Abstract
Replication and transcription activator (RTA), an immediate-early gene product of gamma-2 herpesviruses including Kaposi's sarcoma-associated herpesvirus (KSHV) and murine gamma herpesvirus 68 (MHV-68), plays a critical role in controlling the viral life cycle. RTA acts as a strong transcription activator for several downstream genes of KSHV and MHV-68 through direct DNA binding, as well as via indirect mechanisms. HMGB1 (also called HMG-1) protein is a highly conserved nonhistone chromatin protein with the ability to bind and bend DNA. HMGB1 protein promoted RTA binding to different RTA target sites in vitro, with greater enhancement to low-affinity sites than to high-affinity sites. Box A or box B and homologues of HMGB1 also enhanced RTA binding to DNA. Transient transfection of HMGB1 stimulated RTA transactivation of RTA-responsive promoters from KSHV and MHV-68. Furthermore, MHV-68 viral gene expression, as well as viral replication, was significantly reduced in HMGB1-deficient cells than in the wild type. This abated viral gene expression was partially restored by HMGB1 transfection into HMGB1−/− cells. These results suggest an important function of the DNA architectural protein, HMGB1, in RTA-mediated gene expression, as well as viral replication in gamma-2 herpesviruses.
Gammaherpesviruses, including Epstein-Barr virus (EBV), Kaposi's sarcoma-associated herpesvirus (KSHV), herpesvirus saimiri (HVS), and murine gammaherpesvirus 68 (MHV-68) are characterized by their ability to infect and establish latency in lymphocytes. These viruses have been shown to be associated with many malignancies in lymphocytes, as well as in epithelial and endothelial cells. Although latent infection is thought to be important for tumorigenesis associated with these viruses, it has been suggested that even low-frequency viral reactivation, from latency to lytic replication, plays a significant role in pathogenesis (42, 46).
Replication and transcription activator (RTA) is a well-conserved immediate-early gene product among gammaherpesviruses (24, 45, 52, 54). Especially in gamma-2 herpesviruses, RTA alone functions as a potent molecular switch in controlling the viral life cycle between latency and lytic replication, as well as in de novo infection (16, 17, 45, 54). The N termini of RTA proteins share a highly conserved DNA-binding domain, followed by a leucine-zipper domain, and the C termini contain an activation domain with little homology to other proteins (23, 25, 30-32). The RTA of KSHV has been shown to activate several downstream genes, including PAN RNA, Kaposin (Kpsn or K12), open reading frame 57 (ORF57), viral interferon regulatory factors, thymidine kinase, ORF6, and the viral homolog of interleukin-6 (vIL-6 or K2) (7, 8, 10, 11, 29, 31, 39, 57). DNA binding of RTA is thought to be one of the important mechanisms of RTA transactivation, whereas interactions with cellular proteins are involved in activation of selected groups of target genes. A group of proteins have been shown to interact with KSHV RTA: CBP, HDAC1, Stat3, MGC2663, RBP-Jκ, C/EBPα, the SWI/SNF chromatin remodeling complex, the TRAP/Mediator coactivator, cellular poly(ADP-ribose) polymerase 1, and Ste20-like kinase hKFC (19-22, 27, 49, 50). However, the detailed mechanism by which RTA activates downstream target genes remains to be further elucidated.
Our recent comparative studies on PAN RNA, Kpsn, ORF57, and vIL-6 gene expression indicated that direct binding of RTA to these target sequences makes a significant contribution to activation of these promoters (40). RTA binding affinity for different target sites in vitro showed dramatic differences by up to 100-fold, based on competition analyses. When we examined the promoter strengths, transcription rates, and steady-state transcript levels to measure RTA responsiveness of four target promoters in vivo, the order of RTA responsiveness in vivo was consistent with that of RTA binding affinities in vitro. However, it was noted that variations in RTA responsiveness of these target genes in vivo were in the range of 2- to 10-fold, far less than the results in in vitro binding assays (40). This led us to propose a role of other cellular or viral factors in controlling the expression of RTA target genes in vivo, in addition to direct binding of RTA alone to DNA. These findings suggest that RTA interacts, either directly or indirectly, with distinct promoter sequences or that the RTA recognition is markedly degenerate. Another plausible explanation for the binding of different promoters is the interaction of RTA with cellular transcriptional coactivators that alter the capacity and specificity for DNA recognition. The fact that their TATA boxes differ may also contribute to these results.
High-mobility-group proteins (HMGs) are a large group of heterogeneous chromosomal proteins with little homology at the level of sequence and structure but thought to function as architectural elements that facilitate interactions of neighboring proteins and DNA by creating favorable DNA conformations (5). One of the most abundant HMGs belongs to the HMG box protein subfamily. The HMG box proteins include HMGB1 and HMGB2, which are highly abundant (∼106 molecules per nucleus) and ubiquitously expressed in eukaryotic cells (47, 48). HMGB1 is especially highly conserved in mammals with >95% amino acid identity between rodent and human forms (51). HMGB1 and -2 are extremely versatile in that they are involved in numerous DNA events in the nucleus such as DNA repair, replication, recombination, and transcription (5, 35). HMGB1, as well as HMGB2, is characterized by a tripartite structure consisting of two tandem HMG box domains (A and B) and an acidic C terminus composed of glutamic and aspartic residues. Both HMG boxes A and B share a common three-dimensional fold structure. Three α-helices arranged in the shape of an L form an 80-amino-acid motif, which can bind and bend DNA in part by intercalating hydrophobic amino acids between the base pairs. Interactions between the HMG box and DNA through the minor groove largely determine the DNA-binding properties of HMGB1 (18, 43, 44). HMGB1 has been shown to stimulate the binding of a variety of sequence-specific DNA-binding proteins in vitro. These include p53, Hox domain proteins, steroid hormone receptors, octamer binding factors (Oct1/2), the RAG1/2 recombinase, and the viral transactivators ZEBRA and RTA of EBV (1, 2, 4, 12, 13, 26, 34, 56). Direct interactions between the HMG boxes of HMGB1 and sequence-specific DNA-binding proteins were reported in most cases, whereas there was no evidence of direct contacts in the cases of EBV ZEBRA and RTA (12, 13, 34). Unlike gamma-2 herpesviruses, both ZEBRA and RTA of EBV, a gamma-1 herpesvirus, are critical in controlling the viral life cycle through activation of their target genes (9, 38, 55). The role of HMGB1 in the enhanceosome assembly of these viral transcription activators has been studied in the context of the viral promoter BHLF1 (34). Interestingly, it was found that HMGB1 displays two distinct modes of action in promoting ZEBRA and RTA binding to their cognate sites. In the case of ZEBRA, HMGB1 and ZEBRA form a nucleoprotein complex wherein HMGB1 binds to the DNA segment between two ZEBRA-binding sites in a sequence-dependent manner to promote the cooperative assembly of a stable ZEBRA and HMGB1 nucleoprotein complex (13). In contrast, HMGB1 DNA association is transient and displays little specificity when it promotes RTA-DNA binding (34). Despite apparent dissimilarities in the modes of the action of HMGB1, DNA bending activity rather than specific protein-protein interactions has been proposed to be a main contributing factor to binding enhancement by HMGB1.
Given the differences in molecular switch systems between gamma-1 and gamma-2 herpesviruses, it is intriguing to determine whether the DNA architectural protein, HMGB1, plays a similar role in RTA function in gamma-2 herpesviruses. In addition, it is important to study how the role of HMGB1 applies to the virus life cycle in the context of the viral genome rather than in isolated promoters. We set out to examine the effect of HMGB1 on RTA binding to four different cognate sites and found enhancement of RTA binding by HMGB1. Next, we took advantage of the robust viral replication system of MHV-68, as well as cell lines established from HMGB1 knockout mice. Our results indicate that the DNA architectural protein HMGB1 enhances RTA-mediated viral gene expressions and replication of gamma-2 herpesviruses.
MATERIALS AND METHODS
Cells, virus, and plaque assays.
All cells were cultured at 37°C in the presence of 5% CO2. 293T and BHK-21 cells were cultured in Dulbecco modified Eagle medium (Cellgro) containing 10% fetal bovine serum and antibiotics (50 U of penicillin and 50 μg of streptomycin/ml). Mouse HMGB1−/− (C1) and wild-type (VA1) fibroblast cell lines were kindly provided by M. E. Bianchi (6) and were grown in Dulbecco modified Eagle medium supplemented with 10% fetal bovine serum and antibiotics. Construction of enhanced green fluorescent protein (EGFP)/MHV-68, a recombinant MHV-68 expressing EGFP, was described in detail elsewhere (53). Briefly, the EGFP expression cassette including the cytomegalovirus (CMV) promoter was cloned into TW25 vector containing a region homologous to the left end of the virus genome. The virus titer was determined by plaque assays by using BHK-21 monolayers overlaid with medium containing 1% methylcellulose as previously described (53, 54).
Transfection and flow cytometry analysis.
All reporter plasmids containing a copy of each RRE from the promoters of PAN RNA, Kpsn, ORF57, and vIL-6 have been described (40). mORF57p containing 565-bp fragment, spanning from bp 75218 to 75782 in the MHV68 genome was cloned into pGL3-Basic (Promega) (28). Construction of mM3-2p containing 595-bp fragment upstream of the M3 TATA box was previously described elsewhere (33). For the luciferase reporter assays, 3 × 105 293T cells were transfected with 1 to 100 ng of pcDNA3/RTA or mFLAG/RTA and 25 ng of a reporter plasmid into 12-well plates in the presence or absence of the HMGB1-expressing plasmid (pFLAGHMGB1; 100 ng) by using a calcium-phosphate transfection method. Each transfection for reporter assays was performed in duplicate and contained 1 ng of pRLCMV or pRLSV40 (Promega) as a control for transfection efficiency, as well as 375 ng of pcDNA3. At 24 h posttransfection, cells were washed with 1× phosphate-buffered saline and subjected to reporter assays.
To reconstitute HMGB1 expression in HMGB1 deficient cells, 20 μg of a murine HMGB1-expressing plasmid (pHMGB1) or a vector [pBluescript II KS(−)] was introduced by electroporation (960 μF, 250 V, 200 Ω) with a Genepulser II (Bio-Rad, Hercules, Calif.) into 4 × 106 HMGB1-deficient cells in incomplete medium in the presence of 4 μg of pCMVDsRed2Exp plasmid expressing red fluorescent protein (RFP). Electroporated cells were divided into four wells in a six-well plate. At 24 h posttransfection, the cells were either mock infected or infected with EGFP/MHV-68 at a multiplicity of infection (MOI) equivalent to 0.005, 0.05, or 0.5 PFU/cell. The vector-transfected cells were used as a control for flow cytometry analysis. At day 3 postinfection, the cells transfected with the RFP-expressing plasmid and infected with EGFP/MHV-68 were sorted on a FACTStarPlus (Becton-Dickinson) and counted for EGFP- and RFP-positive cells. To establish cell lines that stably express HMGB1 in HMGB1-deficient cells, pHMGB1 (4 μg), as well as pBabe-puro (0.4 μg), was introduced by using Lipofectamine 2000 (Invitrogen), and the pHMGB1-transfected cells were selected in the presence of puromycin (4 μg/ml) for 4 weeks and examined for HMGB1 expression by Western and real-time PCR analysis.
EMSAs.
The recombinant Rdbd protein (amino acids 1 to 320) tagged with a FLAG peptide at the N terminus and six-histidine residues at the C terminus was expressed in bacteria and purified as previously described (41). Various forms of recombinant HMGB1 were expressed and purified as described in previous studies (34). The recombinant HMGB1 AB protein containing HMG box A and box B was used instead of HMGB1 unless otherwise indicated. A set of double-stranded oligonucleotides, including pan1 (pPAN RRE), Kpsn (pKpsn RRE), K-ORF572 (pORF57 RRE), and K2p25 (pvIL-6 RRE), were used for electrophoretic mobility shift assays (EMSAs), and their sequences are indicated in Fig. 1A. The detailed sequence information is described elsewhere (40). All double-stranded oligonucleotides were end labeled with [γ-32P]ATP, followed by fill-in reaction, and EMSAs were performed as previously described (39). Supershift assays were performed with a monoclonal antibody (2.2 μg) against a FLAG peptide (Sigma) and polyclonal antibody (1 μl) against HMGB1 (BD Pharmingen, San Diego, Calif.).
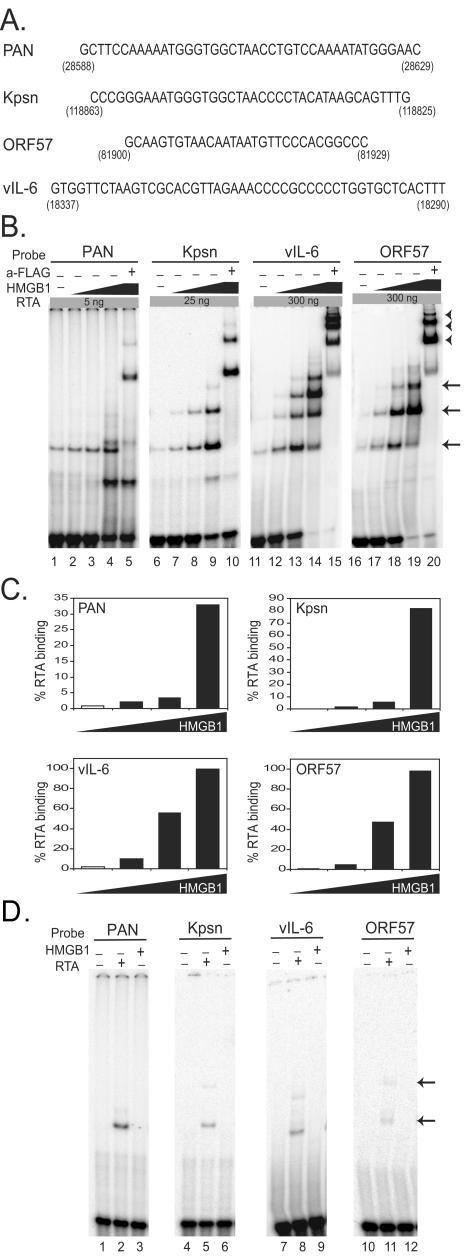
FIG. 1.
In vitro binding of KSHV RTA in the presence or absence of HMGB1. (A) Sequences of previously identified RREs from the promoters of PAN, Kpsn, vIL-6, and ORF57 of KSHV. Numbers indicate the locations of each RRE in the KSHV genome. (B) Dose-dependent enhancement of HMGB1 on RTA binding to RREs. Re-combinant RTA protein (amino acids 1 to 320 out of a total 691 of amino acids), including a putative DNA-binding domain as well as a leucine zipper domain, was tagged with FLAG at the N terminus and six-histidine residues at the C terminus. Recombinant HMGB1 protein contains HMG box A and box B domains without C-terminus tails (HMGB1 AB). A double-stranded probe (1 fmol) encompassing an RRE from pPAN, pKpsn, pvIL-6, or pORF57 with common flanking sequences was 32P end labeled and incubated with RTA protein in the presence of increasing amounts of HMGB1 (0, 80, 120, and 160 ng). These RREs are shown to confer different levels of RTA-binding affinities in the absence of HMGB1. Different amounts of the purified RTA protein (5, 25, or 300 ng) were incubated with end-labeled probes according to their RTA-binding affinities: PAN (5 ng; lanes 1 to 5), Kpsn (25 ng; lanes 6 to 10), vIL-6 (300 ng; lanes 11 to 15), and ORF57 (300 ng; lanes 16 to 20). Arrows indicate the DNA-protein complexes of RTA. Anti-FLAG antibody that recognizes recombinant RTA protein was used to confirm specificity of the RTA protein-DNA complexes (lanes 5, 10, 15, and 20). Arrowheads indicate the DNA-RTA complexes supershifted by anti-FLAG antibody. (C) Quantitative analysis of RTA binding with increasing amounts of HMGB1 protein. RTA-binding affinity to the probe was calculated based on the ratio of the bound probe to the total (bound plus unbound) in the absence (□) or presence (▪) of HMGB1. (D) EMSAs with HMGB1 alone. The 32P-end-labeled probe (1 fmol) encompassing a RRE from pPAN, pKpsn, pvIL-6, or pORF57 was incubated with either RTA protein (5, 25, or 300 ng) or HMGB1 alone (40 ng).
Dual luciferase assays.
The dual luciferase reporter assay system (Promega) was used to test promoter activity. Transfected 293T cells in 12-well plates were washed with 1× phosphate-buffered saline and incubated with 250 μl of 1× passive lysis buffer provided by the manufacturer. Lysates were frozen, thawed once, and centrifuged at top speed in a microcentrifuge for 5 min. Lysates were assayed by using an Optocomp I Luminometer (MGM Instruments, Hamden, Calif.). The reporter assays were carried out according to the manufacturer's protocol for the dual luciferase reporter assay system (Promega). The Renilla luciferase activities from pRLCMV or pRLSV40 plasmid were used as an internal control to normalize the transfection efficiency in different transfection samples.
Western analysis.
To examine the expression of a viral protein, cells were harvested with sample buffer and denatured prior to loading on a 12% sodium dodecyl sulfate-polyacrylamide electrophoresis gel. Western analysis with polyclonal antibody against M9 protein (1:250) was carried out as previously described (41). Antibody to HMGB1 (1:5,000; BD Biosciences Pharmingen, San Diego, Calif.) was used to detect the expression of HMGB1. The same membranes were reprobed with antibody against actin (1:500) as a loading control.
Real-time PCR.
Viral DNAs from supernatants of infected cells were extracted by using DNeasy kit (Qiagen) according to the manufacturer's recommendations. One-tenth of total DNA extracted from supernatants and ORF56-specific primers (nucleotides 75598 to 75783 of the MHV-68 genome; M56F, 5′-GTAACTCGAGACTGAAACCTCGCAGAGGTCC-3′; M56R, 5′-CCGAAGCTTGCACGGTGCAATGTGTCACAG-3′) were mixed with 2× Master mix containing SYBR Green (Applied Biosystems). Real-time PCR was run at 95°C for 15 min, followed by 45 cycles of 95°C for 30 s, 60°C for 30 s, and 72°C for 15 s, with the results analyzed in Opticon II (MJ Research).
RESULTS
HMGB1 facilitates KSHV RTA binding to the target sequences with various RTA binding affinities.
RTA of KSHV is a potent transactivator of several downstream genes. RTA-responsive elements (RREs) were identified in the promoters of some target genes, including PAN RNA, Kpsn, ORF57, and vIL-6 (10, 11, 29, 31, 39, 41). The RREs in the promoters of PAN RNA and Kpsn are highly homologous with 16 consecutive base pairs of identical sequence and 5-bp additional exact matches, all contained within a short 25-bp region of the respective RREs. In contrast, the RREs of ORF57 and vIL-6 have no apparent similarity. The RTA-responsive sequences of these promoters are presented in Fig. 1A, with numbers indicating the genomic locations. RTA was shown to bind to these RREs to various extents: the RREs for PAN and Kpsn appeared to bind with high affinity, whereas those for ORF57 and vIL-6 bound with low affinity (40).
First, we set out to test whether the enhancement of RTA binding by HMGB1 is conserved among gammaherpesviruses. We examined the effect of HMGB1 protein on KSHV RTA binding by using EMSAs. The DNA-binding domain of RTA and a recombinant form of HMGB1 protein containing HMG box A and B were expressed in bacteria and purified near to homogeneity. Increasing amounts of the HMGB1 protein (0, 80, 120, and 160 ng) were incubated with preselected amounts of RTA and 32P-end-labeled oligonucleotides (1 fmol) bearing RRE sequences. The amount of RTA (5, 25, or 300 ng) incubated with each probe was predetermined, based on RTA-binding affinity to the RREs in order to show detectable RTA binding in the absence of HMGB1. Coincubation of HMGB1 resulted in increased formation of RTA/DNA complexes in a dose-dependent manner for each probe (Fig. 1B, lanes 1 to 4, 6 to 9, 11 to 19, and 21 to 24). The specificity of these complexes was confirmed with anti-FLAG antibody that supershifted RTA-containing complexes since RTA contains the FLAG tag at the N terminus (Fig. 1B, lanes 5, 10, 15, and 20). The binding affinity of RTA to its RREs was calculated as a ratio of the bound probe to the total (the bound plus the unbound) and shown in Fig. 1C. EMSA results showed that HMGB1 enhancement of RTA binding has little sequence specificity since it increased RTA binding to quite diverse sequences. In some cases, mobility shift assays of RTA complexed with DNA revealed a series of bands that are believed to be a result of different RTA conformations or oligomeric status. In the presence of HMGB1, there was increased formation of these RTA/DNA complexes. However, these multiple complexes migrated identically to those assembled without HMGB1; no additional shifted complex, which could be attributed to the presence of HMGB1, was observed. As a negative control, HMGB1 alone (40 ng) was incubated in the absence of RTA and did not yield any specific shift of each probe (Fig. 1D). Incubation with a higher dose of HMGB1 (150 ng) also showed no shifted band (data not shown). This is consistent with other studies showing no alteration in mobility due to HMGB1 (4, 34, 56).
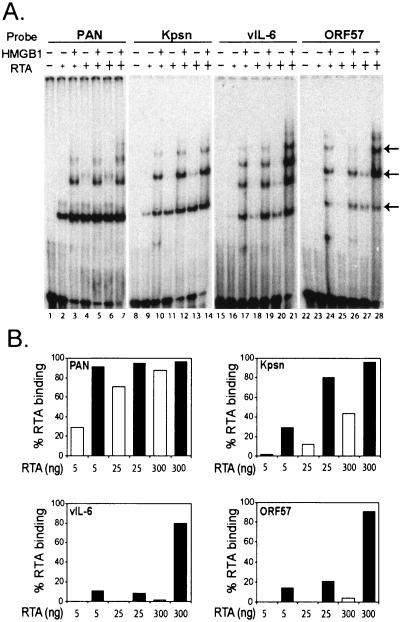
To examine the degree of HMGB1 effect on RTA binding to each RRE, we used increasing amounts of RTA (5, 25, and 300 ng) with or without the fixed amount of HMGB1 (150 ng) (Fig. 2A). A quantitative analysis of RTA binding is shown in Fig. 2B. In agreement with our previous study, EMSA results showed various binding affinities of different RREs; PAN and Kpsn RREs conferred higher binding affinity than the vIL-6 and ORF57 RREs (Fig. 2B). HMGB1 protein facilitated RTA binding to all RREs, notably with greater effect on binding low affinity sites than high-affinity sites. In addition, the higher fold of enhancements by HMGB1 was detected when the lower amounts of RTA were used. For example, at a dose of 300 ng of RTA, vIL-6, and ORF57 RREs displayed ∼5% RTA binding, but this increased to >80% when HMGB1was added to the binding mixtures, manifesting >16-fold increase (Fig. 2B, lower panels). However, HMGB1 stimulated a threefold increase of RTA binding to the highest binding affinity site (PAN RRE) at 5 ng, and little enhancement at 300 ng of RTA. These results are consistent with our notion that HMGB1 may be a cellular factor that contributes to enhanced RTA binding in vivo, to low-affinity sites, which would in turn increase RTA transactivation of these promoters in cells.
FIG. 2.
HMGB1 enhancement of RTA binding was greater to low-affinity binding sites than to high-affinity ones. (A) EMSAs with increasing amounts of RTA protein in the absence or presence of HMGB1 protein. Increasing amounts of RTA protein (5, 25, and 300 ng) were incubated with the end-labeled probes (1 fmol) PAN (lanes 1 to 7), Kpsn (lanes 8 to 14), vIL-6 (lanes 15 to 21), and ORF57 (lanes 22 to 28). Recombinant HMGB1 protein (150 ng) was added to each reaction mixture as indicated. Arrows indicate the DNA-protein complexes of RTA. (B) Quantitative analysis of RTA DNA binding stimulated by HMGB1 protein. RTA-binding affinities were shown in the absence (□) or presence (▪) of HMGB1 with increasing amounts of RTA.
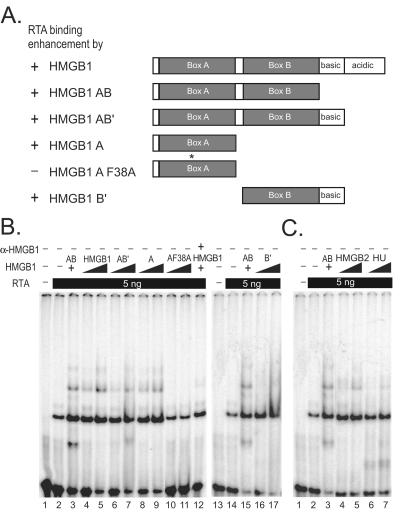
Box A and box B and other functional homologues of HMGB1 were sufficient to promote RTA binding.
HMGB1 consists of two DNA-binding domains, box A and B, followed by basic and acidic tails at the C terminus (Fig. 3A). A set of deletion mutants of HMGB1 were tested for their ability to facilitate RTA binding to the PAN RRE. EMSA results showed that each mutant was capable of promoting RTA binding to the same extent as full-length HMGB1 (Fig. 3B), suggesting that DNA bending caused by either HMG box may be sufficient to facilitate RTA binding. A mutation in the box A was introduced at phenylalanine 38 (HMGB1A F38A) known to intercalate into the minor groove of DNA. Phenylalanine 38 of box A has been shown to be critical for DNA bending and DNA binding (36). This mutant form of HMGB1 box A failed to stimulate RTA binding, reinforcing the notion that DNA bending and binding by HMGB1 is important in facilitating RTA binding (Fig. 3B, lanes 10 and 11). When anti-HMGB1 antibody was coincubated with the full-length HMGB1, enhancement of RTA binding by HMGB1 was significantly reduced, suggesting that increased RTA binding is a HMGB1-specific effect (Fig. 3B, lanes 4, 5, and 12).
FIG. 3.
Effects of individual boxes and homologues of HMGB1 on KSHV RTA binding. (A) The schematic diagram is shown with the summarized effect of each domain of HMGB1 on RTA-binding enhancement in comparison with that of recombinant AB domain of HMGB1. Individual DNA-binding domains of HMGB1 stimulate RTA binding to pPAN RRE. A mutation (F38A) that abolishes its DNA bending and binding was introduced to HMGB1 box A, as indicated with an asterisk. (B) The end-labeled pPANRRE (1 fmol) was incubated with RTA protein (5 ng) with increasing amounts of individual domains of HMGB1: HMGB1 AB (150 ng, lanes 3 and 15), HMGB1 full-length (300 and 600 ng, lanes 4 and 5; 600 ng, lane 12), HMGB1 AB′ (300 and 600 ng, lanes 6 and 7), HMGB1 A (300 and 600 ng, lanes 8 and 9), and HMGB1 AF38A (300 and 600 ng, lanes 10 and 11). HMGB1 B′ (300 and 600 ng, lanes 16 and 17). Purified polyclonal antibody to HMGB1 was coincubated with full-length HMGB1 protein to test the specificity of HMGB1 effect on RTA-binding enhancement (lane 12). (C) Homologues of HMGB1, eukaryotic and prokaryotic architectural proteins, promote RTA binding to pPAN RRE. RTA protein (5 ng) was incubated with the labeled pPAN RRE probe (1 fmol) in the presence of increasing amounts of HMG2 (150 and 300 ng, lanes 4 and 5) and HU (150 and 300 ng, lanes 6 and 7) architectural proteins. HMGB1 AB protein (150 ng, lane 3) was used as a positive control.
Other functional homologues of HMGB1 were tested for their ability to facilitate RTA binding to the pPAN RRE (Fig. 3C). HMG2, a protein highly homologous to HMGB1, elicited similar enhancement to that of HMGB1. HU, a bacterial protein distantly related to HMGB1, was also able to facilitate RTA binding. Taken together, these results argue for the hypothesis that DNA bending induced by DNA architectural proteins plays a significant role in promoting RTA binding, although we cannot completely rule out the possibility of specific transient protein-protein interactions that might occur but not be detected in this experimental system.
HMGB1 stimulated RTA-mediated transactivation of RREs.
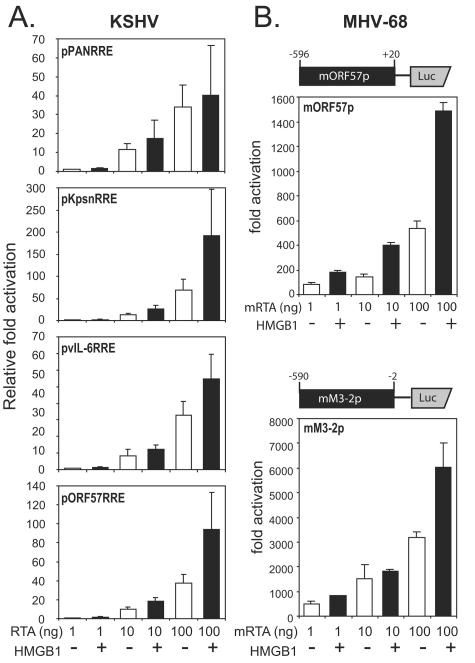
One copy of each RRE from the promoters of PAN, Kpsn, ORF57, and vIL-6 was cloned in front of the luciferase reporter to examine the effect of HMGB1 on RTA transactivation in a transient-transfection assay. Different amounts of the RTA expressing plasmid (1, 10, and 100 ng) were transfected into 293T cells in the presence or absence of the construct expressing HMGB1 (100 ng). HMGB1 alone activated neither the promoters containing RREs nor the CMV promoter used as a control (data not shown). Consistent with our results from in vitro binding assays, HMGB1 facilitated RTA-mediated transactivation of all RREs tested (Fig. 4A). Although less prominent in reporter assays than in binding assays, the extent of enhancement by HMGB1 was comparable to studies related to other sequence-specific transcription activators (4, 34, 56). This might be due to the abundant amount of preexisting HMGB1 and/or redundant functional homologues in cells.
FIG. 4.
HMGB1 effects on RTA-mediated transactivation in gamma-2 herpesviruses. (A) HMGB1 promotes KSHV RTA-mediated transactivation of RREs. One copy of each RRE upstream of the adenovirus E4 core promoter sequences was cloned into a luciferase reporter. Each reporter construct (25 ng) was cotransfected into 293T cells with various amounts (1 to 100 ng) of RTA-expressing plasmid (pcDNA3/gRTA) in the absence (□) or presence (▪) of HMGB1-expressing plasmid (pFLAGHMGB1; 100 ng). A plasmid, pRLCMV (1 ng), constitutively expressing Renilla luciferase was added to each transfection as an internal control. At 24 h posttransfection, the cells were harvested and subjected to dual luciferase assays. Promoter activities were calculated, relative to the fold activation from transfection of 1 ng of pcDNA/gRTA for each reporter construct. The values represent the averages of at least three independent transfections in duplicate, and the standard deviations are shown as error bars. (B) HMGB1 potentiates MHV-68 RTA-mediated transactivation. A schematic diagram of a reporter construct for the ORF57 promoter of MHV-68 (mORF57p, upper panel) or for the ORF M3 promoter (mM3-2p, lower panel) is shown above the graphs. HMGB1 effects on MHV-68 RTA (mRTA)-mediated transactivation of mORF57p (upper panel) or mM3-2p (lower panel). Each reporter construct (25 ng) was cotransfected into 293T cells with various amounts (1 to 100 ng) of mRTA-expressing plasmid (pFLAG/mRTA) in the absence (□) or presence (▪) of HMGB1-expressing plasmid (pFLAGHMGB1; 100 ng). A plasmid, pRLCMV (1 ng), constitutively expressing Renilla luciferase was added to each transfection as an internal control. At 24 h posttransfection, the cells were harvested and subjected to dual luciferase assays. The fold activations by mRTA were calculated relative to luciferase activity from cells transfected with pcDNA3 alone. Transfections were performed in triplicate, and the standard deviation is indicated by error bars.
HMGB1 also facilitates mRTA-mediated transactivation in MHV-68.
MHV-68 belongs to a gamma-2 herpesvirus subfamily that is closely related to KSHV. RTA of MHV-68 (mRTA) is also necessary and sufficient to initiate viral lytic replication. Both promoters from mORF57 and M3 were previously shown to be mRTA responsive (28, 33, 53). Segments (∼600 bp) of the mORF57 and M3 promoters, which contain RTA-responsive elements, were cloned into a reporter plasmid driving luciferase expression (Fig. 4B, upper panel). ORF57 is a well-conserved gene among gammaherpesviruses, whereas M3 is unique to MHV-68. The two reporter constructs driven by each promoter were individually transfected into 293T cells with various amounts of mRTA (1, 10, and 100 ng) in the presence or absence of HMGB1 (100 ng). Although the detailed mechanisms by which these promoters are activated by mRTA are not yet known, cotransfection of HMGB1 enhanced both promoter activities, implying a conserved role of HMGB1 in RTA function (Fig. 4B, lower panel). Transfection of pFLAGHMGB1 (100 ng) in the absence of mRTA did not exhibit activation of the reporters (data not shown).
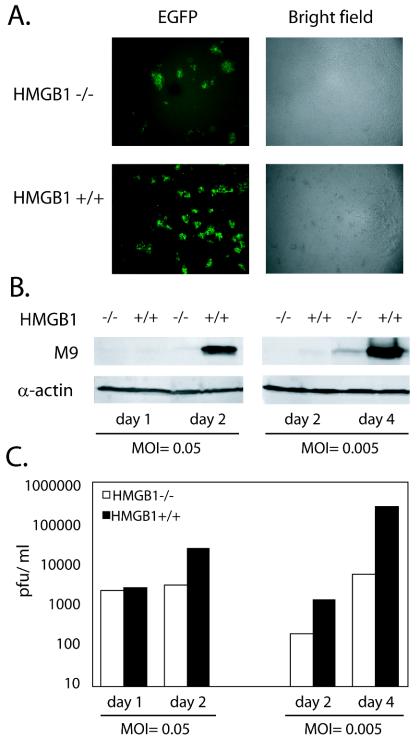
MHV-68 viral replication was reduced in HMGB1−/− cells compared to that in HMGB1+/+ cells.
Our results are consistent with the recent study on HMGB1 and EBV RTA, suggesting that the role of HMGB1 on RTA might be conserved among gammaherpesviruses. However, a limited experimental system for human gammaherpesviruses has impeded understanding of this phenomenon in the context of the viral genome and the ability to test the effect of HMGB1 on viral replication. Unlike human herpesviruses, many permissive cell lines, including fibroblast cell lines, are available to support the robust replication of MHV-68, enabling us to examine the role of HMGB1 on virus replication. To study how HMGB1 may affect virus replication, fibroblast cell lines from HMGB1−/− and wild-type littermates were obtained (courtesy of M. E. Bianchi) (6) and used for functional assays of MHV-68 replication. MHV-68 expressing EGFP (EGFP/MHV-68) was previously generated by inserting an expression cassette of EGFP at the left end of the viral genome (53). EGFP/MHV-68 grows similarly to the wild-type virus in tissue culture and was therefore used to monitor virus replication by monitoring the expression of EGFP. HMGB1-deficient and wild-type fibroblasts were infected with EGFP/MHV-68 at MOIs equivalent to 0.005 or 0.05. MOIs were calculated as the PFU of viruses per cell. One PFU is thought to contain approximately 10 infectious virus particles, based on the results from comparing plaque assays and limiting dilutions of the same virus stock. The cells were harvested at the indicated time points, and three assays were carried out to investigate the role of HMGB1 in virus replication: EGFP expression, Western analysis, and plaque assays. At each time point, pictures were taken for expression of EGFP as a measure of virus replication, and a set of representative pictures for EGFP and the corresponding bright fields are shown in Fig. 5A. At 4 days postinfection, the expression of EGFP was detected in both cell lines. However, MHV-68 exhibited more robust replication in wild-type fibroblasts than in the HMGB1-deficient cell line (Fig. 5A, left panels). This did not appear to be due to different cell growth rates of the two different cell lines since they showed similar density at the time of harvest (Fig. 5A, right panels). Next, the harvested cells were subject to Western analysis. Expression of M9 or ORF65, an abundant capsid protein of MHV-68, has been used successfully to indicate the gene expression of MHV-68 at the levels of transcription and translation and was therefore examined as a sensitive marker for viral gene expression (33). At both high and low MOIs, M9 expression was dramatically reduced in the HMGB1-deficient cell line versus the wild type (Fig. 5B, upper panels). The same membranes were probed for α-actin expression as a loading control (Fig. 5B, lower panels). Third, the supernatants from the infected cell cultures were harvested, and their titers were measured for PFU of infectious virus particles (Fig. 5C). Infectious virus titers from the wild-type cells were up to 50-fold higher than from the HMGB1−/− cells. Moreover, the hampered ability of HMGB1-deficient cells to support virus replication was more apparent in low-MOI infections than in high-MOI infections, which is probably due to accumulated effects of lost HMGB1 protein during multiple rounds of replication at low MOIs.
FIG. 5.
MHV-68 viral replication in HMGB1−/− and HMGB1+/+ cells. (A) Viral replication of a recombinant MHV-68 expressing EGFP (EGFP/MHV-68) in HMGB1−/− and HMGB1+/+ cells. EGFP/MHV-68 was infected into cell lines established from HMGB1−/− and HMGB1+/+ mice at an MOI equivalent to 0.005 or 0.05 PFU/cell in a six-well plate. On day 4 postinfection, green fluorescence images were recorded to measure viral replication. Bright-field pictures were also taken to show cell density. Representative sets of the pictures at an MOI of 0.005 PFU/cell are shown. (B) Viral gene expression in HMGB1−/− and HMGB1+/+ cells. HMGB1−/− and HMGB1+/+ cells were infected with EGFP/MHV-68 at different MOIs of either 0.005 or 0.05 PFU/cell and harvested at the indicated days postinfection. Protein samples were analyzed with polyclonal antibody to M9 (ORF65), a viral capsid protein. The same membranes were reprobed with monoclonal antibody against α-actin as a loading control. (C) Virus titers from HMGB1−/− and HMGB1+/+ cells infected by EGFP/MHV-68. From the same samples described in panel B, the supernatants were collected and subjected to plaque assays to measure the PFU of the viruses produced in HMGB1−/− and HMGB1+/+ cells. Three independent plaque assays were performed in duplicate.
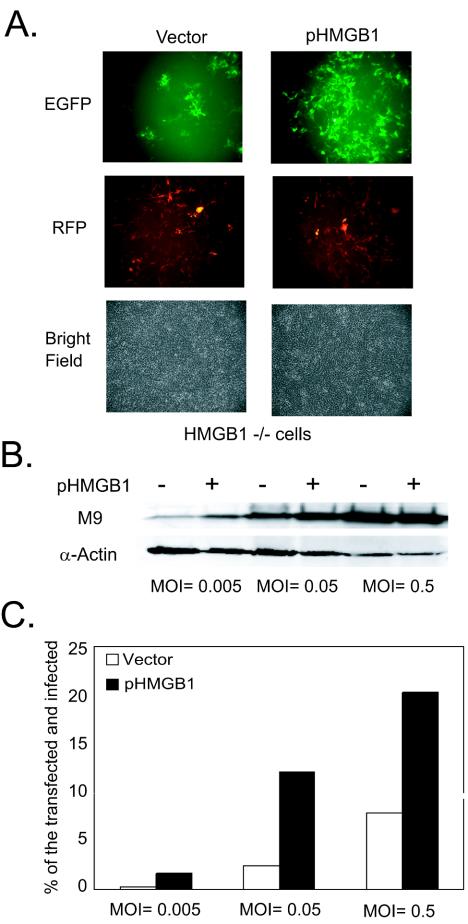
HMGB1 transfection into HMGB1-deficient cells restored their ability to support virus replication.
To verify the contribution of HMGB1 to MHV-68 virus replication in the two cell lines, we performed reconstitution experiments to restore HMGB1 expression in HMGB1-deficient cells. Either a vector alone or a plasmid expressing murine HMGB1 (pHMGB1; 5 μg) (14) was transfected into HMGB1-deficient cells, along with a control plasmid expressing RFP. At 24 h posttransfection, the cells were infected with EGFP/MHV-68 at MOIs of 0.005, 0.05, and 0.5 (PFU/cell). At day 3 postinfection, we examined EFGP expression for virus replication and RFP for transfection efficiency. A set of representative pictures is shown in Fig. 6A. Similar transfection efficiencies were obtained from both transfections, as determined by RFP expression. Transfection of the HMGB1-expressing plasmid enhanced virus replication in the HMGB1−/− cells compared to that in the vector-transfected cells (Fig. 6A). In addition, the HMGB1−/− cells were subjected to flow cytometry analysis and scored for transfected (red) and infected (green) cells (Fig. 6C). When the percentages of the transfected and infected cells were examined and counted as double positive for green and red fluorescence, the number of double-positive cells was significantly increased (three- to fivefold) in HMGB1-transfected cells at all of the MOIs tested, suggesting that more vigorous virus replication occurred in the cells that restored HMGB1 expression. The results from Western analysis with M9 antibody were also consistent with these findings in that M9 protein expression was enhanced in the HMGB1-transfected cells (Fig. 6B). The differences were more apparent when the cells were infected with virus at lower MOIs. Due to relatively low transfection efficiency (20 to 30%) into the HMGB1-deficient cells, as determined by flow cytometry analysis, it would be more difficult to discriminate between the portion of virus replication contributed by the transfected cell population and the portion contributed by the untransfected at higher MOIs. A large number of untransfected cells stemming from a low transfection efficiency may account for a moderate increase (two- to threefold) in the virus titers from the HMGB1-transfected cells compared to those from the vector-transfected cells (data not shown).
FIG. 6.
Effects of HMGB1 supplementation in HMGB1-deficient cells on MHV-68 viral replication. (A) Viral replication of EGFP/MHV-68 in HMGB1−/− transfected with a vector alone or a murine HMGB1-expressing plasmid (pHMGB1). HMGB1−/− cells were transfected with 5 μg of either vector alone or pHMGB1 in addition to a control plasmid expressing RFP (pCMVDsRed2Exp) and then infected with EGFP/MHV-68 at various MOIs from 0.005 to 0.5 PFU/cell at 24 h posttransfection. Green and red fluorescence images were recorded at 3 days postinfection. A set of representative pictures from infection at an MOI of 0.05 PFU/cell are shown for viral replication (EGFP), transfection efficiency (RFP), and cell density (bright field). (B) Viral gene expression in HMGB1−/− transfected with a vector alone or pHMGB1 (5 μg). The cells were harvested at 3 days postinfection after transfection as described in panel A. Western analyses were performed with anti-M9 antibody. The same membranes were stripped and reprobed with α-actin antibody as a loading control. (C) Flow cytometry analysis of the transfected and infected HMGB1−/− cells. As described in panel A, the HMGB1−/− cells were harvested at 3 days postinfection and subjected to flow cytometry analysis. The cells were assayed for RFP expression of transfected cell population and for EGFP expression of virus-infected cell population.The percentage of double-positive cells (RFP+EGFP) was shown to be the percentage of the transfected and infected population in HMGB1−/− cells (□) and HMGB1−/− cells transfected with pHMGB1 (▪).
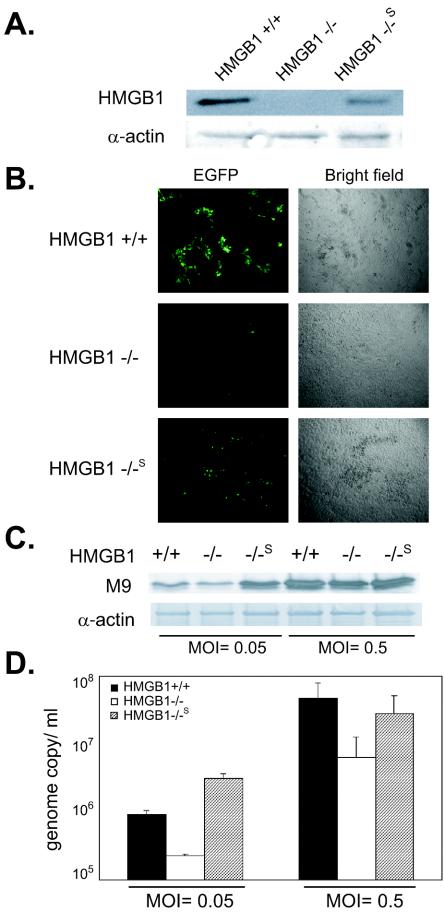
Due to the heterogeneous cell population in our transient transfection, we set to establish a reconstituted clone (HMGB1−/−S) of HMGB1-deficient cells. The results of Western analysis with anti-HMGB1 antibody showed that the level of HMGB1 expression in HMGB1−/−S was partially restored (Fig. 7A). To compare the ability of these cells to support virus replication, EGFP/MHV-68 was infected at MOIs of 0.05 and 0.5 (PFU/cell). At day 3 postinfection, we examined EFGP expression for virus replication (Fig. 7B). There was a substantial increase in viral replication in the stable transfectants. M9 expression for viral gene expression was also increased to a level comparable to that of HMGB1+/+ cells (Fig. 7C). Moreover, we determined the viral genome copy number from the supernatants of infected cells by using real-time PCR (Fig. 7D). Consistent with previous results, HMGB1−/−S cells showed substantial restoration of viral replication compared to parental cells. Notably, the virus appeared to replicate better in HMGB1−/−S cells at a lower MOI than in HMGB1+/+ cells. Our results indicated that partial complementation of HMGB1 expression in HMGB1−/−S was sufficient to reverse the defect of HMGB1-deficient cells in supporting MHV-68 virus replication. Taken together, all of our results reinforce our hypothesis that HMGB1 plays a significant role in viral gene expression, as well as virus replication.
FIG. 7.
MHV-68 viral replication in a reconstituted clone of HMGB1-deficient cells (HMGB1−/−S). (A) HMGB1 expression in HMGB1−/−S. A cell line stably expressing HMGB1 in HMGB1−/− cells was established and examined for HMGB1 expressing by using anti-HMGB1 antibody. Cell lysates from HMGB1+/+ cells and HMGB1−/− cells were also loaded as positive and negative controls, respectively. Actin was used as a loading control. (B) Viral replication of EGFP/MHV-68 in HMGB1−/− cells. HMGB1+/+, HMGB1−/−, and HMGB1−/−S cells were infected with EGFP/MHV-68 at MOIs of 0.05 and 0.5. At 3 days postinfection, the infected cells were monitored as described in Fig. 5. A set of representative images from cells infected at an MOI of 0.05 PFU/cell are shown. (C) Viral gene expression in HMGB1−/− and HMGB1+/+ cells. The infected cells were harvested at 3 days postinfection and examined for M9 expression by Western analysis. The same membranes were stripped and reprobed with α-actin antibody as a loading control. (D) Viral genome copy in the supernatants of infected HMGB1+/+, HMGB1−/−, and HMGB1−/−S cells. Viral DNA from supernatants of infected cells were extracted and subjected to real-time PCR with a set of viral genome-specific primers to determine the genome copy number in the supernatants. The numbers of viral genome copy per ml of supernatants are shown for HMGB+/+ (▪), HMGB1−/− (□), and HMGB1−/−S ( ) cells with error bars (standard error of the mean).
) cells with error bars (standard error of the mean).
DISCUSSION
The ability of HMGB1 to bind and bend DNA appears to make it a versatile component as an architectural facilitator in a wide variety of nuclear processes. We have tested the role of HMGB1 as it applies to the functions of RTA, a potent molecular switch of gamma-2 herpesviruses. HMGB1 facilitated RTA binding in vitro to its different cognate sites of KSHV, with greater effects on binding sites with low affinity than those with high affinity. Transient-transfection experiments showed that HMGB1 augments transactivation on RTA-responsive promoters from both KSHV and MHV-68. Next, we took advantage of fibroblasts established from HMGB1-deficient mice and explored the functional significance in gene expression and replication of MHV-68. Our results with transient and stable transfectants indicate that HMGB1 plays an important role in RTA-mediated gene expression and viral replication. Given that RTA plays a key role in viral gene expression and replication of gamma-2 herpesviruses and that HMGB1 makes a significant contribution to RTA-mediated transactivation, as well as RTA binding, HMGB1 may facilitate viral replication in gamma-2 herpesviruses.
We have used four different RTA-responsive sequences from KSHV and found that HMGB1 increased in vitro RTA binding to all four sequences. Our results indicate that HMGB1 enhancement of RTA binding has little sequence specificity. This is consistent with the hypothesis that HMGB1 recognizes DNA structure rather than the sequence per se, although the detailed mechanism by which HMGB1 enhances RTA binding is yet to be elucidated. Although HMGB1 protein is rather abundant in cells, our results showing increased RTA-mediated transactivation by transfected HMGB1 suggest that HMGB1 freely available in the cells can be limiting when RTA is overexpressed (3). Thus, ectopic expression of HMGB1 may still have a positive effect on RTA function, although the extent of the HMGB1 effect on RTA-mediated transactivation was less dramatic than those on RTA binding in vitro.
Considering the critical role of RTA in the life cycle of gammaherpesviruses, our initial results on HMGB1 enhancement of KSHV RTA binding led us to hypothesize that HMGB1 may play a significant role in the virus life cycle. However, this is difficult to test with human gammaherpesviruses, mainly due to the limited experimental systems of lytic replication. We have used a closely related murine gammaherpesvirus, MHV-68, which exhibits robust virus replication in cell culture. We found lowered viral gene expression and less replication in HMGB1−/− cells than in wild-type cells. Initial stages of viral infection such as virus-cell fusion or virus entry did not seem to account for this discrepancy due to the following observation. Upon infection at a high MOI, virus titers were similar in both cell lines at 1 day postinfection, suggesting initial success of virus infection in both cell lines.
Herpesviruses and HMG proteins.
We tried to calculate and link functional surface pockets of eight RTA proteins from gammaherpesviruses with known structures and functions by using a computer program that recognizes the surface of protein. Our effort in this computer-based functional domain search of RTA revealed a remarkable identity (13 of 15 amino acids) of the AT-hook domain of HMG I(Y) to RTA of HVS (amino acids 402 to 416). However, since this domain is only moderately conserved with other gammaherpesviruses, it is plausible to think that RTA of herpes simplex virus might have evolved to pirate this motif from the architectural proteins, whereas other herpesviruses utilize cellular DNA architectural proteins to fulfill a similar need. The role of this motif in HVS RTA has been tested and shown to be important in RTA function (Adrian Whitehouse, unpublished data). HMG family proteins such as HMGB1 and HMG I(Y) have been shown to be involved in the transcription regulation of other herpesviruses, such as herpes simplex virus type 1 on the cognate sites of infected cell protein 4 and on the latency-active promoter 2 (15, 37). In both cases, it was suggested that HMG family proteins facilitate the formation of higher-order DNA-protein complexes. However, it is unclear how these findings are related to the virus life cycle. We extended our findings to explore the functional significance of HMGB1 in the virus life cycle and showed that HMGB1 plays an important role in supporting RTA-mediated DNA binding and gene expression, as well as viral replication.
Although HMGB1 is involved in supporting lytic replication of MHV-68, we did not prove that HMGB1 is an essential factor for MHV-68 virus replication. MHV-68 was able to replicate over time from day 2 to day 4 postinfection at low MOIs in HMGB1-deficient cells, although to a lesser extent than in the wild type. However, we do not rule out the possibility that other functional homologues such as HMGB2 in mammalian cells might complement the role of HMGB1 in virus replication since we demonstrated this complementation in facilitation of RTA-binding assays in vitro. Furthermore, HU, a bacterial architectural protein, can also functionally substitute for HMGB1 to stimulate RTA binding. Moreover, the mutant form of HMG box A (F38A) on a residue critical for DNA bending by HMGB1, failed to stimulate RTA binding. These findings, taken collectively, are consistent with our interpretation in that it appears to be the DNA bending property of HMGB1 that contributes to the enhancement of RTA binding and function.
Conserved mechanism by which HMGB1 stimulated RTA binding of gammaherpesviruses.
Our results are consistent with previous studies of RTA of EBV, another human gammaherpesvirus: (i) functional substitution of HMGB1-stimulated RTA binding by individual domains and homologues of HMGB1, such as HMGB2 and HU; (ii) no apparent ternary complex containing RTA and HMGB1; and (iii) no enhancement of RTA binding by the mutant in HMG box A with substitution of F38A at a critical bending residue. The bacterial architectural protein HU is known to function as a DNA chaperone that can induce and stabilize a DNA conformation to be more favorable for the assembly of nucleoprotein complexes. It has been suggested that a “DNA chaperone function” (47) without specific protein-protein interactions might be one of the mechanisms by which HMGB1 stimulated EBV RTA, based on the ability of the completely unrelated bacterial factor HU to functionally substitute HMGB1 (34). Thus, the mechanisms by which HMGB1 promotes RTA binding are likely to be conserved among gammaherpesviruses. However, the detailed mechanisms of how HMGB1 functions with RTA are not entirely clear and remain to be determined. In conclusion, our study highlights the importance of DNA architectural proteins in RTA functions, as well as in the life cycle of gammaherpesviruses.
Acknowledgments
We thank L. Ronfani and M. E. Bianchi for the gift of HMGB1−/− and HMGB1+/+ cell lines and the construct expressing murine HMGB1.
This study was supported by NIH grants CA83525, CA91791, and DE14153; the Stop Cancer Foundation (R.S.); and the California Cancer Research Committee.
REFERENCES
- 1.Agrawal, A., and D. G. Schatz. 1997. RAG1 and RAG2 form a stable postcleavage synaptic complex with DNA containing signal ends in V(D)J recombination. Cell 89:43-53. [DOI] [PubMed] [Google Scholar]
- 2.Aidinis, V., T. Bonaldi, M. Beltrame, S. Santagata, M. E. Bianchi, and E. Spanopoulou. 1999. The RAG1 homeodomain recruits HMG1 and HMG2 to facilitate recombination signal sequence binding and to enhance the intrinsic DNA-bending activity of RAG1-RAG2. Mol. Cell. Biol. 19:6532-6542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bianchi, M. E., and M. Beltrame. 1998. Flexing DNA: HMG-box proteins and their partners. Am. J. Hum. Genet. 63:1573-1577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Boonyaratanakornkit, V., V. Melvin, P. Prendergast, M. Altmann, L. Ronfani, M. E. Bianchi, L. Taraseviciene, S. K. Nordeen, E. A. Allegretto, and D. P. Edwards. 1998. High-mobility group chromatin proteins 1 and 2 functionally interact with steroid hormone receptors to enhance their DNA binding in vitro and transcriptional activity in mammalian cells. Mol. Cell. Biol. 18:4471-4487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bustin, M. 1999. Regulation of DNA-dependent activities by the functional motifs of the high-mobility-group chromosomal proteins. Mol. Cell. Biol. 19:5237-5246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Calogero, S., F. Grassi, A. Aguzzi, T. Voigtlander, P. Ferrier, S. Ferrari, and M. E. Bianchi. 1999. The lack of chromosomal protein Hmg1 does not disrupt cell growth but causes lethal hypoglycemia in newborn mice. Nat. Genet. 22:276-280. [DOI] [PubMed] [Google Scholar]
- 7.Chang, P. J., D. Shedd, L. Gradoville, M. S. Cho, L. W. Chen, J. Chang, and G. Miller. 2002. Open reading frame 50 protein of Kaposi's sarcoma-associated herpesvirus directly activates the viral PAN and K-12 genes by binding to related response elements. J. Virol. 76:3168-3178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chen, J., K. Ueda, S. Sakakibara, T. Okuno, and K. Yamanishi. 2000. Transcriptional regulation of the Kaposi's sarcoma-associated herpesvirus viral interferon regulatory factor gene. J. Virol. 74:8623-8634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cox, M. A., J. Leahy, and J. M. Hardwick. 1990. An enhancer within the divergent promoter of Epstein-Barr virus responds synergistically to the R and Z transactivators. J. Virol. 64:313-321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Deng, H., M. J. Song, J. T. Chu, and R. Sun. 2002. Transcriptional regulation of the interleukin-6 gene of human herpesvirus 8 (Kaposi's sarcoma-associated herpesvirus). J. Virol. 76:8252-8264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Duan, W., S. Wang, S. Liu, and C. Wood. 2001. Characterization of Kaposi's sarcoma-associated herpesvirus/human herpesvirus-8 ORF57 promoter. Arch. Virol. 146:403-413. [DOI] [PubMed] [Google Scholar]
- 12.Ellwood, K., W. Huang, R. Johnson, and M. Carey. 1999. Multiple layers of cooperativity regulate enhanceosome-responsive RNA polymerase II transcription complex assembly. Mol. Cell. Biol. 19:2613-2623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ellwood, K. B., Y. M. Yen, R. C. Johnson, and M. Carey. 2000. Mechanism for specificity by HMG-1 in enhanceosome assembly. Mol. Cell. Biol. 20:4359-4370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Falciola, L., F. Spada, S. Calogero, G. Langst, R. Voit, I. Grummt, and M. E. Bianchi. 1997. High mobility group 1 protein is not stably associated with the chromosomes of somatic cells. J. Cell Biol. 137:19-26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.French, S. W., M. C. Schmidt, and J. C. Glorioso. 1996. Involvement of a high-mobility-group protein in the transcriptional activity of herpes simplex virus latency-active promoter 2. Mol. Cell. Biol. 16:5393-5399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Goodwin, D. J., M. S. Walters, P. G. Smith, M. Thurau, H. Fickenscher, and A. Whitehouse. 2001. Herpesvirus saimiri open reading frame 50 (Rta) protein reactivates the lytic replication cycle in a persistently infected A549 cell line. J. Virol. 75:4008-4013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gradoville, L., J. Gerlach, E. Grogan, D. Shedd, S. Nikiforow, C. Metroka, and G. Miller. 2000. Kaposi's sarcoma-associated herpesvirus open reading frame 50/Rta protein activates the entire viral lytic cycle in the HH-B2 primary effusion lymphoma cell line. J. Virol. 74:6207-6212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Grasser, K. D., S. H. Teo, K. B. Lee, R. W. Broadhurst, C. Rees, C. H. Hardman, and J. O. Thomas. 1998. DNA-binding properties of the tandem HMG boxes of high-mobility-group protein 1 (HMG1). Eur. J. Biochem. 253:787-795. [DOI] [PubMed] [Google Scholar]
- 19.Gwack, Y., H. J. Baek, H. Nakamura, S. H. Lee, M. Meisterernst, R. G. Roeder, and J. U. Jung. 2003. Principal role of TRAP/mediator and SWI/SNF complexes in Kaposi's sarcoma-associated herpesvirus RTA-mediated lytic reactivation. Mol. Cell. Biol. 23:2055-2067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gwack, Y., H. Byun, S. Hwang, C. Lim, and J. Choe. 2001. CREB-binding protein and histone deacetylase regulate the transcriptional activity of Kaposi's sarcoma-associated herpesvirus open reading frame 50. J. Virol. 75:1909-1917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gwack, Y., S. Hwang, C. Lim, Y. S. Won, C. H. Lee, and J. Choe. 2002. Kaposi's Sarcoma-associated herpesvirus open reading frame 50 stimulates the transcriptional activity of STAT3. J. Biol. Chem. 277:6438-6442. [DOI] [PubMed] [Google Scholar]
- 22.Gwack, Y., H. Nakamura, S. H. Lee, J. Souvlis, J. T. Yustein, S. Gygi, H. J. Kung, and J. U. Jung. 2003. Poly(ADP-ribose) polymerase 1 and Ste20-like kinase hKFC act as transcriptional repressors for gamma-2 herpesvirus lytic replication. Mol. Cell. Biol. 23:8282-8294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hall, K. T., A. J. Stevenson, D. J. Goodwin, P. C. Gibson, A. F. Markham, and A. Whitehouse. 1999. The activation domain of herpesvirus saimiri R protein interacts with the TATA-binding protein. J. Virol. 73:9756-9763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hardwick, J. M., P. M. Lieberman, and S. D. Hayward. 1988. A new Epstein-Barr virus transactivator, R, induces expression of a cytoplasmic early antigen. J. Virol. 62:2274-2284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hardwick, J. M., L. Tse, N. Applegren, J. Nicholas, and M. A. Veliuona. 1992. The Epstein-Barr virus R transactivator (Rta) contains a complex, potent activation domain with properties different from those of VP16. J. Virol. 66:5500-5508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Jayaraman, L., N. C. Moorthy, K. G. Murthy, J. L. Manley, M. Bustin, and C. Prives. 1998. High mobility group protein-1 (HMG-1) is a unique activator of p53. Genes Dev. 12:462-472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Liang, Y., J. Chang, S. J. Lynch, D. M. Lukac, and D. Ganem. 2002. The lytic switch protein of KSHV activates gene expression via functional interaction with RBP-Jκ (CSL), the target of the Notch signaling pathway. Genes Dev. 16:1977-1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Liu, S., I. V. Pavlova, H. W. T. Virgin, and S. H. Speck. 2000. Characterization of gammaherpesvirus 68 gene 50 transcription. J. Virol. 74:2029-2037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lukac, D. M., L. Garibyan, J. R. Kirshner, D. Palmeri, and D. Ganem. 2001. DNA binding by Kaposi's sarcoma-associated herpesvirus lytic switch protein is necessary for transcriptional activation of two viral delayed early promoters. J. Virol. 75:6786-6799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lukac, D. M., J. R. Kirshner, and D. Ganem. 1999. Transcriptional activation by the product of open reading frame 50 of Kaposi's sarcoma-associated herpesvirus is required for lytic viral reactivation in B cells. J. Virol. 73:9348-9361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lukac, D. M., R. Renne, J. R. Kirshner, and D. Ganem. 1998. Reactivation of Kaposi's sarcoma-associated herpesvirus infection from latency by expression of the ORF 50 transactivator, a homolog of the EBV R protein. Virology 252:304-312. [DOI] [PubMed] [Google Scholar]
- 32.Manet, E., A. Rigolet, H. Gruffat, J. F. Giot, and A. Sergeant. 1991. Domains of the Epstein-Barr virus (EBV) transcription factor R required for dimerization, DNA binding and activation. Nucleic Acids Res. 19:2661-2667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Martinez-Guzman, D., T. Rickabaugh, T. T. Wu, H. Brown, S. Cole, M. J. Song, L. Tong, and R. Sun. 2003. Transcription program of murine gammaherpesvirus 68. J. Virol. 77:10488-10503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Mitsouras, K., B. Wong, C. Arayata, R. C. Johnson, and M. Carey. 2002. The DNA architectural protein HMGB1 displays two distinct modes of action that promote enhanceosome assembly. Mol. Cell. Biol. 22:4390-4401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Muller, S., P. Scaffidi, B. Degryse, T. Bonaldi, L. Ronfani, A. Agresti, M. Beltrame, and M. E. Bianchi. 2001. New EMBO members' review: the double life of HMGB1 chromatin protein: architectural factor and extracellular signal. EMBO J. 20:4337-4340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ohndorf, U. M., M. A. Rould, Q. He, C. O. Pabo, and S. J. Lippard. 1999. Basis for recognition of cisplatin-modified DNA by high-mobility-group proteins. Nature 399:708-712. [DOI] [PubMed] [Google Scholar]
- 37.Panagiotidis, C. A., and S. J. Silverstein. 1999. The host-cell architectural protein HMG I(Y) modulates binding of herpes simplex virus type 1 ICP4 to its cognate promoter. Virology 256:64-74. [DOI] [PubMed] [Google Scholar]
- 38.Quinlivan, E. B., E. A. Holley-Guthrie, M. Norris, D. Gutsch, S. L. Bachenheimer, and S. C. Kenney. 1993. Direct BRLF1 binding is required for cooperative BZLF1/BRLF1 activation of the Epstein-Barr virus early promoter, BMRF1. Nucleic Acids Res. 21:1999-2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Song, M. J., H. J. Brown, T. T. Wu, and R. Sun. 2001. Transcription activation of polyadenylated nuclear rna by rta in human herpesvirus 8/Kaposi's sarcoma-associated herpesvirus. J. Virol. 75:3129-3140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Song, M. J., H. Deng, and R. Sun. 2003. Comparative study of regulation of RTA-responsive genes in Kaposi's sarcoma-associated herpesvirus/human herpesvirus 8. J. Virol. 77:9451-9462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Song, M. J., X. Li, H. J. Brown, and R. Sun. 2002. Characterization of interactions between RTA and the promoter of polyadenylated nuclear RNA in Kaposi's sarcoma-associated herpesvirus/human herpesvirus 8. J. Virol. 76:5000-5013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Staskus, K. A., W. Zhong, K. Gebhard, B. Herndier, H. Wang, R. Renne, J. Beneke, J. Pudney, D. J. Anderson, D. Ganem, and A. T. Haase. 1997. Kaposi's sarcoma-associated herpesvirus gene expression in endothelial (spindle) tumor cells. J. Virol. 71:715-719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Stros, M. 1998. DNA bending by the chromosomal protein HMG1 and its high mobility group box domains. Effect of flanking sequences. J. Biol. Chem. 273:10355-10361. [PubMed] [Google Scholar]
- 44.Stros, M. 2001. Two mutations of basic residues within the N terminus of HMG-1 B domain with different effects on DNA supercoiling and binding to bent DNA. Biochemistry 40:4769-4779. [DOI] [PubMed] [Google Scholar]
- 45.Sun, R., S. F. Lin, L. Gradoville, Y. Yuan, F. Zhu, and G. Miller. 1998. A viral gene that activates lytic cycle expression of Kaposi's sarcoma-associated herpesvirus. Proc. Natl. Acad. Sci. USA 95:10866-10871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Sun, R., S. F. Lin, K. Staskus, L. Gradoville, E. Grogan, A. Haase, and G. Miller. 1999. Kinetics of Kaposi's sarcoma-associated herpesvirus gene expression. J. Virol. 73:2232-2242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Thomas, J. O. 2001. HMG1 and 2: architectural DNA-binding proteins. Biochem. Soc. Trans. 29:395-401. [DOI] [PubMed] [Google Scholar]
- 48.Thomas, J. O., and A. A. Travers. 2001. HMG1 and 2, and related “architectural” DNA-binding proteins. Trends Biochem. Sci. 26:167-174. [DOI] [PubMed] [Google Scholar]
- 49.Wang, S., S. Liu, M. H. Wu, Y. Geng, and C. Wood. 2001. Identification of a cellular protein that interacts and synergizes with the RTA (ORF50) protein of Kaposi's sarcoma-associated herpesvirus in transcriptional activation. J. Virol. 75:11961-11973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Wang, S. E., F. Y. Wu, Y. Yu, and G. S. Hayward. 2003. CCAAT/enhancer-binding protein-alpha is induced during the early stages of Kaposi's sarcoma-associated herpesvirus (KSHV) lytic cycle reactivation and together with the KSHV replication and transcription activator (RTA) cooperatively stimulates the viral RTA, MTA, and PAN promoters. J. Virol. 77:9590-9612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Wei, M., O. Burenkova, and S. J. Lippard. 2003. Cisplatin sensitivity in Hmbg1−/− and Hmbg1+/+ mouse cells. J. Biol. Chem. 278:1769-1773. [DOI] [PubMed] [Google Scholar]
- 52.Whitehouse, A., I. M. Carr, J. C. Griffiths, and D. M. Meredith. 1997. The herpesvirus saimiri ORF50 gene, encoding a transcriptional activator homologous to the Epstein-Barr virus R protein, is transcribed from two distinct promoters of different temporal phases. J. Virol. 71:2550-2554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Wu, T. T., L. Tong, T. Rickabaugh, S. Speck, and R. Sun. 2001. Function of Rta is essential for lytic replication of murine gammaherpesvirus 68. J. Virol. 75:9262-9273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Wu, T. T., E. J. Usherwood, J. P. Stewart, A. A. Nash, and R. Sun. 2000. Rta of murine gammaherpesvirus 68 reactivates the complete lytic cycle from latency. J. Virol. 74:3659-3667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Zalani, S., E. Holley-Guthrie, and S. Kenney. 1996. Epstein-Barr viral latency is disrupted by the immediate-early BRLF1 protein through a cell-specific mechanism. Proc. Natl. Acad. Sci. USA 93:9194-9199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Zappavigna, V., L. Falciola, M. Helmer-Citterich, F. Mavilio, and M. E. Bianchi. 1996. HMG1 interacts with HOX proteins and enhances their DNA binding and transcriptional activation. EMBO J. 15:4981-4991. [PMC free article] [PubMed] [Google Scholar]
- 57.Zhang, L., J. Chiu, and J. C. Lin. 1998. Activation of human herpesvirus 8 (HHV-8) thymidine kinase (TK) TATAA-less promoter by HHV-8 ORF50 gene product is SP1 dependent. DNA Cell Biol. 17:735-742. [DOI] [PubMed] [Google Scholar]