Summary
Salmonella Typhimurium is an intracellular bacterial pathogen that infects both epithelial cells and macrophages. Salmonella effector proteins, which are translocated into the host cell and manipulate host cell components, control the ability to replicate and/or survive in host cells. Due to the complexity and heterogeneity of Salmonella infections, there is growing recognition of the need for single cell and live-cell imaging approaches to identify and characterize the diversity of cellular phenotypes and how they evolve over time. Here we establish a pipeline for long-term (16 hours) live-cell imaging of infected cells and subsequent image analysis methods. We apply this pipeline to track bacterial replication within the Salmonella-containing vacuole in epithelial cells, quantify vacuolar replication versus survival in macrophages, and investigate the role of individual effector proteins in mediating these parameters. This approach revealed that dispersed bacteria can coalesce at later stages of infection, that the effector protein SseG influences the propensity for cytosolic hyperreplication in epithelial cells, and that while SteA only has a subtle effect on vacuolar replication in epithelial cells, it has a profound impact on infection parameters in immunocompetent macrophages, suggesting differential roles for effector proteins in different infection models.
INTRODUCTION
There is growing recognition of the importance of heterogeneity in biological systems. It is now apparent that even genetically identical populations of cells have substantial variability in gene expression, protein expression, and growth rate (Spudich and Koshland, 1976), as well as physiological heterogeneity such as response to environmental stressors, sensitivity to drugs, and propensity for cell death (Perkins and Swain, 2009). This heterogeneity is particularly pertinent to microbial pathogenesis because growth rate, replicative ability, responsiveness to stress, and antimicrobial resistance profoundly influence key features of disease, such as infectivity, spread, the existence of non-replicating states, establishment of chronic infection, and sensitivity to therapeutic intervention (Booth, 2002; Avery, 2006; Lidstrom and Konopka, 2010).
Salmonella enterica subsp. enterica are intracellular bacterial pathogens capable of infecting a wide range of host organisms. While there are over 2500 serovars, the Typhimurium serovar is widely used as a model strain because it is a major contributor to human enteric disease (Majowicz et al., 2010), and it gives rise to a systemic typhoid-like disease (Haraga et al., 2008) in mice. To establish infection, S. Typhimurium uses two type III secretion systems (T3SSs) to translocate bacterial effector proteins into the host cell (Moraes et al., 2008; Marlovits and Stebbins, 2010). In the gastroenteritis model of infection, the first secretion system (T3SS1) delivers effector proteins into the host cell to induce bacterial uptake by intestinal epithelial cells, whereas in systemic disease, bacteria are phagocytosed by resident macrophages (McGhie et al., 2009; Srikanth et al., 2011; Moest and Méresse, 2013). Once inside either cell type, S. Typhimurium uses a second secretion system (T3SS2) to deliver additional effector proteins that work in concert to establish a replicative niche within the cell (Jones et al., 1998; McGhie et al., 2001; Schlumberger and Hardt, 2006; Srikanth et al., 2011). Salmonella has long been described as a vacuolar pathogen, as infection of multiple cell types is characterized by encapsulation of bacteria within a membrane-bound vacuole (Creasey and Isberg, 2014). However, recent reports establish that there are distinct replicative populations within epithelial cells, where Salmonella can escape from the Salmonella containing vacuole (SCV) in some cells and hyperreplicate in the cytosol, a phenomenon which contributes to extrusion of cells from the gut epithelium, possibly promoting bacterial dissemination along the gastrointestinal tract (Knodler et al., 2010). Cytosolic bacteria undergo transcriptional reprogramming so that they are motile and primed for infection (Knodler et al., 2010), whereas vacuolar bacteria express virulence genes that mediate the establishment and maintenance of the vacuole to promote controlled replication (Abrahams and Hensel, 2006; Adam C Smith et al., 2007; Steele-Mortimer, 2008; Bakowski et al., 2008; Sherwood and Craig R Roy, 2013; Creasey and Isberg, 2014). Such heterogeneity is not unique to epithelial cells as multiple replication phenotypes have been observed in macrophages, including proliferative bacteria and bacteria in a dormant-like state (Helaine et al., 2010; Claudi et al., 2014; Avraham et al., 2015).
A major unanswered question is how individual effector proteins modulate the intracellular environment for the different subpopulations in epithelial cells or macrophages, and whether different effector proteins serve different roles during the evolution of infection in different model systems (epithelial cells versus macrophages). Characterizing and quantifying the survival and replication of bacterial pathogens within host cells, as well as examining phenotypes such as the appearance, dynamics, and location of bacteria, are important steps in defining the infection context and identifying factors that perturb the intracellular niche. Yet it is now clear that there is abundant heterogeneity in infection, replication, and infection phenotypes. To complicate matters further, there is growing recognition for the need to define infection parameters in living cells due to the dynamic nature of the host-pathogen interface, the potential introduction of artifacts upon fixation (Rajashekar et al., 2014), and the drive to define how phenotypes evolve over time. Thus, there is a need for a broader repertoire of experimental approaches that interrogate the role of individual effector proteins in establishing infection while preserving the full complexity and heterogeneity of the system, enabling quantitative evaluation of many cells to provide statistical power, and discern common from rare phenotypes.
In this study we set out to use long-term live-cell imaging of infected cells to monitor Salmonella survival and replication at the single cell level in both epithelial cells and macrophages, as well as characterize the appearance and location of the SCV and intracellular bacteria. We were particularly interested in investigating T3SS2 effector proteins that mediate maturation of the SCV and whether these effector proteins play distinct roles in epithelial cells versus macrophages. SCV maturation is a complex process involving positioning near the nucleus, interception of membrane trafficking pathways, and control of membrane dynamics (Steele-Mortimer, 2008). Numerous effector proteins have been implicated in this process, including SifA, PipB2, SopD2, SseF, SseG, and SseJ (McGhie et al., 2009; Agbor and McCormick, 2011; Ramos-Morales, 2012). More recently it has been proposed that SteA may also contribute to SCV membrane dynamics (Domingues et al., 2014; Domingues et al., 2015). However, the precise cellular role of SteA is not well characterized and how it contributes to vacuolar replication in epithelial cells or survival versus replication within macrophages has not been defined. It was recently suggested that in infected epithelial cells, SteA may be functionally linked with another effector protein involved in SCV maturation, SseG (Domingues et al., 2014). SseG, along with a related protein SseF, has been shown to be important for localization of the SCV near the Golgi by controlling molecular motors, as well as for SCV maintenance by recruiting host cell membrane components and promoting microtubule bundling at the SCV (Kuhle and Hensel, 2002; Salcedo and Holden, 2003; Kuhle et al., 2004; Kuhle et al., 2006; Deiwick et al., 2006; Ramsden et al., 2007). In this study we reveal that SteA is important for Salmonella replication within the SCV in epithelial cells as well as vacuolar replication and survival in macrophages. Additionally, we uncover unexpected and previously undocumented roles for SseG in epithelial cells and macrophages. In epithelial cells, dispersed bacteria can coalesce at later stages of infection and reestablish a compact structure resembling an SCV. Intriguingly, the double deletion of sseG and sseF affects the coalescence/dispersal ratio of these bacteria. In macrophages, we found that SseG plays a role in preventing bacterial clearance. Finally, we found that T3SS2 effector proteins can influence the propensity for cytosolic hyperreplication.
RESULTS
Long-term live cell imaging reveals diverse bacterial growth phenotypes in epithelial cells
Characterizing and quantifying the survival and replication of bacterial pathogens within host cells is an important step in defining the intracellular niche and identifying factors that perturb this niche. Given documented heterogeneity in intracellular bacterial growth rates (Avery, 2006; Lidstrom and Konopka, 2010) and replicative states (Helaine et al., 2010; Malik-Kale et al., 2012), these experiments need to be performed at the single cell level. Here we utilize a quantitative approach for tracking bacterial replication in individual host cells for hundreds of cells over long time periods (16 hours) to examine the role of effector proteins in regulating this niche. Similar to the approach pioneered by Malik-Kale et al (Malik-Kale et al., 2012), two criteria were used to distinguish infected cells with exclusively vacuolar bacteria from those containing cytosolic hyperreplicating bacteria. The first criterion was colocalization of bacteria with a fluorescently-tagged Dextran that was added to cells immediately before infection so that it was engulfed with bacteria and labeled the SCV. We chose Dextran to label the SCV instead of a host cell marker, such as LAMP1, because Dextran does not require active recruitment after infection and was previously documented as a robust marker of the SCV (Rajashekar et al., 2008; Malik-Kale et al., 2012). The second criterion was that bacteria exposed to the cytosol of epithelial cells hyperreplicate such that at late time points the cytosol becomes filled with bacteria, frequently causing the infected cell to die or slough off after ~ 9 hrs (Knodler et al., 2010; Malik-Kale et al., 2012). On the contrary, infected cells with exclusively vacuolar bacteria tended to survive for the duration of the experiment (~ 17 hours). Infection of HeLa cells treated with Alexa Fluor 488 Dextran with a Salmonella strain constitutively expressing the mCherry fluorescent protein led to engulfment of the fluorescently-tagged Dextran in the SCV and enabled simultaneous tracking of the fate of Salmonella and the vacuole over time. Dextran was routinely visible for up to 17 hours post infection and the combination of Dextran colocalization and monitoring of fluorescently-tagged bacteria allowed for clear differentiation of vacuolar and cytosolic bacteria (Figure 1, Supplementary Figure S1). Some infected cells contained both vacuolar and cytosolic bacteria at early time points, but at later times, the cytosolic bacteria typically overran the cytoplasm; the bacteria in these cells were classified as cytosolic (Supplementary Figure S1). In order to categorize intracellular bacterial growth at the single cell level, the bioimaging analysis program ICY was used to extract the number of pixels corresponding to fluorescent bacteria. To establish whether there was a direct correlation between the number of bacteria and the number of pixels reported by ICY, we counted individual bacteria manually across 10 experiments from four different conditions and compared our results to the ICY output (Supplementary Figure S2). The pixel area linearly correlated (R2 > 0.9) with the number of bacteria until the bacteria reached a density that was too difficult to count manually (~50 bacteria, correlating to 200-250 pixels). Based on this correlation, the total pixel area reported by ICY was used as a measure of bacterial load and hence bacterial replication over time.
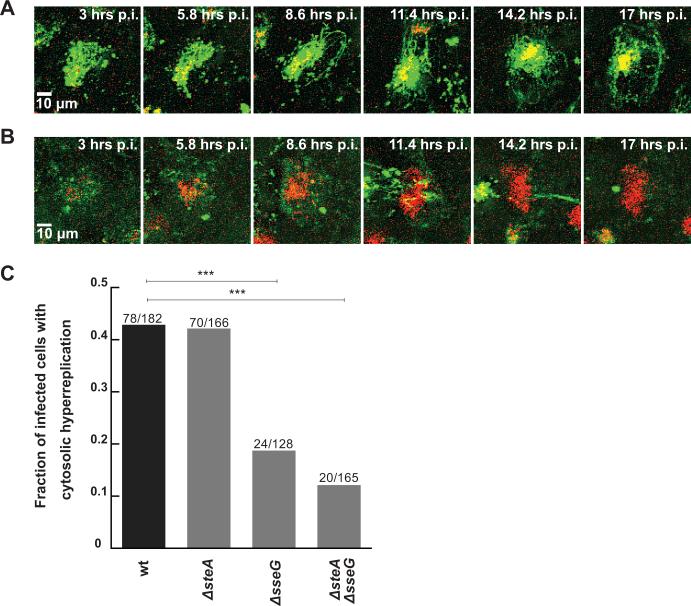
Figure 1.
Live cell imaging distinguishes cytosolic hyperreplication from vacuolar replication in epithelial cells. Representative fluorescence images of an infected cell with (A) vacuolar bacteria or (B) cytosolic hyperreplicating bacteria at distinct time points post infection (wt mCherry Salmonella in red, AlexaFluor 488 Dextran in green, colocalization in yellow). (C) Fraction of cells infected with wt or deletion Salmonella strains that contain cytosolic hyperreplicating bacteria. ΔsseG causes a decrease in cytosolic replication events. Values (wt n= 182 cells, ΔsteA n= 166 cells, ΔsseG n=128 cells, ΔsteA/ΔsseG n=165 cells) are a combination of two (for ΔsseG) or three independent experiments (all other strains). p values labeled with *** are <0.0001. All p values were calculated using a χ2 test (MVPstats).
The existence of sub-populations of replicating Salmonella in epithelial cells has only recently become widely recognized (Knodler, 2015) and the repertoire of host and bacterial factors that give rise to these different physiologies remain to be elucidated. It has been shown that a ΔssaR mutant, defective for the translocation of T3SS2 effector proteins, did not alter the percentage of cytosolic bacteria in Caco-2 C2Bbe1 cells (Knodler et al., 2014), suggesting that perhaps SPI2 effector proteins do not play a role in cytosolic replication. However, the role of individual effector proteins has not been explored and it is possible that these proteins could differentially impact the distribution amongst these niches such that the deletion of the entire effector protein cohort yields no net effect. Therefore, we examined whether individual SPI-2 effector proteins could influence the distribution of cytosolic hyperreplication versus vacuolar replication. These experiments focused on two Salmonella effector proteins: SteA and SseG, which localize to the SCV upon translocation (Kuhle et al., 2004; Van Engelenburg and Palmer, 2010), play a role in maintaining the SCV, and have been shown to be functionally linked (Domingues et al., 2014). HeLa cells were infected with ΔsteA, ΔsseG, and ΔsteA/ΔsseG mutant strains as described above and both bacteria and Dextran were monitored (n ~ 100 cells for each condition). There was no significant difference in the overnight growth of these strains in LB media (Supplementary Figure S3). Within HeLa cells, a significant shift in the distribution of vacuolar versus cytosolic replication was observed. Cells with cytosolic hyperreplicating bacteria corresponded to 43%, 42%, 19%, and 12% of wt, ΔsteA, ΔsseG, and ΔsteA/ΔsseG infected cells, respectively (Figure 1C). These results indicate that strains lacking sseG gave rise to significantly less cytosolic hyperreplication, suggesting that SseG, but not SteA, contributes to escape from the vacuole and/or promotes cytosolic replication.
The primary advantage of this single cell assay is the ability to monitor changes in bacterial load inside an individual host cell and to determine how effector proteins perturb this process. While heterogeneity in bacterial growth rates in vitro (Kelly and Rahn, 1932; Elfwing et al., 2004) and in SCVs within infected epithelial cells (Malik-Kale et al., 2012) and macrophages (Helaine et al., 2010) have been documented, the possibility that there are different replicative sub-populations within the SCVs of distinct epithelial cells has not been described. Bacterial growth in vitro in a closed system such as batch culture is typically characterized by a lag phase, followed by exponential growth in which the bacterial population doubles each generation, and finally a stationary phase (Baty et al., 2002). Intracellular replication is likely to be more complex given exchange of nutrients with the host cell and replication that is regulated by both host and bacterial factors. Analysis of the change in bacterial load over time at the single cell level (i.e. replication curves) has the potential to reveal different kinds of replication defects such as a pronounced lag phase that could indicate some factor is not yet optimal for growth, a change in the doubling time indicating slow growth, or increased bacterial death. The change in pixel area over time enabled us to generate an in situ replication curve for each epithelial cell in which bacteria replicated within a vacuole (Figure 2A). In situ replication curves generally exhibited features similar to in vitro growth curves; however, there was substantial heterogeneity in the shapes of these curves, including steady growth, a pronounced lag phase followed by steady growth, growth followed by a plateau, no growth (i.e. a flat line), or growth followed by a crash (decrease in bacterial area) (Figure 2B). The wild type (wt) and ΔsteA strains show a similar distribution of replication curve phenotypes, whereas ΔsseG strains show a marked reduction in the number of curves with steady growth and an increase in infections with a pronounced lag phase prior to steady growth, indicating that in the absence of SseG the SCV environment is compromised for replication. Finally ΔsteA/ΔsseG infections had a substantial number of curves with no growth or with steady growth followed by a crash, perhaps suggestive of increased bacterial cell death.
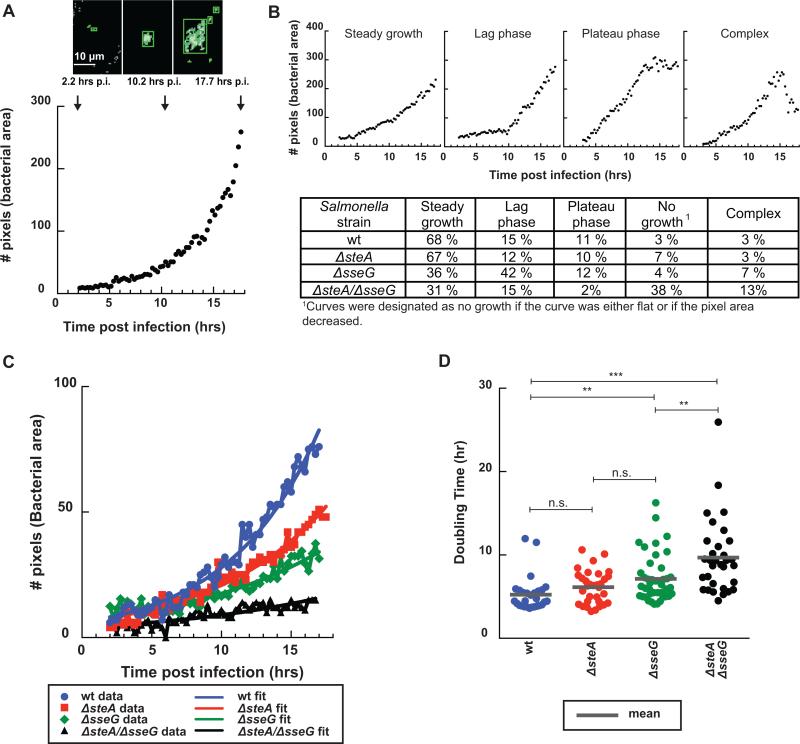
Figure 2.
Comparison of replication curves between wt and deletion Salmonella strains. (A) Representative images showing ICY analysis of the bacterial channel at three time points post infection. Green boxes mark the detected bacterial areas at each time point (arrows). Images were processed using a threshold plugin and pixels were quantified over time to generate a replication curve (bacterial area vs. time post infection in hours). (B) Examples of replication curve phenotypes (top) and the frequency for each strain (bottom). Curves were categorized by visual inspection, wt n= 103 cells, ΔsteA n= 99 cells, ΔsseG n= 103 cells, and ΔsteA/ΔsseG n= 116 cells. (C) Representative “Steady growth” replication curves and their corresponding fits to a mathematical function representing bacterial growth (Y=A*2t/tau) for wt (blue), ΔsteA (red), ΔsseG (green), and ΔsteA/ΔsseG (black) strains. (D) Doubling times for infections categorized as “Steady growth” for ΔsteA (red), ΔsseG (green), and ΔsteA/ΔsseG (black) strains compared to wt Salmonella (blue). ΔsteA, ΔsseG, and ΔsteA/ΔsseG infections have longer average doubling times compared to wt Salmonella within infected epithelial cells (wt n= 28 cells, ΔsteA n= 32 cells, ΔsseG n= 43 cells, and ΔsteA/ΔsseG n= 32 cells). Each dot represents the doubling time for an infection within a single cell. The mean for each strain is marked with a gray line. All p values were calculated using a one-way ANOVA test (KaleidaGraph), *** denotes p <0.0001, ** denotes p = 0.003, and n.s. denotes p=0.09.
To examine whether the rate of replication differed among the mutant strains, replication curves that were characterized as “steady growth” were fit to a mathematical expression for bacterial growth (Y = A*bt/tau, where A represents the amount of bacteria at t = 0, b = 2 due to doubling each generation, and tau is the time constant that represents the doubling time). Figures 2C and D present representative fits and the doubling times compared across the different strains. There was no difference between the average doubling times for wild type Salmonella and ΔsteA (5 hours and 6 hours, respectively). However there were substantial increases in the doubling times for ΔsseG (7 hours) and ΔsteA/ΔsseG (10 hours), revealing that in addition to altered replication curve phenotypes, the mutant strains also had overall lower replication rates than the wt strain.
Comparison of bacterial growth in single host cells versus in bulk assays
Quantification of vacuolar replication in individual cells enabled determination of fold replication at the single cell level, where fold replication was calculated by dividing the number of pixels corresponding to bacterial fluorescence at discrete time points by the number of pixels at three hours post infection. A comparison of wt and ΔsteA Salmonella demonstrates that the overall fold replication of the two strains is similar until late time points (> 16 hours) at which point the difference in the mean (21%) becomes statistically significant (p =0.02) (Figure 3A). This difference was complemented by the addition of SteA-GFP11 on a plasmid (Supplementary Figure S4). Intriguingly, examination of fold replication at the single cell level reveals that at later time points, the majority of ΔsteA infections show a replication defect, but there are 7 infected cells that exhibit high replication, comparable to wt, perhaps indicative of subpopulations of vacuolar Salmonella.
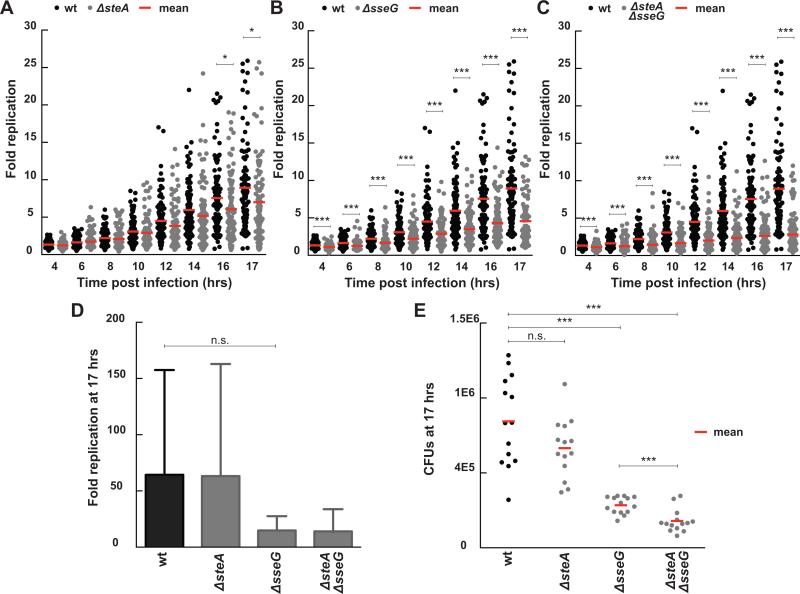
Figure 3.
Fold change in bacterial load for infections with ΔsteA, ΔsseG, and ΔsteA/ΔsseG strains compared to wt Salmonella. Results compare single cell long term imaging assays (A-C) with CFU assays (D-E). For the single cell imaging assays, fold replication was calculated by dividing the bacterial area in pixels at discrete time points by the bacterial area at three hours post infection. Each dot represents the fold replication within a single cell. (A) ΔsteA infections exhibit decreased fold replication compared to wt infections at late time points (wt n= 100 cells, ΔsteA n= 99 cells, 3 independent experiments, * indicates p <0.02). (B) ΔsseG infections exhibit decreased fold replication compared to wt infections at all time points (ΔsseG n= 98 cells, 2 independent experiments, *** indicates p <0.0002). (C) ΔsteA/ΔsseG infections exhibit decreased fold replication that is additive between the two single knock out strains (ΔsteA/ΔsseG n= 116 cells, 3 independent experiments, *** indicates p <0.0003). (D) Fold replication as measured by a CFU assay (n = 3 independent experiments, n.s. signifies p = 0.09). Fold replication for the CFU assay was calculated by dividing the CFUs at 17 hours post infection by the CFUs at 3 hours post infection. Values are the mean + 95% confidence interval. (E) CFUs at 17 hours post infection show decreased bacterial load for ΔsseG and ΔsteA/ΔsseG. Each dot represents CFUs from a single infection (n=14 for all strains; data from three independent experiments, *** signifies p=0.0003, n.s. signifies p = 0.07). All p values were calculated using a one-way ANOVA test (KaleidaGraph).
Our single cell replication results are consistent with a previous study using a CFU assay that found no replication requirement for SteA in HeLa or Caco-2 cells at six hours post-infection (Buckner et al., 2011), but we show that a replication defect begins to emerge at later time points. Our results differ from previous reports that deletion of steA did not significantly impair replication in a model mouse macrophage cell line (Raw264.7 cells) (Domingues et al., 2014) or bone marrow derived macrophage from susceptible mice (Figueira et al., 2013) at 16 or 17 hours post infection, respectively. However, this difference could result from the use of different cell types for these experiments, the ability of our assay to track replication within individual cells, or the fact that our assay enables us to explicitly define replication within the SCV, excluding the potentially confounding contribution of cytosolic replication.
Deletion of sseG led to a more profound replication defect. Decreased replication was observed at all time points, resulting in a 48% decrease in mean fold replication compared to wt at 17 hours post infection (Figure 3B), consistent with previous results using a CFU assay to quantify replication between 2 and 16 hours post infection in HeLa cells (Salcedo and Holden, 2003). The ΔsteA/ΔsseG strain also had a replication defect throughout the experiment resulting in a 68% decrease in the mean fold replication compared to wt at 17 hours post infection (Figure 3C). The fold replication for the ΔsseG and ΔsteA/ΔsseG strains were significantly different by 17 hours post infection, with deletion of steA causing an additional 20% decrease in the mean fold replication compared to deletion of sseG alone, suggesting the contributions of SteA and SseG to vacuolar replication may be additive. Intriguingly, for both ΔsseG and ΔsteA/ΔsseG, there was substantially less heterogeneity (variance wt: 31.58, ΔsseG: 8.89, ΔsteA/ΔsseG: 6.33, P < 0.0001) in the fold replication than wt. Additionally, for the ΔsteA/ΔsseG strain there appears to be a subpopulation that doesn't replicate at all and a subpopulation that replicates but to a lower level than wt.
We next compared the fold replication of infections at the single cell level and the total bacterial load with a CFU assay (Figure 3D, E). Not surprisingly, the CFU assay yielded much higher values for fold replication likely because it combines cytosolic hyperreplication with vacuolar replication. There was a high degree of heterogeneity in the CFU assay resulting in very large error bars, likely due to variability in bacterial load at 3 hours, the presence of both vacuolar and hyperreplicating bacteria, and variable numbers of recoverable bacteria due to cell death. Overall, the single host cell assay provides significantly more information than the population-based CFU assay, allowing us to separate vacuolar from cytosolic replication, identify heterogeneity and subpopulations, measure replication curves and replication rates at the single host cell level, and discern subtle differences in replication that are masked in a bulk population assay.
A new infection phenotype in epithelial cells for ΔsseG strains
Establishing an intracellular niche is essential for efficient Salmonella replication (Steele-Mortimer, 2008; Bakowski et al., 2008; Creasey and Isberg, 2014), and effector proteins are key to this process due to their ability to modify the nature of the SCV. Examination of phenotypes such as appearance, location, and dynamics of bacteria in infected cells has led to identification of important features of the vacuolar environment and the role of effector proteins in directing this niche (Salcedo and Holden, 2003; Brawn et al., 2007; Ramsden et al., 2007). Previous studies have revealed that deletion of sseG alters the directional movement of the SCV and leads to more dispersed, non-Golgi localized bacteria (Salcedo and Holden, 2003). For example, only 30% of SCVs associated with the Golgi marker Giantin at 8 hours post infection (Salcedo and Holden, 2003). By tracking bacteria over long periods of time (17 hours) in many cells (~ 100 cells per strain), three different localization phenotypes emerged for ΔsseG and ΔsteA/ΔsseG strains (Figure 4 and Supplementary Figure S5). In 50% of host cells infected with ΔsseG or ΔsteA/ΔsseG Salmonella, bacteria remained clustered together in a compact structure, similar to the SCV in wt and ΔsteA Salmonella infections. In the other 50% of infected cells, bacteria spread out across the cell between 3 and 12 hours post-infection, reminiscent of the dispersed phenotype first described by Salcedo and Holden (Salcedo and Holden, 2003). Surprisingly, for ΔsseG infections, in 20% of infected cells, the bacteria re-established a compact structure within the next 5 hours, while in 30% of cells Salmonella remained dispersed (Figure 4). The ΔsteA/ΔsseG strain showed a similar trend with 18% of cells exhibiting Salmonella dispersion followed by coalescence, and 32% of cells remaining dispersed (Supplementary Figure S5). Intriguingly, we found that all ΔsseG infections had similar replication rates, regardless of whether or not the bacteria were dispersed (Supplementary Figure S6), indicating that bacteria replicate equally as well as a microcolony or dispersed across the cell, despite a previous study suggesting that Salmonella fail to replicate when they are not near the Golgi (Salcedo and Holden, 2003). The dispersion of ΔsseG bacteria was complemented by the addition of SseG-GFP11 on a plasmid (4% of wt and ΔsseG pSseG-GFP11 Salmonella demonstrate a dispersion phenotype over the course of infection, Supplementary Figure S7). This coalescence phenotype would have been completely masked by a fixed time point assay, as live cell imaging was necessary to reveal that within an individual cell, bacteria could disperse and then re-cluster.
Figure 4.
Intracellular replication for wt versus mutant strains. Representative fluorescence images showing mCherry wt or the indicated mutant Salmonella strains at 3, 12, and 17 hrs post infection. Individual replication curves were generated using ICY to measure the area of bacterial pixels for each infection. Arrowheads on the replication curves denote the vacuole size at the time points shown in the image on the left. Percentages for ΔsseG infection phenotypes were calculated by visually inspecting every replication event.
As SseG and SseF are hypothesized to have similar roles in establishing the SCV (Kuhle and Hensel, 2002; Kuhle et al., 2004; Kuhle et al., 2006; Abrahams et al., 2006; Deiwick et al., 2006), we tested whether the dispersion/coalescence phenotype depended on both SseG and SseF. We found that in ΔsseG/ΔsseF infected cells, there was a significant increase in the % of Salmonella that dispersed (72% for ΔsseG/ΔsseF vs. 50% for ΔsseG alone) and a decrease in the fraction of infected cells in which dispersed Salmonella coalesced into a microcolony at later time points (7% vs. 20% for ΔsseG, Supplementary Figure S8). These results suggest that like SseG, SseF also contributes to maintenance of the SCV as a microcolony near the nucleus.
We next examined whether dispersed bacteria were replicating in a membrane-enclosed compartment. These bacteria clearly lacked the hyperreplication phenotype observed for cytosolic bacteria, but they also exhibited decreased colocalization with the fluid-phase Dextran marker used to define the SCV (44% and 12% of non-hyperreplicating infections colocalized with Dextran for ΔsseG and ΔsteA/ΔsseG, respectively, as opposed to > 70% for wt and ΔsteA). To test whether dispersed bacteria were enclosed in a membrane-bound compartment, we used the Split GFP system previously developed in our lab to visualize translocated effector proteins (Van Engelenburg and Palmer, 2010). Briefly, a fragment of GFP (GFP11) was genetically fused to an effector protein expressed in Salmonella, and the complementary fragment (GFP1-10) was transfected into the host cell. Upon translocation of the effector protein, the two fragments of GFP recombine and reconstitute fluorescence, illuminating the effector protein within the host cell. In this work, we tagged SteA, which upon translocation is membrane associated and localized to the SCV and membrane tubules, as previously described (Van Engelenburg and Palmer, 2010; Domingues et al., 2015). SteA colocalized with ΔsseG bacteria at all time points post infection, even when these bacteria were dispersed (Supplementary Figure S9), suggesting that ΔsseG bacteria are indeed enclosed within a membrane compartment, one that is marked by the effector protein SteA.
Comparison of wt and ΔsteA infections revealed a small difference in the appearance of the SCV (Figure 4). At 12 or more hours post-infection, the vacuole containing ΔsteA Salmonella appeared to contain fewer bacteria and was more compact, consistent with previously reported immunofluorescence results at 14 hours post infection (Domingues et al., 2014) and our replication data suggesting a mild replication defect at later stages of infection.
SteA plays a significant role in Salmonella survival and replication within macrophages
In the murine typhoid model of S. Typhimurium infection, Salmonella infect macrophages as well as epithelial cells (Haraga et al., 2008). This infection process has both acute and systemic phases of infection. The acute phase of infection is studied in susceptible murine strains (e.g. C57BL/6 or BALB/c) whereas systemic infection is studied in immunocompetent murine strains (e.g. SV129S6) (Vidal et al., 1996; Monack et al., 2004; Brown et al., 2013). Macrophages derived from immunocompetent mice express the metal transport protein NRAMP1 (Slc11a1) which confers resistance to pathogens that reside within vacuoles by restricting pathogen access to iron, an essential nutrient (Gruenheid et al., 1997; Cuellar-Mata et al., 2002; Nairz et al., 2009). Nramp1+/+ mice allow Salmonella to persist within macrophages in a chronic infection state for up to one year (M-F Roy and Malo, 2002; Monack et al., 2004). Although there are studies that explore the roles of effector proteins in bacterial replication in primary macrophages derived from susceptible mice (Figueira et al., 2013), to our knowledge, no study has addressed the roles of effector proteins in determining whether Salmonella survive, replicate, or are cleared in primary macrophages derived from immunocompetent mice at the single cell level. The balance of Salmonella replication, survival, or clearance within NRAMP1+/+ macrophages may be particularly important for establishing a systemic infection state. This part of the study focuses on the roles of two effector proteins in determining this balance within infected bone marrow derived macrophages (BMDMs) from immunocompetent SV129S6 mice.
Replication is most commonly assessed by quantifying the increase in bacterial load at distinct time points via a CFU assay. However, the net bacterial load reflects a combination of bacterial replication, survival, and clearance by the host, which may be differentially regulated by effector proteins. Therefore, we utilized long-term time lapse imaging to define infection parameters at the single cell level. BMDMs from SV129S6 mice were infected with opsonized mCherry-expressing Salmonella grown to stationary phase. Salmonella grown in this way exhibit less SPI-1 expression compared to Salmonella grown to late log phase (Kröger et al., 2013). We readily distinguished diverse phenotypes including bacterial replication, non-replicating bacteria, and clearance of bacteria by the host cells (Figure 5A). There was substantial variability for preparations of BMDMs from different mice, similar to what was observed for immunocompetent human macrophages from different donors (Lathrop et al., 2015). Nevertheless, because we tracked individual cells, we could exclude from the analysis macrophages that underwent cell death or were engulfed by other macrophages during the course of the experiment, and hence we were able to identify individual macrophages clearing bacteria.
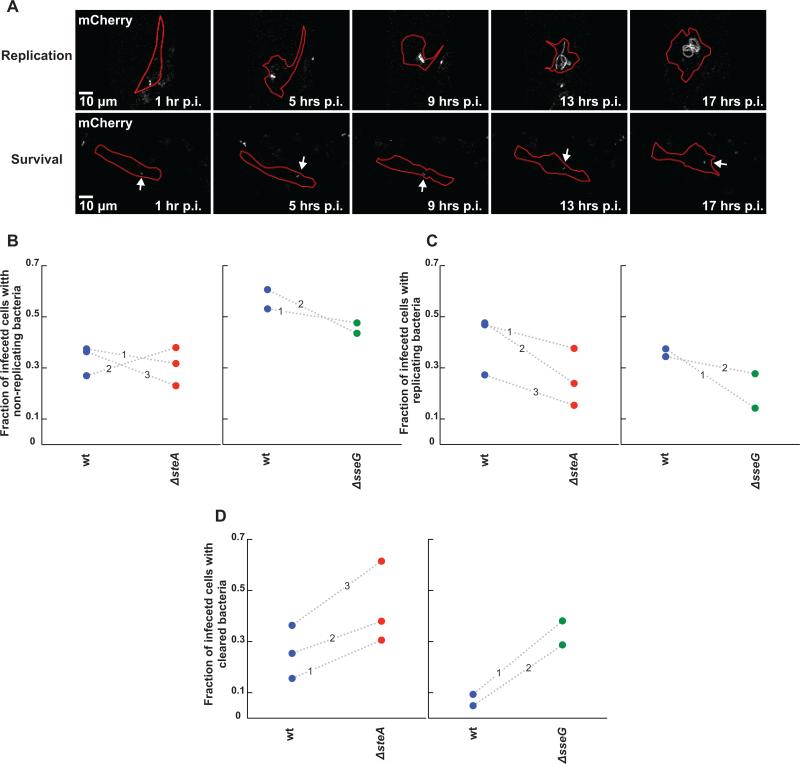
Figure 5.
Assay for monitoring the roles of SteA and SseG in Salmonella persistence, replication, or clearance in single primary bone marrow derived macrophages (BMDMs). (A) Representative fluorescence images of mCherry Salmonella replication (top panel) or survival (non-replicating state, bottom panel, labeled with an arrow) at distinct time points post infection. Salmonella is in gray and cell outlines are marked in red. (B-D) Comparison of ΔsteA (red circles) or ΔsseG (green circles) to wt Salmonella (blue circles) in altering the fraction of infected cells containing persisting (B), replicating (C), or cleared bacteria (D) between 1 and 16 hours post infection. Shown here is the fraction of cells exhibiting each phenotype for each strain. Each circle represents a fraction from a single independent experiment. Wt and mutant fractions from the same experiment are connected by a dotted line marked by the experiment number. (B) Deletion of either steA or sseG does not alter the fraction of cells containing persisting bacteria compared to wt Salmonella. (C) Deletion of both steA and sseG decrease the fraction of cells containing replicating bacteria. (D) ΔsteA or ΔsseG bacteria are more easily cleared by BMDMs compared to wt bacteria, though deletion of sseG causes a much stronger phenotype.
We carried out simultaneous imaging experiments in which the same preparation of BMDMs was infected by wt or deletion Salmonella strains in a 96-well plate to explore the role of effector proteins in shaping the infection. These conditions ensured that mutant strains could be directly compared to wt for each experiment (Supplementary Tables S1 and S2). Deletion of steA caused a 32% decrease in the percentage of cells that contained bacteria at one hour post infection, suggesting that SteA is important for helping to establish infection at early time points (Supplementary Table S1). To define the role of SteA in modifying the balance of replication, survival without replication, and clearance, we monitored infected cells over the next 16 hours. Deletion of steA did not alter the percentage of cells with non-replicating bacteria (4% decrease compared to wt Salmonella, Figure 5B, Supplementary Table S1), but caused a 38% decrease in the percentage of cells with replicating bacteria (Figure 5C, Supplementary Table S1). Finally, there was a significant increase (75%) in the ability of macrophage to clear ΔsteA bacteria compared to wt (Figure 5D, Supplementary Table S1). These results demonstrate that SteA plays a critical role in establishing infection in immunocompetent macrophages, but is not equally involved in promoting replication and survival in these cells.
We next compared the extent of perturbation of infection parameters between ΔsteA and ΔsseG strains, as deletion of sseG has been shown in multiple infection models to affect replication (Kuhle and Hensel, 2002; Salcedo and Holden, 2003; Figueira et al., 2013). Deletion of sseG was comparable to deletion of steA for interfering with replication (Figures 5B-C, Supplementary Table S2). However, the percentage of macrophages that cleared ΔsseG bacteria was much higher than ΔsteA (401% increase compared to wt, Figure 5D, Supplementary Table S2). Taken together, these results show that while SteA and SseG are both important for replication and prevention of clearance, SseG may play a more prominent role than SteA in the prevention of clearance.
Because analysis of replication curves allowed us to compare doubling times for wt and mutant Salmonella strains in epithelial cells, we applied a similar analysis for Salmonella strains within BMDMs. Salmonella replication curves in BMDMs had five phenotypes: steady growth, a lag phase followed by steady growth, an increase followed by a plateau, no growth, or growth followed by a decrease in bacterial area (Figure 6A). The wt strains were more likely to have curves that were considered steady growth, whereas ΔsteA strains had equal fractions of steady growth curves and lag phase curves. The ΔsseG strains had equal fractions of steady growth curves and no growth.
Figure 6.
Comparison of replication curves between wt and deletion Salmonella strains in bone marrow derived macrophages (BMDMs). All Salmonella categorized as “replicating” were analyzed using ICY to generate replication curves (bacterial area vs. time post infection in hours). (A) Examples of replication curve phenotypes (top) and the frequency for each strain (bottom). Curves were categorized by visual inspection, wt n= 116 cells, ΔsteA n= 50 cells, and ΔsseG n= 31 cells. (B) Representative “Steady growth” replication curves and their corresponding fits to a mathematical function representing bacterial growth (Y=A*2t/tau) for wt (blue), ΔsteA (red), and ΔsseG (green) strains. (C) Doubling times for infections categorized as “Steady growth” for ΔsteA (red) and ΔsseG (green) strains compared to wt Salmonella (blue). In bone marrow derived macrophages, wt and ΔsteA strains have similar doubling times while ΔsseG Salmonella has a much larger doubling time (wt n= 46 cells, ΔsteA n= 22 cells, and ΔsseG n= 5 cells). Each dot represents the doubling time for an infection within a single cell. The mean for each strain is marked with a gray line. All p values were calculated using a one-way ANOVA test (KaleidaGraph), ** denotes p < 0.002.
We next fit the curves designated as “steady growth” as described previously (Figure 6B). There was no difference between average doubling times for wild type and ΔsteA Salmonella (9 hours). Salmonella lacking sseG had a larger average doubling time (17 hours, Figure 6C), suggesting that when ΔsseG Salmonella have steady growth curves, they do not replicate at the same rate as wt and ΔsteA strains. These data agree with the replication defects seen with ΔsseG Salmonella in Raw264.7 cells (Kuhle and Hensel, 2002).
DISCUSSION
Infections are by their very nature dynamic, and this dynamism is often key to understanding cause and effect. Live cell imaging permits visualization and quantification of these dynamics and evolution of complex phenotypes over time. Moreover, given the heterogeneity in infection, live cell imaging can help discern whether a particular observation is deterministic of downstream events. In this study, we used long term imaging assays and subsequent data analysis methods, applied these methods to examine vacuolar replication and survival of wt and mutant Salmonella strains in both epithelial cells and immunocompetent BMDMs, and determined whether effector proteins play different roles in these model systems. Our results uncovered new features of infection and the roles of effector proteins in shaping these phenotypes. For example, in both epithelial cells and macrophages we quantified a variety of growth phenotypes that were differentially affected by deletion of effector proteins, observed for the first time that dispersed bacteria can coalesce back into an aggregate resembling a canonical vacuole, discovered that SseG increases the fraction of infections categorized by cytosolic hyperreplication, and observed that SteA is important for replication at late stages of infection. In immunocompetent macrophages, we determined that both SteA and SseG are important for promoting replication and resisting clearance during infection.
Monitoring bacterial replication in epithelial cells at the single cell level revealed a rich variety of replication phenotypes that were differentially impacted by effector proteins. The absence of SseG increased the percent of infections that exhibited a pronounced lag phase and increased the doubling time (Figure 2), indicating that the deletion of sseG causes a defect in the SCV development even at early time points. This observation builds on and extends previous studies (Salcedo and Holden, 2003) by providing more detailed and quantitative information on the diversity of growth phenotypes and how the deletion of sseG affects these parameters. On the other hand, deletion of steA had a more subtle effect, showing only a defect in fold-replication at late time points, suggesting that perhaps SteA is not required for establishing the SCV but rather plays a role in maintaining the SCV environment over time. Our findings lead us to speculate that perhaps loss of effector proteins that help set up the SCV environment lead to substantially delayed replication and an overall decrease in growth rate. These gross defects give rise to pronounced decrease in total bacterial load even at early time points. Such effector proteins are likely to be identified in single time point assays using fold replication. However, effector proteins that play a role at later stages maintaining the SCV environment can still impact the rate of replication, but loss of these effector proteins yields a subtle defect that is only manifested at late stages of infection and captured in single cell assays. This phenomenon could explain why SteA was not previously identified as influencing replication in epithelial cells (Buckner et al., 2011).
Tracking infections over a long time period enabled us to capture a new phenotype associated with deletion of the sseG gene. Previous studies have suggested that SseG plays a role in vacuole construction and placement within epithelial cells, either by physically tethering the SCV to the Golgi or by manipulating molecular motors (Salcedo and Holden, 2003; Kuhle et al., 2004; Kuhle et al., 2006; Deiwick et al., 2006; Ramsden et al., 2007). Consequently, loss of SseG results in dispersion of bacteria away from the Golgi (Salcedo and Holden, 2003). We observed dispersion of SCVs containing ΔsseG bacteria in ~ 50% of infections (Figure 4B). However, much to our surprise, in 20% of infections the dispersed SCV coalesced into what appeared to be a normal SCV in the perinuclear region. Further deletion of sseF, which has been shown to collaborate with SseG, resulted in exacerbation of these phenotypes: an increase the fraction of infected cells with dispersed bacteria and a decrease in the fraction of dispersed bacteria that coalesced. However, we still observed that 27% of infected cells exhibited microcolonies that did not disperse suggesting an additional factor, perhaps another effector protein that contributes to maintenance of compact microcolonies near the Golgi.
A final unexpected observation from our epithelial cell study was that the absence of SseG led to a substantial increase in the proportion of infections that exhibited vacuolar replication and a concomitant decrease in the number of infections in which bacteria hyperreplicated in the cytosol (Figure 1C). This result suggests that either SseG plays a role in promoting escape from the vacuole or that it facilitates survival and replication in the cytosol of epithelial cells. This finding was unanticipated as previous work using a GFP reporter system demonstrated a lack of expression of T3SS2 effector proteins (such as SseG) in cytosolic bacteria at 8 hours post invasion (Knodler et al., 2010). More recently, Knodler and colleagues showed that a ΔssaR mutant, defective for the translocation of T3SS2 effector proteins, did not give rise to a significant change in the percentage of cytosolic bacteria in Caco-2 C2Bbe1 cells, as determined by a chloroquine resistance CFU assay at 2, 4, and 7 hours post-invasion, suggesting that deletion of the T3SS2 translocon does not alter vacuolar escape (Knodler et al., 2014). One possible explanation to reconcile these observations is that T3SS2 effector proteins that are translocated early in infection, while bacteria are still contained within the SCV, may differentially influence vacuolar escape and/or cytosolic replication, such that deletion of the translocon yields no net effect, but deletion of individual effector proteins may reveal effector proteins that alter the balance between these two phenotypes. Indeed, using the Split GFP system to monitor SteA at early time points post infection (~5 hours post infection), we observed SteA expression even in cells containing hyperreplicating bacteria (Supplementary Figure S10), demonstrating that SPI-2 effector proteins can be detected in the cytosol of infected cells dominated by hyperreplication. As the infection progresses and hyperreplicating bacteria take over the cell, SteA signal decreases, consistent with the observation that SteA and other T3SS2 effector proteins are not expressed by cytosolic bacteria (Knodler et al., 2010). However these results raise the intriguing idea that T3SS2 effector proteins could influence the propensity for hyperreplication in the cytosol or replication within the SCV. It is worth noting that the percentage of cytosolic hyperreplicating wt bacteria that we observed (40%) was higher than that observed by Knodler and coworkers (~10%). While the origin of this difference is unclear, there were methodological differences between the two studies. Both studies used the SL1344 strain but these bacteria harbored different plasmids and it has been shown that different plasmids can affect Salmonella invasion and replication ability (Knodler et al., 2005; Clark et al., 2009). In addition, our bacteria were grown under conditions that induce SPI-1 using different methods than those of Knodler and coworkers, and different culturing conditions can affect the “invasiveness” of the bacteria (Ibarra et al., 2010).
In mouse models of acute and systemic infection a ΔsteA mutant shows decreased virulence and persistence, respectively (Geddes et al., 2005; Lawley et al., 2006). Yet the mechanism by which SteA promotes virulence remains elusive, and studies have yielded confounding results with respect to the cellular role of SteA. For example, ΔsteA Salmonella did not show a replication defect in a model macrophage cell line RAW 264.7 or BMDMs from CL57BL/6 mice, both of which lack a functional NRAMP1 (Figueira et al., 2013; Domingues et al., 2014), and although we find a replication defect in epithelial cells at late stages of infection, the effect is subtle and hard to reconcile with the documented virulence defect for ΔsteA in animal models. Therefore we examined the role of SteA in promoting survival in a model of infection involving BMDMs from SV129S6 mice, which possess an intact NRAMP1 (M-F Roy and Malo, 2002). Our results reveal that SteA plays a significant role in establishing infection, promoting replication, and preventing clearance. These results contrast with previous findings that SteA was not important for replication in susceptible macrophages (Figueira et al., 2013), supporting the idea that SteA may be more important during the systemic phase of infection (Lawley et al., 2006). Both wt and ΔsteA infections have a similar percentage of cells containing non-replicating bacteria (~30%), consistent with observations in immunocompetent macrophages (Helaine et al., 2010). Although ΔsteA Salmonella were less likely to replicate in BMDMs, it is important to note that when ΔsteA Salmonella did replicate, they did so with the same replication rate as wt. Thus SteA may be more important for the perpetuation of Salmonella infection throughout the host than playing a direct role in replication. Regardless, these results are particularly intriguing in light of the subtle defect in replication caused by ΔsteA in infected epithelial cells (Figure 3), and lack of effect in susceptible macrophages (Figueira and Holden, 2012), and suggest that effector proteins may play different roles in infection depending on cell type, and in different infection models (acute versus systemic infection).
Although long term imaging of single infected cells provides in depth information about the roles effector proteins play during Salmonella replication or survival in infected cells, these experiments are challenging to design and perform. The ability to determine subtle differences between replication or survival phenotypes is directly related to the size of the data set (Krzywinski and Altman, 2013). Image analysis programs such as ICY allow for the development of pipeline protocols that yield quantitative information about each infection and allow users to conduct analysis on entire datasets at least semi-automatically.
EXPERIMENTAL PROCEDURES
Salmonella strains
All red Salmonella strains were Salmonella enterica serovar Typhimurium SL1344 (B P Smith et al., 1984) constitutively expressing mCherry or mRuby3 from a plasmid under the rpsM promoter (pAmCh or pAmR plasmid, respectively; parent pACYC177). ΔsteA and ΔsseG mCherry Salmonella strains were made as described previously (Datsenko and Wanner, 2000; Uzzau et al., 2001; Van Engelenburg and Palmer, 2010). Briefly, the steA or sseG gene was replaced with a kanamycin resistance gene in the 14028S Salmonella strain (Jarvik et al., 2010) by recombination. This knockout was transferred to wild type SL1344 strains using P22 phage derived from the resistance free strain SL1290. To make the ΔsteA/ΔsseG strain, the endogenous sseG and steA genes were replaced with a kanamycin and chloramphenicol resistance gene, respectively. For the ΔsseF/ΔsseG strain, both sseF and sseG were replaced with a single kanamycin resistance gene. To complement the deletion of steA or sseG genes, we created pSteA-GFP11 and the pSseG-GFP11 plasmids using pAmCh and the pAmR plasmids, respectively. These new plasmids express SteA-GFP11 and SseG-GFP11 under the SteA promoter. We transformed ΔsteA and ΔsseG Salmonella with pSteA-GFP11 and pSseG-GFP11 plasmids to make the SteA-GFP11 mCherry and SseG-GFP11 mRuby3 complement strains.
For infection of HeLa cells, Salmonella strains were grown in SPI-1 media (LB (EMD) supplemented with 300 mM NaCl (Fisher Scientific) and 25 mM MOPS (Sigma) at pH 7.6) with appropriate antibiotics (wt mCherry SL1344 strain: 50 μg/mL ampicillin and 50 μg/mL streptomycin; ΔsteA, ΔsseG, and ΔsseF/ΔsseG mCherry strains, SteA-GFP11 mCherry, SseG-GFP11 mRuby3, or SteA-GFP11 mTagBFP2 strains: 50 μg/mL ampicillin, 50 μg/mL streptomycin, and 50 μg/mL kanamycin; and ΔsteA/ΔsseG mCherry or ΔsseG SteA-GFP11 mTagBFP2 strains: 50 μg/mL ampicillin, 50 μg/mL streptomycin, 50 μg/mL kanamycin, and 10 μg/mL chloramphenicol) overnight at 37°C without aeration. Prior to infection, bacteria were diluted 1:33 in 3 mL SPI-1 media with appropriate antibiotics for 4 hours at 37°C without aeration. For infection of primary Bone Marrow Derived Macrophages (BMDMs), bacteria were grown to stationary phase in LB with appropriate antibiotics at 37°C with aeration. Prior to infection of BMDMs, bacteria were opsonized in a 1:1 solution of mouse serum (Sigma) and cell culture media (Gibco) for 20 minutes at room temperature.
Cell Culture and Salmonella infection
HeLa cells were maintained in Dulbecco's modified Eagle medium (DMEM, Gibco) supplemented with 10% FBS (Gibco), 100 Units/mL penicillin G sodium (Gibco), and 100 μg/mL streptomycin sulfate (Gibco) at 37°C with 5% CO2. For infections, HeLa cells were plated in 35 mm glass-bottom dishes. Primary BMDMs were isolated as previously described (Silva-Herzog and Detweiler, 2010). Briefly, marrow was flushed from the femurs, tibias, and humeri of 2- to 4-month-old SV129S6 mice (Taconic Laboratories, Hudson, NY). Cells were resuspended in DMEM (Sigma-Aldrich, St. Louis, MO) supplemented with fetal bovine serum (10%), L-glutamine (2 mM), sodium pyruvate (1 mM), beta-mercaptoethanol (50 μM), HEPES (10 mM), and penicillin-streptomycin (50 IU/ml penicillin and 50 μg/ml streptomycin). Cells were overlaid onto an equal volume of Histopaque-1083 (Sigma-Aldrich, St. Louis, MO) and centrifuged at 500 × g for 15 min. Monocytes at the interface were harvested and incubated for 6 to 7 days at 37°C in 5% CO2 in supplemented DMEM that also contained 30% MCSF (macrophage colony stimulating factor) obtained from NIH/3T3 cells (acquired from Jeffery Cox, University of California, San Francisco) to promote monocyte differentiation into macrophages. For infections, BMDMs were plated in 96 well glass bottom plates coated with poly-L-lysine (Sigma).
Immediately prior to infection, AlexaFluor 488 labeled Dextran (10,000 MW, Molecular Probes) was added to HeLa cells or BMDMs to a final concentration of 200 μg/mL in order to label the Salmonella containing vacuole. HeLa cells were infected at a multiplicity of infection (MOI) of 30 while BMDMs were infected at an MOI of 10. HeLa infections were allowed to proceed for 30 minutes at 37°C and 5% CO2 before the Salmonella-containing media was exchanged with phenol red free DMEM containing 10% FBS and 100 μg/mL gentamicin to kill any extracellular bacteria. After incubating for 45 minutes at 37°C and 5% CO2, the media was replaced with phenol red free DMEM containing 10% FBS and 10 μg/mL gentamicin for the remainder of the experiment. BMDM infections were allowed to proceed for 30 minutes at 37°C and 5% CO2 before the Salmonella-containing media was exchanged with phenol red free DMEM containing 10% FBS and 10 μg/mL gentamicin for the remainder of the experiment.
Imaging of infected cells
All infections were performed in duplicate or triplicate and imaged on either a Nikon Ti-E wide-field or a Nikon A1R laser scanning confocal microscope. Both microscopes were equipped with the Nikon Elements software platform, Ti-E Perfect Focus system, a motorized XY stage with a Ti Z drive, and an environmental chamber (Pathology Devices) to maintain cells at 37°C, 5% CO2 and 70% humidity. The motorized XY stage enabled us to select and store the locations of multiple fields of view in order to follow the fates of many infected cells over the course of the experiment. The Z drive was used to create z stacks that encompassed the entirety of the cells within each field of view, thus ensuring the complete detection of any bacteria present. Images for all fields of view and Z planes were collected every 15 minutes from 1-2 hours to 17 hours post infection.
Images acquired on the wide-field microscope used a 60x oil objective (NA 1.40), an iXon3 897 EMCCD camera (Andor), and a xenon-arc lamp to image mCherry Salmonella (excitation: 560/40 nm, emission: 630/75 nm, dichroic: 585 nm) and AlexaFluor 488 labeled Dextran (excitation: 472/30 nm, emission: 520/40 nm, dichroic: 490 nm). Confocal imaging experiments used a 40x oil objective (NA 1.30) and the following channels: red (561 nm laser line, PMT gain: 110, emission filter: 600/50 nm), green (488 nm laser line, PMT gain: 90, emission filter: 525/50 nm), and bright field DIC. Imaging was performed with the channel series function turned on (i.e. red and green channels were not acquired simultaneously) to prevent bleedthrough between fluorescence channels. All fields of view were imaged with a pixel dwell time of 2 μs.
ICY Analysis of Salmonella replication in HeLa cells
To streamline image analysis, a semi-automated protocol for quantifying the change in fluorescence pixel area was developed for ICY (de Chaumont et al., 2012). First, each field of view was split into an individual file and z stacks were flattened using the Extended Depth of Field algorithm within the Nikon Elements software platform. Each channel was exported as a series of Tiff files for further analysis. Fiji (http://fiji.sc/Fiji) was used to generate a separate Tiff file for each infected cell containing bacteria colocalized with Dextran (indicating bacteria within a vacuole). The mCherry bacterial channel was imported into ICY and the Thresholder plugin was used to remove noise. Pixels corresponding to fluorescent bacteria were detected automatically using the Connected Components plugin, and a script was written to count the number of pixels for each time point. Data were exported to Excel and used to generate a bacterial replication curve for each infected cell, where the total pixel area was used to represent the amount of bacteria. In order to analyze bacterial replication, individual replication curves were generated for all infections imaged by confocal microscopy. Visual inspection of the replication curves led to the following categories: steady growth; lag phase: a curve containing a lag phase followed by steady growth; plateau phase: a curve containing a substantial stationary phase after growth; no growth: a curve that is flat or has a decrease in area; and complex: a curve with both increases and decreases in bacterial pixels.
For fitting replication curves, a MATLAB script was developed to automatically take the excel file generated as output from ICY, extract pixels in the bacterial channel for each infected cell at each time point, fit these curves to a mathematical function representing bacterial growth (Y = A*bt/tau, where A represents the amount of bacteria at t = 0, b = 2 due to doubling each generation, and tau is the time constant that represents the doubling time). Our script was designed to only accept fits with R2 > 0.7 and a Durbin-Watson score between 1 and 2. For the wt replication curves, 29% met the fit criterion. For mutant strains, 35% of ΔsteA replication curves, 42% of ΔsseG replication curves, and 26% of ΔsteA/ΔsseG replication curves fit this criterion. For ΔsseG and ΔsteA/ΔsseG fewer curves met the fit criterion because these mutants had increased numbers of replication curves with pronounced lag phases, stationary phases, or no growth. For those curves that were characterized by steady growth, the time constant (tau) was extracted from the fit and compared across strains. Statistical significance was measured using a one-way ANOVA test in KaleidaGraph (Synergy Software).
Analysis of Salmonella replication and survival in BMDMs
For each field of view, z stacks were flattened using the Maximum z Projection algorithm in Fiji and fluorescence channels corresponding to mCherry bacteria, green Dextran, and bright field were separated in a series of Tiff files for further analysis. Because we wanted to track bacterial fluorescence intensity (i.e. absence or presence of a bacterial signal), these channels were corrected for noise caused by the PMTs by subtracting a dark image from each channel. Dark images were created by averaging 16 different images acquired for all channels with the lasers turned off. These corrected images were imported into ICY where background was removed from the fluorescence channels using the Thresholder plugin. The channels were merged into one image and each cell was labeled according to its fate over the course of the experiment (infected, uninfected, dead, etc). For infected cells, bacteria were tracked manually and recorded as replicating (increase in the number of rod shaped bacteria), surviving (fluorescence intensity remains constant over the course of the experiment), or cleared (fluorescence intensity disappears).
Analysis of Salmonella replication rates in BMDMs
Bacteria manually recorded as “replicating” were analyzed using the ICY assay discussed above. For fitting replication curves, we used the same MATLAB script as discussed above. For the wt replication curves, 39% met the fit criterion. For mutant strains, 43% of ΔsteA replication curves, and 16% of ΔsseG replication curves. For the ΔsseG strain, fewer curves met the fit criterion because this mutant had increased numbers of replication curves with no growth. Statistical significance was measured using a one-way ANOVA test in KaleidaGraph (Synergy Software).
Colony Forming Unit (CFU) assays
HeLa cells were seeded into 12 well cell culture plates maintained in antibiotic free DMEM supplemented with 10% FBS and were held at 37°C with 5% CO2 throughout the infection process. Infections with Salmonella strains were carried out as described above. At 3 and 17 hours post infection, infected cells were rinsed twice with PBS and incubated with 0.1% Triton in PBS at room temperature for 5 min. A series of dilutions in PBS were generated and plated in quadruplicate on appropriate antibiotic containing LB-Agar plates. The CFU calculated for each infection were normalized to the wt strain. Statistical significance was measured using a one-way ANOVA test.
Supplementary Material
ACKNOWLEDGEMENTS
We would like to acknowledge the following sources for financial support: University of Colorado Signaling and Cell Cycle Regulation Training Grant (T32 GM08759 to SEM and EB), NIH Predoctoral Fellowship (F31 GM106644 to SEM), NSF Career Award (MCB-0950411 to AEP), Public Health Service grant (R01 AI-095395 to CSD) and a Butcher Foundation Seed Grant Award (to CSD and AEP). The laser scanning confocal work was performed in the BioFrontiers Institute Advanced Light Microscopy Core. The Nikon A1R microscope was acquired through the generous support of NIST-CU Cooperative Agreement award number 70NANB15H226.
Footnotes
The authors declare no conflict of interest.
REFERENCES
- Abrahams GL, Hensel M. Manipulating cellular transport and immune responses: dynamic interactions between intracellular Salmonella enterica and its host cells. Cell Microbiol. 2006;8:728–737. doi: 10.1111/j.1462-5822.2006.00706.x. [DOI] [PubMed] [Google Scholar]
- Abrahams GL, Müller P, Hensel M. Functional dissection of SseF, a type III effector protein involved in positioning the salmonella-containing vacuole. Traffic. 2006;7:950–965. doi: 10.1111/j.1600-0854.2006.00454.x. [DOI] [PubMed] [Google Scholar]
- Agbor TA, McCormick BA. Salmonella effectors: important players modulating host cell function during infection. Cell Microbiol. 2011;13:1858–1869. doi: 10.1111/j.1462-5822.2011.01701.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Avery SV. Microbial cell individuality and the underlying sources of heterogeneity. Nat Rev Micro. 2006;4:577–587. doi: 10.1038/nrmicro1460. [DOI] [PubMed] [Google Scholar]
- Avraham R, Haseley N, Brown D, Penaranda C, Jijon HB, Trombetta JJ, et al. Pathogen Cell-to-Cell Variability Drives Heterogeneity in Host Immune Responses. Cell. 2015;162:1309–1321. doi: 10.1016/j.cell.2015.08.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bakowski MA, Braun V, Brumell JH. Salmonella-containing vacuoles: directing traffic and nesting to grow. Traffic. 2008;9:2022–2031. doi: 10.1111/j.1600-0854.2008.00827.x. [DOI] [PubMed] [Google Scholar]
- Baty F, Flandrois JP, Delignette-Muller ML. Modeling the Lag Time of Listeria monocytogenes from Viable Count Enumeration and Optical Density Data. Appl Environ Microbiol. 2002;68:5816–5825. doi: 10.1128/AEM.68.12.5816-5825.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Booth IR. Stress and the single cell: intrapopulation diversity is a mechanism to ensure survival upon exposure to stress. Int J Food Microbiol. 2002;78:19–30. doi: 10.1016/s0168-1605(02)00239-8. [DOI] [PubMed] [Google Scholar]
- Brawn LC, Hayward RD, Koronakis V. Salmonella SPI1 effector SipA persists after entry and cooperates with a SPI2 effector to regulate phagosome maturation and intracellular replication. Cell Host Microbe. 2007;1:63–75. doi: 10.1016/j.chom.2007.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown DE, Libby SJ, Moreland SM, McCoy MW, Brabb T, Stepanek A, et al. Salmonella enterica causes more severe inflammatory disease in C57/BL6 Nramp1G169 mice than Sv129S6 mice. Vet Pathol. 2013;50:867–876. doi: 10.1177/0300985813478213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buckner MMC, Croxen MA, Arena ET, Finlay BB. A comprehensive study of the contribution of Salmonella enterica serovar Typhimurium SPI2 effectors to bacterial colonization, survival, and replication in typhoid fever, macrophage, and epithelial cell infection models. virulence. 2011;2:208–216. doi: 10.4161/viru.2.3.15894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clark L, Martinez-Argudo I, Humphrey TJ, Jepson MA. GFP plasmid-induced defects in Salmonella invasion depend on plasmid architecture, not protein expression. Microbiology (Reading, Engl) 2009;155:461–467. doi: 10.1099/mic.0.025700-0. [DOI] [PubMed] [Google Scholar]
- Claudi B, Spröte P, Chirkova A, Personnic N, Zankl J, Schürmann N, et al. Phenotypic variation of Salmonella in host tissues delays eradication by antimicrobial chemotherapy. Cell. 2014;158:722–733. doi: 10.1016/j.cell.2014.06.045. [DOI] [PubMed] [Google Scholar]
- Creasey EA, Isberg RR. Maintenance of vacuole integrity by bacterial pathogens. Curr Opin Microbiol. 2014;17:46–52. doi: 10.1016/j.mib.2013.11.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cuellar-Mata P, Jabado N, Liu J, Furuya W, Finlay BB, Gros P, Grinstein S. Nramp1 modifies the fusion of Salmonella typhimurium-containing vacuoles with cellular endomembranes in macrophages. J Biol Chem. 2002;277:2258–2265. doi: 10.1074/jbc.M105508200. [DOI] [PubMed] [Google Scholar]
- Datsenko KA, Wanner BL. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc Natl Acad Sci USA. 2000;97:6640–6645. doi: 10.1073/pnas.120163297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Chaumont F, Dallongeville S, Chenouard N, Hervé N, Pop S, Provoost T, et al. Icy: an open bioimage informatics platform for extended reproducible research. Nat Meth. 2012;9:690–696. doi: 10.1038/nmeth.2075. [DOI] [PubMed] [Google Scholar]
- Deiwick J, Salcedo SP, Boucrot E, Gilliland SM, Henry T, Petermann N, et al. The translocated Salmonella effector proteins SseF and SseG interact and are required to establish an intracellular replication niche. Infect Immun. 2006;74:6965–6972. doi: 10.1128/IAI.00648-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Domingues L, Holden DW, Mota LJ. The Salmonella Effector SteA Contributes to the Control of Membrane Dynamics of Salmonella-Containing Vacuoles. Infect Immun. 2014;82:2923–2934. doi: 10.1128/IAI.01385-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Domingues L, Ismail A, Charro N, Rodríguez-Escudero I, Holden DW, Molina M, et al. The Salmonella effector SteA binds phosphatidylinositol 4-phosphate for subcellular targeting within host cells. Cell Microbiol. 2015 doi: 10.1111/cmi.12558. [DOI] [PubMed] [Google Scholar]
- Elfwing A, LeMarc Y, Baranyi J, Ballagi A. Observing growth and division of large numbers of individual bacteria by image analysis. Appl Environ Microbiol. 2004;70:675–678. doi: 10.1128/AEM.70.2.675-678.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Figueira R, Holden DW. Functions of the Salmonella pathogenicity island 2 (SPI- 2) type III secretion system effectors. Microbiology. 2012;158:1147–1161. doi: 10.1099/mic.0.058115-0. [DOI] [PubMed] [Google Scholar]
- Figueira R, Watson KG, Holden DW, Helaine S. Identification of salmonella pathogenicity island-2 type III secretion system effectors involved in intramacrophage replication of S. enterica serovar typhimurium: implications for rational vaccine design. mBio. 2013;4:e00065. doi: 10.1128/mBio.00065-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geddes K, Worley M, Niemann G, Heffron F. Identification of new secreted effectors in Salmonella enterica serovar Typhimurium. Infect Immun. 2005;73:6260–6271. doi: 10.1128/IAI.73.10.6260-6271.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gruenheid S, Pinner E, Desjardins M, Gros P. Natural resistance to infection with intracellular pathogens: the Nramp1 protein is recruited to the membrane of the phagosome. J Exp Med. 1997;185:717–730. doi: 10.1084/jem.185.4.717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haraga A, Ohlson MB, Miller SI. Salmonellae interplay with host cells. Nat Rev Micro. 2008;6:53–66. doi: 10.1038/nrmicro1788. [DOI] [PubMed] [Google Scholar]
- Helaine S, Thompson JA, Watson KG, Liu M, Boyle C, Holden DW. Dynamics of intracellular bacterial replication at the single cell level. Proc Natl Acad Sci USA. 2010;107:3746–3751. doi: 10.1073/pnas.1000041107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ibarra JA, Knodler LA, Sturdevant DE, Virtaneva K, Carmody AB, Fischer ER, et al. Induction of Salmonella pathogenicity island 1 under different growth conditions can affect Salmonella-host cell interactions in vitro. Microbiology. 2010;156:1120–1133. doi: 10.1099/mic.0.032896-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jarvik T, Smillie C, Groisman EA, Ochman H. Short-term signatures of evolutionary change in the Salmonella enterica serovar typhimurium 14028 genome. Journal of Bacteriology. 2010;192:560–567. doi: 10.1128/JB.01233-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones MA, Wood MW, Mullan PB, Watson PR, Wallis TS, Galyov EE. Secreted effector proteins of Salmonella dublin act in concert to induce enteritis. Infect Immun. 1998;66:5799–5804. doi: 10.1128/iai.66.12.5799-5804.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelly CD, Rahn O. The Growth Rate of Individual Bacterial Cells. Journal of Bacteriology. 1932;23:147–153. doi: 10.1128/jb.23.2.147-153.1932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knodler LA. Salmonella enterica: living a double life in epithelial cells. Curr Opin Microbiol. 2015;23:23–31. doi: 10.1016/j.mib.2014.10.010. [DOI] [PubMed] [Google Scholar]
- Knodler LA, Bestor A, Ma C, Hansen-Wester I, Hensel M, Vallance BA, Steele- Mortimer O. Cloning vectors and fluorescent proteins can significantly inhibit Salmonella enterica virulence in both epithelial cells and macrophages: implications for bacterial pathogenesis studies. Infect Immun. 2005;73:7027–7031. doi: 10.1128/IAI.73.10.7027-7031.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knodler LA, Nair V, Steele-Mortimer O. Quantitative assessment of cytosolic Salmonella in epithelial cells. PLoS ONE. 2014;9:e84681. doi: 10.1371/journal.pone.0084681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knodler LA, Vallance BA, Celli J, Winfree S, Hansen B, Montero M, Steele- Mortimer O. Dissemination of invasive Salmonella via bacterial-induced extrusion of mucosal epithelia. Proceedings of the National Academy of Sciences. 2010;107:17733–17738. doi: 10.1073/pnas.1006098107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kröger C, Colgan A, Srikumar S, Händler K, Sivasankaran SK, Hammarlöf DL, et al. An infection-relevant transcriptomic compendium for Salmonella enterica Serovar Typhimurium. Cell Host Microbe. 2013;14:683–695. doi: 10.1016/j.chom.2013.11.010. [DOI] [PubMed] [Google Scholar]
- Krzywinski M, Altman N. Points of significance: Power and sample size. Nat Meth. 2013;10:1139–1140. [Google Scholar]
- Kuhle V, Abrahams GL, Hensel M. Intracellular Salmonella enterica redirect exocytic transport processes in a Salmonella pathogenicity island 2-dependent manner. Traffic. 2006;7:716–730. doi: 10.1111/j.1600-0854.2006.00422.x. [DOI] [PubMed] [Google Scholar]
- Kuhle V, Hensel M. SseF and SseG are translocated effectors of the type III secretion system of Salmonella pathogenicity island 2 that modulate aggregation of endosomal compartments. Cell Microbiol. 2002;4:813–824. doi: 10.1046/j.1462-5822.2002.00234.x. [DOI] [PubMed] [Google Scholar]
- Kuhle V, Jäckel D, Hensel M. Effector proteins encoded by Salmonella pathogenicity island 2 interfere with the microtubule cytoskeleton after translocation into host cells. Traffic. 2004;5:356–370. doi: 10.1111/j.1398-9219.2004.00179.x. [DOI] [PubMed] [Google Scholar]
- Lathrop SK, Binder KA, Starr T, Cooper KG, Chong A, Carmody AB, Steele- Mortimer O. Replication of Salmonella Typhimurium in Human Monocyte-Derived Macrophages. Infect Immun. 2015 doi: 10.1128/IAI.00033-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lawley TD, Chan K, Thompson LJ, Kim CC, Govoni GR, Monack DM. Genome-wide screen for Salmonella genes required for long-term systemic infection of the mouse. PLoS Pathog. 2006;2:e11. doi: 10.1371/journal.ppat.0020011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lidstrom ME, Konopka MC. The role of physiological heterogeneity in microbial population behavior. Nat Chem Biol. 2010;6:705–712. doi: 10.1038/nchembio.436. [DOI] [PubMed] [Google Scholar]
- Majowicz SE, Musto J, Scallan E, Angulo FJ, Kirk M, O'Brien SJ, et al. The global burden of nontyphoidal Salmonella gastroenteritis. Clin Infect Dis. 2010;50:882–889. doi: 10.1086/650733. [DOI] [PubMed] [Google Scholar]
- Malik-Kale P, Winfree S, Steele-Mortimer O. The Bimodal Lifestyle of Intracellular Salmonella in Epithelial Cells: Replication in the Cytosol Obscures Defects in Vacuolar Replication. PLoS ONE. 2012;7:e38732. doi: 10.1371/journal.pone.0038732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marlovits TC, Stebbins CE. Type III secretion systems shape up as they ship out. Curr Opin Microbiol. 2010;13:47–52. doi: 10.1016/j.mib.2009.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGhie EJ, Brawn LC, Hume PJ, Humphreys D, Koronakis V. Salmonella takes control: effector-driven manipulation of the host. Curr Opin Microbiol. 2009;12:117–124. doi: 10.1016/j.mib.2008.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGhie EJ, Hayward RD, Koronakis V. Cooperation between actin-binding proteins of invasive Salmonella: SipA potentiates SipC nucleation and bundling of actin. EMBO J. 2001;20:2131–2139. doi: 10.1093/emboj/20.9.2131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moest TP, Méresse S. Salmonella T3SSs: successful mission of the secret(ion) agents. Curr Opin Microbiol. 2013;16:38–44. doi: 10.1016/j.mib.2012.11.006. [DOI] [PubMed] [Google Scholar]
- Monack DM, Bouley DM, Falkow S. Salmonella typhimurium persists within macrophages in the mesenteric lymph nodes of chronically infected Nramp1+/+ mice and can be reactivated by IFNgamma neutralization. J Exp Med. 2004;199:231–241. doi: 10.1084/jem.20031319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moraes TF, Spreter T, Strynadka NC. Piecing together the type III injectisome of bacterial pathogens. Curr Opin Struct Biol. 2008;18:258–266. doi: 10.1016/j.sbi.2007.12.011. [DOI] [PubMed] [Google Scholar]
- Nairz M, Fritsche G, Crouch M-LV, Barton HC, Fang FC, Weiss G. Slc11a1 limits intracellular growth of Salmonella enterica sv. Typhimurium by promoting macrophage immune effector functions and impairing bacterial iron acquisition. Cell Microbiol. 2009;11:1365–1381. doi: 10.1111/j.1462-5822.2009.01337.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perkins TJ, Swain PS. Strategies for cellular decision-making. Molecular Systems Biology. 2009;5:326. doi: 10.1038/msb.2009.83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rajashekar R, Liebl D, Chikkaballi D, Liss V, Hensel M. Live Cell Imaging Reveals Novel Functions of Salmonella enterica SPI2-T3SS Effector Proteins in Remodeling of the Host Cell Endosomal System. PLoS ONE. 2014 doi: 10.1371/journal.pone.0115423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rajashekar R, Liebl D, Seitz A, Hensel M. Dynamic remodeling of the endosomal system during formation of Salmonella-induced filaments by intracellular Salmonella enterica. Traffic. 2008;9:2100–2116. doi: 10.1111/j.1600-0854.2008.00821.x. [DOI] [PubMed] [Google Scholar]
- Ramos-Morales F. Impact of Salmonella enterica Type III Secretion System Effectors on the Eukaryotic Host Cell. ISRN Cell Biology. 2012;2012:1–36. [Google Scholar]
- Ramsden AE, Mota LJ, Münter S, Shorte SL, Holden DW. The SPI-2 type III secretion system restricts motility of Salmonella-containing vacuoles. Cell Microbiol. 2007;9:2517–2529. doi: 10.1111/j.1462-5822.2007.00977.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roy M-F, Malo D. Genetic regulation of host responses to Salmonella infection in mice. Genes Immun. 2002;3:381–393. doi: 10.1038/sj.gene.6363924. [DOI] [PubMed] [Google Scholar]
- Salcedo SP, Holden DW. SseG, a virulence protein that targets Salmonella to the Golgi network. EMBO J. 2003;22:5003–5014. doi: 10.1093/emboj/cdg517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schlumberger MC, Hardt W-D. Salmonella type III secretion effectors: pulling the host cell's strings. Curr Opin Microbiol. 2006;9:46–54. doi: 10.1016/j.mib.2005.12.006. [DOI] [PubMed] [Google Scholar]
- Sherwood RK, Roy CR. A Rab-centric perspective of bacterial pathogen-occupied vacuoles. Cell Host Microbe. 2013;14:256–268. doi: 10.1016/j.chom.2013.08.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silva-Herzog E, Detweiler CS. Salmonella enterica replication in hemophagocytic macrophages requires two type three secretion systems. Infect Immun. 2010;78:3369–3377. doi: 10.1128/IAI.00292-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith AC, Heo WD, Braun V, Jiang X, Macrae C, Casanova JE, et al. A network of Rab GTPases controls phagosome maturation and is modulated by Salmonella enterica serovar Typhimurium. J Cell Biol. 2007;176:263–268. doi: 10.1083/jcb.200611056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith BP, Reina-Guerra M, Hoiseth SK, Stocker BA, Habasha F, Johnson E, Merritt F. Aromatic-dependent Salmonella typhimurium as modified live vaccines for calves. Am J Vet Res. 1984;45:59–66. [PubMed] [Google Scholar]
- Spudich JL, Koshland DE. Non-genetic individuality: chance in the single cell. Nature. 1976;262:467–471. doi: 10.1038/262467a0. [DOI] [PubMed] [Google Scholar]
- Srikanth CV, Mercado-Lubo R, Hallstrom K, McCormick BA. Salmonella effector proteins and host-cell responses. Cell Mol Life Sci. 2011;68:3687–3697. doi: 10.1007/s00018-011-0841-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steele-Mortimer O. The Salmonella-containing vacuole: moving with the times. Curr Opin Microbiol. 2008;11:38–45. doi: 10.1016/j.mib.2008.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uzzau S, Figueroa-Bossi N, Rubino S, Bossi L. Epitope tagging of chromosomal genes in Salmonella. Proc Natl Acad Sci USA. 2001;98:15264–15269. doi: 10.1073/pnas.261348198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Engelenburg SB, Palmer AE. Imaging type-III secretion reveals dynamics and spatial segregation of Salmonella effectors. Nat Meth. 2010;7:325–330. doi: 10.1038/nmeth.1437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vidal SM, Pinner E, Lepage P, Gauthier S, Gros P. Natural resistance to intracellular infections: Nramp1 encodes a membrane phosphoglycoprotein absent in macrophages from susceptible (Nramp1 D169) mouse strains. J Immunol. 1996;157:3559–3568. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.