Abstract
The 37/67-kDa laminin receptor (LAMR/RPSA) was originally identified as a 67-kDa binding protein for laminin, an extracellular matrix glycoprotein that provides cellular adhesion to the basement membrane. LAMR has evolutionary origins, however, as a 37-kDa RPS2 family ribosomal component. Expressed in all domains of life, RPS2 proteins have been shown to have remarkably diverse physiological roles that vary across species. Contributing to laminin binding, ribosome biogenesis, cytoskeletal organization, and nuclear functions, this protein governs critical cellular processes including growth, survival, migration, protein synthesis, development, and differentiation. Unsurprisingly given its purview, LAMR has been associated with metastatic cancer, neurodegenerative disease and developmental abnormalities. Functioning in a receptor capacity, this protein also confers susceptibility to bacterial and viral infection. LAMR is clearly a molecule of consequence in human disease, directly mediating pathological events that make it a prime target for therapeutic interventions. Despite decades of research, there are still a large number of open questions regarding the cellular biology of LAMR, the nature of its ability to bind laminin, the function of its intrinsically disordered C-terminal region and its conversion from 37 to 67 kDa. This review attempts to convey an in-depth description of the complexity surrounding this multifaceted protein across functional, structural and pathological aspects.
Keywords: 37 67 kDa laminin receptor, RPSA, 37LRP, 67LR, YIGSR, p40, 37LBP
I. INTRODUCTION
Laminins are a family of large (400–900 kDa) heterotrimeric glycoproteins typically found within the basal lamina region of the extracellular matrix. There are currently 15 known laminin proteins, each consisting of an α, β and γ-chain organized into a complex cruciform structure. Laminins have crucial and diverse rolls in physiology, influencing cellular adhesion, migration, homing, proliferation, polarity, differentiation, tissue development and cellular communication with the extracellular environment (Miner, 2008; Ryan et al., 1996; Yurchenco, 2011).
Shortly after the discovery of laminins and observation of their importance in tumour metastasis, the search for cell-surface proteins mediating their effects identified receptors of two main classes: the integrins and the non-integrin laminin receptors. The first to be revealed was the non-integrin 67-kDa laminin receptor, identified via binding to immobilized laminin-1 (α1β1γ1) (Lesot, Kuhl & Mark, 1983; Malinoff & Wicha, 1983; Rao et al., 1983). Further study demonstrated this protein might arise from a 32–33 kDa precursor that migrates at approximately 37 kDa in sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS-PAGE) (Rao et al., 1989; Wewer et al., 1986). Quite separately, a protein called p40 in a different line of investigation was found to be equivalent (Makrides et al., 1988; McCaffery, Neve & Drager, 1990) and later confirmed to be a component of the 40S ribosome now called ribosomal protein SA (RPSA) (Tohgo et al., 1994). Hence, this protein has been called 37/67-kDa laminin receptor (37LR, 67LR, LAMR1), 32 kDa laminin binding protein (LBP), 32 kDa laminin binding protein precursor (LBP-32, 37LRP), p40 and RPSA. This review notes that the most common descriptive term is 37/67-kDa laminin receptor and will refer to the protein as LAMR, designating the relevant species when possible in superscript (i.e. LAMR37/67) even in instances referring to non-laminin binding homologs.
The functions of LAMR have proven to be surprisingly diverse, representing a repertoire that extends far beyond cell anchoring via laminins to include ribosomal biogenesis and translation, pre-rRNA processing, roles in cellular migration, invasion, viability and growth, cytoskeleton (re)organization as well as chromatin and histone binding. These functions reflect the protein’s presence in the plasma membrane, ribosomes, cytosol and the nucleus. The importance of LAMR in development and differentiation has also been reported, and the cell signalling cascades LAMR participates in are beginning to be elucidated.
As proteins homologous to ancestral prokaryotic RPS2 superfamily members – not to be confused with eukaryotic RPS2 of the RPS5 family – RPSA/LAMR homologs are found in all taxonomic domains of life and have been studied specifically in mammals, birds, amphibians, a number of invertebrates, yeast, nematodes, plants, bacteria and archaea either for ribosomal or laminin binding function. Laminin binding appears to be a more recently acquired function particularly concentrated in metazoans, thus not all RPSA homologs have a laminin binding function.
LAMR has also been implicated in several pathological conditions, including neoplastic and metastatic cancers, neurodegenerative diseases, microbial infections and several developmental aberrations. The protein is perhaps most well studied for its overexpression in solid and haematological malignancies and its positive association with invasive and metastatic cell behaviour (Menard, Tagliabue & Colnaghi, 1998). Its role in other maladies appears to be a function of its ability to act as a receptor for several exogenous agents including prion proteins (Vana et al., 2009), viruses (Kielian, Chanel-Vos & Liao, 2010) and bacteria (Huang & Jong, 2009).
To date there are over 500 research publications specifically pertaining to LAMR/RPSA; they have been critically examined for expert commentary in this review. This topic has been reviewed previously by Nelson et al. (2008), Menard et al. (1997) and Rea et al. (2012). Because of the multifaceted nature of LAMR, its controversial role in laminin binding and the complexities of its study, this review emphasizes the strengths and weaknesses of the evidence for each of its putative roles. Additional attention is given to examination of LAMR’s structural features with particular focus on implications for function. Lastly, an attempt is made to piece together various lines of evidence to arrive at probable signal transduction pathways to which LAMR may contribute.
II. 37/67-kDa LAMININ RECEPTOR IDENTITY
(1) Gene and genome
LAMR, officially RPSA, is encoded in humans by the ribosomal protein SA gene (RPSA, NCBI Gene ID: 3921, Accession No.: AC_000135). The gene is a 5833 bp stretch along chromosome location 3p22.2 (3p21.3 by in situ hybridization) consisting of seven exons and six introns (Jackers et al., 1996b). Based on analysis of the avian gene promoter, no TATA or CAAT box was found, however, several specificity protein 1 (Sp1) and activator protein 1/2 (AP1/2) binding sites were noted in addition to response elements for glucocorticoids and consensus motifs for signal transducer and activator of transcription (STAT) proteins (Clausse et al., 1996). Sequence comparison and prediction of transcription factor binding sites using the human promoter are in agreement with this report (Messeguer et al., 2002). The promoter may also be responsive to p53 (Modugno et al., 2002).
Within the introns of RPSA, there are two H/ACA box-type small nucleolar RNA (snoRNA) genes, SNORA6 and SNORA62, understood to be important for rRNA processing as part of ribonucleoparticles (Kiss et al., 2004; Lestrade & Weber, 2006; Ruff et al., 1993).
It is unknown how many functional copies of the gene exist, but there are at least 64 pseudogenes spread throughout several chromosomes (Balasubramanian et al., 2009), many of which appear to be processed retroposons (Jackers et al., 1996a). Interestingly, the avian genome has only one gene copy and contains no pseudogenes, a convenience exploited for early efforts at gene description.
(2) The 37-kDa laminin receptor protein
The human RPSA gene produces a final mRNA of 1155 nucleotides (Accession No.: NM_002295) or a 1109 nt variant with an alternative 5' exon prior to the start codon (NM_001012321) – both encode the same protein. Northern blotting detects a single transcript of approximately 1.2–1.4 kb (Satoh et al., 1992). The open reading frame of 885 bases encodes a 295 amino acid protein.
Of the seven exons in the human RPSA mRNA, the first is non-coding and 2–5 encode the N-terminal RPS2 domain [amino acids (a.a.) 1–209], which shares roughly 20–30% sequence identity with prokaryotic rps2 proteins (>60% among eukaryotes). The latter two exons encode a C-terminal region (a.a. 210–295) absent in prokaryotes. Though highly homologous amongst vertebrates, this C-terminal portion is of variable length and sequence in eukaryotes (~80 a.a. in animals and plants versus ~45 in yeast and excavates, ranges of 18–98% identity); exon splicing structure also varies across kingdoms. The genetic origins of the C-terminal-encoding segments are unknown, but they have been implicated in laminin binding for vertebrates (see Section III.4).
LAMR37 is the species of protein produced upon transfection of mammalian cells with RPSA cDNA (Castronovo et al., 1991a), recombinant expression in bacteria (Jamieson et al., 2008) and with in vitro translation reactions (Grosso, Park & Mecham, 1991; Rao et al., 1989). This species has been found in the cytosol, associated with the cytoskeleton, in the plasma membrane (Venticinque, Jamieson & Meruelo, 2011), in the nucleus (Scheiman et al., 2010a) and associated with ribosomes (McCaffery et al., 1990; Scheiman et al., 2010b). Originally presumed to be simply a precursor to LAMR67, LAMR37 has since been shown to be important for several intracellular roles and is most likely involved in laminin binding (see Section III).
The protein is potentially phosphorylated on Tyr-139 (Davis et al., 1991; K. Kim et al., 2005; Rush et al., 2005), but all other potential post-translational modifications are typically discussed with respect to conversion to the 67-kDa species, LAMR67.
(3) Higher molecular weight species
The first species of LAMR described was the 67-kDa LAMR67, purified from cellular membranes by laminin-1 immobilized on sepharose and confirmed as a laminin binding protein via radiolabelled laminin overlay blots (Huard, Malinoff & Wicha, 1986; Lesot et al., 1983; Malinoff & Wicha, 1983; Rao et al., 1983). The precise identity of this protein remains uncertain, but it is presumed to be a higher molecular weight form of LAMR37. There are several lines of evidence for this. First, following the development of an antibody derived from the purified 67-kDa protein (Liotta et al., 1985), immunoscreening of a cDNA library identified RPSA coding sequences (Wewer et al., 1986). Second, partial protein sequencing of the purified 67-kDa protein revealed a short peptide (MLAREVLR) corresponding to the LAMR37 sequence 177–184 (Wewer et al., 1986) – human protein BLAST searches to date show only LAMR37/RPSA as a direct match with 100% identity and coverage. Lastly, several antibodies raised against different synthetic peptides within the LAMR37 sequence recognize both 37-kDa and 67-kDa bands on immunoblots of cell lysates (Castronovo, Taraboletti & Sobel, 1991b). Indeed, numerous reports demonstrate that different antibodies cross-react between the two species (Buto et al., 1997; Castronovo et al., 1991a; Rao et al., 1989; Wewer et al., 1987). Clearly, it would appear that LAMR37 and LAMR67 are at least immunologically related. Of note, however, is the regularity with which LAMR antibodies recognize either the 37 or the 67-kDa proteins, but not both.
It has long been thought that there is a precursor relationship between the two species, that the 67-kDa protein is somehow derived from LAMR37. This is a matter of some controversy and has been directly addressed by only two studies. The first tracks the conversion of LAMR37 to a higher molecular weight (HMW) 67-kDa form using pulse-chase radiolabelled cells to monitor the transition over time. After immunoprecipitation of LAMR, the appearance of a weak 67-kDa signal begins to occur within 30–90 min of the pulse accompanied by decreased LAMR37 detection (Castronovo et al., 1991a). Complete conversion of LAMR37 to 67-kDa is said to occur, an observation inconsistent with the known presence of LAMR37 in cells. Crucially, in the same report, exogenous introduction of RPSA cDNA increases LAMR37 but not the 67-kDa HMW form.
Though the precursor relationship is largely accepted, no data have emerged showing the production of a HMW species from a tagged LAMR37 construct – only 37-kDa+Tag molecular weights have been observed (see for examples: Scheiman et al., 2010a; Chen et al., 2003; Castronovo et al., 1991a; Gauczynski et al., 2001; Amano et al., 2005b). Anecdotal reports are discussed in Buto et al. (1998), Clausse et al. (1996) and Menard et al. (1997), who reference unpublished data and a conference abstract by Montuori et al. (1995) claiming to have demonstrated production of a 67-kDa protein from a histidine-tagged LAMR37. Accounts claiming overexpression of LAMR67 upon RPSA cDNA introduction are available, but problems with interpretation arise due to ambiguous demonstration of protein size (e.g. use of flow cytometry or failure to distinguish between 37 and 67 species; see Landowski, Uthayakumar & Starkey, 1995b and Tachibana et al., 2004). In the future, the field would benefit from clear molecular weight distinguishers and precise terminology when referring to various LAMR species.
The second attempt to solidify a precursor relationship comes from a study hypothesizing that fatty acid acylation of LAMR37 may be required for transition to the HMW form. In support of this, treatment of cells with cerulenin, an inhibitor of fatty acid synthase, reduces the detection of LAMR67 in cells with a concomitant appearance of LAMR37. Usage of hydroxylamine to strip thio-/oxy-ester bound fatty acids from proteins produces a similar effect (Buto et al., 1998). The presence of palmitate, stearate and oleate fatty acid residues on purified LAMR67 has been detected using gas chromatography–mass spectrometry (Landowski, Dratz & Starkey, 1995a).
Several groups have reported the presence of additional HMW LAMR species. Proteins of 32, 37, 45, 53, 55, 67, 80 and >110-kDa size have been isolated using laminin-1 columns in addition to the integrins (Clement et al., 1990; Davis et al., 1991; Douville, Harvey & Carbonetto, 1988; Kleinman et al., 1988; Landowski et al., 1995a; Languino et al., 1989; Mercurio & Shaw, 1988; Tandon et al., 1991; Weeks et al., 1991). Cross-reactivity of LAMR67 antiserum with some of these proteins has been reported, including a 110-kDa species and galactosidase beta-1 (GLB1, Protein Accession No.: P16278). GLB1 is a natively 67-kDa elastin/laminin binding protein often confused for LAMR67, especially when lectin and elastin are involved (Mecham et al., 1989; Weeks et al., 1991). These reports serve as examples of possible shared epitopes and potential sources of contaminating sequences.
Taking into account the descriptions of known laminin binding proteins and the LAMR literature, it seems likely that reports of 32–45 kDa species correspond to LAMR37. Curiously, 53–55 kDa entities (Graf et al., 1987b; Karpatova et al., 1996; Tandon et al., 1991) may represent another HMW species, LAMR53, distinct from the 67-kDa form – 37, 53 and 67 can be detected in cells with a single antibody. Unfortunately, very little published literature is available to address this possibility.
(4) Identity of LAMR67
Given that LAMR67 is a stable protein species that conserves its molecular weight under denaturing and reducing conditions (Landowski et al., 1995a), it is unclear what event is responsible for the increase in size if it is indeed formed from LAMR37. Efforts to sequence the protein directly have met with little success, typically attributed to a blocked N-terminus and/or limited ability to isolate the protein in sufficient quantities and purity – bovine serum albumin (BSA) especially is reported to be difficult to eliminate upon co-purification (von der Mark & Risse, 1987; Wewer et al., 1986). The amino acid composition of purified LAMR67 has been determined (Landowski et al., 1995a), but efforts to sequence the protein using modern technologies appear to be absent from the literature.
It has been suggested that HMW LAMR species are not produced upon introduction of RPSA cDNA into cells because of the necessity for another protein, hence the hypothesis that LAMR67 is a heteromeric protein. A galectin family protein is the most frequently proposed candidate for a LAMR binding partner owing to reports demonstrating that lectin-targeted antibodies cross-react with LAMR67 (Buto et al., 1998; Castronovo et al., 1992; Mecham et al., 1989). LAMR has been reported to be eluted from laminin with galactosides (Mecham et al., 1989) and lectins do have laminin binding properties (Mercurio & Shaw, 1991). This issue is confused, however, by the presence of the 67-kDa elastin/laminin binding protein (GLB1) which also has lectin properties (Ochieng et al., 1999), thus raising the possibility that identity errors have occurred. Indeed, there are directly contradictory reports that LAMR does not elute from laminin affinity columns with galactosides (Fulcher et al., 1993) nor bind elastin (Grosso et al., 1991). Complicating the matter further, there is evidence of a lectin protein that binds the carbohydrate support of sepharose columns; this protein is a common contaminant of integrin preparations and is reduced to 70-kDa subunits when denatured, introducing another source of uncertainty (Bao, Muschler & Horwitz, 1992; McCaffery et al., 1990).
Amino acid composition analysis performed on purified LAMR67 is in agreement with the predicted composition of LAMR37, which has been cited as evidence for homodimer formation of LAMR67 (Landowski et al., 1995a). A potential galectin-3-LAMR37 heterodimer and GLB1 show significant deviations from this amino acid composition. Evidence for a homodimer is sparse, however. Two-hybrid testing failed to demonstrate LAMR37 could interact with itself or galectin-3 (Hundt et al., 2001), however this trial was performed with a possibly abnormal N-terminally truncated LAMR37(Δ1–43) in yeast where modifications necessary for dimerization may not occur. Bacterially grown recombinant and in vitro translated LAMR37 remains monomeric (Grosso et al., 1991; Jamieson et al., 2008; Rao et al., 1989) and exogenous expression of RPSA cDNA has not been clearly shown to produce LAMR67.
It would appear the oligomeric state of LAMR67 remains unsolved. Alternative splicing or a separate transcript does not provide a satisfactory explanation, as the vast majority of reports detect a single RPSA mRNA of approximately 1.2 kb in several cell lines as well as tissue samples (Satoh et al., 1992). There are sparse examples of larger mRNAs (5.5 kb) being co-detected in low abundance in tumour cells using RPSA probes, but their identity is uncertain (Rabacchi, Neve & Drager, 1990; Yow et al., 1988).
Why a protein–protein complex involving LAMR37 would be stable during denaturing/reducing SDS-PAGE is unclear, but a strong hydrophobic fatty-acid mediated interaction may contribute. Alternatively, covalently attached additions from post-translational modifications are possible. LAMR67 and LAMR37 appear to be acylated, but fatty-acid residues are low in molecular weight and not considered to be in abundance on LAMR (Buto et al., 1998; Landowski et al., 1995a). Glycosyl group addition is another potential low molecular weight modification, but LAMR shows no signs of glycosylation (Castronovo et al., 1991a; Landowski et al., 1995a). There is a singular report of LAMR detection by an antibody that recognizes moieties within glycosphingolipids that also demonstrates LAMR incorporation of labelled galactose and sialic acid during synthesis (Katagiri et al., 2005).
There do exist post-translational modifications that can add significant mass to a protein by covalent attachment. Ubiquitin-like proteins (UBLs) such as ubiquitin, NEDD (neural precursor cell expressed developmentally down-regulated protein) and SUMO (small ubiquitin-like modifier) are within the mass range of 8–14-kDa and are capable of forming oligomeric chains (van der Veen & Ploegh, 2012). This possibility may have significant explanatory power with regard to LAMR67.
Several large-scale proteomics screens for SUMOylated proteins have identified human LAMR as a target for SUMO-1, SUMO-2/3 and SUMO-4 modification (Blomster et al., 2009; Golebiowski et al., 2009; Guo et al., 2005; Manza et al., 2004; Schimmel et al., 2008). SUMOylated RPS0 (the yeast RPSA/LAMR homolog) was also detected in a yeast screen (Wohlschlegel et al., 2004). The primary LAMR37 structure has a consensus SUMOylation site near the N-terminus (Lys-11) and several non-consensus sites (Lys-212 and Lys-224) which happen to overlap with a putative laminin binding region within residues 205–229 (SUMOsp: Ren et al., 2009). It is tempting to speculate that a single SUMOylation event might be responsible for the reported LAMR53 species while multiple modifications or SUMO oligomers might build LAMR67 and larger species; the predicted sizes of LAMR-SUMO species are consistent with this proposal.
SUMOylation is known to positively or negatively influence protein stability, protein trafficking, and protein–protein interactions. The latter influences occur due to creating or masking of a binding interface and/or inducing conformational changes within the SUMOylated protein while protein stability effects occur in conjunction with the ubiquitin/proteasome system (Wilkinson & Henley, 2010).
Definitive identification of the LAMR HMW species would seem to be lacking, leading to justifiable skepticism concerning their characteristics. Nonetheless, there appears to be sufficient evidence supporting that LAMR67 is a genuine entity and unique molecule related to LAMR37, albeit abundance of the protein is likely low and dependent on various experimental and physiological conditions.
(5) LAMR membrane presence
LAMR67 is considered to be primarily located at the plasma membrane, possibly concentrated in lipid rafts (Fujimura, Yamada & Tachibana, 2005). This has been sufficiently demonstrated using fluorescent microscopy, membrane fractionation and FACS on unpermeabilized cells (Abe et al., 1996; Castronovo et al., 1991b; Landowski et al., 1995b; Liotta et al., 1985).
The membrane abundance of both LAMR67 and LAMR37 is significantly less than the overall cytoplasmic content. LAMR67 cytosolic levels in particular appear to be minimal. It is important to note that species abundance data are often difficult to judge because of the varying antibody affinities and the possibility that some epitopes are only exposed (or destroyed) upon protein denaturing.
Intriguingly, it appears that antibodies against epitopes C-terminal to the 104th residue tend to be capable of labelling cell surface LAMR without cell permeabilization, whereas more N-terminal-directed antibodies only detect total LAMR with permeabilization (Castronovo et al., 1991b). This seemingly corroborates the prediction of a short, atypical transmembrane domain within LAMR37 residues 86–101 (Rao et al., 1989), which would result in approximately 100 intracellular N-terminal residues. No transmembrane region is apparent in the LAMR37(9–205) crystal structure, however, nor is the presence of an exposed amphipathic domain consistent with LAMR cytosolic solubility (see Section IV). The N-terminal regions may simply be masked in particular situations, and the targeted epitopes have not been determined for all LAMR antibodies. A transmembrane region need be considered only if clear evidence arises implicating LAMR in direct structural transduction of extracellular-to-cytosolic signals without the need for accessory proteins or translocation events.
LAMR37 has no membrane-targeting signal peptide and a transmembrane domain is unlikely, leaving open the question of how association with the plasma membrane occurs. Electrostatic interactions with the membrane are unlikely as LAMR is predominantly negatively charged. Although fatty acid acylation appears to occur on LAMR, the pattern is inconsistent with that of a glycosylphosphatidylinositol (GPI) anchor (Karpatova et al., 1996; Landowski et al., 1995a). This situation brings up the possibility that membrane association could occur via a hydrophobic segment on LAMR37 or a modification, an adhesion method not uncommon for exoplasmic peripheral membrane proteins. This mode of membrane attachment is supported by numerous reports of LAMR association with intact liposomes (Anilkumar & Sudhakaran, 1993; Lesot et al., 1983; Sengupta et al., 1991) and shedding of intact LAMR67 into cell culture medium (Karpatova et al., 1996), but definitive evidence is lacking.
Peripheral membrane proteins are often known to associate with other proteins for added stability in the membrane. Consistent with this hypothesis, LAMR has been demonstrated to interact with the α6 integrin subunit, presumably as part of a full integrin (Ardini et al., 1997). This may explain how initial membrane targeting occurs; LAMR has been found to co-localize with integrins in cytoplasmic particles that may represent vesicular trafficking to the plasma membrane (Romanov et al., 1994).
A heteromeric or otherwise modified HMW species may also associate with the membrane via the properties of its partner(s). The possibility of a LAMR67 homodimer having a considerably different conformation cannot be entirely ruled out either, but these options do not suffice to explain the conservation of protein properties or the membrane presence of LAMR37. Several possibilities are depicted in Fig. 1.
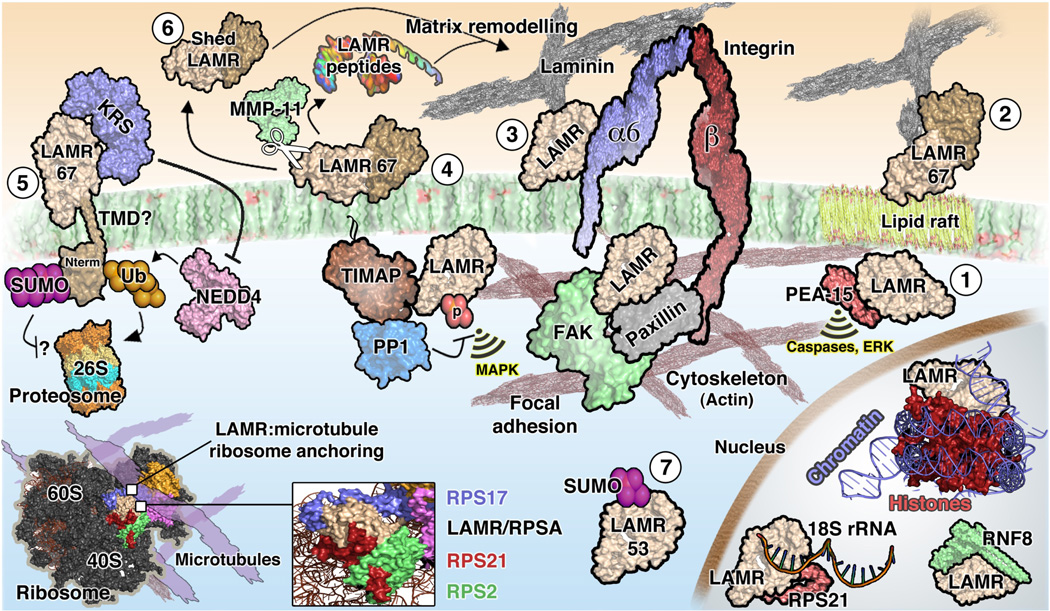
Figure 1.
Theoretical cellular biology of laminin receptor (LAMR/RPSA). Several possible LAMR species are depicted including (1) intracellular LAMR37; (2) extracellular peripheral membrane LAMR67 capable of independently binding laminin; (3) extracellular peripheral membrane LAMR37 supporting integrin laminin binding; (4) LAMR67 membrane association via the properties of a homo- or heteromer; (5) LAMR67 with hypothetical transmembrane domain (TMD). It is unclear how LAMR membrane presence is dynamically regulated by intracellular proteins in the absence of a TMD; (6) shed LAMR67 and (7) intracellular SUMOylated LAMR53. Ribosomal, cytoskeletal and nuclear localizations of LAMR in addition to its presence at focal adhesions are also depicted. All protein–protein contacts represent experimentally demonstrated interactions. Signalling indications (yellow) represent preliminary evidence for signalling cascades involving LAMR. Because of ambiguity in the data, the subcellular locations of each protein are not necessarily accurate and represent only predictions based on available evidence. Similarly, the species of LAMR (i.e. 37/67 kDa) depicted for each function is not meant to suggest exclusivity or a definitive understanding of the relevant molecule(s). p, phosphorylation. Nterm, intracellular N-terminus of LAMR. Ub, ubiquitin. SUMO, small ubiquitin-like modifier. NEDD4, neural precursor cell expressed developmentally down-regulated protein 4. RPS, ribosomal protein small subunit. MMP-11, matrix metalloproteinase-11/stromelysin-3. KRS, lysyl-tRNA synthetase. TIMAP, TGF-inhibited membrane-associated protein. PP1, protein phosphatase-1. FAK, focal adhesion kinase. PEA-15, phosphoprotein enriched in astrocytes-15. RNF8, ring finger protein 8. MAPK, mitogen-activated protein kinase pathways. ERK, extracellular signal-regulated kinase pathways.
The mechanism of membrane targeting also remains unresolved, but it is apparent that there are control systems in place to influence LAMR turnover in the plasma membrane. The anti-metastatic protein HEXIM1 (hexamethylene bis-acetamide inducible 1), for example, is found to serve a role in downregulation of LAMR content in the membrane without affecting total levels (Ketchart et al., 2013). Membranous LAMR67 is also mediated by NEDD4, which can target the protein for ubiquitination and 26S proteasome degradation. Protection from ubiquitination events – and thus LAMR67 stabilization – can be conferred by the LAMR binding protein KRS (lysyl-tRNA synthetase), which is activated by laminin signalling (Kim et al., 2012). It is yet unclear how a protein residing wholly in the outer plasma membrane layer could be regulated in this manner (Scenario 5 in Fig. 1).
SUMOylation is often brought about under conditions of cellular stress; it may also be a response to other signalling stimuli. Reports of specific proteins’ plasma membrane association aided or disrupted by SUMOylation exist (Huang et al., 2012; Martin et al., 2007). SUMO modification could also play a role in regulating the stability of LAMR, whether by targeting it for degradation or as a competitive counter to ubiquitination, as it can for other proteins (Schimmel et al., 2008). It is also possible that SUMO modification changes the laminin binding properties of membranous LAMR (i.e. in the 37- to 67-kDa transition, see Section II.4).
One particularly appealing speculative scenario is the alternating usage of ubiquitin and SUMO modifications on LAMR to regulate the degree of laminin binding when a cell may have need to migrate. This may be affected via changes in LAMR stability, targeting for endocytosis or degradation, or alteration of ligand binding affinity. Ribosomal incorporation (exclusively LAMR37) and nuclear import of LAMR could similarly be regulated by these modifications.
III. 37/67-kDa LAMININ RECEPTOR FUNCTION
(1) Functional importance
Little is conclusively understood about the normal physiological province of LAMR and its various higher molecular weight species in cells. What is clear is that it functions in laminin binding and as a structural component of the ribosome. Further, we can conclude from the evidence that it is important for certain modes of cellular adhesion through laminin, and a factor in cellular migration. Homozygous Rpsa null mice fail to develop past embryonic day 3.5 while heterozygotes suffer only delayed embryonic growth, becoming otherwise phenotypically normal (Han, Logsdon & Ellis, 2008). A comprehensive assessment of the intricacies of LAMR remains elusive, but investigations using methods to reduce expression or modulate function are beginning to shed light on its numerous roles.
Depending on the cell line being investigated and the degree of knockdown, the effects of reduced LAMR are known to include: reduction in proliferation and induction of G1-phase cell cycle arrest (Satoh et al., 1999; Scheiman et al., 2010a, b), attenuation of protein synthesis (Demianova, Formosa & Ellis, 1996; Ford, Randal-Whitis & Ellis, 1999; Scheiman et al., 2010b; Venticinque et al., 2011), disruption of pre-rRNA processing (Ford et al., 1999; O'Donohue et al., 2010), decreases in viability (Demianova et al., 1996; Moodley & Weiss, 2013; Scheiman et al., 2010a; Susantad & Smith, 2008), and reduced adhesive and migratory ability (Chen et al., 2009; Poon et al., 2011; Venticinque et al., 2011). Unsurprisingly given RPS2 family homology, there may be distinctions between ribosomal and extraribosomal functions conferred by the N- and C-terminal regions, respectively (Scheiman et al., 2010a). Antibody blockade studies are generally in agreement with LAMR involvement in adhesion, migration and invasion (Davis et al., 1991; Omar et al., 2012; Tandon et al., 1991; Terranova & Lyall, 1986; Wewer et al., 1987), and thus serve to partially alleviate concern over secondary effects arising from the small interfering RNA (siRNA) approach, which also compromises ribosomal function and protein synthesis. In addition, LAMR interacts with many individual proteins, presumably participating in several cell signalling cascades.
(2) Laminin binding
The most studied and rudimentary aspect of LAMR is its function in binding laminin. If this interaction goes beyond simple promotion of cell adhesion to the basement membrane, LAMR’s involvement in transducing signals from laminin to affect cellular processes has not been clearly demonstrated. This has led to the proposal that cell surface LAMR functions only to stabilize or regulate the binding of laminin to other receptors (Ardini et al., 1997; Pellegrini et al., 1994), that LAMR is only necessary for particular stages of attachment (Basson et al., 1990) or that accessory molecules are necessary for its function (Landowski et al., 1995a).
Several laminin receptors are known to overlap in function (particularly adhesion and migration), although they do not seem to be entirely redundant or compensatory in vitro. With so many identified, there are inevitably cell-type-specific differences in laminin receptor profiles. This varied receptor complement can switch in response to certain stimuli – for example, haematopoietic stem cells have been shown to shift from α6 integrin display to LAMR during signal-induced mobilization for egress and migration (Selleri et al., 2006). Development and differentiation (Hara, Satoh & Ide, 1997; McKenna et al., 2001), malignant transformation (Menard et al., 1998), tissue stimulus responses (Anilkumar & Sudhakaran, 1993; Baloui et al., 2009; Stitt et al., 1998) and other signalling molecules (e.g. cytokines, hormones) have also been noted to induce changes in LAMR expression (Castronovo et al., 1989; Chen et al., 2002; Clausse et al., 1998; Raghunath et al., 1993; Shi et al., 1993). Some of these changes are linked to laminin expression in adjacent tissue and may be due to a need for a change in laminin binding propensity (positive or negative).
Cell membrane purified LAMR67 binds native laminin isolated from Engelbreth Holm Swarm murine sarcoma (predominantly laminin-1) with an affinity (Kd) of 2 nM (Malinoff & Wicha, 1983; Rao et al., 1983). In comparison, Escherichia coli purified recombinant LAMR37 binds laminin-1 with a 700 nM Kd (Jamieson et al., 2008; Zidane et al., 2013). The difference in affinity is likely to reflect differences between the 67- and 37-kDa species, the contribution of any post-translational modifications (to cellular LAMR37) as well as inter-assay and preparation variability. The only comparison of affinity in a consistent system found a similar Kd for both species, 300–400 nM, using surface plasmon resonance (Fatehullah et al., 2010). It is challenging to speculate on the physiological significance – if any – of the differences in the dissociation constants, but a 37/67 expression ratio may factor into a laminin binding fine-tuning system.
(3) Laminin-1 binding sites for LAMR
Not many studies have directly examined LAMR binding to laminin in order to refine our knowledge of the binding site. Most of the information collected is for laminin-1. One low-resolution attempt used the native laminin-1 molecule to investigate the binding region of purified LAMR67, employing rotary shadowing electron microscopy. The result showed the LAMR67 binding site to reside on the laminin long arm just below its intersection with the short arms (Cioce et al., 1993), presumably on domain II, which adopts a triple coiled-coil of all three laminin subunits (Engvall & Wewer, 1996). Large laminin proteolytic fragments, C1 and P1 (Rao et al., 1982; Rohde, Bachinger & Timpl, 1980), were used to substantiate this binding site. Information about the role of laminin domain II in cell binding is limited due to lack of applicable large fragments (Aumailley, 2013; Aumailley et al., 2005), and detailed peptide analysis may be unfeasible because of quaternary structure.
Another approach to identify the LAMR binding region of laminin was taken with the use of antibodies derived against synthetic peptides from the β1 subunit of laminin-1. An antibody that blocked cell adhesion to laminin was identified, and screening of peptides nearby the targeted epitope identified CDPGYIGSR (Peptide 11) as being capable of similarly blocking cell attachment (Graf et al., 1987a). The activity of this synthetic peptide was later refined to the minimal pentapeptide YIGSR on the laminin β1 short arm, a.a. 929–933 (Graf et al., 1987b). CDPGYIGSR and YIGSR can influence cell attachment and migration in a manner generally consistent with that of a laminin analog or decoy (Kikkawa et al., 2013). YIGSR peptides have also been demonstrated to elute LAMR67 from laminin columns (Graf et al., 1987a, b) as well as associate with LAMR67 (Bushkin-Harav, Garty & Littauer, 1995; Hadley et al., 1990; Starkey, Uthayakumar & Berglund, 1999). YIGSR’s interaction with LAMR37 has not been examined in detail, but the peptide failed to co-crystallize with a LAMR37(1–220) recombinant protein (Jamieson, K.V. & Wu, J., unpublished data) – although it may only interact with LAMR67 or require full-length LAMR37.
Skepticism about YIGSR and related peptides does exist in the literature. Some groups have failed to elute LAMR from laminin columns with the peptide, and others still express concern that its properties do not adequately mimic those of laminin binding or display LAMR specificity (Castronovo et al., 1991b; Clement et al., 1990; Maeda, Titani & Sekiguchi, 1994; Yamada et al., 1990). YIGSR is certainly a bioactive peptide (Kikkawa et al., 2013), but whether all its effects are mediated through LAMR or reflective of native laminin (folding state, receptor usage, etc.) is uncertain.
(4) LAMR binding sites for laminin
Several binding sites on LAMR have been suggested to interact with laminin. These sites are typically discussed as if they possess independent ability to bind laminin, although in native binding it is unclear if they function redundantly or in collaboration.
The most common approach of using synthetic peptides derived from the LAMR37 primary sequence has identified two putative laminin binding regions. The first is referred to as Peptide G, residues 161–180 (Castronovo et al., 1991b). The second site, typically referred to as the “direct binding region”, is the stretch of amino acids between 205 and 229 (Landowski et al., 1995b). Both peptides can bind laminin-1 and have bioactive properties in vivo (Landowski et al., 1995b; Taraboletti et al., 1993). These peptides also display heparan sulfate (HS) binding properties (Guo et al., 1992; Kazmin et al., 2000). The functional implications of HS binding are not clear, as laminin-1 preparations are often said to contain HS proteoglycan. Truncated constructs of LAMR are seemingly capable of binding HS and laminin simultaneously, while full-length LAMR37 and LAMR67 may bind HS more weakly or not at all (Fatehullah et al., 2010; Zidane et al., 2013). HS binding may act as a laminin binding stabilization or inhibitory event in these scenarios, but artifactual binding cannot be ruled out.
The palindromic sequence LMWWML (a.a. 173–178) contained within Peptide G has been suggested to be the minimal sequence responsible for laminin binding, the two consecutive tryptophan (W) residues considered suitable for protein–protein interaction (Jaseja et al., 2005). Suspicions arise, however, due to the location of this sequence in the protein crystal structure: it is a helical stretch buried within the interior of the protein without obvious surface accessibility, calling into question its availability for protein–protein interaction while also accounting for the presence of the tryptophan residues. Nonetheless, high sequence conservation in this region was shown to correlate with laminin-expressing organisms while divergence of sequence occurs in non-laminin-expressing species (Ardini et al., 1998). Though the RPS2 family domain (human residues 1–209) is highly conserved as a whole, this palindromic sequence has especially high sequence identity amongst metazoans.
Notably, the presence of both tryptophans does not appear to be required, as laminin binding has been observed in LAMR homologs even with sequences LMLWML (metazoan Echinococcus granulosus: Zhang et al., 1997), LMFWLL (amoebozoa Acanthamoeba healyi: Hong et al., 2004) and LMYWLL (alveolate Toxoplasma gondii: Furtado et al., 1992). Thus, laminin binding exists even with sequence variation in the palindrome and is not limited to metazoans. Interestingly, the second tryptophan in particular (human residue W176) appears to be universally conserved, although it is also conserved in non-laminin-binding archaea (IVYWLL) and plants (CLFWLL).
Ultimately, the significance of Peptide G and the palindrome will have to be verified experimentally within the full protein. Peptide G and its possible availability for laminin binding is further discussed in Section IV.
A separate study using a phage peptide display identified 9-mer mimotopes similar in sequence to Peptide G and Peptide 205–229 as functional in binding laminin-1 (Kazmin et al., 2000). This study also identified sequences similar to the five x(D/E)W(S/T) repeats in the C-terminus of LAMR (a.a. 247–9, 264–8, 273–7, 285–7, 291–5) as potential laminin binding contributors – offering a third laminin binding site. Although the homologous sequence mimotopes could be eluted from laminin-1 by YIGSR peptides, they may not represent actual LAMR functionality.
The only study to attempt a mutational approach to determine the laminin binding site identified three amino acids (Phe-32, Glu-35, Arg-155) in structural proximity to each other using a truncated LAMR37(1–220) recombinant protein; individual or mutations in combination resulted in significant reduction of laminin binding capacity (Jamieson, Hubbard & Meruelo, 2011). However, introduction of these mutations to the full-length LAMR37(1–295) protein did not significantly affect laminin-1 binding (DiGiacomo, V. & Jamieson, K.V., unpublished data).
There are reports that N-terminal portions of LAMR37 (2–209 or 1–220) and C-terminal portions (210–295 or 225–295) can independently bind laminin-1 (Jamieson et al., 2011, 2008; Zidane et al., 2013), suggesting the four putative laminin binding sites work individually. Transference of the human C-terminal region (a.a. 210–295) to a non-laminin-binding archaeal LAMR homolog, Archaeoglobus fulgidus rps2P (Protein Accession No.: AAB90111), however, failed to confer laminin binding (DiGiacomo, V., unpublished data). The presence of multiple binding sites in the same construct does not appear to significantly alter target affinity – the full-length protein (four binding sites) engages laminin-1 with an approximately twofold higher Kd than N- and C-terminally truncated portions that each contain two proposed binding regions (Jamieson et al., 2008; Zidane et al., 2013).
The laminin binding regions will be discussed in detail with respect to the protein structure below, but it would seem as if the laminin binding site(s) is still a matter of debate (see Section IV). It may very well be that peptide analysis is insufficient for determining the true binding mode of LAMR to laminin, as it remains possible the binding site is a non-linear surface presented only by the full structure. It is also possible that LAMR37 interacts with laminin differently than LAMR67.
(5) Ribosomal functions
LAMR is likely to have originated as a ribosomal component. The most highly conserved portion of the protein, the N-terminal bulk (a.a. 1–209), is highly homologous to prokaryotic s2 family ribosomal proteins. Laminin binding functions may have evolved along with the appearance of laminin, although the nature of that development is incompletely understood (Ardini et al., 1998).
The earliest signs of translational function came from an observation of cytosolic p40 being associated with ribosomal particles in mice (Auth & Brawerman, 1992). When that protein was purified from ribosomes and sequenced, it was equated with LAMR37/RPSA (Tohgo et al., 1994). It is now clear that LAMR37 associates with the 40S small subunit of ribosomes (Anger et al., 2013; Ben-Shem et al., 2011; Malygin et al., 2011; Scheiman et al., 2010a, b) and has direct contacts with RPS2 (RPS5 family), RPS17, RPS21 and sparse contact with rRNA in humans, insects, yeast, plants and Tetrahymena (Anger et al., 2013; Armache et al., 2010; Ben-Shem et al., 2011; Rabl et al., 2011). Treatment with RNase does not disassociate LAMR from 40S particles (Auth & Brawerman, 1992).
It appears the N-terminal domain of the protein is sufficient for ribosomal binding, as LAMR37(1–220) is incorporated into ribosomes and can rescue translational defects caused by LAMR siRNA knockdown (Scheiman et al., 2010a). The C-terminus (210–295) may have been co-opted for binding additional proteins, stabilizing binding or interacting with rRNA (Malygin et al., 2011). The wheat (Triticum aestivum) LAMR homolog, RPSA/S2p, for example, has been suggested to bind ribosomal RACK1 (receptor for activated protein C kinase 1) via the C-terminal region (Armache et al., 2010). The C-terminus, encoded by separate exons (see Section II.2), alternatively may have evolved for laminin binding.
The function of LAMR ribosomal incorporation is not known outside the observation that it is important for protein synthesis and ribosome biogenesis (Demianova et al., 1996; Scheiman et al., 2010a, b; Venticinque et al., 2011), although it is alleged LAMR may not be a constitutive component (Malygin et al., 2011). While occurrence of ribosomal LAMR37 may be enhanced in polysomes of actively growing cells (Garcia-Hernandez et al., 1996) or by other stimuli (Fromm-Dornieden et al., 2012), the protein is still found in free 40S subunits and present during initiation of translation (Ford et al., 1999; Scheiman et al., 2010b; Valasek et al., 2003).
In addition to direct ribosomal function, LAMR is crucial for the processing of the 21S pre-rRNA into mature 18S rRNA. This function may involve binding and partial redundancy with RPS21, experimentally demonstrated in both humans and yeast (O'Donohue et al., 2010; Sato et al., 2003, 1999; Tabb-Massey et al., 2003). This appears to represent a ribosomal biogenesis function of LAMR as well as a nuclear/nucleolar role, as pre-40S subunits require LAMR for nucleolar exit (O'Donohue et al., 2010).
(6) Nuclear functions
Although nuclear and nucleolar localization of LAMR37 was noted early, the functional implications were not apparent until recently. There is no nuclear localization signal in LAMR (Sato et al., 1996), and it is not clear if SUMO modifications play a role in nuclear import, a common function of SUMOylation. Part of the LAMR presence in the nucleus is likely explained by its association with nucleolar pre-40S ribosomes and small nucleolar ribonucleoproteins (snoRNPs: O'Donohue et al., 2010). Beyond this, however, LAMR37 potentially interacts with chromatin, directly to some extent and more tightly through histones (Kinoshita et al., 1998; Venticinque & Meruelo, 2012).
LAMR37 may also serve to sequester the DNA damage repair proteins RNF8 (ring finger protein 8) and BRCA1 (breast cancer 1) to a waiting reserve in the nucleolus (Guerra-Rebollo et al., 2012). Intriguingly, RNF8 is an E3 ubiquitin ligase, and in this context, evidence for a polyubiquitinated LAMR species was found. The importance of nuclear LAMR may be more widespread than was originally realized; potential for transcription factor or chromatin modifier activity is a possibility.
(7) Cytoskeletal functions
Soon after the identification of LAMR, its cytosolic presence was shown to co-localize with actin and cytoskeletal stress fibre features (Brown, Malinoff & Wicha, 1983; Keppel & Schaller, 1991; Yannariello-Brown et al., 1988). It was suggested that LAMR may serve as a connection to extracellular laminin and the cytoskeleton in a manner analogous to integrins. In the likely absence of a transmembrane domain, however, it is unclear how LAMR would achieve such a function. Nevertheless, cell laminin binding is known to cause cytoskeletal reorganization events through a variety of mechanisms. LAMR staining patterns appear to react to adhesion and spreading on laminin, concentrating on leading edges of membrane ruffles and podia during migratory and attachment processes (Guirguis et al., 1987; Venticinque et al., 2011; Yannariello-Brown et al., 1988). Because LAMR directly interacts and co-localizes with filamentous actin and focal adhesion complex members (K.J. Kim, Chung & Kim, 2005), it would appear this occurrence is an intracellular event. LAMR may also be separately concentrated to these reacting regions on the extracellular surface (Scenario 3 in Fig. 1).
LAMR interaction with the cytoskeleton also has a prominent intracellular role that likely explains its tight pattern of co-localization and physical binding with tubulin (Venticinque et al., 2011). Critically, it would appear as if LAMR serves as a tether for ribosomes to microtubules, as its reduction results in loss of ribosomal localization to cytoskeletal structure, leading to dysfunction in protein synthesis (Auth & Brawerman, 1992; Venticinque et al., 2011).
(8) Development, differentiation and tissue response
Because laminin has been shown to influence a great many developmental programs and tissue responses to various stimuli such as injury or differentiation signals, it is likely that LAMR’s involvement in such functions is due to its laminin binding. Patterning, homing and establishing polarity may also fall within this purview. Given LAMR’s importance in protein translation and potential nuclear roles, however, it is necessary also to consider the contributions from such functions.
Examination of the literature reveals that LAMR tends to be more highly expressed on the cell surface of undifferentiated and developing tissue with subsequent decline in adulthood (Amano et al., 2005a; Biragyn et al., 2007; Khalfaoui et al., 2013; Laurie et al., 1989; Laurie, Stone & Yamada, 1991; Masuda et al., 2009; McKenna et al., 2001; Rao et al., 1994; Rescan et al., 1990; Weeks et al., 1991). LAMR has been shown to be important for imaginal disc development in Drosophila melanogaster (Melnick, Noll & Perrimon, 1993), for limb bud development in chicks (Hara et al., 1997), cardiomyopathy in mice (Asano et al., 2004) and quite strikingly in the development of the spleen in humans (Bolze et al., 2013). In the latter case, mutations found in the LAMR/RPSA gene are strongly associated with congenital asplenia, representing the only known polymorphisms in LAMR associated with human pathology.
In more well-differentiated cells, LAMR expression is typically confined to its intracellular localization except when a stimulus triggers cell surface upregulation. Upregulation of LAMR has been shown to occur in microglia following traumatic injury to the spinal cord in association with re-expression of laminin (Baloui et al., 2009) and in vasculopathic neovascularization events associated with ischaemic injury in the retina (Stitt et al., 1998). In normal cell behaviour, LAMR upregulation seems to occur in response to hormone or cytokine signalling that triggers chemotaxis (Castronovo et al., 1989; Chen et al., 2002; Poon et al., 2011; Selleri et al., 2006; Shi et al., 1993) while downregulation occurs with anti-tumorigenic-associated cytokines (Clausse et al., 1998). Increases in LAMR were also reported for other cytokine classes (Raghunath et al., 1993; Wenzel et al., 2010). Transforming growth factor (TGF), interferons, tumour necrosis factor (TNF), granulocyte-macrophage colony stimulating factor (GM-CSF) and gonadotropin releasing hormone 2 (GNRH-II) are amongst the effective signalling molecules described. Depending on cell type, differentiation or growth-promoting molecules like retinoids and phorbol esters have also been shown to alter LAMR expression levels (Annabi et al., 2007; Bryant et al., 1986; Hendrix et al., 1990; Montuori et al., 1999; Ross, Ahrens & De Luca, 1994; Tachibana et al., 2004).
(9) Modification of extracellular matrix
A particularly unexpected function of LAMR came to light when it was discovered that the HMW LAMR67 is shed (or secreted) from the cell surface in cell cultures (Karpatova et al., 1996). The function of this shed form (LAMR67-Shed) is unknown, but it may feed into a theorized system of modifying laminin to promote cellular migration.
Synthetic Peptide G, a putative laminin binding segment on LAMR, has been shown to cause cytoskeletal changes in cells and to increase the presence of LAMR on the plasma membrane, especially within filopodia, consequently promoting a more invasive phenotype (Berno et al., 2005). While it is unclear what physiological role this might play, Peptide G has also been shown to increase the rate of laminin degradation by proteolytic enzymes when the extracellular matrix molecule is exposed to the peptide, releasing a laminin fragment that can promote cellular migration (Ardini et al., 2002). This has led to speculation that LAMR contact with laminin modifies the laminin and subsequently alters cell adhesion and mobility.
Stromelysin-3 (MMP-11), a secreted extracellular matrix proteinase, has been shown to interact with and cleave LAMR, presumably liberating LAMR-derived fragments that may function similarly to Peptide G (Amano et al., 2005a, b). This system may act on both membrane anchored LAMR and LAMR67-Shed (Fig. 1). Similarly, LAMR67-Shed has been suggested to alter the profile of angiostatin fragments produced from plasmin, thereby reducing their anti-angiogenic activity; LAMR would thus promote angiogenesis in this manner (Moss et al., 2006). Plasmin normally functions to degrade extracellular matrix components and activate collagenases. LAMR activation has also been proposed to lead to plasmin activation by urokinase-type plasminogen activator (uPA, Soeda et al., 1994) and to upregulate the production of collagenases (Turpeenniemi-Hujanen et al., 1986).
The existence of these intricate systems may serve to promote laminin remodelling when necessary, shifting the environment toward one conducive to migration or, perhaps, invasion in a tumour context where many of these proteins are overexpressed.
(10) Signal transduction
Mounting evidence is beginning to point toward a role for LAMR as a mediator of cell signalling, interconnected with many pathways involved in diverse responses (Fig. 1). A proteomics screen for LAMR binding proteins recently confirmed this, identifying interacting proteins involved not only in cell adhesion and ribosomal biogenesis, but in metabolism, transcription, chromatin functions and signalling (Venticinque & Meruelo, 2012).
The earliest reports suggesting that LAMR might participate in cell responses came with the observation that laminin treatment of cells resulted in the dephosphorylation of a 67-kDa laminin binding protein and that phosphatase inhibitors prevented laminin-induced process formation. Protein kinase C stimulators caused similar process formation blockage (Weeks, DiSalvo & Kleinman, 1990). LAMR has subsequently been confirmed to be a phosphorylated protein (Davis et al., 1991; Rush et al., 2005). Protein phosphatase 1 (PP1) has been shown to affect LAMR phosphorylation after being targeted to the protein via LAMR interactions with the TGF-inhibited membrane-associated protein TIMAP (K. Kim et al., 2005), representing one possible mechanism leading to downstream effects. Laminin treatment of cells has also been reported to decrease the phosphorylation of extracellular signal-regulated kinases (ERKs), c-Jun N-terminal kinases (JNKs) and p38 proteins, an event that can be partially relieved by decreasing LAMR expression — lower LAMR levels result in increases of phosphorylated ERK, JNK and p38 (Givant-Horwitz, Davidson & Reich, 2004). It appears, then, that LAMR is involved in kinase/phosphatase axes and in laminin-induced signalling. It is important to consider, however, that the direct relationship to mitogen-activated protein kinase (MAPK) pathways is unclear because diminishing LAMR potentially triggers cell stress, to which the discussed pathways are known to respond (see Section III.1).
Laminin-induced signals also seem to lead to the phosphorylation of phosphoprotein enriched in astrocytes 15 (PEA-15/PED), an adaptor protein that binds LAMR (Formisano et al., 2012). Phosphorylation events on this protein lead to growth signals through the ERK cascade as well as blockage of caspase activation, linking LAMR to laminin pro-survival signalling.
Since laminin signalling is best known to result in cytoskeletal reorganization associated with migratory and adhesion changes, it is not surprising that LAMR has been implicated in membrane proximal focal adhesions as well. LAMR has been demonstrated to interact with actin, focal adhesion kinase (FAK) and paxillin, all crucial cytoskeletal rearrangement participants (K.J. Kim et al., 2005; Venticinque et al., 2011). Because these components, in addition to LAMR, are known to associate with integrins, involvement in signalling by large macromolecular focal adhesion complexes seems likely. In this case, it is difficult to interpret whether LAMR is required to be extracellular, transmembrane or intracellular, although the latter is at least necessary for binding FAK and actin.
In more specialized conditions, LAMR has also been shown to interact with the GM-CSF receptor (Ando et al., 2011; Chen et al., 2003), insulin-like growth factor receptors I and II (Ku et al., 2012), insulin receptor and insulin receptor substrates (IRS-1/2/4) (Ku et al., 2009), the t-cell specific adaptor protein TSAd (Park et al., 2009). LAMR also potentially serves as a receptor for both the anti-angiogenic agent pigment epithelium-derived factor (PEDF) (Bernard et al., 2009) and the pro-survival pro-angiogenic midkine protein, which may induce nuclear localization of LAMR (Salama et al., 2001).
Functionally, it seems LAMR may play a role in MAPK signalling and participate in events occurring within focal adhesions. This may translate to downstream processes promoting growth and survival as well as the more specific responses that have been described. Difficulty arises in piecing together an accurate understanding of how LAMR mediates or otherwise participates in these events, as there is considerable confusion about the relevant species and locations of LAMR involved, the transduction mechanism, and the potential need for membrane spanning in the explanation of some effects.
IV. STRUCTURE AND IMPLICATIONS
The crystal structure of human LAMR37(1–220) has been solved at resolution of 2.15 Å; amino acids 9–205 were resolved (Jamieson et al., 2008). The structure represents the RPS2 family domain, homologous in sequence and structure to prokaryotic ribosomal s2 proteins. This N-terminal domain (NTD, 1–209) is globular, consisting of a central β-sheet flanked by α-helices, while the C-terminal region (CTR, 210–295) is considered to be intrinsically disordered as demonstrated by circular dichroism spectrometry and several analytical folding assessment methods (Ould-Abeih et al., 2012; Schlessinger et al., 2009). The CTR is also highly negatively charged, incorporating a large content of glutamic and aspartic acid residues (>20%, net −16 charges) particularly concentrated in the 205–229 area suspected to be a laminin binding region (net −5 charges).
For discussion, we have built a hypothetical model of the full-length protein (Fig. 2A) based on known structure, homology modelling, ab initio folding and experimental data (Roy, Kucukural & Zhang, 2010; Xu & Zhang, 2012; Zhang, Liang & Zhang, 2011).
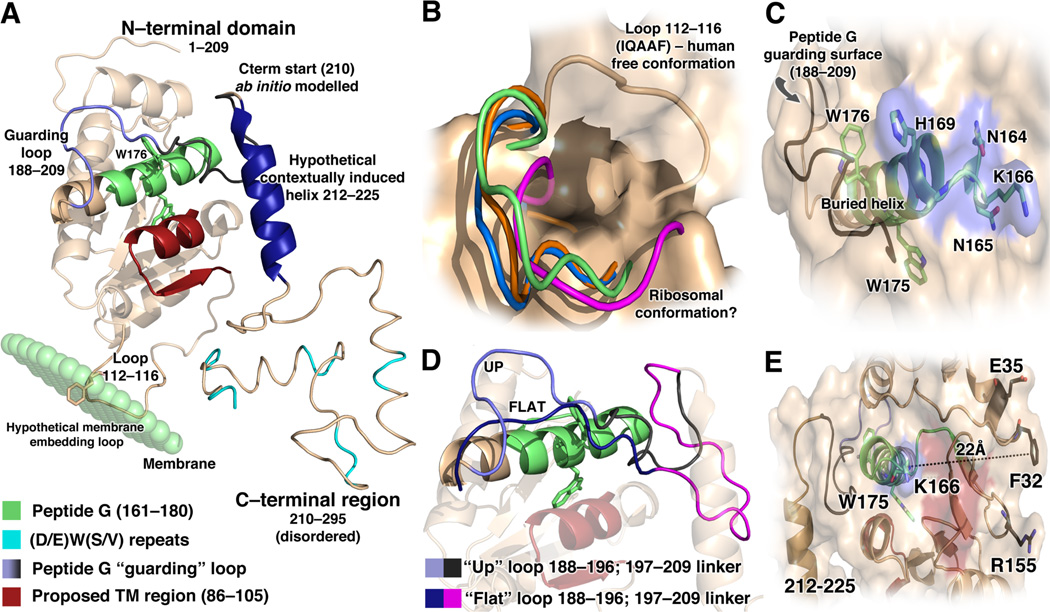
Figure 2.
Model of full-length 37-kDa laminin receptor. (A) Model of LAMR 1–295 built from the structures of human LAMR9–205 (PDB: 3BCH) and T. aestivum RPSA (PDB: 3IZ6) using a combination of homology threading and ab initio folding. The model depicts three proposed laminin binding regions: Peptide G (green), intrinsically disordered Peptide 205–229 in a hypothetical laminin-induced helical structure (blue), and five disordered DWS repeats (cyan). A proposed transmembrane (TM) region is shown packed up against the core of the structure in a conformation not indicative of a transmembrane helix (red). A speculative membrane-embedding mechanism via a hydrophobic loop is also suggested. (B) Structural divergence of the 112–116 loop in human LAMR/RPSA (beige, 3BCH) versus archaea rps2P (green, 1VI6). The ribosomal context conformations of the same loop in human (orange, 3J3A), wheat (magenta, 3IZ6) and Tetrahymena thermophila (blue, 4BPE) RPSA are also shown. Yeast (3U5B) and fly (3J38) ribosomal structures demonstrate loops structurally homologous to ribosomal human RPSA. (C) The buried Peptide G helix shown in green with surface-exposed residues N164–H169 indicated by blue surface overlay. Peptide G guarding loop in black. (D) Highlight of the 188–196 loop in the “up” (light blue) or “flat” (dark blue) conformations. The 197–209 linker could also adopt different conformations: a small turn (black) or an extended protruded loop (magenta). (E) A fourth proposed laminin binding site composed of Phe-32, Glu-35 and Arg-155 shown relative to Peptide G (22 Å from Lys-166 side chain) and a theoretical laminin binding helix (212–225). A hypothetical laminin binding surface is shaded in red.
(1) Membrane association, structural divergence and localization
By examination of the LAMR structure, we can recognize that the proposed transmembrane domain residing approximately within residues 86–105 (Rao et al., 1989) is comprised of an α-helical stretch packed against the central structure and a β-strand participating in the core β-sheet (Fig. 2A). Any conformational change that would occur to enable membrane spanning would likely have drastic consequences for the folding of the protein. A transmembrane LAMR is thus properly considered implausible.
Notably, residues 112–116 (IQAAF) form an outward projected loop with significant hydrophobic burden, unusual for solvent-accessible regions (Fig. 2A,B). This loop is structurally divergent from the corresponding region of the archaeal homolog rps2P, in which it is pinned against the globular core (PDB: 1VI6) and from bacteria (PDB: 2AVY) which possess a larger extended structure for rRNA binding (Badger et al., 2005; Brodersen et al., 2002; Schuwirth et al., 2005; Wimberly et al., 2000).
It is conceivable that hydrophobic loops could serve as membrane-inserted segments capable of attaching LAMR as a peripheral membrane protein. Whether loops such as that containing IQAAF could serve in such a capacity is unclear and highly speculative, as shallow membrane insertion depth would be expected (Lomize et al., 2012). Prokaryotic RPSA/LAMR homologs are purely intracellular and thus have no need for membrane insertion capability, suggesting that divergence in this region could have been an adaptive trait. The hydrophobic nature of the IQAAF loop, however, is not entirely conserved in all species with laminin expression (such as nematodes, IQKTF). As with many peripheral membrane proteins, additional modifications or contacts with other proteins may be required (see Section II.5).
Curiously, the 112–116 loop appears to be in an archaeal-like “pinned back” conformation when human LAMR is within the ribosomal context (PDB: 3J3A). This loop has the same feature in the ribosome structures of several eukaryotes as shown in Fig. 2B (Anger et al., 2013; Armache et al., 2013, 2010; Badger et al., 2005; Ben-Shem et al., 2011; Rabl et al., 2011; Weisser et al., 2013). Since RPSA homolog structures outside of the ribosomal context are not available for invertebrate laminin-expressing species, it is difficult to say if this loop adopts different conformations dependent on its environment. Such a capacity may explain the ability to sustain differentially localized LAMR populations. Admittedly, this loop deviation in the monomeric human structure may be an artifact of crystallization, as it is occupied by the equivalent site in a symmetric partner in the crystal packing. Structural artifacts introduced by resolution inadequacies must also be considered, as cryo-electron microscopy (EM) structures of metazoan ribosomes often use non-metazoan X-ray structures as homology models to fit structural features into EM density maps. Alternatively, this loop may represent a physiological dimerization interface or protein–protein interaction site. Such hypotheses can be tested empirically with simple mutagenesis/deletion experiments in cells or by using synthetic liposomes to determine if they represent a plausible mode of membrane attachment and/or protein interaction (e.g. with exoplasmic integrin segments). Mutation of this region did not dramatically affect the laminin binding of purified LAMR37(1–220) (Jamieson et al., 2011).
(2) Peptide G and its proximal “guarding” loop
Unfortunately, only two of the four proposed laminin binding sites are represented in the available crystal structures. The leading putative laminin binding region, Peptide G (a.a. 161–180), and the palindrome it contains (LMWWML) is buried beneath the protein surface (Fig. 2C). Only Lys-166 and His-169 are reasonably exposed to solvent (>50% surface area). Small portions of residues 164–168 are partially exposed. The consecutive tryptophan residues are buried as well, projecting in opposite directions – Trp-175 towards the protein interior and Trp-176 towards the exterior. Considering its placement within the structure, then, Peptide G becomes an improbable interaction site.
Mutations to this region commonly result in improperly folded recombinant proteins, making confirmation studies difficult. Mutations of surface-exposed Lys-166 only minimally affect laminin binding in LAMR37(1–220) (Jamieson et al., 2011). Trp-175 mutation did not produce protein while a W176A mutant has been purified, although its laminin binding activity is unknown (Zidane et al., 2013).
There is emerging evidence that conformational changes may expose a segment of Peptide G and possibly free it for ligand binding. These conformational changes involve the shift of a segment packed against the Peptide G helix (a.a. 197–209) and a proximal loop (a.a. 188–196, Fig. 2A,D). In molecular dynamics simulations, this “guarding” loop exhibits flexibility, possibly allowing it to flip out from the structure, an event that may further expose Peptide G residues – specifically Arg-180 (Di Giovanni, Grottesi & Lavecchia, 2012).
Further support for conformational shifting was gathered using a combination of techniques to assess folding states of LAMR37. In this analysis, LAMR37 was interpreted to have alternate conformations at biological temperatures (i.e. 37°C) that may not have been reflected in the crystal structure (crystals grown at 17°C). More than half of the LAMR37 population could be in an intermediate state between fully ordered (i.e. crystal structures) and denatured (Ould-Abeih et al., 2012). It is conceivable that true physiological events expose the palindrome for laminin binding by releasing the guarding loop from its interactions with Peptide G.
With the available data, it is not clear how substantial structural changes may be or what region(s) they affect, but the presence and ratio of multiple conformers could explain the diversity of LAMR functions. The equilibrium between different conformers may be skewed toward one species depending on sub-cellular localization or by stabilization of one state by protein–protein binding.
Examination of the aforementioned structures for the LAMR/RPSA homologs of yeast, Tetrahymena, D. melanogaster and T. aestivum as well as several archaea and bacteria hints at variable conformations of the region that guards the Peptide G helix as well. These structures are solved in the ribosomal context, where the flexible 188–196 loop appears to be either stretched out linearly across the posterior helix (“flat”) or gathered as in the free human monomer (“up”). Peptide G is still buried in both scenarios (Fig. 2D).
While archaea and bacteria lack the human-like “up” conformation, it appears as if several eukaryotes may have the pinched loop “up” structure, though there is a limited data set on which to base this comparison. Since non-laminin-expressing tetrahymenidae and yeast also display the “up” loop conformation, its contribution to laminin binding is unclear, though both these groups do have reports of laminin binding capabilities (Furtado et al., 1992; Lopez-Ribot et al., 1994; Narasimhan et al., 1994; Sepulveda et al., 1996). Ribosomal context does not seem to influence the conformation of this region, and in humans, mutation of the guarding loop does not directly affect laminin binding in vitro (Jamieson et al., 2011).
Ultimately, the significance of Peptide G and possible conformational changes to its guarding loop for both laminin binding and other physiological events remains to be experimentally demonstrated.
(3) The Phe-32, Glu-35 and Arg-155 binding face
The second proposed laminin binding site with available structural information is the protein face presented by the structurally proximal Phe-32, Glu-35, Arg-155 residues (FER). Here, there are no obvious physiochemical features or structural grooves indicative of protein binding, but protein–protein interactions can be confined to small surface area contacts. These residues are not conserved by non-laminin binding LAMR/RPSA homologs (Jamieson et al., 2011). Mutations in this region drastically affect laminin binding of the truncated LAMR37(1–220), which also contains Peptide G and possibly the 205–229 receptor site (or a portion). It is not clear, however, how these mutations affect the binding of the full-length protein, which supposedly contains additional binding sites. Interestingly, the FER motif is near surface-exposed Peptide G residues and the model-predicted position of the 205–229 receptor site (Fig. 2E).
(4) The C-terminal tail, DWS repeats and peptide 205–229
The CTR of LAMR and the 5 (D/E)W(S/T) repeats themselves do not seem to adopt significant secondary structure (Ould-Abeih et al., 2012; Schlessinger et al., 2009). Indeed, it is not clear if any ordered regions exist after residue 205 in the free protein’s native conformation – residues 206–220 are not resolved in the crystal structure. Secondary structure in the CTR could conceivably be contextually induced under certain conditions, however (e.g. upon laminin or ribosome binding).
There is some evidence suggesting the NTD and CTR may weakly interact, imparting some order to the random coils of the CTR. Additional α-helicity is reported in the full-length protein over the summated secondary structures of the separated constructs (Ould-Abeih et al., 2012). Indeed, NTD and CTR unification appears to translate to small differences in strength of laminin binding and interaction capacity with other substrates (Fatehullah et al., 2010; Zidane et al., 2013). Intrinsically disordered protein termini (“tails”) frequently contribute to protein interactions by adopting structure in ligand-bound contexts (Uversky, 2013), although fold-upon-binding behaviour is not always necessary for the activity of such proteins (Sigalov, 2011).
The availability of a de novo model of the wheat RPSA C-terminal region fit to a cryo-EM map of the T. aestivum ribosome (taRPSA, PDB: 3IZ6) allowed us to virtually construct the human LAMR CTR210–295 and estimate its position relative to the NTD for representation in our model (Fig. 2A). This model and secondary structure prediction suggests α-helical structure could arise within Peptide 205–229, a putative laminin binding region. This theoretical helix is draped perpendicularly to the anterior face of the linker segment guarding Peptide G and has long been thought to adopt a helical structure (Landowski et al., 1995b). Whether this model accurately represents human LAMR, however, is unclear given that its basis, the wheat RPSA structure, is a non-laminin binding protein in the ribosomal context.
The origins of the LAMR CTR are unknown, but the presence of this region and any function associated with it, was clearly subsequent to the genesis of the ribosomal RPS2 domain represented by the NTD. It is tempting to attribute the acquisition of laminin binding capability with evolutionary gain-of-function arising from the CTR. Indeed, given that LAMR regions 210–295 and 225–295 have independent laminin binding activity, this may be the case. Thus, the C-terminal tail of LAMR is likely a pivotal laminin binding component that may also have implications for localization and nucleic acid binding (e.g. ribosome, membrane, chromatin, RNA). And yet, the isolated NTD (2–209) also binds laminin with similar affinity (Zidane et al., 2013, see Section III.4).
Given the revelations from the crystal structure and the apparent complexities inherent to interpretation of the data available for the laminin binding sites, it seems prudent to conclude the precise nature of the interaction with laminin is poorly understood. The most outstanding questions awaiting clarification are surely: (1) which of the four proposed laminin binding sites can be physiologically verified, and (2) given the presence of multiple binding sites, why can isolated NTD and CTR constructs engage laminin independently and at affinities not dissimilar to the full-length protein?
Based on evidence that truncated proteins and synthetic peptides may behave abnormally, it is necessary to consider the true laminin binding site may not be a purely linear peptide in nature. Binding to a multi-partite face of the N-terminal domain that is subsequently stabilized by involvement of the CTR (or vice versa) represents a promising new explanatory paradigm for laminin interaction. Further research is necessary to clarify this issue with attention paid to using physiologically relevant constructs and conditions. Co-crystal structures or chemical crosslinking studies with laminin peptides – laminin in whole may be too challenging a molecule – and/or structural nuclear magnetic resonance (NMR) data addressing conformational changes would be enormously helpful in resolving the mechanism of laminin engagement by LAMR.
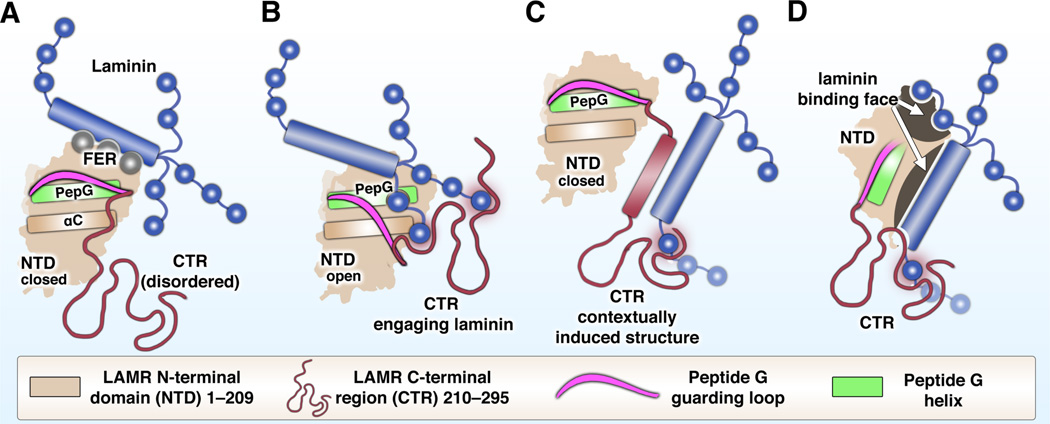
Fig. 3 attempts to summarize the suggested modes of LAMR:laminin interaction.
Figure 3.
LAMR interaction with laminin. Several theoretical modes of LAMR:laminin binding are proposed for discussion. (A) LAMR interaction with laminin through the N-terminal domain (NTD) only. Binding is via the Phe-32, Glu-35, Arg-155 (FER) motif while Peptide G (PepG, green) remains buried by a guarding loop in the “closed” position (magenta) above the C-helix (αC). (B) Guarding loop in the theorized “open” position, allowing laminin interaction with previously buried Peptide G. The C-terminal region (CTR, red) could also cooperatively engage laminin while remaining unfolded. (C) Laminin interaction through the CTR exclusively. The CTR could hypothetically adopt secondary structure in the laminin-bound context. (D) Laminin interaction via a non-linear multipartite protein face on the NTD with involvement of the CTR. The representations of laminin (blue) are not meant to imply particular interaction sites and are not to scale.
V. DISEASE AND THERAPY
(1) Cancer
LAMR is well known to be involved in cancer, where its functions in promoting adhesion and migration likely promote invasive and metastatic phenotypes. It is also possible that LAMR’s ribosomal functions support tumour progression. Indeed, there are many studies examining the expression of LAMR in samples of human cancers. Longstanding investigations examining the use of LAMR as a prognostic indicator in a large variety of both solid and haematological malignancies have compiled over 100 individual reports, although many are small data sets (<20–50 patients). Many of these studies have been reviewed previously (Menard et al., 1997, 1998; Montuori & Sobel, 1996). The larger studies (400+ patients) are focused on breast carcinomas, colorectal carcinomas and lymphomas (Carbone et al., 1995; Colnaghi, 1994; Martignone et al., 1993; Sanjuan et al., 1996; Tagliabue & Pastorino, 1996; Viacava et al., 1997), but gastric, ovarian, lung, thyroid, prostate, laryngeal and renal/adrenal tumours have also been investigated in addition to mesotheliomas, melanomas, leukaemias, astrocytomas, hepatocarcinomas, and the various metastatic sites of these cancers.
It is beyond the scope of this review to attempt a meta-analysis of the available clinical data, but the general trend appears to support a correlation between higher LAMR expression and several disease features. Decreased overall survival, poorer disease-free progression, advanced tumour stage, lesser extent of tumour differentiation, increased metastatic risk and lymph node involvement, higher intratumoral microvessel density, increased presence of invasive tumour fronts and changes in tumour responsiveness to therapy have all been linked to higher LAMR expression (either by mRNA or protein levels in histological sections). It is important to consider that a significant number of studies have found no correlation of LAMR with the previously listed indicators, and that there is likely to be variation of involvement depending on the particular malignancy.
The field awaits a large, prospective study adequately controlled for multiple comparisons, which have been weaknesses in the retrospective studies available to date. Description of membrane versus overall levels may also prove important in using LAMR as a predictive marker of disease course and outcome.
(2) Bacterial, viral and parasite infection
Several meningitis-causing bacteria have been implicated in using LAMR as a target of bacterial surface adhesins. Among them, Streptococcus pneumoniae, Neisseria meningitidis, Haemophilus influenzae and E. coli have been investigated to determine the nature of the tropism (Abouseada et al., 2012; Chung et al., 2003; K.J. Kim et al., 2005; Orihuela et al., 2009). The best-studied system is E. coli interaction via cytotoxic necrotizing factor 1 (CNF1) binding to LAMR (Chung et al., 2003; K.J. Kim et al., 2005). This interaction appears to trigger cytoskeletal rearrangements conducive to bacteria internalization.
Infection has also been shown to be partly mediated by LAMR for viruses capable of using the protein in a receptor capacity. Sindbis, Venezuelan equine encephalitis, Dengue, Adeno-associated, West Nile, Tick-borne encephalitis and Japanese encephalitis viruses have been reported to have LAMR tropism (Akache et al., 2006; Bogachek et al., 2008; Ludwig, Kondig & Smith, 1996; Strauss et al., 1994; Thepparit & Smith, 2004; Thongtan et al., 2012; Wang et al., 1992).
A single report demonstrates the parasitic protozoan Toxoplasma gondii may use LAMR to enhance laminin-mediated cell surface interactions (Furtado et al., 1992).
Importantly, it is acknowledged that LAMR likely does not represent a singular receptor for these infectious species, as many others have been described. LAMR may not be strictly necessary for pathogenesis either, but its presence in lipid rafts may be involved in tropism or enhancing primary receptor interactions (Simons & Ehehalt, 2002). Indeed, for pathogen infectivity as well as for the pathogenic agents described below, it is important to consider that blockade of cell-surface LAMR may be altering cellular uptake mechanisms in a broader context and not necessarily in a ligand-specific manner.
(3) Prion and neurodegenerative diseases
Prions are recently described infectious agents responsible for transmissible spongiform encephalopathies that appear to co-opt LAMR as part of a mechanism of cellular internalization and propagation. This came to light when LAMR was proposed to interact with the PrPc protein, a normally functioning endogenous molecule that can be converted to a protease-resistant, misfolded prion form (PrPSc) (Gauczynski et al., 2001; Rieger et al., 1997; Simoneau et al., 2003). A similar circumstance exists for the yeast Sup35 prion-forming protein (Pampeno, Derkatch & Meruelo, 2014).
The PrPSc prion form is a pathological molecule in bovine spongiform encephalopathy in cattle (BSE, mad cow disease) and Creutzfeldt-Jakob disease in humans (Rieger, Lasmezas & Weiss, 1999). The physiological relevance of LAMR-PrP binding is still being investigated. N-terminally truncated LAMR37(44–295) and smaller LAMR fragments with uncertain folding states have been used to probe the nature of the interaction, revealing similarities to laminin binding. Nonetheless, methods to modulate LAMR function or expression seem to interfere with prion infection and propagation (Gauczynski et al., 2006; Leucht et al., 2003; Zuber et al., 2008a, b).
Because of the commonalities of amyloid beta (Aβ), an etiological agent in Alzheimer’s Disease, with prion proteins and the association of PrPc in Aβ related pathology, it has been suggested LAMR may play a role in Alzheimer’s Disease (Da Costa Dias et al., 2011; Vana et al., 2009). The evidence for this is preliminary but mounting. LAMR has been found to contain irregular oxidative modifications in a small-scale proteomics investigation of the brains of Alzheimer’s model mice (Shin et al., 2004). Modulation of LAMR has also recently been reported to affect Aβ cytotoxicity (Da Costa Dias et al., 2013) and its release from cells (Jovanovic et al., 2013, 2014).
(4) Therapeutic approaches
Given the diverse roles LAMR plays in many key cellular processes, it is not surprising that this protein has been linked to pathology. LAMR’s overexpression in cancer and its receptor behaviour for pathological agents make it a tempting target for therapeutics.
Attempts to exploit LAMR for therapeutic purposes have been frequent endeavours that have met with various degrees of success. Several approaches have been taken. The first efforts attempted to use antibodies intended to block cell-surface LAMR binding to laminin. Indeed, antibodies used in this manner successfully reduce experimental metastasis formation of injected tumour cells in mice (Narumi et al., 1999). Similarly, synthetic peptides designed to mimic laminin (YIGSR peptides) have shown equivalent effects in the same experimental systems (Iwamoto et al., 1987; Nakai et al., 1992; Yamamura et al., 1993). This success led to efforts to derivatize YIGSR and related peptides into more effective forms (e.g. cyclic peptides, extended peptides, tripartite repeats, terminal modifications) and use for tumour targeting of liposomes (Kikkawa et al., 2013). Because YIGSR’s effects have not been conclusively demonstrated to work through LAMR, however, results should be examined with open mindedness for other targets (Maeda et al., 1994).
Other approaches attempting to treat malignancies have focused on a LAMR tumour-associated antigen referred to as oncofetal antigen immature laminin receptor protein (OFA-iLRP). This avenue of research led to tumour vaccine studies encouraging antibodies to develop against OFA-iLRP. The undertaking has produced promising results and recruitment for an early-phase clinical trial is in progress (Barsoum et al., 2009; Biragyn et al., 2007; Friedrichs et al., 2008). Oncolytic viruses carrying LAMR-silencing short hairpin RNA (shRNA) payloads have been used to similar effect (Scheiman et al., 2010b).
Knowledge of LAMR has also been applied in developing treatments for prion diseases (Omar et al., 2011; Sakaguchi, Ishibashi & Matsuda, 2009) and to advance tissue engineering (Gu & Masters, 2010).
Ultimately, small molecule inhibitors are often the most desirable therapeutic interventions. The green-tea-derived polyphenol, (−)-epigallocatechin-3-gallate (EGCG), is one such small molecule that has been reported to affect cells through LAMR67 (Tachibana et al., 2004). EGCG has also been reported to interact with LAMR37 (Zidane et al., 2013). EGCG is often described as having broad health-promoting benefits in a wide range of human pathologies (Chen et al., 2008; Mak, 2012). Unfortunately, EGCG suffers from low bioavailability with poor oral absorption as well as significant stability issues in plasma (Chow et al., 2003). This would suggest EGCG may better serve as a lead compound for aid in designing improved LAMR-targeted small molecules.
Numerous investigations into the mechanisms of LAMR-dependent EGCG actions have been conducted and point toward cytoskeletal involvement (Umeda, Tachibana & Yamada, 2005; Umeda et al., 2008a, b). Curiously, EGCG efficacy appears to be blocked by LAMR antibodies, which do not trigger the same effects, indicating that the polyphenol may be acting agonistically or allosterically. This observation also leads to the conclusion that EGCG may be acting on extracellular LAMR, leaving the upstream mechanism and the signal transduction process unclear. It is also noted that in many of these studies, the adverse effects of LAMR knockdown described herein do not seem to be observed. Because EGCG has been reported to influence a large number of cellular targets, using this compound as a tool for investigating LAMR functions and signal transduction pathways becomes complex and difficult, but the therapeutic implications are clear.
Strikingly, a screen to disrupt the KRS:LAMR interaction, a laminin-induced event that stabilizes the presence of LAMR67 at the plasma membrane (see Section II.5), has led to the discovery of a KRS-targeted small molecule with anti-metastatic activity (Kim et al., 2014).
VI. CONCLUSIONS
The 37/67-kDa laminin receptor/RPSA protein is a demonstrably important biological molecule with roles in some of the most fundamental cellular processes. Alongside its normal physiological functions, LAMR has been strongly linked to neoplastic diseases, bacterial and viral infection, developmental abnormalities and neurodegenerative conditions.
The nature of conversion of the 37-kDa LAMR protein to higher molecular weight species (53- and 67-kDa) remains poorly understood. SUMOylation is a candidate modification, but it is undetermined if other proteins contribute to multimerization. The functional purview of each protein species is similarly incompletely described.
The process by which LAMR is directed to cytosol, ribosomes, nucleus and the outer plasma membrane is unknown. Although ribosomal proteins are often multifunctional, it is unclear how the cell differentially distributes this protein and maintains the particular ratios necessary for homeostatic function.
Native laminin interaction most likely occurs on a protein face apparent only in the full structure of LAMR rather than at discrete linear segments. Binding could be a collaborative event between several binding sites and clearly involves engagement of the intrinsically disordered C-terminal tail. A similar situation may exist for laminin where, again, linear peptides (e.g. YIGSR) may not adequately capture native binding states.
Intracellular functions of LAMR include anchoring ribosomes to microtubules, processing of pre-rRNA, signalling at focal adhesions to laminin and yet undescribed roles in histone interactions and chromatin regulation.
Knowledge of LAMR extracellular functions is considerably more difficult to place into physiological context, especially as it concerns extra- to intracellular signal transduction. Although cell-surface LAMR appears to bind laminin, it is uncertain if this is solely in an adhesion capacity or if LAMR modulates migratory processes directly (i.e. exclusive of intracellular functions). In the probable absence of a transmembrane region, mechanistic progression of signal to response will be a required area of future research.
LAMR cellular biology reflects involvement of translational and transcriptional machinery as well as extracellular signals acting non-exclusively across several sub-cellular locations. This significantly complicates the dissection of the pathways relevant to discrete events. These broad and crucial functions are most likely operating in conjunction and, therefore, highly relevant for understanding the true contribution of LAMR to physiology and disease.
Acknowledgments
We thank Drs. Christine Pampeno, Lisa Venticinque, Tomer Granot and Jonathan Scheiman for critical reading of this manuscript and valuable input concerning its content. This work was supported by US Public Health grant CA100687 from the National Cancer Institute, National Institutes of Health and Department of Health and Human Services as well as Institutional National Research Service Award T32-CA009161 (Levy). Protein structure graphics were generated using the PyMOL Molecular Graphics System, Schrödinger, LLC.
Footnotes
Which has been published in final form at: http://dx.doi.org/10.1111/brv.12170. This article may be used for non-commercial purposes in accordance with Wiley Terms and Conditions for Self-Archiving.
REFERENCES
- Abe K, Murakami S, Mukae N, Mita T, Hashimoto Y, Isemura M, Shimo-Oka T, Ii I, Kimata K, Narumi K, Satoh K, Nukiwa T. Presence of atypical laminin on the surface of mouse Lewis lung carcinoma cells. Tohoku Journal of Experimental Medicine. 1996;180:33–44. doi: 10.1620/tjem.180.33. [DOI] [PubMed] [Google Scholar]
- Abouseada NM, Assafi MS, Mahdavi J, Oldfield NJ, Wheldon LM, Wooldridge KG, Ala'Aldeen DA. Mapping the laminin receptor binding domains of Neisseria meningitidis PorA and Haemophilus influenzae OmpP2. PLoS One. 2012;7:e46233. doi: 10.1371/journal.pone.0046233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Akache B, Grimm D, Pandey K, Yant SR, Xu H, Kay MA. The 37/67-kilodalton laminin receptor is a receptor for adeno-associated virus serotypes 8, 2, 3, and 9. Journal of Virology. 2006;80:9831–9836. doi: 10.1128/JVI.00878-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amano T, Fu L, Marshak A, Kwak O, Shi YB. Spatio-temporal regulation and cleavage by matrix metalloproteinase stromelysin-3 implicate a role for laminin receptor in intestinal remodeling during Xenopus laevis metamorphosis. Developmental Dynamics. 2005a;234:190–200. doi: 10.1002/dvdy.20511. [DOI] [PubMed] [Google Scholar]
- Amano T, Kwak O, Fu L, Marshak A, Shi YB. The matrix metalloproteinase stromelysin-3 cleaves laminin receptor at two distinct sites between the transmembrane domain and laminin binding sequence within the extracellular domain. Cell Research. 2005b;15:150–159. doi: 10.1038/sj.cr.7290280. [DOI] [PubMed] [Google Scholar]
- Ando K, Miyazaki Y, Sawayama Y, Tominaga S, Matsuo E, Yamasaki R, Inoue Y, Iwanaga M, Imanishi D, Tsushima H, Fukushima T, Imaizumi Y, Taguchi J, Yoshida S, Hata T, Tomonaga M. High expression of 67-kDa laminin receptor relates to the proliferation of leukemia cells and increases expression of GM-CSF receptor. Experimental Hematology. 2011;39:179–186. e4. doi: 10.1016/j.exphem.2010.11.001. [DOI] [PubMed] [Google Scholar]
- Anger AM, Armache JP, Berninghausen O, Habeck M, Subklewe M, Wilson DN, Beckmann R. Structures of the human and Drosophila 80S ribosome. Nature. 2013;497:80–85. doi: 10.1038/nature12104. [DOI] [PubMed] [Google Scholar]
- Anilkumar N, Sudhakaran PR. Isolation and characterization of laminin binding protein from regenerating rat liver plasma membrane. Biochemistry and Molecular Biology International. 1993;31:201–209. [PubMed] [Google Scholar]
- Annabi B, Currie JC, Moghrabi A, Beliveau R. Inhibition of HuR and MMP-9 expression in macrophage-differentiated HL-60 myeloid leukemia cells by green tea polyphenol EGCg. Leukemia Research. 2007;31:1277–1284. doi: 10.1016/j.leukres.2006.10.001. [DOI] [PubMed] [Google Scholar]
- Ardini E, Pesole G, Tagliabue E, Magnifico A, Castronovo V, Sobel ME, Colnaghi MI, Menard S. The 67-kDa laminin receptor originated from a ribosomal protein that acquired a dual function during evolution. Molecular Biology and Evolution. 1998;15:1017–1025. doi: 10.1093/oxfordjournals.molbev.a026000. [DOI] [PubMed] [Google Scholar]
- Ardini E, Sporchia B, Pollegioni L, Modugno M, Ghirelli C, Castiglioni F, Tagliabue E, Menard S. Identification of a novel function for 67-kDa laminin receptor: increase in laminin degradation rate and release of motility fragments. Cancer Research. 2002;62:1321–1325. [PubMed] [Google Scholar]
- Ardini E, Tagliabue E, Magnifico A, Buto S, Castronovo V, Colnaghi MI, Menard S. Co-regulation and physical association of the 67-kDa monomeric laminin receptor and the alpha6beta4 integrin. Journal of Biological Chemistry. 1997;272:2342–2345. doi: 10.1074/jbc.272.4.2342. [DOI] [PubMed] [Google Scholar]
- Armache JP, Anger AM, Marquez V, Franckenberg S, Frohlich T, Villa E, Berninghausen O, Thomm M, Arnold GJ, Beckmann R, Wilson DN. Promiscuous behaviour of archaeal ribosomal proteins: implications for eukaryotic ribosome evolution. Nucleic Acids Research. 2013;41:1284–1293. doi: 10.1093/nar/gks1259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Armache JP, Jarasch A, Anger AM, Villa E, Becker T, Bhushan S, Jossinet F, Habeck M, Dindar G, Franckenberg S, Marquez V, Mielke T, Thomm M, Berninghausen O, Beatrix B, Soding J, Westhof E, Wilson DN, Beckmann R. Localization of eukaryote-specific ribosomal proteins in a 5.5-A cryo-EM map of the 80S eukaryotic ribosome. Proceedings of the National Academy of Sciences of the United States of America. 2010;107:19754–19759. doi: 10.1073/pnas.1010005107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Asano Y, Takashima S, Asakura M, Shintani Y, Liao Y, Minamino T, Asanuma H, Sanada S, Kim J, Ogai A, Fukushima T, Oikawa Y, Okazaki Y, Kaneda Y, Sato M, Miyazaki J, Kitamura S, Tomoike H, Kitakaze M, Hori M. Lamr1 functional retroposon causes right ventricular dysplasia in mice. Nature Genetics. 2004;36:123–130. doi: 10.1038/ng1294. [DOI] [PubMed] [Google Scholar]
- Aumailley M. The laminin family. Cell Adhesion & Migration. 2013;7:48–55. doi: 10.4161/cam.22826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aumailley M, Bruckner-Tuderman L, Carter WG, Deutzmann R, Edgar D, Ekblom P, Engel J, Engvall E, Hohenester E, Jones JC, Kleinman HK, Marinkovich MP, Martin GR, Mayer U, Meneguzzi G, Miner JH, Miyazaki K, Patarroyo M, Paulsson M, Quaranta V, Sanes JR, Sasaki T, Sekiguchi K, Sorokin LM, Talts JF, Tryggvason K, Uitto J, Virtanen I, von der Mark K, Wewer UM, Yamada Y, Yurchenco PD. A simplified laminin nomenclature. Matrix Biology. 2005;24:326–332. doi: 10.1016/j.matbio.2005.05.006. [DOI] [PubMed] [Google Scholar]
- Auth D, Brawerman G. A 33-kDa polypeptide with homology to the laminin receptor: component of translation machinery. Proceedings of the National Academy of Sciences of the United States of America. 1992;89:4368–4372. doi: 10.1073/pnas.89.10.4368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Badger J, Sauder JM, Adams JM, Antonysamy S, Bain K, Bergseid MG, Buchanan SG, Buchanan MD, Batiyenko Y, Christopher JA, Emtage S, Eroshkina A, Feil I, Furlong EB, Gajiwala KS, Gao X, He D, Hendle J, Huber A, Hoda K, Kearins P, Kissinger C, Laubert B, Lewis HA, Lin J, Loomis K, Lorimer D, Louie G, Maletic M, Marsh CD, Miller I, Molinari J, Muller-Dieckmann HJ, Newman JM, Noland BW, Pagarigan B, Park F, Peat TS, Post KW, Radojicic S, Ramos A, Romero R, Rutter ME, Sanderson WE, Schwinn KD, Tresser J, Winhoven J, Wright TA, Wu L, Xu J, Harris TJ. Structural analysis of a set of proteins resulting from a bacterial genomics project. Proteins. 2005;60:787–796. doi: 10.1002/prot.20541. [DOI] [PubMed] [Google Scholar]
- Balasubramanian S, Zheng D, Liu YJ, Fang G, Frankish A, Carriero N, Robilotto R, Cayting P, Gerstein M. Comparative analysis of processed ribosomal protein pseudogenes in four mammalian genomes. Genome Biology. 2009;10:R2. doi: 10.1186/gb-2009-10-1-r2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baloui H, Stettler O, Weiss S, Nothias F, von Boxberg Y. Upregulation in rat spinal cord microglia of the nonintegrin laminin receptor 37 kDa-LRP following activation by a traumatic lesion or peripheral injury. Journal of Neurotrauma. 2009;26:195–207. doi: 10.1089/neu.2008.0677. [DOI] [PubMed] [Google Scholar]
- Bao ZZ, Muschler J, Horwitz AF. LBL, a novel, developmentally regulated, laminin-binding lectin. Journal of Biological Chemistry. 1992;267:4974–4980. [PubMed] [Google Scholar]
- Barsoum AL, Liu B, Rohrer JW, Coggin JH, Jr, Tucker JA, Pannell LK, Schwarzenberger PO. Production, safety and antitumor efficacy of recombinant Oncofetal Antigen/immature laminin receptor protein. Biomaterials. 2009;30:3091–3099. doi: 10.1016/j.biomaterials.2009.02.022. [DOI] [PubMed] [Google Scholar]
- Basson CT, Knowles WJ, Bell L, Albelda SM, Castronovo V, Liotta LA, Madri JA. Spatiotemporal segregation of endothelial cell integrin and nonintegrin extracellular matrix-binding proteins during adhesion events. Journal of Cell Biology. 1990;110:789–801. doi: 10.1083/jcb.110.3.789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ben-Shem A, Garreau de Loubresse N, Melnikov S, Jenner L, Yusupova G, Yusupov M. The structure of the eukaryotic ribosome at 3.0 A resolution. Science. 2011;334:1524–1529. doi: 10.1126/science.1212642. [DOI] [PubMed] [Google Scholar]
- Bernard A, Gao-Li J, Franco CA, Bouceba T, Huet A, Li Z. Laminin receptor involvement in the anti-angiogenic activity of pigment epithelium-derived factor. Journal of Biological Chemistry. 2009;284:10480–10490. doi: 10.1074/jbc.M809259200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berno V, Porrini D, Castiglioni F, Campiglio M, Casalini P, Pupa SM, Balsari A, Menard S, Tagliabue E. The 67 kDa laminin receptor increases tumor aggressiveness by remodeling laminin-1. Endocrine Related Cancer. 2005;12:393–406. doi: 10.1677/erc.1.00870. [DOI] [PubMed] [Google Scholar]
- Biragyn A, Schiavo R, Olkhanud P, Sumitomo K, King A, McCain M, Indig FE, Almanzar G, Baatar D. Tumor-associated embryonic antigen-expressing vaccines that target CCR6 elicit potent CD8+ T cell-mediated protective and therapeutic antitumor immunity. Journal of Immunology. 2007;179:1381–1388. doi: 10.4049/jimmunol.179.2.1381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blomster HA, Hietakangas V, Wu J, Kouvonen P, Hautaniemi S, Sistonen L. Novel proteomics strategy brings insight into the prevalence of SUMO-2 target sites. Molecular and Cellular Proteomics. 2009;8:1382–1390. doi: 10.1074/mcp.M800551-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bogachek MV, Protopopova EV, Loktev VB, Zaitsev BN, Favre M, Sekatskii SK, Dietler G. Immunochemical and single molecule force spectroscopy studies of specific interaction between the laminin binding protein and the West Nile virus surface glycoprotein E domain II. Journal of Molecular Recognition. 2008;21:55–62. doi: 10.1002/jmr.866. [DOI] [PubMed] [Google Scholar]
- Bolze A, Mahlaoui N, Byun M, Turner B, Trede N, Ellis SR, Abhyankar A, Itan Y, Patin E, Brebner S, Sackstein P, Puel A, Picard C, Abel L, Quintana-Murci L, Faust SN, Williams AP, Baretto R, Duddridge M, Kini U, Pollard AJ, Gaud C, Frange P, Orbach D, Emile JF, Stephan JL, Sorensen R, Plebani A, Hammarstrom L, Conley ME, Selleri L, Casanova JL. Ribosomal protein SA haploinsufficiency in humans with isolated congenital asplenia. Science. 2013;340:976–978. doi: 10.1126/science.1234864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brodersen DE, Clemons WM, Jr, Carter AP, Wimberly BT, Ramakrishnan V. Crystal structure of the 30 S ribosomal subunit from Thermus thermophilus: structure of the proteins and their interactions with 16 S RNA. Journal of Molecular Biology. 2002;316:725–768. doi: 10.1006/jmbi.2001.5359. [DOI] [PubMed] [Google Scholar]
- Brown SS, Malinoff HL, Wicha MS. Connectin: cell surface protein that binds both laminin and actin. Proceedings of the National Academy of Sciences of the United States of America. 1983;80:5927–5930. doi: 10.1073/pnas.80.19.5927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bryant G, Haberern C, Rao CN, Liotta LA. Butyrate induced reduction of tumor cell laminin receptors. Cancer Research. 1986;46:807–811. [PubMed] [Google Scholar]
- Bushkin-Harav I, Garty NB, Littauer UZ. Down-regulation of a 67-kDa YIGSR-binding protein upon differentiation of human neuroblastoma cells. Journal of Biological Chemistry. 1995;270:13422–13428. doi: 10.1074/jbc.270.22.13422. [DOI] [PubMed] [Google Scholar]
- Buto S, Ghirelli C, Aiello P, Tagliabue E, Ardini E, Magnifico A, Montuori N, Sobel ME, Colnaghi MI, Menard S. Production and characterization of monoclonal antibodies directed against the laminin receptor precursor. International Journal of Biological Markers. 1997;12:1–5. doi: 10.1177/172460089701200101. [DOI] [PubMed] [Google Scholar]
- Buto S, Tagliabue E, Ardini E, Magnifico A, Ghirelli C, van den Brule F, Castronovo V, Colnaghi MI, Sobel ME, Menard S. Formation of the 67-kDa laminin receptor by acylation of the precursor. Journal of Cellular Biochemistry. 1998;69:244–251. doi: 10.1002/(sici)1097-4644(19980601)69:3<244::aid-jcb2>3.0.co;2-r. [DOI] [PubMed] [Google Scholar]
- Carbone A, Gloghini A, Colombatti A, Castronovo V, Menard S. Expression of the monomeric 67-kd laminin-binding protein in human lymphomas as defined by MLuC5 monoclonal antibody and paraffin section immunohistochemistry. Human Pathology. 1995;26:541–546. doi: 10.1016/0046-8177(95)90251-1. [DOI] [PubMed] [Google Scholar]
- Castronovo V, Claysmith AP, Barker KT, Cioce V, Krutzsch HC, Sobel ME. Biosynthesis of the 67 kDa high affinity laminin receptor. Biochemical and Biophysical Research Communications. 1991a;177:177–183. doi: 10.1016/0006-291x(91)91965-f. [DOI] [PubMed] [Google Scholar]
- Castronovo V, Luyten F, van den Brule F, Sobel ME. Identification of a 14-kDa laminin binding protein (HLBP14) in human melanoma cells that is identical to the 14-kDa galactoside binding lectin. Archives of Biochemistry and Biophysics. 1992;297:132–138. doi: 10.1016/0003-9861(92)90650-l. [DOI] [PubMed] [Google Scholar]
- Castronovo V, Taraboletti G, Liotta LA, Sobel ME. Modulation of laminin receptor expression by estrogen and progestins in human breast cancer cell lines. Journal of the National Cancer Institute. 1989;81:781–788. doi: 10.1093/jnci/81.10.781. [DOI] [PubMed] [Google Scholar]
- Castronovo V, Taraboletti G, Sobel ME. Functional domains of the 67-kDa laminin receptor precursor. Journal of Biological Chemistry. 1991b;266:20440–20446. [PubMed] [Google Scholar]
- Chen A, Ganor Y, Rahimipour S, Ben-Aroya N, Koch Y, Levite M. The neuropeptides GnRH-II and GnRH-I are produced by human T cells and trigger laminin receptor gene expression, adhesion, chemotaxis and homing to specific organs. Nature Medicine. 2002;8:1421–1426. doi: 10.1038/nm1202-801. [DOI] [PubMed] [Google Scholar]
- Chen D, Milacic V, Chen MS, Wan SB, Lam WH, Huo C, Landis-Piwowar KR, Cui QC, Wali A, Chan TH, Dou QP. Tea polyphenols, their biological effects and potential molecular targets. Histology and Histopathology. 2008;23:487–496. doi: 10.14670/hh-23.487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen FX, Qian YR, Duan YH, Ren WW, Yang Y, Zhang CC, Qiu YM, Ji YH. Down-regulation of 67LR reduces the migratory activity of human glioma cells in vitro. Brain Research Bulletin. 2009;79:402–408. doi: 10.1016/j.brainresbull.2009.04.019. [DOI] [PubMed] [Google Scholar]
- Chen J, Carcamo JM, Borquez-Ojeda O, Erdjument-Bromage H, Tempst P, Golde DW. The laminin receptor modulates granulocyte-macrophage colony-stimulating factor receptor complex formation and modulates its signaling. Proceedings of the National Academy of Sciences of the United States of America. 2003;100:14000–14005. doi: 10.1073/pnas.2334584100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chow HH, Cai Y, Hakim IA, Crowell JA, Shahi F, Brooks CA, Dorr RT, Hara Y, Alberts DS. Pharmacokinetics and safety of green tea polyphenols after multiple-dose administration of epigallocatechin gallate and polyphenon E in healthy individuals. Clinical Cancer Research. 2003;9:3312–3319. [PubMed] [Google Scholar]
- Chung JW, Hong SJ, Kim KJ, Goti D, Stins MF, Shin S, Dawson VL, Dawson TM, Kim KS. 37-kDa laminin receptor precursor modulates cytotoxic necrotizing factor 1-mediated RhoA activation and bacterial uptake. Journal of Biological Chemistry. 2003;278:16857–16862. doi: 10.1074/jbc.M301028200. [DOI] [PubMed] [Google Scholar]
- Cioce V, Margulies IM, Sobel ME, Castronovo V. Interaction between the 67 kilodalton metastasis-associated laminin receptor and laminin. Kidney International. 1993;43:30–37. doi: 10.1038/ki.1993.7. [DOI] [PubMed] [Google Scholar]
- Clausse N, Jackers P, Jares P, Joris B, Sobel ME, Castronovo V. Identification of the active gene coding for the metastasis-associated 37LRP/p40 multifunctional protein. DNA and Cell Biology. 1996;15:1009–1023. doi: 10.1089/dna.1996.15.1009. [DOI] [PubMed] [Google Scholar]
- Clausse N, van den Brule F, Delvenne P, Jacobs N, Franzen-Detrooz E, Jackers P, Castronovo V. TNF-alpha and IFN-gamma down-regulate the expression of the metastasis-associated bi-functional 37LRP/p40 gene and protein in transformed keratinocytes. Biochemical and Biophysical Research Communications. 1998;251:564–569. doi: 10.1006/bbrc.1998.9431. [DOI] [PubMed] [Google Scholar]
- Clement B, Segui-Real B, Savagner P, Kleinman HK, Yamada Y. Hepatocyte attachment to laminin is mediated through multiple receptors. Journal of Cell Biology. 1990;110:185–192. doi: 10.1083/jcb.110.1.185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colnaghi MI. The simultaneous expression of c-erbB-2 oncoprotein and laminin receptor on primary breast tumors has a predicting potential analogous to that of the lymph node status. Advances in Experimental Medicine and Biology. 1994;353:149–154. doi: 10.1007/978-1-4615-2443-4_14. [DOI] [PubMed] [Google Scholar]
- Da Costa Dias B, Jovanovic K, Gonsalves D, Moodley K, Reusch U, Knackmuss S, Penny C, Weinberg MS, Little M, Weiss SF. Anti-LRP/LR specific antibody IgG1-iS18 and knock-down of LRP/LR by shRNAs rescue cells from Abeta42 induced cytotoxicity. Science Reports. 2013;3:2702. doi: 10.1038/srep02702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Da Costa Dias B, Jovanovic K, Gonsalves D, Weiss SF. Structural and mechanistic commonalities of amyloid-beta and the prion protein. Prion. 2011;5:126–137. doi: 10.4161/pri.5.3.17025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis CM, Papadopoulos V, Jia MC, Yamada Y, Kleinman HK, Dym M. Identification and partial characterization of laminin binding proteins in immature rat Sertoli cells. Experimental Cell Research. 1991;193:262–273. doi: 10.1016/0014-4827(91)90095-c. [DOI] [PubMed] [Google Scholar]
- Demianova M, Formosa TG, Ellis SR. Yeast proteins related to the p40/laminin receptor precursor are essential components of the 40 S ribosomal subunit. Journal of Biological Chemistry. 1996;271:11383–11391. doi: 10.1074/jbc.271.19.11383. [DOI] [PubMed] [Google Scholar]
- Di Giovanni C, Grottesi A, Lavecchia A. Conformational switch of a flexible loop in human laminin receptor determines laminin-1 interaction. European Biophysics Journal. 2012;41:353–358. doi: 10.1007/s00249-012-0793-9. [DOI] [PubMed] [Google Scholar]
- Douville PJ, Harvey WJ, Carbonetto S. Isolation and partial characterization of high affinity laminin receptors in neural cells. Journal of Biological Chemistry. 1988;263:14964–14969. [PubMed] [Google Scholar]
- Engvall E, Wewer UM. Domains of laminin. Journal of Cellular Biochemistry. 1996;61:493–501. doi: 10.1002/(SICI)1097-4644(19960616)61:4%3C493::AID-JCB2%3E3.0.CO;2-J. [DOI] [PubMed] [Google Scholar]
- Fatehullah A, Doherty C, Pivato G, Allen G, Devine L, Nelson J, Timson DJ. Interactions of the 67 kDa laminin receptor and its precursor with laminin. Bioscience Reports. 2010;30:73–79. doi: 10.1042/BSR20090023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ford CL, Randal-Whitis L, Ellis SR. Yeast proteins related to the p40/laminin receptor precursor are required for 20S ribosomal RNA processing and the maturation of 40S ribosomal subunits. Cancer Research. 1999;59:704–710. [PubMed] [Google Scholar]
- Formisano P, Ragno P, Pesapane A, Alfano D, Alberobello AT, Rea VE, Giusto R, Rossi FW, Beguinot F, Rossi G, Montuori N. PED/PEA-15 interacts with the 67 kD laminin receptor and regulates cell adhesion, migration, proliferation and apoptosis. Journal of Cellular and Molecular Medicine. 2012;16:1435–1446. doi: 10.1111/j.1582-4934.2011.01411.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friedrichs B, Siegel S, Kloess M, Barsoum A, Coggin J, Jr, Rohrer J, Jakob I, Tiemann M, Heidorn K, Schulte C, Kabelitz D, Steinmann J, Schmitz N, Zeis M. Humoral immune responses against the immature laminin receptor protein show prognostic significance in patients with chronic lymphocytic leukemia. Journal of Immunology. 2008;180:6374–6384. doi: 10.4049/jimmunol.180.9.6374. [DOI] [PubMed] [Google Scholar]
- Fromm-Dornieden C, von der Heyde S, Lytovchenko O, Salinas-Riester G, Brenig B, Beissbarth T, Baumgartner BG. Novel polysome messages and changes in translational activity appear after induction of adipogenesis in 3T3-L1 cells. BMC Molecular Biology. 2012;13:9. doi: 10.1186/1471-2199-13-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujimura Y, Yamada K, Tachibana H. A lipid raft-associated 67kDa laminin receptor mediates suppressive effect of epigallocatechin-3-O-gallate on FcepsilonRI expression. Biochemical and Biophysical Research Communications. 2005;336:674–681. doi: 10.1016/j.bbrc.2005.08.146. [DOI] [PubMed] [Google Scholar]
- Fulcher KD, Welch JE, Davis CM, O'Brien DA, Eddy EM. Characterization of laminin receptor messenger ribonucleic acid and protein expression in mouse spermatogenic cells. Biology of Reproduction. 1993;48:674–682. doi: 10.1095/biolreprod48.3.674. [DOI] [PubMed] [Google Scholar]
- Furtado GC, Slowik M, Kleinman HK, Joiner KA. Laminin enhances binding of Toxoplasma gondii tachyzoites to J774 murine macrophage cells. Infection and Immunity. 1992;60:2337–2342. doi: 10.1128/iai.60.6.2337-2342.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia-Hernandez M, Davies E, Baskin TI, Staswick PE. Association of Plant p40 Protein with Ribosomes Is Enhanced When Polyribosomes Form during Periods of Active Tissue Growth. Plant Physiology. 1996;111:559–568. doi: 10.1104/pp.111.2.559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gauczynski S, Nikles D, El-Gogo S, Papy-Garcia D, Rey C, Alban S, Barritault D, Lasmezas CI, Weiss S. The 37-kDa/67-kDa laminin receptor acts as a receptor for infectious prions and is inhibited by polysulfated glycanes. Journal of Infectious Diseases. 2006;194:702–709. doi: 10.1086/505914. [DOI] [PubMed] [Google Scholar]
- Gauczynski S, Peyrin JM, Haik S, Leucht C, Hundt C, Rieger R, Krasemann S, Deslys JP, Dormont D, Lasmezas CI, Weiss S. The 37-kDa/67-kDa laminin receptor acts as the cell-surface receptor for the cellular prion protein. EMBO Journal. 2001;20:5863–5875. doi: 10.1093/emboj/20.21.5863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Givant-Horwitz V, Davidson B, Reich R. Laminin-induced signaling in tumor cells: the role of the M(r) 67,000 laminin receptor. Cancer Research. 2004;64:3572–3579. doi: 10.1158/0008-5472.CAN-03-3424. [DOI] [PubMed] [Google Scholar]
- Golebiowski F, Matic I, Tatham MH, Cole C, Yin Y, Nakamura A, Cox J, Barton GJ, Mann M, Hay RT. System-wide changes to SUMO modifications in response to heat shock. Science Signaling. 2009;2:ra24. doi: 10.1126/scisignal.2000282. [DOI] [PubMed] [Google Scholar]
- Graf J, Iwamoto Y, Sasaki M, Martin GR, Kleinman HK, Robey FA, Yamada Y. Identification of an amino acid sequence in laminin mediating cell attachment, chemotaxis, and receptor binding. Cell. 1987a;48:989–996. doi: 10.1016/0092-8674(87)90707-0. [DOI] [PubMed] [Google Scholar]
- Graf J, Ogle RC, Robey FA, Sasaki M, Martin GR, Yamada Y, Kleinman HK. A pentapeptide from the laminin B1 chain mediates cell adhesion and binds the 67,000 laminin receptor. Biochemistry. 1987b;26:6896–6900. doi: 10.1021/bi00396a004. [DOI] [PubMed] [Google Scholar]
- Grosso LE, Park PW, Mecham RP. Characterization of a putative clone for the 67-kilodalton elastin/laminin receptor suggests that it encodes a cytoplasmic protein rather than a cell surface receptor. Biochemistry. 1991;30:3346–3350. doi: 10.1021/bi00227a026. [DOI] [PubMed] [Google Scholar]
- Gu X, Masters KS. Regulation of valvular interstitial cell calcification by adhesive peptide sequences. Journal of Biomedical Materials Research, Part A. 2010;93:1620–1630. doi: 10.1002/jbm.a.32660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guerra-Rebollo M, Mateo F, Franke K, Huen MS, Lopitz-Otsoa F, Rodriguez MS, Plans V, Thomson TM. Nucleolar exit of RNF8 and BRCA1 in response to DNA damage. Experimental Cell Research. 2012;318:2365–2376. doi: 10.1016/j.yexcr.2012.07.003. [DOI] [PubMed] [Google Scholar]
- Guirguis R, Margulies I, Taraboletti G, Schiffmann E, Liotta L. Cytokine-induced pseudopodial protrusion is coupled to tumour cell migration. Nature. 1987;329:261–263. doi: 10.1038/329261a0. [DOI] [PubMed] [Google Scholar]
- Guo D, Han J, Adam BL, Colburn NH, Wang MH, Dong Z, Eizirik DL, She JX, Wang CY. Proteomic analysis of SUMO4 substrates in HEK293 cells under serum starvation-induced stress. Biochemical and Biophysical Research Communications. 2005;337:1308–1318. doi: 10.1016/j.bbrc.2005.09.191. [DOI] [PubMed] [Google Scholar]
- Guo NH, Krutzsch HC, Vogel T, Roberts DD. Interactions of a laminin-binding peptide from a 33-kDa protein related to the 67-kDa laminin receptor with laminin and melanoma cells are heparin-dependent. Journal of Biological Chemistry. 1992;267:17743–17747. [PubMed] [Google Scholar]
- Hadley MA, Weeks BS, Kleinman HK, Dym M. Laminin promotes formation of cord-like structures by Sertoli cells in vitro. Developmental Biology. 1990;140:318–327. doi: 10.1016/0012-1606(90)90082-t. [DOI] [PubMed] [Google Scholar]
- Han J, Logsdon PM, Ellis SR. LAMR1 (+/−) mice as a model for Diamond-Blackfan anemia. Mouse Genome Informatics (MGI) direct data submission. The Jackson Laboratory. 2008 MGI Ref ID J:138267. [Google Scholar]
- Hara K, Satoh K, Ide H. Apical ectodermal ridge-dependent expression of the chick 67 kDa laminin binding protein gene (cLbp) in developing limb bud. Zoological Science. 1997;14:969–978. doi: 10.2108/zsj.14.969. [DOI] [PubMed] [Google Scholar]
- Hendrix MJ, Wood WR, Seftor EA, Lotan D, Nakajima M, Misiorowski RL, Seftor RE, Stetler-Stevenson WG, Bevacqua SJ, Liotta LA, Sobel ME, Raz A, Lotan R. Retinoic acid inhibition of human melanoma cell invasion through a reconstituted basement membrane and its relation to decreases in the expression of proteolytic enzymes and motility factor receptor. Cancer Research. 1990;50:4121–4130. [PubMed] [Google Scholar]
- Hong YC, Lee WM, Kong HH, Jeong HJ, Chung DI. Molecular cloning and characterization of a cDNA encoding a laminin-binding protein (AhLBP) from Acanthamoeba healyi. Experimental Parasitology. 2004;106:95–102. doi: 10.1016/j.exppara.2004.01.011. [DOI] [PubMed] [Google Scholar]
- Huang J, Yan J, Zhang J, Zhu S, Wang Y, Shi T, Zhu C, Chen C, Liu X, Cheng J, Mustelin T, Feng GS, Chen G, Yu J. SUMO1 modification of PTEN regulates tumorigenesis by controlling its association with the plasma membrane. Nature Communications. 2012;3:911. doi: 10.1038/ncomms1919. [DOI] [PubMed] [Google Scholar]
- Huang SH, Jong A. Evolving role of laminin receptors in microbial pathogenesis and therapeutics of CNS infection. Future Microbiol. 2009;4:959–962. doi: 10.2217/fmb.09.67. [DOI] [PubMed] [Google Scholar]
- Huard TK, Malinoff HL, Wicha MS. Macrophages express a plasma membrane receptor for basement membrane laminin. American Journal of Pathology. 1986;123:365–370. [PMC free article] [PubMed] [Google Scholar]
- Hundt C, Peyrin JM, Haik S, Gauczynski S, Leucht C, Rieger R, Riley ML, Deslys JP, Dormont D, Lasmezas CI, Weiss S. Identification of interaction domains of the prion protein with its 37-kDa/67-kDa laminin receptor. EMBO Journal. 2001;20:5876–5886. doi: 10.1093/emboj/20.21.5876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iwamoto Y, Robey FA, Graf J, Sasaki M, Kleinman HK, Yamada Y, Martin GR. YIGSR, a synthetic laminin pentapeptide, inhibits experimental metastasis formation. Science. 1987;238:1132–1134. doi: 10.1126/science.2961059. [DOI] [PubMed] [Google Scholar]
- Jackers P, Clausse N, Fernandez M, Berti A, Princen F, Wewer U, Sobel ME, Castronovo V. Seventeen copies of the human 37 kDa laminin receptor precursor/p40 ribosome-associated protein gene are processed pseudogenes arisen from retropositional events. Biochimica et Biophysica Acta. 1996a;1305:98–104. doi: 10.1016/0167-4781(95)00206-5. [DOI] [PubMed] [Google Scholar]
- Jackers P, Minoletti F, Belotti D, Clausse N, Sozzi G, Sobel ME, Castronovo V. Isolation from a multigene family of the active human gene of the metastasis-associated multifunctional protein 37LRP/p40 at chromosome 3p21.3. Oncogene. 1996b;13:495–503. [PubMed] [Google Scholar]
- Jamieson KV, Hubbard SR, Meruelo D. Structure-guided identification of a laminin binding site on the laminin receptor precursor. Journal of Molecular Biology. 2011;405:24–32. doi: 10.1016/j.jmb.2010.10.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jamieson KV, Wu J, Hubbard SR, Meruelo D. Crystal structure of the human laminin receptor precursor. Journal of Biological Chemistry. 2008;283:3002–3005. doi: 10.1074/jbc.C700206200. [DOI] [PubMed] [Google Scholar]
- Jaseja M, Mergen L, Gillette K, Forbes K, Sehgal I, Copie V. Structure-function studies of the functional and binding epitope of the human 37 kDa laminin receptor precursor protein. Journal of Peptide Research. 2005;66:9–18. doi: 10.1111/j.1399-3011.2005.00267.x. [DOI] [PubMed] [Google Scholar]
- Jovanovic K, Gonsalves D, Da Costa Dias B, Moodley K, Reusch U, Knackmuss S, Penny C, Weinberg MS, Little M, Weiss SF. Anti-LRP/LR specific antibodies and shRNAs impede amyloid beta shedding in Alzheimer's disease. Scientific Reports. 2013;3:2699. doi: 10.1038/srep02699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jovanovic K, Loos B, Da Costa Dias B, Penny C, Weiss SF. High resolution imaging study of interactions between the 37 kDa/67 kDa laminin receptor and APP, beta-secretase and gamma-secretase in Alzheimer's Disease. PLoS One. 2014;9(6):e100373. doi: 10.1371/journal.pone.0100373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karpatova M, Tagliabue E, Castronovo V, Magnifico A, Ardini E, Morelli D, Belotti D, Colnaghi MI, Menard S. Shedding of the 67-kD laminin receptor by human cancer cells. Journal of Cellular Biochemistry. 1996;60:226–234. doi: 10.1002/(SICI)1097-4644(19960201)60:2%3C226::AID-JCB7%3E3.0.CO;2-Z. [DOI] [PubMed] [Google Scholar]
- Katagiri YU, Kiyokawa N, Nakamura K, Takenouchi H, Taguchi T, Okita H, Umezawa A, Fujimoto J. Laminin binding protein, 34/67 laminin receptor, carries stage-specific embryonic antigen-4 epitope defined by monoclonal antibody Raft.2. Biochemical and Biophysical Research Communications. 2005;332:1004–1011. doi: 10.1016/j.bbrc.2005.05.052. [DOI] [PubMed] [Google Scholar]
- Kazmin DA, Hoyt TR, Taubner L, Teintze M, Starkey JR. Phage display mapping for peptide 11 sensitive sequences binding to laminin-1. Journal of Molecular Biology. 2000;298:431–445. doi: 10.1006/jmbi.2000.3680. [DOI] [PubMed] [Google Scholar]
- Keppel E, Schaller HC. A 33 kDa protein with sequence homology to the 'laminin binding protein' is associated with the cytoskeleton in hydra and in mammalian cells. Journal of Cell Science. 1991;100(Pt 4):789–797. doi: 10.1242/jcs.100.4.789. [DOI] [PubMed] [Google Scholar]
- Ketchart W, Smith KM, Krupka T, Wittmann BM, Hu Y, Rayman PA, Doughman YQ, Albert JM, Bai X, Finke JH, Xu Y, Exner AA, Montano MM. Inhibition of metastasis by HEXIM1 through effects on cell invasion and angiogenesis. Oncogene. 2013;32:3829–3839. doi: 10.1038/onc.2012.405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khalfaoui T, Groulx JF, Sabra G, Guezguez A, Basora N, Vermette P, Beaulieu JF. Laminin receptor 37/67LR regulates adhesion and proliferation of normal human intestinal epithelial cells. PLoS One. 2013;8:e74337. doi: 10.1371/journal.pone.0074337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kielian M, Chanel-Vos C, Liao M. Alphavirus entry and membrane fusion. Viruses. 2010;2:796–825. doi: 10.3390/v2040796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kikkawa Y, Hozumi K, Katagiri F, Nomizu M, Kleinman HK, Koblinski JE. Laminin-111-derived peptides and cancer. Cell Adhesion & Migration. 2013;7:150–256. doi: 10.4161/cam.22827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim DG, Choi JW, Lee JY, Kim H, Oh YS, Lee JW, Tak YK, Song JM, Razin E, Yun SH, Kim S. Interaction of two translational components, lysyl-tRNA synthetase and p40/37LRP, in plasma membrane promotes laminin-dependent cell migration. FASEB Journal. 2012;26:4142–4159. doi: 10.1096/fj.12-207639. [DOI] [PubMed] [Google Scholar]
- Kim DG, Lee JY, Kwon NH, Fang P, Zhang Q, Wang J, Young NL, Guo M, Cho HY, Mushtaq AU, Jeon YH, Choi JW, Han JM, Kang HW, Joo JE, Hur Y, Kang W, Yang H, Nam DH, Lee MS, Lee JW, Kim ES, Moon A, Kim K, Kim D, Kang EJ, Moon Y, Rhee KH, Han BW, Yang JS, Han G, Yang WS, Lee C, Wang MW, Kim S. Chemical inhibition of prometastatic lysyl-tRNA synthetase-laminin receptor interaction. Nature Chemical Biology. 2014;10:29–34. doi: 10.1038/nchembio.1381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim K, Li L, Kozlowski K, Suh HS, Cao W, Ballermann BJ. The protein phosphatase-1 targeting subunit TIMAP regulates LAMR1 phosphorylation. Biochemical and Biophysical Research Communications. 2005;338:1327–1334. doi: 10.1016/j.bbrc.2005.10.089. [DOI] [PubMed] [Google Scholar]
- Kim KJ, Chung JW, Kim KS. 67-kDa laminin receptor promotes internalization of cytotoxic necrotizing factor 1-expressing Escherichia coli K1 into human brain microvascular endothelial cells. Journal of Biological Chemistry. 2005;280:1360–1368. doi: 10.1074/jbc.M410176200. [DOI] [PubMed] [Google Scholar]
- Kinoshita K, Kaneda Y, Sato M, Saeki Y, Wataya-Kaneda M, Hoffmann A. LBP-p40 binds DNA tightly through associations with histones H2A, H2B, and H4. Biochemical and Biophysical Research Communications. 1998;253:277–282. doi: 10.1006/bbrc.1998.9699. [DOI] [PubMed] [Google Scholar]
- Kiss AM, Jady BE, Bertrand E, Kiss T. Human box H/ACA pseudouridylation guide RNA machinery. Molecular and Cellular Biology. 2004;24:5797–5807. doi: 10.1128/MCB.24.13.5797-5807.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kleinman HK, Ogle RC, Cannon FB, Little CD, Sweeney TM, Luckenbill-Edds L. Laminin receptors for neurite formation. Proceedings of the National Academy of Sciences of the United States of America. 1988;85:1282–1286. doi: 10.1073/pnas.85.4.1282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ku HC, Chang HH, Liu HC, Hsiao CH, Lee MJ, Hu YJ, Hung PF, Liu CW, Kao YH. Green tea (−)-epigallocatechin gallate inhibits insulin stimulation of 3T3-L1 preadipocyte mitogenesis via the 67-kDa laminin receptor pathway. American Journal of Physiology: Cell Physiology. 2009;297:C121–C132. doi: 10.1152/ajpcell.00272.2008. [DOI] [PubMed] [Google Scholar]
- Ku HC, Liu HS, Hung PF, Chen CL, Liu HC, Chang HH, Tsuei YW, Shih LJ, Lin CL, Lin CM, Kao YH. Green tea (−)-epigallocatechin gallate inhibits IGF-I and IGF-II stimulation of 3T3-L1 preadipocyte mitogenesis via the 67-kDa laminin receptor, but not AMP-activated protein kinase pathway. Molecular Nutrition & Food Research. 2012;56:580–592. doi: 10.1002/mnfr.201100438. [DOI] [PubMed] [Google Scholar]
- Landowski TH, Dratz EA, Starkey JR. Studies of the structure of the metastasis-associated 67 kDa laminin binding protein: fatty acid acylation and evidence supporting dimerization of the 32 kDa gene product to form the mature protein. Biochemistry. 1995a;34:11276–11287. doi: 10.1021/bi00035a037. [DOI] [PubMed] [Google Scholar]
- Landowski TH, Uthayakumar S, Starkey JR. Control pathways of the 67 kDa laminin binding protein: surface expression and activity of a new ligand binding domain. Clinical and Experimental Metastasis. 1995b;13:357–372. doi: 10.1007/BF00121912. [DOI] [PubMed] [Google Scholar]
- Languino LR, Gehlsen KR, Wayner E, Carter WG, Engvall E, Ruoslahti E. Endothelial cells use alpha 2 beta 1 integrin as a laminin receptor. Journal of Cell Biology. 1989;109:2455–2462. doi: 10.1083/jcb.109.5.2455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laurie GW, Horikoshi S, Killen PD, Segui-Real B, Yamada Y. In situ hybridization reveals temporal and spatial changes in cellular expression of mRNA for a laminin receptor, laminin, and basement membrane (type IV) collagen in the developing kidney. Journal of Cell Biology. 1989;109:1351–1362. doi: 10.1083/jcb.109.3.1351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laurie GW, Stone CM, Yamada Y. Elevated 32-kDa LBP and low laminin mRNA expression in developing mouse cerebrum. Differentiation. 1991;46:173–179. doi: 10.1111/j.1432-0436.1991.tb00879.x. [DOI] [PubMed] [Google Scholar]
- Lesot H, Kuhl U, Mark K. Isolation of a laminin-binding protein from muscle cell membranes. EMBO Journal. 1983;2:861–865. doi: 10.1002/j.1460-2075.1983.tb01514.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lestrade L, Weber MJ. snoRNA-LBME-db, a comprehensive database of human H/ACA and C/D box snoRNAs. Nucleic Acids Research. 2006;34:D158–D162. doi: 10.1093/nar/gkj002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leucht C, Simoneau S, Rey C, Vana K, Rieger R, Lasmezas CI, Weiss S. The 37 kDa/67 kDa laminin receptor is required for PrP(Sc) propagation in scrapie-infected neuronal cells. EMBO Reports. 2003;4:290–295. doi: 10.1038/sj.embor.embor768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liotta LA, Horan Hand P, Rao CN, Bryant G, Barsky SH, Schlom J. Monoclonal antibodies to the human laminin receptor recognize structurally distinct sites. Experimental Cell Research. 1985;156:117–126. doi: 10.1016/0014-4827(85)90266-6. [DOI] [PubMed] [Google Scholar]
- Lopez-Ribot JL, Casanova M, Monteagudo C, Sepulveda P, Martinez JP. Evidence for the presence of a high-affinity laminin receptor-like molecule on the surface of Candida albicans yeast cells. Infection and Immunity. 1994;62:742–746. doi: 10.1128/iai.62.2.742-746.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lomize MA, Pogozheva ID, Joo H, Mosberg HI, Lomize AL. OPM database and PPM web server: resources for positioning of proteins in membranes. Nucleic Acids Research. 2012;40:D370–D376. doi: 10.1093/nar/gkr703. (Database Issue) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ludwig GV, Kondig JP, Smith JF. A putative receptor for Venezuelan equine encephalitis virus from mosquito cells. Journal of Virology. 1996;70:5592–5599. doi: 10.1128/jvi.70.8.5592-5599.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maeda T, Titani K, Sekiguchi K. Cell-adhesive activity and receptor-binding specificity of the laminin-derived YIGSR sequence grafted onto Staphylococcal protein A. Journal of Biochemistry. 1994;115:182–189. doi: 10.1093/oxfordjournals.jbchem.a124315. [DOI] [PubMed] [Google Scholar]
- Mak JC. Potential role of green tea catechins in various disease therapies: progress and promise. Clinical and Experimental Pharmacology and Physiology. 2012;39:265–273. doi: 10.1111/j.1440-1681.2012.05673.x. [DOI] [PubMed] [Google Scholar]
- Makrides S, Chitpatima ST, Bandyopadhyay R, Brawerman G. Nucleotide sequence for a major messenger RNA for a 40 kilodalton polypeptide that is under translational control in mouse tumor cells. Nucleic Acids Research. 1988;16:2349. doi: 10.1093/nar/16.5.2349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malinoff HL, Wicha MS. Isolation of a cell surface receptor protein for laminin from murine fibrosarcoma cells. Journal of Cell Biology. 1983;96:1475–1479. doi: 10.1083/jcb.96.5.1475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malygin AA, Babaylova ES, Loktev VB, Karpova GG. A region in the C-terminal domain of ribosomal protein SA required for binding of SA to the human 40S ribosomal subunit. Biochimie. 2011;93:612–617. doi: 10.1016/j.biochi.2010.12.005. [DOI] [PubMed] [Google Scholar]
- Manza LL, Codreanu SG, Stamer SL, Smith DL, Wells KS, Roberts RL, Liebler DC. Global shifts in protein sumoylation in response to electrophile and oxidative stress. Chemical Research in Toxicology. 2004;17:1706–1715. doi: 10.1021/tx049767l. [DOI] [PubMed] [Google Scholar]
- Martignone S, Menard S, Bufalino R, Cascinelli N, Pellegrini R, Tagliabue E, Andreola S, Rilke F, Colnaghi MI. Prognostic significance of the 67-kilodalton laminin receptor expression in human breast carcinomas. Journal of the National Cancer Institute. 1993;85:398–402. doi: 10.1093/jnci/85.5.398. [DOI] [PubMed] [Google Scholar]
- Martin S, Nishimune A, Mellor JR, Henley JM. SUMOylation regulates kainate-receptor-mediated synaptic transmission. Nature. 2007;447:321–325. doi: 10.1038/nature05736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masuda T, Sakuma C, Kobayashi K, Kikuchi K, Soda E, Shiga T, Yaginuma H. Laminin peptide YIGSR and its receptor regulate sensory axonal response to the chemoattractive guidance cue in the chick embryo. Journal of Neuroscience Research. 2009;87:353–359. doi: 10.1002/jnr.21868. [DOI] [PubMed] [Google Scholar]
- McCaffery P, Neve RL, Drager UC. A dorso-ventral asymmetry in the embryonic retina defined by protein conformation. Proceedings of the National Academy of Sciences of the United States of America. 1990;87:8570–8574. doi: 10.1073/pnas.87.21.8570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKenna DJ, Simpson DA, Feeney S, Gardiner TA, Boyle C, Nelson J, Stitt AW. Expression of the 67 kDa laminin receptor (67LR) during retinal development: correlations with angiogenesis. Experimental Eye Research. 2001;73:81–92. doi: 10.1006/exer.2001.1013. [DOI] [PubMed] [Google Scholar]
- Mecham RP, Hinek A, Griffin GL, Senior RM, Liotta LA. The elastin receptor shows structural and functional similarities to the 67-kDa tumor cell laminin receptor. Journal of Biological Chemistry. 1989;264:16652–16657. [PubMed] [Google Scholar]
- Melnick MB, Noll E, Perrimon N. The Drosophila stubarista phenotype is associated with a dosage effect of the putative ribosome-associated protein D-p40 on spineless. Genetics. 1993;135:553–564. doi: 10.1093/genetics/135.2.553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Menard S, Castronovo V, Tagliabue E, Sobel ME. New insights into the metastasis-associated 67 kD laminin receptor. Journal of Cellular Biochemistry. 1997;67:155–165. [PubMed] [Google Scholar]
- Menard S, Tagliabue E, Colnaghi MI. The 67 kDa laminin receptor as a prognostic factor in human cancer. Breast Cancer Research and Treatment. 1998;52:137–145. doi: 10.1023/a:1006171403765. [DOI] [PubMed] [Google Scholar]
- Mercurio AM, Shaw LM. Macrophage interactions with laminin: PMA selectively induces the adherence and spreading of mouse macrophages on a laminin substratum. Journal of Cell Biology. 1988;107:1873–1880. doi: 10.1083/jcb.107.5.1873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mercurio AM, Shaw LM. Laminin binding proteins. Bioessays. 1991;13:469–473. doi: 10.1002/bies.950130907. [DOI] [PubMed] [Google Scholar]
- Messeguer X, Escudero R, Farre D, Nunez O, Martinez J, Alba MM. PROMO: detection of known transcription regulatory elements using species-tailored searches. Bioinformatics. 2002;18:333–334. doi: 10.1093/bioinformatics/18.2.333. [DOI] [PubMed] [Google Scholar]
- Miner JH. Laminins and their roles in mammals. Microscopy Research and Technique. 2008;71:349–356. doi: 10.1002/jemt.20563. [DOI] [PubMed] [Google Scholar]
- Modugno M, Tagliabue E, Ardini E, Berno V, Galmozzi E, De Bortoli M, Castronovo V, Menard S. p53-dependent downregulation of metastasis-associated laminin receptor. Oncogene. 2002;21:7478–7487. doi: 10.1038/sj.onc.1205957. [DOI] [PubMed] [Google Scholar]
- Montuori N, Selleri C, Risitano AM, Raiola AM, Ragno P, Del Vecchio L, Rotoli B, Rossi G. Expression of the 67-kDa laminin receptor in acute myeloid leukemia cells mediates adhesion to laminin and is frequently associated with monocytic differentiation. Clinical Cancer Research. 1999;5:1465–1472. [PubMed] [Google Scholar]
- Montuori N, Siri J, Simmons T, Gillet C, Senterre G, Wrathall L, Sobel ME. Precursor-product relationship between a 37-kDa polypeptide and the 67-kDa laminin receptor. The FASEB Journal. 1995;9(3):A539. Inaccessible meeting abstract (SICI: 0892-6638(1995)9:3<539:PRBA3P>2.0.ZU;2-8) [Google Scholar]
- Montuori N, Sobel ME. The 67-kDa laminin receptor and tumor progression. Current Topics in Microbiology and Immunology. 1996;213(Pt 1):205–214. doi: 10.1007/978-3-642-61107-0_13. [DOI] [PubMed] [Google Scholar]
- Moodley K, Weiss SF. Downregulation of the non-integrin laminin receptor reduces cellular viability by inducing apoptosis in lung and cervical cancer cells. PLoS One. 2013;8:e57409. doi: 10.1371/journal.pone.0057409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moss BL, Taubner L, Sample YK, Kazmin DA, Copie V, Starkey JR. Tumor shedding of laminin binding protein modulates angiostatin production in vitro and interferes with plasmin-derived inhibition of angiogenesis in aortic ring cultures. International Journal of Cancer. 2006;118:2421–2432. doi: 10.1002/ijc.21674. [DOI] [PubMed] [Google Scholar]
- Nakai M, Mundy GR, Williams PJ, Boyce B, Yoneda T. A synthetic antagonist to laminin inhibits the formation of osteolytic metastases by human melanoma cells in nude mice. Cancer Research. 1992;52:5395–5399. [PubMed] [Google Scholar]
- Narasimhan S, Armstrong MY, Rhee K, Edman JC, Richards FF, Spicer E. Gene for an extracellular matrix receptor protein from Pneumocystis carinii. Proceedings of the National Academy of Sciences of the United States of America. 1994;91:7440–7444. doi: 10.1073/pnas.91.16.7440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Narumi K, Inoue A, Tanaka M, Isemura M, Shimo-Oka T, Abe T, Nukiwa T, Satoh K. Inhibition of experimental metastasis of human fibrosarcoma cells by anti-recombinant 37-kDa laminin binding protein antibody. Japanese Journal of Cancer Research. 1999;90:425–431. doi: 10.1111/j.1349-7006.1999.tb00765.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelson J, McFerran NV, Pivato G, Chambers E, Doherty C, Steele D, Timson DJ. The 67 kDa laminin receptor: structure, function and role in disease. Bioscience Reports. 2008;28:33–48. doi: 10.1042/BSR20070004. [DOI] [PubMed] [Google Scholar]
- O'Donohue MF, Choesmel V, Faubladier M, Fichant G, Gleizes PE. Functional dichotomy of ribosomal proteins during the synthesis of mammalian 40S ribosomal subunits. Journal of Cell Biology. 2010;190:853–866. doi: 10.1083/jcb.201005117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ochieng J, Warfield P, Green-Jarvis B, Fentie I. Galectin-3 regulates the adhesive interaction between breast carcinoma cells and elastin. Journal of Cellular Biochemistry. 1999;75:505–514. doi: 10.1002/(sici)1097-4644(19991201)75:3<505::aid-jcb14>3.3.co;2-9. [DOI] [PubMed] [Google Scholar]
- Omar A, Jovanovic K, Da Costa Dias B, Gonsalves D, Moodley K, Caveney R, Mbazima V, Weiss SF. Patented biological approaches for the therapeutic modulation of the 37 kDa/67 kDa laminin receptor. Expert Opinion on Therapeutic Patents. 2011;21:35–53. doi: 10.1517/13543776.2011.539203. [DOI] [PubMed] [Google Scholar]
- Omar A, Reusch U, Knackmuss S, Little M, Weiss SF. Anti-LRP/LR-specific antibody IgG1-iS18 significantly reduces adhesion and invasion of metastatic lung, cervix, colon and prostate cancer cells. Journal of Molecular Biology. 2012;419:102–109. doi: 10.1016/j.jmb.2012.02.035. [DOI] [PubMed] [Google Scholar]
- Orihuela CJ, Mahdavi J, Thornton J, Mann B, Wooldridge KG, Abouseada N, Oldfield NJ, Self T, Ala'Aldeen DA, Tuomanen EI. Laminin receptor initiates bacterial contact with the blood brain barrier in experimental meningitis models. Journal of Clinical Investigation. 2009;119:1638–1646. doi: 10.1172/JCI36759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ould-Abeih MB, Petit-Topin I, Zidane N, Baron B, Bedouelle H. Multiple folding states and disorder of ribosomal protein SA, a membrane receptor for laminin, anticarcinogens, and pathogens. Biochemistry. 2012;51:4807–4821. doi: 10.1021/bi300335r. [DOI] [PubMed] [Google Scholar]
- Pampeno C, Derkatch IL, Meruelo D. Interaction of human laminin receptor with Sup35, the [PSI+] prion-forming protein from S. cerevisiae: a yeast model for studies of LamR interactions with amyloidogenic proteins. PLoS One. 2014;9(1):e86013. doi: 10.1371/journal.pone.0086013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park E, Choi Y, Ahn E, Park I, Yun Y. The adaptor protein LAD/TSAd mediates laminin-dependent T cell migration via association with the 67 kDa laminin binding protein. Experimental and Molecular Medicine. 2009;41:728–736. doi: 10.3858/emm.2009.41.10.079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pellegrini R, Martignone S, Menard S, Colnaghi MI. Laminin receptor expression and function in small-cell lung carcinoma. International Journal of Cancer. Supplement. 1994;8:116–120. doi: 10.1002/ijc.2910570725. [DOI] [PubMed] [Google Scholar]
- Poon SL, Klausen C, Hammond GL, Leung PC. 37-kDa laminin receptor precursor mediates GnRH-II-induced MMP-2 expression and invasiveness in ovarian cancer cells. Molecular Endocrinology. 2011;25:327–338. doi: 10.1210/me.2010-0334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rabacchi SA, Neve RL, Drager UC. A positional marker for the dorsal embryonic retina is homologous to the high-affinity laminin receptor. Development. 1990;109:521–531. doi: 10.1242/dev.109.3.521. [DOI] [PubMed] [Google Scholar]
- Rabl J, Leibundgut M, Ataide SF, Haag A, Ban N. Crystal structure of the eukaryotic 40S ribosomal subunit in complex with initiation factor 1. Science. 2011;331:730–736. doi: 10.1126/science.1198308. [DOI] [PubMed] [Google Scholar]
- Raghunath PN, Sidhu GS, Coon HC, Liu K, Srikantan V, Maheshwari RK. Interferons upregulate the expression of laminin and its receptor LBP-32 in cultured cells. Journal of Biological Regulators and Homeostatic Agents. 1993;7:22–30. [PubMed] [Google Scholar]
- Rao CN, Castronovo V, Schmitt MC, Wewer UM, Claysmith AP, Liotta LA, Sobel ME. Evidence for a precursor of the high-affinity metastasis-associated murine laminin receptor. Biochemistry. 1989;28:7476–7486. doi: 10.1021/bi00444a047. [DOI] [PubMed] [Google Scholar]
- Rao CN, Margulies IM, Goldfarb RH, Madri JA, Woodley DT, Liotta LA. Differential proteolytic susceptibility of laminin alpha and beta subunits. Archives of Biochemistry and Biophysics. 1982;219:65–70. doi: 10.1016/0003-9861(82)90134-5. [DOI] [PubMed] [Google Scholar]
- Rao M, Manishen WJ, Maheshwari Y, Sykes DE, Siyanova EY, Tyner AL, Weiser MM. Laminin receptor expression in rat intestine and liver during development and differentiation. Gastroenterology. 1994;107:764–772. doi: 10.1016/0016-5085(94)90125-2. [DOI] [PubMed] [Google Scholar]
- Rao NC, Barsky SH, Terranova VP, Liotta LA. Isolation of a tumor cell laminin receptor. Biochemical and Biophysical Research Communications. 1983;111:804–808. doi: 10.1016/0006-291x(83)91370-0. [DOI] [PubMed] [Google Scholar]
- Rea VE, Rossi FW, De Paulis A, Ragno P, Selleri C, Montuori N. 67 kDa laminin receptor: structure, function and role in cancer and infection. Le Infezioni in Medicina. 2012;20(Suppl 2):8–12. [PubMed] [Google Scholar]
- Ren J, Gao X, Jin C, Zhu M, Wang X, Shaw A, Wen L, Yao X, Xue Y. Systematic study of protein sumoylation: Development of a site-specific predictor of SUMOsp 2.0. Proteomics. 2009;9:3409–3412. doi: 10.1002/pmic.200800646. [DOI] [PubMed] [Google Scholar]
- Rescan PY, Clement B, Yamada Y, Segui-Real B, Baffet G, Guguen-Guillouzo C, Guillouzo A. Differential expression of laminin chains and receptor (LBP-32) in fetal and neoplastic hepatocytes compared to normal adult hepatocytes in vivo and in culture. American Journal of Pathology. 1990;137:701–709. [PMC free article] [PubMed] [Google Scholar]
- Rieger R, Edenhofer F, Lasmezas CI, Weiss S. The human 37-kDa laminin receptor precursor interacts with the prion protein in eukaryotic cells. Nature Medicine. 1997;3:1383–1388. doi: 10.1038/nm1297-1383. [DOI] [PubMed] [Google Scholar]
- Rieger R, Lasmezas CI, Weiss S. Role of the 37 kDa laminin receptor precursor in the life cycle of prions. Transfusion Clinique et Biologique. 1999;6:7–16. doi: 10.1016/S1246-7820(99)80006-8. [DOI] [PubMed] [Google Scholar]
- Rohde H, Bachinger HP, Timpl R. Characterization of pepsin fragments of laminin in a tumor basement membrane. Evidence for the existence of related proteins. Hoppe-Seylers Zeitschrift fur Physiologische Chemie. 1980;361:1651–1660. doi: 10.1515/bchm2.1980.361.2.1651. [DOI] [PubMed] [Google Scholar]
- Romanov V, Sobel ME, pinto da Silva P, Menard S, Castronovo V. Cell localization and redistribution of the 67 kD laminin receptor and alpha 6 beta 1 integrin subunits in response to laminin stimulation: an immunogold electron microscopy study. Cell Adhesion and Communication. 1994;2:201–209. doi: 10.3109/15419069409004438. [DOI] [PubMed] [Google Scholar]
- Ross SA, Ahrens RA, De Luca LM. Retinoic acid enhances adhesiveness, laminin and integrin beta 1 synthesis, and retinoic acid receptor expression in F9 teratocarcinoma cells. Journal of Cellular Physiology. 1994;159:263–273. doi: 10.1002/jcp.1041590210. [DOI] [PubMed] [Google Scholar]
- Roy A, Kucukural A, Zhang Y. I-TASSER: a unified platform for automated protein structure and function prediction. Nature Protocols. 2010;5:725–738. doi: 10.1038/nprot.2010.5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruff EA, Rimoldi OJ, Raghu B, Eliceiri GL. Three small nucleolar RNAs of unique nucleotide sequences. Proceedings of the National Academy of Sciences of the United States of America. 1993;90:635–638. doi: 10.1073/pnas.90.2.635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rush J, Moritz A, Lee KA, Guo A, Goss VL, Spek EJ, Zhang H, Zha XM, Polakiewicz RD, Comb MJ. Immunoaffinity profiling of tyrosine phosphorylation in cancer cells. Nature Biotechnology. 2005;23:94–101. doi: 10.1038/nbt1046. [DOI] [PubMed] [Google Scholar]
- Ryan MC, Christiano AM, Engvall E, Wewer UM, Miner JH, Sanes JR, Burgeson RE. The functions of laminins: lessons from in vivo studies. Matrix Biology. 1996;15:369–381. doi: 10.1016/s0945-053x(96)90157-2. [DOI] [PubMed] [Google Scholar]
- Sakaguchi S, Ishibashi D, Matsuda H. Antibody-based immunotherapeutic attempts in experimental animal models of prion diseases. Expert Opinion on Therapeutic Patents. 2009;19:907–917. doi: 10.1517/13543770902988530. [DOI] [PubMed] [Google Scholar]
- Salama RH, Muramatsu H, Zou K, Inui T, Kimura T, Muramatsu T. Midkine binds to 37-kDa laminin binding protein precursor, leading to nuclear transport of the complex. Experimental Cell Research. 2001;270:13–20. doi: 10.1006/excr.2001.5341. [DOI] [PubMed] [Google Scholar]
- Sanjuan X, Fernandez PL, Miquel R, Munoz J, Castronovo V, Menard S, Palacin A, Cardesa A, Campo E. Overexpression of the 67-kD laminin receptor correlates with tumour progression in human colorectal carcinoma. Journal of Pathology. 1996;179:376–380. doi: 10.1002/(SICI)1096-9896(199608)179:4<376::AID-PATH591>3.0.CO;2-V. [DOI] [PubMed] [Google Scholar]
- Sato M, Kinoshita K, Kaneda Y, Saeki Y, Iwamatsu A, Tanaka K. Analysis of nuclear localization of laminin binding protein precursor p40 (LBP/p40) Biochemical and Biophysical Research Communications. 1996;229:896–901. doi: 10.1006/bbrc.1996.1899. [DOI] [PubMed] [Google Scholar]
- Sato M, Kong CJ, Yoshida H, Nakamura T, Wada A, Shimoda C, Kaneda Y. Ribosomal proteins S0 and S21 are involved in the stability of 18S rRNA in fission yeast, Schizosaccharomyces pombe. Biochemical and Biophysical Research Communications. 2003;311:942–947. doi: 10.1016/j.bbrc.2003.10.086. [DOI] [PubMed] [Google Scholar]
- Sato M, Saeki Y, Tanaka K, Kaneda Y. Ribosome-associated protein LBP/p40 binds to S21 protein of 40S ribosome: analysis using a yeast two-hybrid system. Biochemical and Biophysical Research Communications. 1999;256:385–390. doi: 10.1006/bbrc.1999.0343. [DOI] [PubMed] [Google Scholar]
- Satoh K, Narumi K, Abe T, Sakai T, Kikuchi T, Tanaka M, Shimo-Oka T, Uchida M, Tezuka F, Isemura M, Nukiwa T. Diminution of 37-kDa laminin binding protein expression reduces tumour formation of murine lung cancer cells. British Journal of Cancer. 1999;80:1115–1122. doi: 10.1038/sj.bjc.6690474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Satoh K, Narumi K, Isemura M, Sakai T, Abe T, Matsushima K, Okuda K, Motomiya M. Increased expression of the 67kDa-laminin receptor gene in human small cell lung cancer. Biochemical and Biophysical Research Communications. 1992;182:746–752. doi: 10.1016/0006-291x(92)91795-r. [DOI] [PubMed] [Google Scholar]
- Scheiman J, Jamieson KV, Ziello J, Tseng JC, Meruelo D. Extraribosomal functions associated with the C terminus of the 37/67 kDa laminin receptor are required for maintaining cell viability. Cell Death & Disease. 2010a;1:e42. doi: 10.1038/cddis.2010.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scheiman J, Tseng JC, Zheng Y, Meruelo D. Multiple functions of the 37/67-kd laminin receptor make it a suitable target for novel cancer gene therapy. Molecular Therapy. 2010b;18:63–74. doi: 10.1038/mt.2009.199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schimmel J, Larsen KM, Matic I, van Hagen M, Cox J, Mann M, Andersen JS, Vertegaal AC. The ubiquitin-proteasome system is a key component of the SUMO-2/3 cycle. Molecular and Cellular Proteomics. 2008;7:2107–2122. doi: 10.1074/mcp.M800025-MCP200. [DOI] [PubMed] [Google Scholar]
- Schlessinger A, Punta M, Yachdav G, Kajan L, Rost B. Improved disorder prediction by combination of orthogonal approaches. PLoS One. 2009;4:e4433. doi: 10.1371/journal.pone.0004433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schuwirth BS, Borovinskaya MA, Hau CW, Zhang W, Vila-Sanjurjo A, Holton JM, Cate JH. Structures of the bacterial ribosome at 3.5 A resolution. Science. 2005;310:827–834. doi: 10.1126/science.1117230. [DOI] [PubMed] [Google Scholar]
- Selleri C, Ragno P, Ricci P, Visconte V, Scarpato N, Carriero MV, Rotoli B, Rossi G, Montuori N. The metastasis-associated 67-kDa laminin receptor is involved in G-CSF-induced hematopoietic stem cell mobilization. Blood. 2006;108:2476–2484. doi: 10.1182/blood-2005-11-012625. [DOI] [PubMed] [Google Scholar]
- Sengupta S, Shovlin FE, Slomiany A, Slomiany BL. Identification of laminin receptor in gingival tissue and its interaction with tooth cementum laminin. International Journal of Biochemistry. 1991;23:115–121. doi: 10.1016/0020-711x(91)90017-h. [DOI] [PubMed] [Google Scholar]
- Sepulveda P, Cervera AM, Lopez-Ribot JL, Chaffin WL, Martinez JP, Gozalbo D. Cloning and characterization of a cDNA coding for Candida albicans polyubiquitin. Journal of Medical & Veterinary Mycology. 1996;34:315–322. [PubMed] [Google Scholar]
- Shi YE, Torri J, Yieh L, Sobel ME, Yamada Y, Lippman ME, Dickson RB, Thompson EW. Expression of 67 kDa laminin receptor in human breast cancer cells: regulation by progestins. Clinical and Experimental Metastasis. 1993;11:251–261. doi: 10.1007/BF00121168. [DOI] [PubMed] [Google Scholar]
- Shin SJ, Lee SE, Boo JH, Kim M, Yoon YD, Kim SI, Mook-Jung I. Profiling proteins related to amyloid deposited brain of Tg2576 mice. Proteomics. 2004;4:3359–3368. doi: 10.1002/pmic.200400961. [DOI] [PubMed] [Google Scholar]
- Sigalov AB. Uncoupled binding and folding of immune signaling-related intrinsically disordered proteins. Progress in Biophysics and Molecular Biology. 2011;106:525–536. doi: 10.1016/j.pbiomolbio.2011.08.005. [DOI] [PubMed] [Google Scholar]
- Simoneau S, Haik S, Leucht C, Dormont D, Deslys JP, Weiss S, Lasmezas C. Different isoforms of the non-integrin laminin receptor are present in mouse brain and bind PrP. Biological Chemistry. 2003;384:243–246. doi: 10.1515/BC.2003.027. [DOI] [PubMed] [Google Scholar]
- Simons K, Ehehalt R. Cholesterol, lipid rafts, and disease. Journal of Clinical Investigation. 2002;110:597–603. doi: 10.1172/JCI16390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soeda S, Ishida S, Honda O, Shimeno H, Nagamatsu A. Aminated fucoidan promotes the invasion of 3 LL cells through reconstituted basement membrane: its possible mechanism of action. Cancer Letters. 1994;85:133–138. doi: 10.1016/0304-3835(94)90249-6. [DOI] [PubMed] [Google Scholar]
- Starkey JR, Uthayakumar S, Berglund DL. Cell surface and substrate distribution of the 67-kDa laminin-binding protein determined by using a ligand photoaffinity probe. Cytometry. 1999;35:37–47. doi: 10.1002/(sici)1097-0320(19990101)35:1<37::aid-cyto6>3.0.co;2-c. [DOI] [PubMed] [Google Scholar]
- Stitt AW, McKenna D, Simpson DA, Gardiner TA, Harriott P, Archer DB, Nelson J. The 67-kd laminin receptor is preferentially expressed by proliferating retinal vessels in a murine model of ischemic retinopathy. American Journal of Pathology. 1998;152:1359–1365. [PMC free article] [PubMed] [Google Scholar]
- Strauss JH, Wang KS, Schmaljohn AL, Kuhn RJ, Strauss EG. Host-cell receptors for Sindbis virus. Archives of Virology. Supplementum. 1994;9:473–484. doi: 10.1007/978-3-7091-9326-6_46. [DOI] [PubMed] [Google Scholar]
- Susantad T, Smith DR. siRNA-mediated silencing of the 37/67-kDa high affinity laminin receptor in Hep3B cells induces apoptosis. Cellular & Molecular Biology Letters. 2008;13:452–464. doi: 10.2478/s11658-008-0017-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tabb-Massey A, Caffrey JM, Logsden P, Taylor S, Trent JO, Ellis SR. Ribosomal proteins Rps0 and Rps21 of Saccharomyces cerevisiae have overlapping functions in the maturation of the 3' end of 18S rRNA. Nucleic Acids Research. 2003;31:6798–6805. doi: 10.1093/nar/gkg899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tachibana H, Koga K, Fujimura Y, Yamada K. A receptor for green tea polyphenol EGCG. Nature Structural & Molecular Biology. 2004;11:380–381. doi: 10.1038/nsmb743. [DOI] [PubMed] [Google Scholar]
- Tagliabue E, Pastorino U. Re: Killing of laminin receptor-positive human lung cancers by tumor-infiltrating lymphocytes bearing gamma delta + T-cell receptors. Journal of the National Cancer Institute. 1996;88:1241–1242. doi: 10.1093/jnci/88.17.1241. [DOI] [PubMed] [Google Scholar]
- Tandon NN, Holland EA, Kralisz U, Kleinman HK, Robey FA, Jamieson GA. Interaction of human platelets with laminin and identification of the 67 kDa laminin receptor on platelets. Biochemical Journal. 1991;274(Pt 2):535–542. doi: 10.1042/bj2740535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taraboletti G, Belotti D, Giavazzi R, Sobel ME, Castronovo V. Enhancement of metastatic potential of murine and human melanoma cells by laminin receptor peptide G: attachment of cancer cells to subendothelial matrix as a pathway for hematogenous metastasis. Journal of the National Cancer Institute. 1993;85:235–240. doi: 10.1093/jnci/85.3.235. [DOI] [PubMed] [Google Scholar]
- Terranova VP, Lyall RM. Chemotaxis of human gingival epithelial cells to laminin. A mechanism for epithelial cell apical migration. Journal of Periodontology. 1986;57:311–317. doi: 10.1902/jop.1986.57.5.311. [DOI] [PubMed] [Google Scholar]
- Thepparit C, Smith DR. Serotype-specific entry of dengue virus into liver cells: identification of the 37-kilodalton/67-kilodalton high-affinity laminin receptor as a dengue virus serotype 1 receptor. Journal of Virology. 2004;78:12647–12656. doi: 10.1128/JVI.78.22.12647-12656.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thongtan T, Wikan N, Wintachai P, Rattanarungsan C, Srisomsap C, Cheepsunthorn P, Smith DR. Characterization of putative Japanese encephalitis virus receptor molecules on microglial cells. Journal of Medical Virology. 2012;84:615–623. doi: 10.1002/jmv.23248. [DOI] [PubMed] [Google Scholar]
- Tohgo A, Takasawa S, Munakata H, Yonekura H, Hayashi N, Okamoto H. Structural determination and characterization of a 40 kDa protein isolated from rat 40 S ribosomal subunit. FEBS Letters. 1994;340:133–138. doi: 10.1016/0014-5793(94)80188-6. [DOI] [PubMed] [Google Scholar]
- Turpeenniemi-Hujanen T, Thorgeirsson UP, Rao CN, Liotta LA. Laminin increases the release of type IV collagenase from malignant cells. Journal of Biological Chemistry. 1986;261:1883–1889. [PubMed] [Google Scholar]
- Umeda D, Tachibana H, Yamada K. Epigallocatechin-3-O-gallate disrupts stress fibers and the contractile ring by reducing myosin regulatory light chain phosphorylation mediated through the target molecule 67 kDa laminin receptor. Biochemical and Biophysical Research Communications. 2005;333:628–635. doi: 10.1016/j.bbrc.2005.05.108. [DOI] [PubMed] [Google Scholar]
- Umeda D, Yano S, Yamada K, Tachibana H. Green tea polyphenol epigallocatechin-3-gallate signaling pathway through 67-kDa laminin receptor. Journal of Biological Chemistry. 2008a;283:3050–3058. doi: 10.1074/jbc.M707892200. [DOI] [PubMed] [Google Scholar]
- Umeda D, Yano S, Yamada K, Tachibana H. Involvement of 67-kDa laminin receptor-mediated myosin phosphatase activation in antiproliferative effect of epigallocatechin-3-O-gallate at a physiological concentration on Caco-2 colon cancer cells. Biochemical and Biophysical Research Communications. 2008b;371:172–176. doi: 10.1016/j.bbrc.2008.04.041. [DOI] [PubMed] [Google Scholar]
- Uversky VN. The most important thing is the tail: multitudinous functionalities of intrinsically disordered protein termini. FEBS Letters. 2013;587:1891–1901. doi: 10.1016/j.febslet.2013.04.042. [DOI] [PubMed] [Google Scholar]
- Valasek L, Mathew AA, Shin BS, Nielsen KH, Szamecz B, Hinnebusch AG. The yeast eIF3 subunits TIF32/a, NIP1/c, and eIF5 make critical connections with the 40S ribosome in vivo. Genes and Development. 2003;17:786–799. doi: 10.1101/gad.1065403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Veen AG, Ploegh HL. Ubiquitin-like proteins. Annual Review of Biochemistry. 2012;81:323–357. doi: 10.1146/annurev-biochem-093010-153308. [DOI] [PubMed] [Google Scholar]
- Vana K, Zuber C, Pflanz H, Kolodziejczak D, Zemora G, Bergmann AK, Weiss S. LRP/LR as an alternative promising target in therapy of prion diseases, Alzheimer's disease and cancer. Infectious Disorders Drug Targets. 2009;9:69–80. doi: 10.2174/1871526510909010069. [DOI] [PubMed] [Google Scholar]
- Venticinque L, Jamieson KV, Meruelo D. Interactions between laminin receptor and the cytoskeleton during translation and cell motility. PLoS One. 2011;6:e15895. doi: 10.1371/journal.pone.0015895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Venticinque L, Meruelo D. Comprehensive proteomic analysis of nonintegrin laminin receptor interacting proteins. Journal of Proteome Research. 2012;11:4863–4872. doi: 10.1021/pr300307h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Viacava P, Naccarato AG, Collecchi P, Menard S, Castronovo V, Bevilacqua G. The spectrum of 67-kD laminin receptor expression in breast carcinoma progression. Journal of Pathology. 1997;182:36–44. doi: 10.1002/(SICI)1096-9896(199705)182:1<36::AID-PATH802>3.0.CO;2-W. [DOI] [PubMed] [Google Scholar]
- von der Mark K, Risse G. Isolation and characterization of laminin receptors. Methods in Enzymology. 1987;144:490–507. doi: 10.1016/0076-6879(87)44197-9. [DOI] [PubMed] [Google Scholar]
- Wang KS, Kuhn RJ, Strauss EG, Ou S, Strauss JH. High-affinity laminin receptor is a receptor for Sindbis virus in mammalian cells. Journal of Virology. 1992;66:4992–5001. doi: 10.1128/jvi.66.8.4992-5001.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weeks BS, DiSalvo J, Kleinman HK. Laminin-mediated process formation in neuronal cells involves protein dephosphorylation. Journal of Neuroscience Research. 1990;27:418–426. doi: 10.1002/jnr.490270321. [DOI] [PubMed] [Google Scholar]
- Weeks BS, Kopp JB, Horikoshi S, Cannon FB, Garrett M, Kleinman HK, Klotman PE. Adult and fetal human mesangial cells interact with specific laminin domains. American Journal of Physiology. 1991;261:F688–F695. doi: 10.1152/ajprenal.1991.261.4.F688. [DOI] [PubMed] [Google Scholar]
- Weisser M, Voigts-Hoffmann F, Rabl J, Leibundgut M, Ban N. The crystal structure of the eukaryotic 40S ribosomal subunit in complex with eIF1 and eIF1A. Nature Structural & Molecular Biology. 2013;20:1015–1017. doi: 10.1038/nsmb.2622. [DOI] [PubMed] [Google Scholar]
- Wenzel S, Henning K, Habbig A, Forst S, Schreckenberg R, Heger J, Maxeiner H, Schluter KD. TGF-beta1 improves cardiac performance via up-regulation of laminin receptor 37/67 in adult ventricular cardiomyocytes. Basic Research in Cardiology. 2010;105:621–629. doi: 10.1007/s00395-010-0108-1. [DOI] [PubMed] [Google Scholar]
- Wewer UM, Liotta LA, Jaye M, Ricca GA, Drohan WN, Claysmith AP, Rao CN, Wirth P, Coligan JE, Albrechtsen R, Mudryj M, Sobel ME. Altered levels of laminin receptor mRNA in various human carcinoma cells that have different abilities to bind laminin. Proceedings of the National Academy of Sciences of the United States of America. 1986;83:7137–7141. doi: 10.1073/pnas.83.19.7137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wewer UM, Taraboletti G, Sobel ME, Albrechtsen R, Liotta LA. Role of laminin receptor in tumor cell migration. Cancer Research. 1987;47:5691–5698. [PubMed] [Google Scholar]
- Wilkinson KA, Henley JM. Mechanisms, regulation and consequences of protein SUMOylation. Biochemical Journal. 2010;428:133–145. doi: 10.1042/BJ20100158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wimberly BT, Brodersen DE, Clemons WM, Jr, Morgan-Warren RJ, Carter AP, Vonrhein C, Hartsch T, Ramakrishnan V. Structure of the 30S ribosomal subunit. Nature. 2000;407:327–339. doi: 10.1038/35030006. [DOI] [PubMed] [Google Scholar]
- Wohlschlegel JA, Johnson ES, Reed SI, Yates JR., 3rd Global analysis of protein sumoylation in Saccharomyces cerevisiae. Journal of Biological Chemistry. 2004;279:45662–45668. doi: 10.1074/jbc.M409203200. [DOI] [PubMed] [Google Scholar]
- Xu D, Zhang Y. Ab initio protein structure assembly using continuous structure fragments and optimized knowledge-based force field. Proteins. 2012;80:1715–1735. doi: 10.1002/prot.24065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamada KM, Kennedy DW, Yamada SS, Gralnick H, Chen WT, Akiyama SK. Monoclonal antibody and synthetic peptide inhibitors of human tumor cell migration. Cancer Research. 1990;50:4485–4496. [PubMed] [Google Scholar]
- Yamamura K, Kibbey MC, Jun SH, Kleinman HK. Effect of Matrigel and laminin peptide YIGSR on tumor growth and metastasis. Seminars in Cancer Biology. 1993;4:259–265. [PubMed] [Google Scholar]
- Yannariello-Brown J, Wewer U, Liotta L, Madri JA. Distribution of a 69-kD laminin-binding protein in aortic and microvascular endothelial cells: modulation during cell attachment, spreading, and migration. Journal of Cell Biology. 1988;106:1773–1786. doi: 10.1083/jcb.106.5.1773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yow HK, Wong JM, Chen HS, Lee CG, Davis S, Steele GD, Jr, Chen LB. Increased mRNA expression of a laminin-binding protein in human colon carcinoma: complete sequence of a full-length cDNA encoding the protein. Proceedings of the National Academy of Sciences of the United States of America. 1988;85:6394–6398. doi: 10.1073/pnas.85.17.6394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yurchenco PD. Basement membranes: cell scaffoldings and signaling platforms. Cold Spring Harbor Perspectives in Biology. 2011;3 doi: 10.1101/cshperspect.a004911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J, Liang Y, Zhang Y. Atomic-level protein structure refinement using fragment-guided molecular dynamics conformation sampling. Structure. 2011;19:1784–1795. doi: 10.1016/j.str.2011.09.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang L, Leggatt GR, Kalinna BH, Piva TJ, McManus DP. Cloning and expression of a cDNA encoding a nonintegrin laminin-binding protein from Echinococcus granulosus with localization of the laminin-binding domain. Molecular and Biochemical Parasitology. 1997;87:183–192. doi: 10.1016/s0166-6851(97)00066-2. [DOI] [PubMed] [Google Scholar]
- Zidane N, Ould-Abeih MB, Petit-Topin I, Bedouelle H. The folded and disordered domains of human ribosomal protein SA have both idiosyncratic and shared functions as membrane receptors. Bioscience Reports. 2013;33:113–124. doi: 10.1042/BSR20120103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuber C, Knackmuss S, Rey C, Reusch U, Rottgen P, Frohlich T, Arnold GJ, Pace C, Mitteregger G, Kretzschmar HA, Little M, Weiss S. Single chain Fv antibodies directed against the 37 kDa/67 kDa laminin receptor as therapeutic tools in prion diseases. Molecular Immunology. 2008a;45:144–151. doi: 10.1016/j.molimm.2007.04.030. [DOI] [PubMed] [Google Scholar]
- Zuber C, Mitteregger G, Schuhmann N, Rey C, Knackmuss S, Rupprecht W, Reusch U, Kretzschmar HA, Hallek M, Buning H, Weiss S. Delivery of single-chain antibodies (scFvs) directed against the 37/67 kDa laminin receptor into mice via recombinant adeno-associated viral vectors for prion disease gene therapy. Journal of General Virology. 2008b;89:2055–2061. doi: 10.1099/vir.0.83670-0. [DOI] [PubMed] [Google Scholar]