Abstract
We and others have shown that infection of dendritic cells with murine cytomegalovirus (MCMV) leads to severe functional impairment of these antigen-presenting cells (D. M. Andrews, C. E. Andoniou, F. Granucci, P. Ricciardi-Castagnoli, and M. A. Degli-Esposti, Nat. Immunol. 2:1077-1084, 2001; S. Mathys, T. Schroeder, J. Ellwart, U. H. Koszinowski, M. Messerle, and U. Just, J. Infect. Dis. 187:988-999, 2003). Phenotypically, reduced surface expression of costimulatory molecules and major histocompatibility complex molecules was detected. In order to identify the molecular basis for the observed effects, we generated MCMV mutants with large deletions of nonessential genes. The study was facilitated by the finding that a monocyte-macrophage cell line displayed similar phenotypic alterations after MCMV infection. By analyzing the expression of cell surface molecules on infected cells, we identified a mutant virus which is no longer able to downmodulate the expression of the costimulatory molecule CD86. Additional mutants with smaller deletions allowed us to pin down the responsible gene to a certain genomic region. RNA analysis led to the identification of the spliced gene m147.5, encoding a protein with 145 amino acids. Experiments with an m147.5 mutant revealed that the protein affects CD86 expression only, suggesting that additional MCMV genes are responsible for downmodulation of the other surface molecules. Identification of viral gene products interfering with functionally important proteins of antigen-presenting cells will provide the basis to dissect the complex interaction of CMV with these important cells and to evaluate the biological importance of these viral genes in vivo.
Human cytomegalovirus (HCMV) is distributed worldwide among the human population (for review, see references 42 and 47). CMV infection is primarily a threat to immunocompromised patients, e.g., transplant recipients or AIDS patients, as well as immunologically immature neonates. The severity of CMV disease correlates with the degree of immunosuppression, underscoring the importance of immune control for containment of the viral infection. However, even in healthy individuals with an intact immune system, primary CMV infection is characterized by viremia and virus shedding that can last for months or even years. The delayed clearance of CMV infection has been attributed to viral gene products, which allow the virus to escape control by various immune effector mechanisms (reviewed in references 36 and 52). Murine CMV (MCMV), which serves as a model system for HCMV, has proven especially useful for dissecting the principles of immune evasion and for defining the biological significance of immunomodulatory CMV genes (reviewed in references 21, 28, and 34).
A series of recent studies suggest that cytomegaloviruses do not just elude the immune effector mechanisms but actively interfere with the initiation of the immune response (4, 8, 22, 23, 29, 38, 44, 45, 48, 49, 55). It is well established that professional antigen-presenting cells, especially monocytes and macrophages, play a central role in the life cycle of CMVs (26, 56; reviewed in references 32 and 33). More recently, evidence was provided that both MCMV and HCMV can also infect dendritic cells in vitro and in vivo (4, 17, 49, 53).
Considering the crucial role of these antigen-presenting cells for induction and maintenance of protective T-cell immunity against viruses (6, 7), one must assume that CMVs were virtually forced to evolve mechanisms that counter the functions of these cells in order to prevent elimination and to guarantee dissemination within the host organism and ultimately transmission to other susceptible individuals. Several different mechanisms have been proposed that may explain the functional impairment of CMV-infected macrophages and dendritic cells, ranging from the blockade of gamma interferon-induced antigen presentation by major histocompatibility complex (MHC) class II molecules (27), the inhibition of monocyte differentiation (22), and affected maturation and migration of immature dendritic cells (44, 45) up to the altered cell surface expression of functionally important molecules such as costimulatory proteins on mature dendritic cells (4, 29, 38, 49). So far, the contribution of the various mechanisms leading to the functional defect of the antigen-presenting cells is not known, and the viral proteins mediating these effects are not yet defined. The dissection and understanding of the complex interactions of CMV with the manifold functions of antigen-presenting cells would certainly benefit from the identification and characterization of the viral genes responsible.
Since many immunomodulatory genes of viruses can only be detected by functional assays, we have recently set up procedures for the generation and screening of libraries of viral mutants (13, 24, 31). This became feasible by the ability to clone the genomes of MCMV and HCMV as bacterial artificial chromosomes (BACs) and the establishment of random and site-directed mutagenesis procedures for manipulation of herpesvirus BACs (9, 25, 41, 58, 62). With these new powerful genetic approaches, several CMV genes interacting with different host functions have been found in the meanwhile (5, 12, 15), implying that these procedures will also help to define the viral genes that impair the functions of antigen-presenting cells.
In the present study we report on the identification of an MCMV gene targeting the expression of the costimulatory molecule CD86 on the surface of antigen-presenting cells. By construction and testing of MCMV deletion mutants, we could consecutively narrow down the genomic region encoding this viral function. RNA analysis led to the identification of a spliced gene that we termed m147.5 or modulator of B7-2 (modB7-2). Experiments with a modB7-2 knockout mutant showed that the gene product exerts a selective effect on CD86 expression, suggesting that additional viral genes are responsible for the observed downmodulation of other functionally important surface molecules on antigen-presenting cells.
MATERIALS AND METHODS
Cells and viruses.
Mouse embryonic fibroblasts obtained from BALB/c mice, bone marrow stromal M2-10B4 cells (ATCC CRL1972), and the monocyte-macrophage cell lines RAW264.7 (ATCC TIB-71) (50) and J774A.1 (ATCC TIB-67) were cultured in Dulbecco's modified Eagle's medium supplemented with 10% fetal calf serum. Dendritic cells were derived from bone marrow cells of BALB/c or C57BL/6 mice by cultivation in granulocyte-macrophage colony-stimulating factor-conditioned RPMI 1640 medium supplemented with 10% heat-inactivated and filtered fetal calf serum essentially following the protocol of Lutz et al. (37). Cell cultures were regularly checked for absence of mycoplasma contamination by use of an enzyme-linked immunosorbent assay-based mycoplasma detection kit (Roche, Mannheim, Germany).
MCMV recombinants expressing green fluorescent protein (GFP) were derived from the bacterial artificial chromosome (BAC)-cloned MCMV-GFP genome pSM3fr-GFP (38). Recombinant viruses were reconstituted by transfecting DNA of the mutated MCMV BACs into M2-10B4 or mouse embryonic fibroblasts by electroporation. Briefly, 2 to 3 μg of BAC DNA was electroporated into 106 cells at 250 V and 1,500 μF with an EasyjecT Optima electroporator (Peqlap, Erlangen, Germany). For preparation of virus stocks, MCMV recombinants were propagated on mouse embryonic fibroblasts and purified as described (11). Titers of virus stocks were determined by standard plaque assay on mouse embryonic fibroblasts. For screening purposes, RAW264.7 cells were subjected to centrifugation-enhanced infection (800 × g for 30 min) (11) with supernatants of MCMV-infected M2-10B4 or mouse embryonic fibroblast cultures, which were harvested when complete cytopathic effect occurred. By following this protocol, usually 20 to 50% of the RAW264.7 cells became infected as determined by GFP expression after 24 h. Results were verified with purified virus preparations.
Plasmids.
Candidate genes for reinsertion into the genome of MCMV-GFPΔ6 were amplified by PCR and cloned between the BamHI and NotI sites of shuttle plasmid pOri6k-AL, which contains a kanamycin resistance marker, an Flp recognition target (FRT) site, and the bacterial origin of replication R6Kγ (35). Open reading frame (ORFs) m147, m148, m149, and m148 plus m149 and approximately 300 bp of upstream sequences representing the putative promoters were amplified by PCR with primers m147ori6K.for (5′-GGA TGC GGC CGC TTT GGC AAG CTT TCC ACC TC-3′ ) and m147ori6K.rev (5′-GAT CGG ATC CAC AAG ACA CAA GTG CTC AAT ATA TAT CAA AAA GGA TTT ATT TGA CCG GAA AGG TTG AAT TTT T-3′ ), m148ori6k.for (5′-GGATGC GGC CGC TCA CGT AGC CCA CCC AAA GCG AGA-3′ ) and m148ori6k.rev (5′-GAT CGG ATC CAC AAG ACA CAA GTG CTC AAT ATA TAT CAA AAA GGA TTT ATT TTC TAG AAC ATC ATG TTT GCT AAT-3′ ), m149ori6k.for (5′-GGATGC GGC CGC GGC GGA ATC CAT TTC AAG AC-3′ ) and m149ori6k.rev (5′-GATCGG ATC CAC AAG ACA CAA GTG CTC AAT ATA TAT CAA AAA GGA TTT ATT TTT CGG ATA CGG TGG CGT G-3′ ), and m148ori6k.for and m149ori6k.rev, respectively. An RNA termination sequence derived from the MCMV ie3 polyadenylation signal (40) was included in the reverse primers.
In order to reinsert the modB-7 gene without intron, the ORF was PCR amplified with primers m147ori6K.rev and Pr-AL13 (5′-ATCGTC GAC CTT TCC ACC TCA GAC CTT TCA G-3′ ) and the cloned cDNA of modB7-2 as a template. The resulting DNA fragment was cloned into the BamHI and SalI sites of pOri6k-AL. Then, a BamHI-SacII fragment at the 3′ end of the insert was replaced by a DNA fragment obtained by PCR with primers m148ori6kBam.for (5′-ATCGGA TCC CAC GTA GCC CAC CCA AAG CGA GA-3′ ) and m147/m148RT.rev (5′-TGGAAT AAG ATG CAA CGA AGA C-3′ ) and viral DNA as a template. Finally, a PCR product generated with primers Pr-AL14 (5′-ATCGTC GAC AAA AAC CGC TTT ATA TAC AAA GTC TAA GC-3′ ) and m149i2.rev (5′-GAG GCG GCC GCG TGT TGA AGG GGG TCT TGT G-3′ ) was cloned at the 5′ end of the insert as a SalI-NotI fragment, resulting in plasmid pOri6k-modB7-2. All cloned DNA fragments were checked by sequencing.
In order to selectively disrupt the modB7-2 ORF, a 4.7-kbp PstI fragment (representing nucleotides 204956 to 209647 of the MCMV genome) (51) was isolated from BAC pSM3fr-GFP and subcloned into a pBluescript derivative with a modified polylinker carrying the recognition sites for BamHI, PstI, and SmaI. An internal KpnI fragment (equivalent to nucleotides 207143 to 207462) (51) was deleted and replaced by a KpnI fragment obtained by PCR with primer Pr-AL17 (5′-TTT TCG GTA CCT aGT CCT aCT aTC aGT CCG CGT CTC TCC GAT CCT-3′ ), which provided the mutation (lowercase letters), and primer m147/m148RT.rev. A tetracycline resistance gene derived from pCP16 (16) was then inserted into the SmaI site of the resulting plasmid adjacent to the cloned PstI fragment, and finally the complete 7.3-kbp insert was transferred as a BamHI-PmeI fragment to shuttle plasmid pST76-KSR (10), resulting in pST76-KSR-AL1.
Mutagenesis of the BAC-cloned MCMV genome.
Deletion mutants were constructed in Escherichia coli by homologous recombination between linear DNA fragments and the MCMV BAC pSM3fr-GFP (38) exploiting the bacteriophage λ recombination genes redα, -β, and -γ provided by plasmid pKD46 (18). Linear fragments carrying a kanamycin resistance gene were generated by PCR with plasmid pGP704-Kan as a template. pGP704-Kan contains the kanamycin resistance gene from transposon Tn903 flanked by minimal Flp recombinase recognition target sites (FRT). The primers contained 20 to 22 nucleotides at their 3′ ends that were specific for the kanamycin resistance template and 50 to 60 nucleotides at their 5′ ends that were homologous to the target region in the MCMV BAC and required for homologous recombination. The ranges of the deletions are given in Table 1. In case of the insertion mutants (see Fig. 3), the kanamycin resistance cassette was excised by Flp-mediated recombination in E. coli as described previously (2, 58). Thus, the ORFs targeted in these mutants are disrupted by small deletions (see Table 1) and by the remaining FRT site only.
TABLE 1.
Deletion mutants and CD86 phenotype of infected RAW264.7 cells
| Mutant | Deletion
|
Phenotype | |
|---|---|---|---|
| Rangea (nt) | ORF(s) | ||
| MCMV-GFPΔ6 | 203002-217799 | m144-m158 | Loss of function |
| MCMV-GFPΔ6S1 | 202746-207298 | m144-m148 | Loss of function |
| MCMV-GFPΔ6S2 | 207354-212803 | m149-m153 | Loss of function |
| MCMV-GFPΔ6S3 | 212946-216883 | m154-m157 | Wild type |
| MCMV-GFPΔm146 | 205646-206774 | m146 | Wild type |
| MCMV-GFPΔm147/m148 | 206866-207265 | m147 plus m148 | Loss of function |
| MCMV-GFPΔm149 | 207354-207566 | m149 | Loss of function |
| MCMV-GFPΔm149/m150 | 207354-208762 | m149 plus m150 | Loss of function |
| MCMV-GFPΔm151 | 208844-209982 | m151 | Wild type |
| RV-AL13 | 207112-207141 | m147 plus m148b | Loss of function |
| RV-AL14 | 207290-207337 | m147 plus m149b | Wild type |
| RV-AL15 | 207467-207506 | m149b | Loss of function |
| RV-AL16 | 207812-207864 | m149 plus m150b | Wild type |
| RV-AL17 | 208529-208628 | m150b | Wild type |
Nucleotide positions refer to reference 51.
These ORFs are disrupted by small deletions and the insertion of an FRT site only.
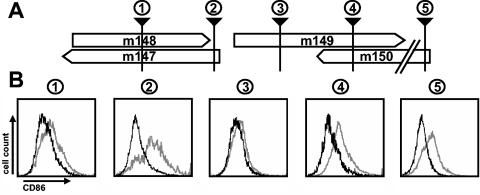
FIG. 3.
Fine mapping of the gene targeting CD86 expression. (A) The ORFs in the genomic region analyzed are represented by open boxes. The black arrowheads indicate the positions of small mutations disrupting the ORFs in the individual mutants. The scheme is not drawn to scale. (B) The recombinant viruses RV-AL13 (box 1), RV-AL14 (box 2), RV-AL15 (box 3), RV-AL16 (box 4), and RV-AL17 (box 5) were used to infect RAW264.7 cells, and their ability to influence the expression of CD86 was determined by flow cytometry 24 h postinfection. Bold lines represent the expression of CD86 on MCMV-infected (GFP-positive) cells, and gray lines represent the expression of CD86 on the uninfected (GFP-negative) population of the same culture.
Tagging of the modB7-2 ORF was also done by homologous recombination in E. coli with a linear DNA fragment, which was obtained by PCR with primers m147HA.for (5′-TAA CAG ATA TGT AGC GGA ACC CTG CGG TCC CGA CGA TGA ATA CCC ATA CGA CGT CCC AGA CTA CGC TTA AAG GAC GAC GAC GAC AAG TAA-3′ ) and m147HA.rev (5′-AGA GAC TAA AGA CAA CAT CAC ACA AGT TTA TTG AGA TGG TGA CAC AGG AAC ACT TAA CGG CTG A-3′ ) and pG704-Kan as a template. The hemagglutinin (HA) epitope sequence is provided by the forward primer (italic). After insertion of the linear fragment into BAC pSM3fr (59), the HA epitope sequence was fused in frame to the 3′ end of the modB7-2 ORF. The kanamycin resistance cassette was subsequently removed by Flp recombinase, and successful tagging was verified by sequencing.
BAC pSM3fr-Δ6-Δkan, obtained from the genome of MCMV-GFPΔ6 by Flp-mediated excision of the kanamycin resistance marker, carries a single FRT site. The pOri6k-based shuttle plasmids described in the section on plasmids were inserted into BAC pSM3fr-Δ6-Δkan by Flp-mediated recombination as described elsewhere (39).
For construction of a BAC with a selective disruption of the modB7-2 gene, first the genomic region from nucleotides 207002 to 207685 was replaced by red α-, β-, and γ-mediated recombination in E. coli with a DNA fragment comprising a pCP16-derived tetracycline resistance marker (16), which was obtained by PCR with primers Pr-AL22 (5′-CGC GGC TAC GGG CGC CAG TCG TAC CGC GTC GGT CCC CCT CTG ATG CCG CCT TCT TCT AGA GGT ACC GCA-3′ ) and Pr-AL23 (5′-AGA CAG ATC AGC GCA CGC TAT GTC TTC GGC AGA CCG CCT GGC GAG TCG AGT GGA ATT CCC GGG AGA GCT-3′ ). The resistance marker was subsequently excised with Flp recombinase. Finally, the DNA sequences carrying the stop codons were reintroduced by a two-step replacement procedure as described (9, 10) with plasmid pST76-KSR-AL1. Successful introduction of the mutation was verified by sequencing.
DNA of mutant BACs was isolated from E. coli cultures by the alkaline lysis procedure (54) and characterized by digestion with appropriate restriction enzymes followed by agarose gel electrophoresis as described (9).
Flow cytometry.
Biotinylated monoclonal antibodies directed against CD40 (no. 553789), CD80 (no. 553767), CD86 (no. 553690), MHC class I (no. 553578), and MHC class II (no. 553546) (all from Becton Dickinson, Heidelberg, Germany) were used for the analysis of phenotypic changes on infected RAW264.7 cells. A streptavidin-allophycocyanin conjugate (no. 554067) was used to visualize primary biotinylated antibodies. Cells were harvested by centrifugation and resuspended in phosphate-buffered saline containing 3% fetal calf serum and Fc-Block (no. 553141) at a dilution of 1:100. After 5 min of incubation at room temperature, an antibody against the respective cell surface antigen or an isotype-matched control antibody was added at a dilution of 1:100. After incubation for 20 min at 4°C in the dark, cells were washed twice and resuspended in phosphate-buffered saline containing 1% fetal calf serum and 1 μg of propidium iodide per ml. Fluorescence-activated cell sorting analysis was done with a FACScalibur machine (Becton Dickinson) equipped with Cell Quest software.
RNA analysis.
Cytoplasmic RNA was isolated from either uninfected or MCMV-GFP-infected RAW264.7 cells 24 h postinfection with the RNeasy mini kit (Qiagen, Hilden, Germany). The RNA was reverse transcribed with random hexamers and Thermoscript reverse transcriptase (Invitrogen, Carlsbad, Calif.) following the instructions of the manufacturer. The resulting cDNA product was PCR amplified with the specific primers m147/m148RT.for (5′-CAG ATA CAC CAG CCG AAA G-3′ ) and Pr-AL10 (5′-ATC ACC CCG GAG GAT GCG TCT TCT TG-3′ ) under the following conditions: one cycle at 95°C for 10 min; 10 cycles of 30 s at 95°C, 45 s at the annealing temperature starting at 65°C and going down 1°C per cycle, and 30 c at 72°C; 23 cycles of 20 s at 95°C, 45 s at 55°C, and 30 s at 72°C; and one cycle at 72°C for 5 min. DNA of the BAC MCMV-GFP served as a template for the control reaction. The amplified products were separated on a 1.2% low-melting-point agarose gel and visualized by ethidium bromide staining. The PCR products were purified with the Qiaquick PCR purification Kit (Qiagen, Hilden, Germany) and sequenced with the primer Pr-AL11 (5′-GGC GGA ATC CAT TTC AAG AC-3′ ). 5′ Rapid amplification of cDNA ends (RACE) and 3′-RACE experiments were performed with the gene-specific primers Pr-AL12 (5′-TTT GGC AAG CTT TCC ACC TC-3′ ) and m147/m148RT.rev, respectively, and the GeneRacer kit (Invitrogen) according to the manufacturer's protocol. The amplified products were separated on a 1.2% low-melt agarose gel and specific bands were eluted and subjected to sequencing reactions with the gene specific primers.
For Northern blot analysis, 5 μg of cytoplasmic RNA of uninfected and MCMV-GFP infected cells was separated on denaturing formaldehyde-1.2% agarose gels and transferred to Hybond XL membranes (Amersham, Freiburg, Germany) by the capillary blot standard procedure (54). The membranes were prehybridized with a prehybridization reagent (Roche) for 2 h at 65°C. Probes 1 and 2 were generated by PCR with primers m147ori6k.rev and m148ori6k.rev and m149RT.for (5′-ACG AGC GGA AAT GTC CAA C-3′ ) and m149i2.rev, respectively. Probes were labeled with [α-32P]dCTP with a random prime labeling kit (Amersham) and filters were hybridized at 65°C overnight. Washing of the filters was done twice, each time for 10 min, in 3× SSC (1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate)-0.1% sodium dodecyl sulfate (SDS), 1× SSC-0.1% SDS, and 0.3× SSC-0.1% SDS. Results were obtained by autoradiography on Kodak Biomax MR films.
Western blot analysis.
RAW264.7 cells were infected with the recombinant virus RV-AL18 at a multiplicity of infection of 1 followed by centrifugal enhancement as described (11). For selective expression of viral immediate-early proteins, cells were incubated from 30 min prior to infection to 3 h postinfection in the presence of cycloheximide (100 μg/ml; Sigma, Munich, Germany), followed by incubation in the presence of actinomycin D (2.5 μg/ml; Sigma) for another 3 h. Selective expression of early genes was achieved by incubation of the cells in the presence of phosphonoacetic acid (250 μg/ml; Sigma) for 24 h. Protein samples were taken from mock-infected cells and from infected cells at the indicated time points by lysis of the cells in protein sample buffer (3% SDS, 2% β-mercaptoethanol, 200 mM Tris [pH 8.8], 0.5 M sucrose, 5 mM EDTA), and subsequently boiled for 5 min. Proteins were separated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) (15% polyacrylamide) and transferred to nitrocellulose filters. Filters were probed with an anti-HA antibody (no. H6908; Sigma). Signals were detected by enhanced chemiluminescence with the ECL detection kit (Amersham).
RESULTS
Reduced surface expression of costimulatory molecules on MCMV-infected RAW264.7 cells.
We and others have recently observed a reduced expression of MHC and costimulatory molecules on the surface of dendritic cells upon infection with MCMV (4, 38). The rationale of our current work is to identify the viral genes mediating these phenotypic alterations of dendritic cells. To this end, a genetic approach was chosen exploiting the techniques for the manipulation of CMV genomes that were established in our laboratory (14).
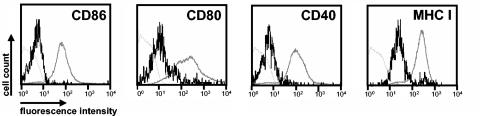
Since screening of hundreds of viral mutants may be required to identify the viral gene responsible for a specific phenotype (12), we sought to design a convenient read-out system for this purpose. Therefore, we asked whether the phenotypic alterations that we found on MCMV-infected dendritic cells (38) may also occur on other antigen-presenting cells. Infection of the macrophage-monocyte cell line RAW264.7 (50) indeed led to alterations in the expression of certain cell surface-expressed molecules (Fig. 1). Cells were infected with the recombinant virus MCMV-GFP (38), expressing green fluorescent protein (GFP), in order to differentiate infected from uninfected cells with flow cytometry by gating for GFP-positive cells. At 24 h postinfection, significantly reduced expression of the costimulatory molecules CD86, CD80, and CD40 as well as of MHC class I molecules was measured on infected (GFP-positive) cells in comparison to noninfected (GFP-negative) cells of the same cultures. There was very little MHC II expression on the surface of RAW264.7 cells (data not shown), preventing analysis of the viral effect on MHC II in these cells. Similar observations were made after infection of the monocyte-macrophage cell line J774A.1 (data not shown). The results suggested that the cell lines examined may facilitate the identification of at least some of the MCMV genes which interfere with the expression of functionally important surface molecules on antigen-presenting cells.
FIG. 1.
Phenotypic changes of MCMV-infected RAW264.7 monocyte-macrophage cells. RAW264.7 cells were infected with the recombinant virus MCMV-GFP expressing green fluorescent protein and analyzed by flow cytometry 24 h postinfection. Surface expression of the indicated markers on MCMV-infected (GFP-positive) cells is represented by bold black lines and on uninfected (GFP-negative) cells of the same culture by bold gray lines. Thin gray lines, isotype control.
Generation and screening of MCMV deletion mutants.
The immunomodulatory genes of murine and human cytomegaloviruses which have been characterized so far are nonessential for replication of the viruses in cell culture (3, 24, 58). Most of these genes are located at the termini of the MCMV genome and at the termini of the UL and US regions of the HCMV genome. Assuming that the viral genes interfering with the expression of surface molecules on antigen-presenting cells belong to the same class of genes, we decided to generate a series of MCMV mutants each carrying a different deletion of approximately 10 to 15 kbp within the terminal regions of their genomes. The mutant genomes were based on the bacterial artificial chromosome (BAC)-cloned MCMV genome pSM3fr-GFP, which contains a GFP gene inserted within the ie2 locus (38). Thus, all of the mutants expressed GFP, easily allowing differentiation between infected and uninfected cells by flow cytometry.
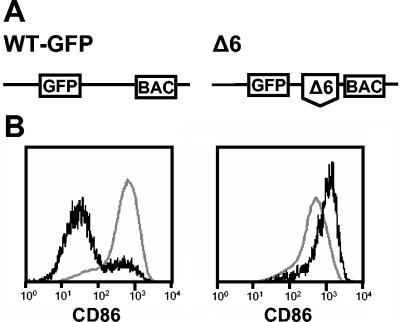
The deletions were introduced into the BAC-cloned MCMV genome by utilizing the ET-mutagenesis technique (58, 64), and mutant viruses were reconstituted by transfecting the mutated genomes into murine fibroblasts (for details, see Materials and Methods). Then, the ability of the mutants to interfere with the expression of the various surface expression markers on RAW264.7 cells was tested by flow cytometry. One of the mutants, MCMV-GFPΔ6, was no longer able to influence the expression of CD86. At 24 h after infection with MCMV-GFPΔ6, GFP-positive (infected) cells and uninfected cells expressed similar levels of CD86 (Fig. 2B, right panel). Even at later times after infection (48 h), expression of CD86 remained at high levels, and there was no significant difference between infected and noninfected cells. For comparison, cells infected with the parental virus MCMV-GFP displayed a clearly reduced expression of CD86 (Fig. 2B, left panel). This result led us to conclude that the mutant MCMV-GFPΔ6 had lost this function. The mutant carries a deletion spanning from nucleotides 203002 to 217799 of the MCMV genome (51), affecting ORFs m144 to m158 (Fig. 2A). Thus, the viral function targeting CD86 expression on antigen-presenting cells is encoded by one or several genes located in this genomic region.
FIG. 2.
Deletion mutant MCMV-GFPΔ6 lost the ability to downmodulate CD86 expression. (A) Genome structure of the recombinant viruses MCMV-GFP and MCMV-GFPΔ6. Boxes represent the GFP gene and the BAC vector sequences, and the broken line and the delta (Δ) indicate the deletion (ORFs m144 to m158). The illustration is not drawn to scale. (B) RAW264.7 cells were infected with MCMV-GFP (left panel) or MCMV-GFPΔ6 (right panel) and analyzed for surface expression of CD86 by flow cytometry as described for Fig. 1. Black lines represent expression of CD86 on MCMV-infected (GFP-positive) cells, and gray lines represent expression of CD86 on the uninfected (GFP-negative) population of the same culture.
Mapping of the gene interfering with CD86 expression.
In order to narrow down the genomic region encoding the viral function, three new mutants with smaller deletions were constructed and tested for their ability to interfere with CD86 expression. Two of the mutants, MCMV-GFPΔ6S1 and MCMV-GFPΔ6S2, were unable to downregulate CD86 expression in RAW264.7 cells (Table 1). Accordingly, we had to consider that two or more proteins that work together (encoded by m144 to m148 and m149 to m153) are needed to accomplish this viral function. Another possibility was that the gene responsible spans the boundary that was defined by the deletions of mutants MCMV-GFPΔ6S1 and MCMV-GFPΔ6S2. A number of additional mutants that carried deletions of single ORFs in this region were generated in order to ascribe the function to a certain viral gene. Since some ORFs are located at the same position but on opposite DNA strands or overlap (see Fig. 3), more than one ORF was affected in some of the mutants. Three of the mutants, carrying deletions of m147 plus m148, m149, and m149 plus m150, respectively, exhibited a loss-of-function phenotype (Table 1). Possible reasons for the observed phenotype can be that several ORFs encode the function or that the deletion of an ORF impaired the promoter and therefore the expression of a neighboring gene because the ORFs are narrowly clustered in this region (51).
Gain of function by reinsertion of genes into the genome of deletion mutant MCMV-GFPΔ6.
In order to assign the function to a specific gene, we reinserted the candidate genes into the genome of mutant MCMV-GFPΔ6 and tested whether the resulting mutants had regained the ability to downmodulate CD86 in infected cells. The ORFs plus approximately 300 bp of upstream sequences representing the putative promoter region were amplified by PCR and cloned into a shuttle plasmid that also carried an FRT site. Reinsertion of the genes into the backbone of MCMV-GFPΔ6 was mediated by Flp recombination (for technical details, see Materials and Methods). Since the results obtained with the deletion mutants strongly suggested that ORFs m147, m148, and m149 may be involved in CD86 downmodulation, we started with a recombinant virus carrying a reinsertion of this genomic region. Indeed, the virus RV-AL4 possessed the function (Table 2), confirming that one or several of the ORFs located on the reinserted fragment encoded the protein(s) interfering with CD86 expression. ORF m150 could definitely be excluded because it was not completely enclosed in the inserted DNA fragment. Recombinant viruses carrying the putative m147, m148, and m149 genes still each showed the same loss-of-function phenotype as the parental virus MCMV-GFPΔ6 (Table 2). Accordingly, the function could not be encoded solely by one of these ORFs.
TABLE 2.
Reinsertion mutants and their CD86 phenotypes on RAW264.7 cells
| Mutant | Gene(s) reinserteda | Phenotype |
|---|---|---|
| RV-AL1 | m147 | Loss of function |
| RV-AL2 | m148 | Loss of function |
| RV-AL3 | m149 | Loss of function |
| RV-AL4 | m147, m148, m149 | Gain of function |
| RV-AL24 | modB7-2 (without intron) | Gain of function |
The indicated genes were reinserted into the genome of deletion mutant MCMV-GFPΔ6 lacking ORFs m144 to m158.
If two gene products are needed to accomplish the function, double infection of cells with reinsertion mutants that carry the responsible genes should restore the function. However, in double infection experiments performed with mutants RV-AL1 and RV-AL2, RV-AL1 and RV-AL3, and RV-AL2 and RV-AL3, downmodulation of CD86 was not observed, not even in a minor population of the infected cells. The results of these experiments excluded that the proteins encoded by the recombinant viruses tested work together to mediate the effect on CD86.
Fine mapping of the gene.
Another set of mutants in which the ORFs were disrupted by small mutations rather than by complete removal of an ORF were constructed in order to minimize potential negative effects on neighboring promoters or genes (Fig. 3). This was accomplished by targeted insertion of an FRT-flanked kanamycin resistance gene into the BAC-cloned MCMV genome by targeted mutagenesis followed by excision of the kanamycin resistance marker by Flp-mediated recombination, leaving one FRT site that disrupted the ORF (for details, see Materials and Methods). The mutant virus RV-AL13 displayed a loss-of-function phenotype (Fig. 3B), indicating that ORF m147 or m148 encodes at least part of the viral protein targeting CD86 expression. Mutant RV-AL17 was still able to downregulate CD86 in infected cells, confirming the results obtained with the reinsertion mutants, excluding ORF m150. Disruption of ORF m149 in mutant RV-AL16 was compatible with the wild-type phenotype, indicating that ORF m149 is not involved either.
Surprisingly, however, an insertion located within the first third of ORF m149 led to a loss of the function (mutant RV-AL15; Fig. 3B). Since several small ORFs are located on both DNA strands in this region, the result strongly suggested that one of these ORFs represents either one of the genes involved or at least a part of the gene. Another insertion affecting the very end of ORF m147 and the 5′ end of ORF m149 only (mutant RV-AL14) had no effect on the phenotype (Fig. 3B). Altogether, these results pointed to the possibility that the gene is composed of at least two exons represented by either ORF m147 or m148 and another exon located in the region which is affected in mutant RV-AL15.
Identification of the gene m147.5.
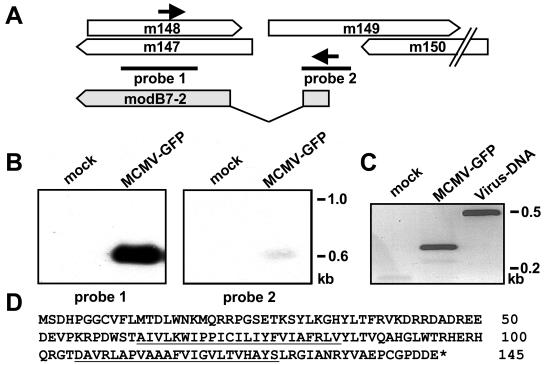
Northern blot experiments were performed to identify transcripts arising from the indicated genomic region. RNA was isolated from RAW264.7 cells that were either mock infected or infected with MCMV-GFP and analyzed by hybridization with probes that were specific for regions proven to be essential for the function by the previous experiment (Fig. 4A). A strong signal corresponding to a transcript about 700 nucleotides was detected by probe 1 (Fig. 4B). Probe 2 revealed a band of similar size, suggesting that a single spliced gene is located in the region examined. The weaker signal observed with probe 2 may be the result of hybridization to a small exon.
FIG. 4.
Detection of viral transcripts. (A) Locations of probes (black bars) and primers (arrows) used for Northern blot analysis and reverse transcription-PCR. The structure of the identified gene modB7-2 is indicated at the bottom. Gray boxes represent exons, and the intron is marked by lines. (B) RAW264.7 cells were mock infected (lane 1) or infected with MCMV-GFP (lane 2). Cytoplasmic RNA was isolated 24 h postinfection and subjected to Northern blot analysis with 32P-labeled probes. The positions of RNA marker bands are indicated at the right margin. (C) The RNA isolated from mock-infected (lane 1) and MCMV-GFP-infected (lane 2) cells was reverse transcribed with oligo(dT) priming, and the resulting cDNA was used as a template for PCR with the primers indicated in panel A. For comparison, viral DNA (lane 3) was used as the template for PCR. Amplified products were separated on a 1.2% agarose gel and visualized by ethidium bromide staining. DNA size markers are shown at the right margin. (D) Deduced amino acid sequence of the ModB7-2 protein. Putative transmembrane regions are underlined.
In order to substantiate these results, cDNA was generated by reverse transcription and PCR amplified with primers m148.for and m149.rev, which were specific for ORFs m147 and m148 and the region affected in mutant RV-AL15 (arrows in Fig. 4A). A PCR product with a size of 250 bp was obtained from the cDNA derived from RNA of MCMV-GFP infected cells (Fig. 4C, lane 2). In comparison, when viral DNA was used as a template, a product of 450 bp was generated (Fig. 4C, lane 3). This result indicated that an intron sequence of about 200 bp was missing in the cDNA. Sequence analysis of the PCR product led to the identification of exon-intron boundaries at nucleotide positions 207466 and 207263 in the lower strand of the MCMV genome (51). The sequences GTGGGT and TTTTCACATACAG at the start and end point of the intron represent consensus splice donor and splice acceptor sites, respectively (43).
5′- and 3′-RACE experiments were carried out to define the start and end point of the transcript. Sequence analysis of the amplified cDNA products indicated a subtle heterogeneity at the transcription start site, locating the 5′ end of the transcript at nucleotides 207525 and 207523 of the MCMV genome. A bona fide TATA box (TATATA) was found another 24 bp upstream (nucleotides 207554 to 207549). The 3′ end of the transcript was mapped to position 206834. A classical polyadenylation signal, AATAAA (nucleotides 206866 to 206861), and a sequence displaying homology to the typical 3′-end consensus sequence YGTTGTTYY (Y, pyrimidine) (63) (TGTGTGATGTTGTCTT; nucleotides 206825 to 206810) were found immediately up- and downstream of the determined 3′ end, respectively. Taken together, an exon with a size of 59 nucleotides spanning from nucleotides 207525 to 207467 is spliced to a second exon starting at position 207263 (Fig. 4A). The second exon, which is mainly identical to the previously described ORF m147 (51), has a size of 430 nucleotides. An ORF starts at position 207502 within exon 1, which contributes 12 codons of a total of 145 codons. Computer-assisted analysis of the amino acid sequence (30) led to the prediction of two putative transmembrane regions (amino acids 62 to 84 and 105 to 127) (Fig. 4D) and suggested a type IIIb topology of the protein.
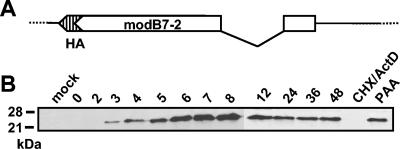
In order to prove that a protein encoded by the identified gene is expressed in infected cells, a short DNA sequence encoding an influenza virus hemagglutinin-derived epitope (HA tag) was inserted at the 3′ end of the ORF in the BAC-cloned MCMV genome (Fig. 5A). RAW264.7 cells were infected with the resulting virus, RV-AL8, and cell lysates were prepared at different time points postinfection and subjected to Western blot analysis with an anti-HA antibody (Fig. 5B). A protein of approximately 23 kDa could be detected at 3 h postinfection. Strongest expression was observed between 6 and 8 h postinfection, and expression lasted throughout the time period examined, until 48 h postinfection. When protein expression in the infected cells was restricted to immediate-early proteins, no signal was found (Fig. 5B, lane CHX/ActD). In contrast, when expression of late proteins was prevented by addition of phosphonoacetic acid to the cell culture medium, the protein could clearly be detected, indicating that it belongs to the class of early viral proteins.
FIG. 5.
Identification of the protein encoded by the modB7-2 gene. (A) A recombinant virus that carries a modB7-2 ORF tagged with an HA epitope sequence at its 3′ end was constructed. (B) Expression of the HA-tagged ModB7-2 protein in RAW264.7 cells infected with RV-AL18. RAW264.7 cells were mock infected or infected at a multiplicity of infection of 1, and cell lysates were harvested at the indicated time points (hours) postinfection. In order to achieve selective expression of immediate-early or early proteins, cells were incubated from 30 min prior to infection to 3 h postinfection in the presence of cycloheximide (CHX) followed by incubation in the presence of actinomycin D (ActD) for another 3 h or incubated in the presence of phosphonoacetic acid (PAA) for 24 h. Protein samples were separated on a 15% polyacrylamide gel, and Western blot analysis was performed with an anti-HA antibody. Molecular mass standards are indicated on the left.
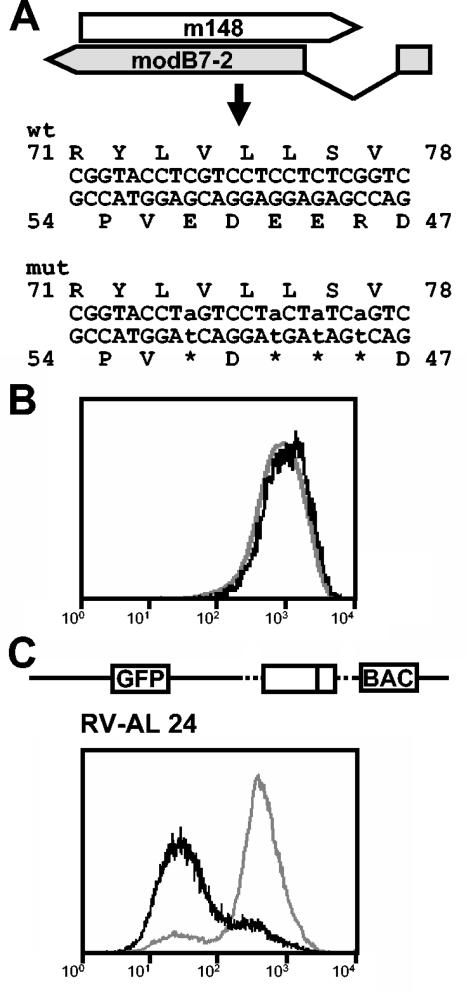
By selectively disrupting the expression of the identified gene without affecting any other viral gene, we wanted to provide definite evidence that the gene is involved in the downmodulation of CD86 surface expression. To this end, we introduced four stop codons at positions 48, 49, 50, and 52 into the open reading frame of the gene (Fig. 6A) with a two-step replacement procedure for mutagenesis of the BAC MCMV-GFP (for details, see Materials and Methods). Please note that the amino acid sequence encoded by ORF m148, which is located on the opposite DNA strand, is not affected by the mutations. Successful mutagenesis was verified by DNA sequencing of the mutated genome. Recombinant virus RV-AL22 was reconstituted from the modified BAC genome and tested for its ability to influence the expression of CD86 on RAW264.7 cells. As can be seen in Fig. 6B, RV-AL22 is missing this function, clearly indicating that the disrupted gene is responsible for interference with CD86 expression.
FIG. 6.
Phenotypic analysis of mutant viruses with either a selective disruption or a reinsertion of the modB7-2 gene. (A) Four stop codons were introduced into the modB7-2 ORF of the BAC-cloned MCMV genome by site-directed mutagenesis to prevent the generation of a functional protein. Indicated are the lower and upper strands of the genomic regions and the resulting amino acid sequences of m148 and modB7-2 before and after mutagenesis. Please note that the amino acid sequence of m148 is not affected by the mutations. (B) The ability of the resulting viral mutant RV-AL22 to influence expression of CD86 was assessed by flow cytometry. (C) Genome structure of mutant RV-AL24, indicating reinsertion of the modB7-2 gene without an intron into the backbone of MCMV-GFPΔ6 and flow cytometric analysis of CD86 expression on the surface of RV-AL24-infected cells. For panels B and C, the bond lines represent CD86 expression in infected (GFP-positive) cells and the gray lines represent CD86 expression in uninfected (GFP-negative) cells of the same culture.
As a second proof, we constructed viral mutant RV-AL24 (Fig. 6C; Table 2) by reinsertion of the identified gene without the intron sequence into the backbone of the deletion mutant MCMV-GFPΔ6 lacking ORFs m144 to m158 (construction of the mutant is described in Materials and Methods). Analysis of CD86 expression on the surface of RV-AL24-infected cells revealed that RV-AL24 possessed the function, confirming that the inserted ORF encodes the gene product mediating the effect. We propose to name the identified gene m147.5 according to its position in the MCMV genome. In addition, we would like to give it the designation modulator of B7-2 (modB7-2) to indicate its modulating effect on CD86 surface expression.
Selective targeting of CD86 expression by the modB7-2 gene.
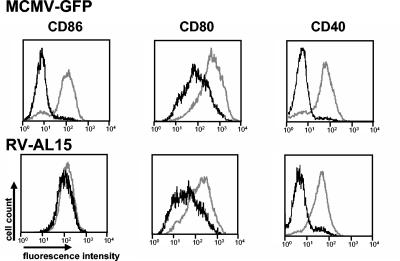
CD86 and other costimulatory molecules as well as MHC class II molecules share the secretory transport pathway to the plasma membrane, and in immature dendritic cells they are stored at least in part together in the same compartment (57). Since CD40 and CD80 are also downmodulated on MCMV-infected antigen-presenting cells, one may expect that MCMV has invented a common principle that affects the transport and/or the expression of all of these molecules. To test this hypothesis, RAW264.7 cells were infected with the recombinant virus RV-AL15 and, for comparison, with the parental virus MCMV-GFP. In RV-AL15, the modB7-2 gene is specifically disrupted by a mutation within the first exon (see the position of the mutation in Fig. 3, arrowhead 3). Accordingly, the virus was unable to influence the expression of CD86 on the surface of infected cells (Fig. 7, lower panel, histogram 1). Downmodulation of the other costimulatory molecules tested was unaffected, as they showed reduced expression levels similar to those on cells infected with the parental virus MCMV-GFP (Fig. 7, compare histograms 2 to 4, upper and lower panels). Virtually identical results were found when infected dendritic cells were examined. In addition, downregulation of MHC class I and class II molecules was still observed on dendritic cells infected with modB7-2 mutants (data not shown). Thus, the effect of modB7-2 is specific for CD86, and there is no impact on the other surface molecules tested.
FIG. 7.
Selectivity of the modB7-2 gene function. RAW264.7 cells were infected with MCMV-GFP or with the mutant virus RV-AL15, which carries a mutation in exon 1 of modB7-2 (compare Fig. 3A). Surface expression of the indicated markers on infected (GFP-positive) (black lines) and uninfected (GFP-negative) cells (gray lines) of the same cultures was determined by flow cytometry.
DISCUSSION
In this study, we report the identification of an MCMV gene which is required for downmodulation of the cell surface expression of the costimulatory molecule CD86 on infected antigen-presenting cells. By taking a genetic approach in combination with screening of mutants for a loss of the viral function, we were able to successively narrow down the genomic region encoding the function and finally to identify the gene responsible. The gene, which has not been described before, is composed of two exons and encodes a 145-amino-acid protein. Selective disruption of the modB7-2 ORF as well as rescue of the phenotype by reinsertion of the gene into the genome of a mutant lacking the function provided definitive evidence that the gene is involved in the downregulation of CD86 surface expression. Since a viral mutant with a mutation in the modB7-2 gene retained its ability to influence several other surface molecules, we conclude that additional immunomodulatory MCMV genes are responsible for interference with these functionally important molecules of antigen-presenting cells.
The procedure that we chose to find the modB7-2 gene was enabled by the availability of the BAC-cloned MCMV genome (41, 59) and of techniques for rapid and specific manipulation of BAC-cloned herpesviruses genomes (1, 60). A previously described approach, the combination of random transposon mutagenesis of the MCMV genome with phenotypic screening procedures, opened the avenue to identify viral genes that are responsible for specific properties of cytomegaloviruses (12, 13, 15). A similar genetic strategy, the systematic knockout of every HCMV ORF encoding a putative glycoprotein, was successfully used to map the HCMV genes encoding the viral Fcγ receptors (5).
Our approach extends the previous possibilities and further speeds up the screening procedure. First, there was no need to know the exact properties of the desired viral protein, and the strategy depended on a stable phenotype and a convenient read-out system only. The observation that monocyte-macrophage cell lines displayed downmodulation of surface molecules upon MCMV infection like that of dendritic cells was especially helpful in this respect. Following the identification of the modB7-2 gene, we could confirm that modB7-2 mutants showed an identical loss of the function in dendritic cells, implying that the ModB7-2 protein affects the same target(s) in both cell types.
Second, the generation and analysis of mutants with large deletions reduced the screening effort. The idea to construct mutants with large deletions was driven by the hypothesis that immunomodulatory functions are encoded by the “luxury” class of CMV genes that are nonessential for replication of the virus in cell culture and that such genes are preferentially located at the termini of the CMV genomes. Indeed, all mutant genomes generated gave rise to viable mutants. By constructing mutants with successively smaller deletions, we could rapidly locate the desired gene. We admit that by following this strategy, one may miss a nonessential gene if it is dispersed between essential genes within the central region of the CMV genomes. Although different, the previously used transposon approach was also afflicted with a few shortcomings. Recent work revealed, for example, that the distribution of transposon insertions in a virus library is not completely random (39). Thus, screening of a transposon library will sometimes have to encompass severalfold coverage of all possible mutants without guaranteeing that the desired mutant is really present. Such potential problems can eventually be circumvented by applying a more systematic mutational analysis, which has already been done for HCMV laboratory strains (20, 61). Such libraries of defined mutants will provide a good basis for further phenotypic screens. But even when such libraries become available, we think it will be worthwhile to generate and to include mutants with large deletions into a screen in order to speed up the genetic analysis of CMV.
We provide several lines of evidence that the modB7-2 gene is involved in the downmodulation of CD86. Every virus carrying a mutation that affected the gene displayed a loss of the function. Interestingly, a mutant with a mutation in the intron sequence retained the ability to influence CD86 surface expression, suggesting that splicing of the primary transcript is not disturbed by the mutation. Reinsertion of a DNA fragment carrying the gene into the genome of the deletion mutant MCMV-GFPΔ6 was sufficient to restore the function. Final assignment of the function to the identified gene was hampered by the fact that both DNA strands in the genomic region analyzed carry ORFs which overlap either partially or even completely. The RNA analysis led to the detection of one transcript only, and the identified splice donor and acceptor sites represent consensus sites. We did not find evidence that other transcripts originate from this area in MCMV-infected RAW267.4 cells. Thus, either some of the ORFs are expressed after infection of other cell types only or they do not represent genuine genes. In fact, recent reevaluation of the coding potential of the HCMV genome led to the discarding of several previously annotated ORFs (19, 46), and the same may apply to some ORFs of MCMV.
Definitive proof that the modB7-2 gene is responsible for CD86 downmodulation was provided by introduction of stop codons into the modB7-2 ORF in mutant RV-AL22, which exclusively disrupted expression of the ModB7-2 protein but retained the integrity of ORF m148. Reinsertion of the modB7-2 gene lacking the intron sequence into the genome of the deletion mutant MCMV-GFPΔ6 led to a rescue of the phenotype, providing further evidence that the identified ORF encodes the function.
An interesting question is whether the modB7-2 gene product mediates the effect solely by itself or has to cooperate with other viral proteins. Analysis of mutants with large deletions elsewhere in the genome did not show a loss of the function (data not shown). The phenotypes of mutant RV-AL4 and of the mutants with disruptions of single ORFs also suggest that no other gene is involved and that the modB7-2 gene exerts the effect alone. However, as mentioned before, we cannot completely rule out that a gene encoding a putative cofactor may have escaped our detection. An answer to the question will be provided by expression of the isolated gene in macrophages or dendritic cells. This kind of experiment is under way. In any case, modB7-2 is an essential factor required for downmodulating CD86.
The observation that ModB7-2 specifically targets CD86 and does not seem to influence the expression of other surface molecules may give us a hint about its mode of action. Although the observation cannot be generalized because we only analyzed a few surface molecules, one can assume that ModB7-2 either interacts with CD86 directly or targets a pathway that is used exclusively by CD86. Because CD86 shares the secretory pathway with other costimulatory molecules as well as with MHC molecules and because these molecules are stored at least partially in the same vesicles in antigen-presenting cells (57), one might expect a common effect on their expression if ModB7-2 acted in a more general way, e.g., by disturbing the transport pathway or by targeting the vesicles. This seems rather unlikely. In fact, we got preliminary results that other individual MCMV genes are responsible for specific effects on the different surface molecules. Analysis of the amino acid sequence suggested that the protein is inserted into a membrane of the cell via its two transmembrane regions. Examination of the subcellular localization of the ModB7-2 protein and of its interaction with other cellular and viral proteins has to await the generation of specific antisera and will be the subject of further work.
Since the description of the functional impairment of MCMV-infected antigen-presenting cells (4, 38), this is the first study that reports on the identification of an MCMV gene that may contribute to the defect in these cells. The genetic approach taken here will allow us to find the other MCMV genes interfering with the functions of dendritic cells and could also be used to define HCMV genes mediating comparable effects. Definition of the molecular basis underlying the observed viral effects is a prerequisite to dissect and to understand the complex interaction of CMV with these important antigen-presenting cells. Identification of the viral genes and the availability of corresponding MCMV mutants will also allow us to evaluate the biological significance of these viral gene functions in vivo.
Acknowledgments
We thank Sabine Herrmann for excellent technical assistance. For generous access to the FACSCalibur machine we are indebted to Andreas Simm. We thank B. L. Wanner and M. Berlyn (E. coli Stock Center, Yale University) for providing plasmid pKD46 and H. Rüssmann for pGP704.
This work was supported by grant ME1102/2-1 of the Deutsche Forschungsgemeinschaft and in part by the NBL3 Program of the Bundesministerium für Bildung und Forschung (BMBF).
REFERENCES
- 1.Adler, H., M. Messerle, and U. H. Koszinowski. 2003. Cloning of herpesviral genomes as bacterial artificial chromosomes. Rev. Med. Virol. 13:111-121. [DOI] [PubMed] [Google Scholar]
- 2.Adler, H., M. Messerle, M. Wagner, and U. H. Koszinowski. 2000. Cloning and mutagenesis of the murine gammaherpesvirus 68 genome as an infectious bacterial artificial chromosome. J. Virol. 74:6964-6974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Alcami, A., and U. H. Koszinowski. 2000. Viral mechanisms of immune evasion. Trends Microbiol. 8:410-418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Andrews, D. M., C. E. Andoniou, F. Granucci, P. Ricciardi-Castagnoli, and M. A. Degli-Esposti. 2001. Infection of dendritic cells by murine cytomegalovirus induces functional paralysis. Nat. Immunol. 2:1077-1084. [DOI] [PubMed] [Google Scholar]
- 5.Atalay, R., A. Zimmermann, M. Wagner, E. Borst, C. Benz, M. Messerle, and H. Hengel. 2002. Identification and expression of human cytomegalovirus transcription units coding for two distinct Fcgamma receptor homologs. J. Virol. 76:8596-8608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Banchereau, J., F. Briere, C. Caux, J. Davoust, S. Lebecque, Y. J. Liu, B. Pulendran, and K. Palucka. 2000. Immunobiology of dendritic cells. Annu. Rev. Immunol. 18:767-811. [DOI] [PubMed] [Google Scholar]
- 7.Banchereau, J., and R. M. Steinman. 1998. Dendritic cells and the control of immunity. Nature 392:245-252. [DOI] [PubMed] [Google Scholar]
- 8.Beck, K., U. Meyer-Konig, M. Weidmann, C. Nern, and F. T. Hufert. 2003. Human cytomegalovirus impairs dendritic cell function: a novel mechanism of human cytomegalovirus immune escape. Eur. J. Immunol. 33:1528-1538. [DOI] [PubMed] [Google Scholar]
- 9.Borst, E. M., G. Hahn, U. H. Koszinowski, and M. Messerle. 1999. Cloning of the human cytomegalovirus (HCMV) genome as an infectious bacterial artificial chromosome in Escherichia coli: a new approach for construction of HCMV mutants. J. Virol. 73:8320-8329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Borst, E. M., G. Posfai, F. Pogoda, and M. Messerle. 2004. Mutagenesis of herpesvirus BACs by allele replacement, p. 269-280. In S. Zhao and M. Stodolsky (ed.), Bacterial artificial chromosomes, vol. 2: functional studies. Humana Press Inc., Totowa, N.J. [DOI] [PubMed]
- 11.Brune, W., H. Hengel, and U. H. Koszinowski. 1999. Current protocols in immunology, p. 19.7.1-19.7.13. John Wiley & Sons, New York, N.Y. [DOI] [PubMed]
- 12.Brune, W., C. Menard, J. Heesemann, and U. H. Koszinowski. 2001. A ribonucleotide reductase homolog of cytomegalovirus and endothelial cell tropism. Science 291:303-305. [DOI] [PubMed] [Google Scholar]
- 13.Brune, W., C. Menard, U. Hobom, S. Odenbreit, M. Messerle, and U. H. Koszinowski. 1999. Rapid identification of essential and nonessential herpesvirus genes by direct transposon mutagenesis. Nat. Biotechnol. 17:360-364. [DOI] [PubMed] [Google Scholar]
- 14.Brune, W., M. Messerle, and U. H. Koszinowski. 2000. Forward with BACs: new tools for herpesvirus genomics. Trends Genet. 16:254-259. [DOI] [PubMed] [Google Scholar]
- 15.Brune, W., M. Nevels, and T. Shenk. 2003. Murine cytomegalovirus m41 open reading frame encodes a Golgi-localized antiapoptotic protein. J. Virol. 77:11633-11643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cherepanov, P. P., and W. Wackernagel. 1995. Gene disruption in Escherichia coli: TcR and KmR cassettes with the option of Flp-catalyzed excision of the antibiotic-resistance determinant. Gene 158:9-14. [DOI] [PubMed] [Google Scholar]
- 17.Dalod, M., T. Hamilton, R. Salomon, T. P. Salazar-Mather, S. C. Henry, J. D. Hamilton, and C. A. Biron. 2003. Dendritic cell responses to early murine cytomegalovirus infection: subset functional specialization and differential regulation by interferon alpha/beta. J. Exp. Med. 197:885-898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Datsenko, K. A., and B. L. Wanner. 2000. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc. Natl. Acad. Sci. USA 97:6640-6645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Davison, A. J., A. Dolan, P. Akter, C. Addison, D. J. Dargan, D. J. Alcendor, D. J. McGeoch, and G. S. Hayward. 2003. The human cytomegalovirus genome revisited: comparison with the chimpanzee cytomegalovirus genome. J. Gen. Virol. 84:17-28. [DOI] [PubMed] [Google Scholar]
- 20.Dunn, W., C. Chou, H. Li, R. Hai, D. Patterson, V. Stolc, H. Zhu, and F. Liu. 2003. Functional profiling of a human cytomegalovirus genome. Proc. Natl. Acad. Sci. USA 100:14223-14228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Farrell, H. E., M. A. Degli-Esposti, and N. J. Davis-Poynter. 1999. Cytomegalovirus evasion of natural killer cell responses. Immunol. Rev. 168:187-197. [DOI] [PubMed] [Google Scholar]
- 22.Gredmark, S., and C. Soderberg-Naucler. 2003. Human cytomegalovirus inhibits differentiation of monocytes into dendritic cells with the consequence of depressed immunological functions. J. Virol. 77:10943-10956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Grigoleit, U., S. Riegler, H. Einsele, S. K. Laib, G. Jahn, H. Hebart, P. Brossart, F. Frank, and C. Sinzger. 2002. Human cytomegalovirus induces a direct inhibitory effect on antigen presentation by monocyte-derived immature dendritic cells. Br. J. Haematol. 119:189-198. [DOI] [PubMed] [Google Scholar]
- 24.Gutermann, A., A. Bubeck, M. Wagner, U. Reusch, C. Menard, and U. H. Koszinowski. 2002. Strategies for the identification and analysis of viral immune-evasive genes—cytomegalovirus as an example. Curr. Top. Microbiol. Immunol. 269:1-22. [DOI] [PubMed] [Google Scholar]
- 25.Hahn, G., H. Khan, F. Baldanti, U. H. Koszinowski, M. G. Revello, and G. Gerna. 2002. The human cytomegalovirus ribonucleotide reductase homolog UL45 is dispensable for growth in endothelial cells, as determined by a BAC-cloned clinical isolate of human cytomegalovirus with preserved wild-type characteristics. J. Virol. 76:9551-9555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hanson, L. K., J. S. Slater, Z. Karabekian, H. W. Virgin, C. A. Biron, M. C. Ruzek, N. van Rooijen, R. P. Ciavarra, R. M. Stenberg, and A. E. Campbell. 1999. Replication of murine cytomegalovirus in differentiated macrophages as a determinant of viral pathogenesis. J. Virol. 73:5970-5980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Heise, M. T., M. Connick, and H. W. Virgin. 1998. Murine cytomegalovirus inhibits interferon gamma-induced antigen presentation to CD4 T cells by macrophages via regulation of expression of major histocompatibility complex class II-associated genes. J. Exp. Med. 187:1037-1046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hengel, H., W. Brune, and U. H. Koszinowski. 1998. Immune evasion by cytomegalovirus—survival strategies of a highly adapted opportunist. Trends Microbiol. 6:190-197. [DOI] [PubMed] [Google Scholar]
- 29.Hertel, L., V. G. Lacaille, H. Strobl, E. D. Mellins, and E. S. Mocarski. 2003. Susceptibility of immature and mature Langerhans cell-type dendritic cells to infection and immunomodulation by human cytomegalovirus. J. Virol. 77:7563-7574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hirokawa, T., S. Boon-Chieng, and S. Mitaku. 1998. SOSUI: Classification and secondary structure prediction system for membrane proteins. Bioinformatics 14:378-379. [DOI] [PubMed] [Google Scholar]
- 31.Hobom, U., W. Brune, M. Messerle, G. Hahn, and U. H. Koszinowski. 2000. Fast screening procedures for random transposon libraries of cloned herpesvirus genomes: mutational analysis of human cytomegalovirus envelope glycoprotein genes. J. Virol. 74:7720-7729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Jahn, G., S. Stenglein, S. Riegler, H. Einsele, and C. Sinzger. 1999. Human cytomegalovirus infection of immature dendritic cells and macrophages. Intervirology 42:365-372. [DOI] [PubMed] [Google Scholar]
- 33.Jarvis, M. A., and J. A. Nelson. 2002. Human cytomegalovirus persistence and latency in endothelial cells and macrophages. Curr. Opin. Microbiol. 5:403-407. [DOI] [PubMed] [Google Scholar]
- 34.Kavanagh, D. G., and A. B. Hill. 2001. Evasion of cytotoxic T lymphocytes by murine cytomegalovirus. Semin. Immunol. 13:19-26. [DOI] [PubMed] [Google Scholar]
- 35.Kolter, R., M. Inuzuka, and D. P. Helinski. 1978. Trans-complementation-dependent replication of a low molecular weight origin fragment from plasmid R6K. Cell 15:1199-1208. [DOI] [PubMed] [Google Scholar]
- 36.Loenen, W. A., C. A. Bruggeman, and E. J. Wiertz. 2001. Immune evasion by human cytomegalovirus: lessons in immunology and cell biology. Semin. Immunol. 13:41-49. [DOI] [PubMed] [Google Scholar]
- 37.Lutz, M. B., N. Kukutsch, A. L. Ogilvie, S. Rossner, F. Koch, N. Romani, and G. Schuler. 1999. An advanced culture method for generating large quantities of highly pure dendritic cells from mouse bone marrow. J. Immunol. Methods 223:77-92. [DOI] [PubMed] [Google Scholar]
- 38.Mathys, S., T. Schroeder, J. Ellwart, U. H. Koszinowski, M. Messerle, and U. Just. 2003. Dendritic cells under influence of mouse cytomegalovirus have a physiologic dual role: to initiate and to restrict T cell activation. J. Infect. Dis. 187:988-999. [DOI] [PubMed] [Google Scholar]
- 39.Menard, C., M. Wagner, Z. Ruzsics, K. Holak, W. Brune, A. E. Campbell, and U. H. Koszinowski. 2003. Role of murine cytomegalovirus US22 gene family members in replication in macrophages. J. Virol. 77:5557-5570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Messerle, M., B. Bühler, G. M. Keil, and U. H. Koszinowski. 1992. Structural organization, expression, and functional characterization of the murine cytomegalovirus immediate-early gene 3. J. Virol. 66:27-36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Messerle, M., I. Crnkovic, W. Hammerschmidt, H. Ziegler, and U. H. Koszinowski. 1997. Cloning and mutagenesis of a herpesvirus genome as an infectious bacterial artificial chromosome. Proc. Natl. Acad. Sci. USA 94:14759-14763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Mocarski, E. S., and C. T. Courcelle. 2001. Cytomegaloviruses and their replication, p. 2629-2674. In D. M. Knipe and P. M. Howley (ed.), Fields virology. Lippincott Williams & Wilkins-Raven Publishers, New York, N.Y.
- 43.Mount, S. M. 1982. A catalogue of splice junction sequences. Nucleic Acids Res. 10:459-472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Moutaftsi, M., P. Brennan, S. A. Spector, and Z. Tabi. 2004. Impaired lymphoid chemokine-mediated migration due to a block on the chemokine receptor switch in human cytomegalovirus-infected dendritic cells. J. Virol. 78:3046-3054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Moutaftsi, M., A. M. Mehl, L. K. Borysiewicz, and Z. Tabi. 2002. Human cytomegalovirus inhibits maturation and impairs function of monocyte-derived dendritic cells. Blood 99:2913-2921. [DOI] [PubMed] [Google Scholar]
- 46.Murphy, E., I. Rigoutsos, T. Shibuya, and T. E. Shenk. 2003. Reevaluation of human cytomegalovirus coding potential. Proc. Natl. Acad. Sci. USA 100:13585-13590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Pass, R. F. 2001. Cytomegalovirus, p. 2675-2705. In D. M. Knipe and P. M. Howley (ed.), Fields virology. Lippincott Williams & Wilkins, Philadelphia, Pa.
- 48.Popkin, D. L., M. A. Watson, E. Karaskov, G. P. Dunn, R. Bremner, and H. W. Virgin. 2003. Murine cytomegalovirus paralyzes macrophages by blocking IFN gamma-induced promoter assembly. Proc. Natl. Acad. Sci. USA 100:14309-14314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Raftery, M. J., M. Schwab, S. M. Eibert, Y. Samstag, H. Walczak, and G. Schonrich. 2001. Targeting the function of mature dendritic cells by human cytomegalovirus: a multilayered viral defense strategy. Immunity 15:997-1009. [DOI] [PubMed] [Google Scholar]
- 50.Raschke, W. C., S. Baird, P. Ralph, and I. Nakoinz. 1978. Functional macrophage cell lines transformed by Abelson leukemia virus. Cell 15:261-267. [DOI] [PubMed] [Google Scholar]
- 51.Rawlinson, W. D., H. E. Farrell, and B. G. Barrell. 1996. Analysis of the complete DNA sequence of murine cytomegalovirus. J. Virol. 70:8833-8849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Reddehase, M. J. 2002. Antigens and immunoevasins: opponents in cytomegalovirus immune surveillance. Nat. Rev. Immunol. 2:831-844. [DOI] [PubMed] [Google Scholar]
- 53.Riegler, S., H. Hebart, H. Einsele, P. Brossart, G. Jahn, and C. Sinzger. 2000. Monocyte-derived dendritic cells are permissive to the complete replicative cycle of human cytomegalovirus. J. Gen. Virol. 81:393-399. [DOI] [PubMed] [Google Scholar]
- 54.Sambrook, J. F., and D. W. Russell. 2000. Molecular cloning: a laboratory manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 55.Senechal, B., A. M. Boruchov, J. L. Reagan, D. N. Hart, and J. W. Young. 2004. Infection of mature monocyte-derived dendritic cells with human cytomegalovirus inhibits stimulation of T-cell proliferation via the release of soluble CD83. Blood 103:4207-4215. [DOI] [PubMed] [Google Scholar]
- 56.Stoddart, C. A., R. D. Cardin, J. M. Boname, W. C. Manning, G. B. Abenes, and E. S. Mocarski. 1994. Peripheral blood mononuclear phagocytes mediate dissemination of murine cytomegalovirus. J. Virol. 68:6243-6253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Turley, S. J., K. Inaba, W. S. Garrett, M. Ebersold, J. Unternaehrer, R. M. Steinman, and I. Mellman. 2000. Transport of peptide-MHC class II complexes in developing dendritic cells. Science 288:522-527. [DOI] [PubMed] [Google Scholar]
- 58.Wagner, M., A. Gutermann, J. Podlech, M. J. Reddehase, and U. H. Koszinowski. 2002. Major histocompatibility complex class I allele-specific cooperative and competitive interactions between immune evasion proteins of cytomegalovirus. J. Exp. Med. 196:805-816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Wagner, M., S. Jonjic, U. H. Koszinowski, and M. Messerle. 1999. Syst. excision of vector sequences from the BAC-cloned herpesvirus genome during virus reconstitution. J. Virol. 73:7056-7060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Wagner, M., Z. Ruzsics, and U. H. Koszinowski. 2002. Herpesvirus genetics has come of age. Trends Microbiol. 10:318-324. [DOI] [PubMed] [Google Scholar]
- 61.Yu, D., M. C. Silva, and T. Shenk. 2003. Functional map of human cytomegalovirus AD169 defined by global mutational analysis. Proc. Natl. Acad. Sci. USA 100:12396-12401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Yu, D., G. A. Smith, L. W. Enquist, and T. Shenk. 2002. Construction of a self-excisable bacterial artificial chromosome containing the human cytomegalovirus genome and mutagenesis of the diploid TRL/IRL13 gene. J. Virol. 76:2316-2328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Zarudnaya, M. I., I. M. Kolomiets, A. L. Potyahaylo, and D. M. Hovorun. 2003. Downstream elements of mammalian pre-mRNA polyadenylation signals: primary, secondary and higher-order structures. Nucleic Acids Res. 31:1375-1386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Zhang, Y., F. Buchholz, J. P. Muyrers, and A. F. Stewart. 1998. A new logic for DNA engineering using recombination in Escherichia coli. Nat. Genet. 20:123-128. [DOI] [PubMed] [Google Scholar]