones, and F. S. L. Brinkman. 2003. IslandPath: aiding detection of genomic islands in prokaryotes. Bioinformatics 19:418-420. [DOI] [PubMed] [Google Scholar]
- 30.Kalkus, J., C. Dorrie, D. Fischer, M. Reh, and H. G. Schlegel. 1993. The giant linear plasmid pHG207 from Rhodococcus sp. encoding hydrogen autotrophy: characterization of the plasmid and its termini. J. Gen. Microbiol. 139:2055-2065. [DOI] [PubMed] [Google Scholar]
- 31.Kalkus, J., R. Menne, M. Reh, and H. G. Schlegel. 1998. The terminal structures of linear plasmids from Rhodococcus opacus. Microbiology 144:1271-1279. [DOI] [PubMed] [Google Scholar]
- 32.Kitagawa, W., K. Miyauchi, E. Masai, and M. Fukuda. 2001. Cloning and characterization of benzoate catabolic genes in the gram-positive polychlorinated biphenyl degrader Rhodococcus sp. strain RHA1. J. Bacteriol. 183:6598-6606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kitagawa, W., A. Suzuki, T. Hoaki, E. Masai, and M. Fukuda. 2001. Multiplicity of aromatic ring hydroxylation dioxygenase genes in a strong PCB degrader, Rhodococcus sp. strain RHA1 demonstrated by denaturing gradient gel electrophoresis. Biosci. Biotechnol. Biochem. 65:1907-1911. [DOI] [PubMed] [Google Scholar]
- 34.Kupfer, K., M. Smith, J. Quackenbush, and G. Evans. 1995. Physical mapping of complex genomes by sampled sequencing: a theoretical analysis. Genomics 27:90-100. [DOI] [PubMed] [Google Scholar]
- 35.Kurtz, S., J. V. Choudhuri, E. Ohlebusch, C. Schleiermacher, J. Stoye, and R. Giegerich. 2001. REPuter: the manifold applications of repeat analysis on a genomic scale. Nucleic Acids Res. 29:4633-4642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kurtz, S., A. Phillippy, A. L. Delcher, M. Smoot, M. Shumway, C. Antonescu, and S. L. Salzberg. 2004. Versatile and open software for comparing large genomes. Genome Biol. 5:R12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lamb, D. C., H. Ikeda, D. R. Nelson, J. Ishikawa, T. Skaug, C. Jackson, S. Omura, M. R. Waterman, and S. L. Kelly. 2003. Cytochrome P450 complement (CYPome) of the avermectin-producer Streptomyces avermitilis and comparison to that of Streptomyces coelicolor A3(2). Biochem. Biophys. Res. Commun. 307:610-619. [DOI] [PubMed] [Google Scholar]
- 38.Le Dantec, C., N. Winter, B. Gicquel, V. Vincent, and M. Picardeau. 2001. Genomic sequence and transcriptional analysis of a 23-kilobase mycobacterial linear plasmid: evidence for horizontal transfer and identification of plasmid maintenance systems. J. Bacteriol. 183:2157-2164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Marmur, J. 1961. A procedure for the isolation of deoxyribonucleic acids from microorganisms. J. Mol. Biol. 3:208-218. [Google Scholar]
- 40.Marra, M. A., T. A. Kucaba, N. L. Dietrich, E. D. Green, B. Brownstein, R. K. Wilson, K. M. McDonald, L. W. Hillier, J. D. McPherson, and R. H. Waterston,. 1997. High throughput fingerprint analysis of large-insert clones. Genome Res. 7:1072-1084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Nagy, I., F. Compernolle, K. Ghys, J. Vanderleyden, and R. De Mot. 1995. A single cytochrome P-450 system is involved in degradation of the herbicide EPTC (S-ethyl dipropylthiocarbamate) and atrazine by Rhodococcus sp. strain NI86/21. Appl. Environ. Microbiol. 61:2056-2060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ness, S. R., W. Terpstra, M. Krzywinski, M. A. Marra, and S. J. Jones. 2002. Assembly of fingerprint contigs: parallelized FPC. Bioinformatics 18:484-485. [DOI] [PubMed] [Google Scholar]
- 43.Page, R. D. M. 1996. TREEVIEW: an application to display phylogenetic trees on personal computers. Comput. Appl. Biosci. 12:357-358. [DOI] [PubMed] [Google Scholar]
- 44.Pandza, S., G. Biukovic, A. Paravic, A. Dadbin, J. Cullum, and D. Hranueli. 1998. Recombination between the linear plasmid pPZG101 and the linear chromosome of Streptomyces rimosus can lead to exchange of ends. Mol. Microbiol. 28:1165-1176. [DOI] [PubMed] [Google Scholar]
- 45.Parsons, J. D. 1995. Miropeats: graphical DNA sequence comparisons. Comput. Appl. Biosci. 11:615-619. [DOI] [PubMed] [Google Scholar]
- 46.Picardeau, M., C. Le Dantec, and V. Vincent. 2000. Analysis of the internal replication region of a mycobacterial linear plasmid. Microbiology 146:305-313. [DOI] [PubMed] [Google Scholar]
- 47.Polo, S., O. Guerini, M. Sosio, and G. Deho. 1998. Identification of two linear plasmids in the actinomycete Planobispora rosea. Microbiology 144:2819-2825. [DOI] [PubMed] [Google Scholar]
- 48.Roberts, G. A., G. Grogan, A. Greter, S. L. Flitsch, and N. J. Turner. 2002. Identification of a new class of cytochrome P450 from a Rhodococcus sp. J . Bacteriol. 184:3898-3908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Rowe-Magnus, D. A., and D. Mazel. 2001. Integrons: natural tools for bacterial genome evolution. Curr. Opin. Microbiol. 4:565-569. [DOI] [PubMed] [Google Scholar]
- 50.Sakaguchi, K. 1990. Invertrons, a class of structurally and functionally related genetic elements that includes linear DNA plasmids, transposable elements, and genomes of adeno-type viruses. Microbiol. Rev. 54:66-74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Sakai, M., E. Masai, H. Asami, K. Sugiyama, K. Kimbara, and M. Fukuda. 2002. Diversity of 2,3-dihydroxybiphenyl dioxygenase genes in a strong PCB degrader, Rhodococcus sp. strain RHA1. J. Biosci. Bioeng. 93:421-427. [DOI] [PubMed] [Google Scholar]
- 52.Sanger, F., S. Nicklen, and A. R. Coulson. 1977. DNA sequencing with chain-terminating inhibitors. Proc. Natl. Acad. Sci. USA 74:5463-5467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52a.Schein, J., T. Kucaba, M. Sekhon, D. Smailus, R. Waterston, and M. Marra. 2004. High-throughput BAC fingerprinting. Methods Mol. Biol. 255:143-156. [DOI] [PubMed] [Google Scholar]
- 53.Seto, M., K. Kimbara, M. Shimura, T. Hatta, M. Fukuda, and K. Yano. 1995. A novel transformation of polychlorinated biphenyls by Rhodococcus sp. strain RHA1. Appl. Environ. Microbiol. 61:3353-3358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Sharp, P. M., J. E. Kelleher, A. S. Daniel, G. M. Cowan, and N. E. Murray. 1992. Roles of selection and recombination in the evolution of type I restriction-modification systems in enterobacteria. Proc. Natl. Acad. Sci. USA 89:9836-9840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Shiffman, D., and S. N. Cohen. 1992. Reconstruction of a Streptomyces linear replicon from separately cloned DNA fragments: existence of a cryptic origin of circular replication within the linear plasmid. Proc. Natl. Acad. Sci. USA 89:6129-6133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Shimizu, S., H. Kobayashi, E. Masai, and M. Fukuda. 2001. Characterization of the 450-kb linear plasmid in a polychlorinated biphenyl degrader, Rhodococcus sp. strain RHA1. Appl. Environ. Microbiol. 67:2021-2028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Soderlund, C., S. Humphray, A. Dunham, and L. French. 2000. Contigs built with fingerprints, markers, and FPC V4.7. Genome Res. 10:1772-17787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Soderlund, C., I. Longden, and R. Mott. 1997. FPC: a system for building contigs from restriction fingerprinted clones. Comput. Appl. Biosci. 13:523-535. [DOI] [PubMed] [Google Scholar]
- 59.Spatz, K., H. Kohn, and M. Redenbach. 2002. Characterization of the Streptomyces violaceoruber SANK95570 plasmids pSV1 and pSV2. FEMS Microbiol. Lett. 213:87-92. [DOI] [PubMed] [Google Scholar]
- 60.Stecker, C., A. Johann, C. Herzberg, B. Averhoff, and G. Gottschalk. 2003. Complete nucleotide sequence and genetic organization of the 210-kilobase linear plasmid of Rhodococcus erythropolis BD2. J. Bacteriol. 185:5269-5274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Thompson, J. D., D. G. Higgins, and T. J. Gibson. 1994. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position specific gap penalties and weight matrix choice. Nucleic Acids Res. 22:4673-4680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.van der Werf, M. J., C. van der Ven, F. Barbirato, M. H. Eppink, J. A. de Bont, and W. J. van Berkel. 1999. Stereoselective carveol dehydrogenase from Rhodococcus erythropolis DCL14: a novel nicotinoprotein belonging to the short chain dehydrogenase/reductase superfamily. J. Biol. Chem. 274:26296-26304. [DOI] [PubMed] [Google Scholar]
- 63.Volff, J. N., and J. Altenbuchner. 2000. A new beginning with new ends: linearization of circular chromosomes during bacterial evolution. FEMS Microbiol. Lett. 186:143-150. [DOI] [PubMed] [Google Scholar]
- 64.Williams, P. A., R. M. Jones, and G. Zylstra. 2003. Genomics of catabolic plasmids, p. 165-195. In J. L. Ramos (ed.), The pseudomonads, vol. 1: genomics, lifestyle, and molecular architecture. Kluwer Academic/Plenum Publishers, New York, N.Y.
- 65.Wu, X., and K. L. Roy. 1993. Complete nucleotide sequence of a linear plasmid from Streptomyces clavuligerus and characterization of its RNA transcripts. J. Bacteriol. 175:37-52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Yamada, A., H. Kishi, K. Sugiyama, T. Hatta, K. Nakamura, E. Masai, and M. Fukuda. 1998. Two nearly identical aromatic compound hydrolase genes in a strong polychlorinated biphenyl degrader, Rhodococcus sp. strain RHA1. Appl. Environ. Microbiol. 64:2006-2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.