Abstract
Efficient replication of hepatitis C virus (HCV) subgenomic RNA in cell culture requires the introduction of adaptive mutations. In this report we describe a system which enables efficient replication of the Con1 subgenomic replicon in Huh7 cells without the introduction of adaptive mutations. The starting hypothesis was that high amounts of the NS5A hyperphosphorylated form, p58, inhibit replication and that reduction of p58 by inhibition of specific kinase(s) below a certain threshold enables HCV replication. Upon screening of a panel of kinase inhibitors, we selected three compounds which inhibited NS5A phosphorylation in vitro and the formation of NS5A p58 in cell culture. Cells, transfected with the HCV Con1 wild-type sequence, support HCV RNA replication upon addition of any of the three compounds. The effect of the kinase inhibitors was found to be synergistic with coadaptive mutations in NS3. This is the first direct demonstration that the presence of high amounts of NS5A-p58 causes inhibition of HCV RNA replication in cell culture and that this inhibition can be relieved by kinase inhibitors.
The hepatitis C virus (HCV) has been identified as one of the major causes of chronic liver disease, and neither vaccines nor broadly effective therapeutic agents are available (7). HCV contains a plus-strand RNA genome which encodes a single polyprotein precursor that is cleaved by cellular and viral proteases (1, 23). The structural proteins core, E1, and E2 are located in the amino terminus of the polyprotein (11), followed by p7, a hydrophobic peptide capable of forming an ion channel (10, 21), and the nonstructural (NS) proteins NS2, NS3, NS4A, NS4B, NS5A, and NS5B, which are putative components of the RNA replication machinery (6). NS5A is a 446-amino-acid phosphoprotein which is phosphorylated on many serine residues and exists in two distinct species, termed p56 (phosphorylated) and p58 (hyperphosphorylated). In p56, two of the modified serine residues have been identified as serine residue 2194 in strain 1B (13) and serine residue 2321 in strain 1A (22). In addition to these basal phosphorylation sites, three serine residues, S2197, S2201, and S2204, have been reported to be important for hyperphosphorylation (25). The hyperphosphorylation of NS5A is a highly regulated process which requires the expression of an intact NS3-5A polyprotein and correct polyprotein processing (14, 20). The family of cellular kinase(s) responsible for NS5A phosphorylation has been identified as the CMGC group of serine-threonine kinases (24).
Studies of HCV biology and the elucidation of the function of the single viral protein has been slowed down for a long time by the lack of a permissive cell culture system supporting the efficient replication of the virus. Some years ago, however, Lohmann and colleagues reported efficient HCV replication in the human hepatoma cell line Huh7 after transfection of a bicistronic subgenomic replicon expressing a selectable marker (18). Replication of this replicon dramatically increases after the occurrence of adaptive mutations, which map predominantly in NS5A and, in some cases, affect its phosphorylation status (2, 16).
The fact that many adaptive mutations reside in NS5A suggests a role for NS5A or its different phosphorylated forms in RNA replication, even though the exact mechanism is still obscure. It is interesting that the most effective adaptive mutations, with the exception of those in NS4B, map exactly at serine residues, which have been implicated in NS5A hyperphosphorylation (2). Adaptive mutations at these sites result in a significant reduction of NS5A p58 formation. This observation suggests not only that NS5A hyperphosphorylation is not necessary for HCV replication in cell culture but that it seems that it may be deleterious.
The replicon system would be an ideal system for the study of HCV replication mechanisms. It has been noted, however, that mutations that are adaptive for replication of HCV in cell culture are highly attenuated in vivo (4). The HCV-N strain seems to be an exception. It has been reported that HCV-N is infectious in the chimpanzee model and that it is able to replicate efficiently in cell culture without any adaptive mutation (12). However, NS5A derived from this HCV strain contains a natural insertion of 4 amino acids, which is unique to this isolate compared to all other viral strains. This 4-amino-acid insertion renders the corresponding replicon highly efficient for replication in cell culture, and at the same time, infectivity in the chimpanzee model is maintained. Viremia in the infected chimpanzee, however, is at a low level, and subsequent infection of two other chimpanzees with RNA derived from the primary infected chimpanzee failed (12). Thus, it seems that the 4-amino-acid insertion significantly attenuates infectivity in the chimpanzee and functions as a naturally occurring cell culture-permissive variant.
It is of great importance to develop a cell culture system in which HCV replicates without losing its in vivo infectivity potential. In this case, the cell culture system could be considered a better mimic of in vivo replication.
In this work, we show that compounds that inhibit the formation of NS5A p58 by inhibition of the putative kinase(s) render a nonadapted HCV replicon capable of replication in cell culture.
MATERIALS AND METHODS
Plasmid construction.
DNA plasmids used in this work were constructed by standard recombinant DNA technology, and the correct sequence was confirmed by automated nucleotide sequencing. Amino acid numbers refer to the location of the Con1 full-length HCV genome (EMBL accession number AJ238799) starting from the core coding region. The DNA plasmids wild type (wt) and m17 have been previously described as pHCVNeo17.wt and pHCVNeo17.C, respectively (27). The other plasmids are identical to pHCVNeo17.wt but contain the following mutations: SA, S2204A in NS5A; m17/SA, S2204A in NS5A and E1202G in NS3; m17-GAA, E1202G in NS3 and D2737A/D2738A in NS5B; wt-GAA, D2737A/D2738A in NS5B. For the experiment for which results are shown in Fig. 3, the neomycin phosphotransferase (neo) gene of plasmid wt and SA was replaced by the β-lactamase (BLA) gene as described previously (19), resulting in plasmids wt-BLA and SA-BLA, respectively.
FIG. 3.
Induction of replication of the Con1 wt subgenomic replicon in the presence of kinase inhibitors. RNA was transcribed from plasmid wt-BLA or SA-BLA, and 10 μg of RNA was transfected into 10A-IFN cells as described in Materials and Methods. Cells were incubated for 4 days with or without 8 μM compound (left column) or with or without compound plus 30 μM RdRP inhibitor RdRP-I (right column). The results shown are from the BLA assay, in which blue cell staining indicates active HCV replication.
In vitro kinase assay.
Huh7 cells containing the Con1 subgenomic replicon with the adaptive mutation A2199T (Huh7-68) were grown to 50% confluence, and the protein extract was prepared as follows. Cells from a 15-cm-diameter dish were lysed with 500 μl of NETN buffer (50 mM Tris-HCl [pH 7.5], 120 mM NaCl, 1 mM EDTA, 0.5% NP-40) supplemented with 5 mM dithiothreitol (DTT), 2 mM phenylmethylsulfonyl fluoride, 100 mM NaF, 20% glycerol, and the protease inhibitor cocktail Complete (Roche) as described previously (24). For immunoprecipitation, 2.5 μl of NS5A-specific antibody (20) was bound to 12 μl of protein A-Sepharose in 300 μl of NETN for 1 h at 4°C. After the incubation, protein A-Sepharose was washed once with NETN and 30 μg of total protein was immunoprecipitated in 300 μl of NETN for 1 h at 4°C. After binding, the resin was washed once with NETN supplemented with 5 mM DTT and once with kinase buffer (50 mM Tris-HCl [pH 7.5], 5 mM MnCl, 5 mM DTT, and 50 mM NaF). For the in vitro kinase reaction, 50 μl of kinase buffer supplemented with 2.5 μCi of [γ-33P]ATP and 2.5 μl of either 100 μM compound dissolved in dimethyl sulfoxide (DMSO; final concentration, 5 μM) or DMSO alone was added to the immunoprecipitate. Reaction mixtures were incubated for 45 min at 37°C, and the reaction was terminated by the addition of 25 μl of 3× protein sample buffer. Proteins were loaded onto a sodium dodecyl sulfate (SDS)-7.5% polyacrylamide gel electrophoresis (PAGE) gel and autoradiographed.
Cell culture and BLA assay.
Huh-7 and Huh7-68 were cultured in Dulbecco's modified Eagle medium supplemented with 10% fetal bovine serum. For cells supporting subgenomic replicons, 0.8 mg of G418 (Geneticin; Gibco-BRL)/ml was added. Cells highly competent for HCV replication (10A-IFN) were obtained by curing HBI10A cells from the endogenous replicon with human alpha-2b interferon as described previously (27). The BLA assay was performed as described previously (19). For microscopy, cell fluorescence was observed by excitation of CCF2 through a 405-nm-pore-size filter with emission at 460 nm (blue fluorescence) or 515 nm (green fluorescence). Fluorescence microscopy was conducted on live 10A-IFN cells with a Leica DMIL microscope, and images were taken with a digital camera (Nikon CoolPix 995).
RNA transfection and protein expression with the vaccinia infection-transfection system.
RNA transcripts were prepared by using pHCVNeo17 plasmids digested with ScaI and the Megascript kit (Ambion) exactly as described previously (27). Ten micrograms of RNA was electroporated in 106 10A-IFN cells with the GenePulser XCell (Bio-Rad) at 110 V for 25 ms. After electroporation, 2.5 × 105 cells (5 × 105 when incubated with compound F495) were plated in a 35-mm-diameter dish and incubated for 4 days in the presence or absence of 8 μM compound as described in the figure legend. The RdRP inhibitor RdRP-I corresponds to compound 1 described elsewhere (26). Protein expression with the vaccinia infection-transfection system was performed as described previously (20). Cells (3.5 × 105 10A-IFN cells/35-mm-diameter dish) were plated the day before the experiment. Two micrograms of pHCVNeo17 plasmid was transfected by using Fugene6 (Roche) as the transfection reagent.
Metabolic labeling of proteins and immunoprecipitation.
Metabolic labeling of proteins and immunoprecipitation was performed as described previously (20). To assess the inhibitory activity of the compounds, cells were preincubated with 5 μM concentrations of the indicated compounds for 1 h before starvation and labeling and were present during the whole time of labeling. Cell extraction and denaturing immunoprecipitation were performed as described previously (20).
Quantitative TaqMan RT-PCR.
RNA was prepared by using the RNeasy mini kit (QIAGEN) as described by the manufacturer. The final volume was 100 μl of RNA/35-mm-diameter dish. Two microliters of RNA was used for quantitative reverse transcription (RT)-PCR by using an ABI PRISM 7900 HT sequence detector (Applied Biosystems). Amplifications were performed in duplicate with the TaqMan one-step RT-PCR master mix reagent kit (Applied Biosystems). Oligonucleotides and probe annealed in the 5′ untranslated region of HCV. The primers (450 nM) are GP5, 5′-CGGGAGAGCCATAGTGG-3′, and GP6, 5′-AGTACCACAAGGCCTTTCG-3′. The probe contains the 6-FAM dye at the 5′ end and the TAMRA quencher at its 3′ end. The HC2 probe (150 nM) is 5′-6-FAM-CTGCGGAACCGGTGAGTACAC-TAMRA-3′. As an endogenous standard, we used the β-actin probe, containing the VIC dye at its 5′ end (Applied Biosystems). Reactions were conducted in three stages under the following conditions: stage 1, 30 min at 48°C; stage 2, 10 min at 95°C; stage 3, 15 s at 95°C and 1 min at 60°C for 40 cycles. The total volume of the reaction mixture is 50 μl.
Compounds.
Compound SB203580 was purchased from Calbiochem. Compounds H479, A852, and F495 were synthesized in-house and are available for research purposes upon request.
RESULTS
In vitro inhibition of NS5A phosphorylation by protein kinase inhibitors.
To identify compounds that inhibit the formation of NS5A p58, we set up an in vitro assay which allowed the screening of several hundred compounds for their inhibitory activity on NS5A phosphorylation. As a source of compounds, we used a collection of proprietary kinase inhibitors with the general structure of ATP-competitive 2,4,5-trisubstituted imidazole inhibitors.
NS5A was expressed in Huh7-68 cells, and cell extract was prepared as described in Materials and Methods. It has previously been reported that cellular kinases remain associated with NS5A after immunoprecipitation with specific anti-NS5A antibodies (24). In this study, we incubated the cell extract with an NS5A-specific antibody and immunoprecipitated the associated protein complex under native conditions. The immunoprecipitated proteins were then incubated with [γ-33P]ATP in the presence or absence of 5 μM compound. After the reaction, phosphorylated proteins were resolved by SDS-PAGE, and the results are shown in Fig. 1. Comparison of the nonspecific phosphoproteins present in Huh7 cells not expressing the replicon (lane 2) with those obtained from the replicon cells Huh7-68 (lane 3) clearly indicated the production of phosphorylated NS5A. Most of the phosphates were incorporated into p56, even though a small quantity was visible also in p58 (lanes 3, 4, 6, and 7). Many compounds of the collection had no or only little effect on any of the kinases present in the immunoprecipitation (lanes 4 and 6). A large number of compounds, around 40%, showed 50% or more inhibition. Examples of compounds that inhibited more than 80 to 90% are shown in lanes 5, 8, and 11 to 13, whereas compounds with intermediate inhibition of NS5A phosphorylation are shown in lanes 7, 9, and 10. Besides NS5A-specific inhibition of phosphorylation, other proteins present in the immunoprecipitate were inhibited to different extents by different compounds, pointing to the presence of more than one protein kinase in the immunoprecipitate. For example, SB203580, a specific inhibitor of the mitogen-actived protein kinase p38 (5), inhibited the phosphorylation of all proteins, including NS5A, by more than 50%. SB203580 was thus used throughout the study as a nonspecific control protein kinase inhibitor.
FIG. 1.
Inhibition of NS5A phosphorylation in vitro. NS5A was immunoprecipitated from Huh7-68 cells and incubated with [γ-33P]ATP as described in Materials and Methods. The phosphorylation of NS5A in the absence (lane 3) or presence of 5 μM concentrations of different compounds (lanes 4 to 13) is shown. As a control, proteins were immunoprecipitated from Huh7 cells (lane 2); the sizes of molecular mass marker proteins are indicated in lane 1. Proteins were loaded on SDS-PAGE gels, and the autoradiogram is shown.
We selected all compounds which inhibited NS5A phosphorylation by more than 50% and monitored their activity in a cell-based assay described below.
Inhibition of NS5A phosphorylation in cell culture by selected compounds.
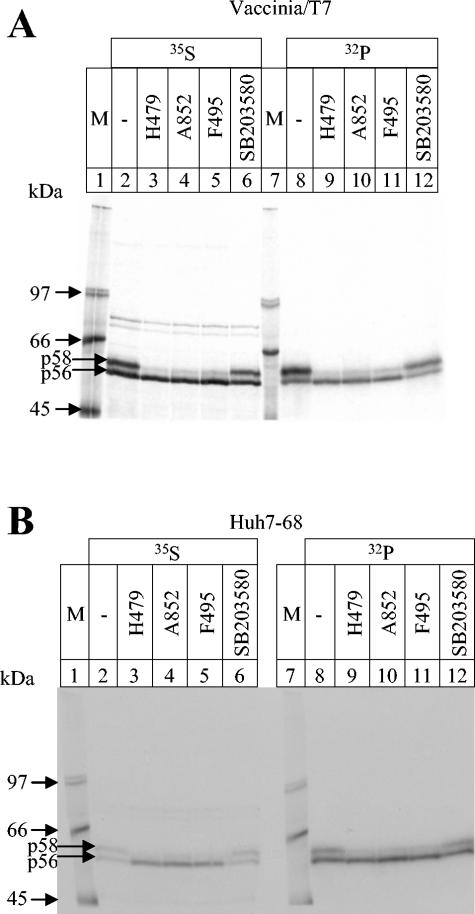
All selected compounds were evaluated in cell culture to assess their effects on NS5A phosphorylation in the context of live cells and active HCV polyprotein processing by using the vaccinia T7 infection-transfection system. DNA plasmid coding for the wt Con1 HCV replicon was transfected into Huh7 cells, and viral proteins were labeled with [35S]methionine, to monitor protein expression, or with [32P]orthophosphate, to investigate phosphorylation efficiency (Fig. 2A). Most of the selected compounds had no effect on NS5A phosphorylation in cell culture or affected the expression of NS5A and the other viral proteins without changing the phosphorylation pattern (data not shown). Three compounds selectively inhibited the formation of the p58 form of NS5A (Fig. 2A, lanes 3 to 5 and 9 to 11). Conversely, no compound was identified which inhibited basal NS5A phosphorylation without affecting NS5A expression. Like many other compounds, the control compound SB203580 inhibited the in vitro phosphorylation of NS5A by more than 50% (Fig. 1) but had no effect either on protein expression or on protein phosphorylation in cells.
FIG. 2.
Inhibition of NS5A p58 formation in cell culture. (A) NS5A was expressed by using the vaccinia infection-transfection system. The plasmid wt-BLA was transfected for 4 h in 10A-IFN cells, and proteins were labeled either with [35S]methionine (lanes 2 to 6) or with [32P]orthophosphate (lanes 8 to 12) in the presence of DMSO (lanes 2 and 8) or with a 5 μM concentration of compound H479 (lanes 3 and 9), A852 (lanes 4 and 10), F495 (lanes 5 and 11), or SB203580 (lanes 6 and 12). After radiolabeling, the protein extract was prepared, NS5A was immunoprecipitated, and proteins were loaded onto an SDS-7.5% PAGE gel and autoradiographed as described in Materials and Methods. (B) The same type of experiment was performed with Huh7-68 cells. The sizes of molecular mass marker proteins (M) are indicated in lanes 1 and 7.
The vaccinia T7 infection-transfection system can be used to study protein phosphorylation during active HCV polyprotein expression and processing. During active HCV replication, however, localization of viral proteins and protein-protein interactions may differ from the vaccinia T7 infection-transfection system, and therefore, the susceptibility of the specific kinase for the inhibitors may change. We incubated Huh7-68 cells with the three indicated compounds and labeled the proteins either with [35S]methionine or with [32P]orthophosphate (Fig. 2B). The results obtained were identical to those already observed with the vaccinia T7 infection-transfection system. Inhibition of NS5A hyperphosphorylation by the three selected kinase inhibitors was further confirmed to not depend on a specific replicon sequence by using cells carrying a replicon that contained an insertion of a lysine after valine 2039 (K@2039) (27), i.e., more than 150 amino acids away from the hyperphosphorylation sites. Also, in this case, the formation of p58 was efficiently inhibited (data not shown).
Activation of replication of wt Con1 replicon in the presence of kinase inhibitors.
Having identified compounds that specifically inhibit NS5A hyperphosphorylation, we tested their effects on HCV replication in cell culture.
To this aim, we used a subgenomic replicon in which the original neomycin phosphotransferase (neo) gene was replaced by the BLA gene (bla) (19). Cells actively replicating HCV express BLA and show a blue staining after incubation with a diffusible BLA substrate (BLA assay). Replicon RNA was electroporated in 10A-IFN cells, and compounds were added at a concentration of 8 μM 2 h after electroporation. The BLA assay was performed after 4 days of incubation, and results are shown in Fig. 3. As a positive control, we used a replicon containing the adaptive mutation S2204A (SA-BLA). This adaptive mutation reduces the formation of NS5A p58, similar to the already published mutation S2204I (Fig. 4A).
FIG. 4.
Detection of HCV-specific proteins or HCV RNA after induction or inhibition with the specific kinase inhibitors. RNA was transcribed from the plasmids wt, wt-GAA, m17, SA, m17/SA, and m17-GAA and electroporated into 10A-IFN cells as described in Materials and Methods. Cells were incubated for 4 days without (red bar) or with 8 μM concentrations of compound H479 (blue bar), compound A852 (green bar), compound F495 (black bar) or SB203580 (pink bar). (A) Western blot analysis of total protein extracts. Cell extract was prepared as described in Materials and Methods, and 50 μg of protein was loaded for each lane. Specific anti-NS5A (lanes 2 to 19) or anti-NS3 (lanes 20 to 25) antibodies were used as primary antibodies, and a peroxidase-conjugated antibody (Pierce) was used as a secondary antibody. The Western blot was developed by using the SuperSignal West Pico chemiluminescent substrate (Pierce). (B) HCV-specific RNA analysis by quantitative RT-PCR. Total cellular RNA was extracted, and HCV-specific RNA was quantified as described in Materials and Methods. Induction with respect to the DMSO control is shown on the y axis. Replicon RNA is indicated at the bottom of the figure.
Electroporation of the wt Con1 replicon did not generate any blue cells (Fig. 3, wt-BLA), whereas the addition of any of the three compounds resulted in the production of blue cells as a consequence of HCV replication (Fig. 3, wt-BLA + compound). The three selected kinase inhibitors, however, did not induce replication to equal efficiencies. Whereas compound H479 produced more than 50% blue cells, which is comparable to the positive control, the other two compounds generated fewer blue cells. The control compound SB203580 had no effect on HCV replication. To demonstrate that the blue staining is a result of HCV replication and not a result of a longer half life of the electroporated HCV RNA or BLA enzyme, we incubated the cells, in addition to the compounds, with an inhibitor of the HCV RNA-dependent RNA polymerase (RdRP) (+ RdRP-I, right column) (8, 26). In all cases, the numbers of blue cells are significantly reduced. In the case of compound F495, some blue cells remain even in the presence of the RdRP inhibitor. It would be possible that compound F495 stabilizes the HCV RNA and consequently BLA expression to some extent (Fig. 3 and 4B).
Synergistic effect between mutation in NS3 and identified kinase inhibitors.
The advantage of the BLA assay is its detection of single cells in which active HCV replication takes place. As a result, HCV replication can be observed also in cases in which only a small percentage of the overall cell population is capable of replication. The previous experiment shows that all three compounds induce the replication of the wt Con1 replicon. We next investigated whether replication is sufficient to allow the detection of viral proteins or viral RNA in the total cell population. RNA of wt Con1 replicon was electroporated into 10A-IFN cells, and compounds were added 2 h after electroporation. After 4 days of incubation, cells were collected and cellular extracts were assayed for the presence of NS5A by immunoblotting (Fig. 4A) or for HCV RNA by quantitative PCR (Fig. 4B). As expected, no NS5A was visible in untreated cells or in cells incubated with the control inhibitor SB203580 (Fig. 4A, lanes 15 and 19). Interestingly, NS5A could be detected only in the presence of compound H479, whereas no or little protein was visible in cells treated with compound A852 or F495 (compare lane 16 with lanes 17 and 18), even though compounds A852 and F495 induced replication (Fig. 3). A possible explanation for this finding is that the replication efficiency, and thus the quantity of viral proteins induced by A852 and F495, is lower than that for H479. Nonetheless, due to the extremely high sensitivity of the BLA assay, sustained low-level replication could be revealed by blue cell staining after prolonged incubation of the cell with the BLA substrate.
During the characterization of the replicon, several mutations have been identified that are synergistic to adaptive mutations, thus increasing the replication efficiency (15, 27). One of these mutations is E1202G, which maps in NS3 (in this work described as m17). By itself, this mutation has little, if any, effect in promoting replication of the Con1 replicon (Fig. 4A, lanes 2 and 5). However, it has a strong synergistic effect on replication when combined with the S2204A mutation (Fig. 4A, compare lanes 3 and 4).
We thus investigated whether the mutation E1202G in NS3 acted synergistically with the kinase inhibitors in a similar way to what was observed with the adaptive mutation (lanes 5 to 9). All three compounds activated replication of the replicon m17, with compound H479 being the most potent activator and compound F495 being the least potent activator. The synergistic effect of the mutation in NS3 with the adaptive mutation in NS5A (lane 3 and 4) is comparable to its synergistic effect with the kinase inhibitors (compare lanes 16 to 18 with lanes 6 to 8). The presence of NS5A is clearly due to active HCV replication and not to protein stabilization because replicons containing the RdRP-inactivating mutation GAA (17) do not show any detectable NS5A protein (lanes 10 to 14).
Compounds which induce replication of wt replicons or m17 have an opposite effect on the replication of already adapted replicons. Replication of the replicon SA is completely inhibited at a 2 μM concentration of compound A852 (Fig. 4A, lanes 20 to 25) (also see Discussion).
The production of HCV-specific RNA was detected by real-time RT-PCR amplification (Fig. 4B). The expression of β-actin was used as an internal control to standardize for the total amount of RNA. The induction of HCV-RNA with respect to the DMSO control is shown. Compound H479 induced the replication of the wt Con1 replicon sixfold, as measured by quantitative PCR, whereas induction of replication by the other compounds was below the background (that of the wt). As already observed in Fig. 4A for protein expression, cells supporting subgenomic replicons with the synergistic mutation in NS3 (m17) contain significantly more HCV RNA upon incubation with the kinase inhibitors than those expressing the wt Con1 replicon. Replication is induced 312-fold for compound H479, 58-fold for compound A852, and 30-fold for compound F495. The presence of HCV RNA is due to active replication, as the HCV RNA polymerase-minus mutants (wt-GAA and m17-GAA) do not show any induced amount of RNA. One exception is compound F495, which induces a slight increase of RNA even without active HCV replication. The same observation was already made by using the BLA assay (Fig. 3).
DISCUSSION
Since the identification of adaptive mutations in the HCV subgenomic replicon, it has been noted by several groups that the hyperphosphorylated form of NS5A, p58, is not necessary for replication in cell culture. In this report, we go one step further and demonstrate that high quantity of p58 is not only not necessary but also deleterious for the replication of the Con1 replicon. To demonstrate this hypothesis, we prevent the formation of NS5A p58 by chemical means, without altering the wt sequence of the Con1 HCV genome.
An in vitro assay was set up which enabled us to screen a collection of small molecules for their inhibitory activity on kinase(s) responsible for NS5A phosphorylation. The principle of this in vitro assay is based on the fact that the specific kinase(s) remain stably associated with NS5A after immunoprecipitation. Incubation of this protein complex with [γ-33P]ATP results in the production of many phosphoproteins (Fig. 1), indicating that the polyclonal antibody nonspecifically binds many cellular proteins and kinases under the conditions we used for the immunoprecipitation. However, the phosphorylation of NS5A is the most efficient reaction in the immunoprecipitate prepared from extracts expressing the replicon (Fig. 1, compare lanes 2 and 3). In replicons containing the A2199T adaptive mutation, p56 and p58 are present in equal amounts (Fig. 2B, lane 2). Nevertheless, in the in vitro assay, radiolabeled phosphate is incorporated predominantly in p56. The formation of p58 depends on the presence of the HCV polyprotein coding from NS3 to NS5A and requires active polyprotein processing (14, 20). For this reason, the formation of p58 during in vitro labeling is not expected. In addition, one can imagine that the conformation of p58 is different from that of p56 and that phosphorylation sites present in p56 are not exposed or masked by other associated proteins in p58. As a consequence, the in vitro phosphorylation of serine residues in p56 may be more efficient than in p58.
The low stringency of the assay produced a hit rate of about 40%, with a threshold of at least 50% inhibition of NS5A phosphorylation. As a consequence, a large number of compounds showed the same in vitro activity as the three selected compounds but were inactive in cell culture. There are several explanations for the difference between in vitro activity and activity in cell culture. One possibility could be limited penetration of the compounds into the cells. Alternatively, a high potential of the compounds for protein binding or instability in the culture medium or within the cell could limit the access to the compound. All compounds tested were designed to be active-site inhibitors and competitors of ATP. Another possible explanation would be that the affinity of the compound for the kinase is too low to compete with the relatively high concentrations of ATP within the cell.
Retesting the selected compounds in cell culture with the vaccinia T7 infection-transfection system as well as Huh7-68 cells identified three active compounds. The formation of p58 is inhibited in both systems, whereas the basal phosphorylation of p56 remained unchanged (Fig. 2A and B, lanes 8 to 12). Interestingly, we could not identify any compound that inhibited the basal phosphorylation without affecting the expression of NS5A. NS5A is proteolytically processed from the polyprotein within 15 min (20). The prevention of basal phosphorylation may destabilize the protein and lead to protein degradation. In this case, any compound which inhibits basal phosphorylation would reduce the total protein amount of NS5A and would have been excluded in our second assay in cell culture.
The identification of three compounds that selectively inhibited NS5A hyperphosphorylation, but not basal phosphorylation, allowed us to verify whether a reduction of the amount of NS5A p58 in the context of the Con1 wt sequence could be sufficient for the activation of subgenomic replication in cell culture. It turned out, in fact, that all three active kinase inhibitors were also active replication inducers (Fig. 3).
Compound H479 is the best inhibitor, as it activates replication in more than 50% of the cells. This efficiency is comparable to the S2204A adaptive mutation, resulting in the expression of HCV proteins at levels detectable by Western blotting. Consistent with the inhibition of NS5A hyperphosphorylation, the formation of p58 is significantly reduced (Fig. 4A, lane 16). It is important to stress the point that the only difference which renders the replicon capable of replication is the reduction of p58 formation by a kinase inhibitor without changing the genomic sequence. Thus, an excess amount of p58 seems to be the causative agent responsible for the inhibition of HCV replication in cell culture.
The other two compounds also activated replication (Fig. 3); however, HCV proteins and HCV RNA were detectable only in the presence of a synergistic mutation in NS3 (Fig. 4). The introduction of mutations may be critical for infectivity, as shown in the case of adaptive mutations. These mutations increase HCV replication efficiency in cell culture, but at the same time, they destroy or significantly attenuate infectivity in vivo (4). One, therefore, has to carefully choose any change in the wt sequence. In this work, we decided to introduce the mutation E1202G in NS3. This mutation has been shown to increase the replication efficiency of the Con1 subgenome when placed together with the adaptive mutations S2197P, K@2039, S2204I (15, 16, 27), and S2204A (Fig. 4) in NS5A. The advantage of the E1202G mutation is the fact that it has been found as a naturally occurring mutation in an infective clone of the 1A strain, thus suggesting that this mutation does not destroy infectivity in vivo (28). The E1202G mutation is synergistic when placed together with adaptive mutations at the hyperphosphorylation site of NS5A and is therefore an ideal candidate for synergistic effects in the presence of the kinase inhibitors.
Replicons containing the E1202G substitution in NS3 (m17) do replicate with low efficiency, and HCV proteins cannot be detected by Western blotting (Fig. 4A). In this genetic background, all three compounds induce replication sufficiently to be detectable by Western blotting and by TaqMan (Fig. 4).
The different degrees of replication efficiency induced by compound H479 and compounds A852 and F495 are probably not a consequence of different potency in the inhibition of p58 formation, which seems to be comparable at 5 μM (Fig. 2). This apparent difference could be explained by considering the cytotoxicity displayed by compound A852 and especially by compound F495 at concentrations of 8 μM or more (data not shown). During the experiments designed for the detection of inhibition of p58 formation, the cells are exposed to 5 μM compound for several hours, whereas they are exposed for 4 days at 8 μM during the induction of replication. It is therefore possible that inhibition of NS5A hyperphosphorylation is necessary for the induction of replication, but different side effects of the compounds may induce or block other pathways which may have antagonistic effects on the induction of replication.
Interestingly, the compounds we identified potently inhibit phosphorylation of NS5A in vitro (Fig. 2, lanes 11 to 13), suggesting that they may directly inhibit NS5A-specific kinase(s). However, we cannot exclude the possibility that these compounds act via the inhibition of upstream kinase(s) that may also be present in the immunoprecipitate. The spectrum of cellular kinases inhibited by these agents and whether any of these kinases is directly or indirectly responsible for NS5A hyperphosphorylation needs to be clarified by future work.
Our findings support a role for NS5A p58 in down-regulating HCV RNA replication in cell culture. Based on the present data, we cannot exclude that the compound(s) we identify act via the inhibition of the phosphorylation of cellular protein(s) regulating HCV RNA replication and that inhibition of p58 is not the actual cause of increased replication. However, we believe that the present data are supportive of a direct, causal connection between NS5A hyperphosphorylation and the inability of hepatitis C subgenomic RNA to replicate in cell culture. This is based on two considerations. First, all of the kinase inhibitors that we selected for specific inhibition of NS5A hyperphosphorylation in cell culture were able to activate RNA replication. Second, it has been previously reported that several adaptive mutations found in the NS5A sequence occur at conserved serine residues and greatly impair the hyperphosphorylation of NS5A (2).
Figure 5 shows two possible models which try to explain the inhibitory effect of NS5A p58 on HCV replication in cell culture. According to model A, p56 is necessary for active replication. In cell culture, p56 is converted to p58 by cellular kinases and recruits a repressor protein (R) to the replication complex, thereby inhibiting replication. In the presence of the specific kinase inhibitors the conversion to p58 is prevented and the repressor protein R will not be recruited to the replication complex (upper part of Fig. 5A). There are three types of adaptive mutations which result in active replication in cell culture, and the first occurs exactly at the sites of NS5A hyperphosphorylation (S2197, S2201, and S2204). These mutations prevent the production of p58 and thus result in replication in cell culture. This situation would result in a p56-containing active replication complex, similar to that formed in the presence of the NS5A hyperphosphorylation inhibitor (Fig. 5A, top panel). The second type of adaptive mutations occurs in NS5A, however, at sites different from the hyperphosphorylation sites. As a consequence, p58 is still expressed (as in the case of Huh7-68). This example is shown in the bottom part of Fig. 5, to the left of each model. The third type of adaptive mutations does not occur in NS5A, but it occurs in other proteins which participate in viral replication, e.g., NS3 or NS5B. This example is shown in the bottom part of Fig. 5, to the right of each model.
FIG. 5.
Working model. Replication in cell culture is inhibited in the presence of p58. (A) p58 recruits a repressor protein R to the replication complex. Upper panel: p56 is converted to the hyperphosphorylated form p58 by cellular kinases, which then binds a repressor protein R. The presence of this protein complex p58-R results in inactivation of HCV RNA replication. Our selected kinase inhibitors prevent the conversion of p56 to p58. Lower panel: the presence of adaptive mutations either in NS5A or in other parts of the replication complex prevents the association of the repressor molecule. (B) p58 is unable to associate with an activator protein. Upper panel: p56 binds an activator protein A and activates the replication complex. In cell culture, p56 is converted to the hyperphosphorylated form p58 by cellular kinases, which is incapable of association with the activator protein A. Our selected kinase inhibitors prevent the conversion of p56 to p58. Lower panel: the presence of adaptive mutations either in NS5A or in other parts of the replication complex enables the association with the activator protein A even in the presence of hyperphosphorylation. p56, NS5A p56; p58, NS5A p58; R, repressor protein; A, activator protein. Stars represent adaptive mutations.
In the latter two cases, adaptive mutations prevent the association of the repressor molecule and replication can take place even in the presence of p58 (Fig. 5A, bottom part). This model is supported by recent data obtained by Graziani et al. (9), who demonstrate that coexpression of wt HCV subgenomic replicons together with adapted replicons results in transdominant inhibition of HCV replication in cell culture. Inhibitory cellular proteins may be expressed only in transformed cell lines and be absent in the liver, thus explaining the strict dependence on adaptive mutations for replication in cell culture.
In model B, p58 is not able to interact with an activator protein (A), which is necessary for replication. Also in this model, the kinase inhibitors prevent the conversion of p56 into p58 and binding of NS5A to the activator protein A is maintained (upper part of Fig. 5B). In this model, one has to assume that the latter two types of adaptive mutations restore protein-protein interactions between NS5A and A, even if NS5A exists as p58. The latter model is supported by recent data obtained by Evans et al. (8a).
Production of high levels of p58 may be an artifact of the subgenomic construct. The expression of the nonstructural proteins is driven by the strong encephalomyocarditis virus internal ribosome entry site, and the amount of NS5A p58 during infection in vivo could be significantly lower. Alternatively, the specific kinases and, thus, the formation of p58 are expressed or regulated differently in cell lines with respect to the liver, thus disturbing the correct equilibrium between p56 and p58. Different expression of cellular factors could also explain the strict host dependence of HCV for replication in cell culture. It would be of great interest to identify the kinases which are responsible for the formation of p58. A cellular system in which the expression of this kinase can be regulated would be an ideal system for the study of the replication of HCV containing the original, infective wt sequence.
There is an additional interesting observation we would like to discuss. Figure 4A clearly shows that treatment of cells with compound H479, A852, or F495 results in a substantial reduction of p58 but is not completely abolished. Two lines of evidence indicate that complete prevention of p58 formation eliminates replication. (i) Blight et al. (3) demonstrated that the mutation S2204I is a very efficient adaptive mutation, whereas the combination of this mutation together with a second adaptive mutation within the hyperphosphorylation sites of NS5A (S2197P) completely abolished replication. (ii) Incubation of cells containing a replicon with the adaptive mutation S2204A with one of our kinase inhibitors also abolishes HCV replication (Fig. 4A, lanes 20 to 25). In both cases, mutation at one hyperphosphorylation site significantly reduces the quantity of p58 and the second event, either a second mutation or inhibition by our compounds, further reduces p58 formation and results in a complete block of replication. This observation suggests, contrary to what has been thought to date, that low amounts of p58 may in fact be necessary for replication and that compounds that completely inhibit NS5A hyperphosphorylation could have antiviral activity in vivo.
In conclusion, we have identified compounds that, by inhibiting protein kinase(s) implicated in NS5A hyperphosphorylation, promote replication of nonadapted HCV sequences in cell culture. This finding points to a role for NS5A p58 in down-regulating HCV RNA replication in cell culture. To date, it is not known whether NS5A p58 has a function in viral replication in vivo or what that function may be. However, compounds that selectively inhibit NS5A hyperphosphorylation could be useful tools for the establishment of a bona fide infection system for HCV in cell culture and could possibly be a starting point for the development of novel antiviral agents.
Acknowledgments
We thank Licia Tomei for helpful discussions throughout the whole work and for critical reading of the manuscript. We also thank Giovanni Migliaccio for critical reading of the manuscript.
REFERENCES
- 1.Bartenschlager, R., and V. Lohmann. 2000. Replication of hepatitis C virus. J. Gen. Virol. 81:1631-1648. [DOI] [PubMed] [Google Scholar]
- 2.Blight, K. J., A. A. Kolykhalov, and C. M. Rice. 2000. Efficient initiation of HCV RNA replication in cell culture. Science 290:1972-1974. [DOI] [PubMed] [Google Scholar]
- 3.Blight, K. J., J. A. McKeating, and C. M. Rice. 2002. Highly permissive cell lines for subgenomic and genomic hepatitis C virus RNA replication. J. Virol. 76:13001-13014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bukh, J., T. Pietschmann, V. Lohmann, N. Krieger, K. Faulk, R. E. Engle, S. Govindarajan, M. Shapiro, M. St. Claire, and R. Bartenschlager. 2002. Mutations that permit efficient replication of hepatitis C virus RNA in Huh-7 cells prevent productive replication in chimpanzees. Proc. Natl. Acad. Sci. USA 99:14416-14421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cuenda, A., J. Rouse, Y. N. Doza, R. Meier, P. Cohen, T. F. Gallagher, P. R. Young, and J. C. Lee. 1995. SB 203580 is a specific inhibitor of a MAP kinase homologue which is stimulated by cellular stresses and interleukin-1. FEBS Lett. 364:229-233. [DOI] [PubMed] [Google Scholar]
- 6.De Francesco, R., P. Neddermann, L. Tomei, C. Steinkuhler, P. Gallinari, and A. Folgori. 2000. Biochemical and immunologic properties of the nonstructural proteins of the hepatitis C virus: implications for development of antiviral agents and vaccines. Semin. Liver Dis. 20:69-83. [DOI] [PubMed] [Google Scholar]
- 7.De Francesco, R., and C. M. Rice. 2003. New therapies on the horizon for hepatitis C: are we close? Clin. Liver Dis 7:211-242, xi. [DOI] [PubMed] [Google Scholar]
- 8.Dhanak, D., A. C. Kaura, and A. Shaw. November 2001. Preparation of 3-(1,1-dioxo-2H-benzo-1,2,4-thiadiazin-3-yl)-2-quinolones as novel anti-infectives. WO patent 01/85172.
- 8a.Evans, M. J., C. M. Rice, and S. P. Goff. 2004. Phosphorylation of hepatitis C virus nonstructural protein 5A modulates its protein interactions and viral RNA replication. Proc. Natl. Acad. Sci. USA 101:13038-13043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Graziani, R., and G. Paonessa. 2004. Dominant negative effect of wild-type NS5A on NS5A-adapted subgenomic hepatitis C virus RNA replicon. J. Gen. Virol. 85:1867-1875. [DOI] [PubMed] [Google Scholar]
- 10.Griffin, S. D., L. P. Beales, D. S. Clarke, O. Worsfold, S. D. Evans, J. Jaeger, M. P. Harris, and D. J. Rowlands. 2003. The p7 protein of hepatitis C virus forms an ion channel that is blocked by the antiviral drug, Amantadine. FEBS Lett. 535:34-38. [DOI] [PubMed] [Google Scholar]
- 11.Hijikata, M., N. Kato, Y. Ootsuyama, M. Nakagawa, and K. Shimotohno. 1991. Gene mapping of the putative structural region of the hepatitis C virus genome by in vitro processing analysis. Proc. Natl. Acad. Sci. USA 88:5547-5551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ikeda, M., M. Yi, K. Li, and S. M. Lemon. 2002. Selectable subgenomic and genome-length dicistronic RNAs derived from an infectious molecular clone of the HCV-N strain of hepatitis C virus replicate efficiently in cultured Huh7 cells. J. Virol. 76:2997-3006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Katze, M. G., B. Kwieciszewski, D. R. Goodlett, C. M. Blakely, P. Neddermann, S. L. Tan, and R. Aebersold. 2000. Ser(2194) is a highly conserved major phosphorylation site of the hepatitis C virus nonstructural protein NS5A. Virology 278:501-513. [DOI] [PubMed] [Google Scholar]
- 14.Koch, J. O., and R. Bartenschlager. 1999. Modulation of hepatitis C virus NS5A hyperphosphorylation by nonstructural proteins NS3, NS4A, and NS4B. J. Virol. 73:7138-7146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Krieger, N., V. Lohmann, and R. Bartenschlager. 2001. Enhancement of hepatitis C virus RNA replication by cell culture-adaptive mutations. J. Virol. 75:4614-4624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lohmann, V., S. Hoffmann, U. Herian, F. Penin, and R. Bartenschlager. 2003. Viral and cellular determinants of hepatitis C virus RNA replication in cell culture. J. Virol. 77:3007-3019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lohmann, V., F. Korner, U. Herian, and R. Bartenschlager. 1997. Biochemical properties of hepatitis C virus NS5B RNA-dependent RNA polymerase and identification of amino acid sequence motifs essential for enzymatic activity. J. Virol. 71:8416-8428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lohmann, V., F. Korner, J. Koch, U. Herian, L. Theilmann, and R. Bartenschlager. 1999. Replication of subgenomic hepatitis C virus RNAs in a hepatoma cell line. Science 285:110-113. [DOI] [PubMed] [Google Scholar]
- 19.Murray, E. M., J. A. Grobler, E. J. Markel, M. F. Pagnoni, G. Paonessa, A. J. Simon, and O. A. Flores. 2003. Persistent replication of hepatitis C virus replicons expressing the beta-lactamase reporter in subpopulations of highly permissive Huh7 cells. J. Virol. 77:2928-2935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Neddermann, P., A. Clementi, and R. De Francesco. 1999. Hyperphosphorylation of the hepatitis C virus NS5A protein requires an active NS3 protease, NS4A, NS4B, and NS5A encoded on the same polyprotein. J. Virol. 73:9984-9991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Pavlovic, D., D. C. Neville, O. Argaud, B. Blumberg, R. A. Dwek, W. B. Fischer, and N. Zitzmann. 2003. The hepatitis C virus p7 protein forms an ion channel that is inhibited by long-alkyl-chain iminosugar derivatives. Proc. Natl. Acad. Sci. USA 100:6104-6108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Reed, K. E., and C. M. Rice. 1999. Identification of the major phosphorylation site of the hepatitis C virus H strain NS5A protein as serine 2321. J. Biol. Chem. 274:28011-28018. [DOI] [PubMed] [Google Scholar]
- 23.Reed, K. E., and C. M. Rice. 2000. Overview of hepatitis C virus genome structure, polyprotein processing, and protein properties. Curr. Top. Microbiol. Immunol. 242:55-84. [DOI] [PubMed] [Google Scholar]
- 24.Reed, K. E., J. Xu, and C. M. Rice. 1997. Phosphorylation of the hepatitis C virus NS5A protein in vitro and in vivo: properties of the NS5A-associated kinase. J. Virol. 71:7187-7197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Tanji, Y., T. Kaneko, S. Satoh, and K. Shimotohno. 1995. Phosphorylation of hepatitis C virus-encoded nonstructural protein NS5A. J. Virol. 69:3980-3986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Tomei, L., S. Altamura, L. Bartholomew, M. Bisbocci, C. Bailey, M. Bosserman, A. Cellucci, E. Forte, I. Incitti, L. Orsatti, U. Koch, R. D. Francesco, D. B. Olsen, S. S. Carroll, and G. Migliaccio. 2004. Characterization of the inhibition of hepatitis C virus RNA replication by nonnucleosides. J. Virol. 78:938-946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Trozzi, C., L. Bartholomew, A. Ceccacci, G. Biasiol, L. Pacini, S. Altamura, F. Narjes, E. Muraglia, G. Paonessa, U. Koch, R. De Francesco, C. Steinkuhler, and G. Migliaccio. 2003. In vitro selection and characterization of hepatitis C virus serine protease variants resistant to an active-site peptide inhibitor. J. Virol. 77:3669-3679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Yanagi, M., R. H. Purcell, S. U. Emerson, and J. Bukh. 1997. Transcripts from a single full-length cDNA clone of hepatitis C virus are infectious when directly transfected into the liver of a chimpanzee. Proc. Natl. Acad. Sci. USA 94:8738-8743. [DOI] [PMC free article] [PubMed] [Google Scholar]