Abstract
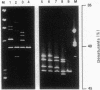
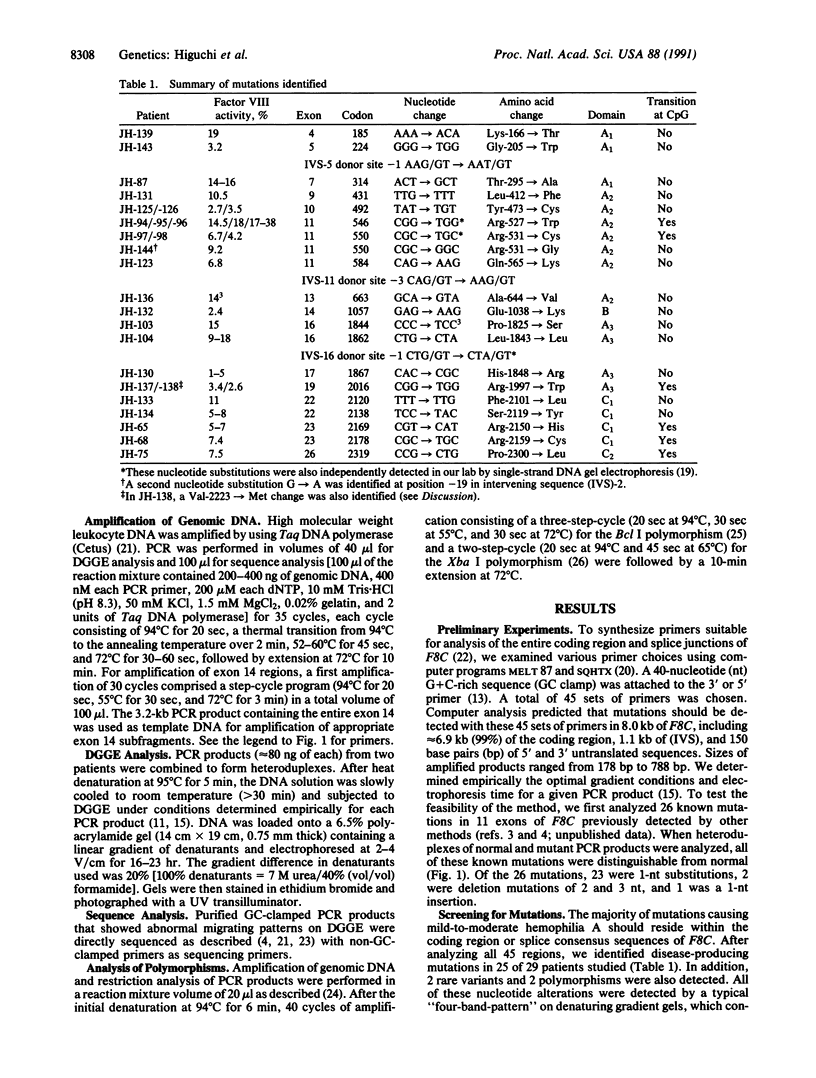
To date it has been difficult to characterize completely a genetic disorder, such as hemophilia A, in which the involved gene is large and unrelated affected individuals have different mutations, most of which are point mutations. Toward this end, we analyzed the DNA of 29 patients with mild-to-moderate hemophilia A in which the causative mutation is likely to be a missense mutation. Using computer analysis, we determined the melting properties of factor VIII gene sequences to design primer sets for PCR amplification and subsequent denaturing gradient gel electrophoresis (DGGE). A total of 45 primer sets was chosen to amplify 99% of the coding region of the gene and 41 of 50 splice junctions. To facilitate detection of point mutations, we mixed DNA from two male patients, and both homoduplexes and heteroduplexes were analyzed. With these 45 primer sets, 26 DNAs containing previously identified point mutations in the factor VIII gene were studied, and all 26 mutations were easily distinguishable from normal. After analyzing the 29 patients with unknown mutations, we identified the disease-producing mutation in 25 (86%). Two polymorphisms and two rare normal variants were also found. Therefore, DGGE after computer analysis is a powerful method for nearly complete characterization of disease-producing mutations and polymorphisms in large genes such as that for factor VIII.
Full text
PDF



Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Antonarakis S. E., Kazazian H. H., Jr The molecular basis of hemophilia A in man. Trends Genet. 1988 Aug;4(8):233–237. doi: 10.1016/0168-9525(88)90156-4. [DOI] [PubMed] [Google Scholar]
- Arai M., Higuchi M., Antonarakis S. E., Kazazian H. H., Jr, Phillips J. A., 3rd, Janco R. L., Hoyer L. W. Characterization of a thrombin cleavage site mutation (Arg 1689 to Cys) in the factor VIII gene of two unrelated patients with cross-reacting material-positive hemophilia A. Blood. 1990 Jan 15;75(2):384–389. [PubMed] [Google Scholar]
- Arai M., Inaba H., Higuchi M., Antonarakis S. E., Kazazian H. H., Jr, Fujimaki M., Hoyer L. W. Direct characterization of factor VIII in plasma: detection of a mutation altering a thrombin cleavage site (arginine-372----histidine). Proc Natl Acad Sci U S A. 1989 Jun;86(11):4277–4281. doi: 10.1073/pnas.86.11.4277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arai M., Scandella D., Hoyer L. W. Molecular basis of factor VIII inhibition by human antibodies. Antibodies that bind to the factor VIII light chain prevent the interaction of factor VIII with phospholipid. J Clin Invest. 1989 Jun;83(6):1978–1984. doi: 10.1172/JCI114107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eaton D. L., Wood W. I., Eaton D., Hass P. E., Hollingshead P., Wion K., Mather J., Lawn R. M., Vehar G. A., Gorman C. Construction and characterization of an active factor VIII variant lacking the central one-third of the molecule. Biochemistry. 1986 Dec 30;25(26):8343–8347. doi: 10.1021/bi00374a001. [DOI] [PubMed] [Google Scholar]
- Fay P. J. Reconstitution of human factor VIII from isolated subunits. Arch Biochem Biophys. 1988 May 1;262(2):525–531. doi: 10.1016/0003-9861(88)90404-3. [DOI] [PubMed] [Google Scholar]
- Fischer S. G., Lerman L. S. DNA fragments differing by single base-pair substitutions are separated in denaturing gradient gels: correspondence with melting theory. Proc Natl Acad Sci U S A. 1983 Mar;80(6):1579–1583. doi: 10.1073/pnas.80.6.1579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foster P. A., Fulcher C. A., Houghten R. A., Zimmerman T. S. Synthetic factor VIII peptides with amino acid sequences contained within the C2 domain of factor VIII inhibit factor VIII binding to phosphatidylserine. Blood. 1990 May 15;75(10):1999–2004. [PubMed] [Google Scholar]
- Fulcher C. A., de Graaf Mahoney S., Roberts J. R., Kasper C. K., Zimmerman T. S. Localization of human factor FVIII inhibitor epitopes to two polypeptide fragments. Proc Natl Acad Sci U S A. 1985 Nov;82(22):7728–7732. doi: 10.1073/pnas.82.22.7728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giannelli F., Green P. M., High K. A., Lozier J. N., Lillicrap D. P., Ludwig M., Olek K., Reitsma P. H., Goossens M., Yoshioka A. Haemophilia B: database of point mutations and short additions and deletions. Nucleic Acids Res. 1990 Jul 25;18(14):4053–4059. doi: 10.1093/nar/18.14.4053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gitschier J., Drayna D., Tuddenham E. G., White R. L., Lawn R. M. Genetic mapping and diagnosis of haemophilia A achieved through a BclI polymorphism in the factor VIII gene. 1985 Apr 25-May 1Nature. 314(6013):738–740. doi: 10.1038/314738a0. [DOI] [PubMed] [Google Scholar]
- Gitschier J., Kogan S., Levinson B., Tuddenham E. G. Mutations of factor VIII cleavage sites in hemophilia A. Blood. 1988 Sep;72(3):1022–1028. [PubMed] [Google Scholar]
- Gitschier J., Wood W. I., Goralka T. M., Wion K. L., Chen E. Y., Eaton D. H., Vehar G. A., Capon D. J., Lawn R. M. Characterization of the human factor VIII gene. Nature. 1984 Nov 22;312(5992):326–330. doi: 10.1038/312326a0. [DOI] [PubMed] [Google Scholar]
- Higuchi M., Kochhan L., Schwaab R., Egli H., Brackmann H. H., Horst J., Olek K. Molecular defects in hemophilia A: identification and characterization of mutations in the factor VIII gene and family analysis. Blood. 1989 Aug 15;74(3):1045–1051. [PubMed] [Google Scholar]
- Higuchi M., Wong C., Kochhan L., Olek K., Aronis S., Kasper C. K., Kazazian H. H., Jr, Antonarakis S. E. Characterization of mutations in the factor VIII gene by direct sequencing of amplified genomic DNA. Genomics. 1990 Jan;6(1):65–71. doi: 10.1016/0888-7543(90)90448-4. [DOI] [PubMed] [Google Scholar]
- Hobbs H. H., Russell D. W., Brown M. S., Goldstein J. L. The LDL receptor locus in familial hypercholesterolemia: mutational analysis of a membrane protein. Annu Rev Genet. 1990;24:133–170. doi: 10.1146/annurev.ge.24.120190.001025. [DOI] [PubMed] [Google Scholar]
- Inaba H., Fujimaki M., Kazazian H. H., Jr, Antonarakis S. E. Mild hemophilia A resulting from Arg-to-Leu substitution in exon 26 of the factor VIII gene. Hum Genet. 1989 Mar;81(4):335–338. doi: 10.1007/BF00283686. [DOI] [PubMed] [Google Scholar]
- Kazazian H. H., Jr The thalassemia syndromes: molecular basis and prenatal diagnosis in 1990. Semin Hematol. 1990 Jul;27(3):209–228. [PubMed] [Google Scholar]
- Kogan S. C., Doherty M., Gitschier J. An improved method for prenatal diagnosis of genetic diseases by analysis of amplified DNA sequences. Application to hemophilia A. N Engl J Med. 1987 Oct 15;317(16):985–990. doi: 10.1056/NEJM198710153171603. [DOI] [PubMed] [Google Scholar]
- Kogan S., Gitschier J. Mutations and a polymorphism in the factor VIII gene discovered by denaturing gradient gel electrophoresis. Proc Natl Acad Sci U S A. 1990 Mar;87(6):2092–2096. doi: 10.1073/pnas.87.6.2092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lerman L. S., Silverstein K. Computational simulation of DNA melting and its application to denaturing gradient gel electrophoresis. Methods Enzymol. 1987;155:482–501. doi: 10.1016/0076-6879(87)55032-7. [DOI] [PubMed] [Google Scholar]
- Mann K. G. Membrane-bound enzyme complexes in blood coagulation. Prog Hemost Thromb. 1984;7:1–23. [PubMed] [Google Scholar]
- Myers R. M., Fischer S. G., Lerman L. S., Maniatis T. Nearly all single base substitutions in DNA fragments joined to a GC-clamp can be detected by denaturing gradient gel electrophoresis. Nucleic Acids Res. 1985 May 10;13(9):3131–3145. doi: 10.1093/nar/13.9.3131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Myers R. M., Lumelsky N., Lerman L. S., Maniatis T. Detection of single base substitutions in total genomic DNA. Nature. 1985 Feb 7;313(6002):495–498. doi: 10.1038/313495a0. [DOI] [PubMed] [Google Scholar]
- Myers R. M., Maniatis T., Lerman L. S. Detection and localization of single base changes by denaturing gradient gel electrophoresis. Methods Enzymol. 1987;155:501–527. doi: 10.1016/0076-6879(87)55033-9. [DOI] [PubMed] [Google Scholar]
- Orita M., Iwahana H., Kanazawa H., Hayashi K., Sekiya T. Detection of polymorphisms of human DNA by gel electrophoresis as single-strand conformation polymorphisms. Proc Natl Acad Sci U S A. 1989 Apr;86(8):2766–2770. doi: 10.1073/pnas.86.8.2766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pattinson J. K., McVey J. H., Boon M., Ajani A., Tuddenham E. G. CRM+ haemophilia A due to a missense mutation (372----Cys) at the internal heavy chain thrombin cleavage site. Br J Haematol. 1990 May;75(1):73–77. doi: 10.1111/j.1365-2141.1990.tb02618.x. [DOI] [PubMed] [Google Scholar]
- Pittman D. D., Kaufman R. J. Proteolytic requirements for thrombin activation of anti-hemophilic factor (factor VIII). Proc Natl Acad Sci U S A. 1988 Apr;85(8):2429–2433. doi: 10.1073/pnas.85.8.2429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saiki R. K., Gelfand D. H., Stoffel S., Scharf S. J., Higuchi R., Horn G. T., Mullis K. B., Erlich H. A. Primer-directed enzymatic amplification of DNA with a thermostable DNA polymerase. Science. 1988 Jan 29;239(4839):487–491. doi: 10.1126/science.2448875. [DOI] [PubMed] [Google Scholar]
- Shapiro M. B., Senapathy P. RNA splice junctions of different classes of eukaryotes: sequence statistics and functional implications in gene expression. Nucleic Acids Res. 1987 Sep 11;15(17):7155–7174. doi: 10.1093/nar/15.17.7155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheffield V. C., Cox D. R., Lerman L. S., Myers R. M. Attachment of a 40-base-pair G + C-rich sequence (GC-clamp) to genomic DNA fragments by the polymerase chain reaction results in improved detection of single-base changes. Proc Natl Acad Sci U S A. 1989 Jan;86(1):232–236. doi: 10.1073/pnas.86.1.232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shima M., Ware J., Yoshioka A., Fukui H., Fulcher C. A. An arginine to cysteine amino acid substitution at a critical thrombin cleavage site in a dysfunctional factor VIII molecule. Blood. 1989 Oct;74(5):1612–1617. [PubMed] [Google Scholar]
- Toole J. J., Knopf J. L., Wozney J. M., Sultzman L. A., Buecker J. L., Pittman D. D., Kaufman R. J., Brown E., Shoemaker C., Orr E. C. Molecular cloning of a cDNA encoding human antihaemophilic factor. Nature. 1984 Nov 22;312(5992):342–347. doi: 10.1038/312342a0. [DOI] [PubMed] [Google Scholar]
- Toole J. J., Pittman D. D., Orr E. C., Murtha P., Wasley L. C., Kaufman R. J. A large region (approximately equal to 95 kDa) of human factor VIII is dispensable for in vitro procoagulant activity. Proc Natl Acad Sci U S A. 1986 Aug;83(16):5939–5942. doi: 10.1073/pnas.83.16.5939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Traystman M. D., Higuchi M., Kasper C. K., Antonarakis S. E., Kazazian H. H., Jr Use of denaturing gradient gel electrophoresis to detect point mutations in the factor VIII gene. Genomics. 1990 Feb;6(2):293–301. doi: 10.1016/0888-7543(90)90569-g. [DOI] [PubMed] [Google Scholar]
- Wion K. L., Tuddenham E. G., Lawn R. M. A new polymorphism in the factor VIII gene for prenatal diagnosis of hemophilia A. Nucleic Acids Res. 1986 Jun 11;14(11):4535–4542. doi: 10.1093/nar/14.11.4535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong C., Dowling C. E., Saiki R. K., Higuchi R. G., Erlich H. A., Kazazian H. H., Jr Characterization of beta-thalassaemia mutations using direct genomic sequencing of amplified single copy DNA. 1987 Nov 26-Dec 2Nature. 330(6146):384–386. doi: 10.1038/330384a0. [DOI] [PubMed] [Google Scholar]
- Yoshitake S., Schach B. G., Foster D. C., Davie E. W., Kurachi K. Nucleotide sequence of the gene for human factor IX (antihemophilic factor B). Biochemistry. 1985 Jul 2;24(14):3736–3750. doi: 10.1021/bi00335a049. [DOI] [PubMed] [Google Scholar]
- Youssoufian H., Antonarakis S. E., Bell W., Griffin A. M., Kazazian H. H., Jr Nonsense and missense mutations in hemophilia A: estimate of the relative mutation rate at CG dinucleotides. Am J Hum Genet. 1988 May;42(5):718–725. [PMC free article] [PubMed] [Google Scholar]
- Youssoufian H., Kazazian H. H., Jr, Phillips D. G., Aronis S., Tsiftis G., Brown V. A., Antonarakis S. E. Recurrent mutations in haemophilia A give evidence for CpG mutation hotspots. 1986 Nov 27-Dec 3Nature. 324(6095):380–382. doi: 10.1038/324380a0. [DOI] [PubMed] [Google Scholar]