Abstract
Ribozymes are small, catalytic RNA molecules that can be engineered to down-regulate gene expression by cleaving specific mRNA. Here we report the selection of hairpin ribozymes that inhibit human immunodeficiency virus (HIV) replication from a combinatorial ribozyme library. We identified a total of 17 effective ribozymes, each capable of inhibiting HIV infection of human CD4+ cells. These ribozymes target diverse steps of the viral replication cycle, ranging from entry to transcription. One ribozyme suppressed HIV integration and transcription by inhibiting the expression of the Ku80 subunit of the DNA-activated protein kinase. Another ribozyme specifically inhibited long terminal repeat transactivation, while two additional ones blocked a step that can be bypassed by vesicular stomatitis virus G-protein pseudotyping. The function of Ku80 in HIV replication and its mechanism of action were further confirmed using short interfering RNA. Identification of the gene targets of these and other selected ribozymes may reveal novel therapeutic targets for combating HIV infection.
Ribozymes (Rzs) are short RNA molecules that can cleave target RNA containing a suitable sequence via their intrinsic enzymatic activities (5). The target specificity of a ribozyme is determined by complementary base pairing between the ribozyme and its target RNA. Cleavage occurs at a required triplet sequence motif in the target RNA: GUC for hairpin ribozymes and NUH (H is an A, C, or T) for hammerhead ribozymes (20). The catalytic activity and the target specificity of ribozymes make them attractive molecular tools for the analysis of RNA-mediated cellular and viral events (6, 32).
Recently, ribozyme libraries have been used in forward genetics strategies to identify genes based on their function (4, 14-16, 19, 28, 29, 33). For example, randomization of the 12-nucleotide DNA sequence encoding the two binding arms of the hairpin ribozyme generates a library of 412 or ∼1.7 × 107 distinct RNA ribozyme genes (16). Following delivery of the library into target cells via retroviral transduction and selection for a desired phenotype, ribozymes that target cellular genes involved in several important viral and oncogenic pathways have been identified (4, 16, 19, 33).
We report here the identification of hairpin ribozymes that suppress human immunodeficiency virus (HIV) in cell culture by using an in vivo selection scheme based on the Rz library and an HIV indicator cell line, CEM-GFP (10). CEM-GFP cells are T cells that contain an integrated copy of the green fluorescent protein (GFP) gene under the control of the HIV long terminal repeat (LTR), such that the cells express GFP upon infection with HIV (10). We identified individual ribozymes that target many different steps of the HIV replication cycle. Three ribozymes, IVRz-1, -9, and -12, were selected for further analysis. Here we show that IVRz-12 blocks a step at or immediately after viral entry, IVRz-9 inhibits the transactivation of the HIV LTR, and IVRz-1 plays a dual role in HIV replication, inhibiting both tat transactivation of the HIV LTR and integration of viral cDNA. Ribozyme IVRz-1 targets Ku80, a component of the nonhomologous end-joining (NHEJ) pathway (8, 27, 31). Involvement of Ku80 in HIV replication was further confirmed using short interfering RNA (siRNA).
MATERIALS AND METHODS
Ribozyme library production and transduction.
We constructed a library for expression of ribozymes with eight random nucleotides in helix 1 and four random nucleotides in helix 2 as described previously (16). We ligated the library into pLHPM, a Moloney virus-based vector with a tRNAval promoter to express the ribozyme RNA. Retroviral vector was prepared by transient transfection of 293-T cells. Vector was harvested at 72 h posttransfection and concentrated by centrifugation at 32,000 rpm, 4°C, for 2 h with a Ti45 rotor in a Beckman L8-80 M preparative ultracentrifuge. CEM-GFP cells were transduced in the presence of 4 μg of Polybrene/ml and selected for 10 days in 0.6 μg of puromycin/ml.
Infection and analysis of viral protection.
HIVLAI and an integrase-deficient mutant (HXB-2 with IN E152A) were used in this study. In large-scale infections, where GFP-negative cells were sorted back for DNA extraction, 2 × 107 cells were infected in the first round of selection and 2 × 106 cells were infected in subsequent rounds. For small-scale infections of CEM-GFP cells, 0.5 × 106 to 1 × 106 cells were infected. Cells were fixed for flow cytometry analysis 6 days postinfection. For CD4-positive cell isolation, cells were labeled with a CD4 monoclonal antibody, OKT4, and purified using microbeads (Miltenyi Biotech, Auburn, Calif.). Single-cell clones were generated by limited dilution. For analysis of intracellular p24 levels, cells were permeabilized with a mild detergent (Intrastain kit; DAKO, Glostrup, Denmark) and stained with a monoclonal antibody against HIV type 1 (HIV-1) p24 followed by detection with an anti-mouse antibody conjugated to Cy-5.
Ribozyme rescue and mini-library production.
We used PCR to amplify an approximately 100-bp region that contains the ribozyme expression cassette. This PCR product was digested with BamHI and Csp45I and then cloned into pLHPM digested with the same enzymes. For mini-library production, colonies grown on bacterial plates were scraped and pooled for plasmid preparations that were then used for retroviral vector production as described above.
MAGI cell transfection and assay.
MAGI cells were transfected with pLHPM containing individual ribozymes by using FuGENE 6 (Roche Diagnostics, Basel, Switzerland) and selected with 0.6 μg of puromycin/ml. Selected cells were infected with HIV-1LAI or vesicular stomatitis virus G-protein (VSV-G)-pseudotyped HIV at a multiplicity of infection (MOI) of 0.5 in the presence of 4 μg of Polybrene/ml and incubated at 37°C in 5% CO2 for 48 h. Cells were stained with 5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside (0.4 mg/ml), and blue cells were scored.
Construction and transfection of siRNAs.
The three siRNAs targeting nucleotides 1224 to 1244, 1691 to 1711, and 2601 to 2621 of the Ku mRNA (M30938) were designated Ku-1224, Ku-1691, and Ku-2601, respectively. The control, scrambled siRNA has a target sequence of 5′-GCGCGCTTTGTAGGATTCG-3′. Double-stranded siRNAs were constructed by in vitro transcription with a Silencer siRNA construction kit (Ambion, Austin, Tex.). Transfection of siRNA was performed using Oligofectamine (Invitrogen, Carlsbad, Calif.). Two days after transfection of siRNA, the cells were replated for infection with HIV as described for the ribozyme-expressing cells.
Stable expression of siRNA by using a lentiviral vector.
The pHIV-7 vector was a gift from M. Li, J.-K. Yee, and J. Rossi. This vector was constructed from pHIV-7-GFP (3, 35) by digestion with PstI to remove the cytomegalovirus promoter-enhanced GFP cDNA cassette and religation of the 530-bp and 5.7-kb fragments. A 1.4-kb Pvu II/BamHI fragment comprising the simian virus 40 promoter/puror cassette was isolated from the pPUR vector (Clontech, Palo Alto, Calif.) and inserted into the SmaI site of pHIV-7 to yield pHIV-7-Puro. The target sequences for Ku-1224 and luciferase siRNA were 5′-AAGAGCTAATCCTCAAGTCGG-3′ and 5′-GTGCGCTGCTGGTGCCAACCC-3′, respectively. U6 promoter/hairpin siRNA expression cassettes were constructed by PCR using the U6 promoter in pSilencer (Ambion) or pTZ U6+1 (17) as the templates for the Ku-1224 and luciferase siRNA expression cassettes, respectively. The primers for the Ku-1224 siRNA were 5′-GAACTAGTGGATCCGACGCC-3′ and 5′-ggcGGATCCAAAAAAgagctaatcctcaagtcggTCTCTTGAAccgacttgaggattagctcAAACAAGGCTTTTCTCCAAGGG-3′. (Underlined sequences indicate the Ku mRNA target sequence, and lowercase letters indicate the siRNA sequence.) The primers for the luciferase siRNA were 5′-GGATCCAAGGTCGGGCAGGAAGAGGG-3′ and 5′-GGATCCAAAAAAGTGCGCTGCTGGTGCCAACCCTCTCTTGAAGGGTTGGCACCAGCAGCGCACGGTGTTTCGTCCTTTCCAC-3′.PCR was performed with Platinum PCR SuperMix High Fidelity (Invitrogen). The PCR products were ligated into the pCR Blunt II-TOPO vector (Invitrogen) and sequenced before being cloned into BamHI-digested pHIV-7-Puro. The reverse orientation (i.e., U6 and simian virus 40 promoters facing in opposite directions) was selected. VSV-G-pseudotyped lentivirus was packaged using the lentivirus support kit (Invitrogen). MAGI and CEM cells were transduced by standard methods (30) and subjected to selection with puromycin (0.6 μg/ml) for 10 days.
PCR amplification of total viral DNA and integrated viral DNA.
Genomic DNA was prepared from infected or mock-infected MAGI cells by using DNAzol (Invitrogen). Total viral DNA PCR was performed using the following primers, which detected the nucleocapsid region of the HIV-1 Gag gene: NC5′ (5′-GCCGGATCCATGCAGAGAGGCAATTTTA-3′) and NC3′ (5′-GCCGAATTCATTAGCCTGTCTCTCAGTAC-3′). Alu-nested PCR was performed with the following primers: SB704 (5′-TGTGGGATTACAGGCGTGAG-3′) and Rc (5′-TAGACCAGATCTGAGCCTGGGA-3′) for first-round PCR and M667 (5′-GGCTAACTAGGGAACCCACTG-3′) and AA55 (5′-CTGCTAGAGATTTTCCACACTGAC-3′) for nested PCR. Primers for the β-globin control were 5′bglo, 5′-AAAGAACAAAGCTGGAGGCATC-3′, and 3′bglo, 5′-GAATCCTTTCCCCATTGCTTG-3′. Aliquots of 250 ng of total genomic DNA were used for all PCRs.
RESULTS
Selection of ribozymes that protect CEM-GFP cells from HIV infection.
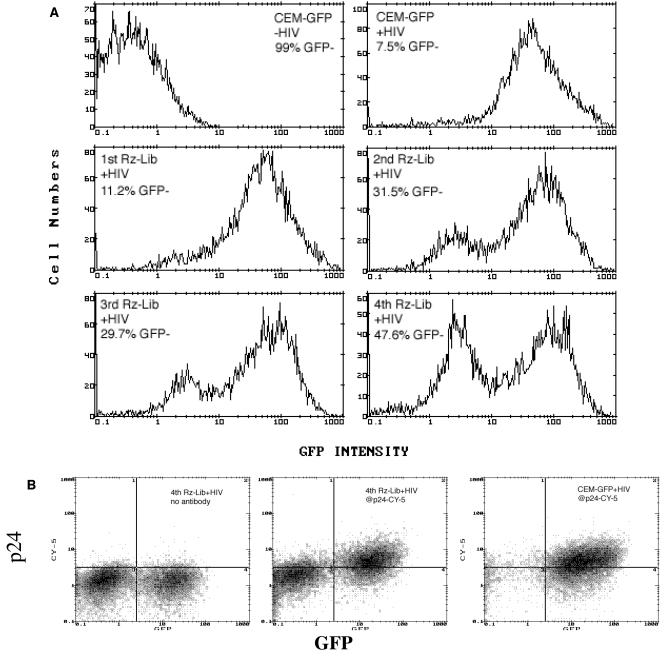
We transduced CEM-GFP cells with the retroviral ribozyme library and then challenged them with HIVLAI at an MOI of 0.02. Infection of the parental CEM-GFP cells induced GFP expression in >90% of the cells at day 6 (Fig. 1A). Prior transduction with the original ribozyme library (1st Rz-Lib) decreased the number of GFP-positive (infected) cells only slightly (88 to 90%, versus 90 to 93% in a typical experiment). Nevertheless, we extracted genomic DNA from the GFP-negative cells, rescued the ribozymes by PCR, and cloned them back into the retroviral vector to generate a mini-library (2nd Rz-Lib). The transduction and HIV challenge cycle were then repeated with naive CEM-GFP cells. This resulted in a significant enrichment of the protective ribozymes, as infection of the transduced cells now produced an average of 30% GFP-low cells. The cycle was repeated two more times, and the percentage of GFP-low cells increased to 41 to 65% in the fourth round. Individual effective ribozymes were identified from the fourth mini-library (4th Rz-Lib) (see below). To confirm that the reduced level of GFP expression in the protected cells was indeed a result of inhibition of HIV replication, we stained the infected cells for HIV p24gag. The GFP-low cells also expressed much lower levels of p24gag than the GFP-positive cells (Fig. 1B).
FIG. 1.
Enrichment of hairpin ribozymes that inhibit HIV infection. (A) Flow cytometry analysis of GFP expression in HIV-1LAI-infected CEM-GFP cells expressing Rz-Lib. CEM-GFP cells in the absence of HIV-1LAI (top left) and CEM-GFP minus Rz-Lib plus HIV-1LAI (top right) are controls. The 1st Rz-Lib is the original ribozyme library, while the 2nd to 4th Rz-Libs represent the mini-libraries generated in subsequent rounds of selection (see Materials and Methods for the construction of mini-libraries). The percentages of GFP-negative cells in each round of selection are shown. (B) Two-color fluorescence-activated cell sorter analysis of intracellular p24 (Cy-5) and GFP expression in HIV-1LAI-infected cells transfected with the 4th Rz-Lib (middle panel) or no ribozyme (right panel). Decrease in GFP+ Cy-5+ cells indicates silencing of HIV-1 gene expression and inhibition of HIV-1 replication.
Identification of individual effective ribozymes that render human CD4+ cells resistant to HIV infection.
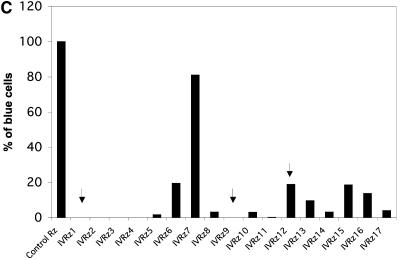
Individual ribozymes were recovered from the fourth mini-library by two independent approaches. In the first approach, we sequenced 100 random bacterial clones of the mini-library and selected ribozymes that were represented more than once in this pool for further confirmation. In the second approach, we took advantage of the fact that HIV infection down-regulates CD4 expression on the cell surface. We challenged CEM-GFP cells harboring the fourth mini-library with HIV and then performed a double selection procedure 6 days later to isolate cells that were both CD4+ and GFP negative (see Materials and Methods). Single-cell clones were then obtained from this population by limited dilution, and ribozymes contained in them were recovered by PCR. Individual ribozymes that are capable of inhibiting HIV replication were identified. An example of such an individual ribozyme (IVRz-1) is given in Fig. 2A and B. Introduction of IVRz-1 into naive CEM-GFP cells inhibited HIV infection by more than 50% as measured by both end-point GFP and kinetic p24 results. All together, we examined the viral inhibitory effects of a total of 30 ribozymes in the parental CEM-GFP cells as well as in MAGI cells (HeLa CD4+ cells containing an LTR-LacZ cassette) (23). For the MAGI assay, we stably transfected MAGI cells with expression vectors for individual ribozymes and then challenged them with HIV. After 24 to 48 h, β-galactosidase-positive cells were scored as a measure of infection. A total of 17 individual ribozymes that conferred various degrees of resistance to HIV in the MAGI assay were identified and designated IVRz-1 through IVRz-17 (Fig. 2C). We also challenged these stably transfected MAGI cells with VSV-G-pseudotyped HIV, which bypasses the normal route of virus entry (Fig. 2D). Only two ribozymes, IVRz-6 and IVRz-12, failed to confer resistance to the pseudotyped virus, indicating that the majority of the ribozymes inhibit HIV infection at a postentry step.
FIG. 2.
Confirmation of individual protective ribozymes. (A) Protection of CEM-GFP from HIV infection by IVRz-1. CEM-GFP cells expressing IVRz-1 were infected by HIV for 6 days and then analyzed for GFP expression. (B) IVRz-1 inhibits HIV spread in CEM-GFP cells. Culture media of the infected cells in panel A were collected at various time points postinfection and assayed for p24. A 2 μM concentration of zidovudine was included as a positive control for viral inhibition. (C) MAGI cells transfected with 17 individual ribozymes and a control ribozyme were infected with HIV. The percentage of infected (blue) cells relative to the control ribozymes is shown. (D) The same cells as shown in panel A were infected with VSV-G-pseudotyped HIV and similarly analyzed. The infection with either the wild-type or the pseudotyped virus was performed at least three times, and the result of a representative experiment is shown.
Selected ribozymes target diverse steps of HIV replication.
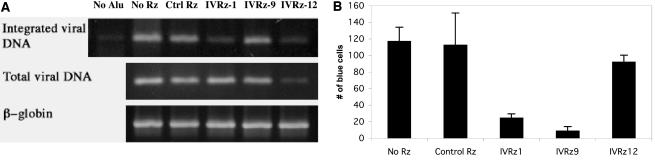
We further explored the mechanism by which three of the ribozymes (IVRz-1, IVRz-9, and IVRz-12) may function to block HIV infection. We first examined the total viral DNA levels 24 h postinfection to look for possible defects in steps leading to successful reverse transcription. Neither IVRz-1 nor IVRz-9 affected total intracellular viral DNA production (Fig. 3A, middle panel). However, when we examined viral DNA integration by an Alu-PCR method, we observed that IVRz-1 caused a severe defect in integration. As expected, both total and integrated HIV DNAs were diminished in cells expressing IVRz-12 due to a block at an earlier step (entry or immediately postentry) as suggested by the VSV-G data (Fig. 3A). Next, we examined the transcriptional activation of Tat/LTR in transient-transfection assays. We transfected an HIV-1 Tat expression plasmid into the ribozyme-expressing MAGI cells and scored β-galactosidase-positive cells 48 h later. We found that IVRz-9 and, to a lesser extent, IVRz-1 inhibited Tat-induced LTR transcription (Fig. 3B). Tat/LTR function was not inhibited in IVRz-12-expressing or control ribozyme-expressing cells. These results suggest that IVRz-9 inhibits HIV transcription while IVRz-1 inhibits viral replication at two steps in the HIV life cycle, integration and transcription.
FIG. 3.
Protective mechanisms of the effective ribozymes. (A) IVRz-1 reduces integrated HIV provirus in infected cells. Shown are results for the Alu-nested PCR of genomic DNA prepared from HIV-1LAI-infected cells expressing Rz (upper panel), total viral DNA PCR amplified using two primers binding to the Gag sequence (middle panel), or the β-globin control PCR (lower panel). PCR without the Alu primer served as a control for total DNA carry-over. Lanes are in the order shown above the gels. (B) IVRz-9 blocks Tat transactivation of HIV LTR. HIV Tat protein was introduced by transfecting MAGI cells with a plasmid encoding the Tat cDNA.
IVRz-1 targets Ku80.
Based on the substrate recognition sequences of ribozyme IVRz-1 (5′-AAUACAAUagaaCUUU-3′), we attempted to identify candidate gene targets in the GenBank database. We searched for targets containing a GUC triplet flanked by four nucleotides complementary to helix 2 and at least six nucleotides complementary to helix 1 of the IVRz-1 hairpin ribozyme (33). Typically, several potential targets can be identified for a single ribozyme based on sequence complementarity. We then used real-time quantitative reverse transcription-PCR (Taqman) to examine the mRNA levels corresponding to the candidate targets and eliminated candidates that were not affected by the cognate ribozyme (data not shown). This process identified a single target for IVRz-1, Ku80. Ku80 forms a heterodimer with Ku70 and is a component of the DNA-dependent protein kinase (31). The target site for IVRz-1 is from nucleotide 2434 to 2450 in Ku mRNA (accession number M30938). The targets for IVRz-9 and -12 remain unidentified at this time.
Confirmation of Ku80 target and mechanism using RNAi.
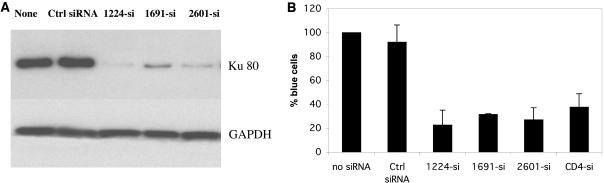
We used RNA interference (RNAi) as an alternative method to silence Ku80 expression and confirm its role in HIV infection. Three siRNAs targeting different sites within the Ku mRNA were produced by in vitro transcription and transiently transfected into MAGI cells. Controls included an siRNA targeting CD4 previously reported to down-regulate CD4 expression and inhibit HIV replication (11) and an siRNA with a scrambled sequence that does not target any known sequence in GenBank. Compared to the controls, all three anti-Ku siRNAs significantly reduced Ku protein expression in transfected MAGI cells, with the Ku-1224 siRNA causing the most dramatic reduction at both the protein and the mRNA levels (Fig. 4A and data not shown). Correspondingly, all three siRNAs inhibited HIV infection in MAGI cells, with the Ku-1224 siRNA conferring the most inhibition. The anti-CD4 siRNA similarly inhibited HIV infection (Fig. 4B), as previously reported.
FIG. 4.
Ku80 is important for HIV replication. (A) Reduction of Ku protein expression by transient siRNA against Ku mRNA. Ku80 and glyceraldehyde-3-phosphate dehydrogenase (GAPDH) protein expression was examined by Western blotting. (B) Inhibition of HIV infection of MAGI cells by Ku siRNA.
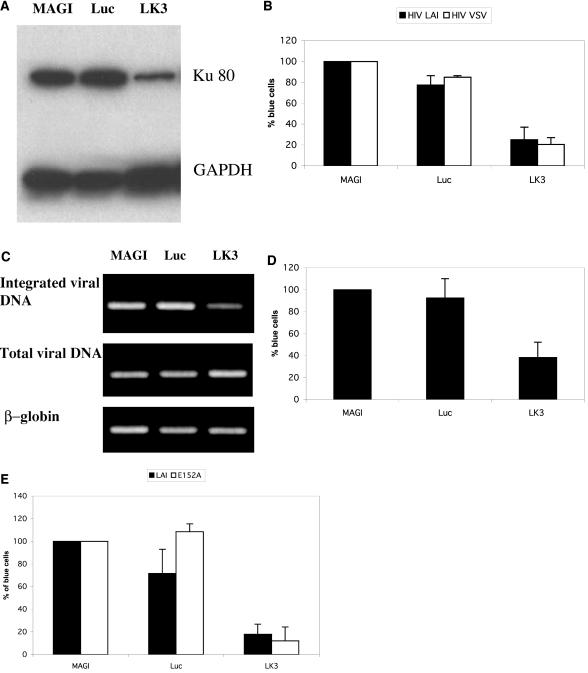
We next constructed lentiviral vectors for expression of hairpin siRNAs corresponding to the Ku-1224 siRNA (LK3) and to a firefly luciferase siRNA (Luc) for use as a control. We transduced MAGI and CEM cells with the LK3 and Luc siRNA expression vectors and selected for puromycin resistance for approximately 2 weeks. As with transiently transfected cells, MAGI cells transduced with the LK3 siRNA vector exhibited reduced Ku80 expression compared to cells transduced with the Luc siRNA vector (Fig. 5A). Transduction of MAGI cells with the LK3 siRNA vector conferred resistance against both wild-type HIV (HIVLAI) and VSV-G-pseudotyped HIV (Fig. 5B), confirming that inhibition of Ku expression blocks HIV replication at a step subsequent to virus entry.
FIG. 5.
Stably transduced cells confirmed the role of Ku80 in HIV replication. (A) Reduction of Ku protein expression by stable siRNA against Ku mRNA. Ku80 and glyceraldehyde-3-phosphate dehydrogenase (GAPDH) protein expression was examined by Western blotting. (B) Inhibition of HIVLAI and VSV-G-pseudotyped HIV infection of MAGI cells by stable Ku siRNA.(C) Stable Ku80 siRNA reduces integrated HIV provirus in infected cells. Shown are results of Alu-nested PCR of genomic DNA prepared from HIV-1LAI-infected cells expressing Ku siRNA (upper panel), total viral DNA PCR amplified using two primers binding to the Gag sequence (middle panel), or the β-globin control PCR (lower panel). (D) Ku80 siRNA blocks Tat transactivation of HIV LTR. HIV Tat protein was introduced by transfecting MAGI cells with a plasmid encoding the Tat cDNA. (E) Ku80 siRNA inhibits an integrase-deficient HIV (E152A) to the same extent as it does the wild-type virus. HIV E152A infection of MAGI cells resulted in around 10% of the number of β-galactosidase-positive cells as with the wild-type infection. The percentage of suppression by Ku80 siRNA, however, was comparable.
Transduction of CEM cells with the LK3 siRNA vector further supported our proposed mechanism for Ku80 in HIV replication. Following transduction of CEM cells with the LK3 siRNA vector, we challenged them with HIVLAI and extracted DNA 24 h postinfection for detection of total and integrated viral DNA by PCR. Neither the Ku siRNA nor the control siRNA vectors affected total viral DNA levels; however, the Ku siRNA significantly reduced the level of integrated viral DNA (Fig. 5C). When transiently transfected with a Tat expression vector, MAGI cells expressing the LK3 siRNA also exhibited a defect in Tat/LTR transcriptional activation (Fig. 5D), which is consistent with the ribozyme data. In addition, an siRNA against Ku80 also inhibited infection of MAGI cells by an integrase-deficient (IN−) virus (Fig. 5E). Several recent studies have suggested that unintegrated HIV-1 DNA can be transcriptionally active (24, 25). Detection of inhibition of HIV replication in MAGI cells is therefore likely due to a block in Tat/LTR transactivation and not integration. These results confirm the dual role of Ku80 in HIV-1 integration and Tat transactivation.
Ku80 knock-down in T cells with siRNA does not sensitize cells for apoptosis induced by various agents or HIV infection.
Many of our functional studies were carried out with stable cells harboring a Ku80-targeting ribozyme or siRNA. We did not observe any defects in cell viability and growth (data not shown) in these cells. We next examined if the Ku80 knock-down would sensitize cells to apoptosis induction. We treated CEM-derived cells harboring an empty lentiviral vector (vector) and a luciferase siRNA (Luc), as well as the Ku80 siRNA (LK-3), with either Brefeldin A or etoposide. Both agents efficiently induced apoptosis in all the treated cells. However, LK-3 cells were not further sensitized for apoptosis induced by Brefeldin A and were only marginally more sensitive to apoptosis induction by etoposide (Fig. 6A and B). We also directly measured apoptosis induction by HIV infection, using the same range of MOIs used in our viral inhibition study. As shown in Fig. 6C, infection of CEM cells with HIVLAI at MOIs of 0.4 and 1.0 did not induce any detectable apoptosis above background. Furthermore, Ku80 knock-down did not sensitive cells for apoptosis by HIV infection under these conditions. Taken together, these results indicate the suppression of HIV integration and transcription by Ku80 ribozyme/siRNA was not a result of loss of infected cells.
FIG. 6.
Knock-down of Ku80 in CEM cells does not sensitize the cells for apoptosis. CEM cells stably transduced with the backbone vector, the luciferase siRNA, and the Ku80 siRNA were treated with either 0.2 μg of Brefeldin A/ml (A) or 10 μg of etoposide/ml (B) for 24 h before the caspase-3 activity was measured. (C) CEM and the derivative cells were infected with HIVLAI for 3 days at the indicated MOIs and then measured for caspase-3 activity.
DISCUSSION
Anti-HIV therapy has so far largely focused on targeting viral genes, e.g., the reverse transcriptase and the protease. The discovery of CCR5 as a coreceptor for HIV and the finding that individuals with a homozygous mutation in CCR5 were healthy and relatively resistant to HIV infection provided a new paradigm for targeting essential cellular factors for HIV infection. This new paradigm not only expands the potential of finding new drug targets but also offers hope that drugs directed against these targets may be less susceptible to virus resistance. Both hammerhead and hairpin ribozymes designed to cleave HIV RNA have been extensively studied as potential anti-HIV agents in the context of gene therapy (26, 34). Ribozymes targeting CCR5 have also been reported to inhibit R5-tropic HIV replication (2). However, ribozymes are in general relatively inefficient gene inactivation tools. Furthermore, it is difficult to design ribozymes that predictably and reproducibly silence gene expression. The key in vivo parameters, such as target site accessibility, on-and-off rates of the ribozymes, proximity of ribozyme and target, and local concentrations of each in a given cellular compartment, cannot be accurately predicted by computer modeling or in vitro cleavage assays (9). In practice, most ribozymes designed against a target gene down-regulate the expression of that gene but rarely knock out its expression completely. For ribozymes targeting essential cellular proteins, a partial knock-down may actually be desirable, as a complete knock-out would likely kill the cells. A unique advantage of our ribozyme library-based approach is the ability to select ribozymes based only on their function (i.e., their ability to confer a defined phenotype), regardless of how effectively they suppress gene expression. The resulting inherent bias towards identification of genes with critical thresholds for conferring a desired phenotype may in fact be favorable from a drug development perspective.
In this report, we applied this in vivo selection system to identify hairpin ribozymes from a randomized ribozyme library that suppress HIV replication in human T-cell cultures. The overexpression of GFP in CEM-GFP cells depends on the introduction of functional Tat proteins into these cells. In HIV challenge experiments, Tat expression culminates from a series of early events, including successful viral entry, reverse transcription, integration, and expression of the early genes from the provirus. Thus, although ribozymes that inhibit later steps in viral replication should also be present in the library and contribute to the phenotype, screening for GFP-low-expressing cells will only recover ribozymes that block upstream of Tat expression. We obtained an increasing percentage of GFP-negative cells in successive rounds of selection, indicating an enrichment of protective ribozymes. The individual selected ribozymes were able to protect the CEM-GFP cells from HIV infection at an MOI of up to 1.0 (data not shown), suggesting that these ribozymes are far more potent than previously reported ribozymes directed against the HIV genome (21, 34). Nevertheless, the inhibition by one of our most potent ribozymes was still not as good as the small-molecule drug zidovudine (Fig. 2B), limiting the potentials of these ribozymes to be therapeutic agents by themselves.
The introduction of a single protective ribozyme was not sufficient to protect 100% of the transduced population from HIV infection in some cases. This may be due to the heterogeneous levels of ribozyme expression in the cell population. Alternatively, the efficiency of ribozyme cleavage of a particular target may also vary in individual cells, depending on the state of the cells. Consistent with either hypothesis, when single-cell clones were isolated from the CEM-GFP cells transduced with IVRz-1, complete protection was observed in some clones (data not shown).
None of the 17 selected ribozymes targeted the HIV genome. One obvious possibility is that our screening was not exhaustive and there are other active ribozymes to be discovered. The 10-kb genome of HIV is a relatively small target compared to the mRNA sequences of all the potential cellular factors important for HIV replication; it also remains possible that HIV RNA is not as readily accessible as cellular mRNA due to the tight association with viral proteins at many stages of the life cycle. One of the ribozymes targets Ku80, a subunit of a heterodimeric protein originally identified as an autoantigen recognized by sera of certain patients with autoimmune diseases (22, 23). This protein was later identified as a dimer of two related proteins, Ku80 and Ku70. This dimeric protein, Ku80/Ku70, further complexes with a 490-kDa kinase to form DNA-dependent protein kinase (31). Ku80 itself has DNA binding and ATPase activities. In addition, Ku80 regulates DNA-dependent protein kinase activity. The latter is an essential component of the NHEJ pathway, which is involved in DNA damage signaling and repair (27). A direct role of this pathway in retroviral integration was studied in rodent cells, and results were controversial but pointed to the possibility that cells deficient in the NHEJ pathway are more prone to apoptosis induced by HIV infection at very high MOIs (1, 7, 18). We did not observe any sensitization to apoptosis induction by several inducing agents or HIV infection at MOIs of 0.4 and 1. The reason for this could be severalfold: our suppression of Ku80 expression is not complete, even with the most potent siRNA; the MOIs used in this study are modest compared to those in some of the studies where massive toxicity was seen; there also could be differences between human and rodent cells in terms of sensitivity to the damage of the NHEJ pathway. Our results do argue that inhibition of Ku80 expression in human cells can suppress HIV infection independent of cell killing, at least at the MOIs that we used. A recent study using human cells reported that depletion of Ku80 with antisense RNA decreased the level of integrated provirus in HIV-infected cells in addition to reduced formation of 2-LTR circles (13). Here, we show that IVRz-1, the selected ribozyme that targets Ku80, inhibited HIV integration in MAGI cells. In addition, siRNAs designed against Ku mRNA also inhibited HIV integration. Taken together, these results confirm a role of Ku80 in HIV integration and validate our ribozyme-based approach for identifying cellular genes involved in the HIV life cycle. Interestingly, HIV transcription was also impaired in cells expressing IVRz-1 or siRNA targeting Ku80, indicating a role of this protein in HIV-1 transcription. An additional inhibition, other than just integration, was confirmed by showing that IVRz-1 also inhibited transient infection of MAGI cells by an IN− mutant HIV. In transient-transfection assays, Jeanson et al. observed enhanced HIV-1 LTR/Tat-mediated reporter expression in rodent cells lacking Ku80, suggesting a negative regulatory role of Ku80 in HIV transcription (12). However, when the same group depleted Ku80 from human cells by using antisense technology, reduced HIV infection was observed instead (13). Our results agree with the latter observation and support a positive role of Ku80 in the HIV replication cycle.
During the course of this study, RNAi emerged as a powerful tool for studying gene function in mammalian cells. Here, we extended our findings with ribozymes to RNAi for validation of our candidate gene targets. All siRNAs we designed reduced expression of their cognate mRNA targets in transient-transfection experiments. However, the degree of knock-down varied between siRNAs targeting different sites within the same mRNA, possibly reflecting the different degrees of in vivo accessibility of these sites.
Although we have limited information on the targets of the other effective ribozymes at this time, these ribozymes likely inhibit HIV replication by targeting mRNAs of cellular proteins involved in diverse steps of viral replication, such as the ones exemplified here. An advantage of therapeutics targeting essential cellular cofactors would be the reduced chance of viral escape. Thus, future studies to identify these genes and their functions in the HIV replication cycle will not only improve our understanding of the virus-host cell interaction but also may reveal additional therapeutically superior drug targets.
Acknowledgments
We acknowledge the UCSD Center for AIDS Research for core services of sequencing and flow cytometry; Rick Bushman for the HIV IN− mutant; John Rossi, Mingjie Li, and Jing-Kuan Yee for kindly providing the pHIV-7 lentiviral vector; and Susan Q. He, Angelica Romero, Maureen Ibanez, and Ky Truong for excellent technical assistance.
REFERENCES
- 1.Baekelandt, V., A. Claeys, P. Cherepanov, E. De Clercq, B. De Strooper, B. Nuttin, and Z. Debyser. 2000. DNA-dependent protein kinase is not required for efficient lentivirus integration. J. Virol. 74:11278-11285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bai, J., S. Gorantla, N. Banda, L. Cagnon, J. Rossi, and R. Akkina. 2000. Characterization of anti-CCR5 ribozyme-transduced CD34+ hematopoietic progenitor cells in vitro and in a SCID-hu mouse model in vivo. Mol. Ther. 1:244-254. [DOI] [PubMed] [Google Scholar]
- 3.Banerjea, A., M. J. Li, G. Bauer, L. Remling, N. S. Lee, J. Rossi, and R. Akkina. 2003. Inhibition of HIV-1 by lentiviral vector-transduced siRNAs in T lymphocytes differentiated in SCID-hu mice and CD34+ progenitor cell-derived macrophages. Mol. Ther. 8:62-71. [DOI] [PubMed] [Google Scholar]
- 4.Beger, C., L. N. Pierce, M. Kruger, E. G. Marcusson, J. M. Robbins, P. Welcsh, P. J. Welch, K. Welte, M. C. King, J. R. Barber, and F. Wong-Staal. 2001. Identification of Id4 as a regulator of BRCA1 expression by using a ribozyme-library-based inverse genomics approach. Proc. Natl. Acad. Sci. USA 98:130-135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cech, T. R., and B. L. Bass. 1986. Biological catalysis by RNA. Annu. Rev. Biochem. 55:599-629. [DOI] [PubMed] [Google Scholar]
- 6.Chatterton, J. E., X. Hu, and F. Wong-Staal. 2004. Ribozymes in gene identification, target validation and drug discovery. Drug Discovery Today: Targets 3:10-17. [Google Scholar]
- 7.Daniel, R., R. A. Katz, and A. M. Skalka. 1999. A role for DNA-PK in retroviral DNA integration. Science 284:644-647. [DOI] [PubMed] [Google Scholar]
- 8.Doherty, A. J., and S. P. Jackson. 2001. DNA repair: how Ku makes ends meet. Curr. Biol. 11:R920-R924. [DOI] [PubMed] [Google Scholar]
- 9.Feng, Y., M. Leavitt, R. Tritz, E. Duarte, D. Kang, M. Mamounas, P. Gilles, F. Wong-Staal, S. Kennedy, J. Merson, M. Yu, and J. R. Barber. 2000. Inhibition of CCR5-dependent HIV-1 infection by hairpin ribozyme gene therapy against CC-chemokine receptor 5. Virology 276:271-278. [DOI] [PubMed] [Google Scholar]
- 10.Gervaix, A., D. West, L. M. Leoni, D. D. Richman, F. Wong-Staal, and J. Corbeil. 1997. A new reporter cell line to monitor HIV infection and drug susceptibility in vitro. Proc. Natl. Acad. Sci. USA 94:4653-4658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Jacque, J., K. Triques, and M. Stevenson. 2002. Modulation of HIV-1 replication by RNA interference. Nature 418:435-438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Jeanson, L., and J. F. Mouscadet. 2002. Ku represses the HIV-1 transcription: identification of a putative Ku binding site homologous to the mouse mammary tumor virus NRE1 sequence in the HIV-1 long terminal repeat. J. Biol. Chem. 277:4918-4924. [DOI] [PubMed] [Google Scholar]
- 13.Jeanson, L., F. Subra, S. Vaganay, M. Hervy, E. Marangoni, J. Bourhis, and J. F. Mouscadet. 2002. Effect of Ku80 depletion on the preintegrative steps of HIV-1 replication in human cells. Virology 300:100-108. [DOI] [PubMed] [Google Scholar]
- 14.Kawasaki, H., R. Onuki, E. Suyama, and K. Taira. 2002. Identification of genes that function in the TNF-alpha-mediated apoptotic pathway using randomized hybrid ribozyme libraries. Nat. Biotechnol. 20:376-380. [DOI] [PubMed] [Google Scholar]
- 15.Kawasaki, H., and K. Taira. 2002. Identification of genes by hybrid ribozymes that couple cleavage activity with the unwinding activity of an endogenous RNA helicase. EMBO Rep. 3:443-450. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 16.Kruger, M., C. Beger, Q. X. Li, P. J. Welch, R. Tritz, M. Leavitt, J. R. Barber, and F. Wong-Staal. 2000. Identification of eIF2Bγ and eIF2γ as cofactors of hepatitis C virus internal ribosome entry site-mediated translation using a functional genomics approach. Proc. Natl. Acad. Sci. USA 97:8566-8571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lee, N. S., T. Dohjima, G. Bauer, H. Li, M. J. Li, A. Ehsani, P. Salvaterra, and J. Rossi. 2002. Expression of small interfering RNAs targeted against HIV-1 rev transcripts in human cells. Nat. Biotechnol. 20:500-505. [DOI] [PubMed] [Google Scholar]
- 18.Li, L., J. M. Olvera, K. E. Yoder, R. S. Mitchell, S. L. Butler, M. Lieber, S. L. Martin, and F. D. Bushman. 2001. Role of the non-homologous DNA end joining pathway in the early steps of retroviral infection. EMBO J. 20:3272-3281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Li, Q. X., J. M. Robbins, P. J. Welch, F. Wong-Staal, and J. R. Barber. 2000. A novel functional genomics approach identifies mTERT as a suppressor of fibroblast transformation. Nucleic Acids Res. 28:2605-2612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lieber, A., and M. Strauss. 1995. Selection of efficient cleavage sites in target RNAs by using a ribozyme expression library. Mol. Cell. Biol. 15:540-551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Macpherson, J. L., J. A. Ely, L. Q. Sun, and G. P. Symonds. 1999. Ribozymes in gene therapy of HIV-1. Front. Biosci. 4:D497-D505. [DOI] [PubMed] [Google Scholar]
- 22.Mimori, T., M. Akizuki, H. Yamagata, S. Inada, S. Yoshida, and M. Homma. 1981. Characterization of a high molecular weight acidic nuclear protein recognized by autoantibodies in sera from patients with polymyositis-scleroderma overlap. J. Clin. Investig. 68:611-620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mimori, T., J. A. Hardin, and J. A. Steitz. 1986. Characterization of the DNA-binding protein antigen Ku recognized by autoantibodies from patients with rheumatic disorders. J. Biol. Chem. 261:2274-2278. [PubMed] [Google Scholar]
- 24.Nakajima, N., R. Lu, and A. Engelman. 2001. Human immunodeficiency virus type 1 replication in the absence of integrase-mediated DNA recombination: definition of permissive and nonpermissive T-cell lines. J. Virol. 75:7944-7955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Poon, B., and I. S. Chen. 2003. Human immunodeficiency virus type 1 (HIV-1) Vpr enhances expression from unintegrated HIV-1 DNA. J. Virol. 77:3962-3972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sarver, N., E. M. Cantin, P. S. Chang, J. A. Zaia, P. A. Ladne, D. A. Stephens, and J. J. Rossi. 1990. Ribozymes as potential anti-HIV-1 therapeutic agents. Science 247:1222-1225. [DOI] [PubMed] [Google Scholar]
- 27.Smith, G. C., and S. P. Jackson. 1999. The DNA-dependent protein kinase. Genes Dev. 13:916-934. [DOI] [PubMed] [Google Scholar]
- 28.Suyama, E., H. Kawasaki, M. Nakajima, and K. Taira. 2003. Identification of genes involved in cell invasion by using a library of randomized hybrid ribozymes. Proc. Natl. Acad. Sci. USA 100:5616-5621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Suyama, E., H. Kawasaki, and K. Taira. 2001. Characterization of pro-apoptotic gene Bak using hammerhead ribozymes. Nucleic Acids Res. Suppl. 2001:207-208. [DOI] [PubMed] [Google Scholar]
- 30.Tiscornia, G., O. Singer, M. Ikawa, and I. M. Verma. 2003. A general method for gene knockdown in mice by using lentiviral vectors expressing small interfering RNA. Proc. Natl. Acad. Sci. USA 100:1844-1848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Tuteja, R., and N. Tuteja. 2000. Ku autoantigen: a multifunctional DNA-binding protein. Crit. Rev. Biochem. Mol. Biol. 35:1-33. [DOI] [PubMed] [Google Scholar]
- 32.Welch, P. J., J. R. Barber, and F. Wong-Staal. 1998. Expression of ribozymes in gene transfer systems to modulate target RNA levels. Curr. Opin. Biotechnol. 9:486-496. [DOI] [PubMed] [Google Scholar]
- 33.Welch, P. J., E. G. Marcusson, Q. X. Li, C. Beger, M. Kruger, C. Zhou, M. Leavitt, F. Wong-Staal, and J. R. Barber. 2000. Identification and validation of a gene involved in anchorage-independent cell growth control using a library of randomized hairpin ribozymes. Genomics 66:274-283. [DOI] [PubMed] [Google Scholar]
- 34.Wong-Staal, F. 1996. Development of ribozyme gene therapy for HIV infection. Antibiot. Chemother. 48:226-232. [DOI] [PubMed] [Google Scholar]
- 35.Yam, P. Y., S. Li, J. Wu, J. Hu, J. A. Zaia, and J. K. Yee. 2002. Design of HIV vectors for efficient gene delivery into human hematopoietic cells. Mol. Ther. 5:479-484. [DOI] [PubMed] [Google Scholar]