Abstract
The RNase H cleavages that generate and remove the polypurine tract (PPT) primer during retroviral reverse transcription must be specific in order to create a linear viral DNA that is suitable for integration. Lentiviruses contain a highly conserved sequence consisting of six guanine residues at the 3′ end of the PPT (hereafter referred to as the G tract). We introduced mutations into the G tract of a human immunodeficiency virus type 1-based vector and determined the effects on the virus titer and RNase H cleavage specificity. Most mutations in the G tract had little or no effect on the virus titer. Mutations at the second and fifth positions of the G tract increased the proportion of two-long-terminal-repeat (2-LTR) circle junctions with one or two nucleotide insertions. The second and fifth positions of the G tract make specific contacts with amino acids in the RNase H domain that are important for RNase H cleavage specificity. These complementary data define protein-nucleic acid interactions that help control the specificity of RNase H cleavage. When the G-tract mutants were analyzed in a viral background that was deficient in integrase, in most cases the proportion of consensus 2-LTR circle junctions increased. However, in the case of a mutant with Ts at the second and fifth positions of the G tract, the proportion of 2-LTR circle junctions containing the one-nucleotide insertion increased, suggesting that linear viral DNAs containing an extra base are substrates for integration. This result is consistent with the idea that the 3′ end-processing reactions of retroviral integrases may help to generate defined ends from a heterogenous population of linear viral DNAs.
Human immunodeficiency virus type 1 (HIV-1) reverse transcriptase (RT) is the virally encoded enzyme that converts the viral genome from single-stranded RNA into double-stranded DNA (reviewed in references 4 and 37). This process involves the collaboration of the two enzymatic activities of RT: it can act as a DNA polymerase that can use either RNA or DNA as a template and as an RNase H that cleaves RNA if (and only if) it is present in an RNA-DNA duplex. Both the polymerase and the RNase H activities are required for conversion of the viral RNA genome into DNA; mutations that inactivate either the polymerase or RNase H activity block virus replication (30, 32, 34, 35).
HIV-1 RT is a heterodimeric protein consisting of p66 and p51 subunits that are derived from the Gag-Pol polyprotein by cleavage with viral protease (6). The p66 subunit is 560 amino acids long, and the p51 subunit contains the first 440 amino acids of the p66 subunit. The p66 subunit is composed of two domains, the polymerase and RNase H domains, with the C-terminal portion of p66 forming the RNase H domain. Based on its crystal structure, the polymerase domain of the p66 subunit of HIV-1 RT has been likened to a right hand, composed of fingers, palm, thumb, and connection subdomains (16). The p51 subunit corresponds closely, but not exactly, to the polymerase domain of p66. Although the polymerase domain of the p66 subunit and the p51 subunit fold into similar subdomains, the arrangements of the subdomains are different for the two subunits (13, 16). The p51 subunit plays a structural role and does not contain a functional polymerase active site (19). Based on crystallographic analysis, the RNase H active site is located approximately 17 to 18 nucleotides away from the polymerase active site; this distance is supported by biochemical experiments (9, 10, 13, 23, 38).
HIV-1 reverse transcription is initiated from a tRNALys3 that is base paired near the 5′ end of the plus-stranded viral RNA genome at the primer-binding site (PBS) (reviewed in references 4 and 37). RT copies the viral RNA into a minus-strand DNA, creating an RNA-DNA duplex that is a substrate for RNase H. There is a terminal redundancy at the ends of the RNA genome that permits the transfer of minus-strand DNA to the 3′ end of the viral RNA, allowing minus-strand DNA synthesis (and RNase H cleavage) to continue. Although most of the degradation of the viral RNA is not sequence specific and a range of differently sized products are generated, the RNase H activity of HIV-1 RT does make specific cleavages that define the ends of the linear viral DNA. One of these specific cleavages removes the tRNA primer used to initiate minus-strand DNA synthesis. In the case of HIV-1, removal of the tRNA primer occurs one nucleotide from the RNA-DNA junction (8, 26, 33, 36); this defines the right end of the unintegrated viral DNA. A second set of specific cleavages generates (and removes) the polypurine tract (PPT) primer used to initiate plus-strand DNA synthesis (25). The PPT primer is generated by two specific cleavages, one at the U tract-PPT junction and another at the PPT-U3 junction (reviewed in reference 4). The resistance of the PPT to RNase H cleavage allows it to serve as the primer for plus-strand DNA synthesis. A specific cleavage removes the PPT primer from the viral DNA; this cleavage occurs at the PPT-U3 junction (RNA-DNA junction) and defines the left end of the unintegrated viral DNA (25). The specificity of the removal of the RNA primers (tRNA and PPT) is important because the ends of the linear viral DNA serve as substrates for the virally encoded enzyme IN: IN inserts the viral DNA into the host cell genome to generate the provirus.
The specificity of the cleavages at the PPT is part of a larger problem: RNase H must be able to specifically cleave the RNA strand of an RNA-DNA duplex. This means that the enzyme must be able to distinguish an RNA-DNA duplex from both an RNA-RNA duplex and a DNA-DNA duplex. RNase H must also be able to recognize (and not cleave) the PPT within the larger RNA-DNA duplex. The first part of this problem, distinguishing an RNA-DNA duplex from both DNA-DNA and RNA-RNA duplexes, is simplified by the fact that the three duplexes have different structures in solution. DNA-DNA duplexes preferentially adopt B-form structures, and RNA-RNA duplexes preferentially adopt A-form structures. An RNA-DNA duplex adopts a structure that is in some ways intermediate between the A form and the B form; this is called the hybrid form, or H form (7). H-form nucleic acids have a minor groove width that falls between those of A-form and B-form nucleic acids; H-form RNA-DNA duplexes also possess altered sugar conformations in the DNA strand compared to DNA-DNA duplexes. All three types of structures are affected by the sequence: specific sequences can cause nucleic acid duplexes to depart significantly from the canonical A-, B-, and H-form structures. In addition, the structures of the nucleic acid duplexes can be profoundly altered by interactions with proteins. Ultimately, it is the structure of the PPT-containing segment of the RNA-DNA duplex when it is bound to HIV-1 RT that determines the specificity of cleavage. Fortunately, we have considerable information about the structure of the PPT both in the absence of HIV-1 RT and in the bound state. Studies of an RNA-DNA hybrid composed of the first 10 bases of the PPT in the absence of HIV-1 RT showed that the PPT has unusual structural features (12). Circular dichromism revealed that the DNA duplex derived from this sequence was in the B form (as expected), and spectra from the RNA-DNA duplex suggested that it had elements of both A-form and B-form structures, which is consistent with an H-form structure (12). The crystal structure of the DNA-DNA duplex revealed substantial base-stacking interactions involving the purines (11); however, in the RNA-DNA hybrid there was discontinuity in the base stacking (17). This deformed nucleic acid structure occurs in the A-G-A steps that are highly conserved in retroviral PPTs. The structure of this segment changes when RT is bound to the nucleic acid; although there are no unpaired bases in the unbound RNA-DNA duplex, there are unpaired bases when the RNA-DNA duplex that contains the PPT is bound to HIV-1 RT (31). Another feature of the unbound nucleic acid structure that may be important is an unusual sugar switch on the second sugar in the RNA strand from a C3′-endo sugar to a C2′-endo sugar (this corresponds to the 5′ end of the PPT); it was suggested that the highly conserved U tract found 5′ of retroviral PPTs may contribute to the maintenance of the unusual sugar configuration (17). Ultimately, it is the structure of the PPT when bound to its cognate RT that allows the RNase H cleavages to occur with specificity.
Many groups have used in vitro assays to study the RNase H cleavage reactions that generate and remove the PPT (18, 24, 25, 28, 29). Mutations in the PPT can affect the specificity of RNase H cleavage in vitro; however, the data are complex (18, 24, 25, 28, 29). In some in vitro experiments, the specificity of PPT-U3 cleavage by HIV-1 RT was affected by mutations in the 3′ end of the HIV-1 PPT (18). Another group reported that mutations in the 5′ end of the PPT are well tolerated and that the sequence of the 3′ end is crucial (24). Because the different studies used different assays and the mutants studied were not the same, the data are difficult to compare directly. These mutations affected the generation and/or removal of the PPT primer and increased the percentage of linear DNAs with aberrant ends. Linear DNAs are substrates for nonhomologous end-joining reactions that generate two-long-terminal-repeat (2-LTR) circles (20). We have used the sequences of 2-LTR circle junctions to show that mutations in the 5′ end of the PPT alter the specificity of RNase H cleavage in vivo (21). We wanted to determine if mutations in the G tract also affect the specificity of RNase H cleavage in vivo. The fact that the G tract is highly conserved between human and simian retroviruses and is moderately well conserved among other retroviruses suggests that it may be important (Table 1). Our results indicated that some G-tract mutations affect viral replication; however, most single point mutations were well tolerated and had modest effects on the viral titer and the specificity of RNase H cleavage. In general, double substitution mutations in the G tract had larger effects and appeared to affect either the generation of the 3′ end of the PPT or its removal by RNase H.
TABLE 1.
Sequences of different retroviral PPT and flanking sequences
| Retrovirusa | Sequence of PPT plus flanking sequencesb |
|---|---|
| HIV-1 (NL4-3) | UUUUUAAAAGAAAAGGGGGGACTGGAAG |
| HIV-1 (SF2) | UUUUUAAAAGAAAAGGGGGGACTGGAAG |
| HIV-2 (ROD) | UUUAAUAAAAACAAGGGGGGGACTGGAAG |
| HIV-2 (BEN) | UUUUAUAAAGAAAAAGGGGGACTGCAA |
| SIV (MND) | UUUUAUAAAAGAAAAGGGGGGACTGGGAG |
| SIV (AGM) | UUUUUAAAAGAAAAGGGAGGACTGGATG |
| EIAV | UAUGUUUAGAAAAACAAGGGGGGAACTGTGGG |
| FIV | UCCUAAAAAAGAAAAAAGGGTGGACTGGGAT |
| BIV | UAUUUUAAACTTAAAAGGGTGGACTGTGGGGC |
| Mo-MLV | UCUCCAGAAAAAGGGGGGAATGAAAGACC |
| Fr-MuLV | UUUCCAGAAAAAGGGGGAATGAAAGA |
| MMTV | UUUUAAAAAAGAAAAAAGGGGGAAATGCCGCGC |
SIV, simian immunodeficiency virus; EIAV, equine infectious anemia virus; FIV, feline immunodeficiency virus; BIV, bovine immunodeficiency virus; Mo-MLV, Moloney murine leukemia virus; Fr-MuLV, Friend murine leukemia virus; MMTV, mouse mammary tumor virus.
PPTs are shown in bold.
MATERIALS AND METHODS
Construction of HIV-1 mutants.
Mutations were generated in the PPT of the HIV-1-based vector pNLNgoMIVR-E-.HSA by a BspMI cassette strategy as previously described (21). The plasmids were analyzed by restriction endonuclease digestion and DNA sequencing to confirm that (only) the desired mutations were present in the vectors. Vectors containing the D116N mutation in the integrase coding region were generated by site-directed mutagenesis with a Quick-Change kit (Stratagene). Briefly, DNA oligonucleotides (Biosource International) containing the necessary mutations were synthesized and used in the site-directed mutagenesis protocol. The mutagenesis substrate was pKS containing the Asp718-to-EcoRI fragment of pNLNgoMIVR-E-.HSA. The resulting plasmid was analyzed by restriction endonuclease digestion, and DNA sequence analysis was used to confirm that only the desired mutation in IN was present; this plasmid was called pKSINTD116N. The Asp718-to-SalI fragment from pKSINTD116N was cloned into pNLNgoMIVR-E-.HSA vectors containing the desired PPT mutations by the use of Asp718 and SalI (these sites are each present once in the plasmid that carries the vector). The resulting plasmids were analyzed by restriction endonuclease digestion and DNA sequence analysis.
Cells.
The human embryonal kidney cell line 293 was obtained from the American Type Culture Collection. The human osteosarcoma cell line HOS was obtained from Richard Schwartz (Michigan State University, Lansing). 293 and HOS cells were maintained in Dulbecco's modified Eagle's medium (Invitrogen) supplemented with 5% fetal bovine serum, 5% newborn calf serum, and penicillin (50 U/ml) plus streptomycin (50 μg/ml) (Quality Biological).
Transfection, infection, and phenotyping protocol.
293 cells were transfected with 3 μg of pNLNgoMIVR-E-.HSA and 1.5 μg of pHCMV-g (obtained from Jane Burns, University of California, San Diego) by the calcium phosphate method. 293 cells were plated in 100-mm-diameter dishes at a density of 106 cells per plate on the day prior to transfection. At this plating density, the cells were approximately 25% confluent on the day of transfection. The precipitate was added dropwise to the 293 cells. Fresh medium was added 16 h after transfection. The supernatants were harvested after 48 h and clarified by low-speed centrifugation, and an aliquot was used to infect HOS cells. The amount of p24 in the supernatant was determined by the Coulter HIV-1 p24 antigen assay (Beckman Coulter); the p24 concentration was used to control for the amount of virus in the samples. HOS cells were plated in 60-mm-diameter dishes at a density of 1.5 × 105 cells per plate on the day prior to infection. Cells were infected with undiluted virus and with viruses diluted 5-fold, 25-fold, and 125-fold in medium. Four milliliters of virus (or diluted virus) was allowed to absorb to the cells for 4 h, the virus was removed by aspiration, and then fresh medium was added. Forty-eight hours after infection, the cells were harvested from the plate by treatment with 1.0 ml of EDTA (Invitrogen), an additional 3 ml of phosphate-buffered saline was added, and then the cells were collected by centrifugation, washed, and resuspended in 200 μl of phosphate-buffered saline. The cells were labeled with a phycoerythrin-conjugated rat anti-mouse CD24 monoclonal antibody (Pharmingen) by standard procedures, fixed with paraformaldehyde, and subjected to florescence-activated cell sorting to determine the viral titer. Virus titers were calculated from the linear range of virus dilution, which included the 1-to-25 and 1-to-125 dilutions of virus.
Transfections, infections, and nucleic acid extraction for 2-LTR circle junction analysis.
293 cells were transfected with 5 μg of plasmid DNA encoding the pNLNgoMIVR-E-.HSA vector and 3 μg of pHCMV-g by the calcium phosphate method. The medium on the cells was changed 16 h after infection. The supernatants were harvested after 48 h and clarified by centrifugation, and 4 ml of virus-containing supernatant was used to infect HOS cells. The supernatants were left on the cells for 4 h, and then fresh medium was added. The total DNA was isolated from the HOS cells approximately 24 h after infection by use of a viral blood DNA kit (Qiagen).
PCR amplification, cloning, and sequencing of 2-LTR circle junctions.
The 2-LTR circle junctions were amplified in 100-μl reactions by use of an upstream PCR primer that anneals near the RU5 junctions and a downstream primer that anneals in the U3 region of the LTR. The sequence of the upstream primer was 5′-CGATGAATTCGCTAACTAGGGAACCCACTGCT-3′, and the sequence of the downstream primer was 5′-GCCATTCTAGAGTTCTCTCCTTTATTGGCCTC-3′. Ten microliters of DNA from cells infected with the PPT mutants and 0.25 μl each of the forward and reverse primers (100 nM final concentration) were used with 90 μl of Platinum PCR Supermix (Invitrogen) for each PCR. The expected product was approximately 350 bp long and had EcoRI and XbaI cleavage sites introduced by the primers. The PCR products were digested with EcoRI and XbaI and cloned into the SK vector (Stratagene). The 2-LTR circle junction clones were analyzed by restriction enzyme digestion and DNA sequence analysis. Approximately 90 2-LTR circle junctions were characterized for each mutant.
Determination of consensus 2-LTR circle junctions in an IN− background.
Transfections, infections, DNA isolation, PCR amplification, and cloning of the 2-LTR circle junction products were done identically for the IN− viruses (containing the D116N mutation) and for the integration-competent (IN+) viruses. A consensus 2-LTR circle junction contains a ScaI recognition sequence. The percentage of consensus 2-LTR circle junctions was determined by digesting the individual clones with ScaI and fractionating the digestion products by agarose gel electrophoresis. The SK plasmid that the PCR products were cloned into also contained a ScaI recognition sequence; plasmids containing a consensus 2-LTR circle junction would produce two bands upon ScaI digestion and agarose gel electrophoresis. Approximately 90 2-LTR circle junctions were characterized for each mutant.
Statistical methods.
The data were analyzed by log linear categorical analysis, contingency table analysis, and related methods. I-by-J circle junction-by-mutant tables were decomposed through the use of likelihood ratio chi-square statistics into independent partitions to show associations between circle junction and/or mutant groupings and categories. Subsets of pertinent groupings were followed up with traditional two-by-two chi-square analyses and Fisher's exact tests.
RESULTS
Effect of mutations in the HIV-1 PPT G tract on virus titer.
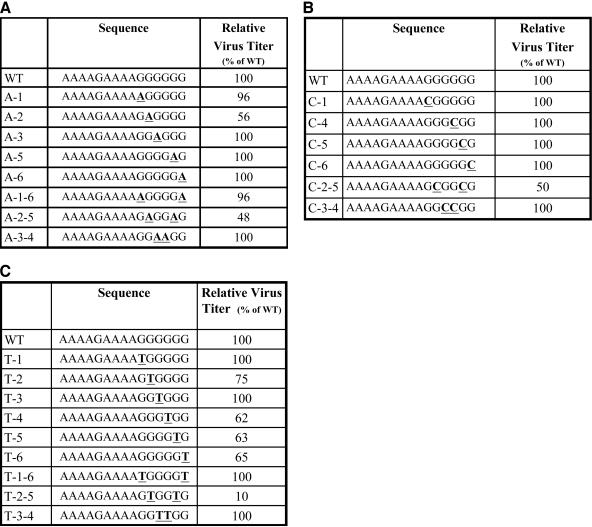
The sequence of the PPT of HIV-1 is 5′-AAAAGAAAAGGGGGG-3′ (Fig. 1) (3). The six G residues at the 3′ end of the PPT are referred to as the G tract. Mutations were introduced into the G tract of the PPT in the HIV-1 vector pNLNgoMIVR-E-.HSA (14). This vector expresses the retroviral gag-pol gene; the murine cell surface marker CD24 (heat-stable antigen or Hsa) is expressed from the nef reading frame. The envelope gene is inactivated in this vector. The DNA sequences of mutant PPTs containing G-to-A transitions, G-to-C transversions, and G-to-T transversions are shown in Fig. 1. Virions were generated by cotransfecting 293 cells with plasmids encoding the vectors and pHCMV-g (expresses the vesicular stomatitis virus envelope glycoprotein [see references 1 and 39]). The viruses were harvested and used to infect human osteosarcoma cells (HOS cells). These vectors were limited to a single cycle of replication. Virus titers were measured by labeling cells exposed to the virus-containing supernatants with an antibody directed against CD24, followed by fluorescence-activated cell sorting. The virus titers were normalized to the amount of p24 antigen present in the virus-containing supernatants.
FIG. 1.
Effects of mutations introduced in the PPT of an HIV-1-based retroviral vector on viral titer. Viruses containing mutations in the PPT were generated as described in Materials and Methods. Left columns, the names of the mutants (the wild-type control is labeled WT and is at the top row of each panel); middle columns, DNA sequences of the mutations in the PPT (mutated bases are underlined and shown in bold); right columns, virus titers (normalized to the amount of p24 in the virus-containing supernatants). The relative virus titers were determined from the linear range of dilution, as described in Materials and Methods. (A) Effects of G-to-A transition mutations on virus titer. (B) Effects of G-to-C transversion mutations on virus titer. (C) Effects of G-to-T transversion mutations on virus titer.
G-to-A transition mutations were introduced at several of the guanine residues at the 3′ end of the HIV-1 PPT (Fig. 1A). The mutants were named according to the position of the G residue that was mutated. The first G is at the 5′ end of the G tract; the A substitutions were named A-1, A-2, A-3, A-5, and A-6. Mutants containing double substitution mutations were also generated; these mutants were named A-1-6, A-2-5, and A-3-4. Most of these mutations did not measurably affect the virus titer in a single cycle of replication. The A-2 single mutant and the A-2-5 double mutant both caused a decrease in virus titer, to about 50% that of the wild-type virus.
G-to-C transversion mutations were introduced at several of the G-tract residues at the 3′ end of the HIV-1 PPT (Fig. 1B). The single mutants C-1, C-4, C-5, and C-6 and the double mutants C-2-5 and C-3-4 were prepared. The only mutation in this series that had a measurable effect on the virus titer was the C-2-5 double mutant; this virus had a titer of about 50% that of the wild type. G-to-T transversion mutations were introduced at each of the G-tract guanine residues (Fig. 1C). In addition to the single mutants T-1, T-2, T-3, T-4, T-5, and T-6, the double mutants T-1-6, T-2-5, and T-3-4 were generated. The T-4, T-5, and T-6 mutants had titers of about 65% that of the wild type, and the T-2-5 double mutant had a titer of about 10% that of the wild type.
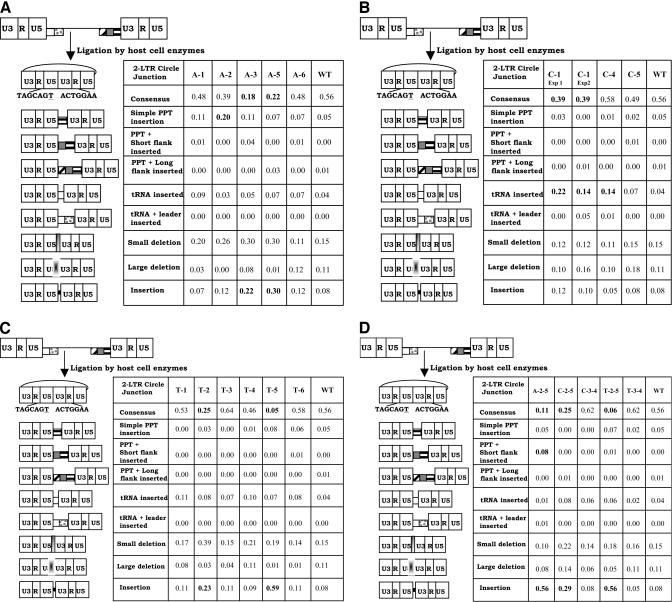
Effects of mutations in the G tract of the HIV-1 PPT on the 2-LTR circle junction.
We previously used the sequence of the 2-LTR circle junction as a surrogate for the ends of the linear viral DNA. The effects of mutations in the G tract of the HIV-1 PPT on the 2-LTR circle junctions are shown in Fig. 2. When cells were infected with a virus containing the wild-type PPT, the percentages of consensus and aberrant 2-LTR circle junction sequences were very similar to previously reported results (14, 21). Fisher's exact tests were performed to determine if mutations caused statistically significant differences in the 2-LTR circle junction sequences. For the G-to-A single mutations (Fig. 2A), the A-3 and A-5 mutants had a statistically significant decrease in the proportion of consensus 2-LTR circle junctions (P < 0.01). These mutants (A-3 and A-5) also had significant increases in the proportion of 2-LTR circle junctions containing insertions at the junction (P < 0.02). The A-2 mutant had a significant increase in the proportion of 2-LTR circle junctions containing simple PPT insertions (P < 0.05). The C-1 transversion mutation (Fig. 2B) had a significant decrease in the proportion of 2-LTR circle junctions with a consensus sequence (P < 0.05). C-1 and C-4 both had significant increases in the proportion of 2-LTR circle junctions containing tRNA insertions (P < 0.05). Of the G-to-T transversion mutations (Fig. 2C), the T-2 and T-5 mutants had significant decreases in the proportion of consensus 2-LTR circle junctions (P < 0.05) and significant increases in the proportion of 2-LTR circle junctions containing insertions other than tRNA- and longer (three nucleotides or more) PPT-derived sequences (P < 0.05). The double mutations A-2-5, C-2-5, and T-2-5 all had significant decreases in the proportion of 2-LTR circle junctions containing consensus sequences (P < 0.05) and significant increases in the proportion of 2-LTR circle junctions containing insertions that were not derived from tRNA or the PPT (P < 0.05). The A-2-5 mutant also had a significant increase in the proportion of 2-LTR circle junctions containing PPT-plus-short-flank insertions (P < 0.05).
FIG. 2.
2-LTR circle junctions derived by infecting cells with PPT mutants. The PBS is indicated by a white box, the leader sequence downstream of the PBS is shown with gray dots, the PPT is shown by a box with black horizontal bars, the U tract is shown by a gray box, and the sequences immediately upstream of the U tract are shown by the box with diagonal bars. The linear viral DNA can be ligated by cellular enzymes in the nucleus of an infected cell to form a 2-LTR circle. A consensus circle junction results from the ligation of linear viral DNAs with complete ends. A consensus 2-LTR circle is shown at the top left of each panel. Different categories of aberrant 2-LTR circle junctions are shown below the consensus. The schematics on the left depict the portions of the viral genome that are retained or deleted at the 2-LTR circle junction. Consensus 2-LTR circle junctions contain complete ends. If PPT-derived sequences were inserted at the 2-LTR circle junction, the result was tallied in the three PPT categories. If a tRNA was inserted at the 2-LTR circle junction, the result was tallied in either of the two tRNA categories. Small deletions are deletions of 5 bp or less at the 2-LTR circle junction, and large deletions are larger than 5 bp. Insertions are insertions at the 2-LTR circle junction that are not of a PPT or tRNA origin. These sequences were typically short 1- to 2-bp insertions. Statistically significant differences between mutants and wild-type are shown in bold. (A) Effects of single G-to-A transition mutations on 2-LTR circle junctions. (B) Effects of single G-to-C transversion mutations on 2-LTR circle junctions. (C) Effects of single G-to-T transversion mutations on 2-LTR circle junctions. (D) Effects of double mutations (both transitions and transversions) on 2-LTR circle junctions.
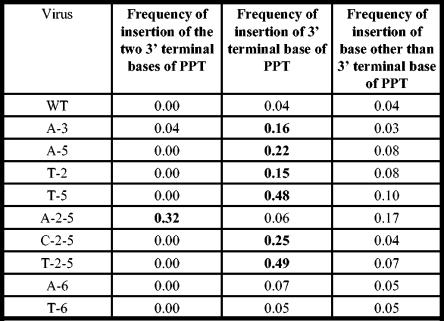
Frequency of insertion of one or two nucleotides, potentially from the PPT.
In order to determine if mutations in the G tract caused aberrant RNase H cleavages that led to small (one or two nucleotide) insertions at the 2-LTR circle junction, we analyzed the frequency of insertions of one or two nucleotides that were potentially derived from the 3′ end of the PPT at the 2-LTR circle junction (Fig. 3). For the wild-type (WT) PPT, the frequency of insertion of a G residue at the 2-LTR circle junction was 0.04. The A-3, A-5, T-2, and T-5 single mutants all had statistically significant increases in the frequency of insertions at the 2-LTR circle junction (P < 0.05). In each of these cases, there was a corresponding increase in the frequency of the terminal nucleotide of the PPT (a G) at the 2-LTR circle junction compared to the wild-type control. The A-6 and T-6 mutants did not cause statistically significant increases in the frequency of insertion of the respective terminal nucleotide (A for A-6 and T for T-6). Insertions of two nucleotides were also analyzed. There were no insertions of GG at the 2-LTR circle junction for the WT PPT. The A-3 mutant and A-2-5 mutants both had 2-bp insertions that appeared to be derived from the PPT. Insertions of GG occurred at a frequency of 0.04 for the A-3 mutant; the A-2-5 mutant had a dramatic increase in frequency, to 0.32 (P ≪ 0.0001) (insertion of AG). The 2-LTR circle junctions derived by infecting cells with the T-2-5 mutant contained insertions of the 3′-terminal base at a frequency of 0.49 (P ≪ 0.0001). The 2-LTR circle junctions that we analyzed from the T-2-5 mutant did not contain any two-nucleotide insertions at the 2-LTR circle junction.
FIG. 3.
Frequencies of insertion of the terminal base and terminal two bases of the PPTs. The mutants chosen for this analysis either showed significant increases in the frequency of insertion or had a modification in the terminal nucleotide of the PPT. The viruses are shown in the left column, the frequencies of insertion of the two terminal nucleotides are shown in the second column, the frequencies of insertion of the 3′ terminal nucleotide are shown in the third column, and the frequencies of insertion of nucleotides other than the terminal nucleotide of the PPT for each mutant are shown in the right column. The A-6 and T-6 mutants did not have statistically significant increases in the frequency of insertion of one or two nucleotides, but the other mutants in the figure did have statistically significant increases in the frequency of insertions at the 2-LTR circle junctions.
G-tract mutations do not increase the frequency of deletions in the U3 domain.
If RNase H were to generate the PPT primer by cleavage downstream (3′) of the PPT-U3 junction, there would be an increase in small deletions in U3. No significant increases in the frequency of U3 deletions were observed (data not shown).
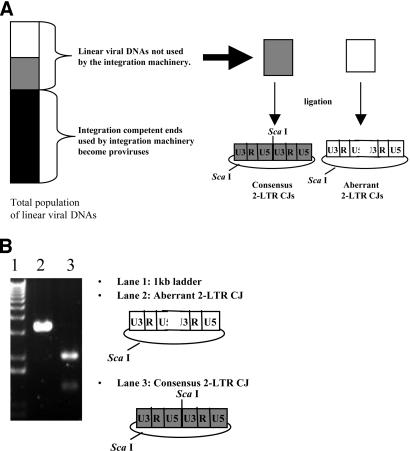
Analysis of the entire linear viral DNA population.
The integration machinery will use linear viral DNAs containing correct ends and may be able to use linear DNAs that are nearly correct. As a consequence, the population of linear viral DNAs that become ligated to form 2-LTR circles preferentially contains aberrant ends. In order to analyze the entire population of linear viral DNAs, we inactivated the viral IN via the D116N mutation. The sequences of 2-LTR circle junctions from cells infected with integration-defective viruses provide information about the population of linear viral DNAs that have not been selected based upon their suitability as substrates for integration. Consequently, all DNAs can be recovered as 2-LTR circles, whether they are suitable for integration or not. However, there may be some residual bias in terms of whether all linear DNAs are equivalently good substrates for the ligation reaction. The proportions of consensus 2-LTR circle junctions in integration-competent and integration-defective backgrounds are shown in Table 2. The proportions of viruses containing a consensus 2-LTR circle junction in the IN− background were determined by use of a ScaI cleavage assay (see Materials and Methods) (Fig. 4). When cells were infected with viruses containing a wild-type PPT and active IN, the proportion of 2-LTR circle junctions containing a consensus sequence was 56%; the percentage of consensus circles was 95% in the absence of integration. When cells were infected with the A-3 and A-5 single mutants in an integration-competent background, 18 and 22% of the 2-LTR circle junctions, respectively, had a consensus sequence; in the absence of integration, these proportions increased to 76 and 70%, respectively. When cells were infected with the T-2 and T-5 single mutants, the proportions of consensus 2-LTR circle junctions were 25 and 5%, respectively, in the presence of integration; these proportions increased to 64 and 49%, respectively, in the absence of integration. The 2-LTR circle junctions arising after cells were infected with the A-2-5 and C-2-5 double mutants also displayed similar increases in the proportions of consensus 2-LTR circle junctions in the absence of integration. For each of the previously discussed mutants, the increase in the proportion of consensus 2-LTR circle junctions in the IN− background was highly significant (P ≪ 0.0001). The T-2-5 double mutant was the only PPT mutant that had similar fractions of consensus 2-LTR circle junctions in the presence and absence of integration. When cells were infected with the T-2-5 mutant, 6% of the 2-LTR circle junctions had a consensus sequence in the presence of integration (based on direct sequencing) and 3% had a consensus sequence in the absence of integration (based on the ScaI assay). In order to validate the ScaI assay and to determine if the aberrant 2-LTR circle junction populations were similar in the presence and absence of integration, we sequenced 2-LTR circle junctions from the T-2-5 IN− virus. The results showed that 3% of the junctions had a consensus sequence, 1% had a PPT insertion, 2% had a tRNA insertion, 8% had small deletions, and 3% had a large deletion. Similar frequencies of these types of aberrant 2-LTR circle junctions were observed in the IN+ background. The proportion of 2-LTR circle junctions containing one-nucleotide insertions at the 3′ end of the PPT was 82% for the IN− background, which was higher than the 56% that was measured for the IN+ background.
TABLE 2.
Percentages of consensus 2-LTR circle junctions in integration-competent and -defective backgrounds
| Virus | % of consensus 2-LTR circle junctions
|
Titer (%) | |
|---|---|---|---|
| IN+ | IN− | ||
| WT | 56 | 95 | 100 |
| A-3 | 18 | 76 | 100 |
| A-5 | 22 | 70 | 100 |
| T-2 | 25 | 64 | 75 |
| T-5 | 5 | 49 | 63 |
| A-2-5 | 11 | 73 | 48 |
| C-2-5 | 25 | 70 | 50 |
| T-2-5 | 6 | 3 | 10 |
FIG. 4.
Enrichment of aberrant ends during and after integration and restriction endonuclease cleavage assay to screen for consensus 2-LTR circle junctions. (A) Integration of correct linear viral DNAs enriches for linear DNAs with aberrant ends, which can then be ligated to form 2-LTR circles. The vertical bar on the left represents the total population of linear viral DNAs. Integration-competent ends are represented by the shaded portions of the bar, and integration-defective ends are represented by the white portion. Integration is efficient (see Discussion), and the majority of linear viral DNAs containing correct ends become integrated to form proviruses. The consensus 2-LTR circle junctions that contain complete ends have a ScaI recognition sequence at the 2-LTR circle junction. (B) Representative picture of a gel in which ScaI endonuclease cleavage was used to screen for consensus 2-LTR circle junctions. Lane 1 contains the 1-kb ladder (Invitrogen), lane 2 contains an aberrant 2-LTR circle junction, and lane 3 contains a consensus 2-LTR circle junction.
DISCUSSION
HIV-1 vectors were used to determine the effects of mutations in the G tract at the 3′ end of the HIV-1 PPT. We are interested in this portion of the PPT because it is highly conserved between different human and simian lentiviruses and is moderately well conserved among other retroviruses as well (Table 1). An analysis of the effects of mutations in the G tract on the virus titer and the sequences of the 2-LTR circle junctions demonstrated that single point mutations in this sequence are generally well tolerated. This result was somewhat surprising because the G tract is conserved. The infections were limited to a single cycle of retroviral replication. If these mutations were present in a replication-competent virus and the virus was allowed to undergo multiple cycles of replication, the effects of relatively small differences in replication capacity would increase in importance.
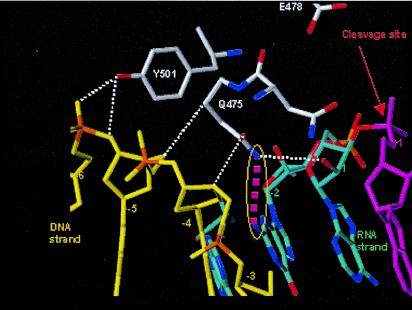
The crystal structure of HIV-1 RT in complex with a DNA-RNA template/primer whose sequence is derived from the PPT has provided valuable insight into features of the PPT and RT that are important for RNase H cleavage specificity (Fig. 5) (31). Mutations of amino acids in the RNase H primer grip and in the amino acids that contact the RNA template PPT affect the specificity of RNase H cleavage (15, 29). The 2-LTR circle junctions that arise in cells infected with viruses containing mutations at Y501 and N474A+Q475A in RT have significant increases in the proportions of circle junctions containing PPT insertions and in the numbers of deletions that arise from miscleavages in U3 (15). These amino acids help to position the nucleic acid in the vicinity of the RNase H active site. Mutations in these amino acids reposition the nucleic acid substrate and alter the RNase H cleavage specificity. Y501 contacts the nucleic acid at position −5 relative to the RNase H active site, and Q475 contacts the nucleic acid at position −2 relative to the active site. If RNase H were aligned so that it would cleave at the 3′ end of the PPT, Y501 would contact the ribose of G2 and the phosphate between G1 and G2; Q475 would contact G5. When we generated mutations in the nucleic acid, the specificities of the RNase H cleavages that generated or removed the PPT were most affected when the mutations were present at the 2nd and 5th positions of the G tract at the 3′ end of the PPT. The A-5, T-5, C-2-5, and T-2-5 mutants all caused a significant increase in the proportion of 2-LTR circle junctions with a one- or two-nucleotide insertion. The greatest effects were seen with the T-2-5 mutant. The data show that changing either the nucleic acid or the amino acid involved in these contacts affects RNase H cleavage. Y501 interacts with the sugar-phosphate backbone between positions −5 and −6; based on the effects of mutations in the G tract, the −5 position (G2) appears to be more important. This interaction involves the hydroxyl group of the tyrosine and the sugar-phosphate backbone of the nucleic acid, and mutations in either the nucleic acid or the protein affect the RNase H specificity. The only major groove contact that was present in the structure involved Q475 and the amino group on the guanine residue at position −2 (G5). A mutation in this amino acid or in the nucleotide involved in this contact affected the RNase H cleavage specificity. These contacts are close to the RNase H cleavage site but are not adjacent to it. This indicates that it is the larger structure that the nucleic acid adopts when in complex with RT that allows the specific cleavages to occur. Mutating the G residues at positions −2 and −5 relative to the RNase H cleavage site had the largest effect on RNase H cleavage. G-to-T mutations at these positions (T-2-5) affected the cleavage specificity in a very defined fashion: the proportion of DNAs containing a 1-bp insertion derived from the PPT increased. It is plausible that when the contacts of the nucleic acid at positions −2 and −5 with Q475 and Y501 of RT, respectively, are disrupted by mutation of the G's that are normally found at these positions to Ts, RT repositions itself and interacts with the sugar-phosphate backbone at position −6 and with the G at position −3. The C-2-5 mutation also caused a significant increase in the number of one-nucleotide insertions; however, the A-2-5 mutation caused two-nucleotide insertions. Repositioning the RT would cause RNase H to cleave the substrate one (or two) nucleotide(s) from the normal cleavage site, causing a one (or two)-nucleotide insertion. If the guanine-RT interactions are important for cleavage specificity, it is possible that the conservation of the G tract (a run of six G's) occurs because local contributions of base stacking help to maintain an appropriate structure for RNase H. Sequence comparisons of several retroviral PPTs showed that G's are conserved at positions 2, 3, and 5 in the G tract. This suggests that other retroviral RTs may make similar contacts with their cognate PPTs.
FIG. 5.
Structure of the PPT in complex with HIV-1 RT. The DNA primer is shown in yellow, and the RNA template is shown in blue and magenta. The red arrow designates the site of RNase H cleavage. Interactions between RT and the sugar-phosphate backbone of the nucleic acid are shown by white dotted lines. The interaction of Y501 with position −5 (G2) is important for the specificity of RNase H cleavage (see Discussion). The major groove contact between Q475 and the guanine base at position −2 (G5) is highlighted in magenta; this interaction is also important for RNase H cleavage specificity.
We previously characterized the effects of mutations in the 5′ end of the PPT on virus replication (21). Single and double mutations had small effects on the virus titer; a complex mutation involving seven of the first nine nucleotides from the 5′ end of the PPT had a large effect on the titer and on the 2-LTR circle junction populations. Mutations at the 5′ end of the PPT primarily affected the generation or removal of the PPT. These mutations caused an increase in the number of 2-LTR circle junctions containing a PPT sequence. In contrast, the G-tract mutations specifically increased the proportion of small insertions in the 2-LTR circle junction. These data suggest that G-tract mutations affect the precise alignment of the RNase H active site at the U3-PPT junction, either when the PPT is generated or when it is removed after its use as the primer for the initiation of plus-strand DNA synthesis.
Many of the mutations had no measurable effect on virus titer in one cycle of replication, some mutations decreased the titer about 50%, and the T-2-5 mutant had a titer of 10% that of the wild type. When we compared the proportions of aberrant 2-LTR circle junctions obtained from infections with viruses that had a functional IN gene to those that were IN−, there was a more dramatic effect on the percentages of aberrant 2-LTR circle junctions than on the viral titers. When the C-1, T-2, T-5, A-3, and A-5 mutants were analyzed in an integrase-proficient background, all had statistically significant decreases in the proportion of consensus 2-LTR circle junctions; however, these mutant viruses all had titers that were similar to that of the wild type. The most dramatic effect was seen with the T-5 mutant: its titer was 63% that of the wild type, but in the presence of active IN only 5% of the 2-LTR circle junctions had the consensus sequence. Many of the double mutants also displayed more dramatic decreases in the proportion of consensus 2-LTR circle junctions than one might have expected based on the titer. The A-2-5 and C-2-5 mutants had consensus circle junctions in only 11 and 25% of the samples, but they had titers of 50% that of the wild type. These data indicate that the integration process enriches 2-LTR circle junctions with aberrant ends; relatively small changes in titer can correlate with dramatic alterations in the 2-LTR circle junction population.
The integration process enriches the pool of linear viral DNAs with aberrant ends. In order to eliminate the effects of integration on the pool of linear DNAs that gave rise to the 2-LTR circle junctions, we repeated some of the experiments with IN− viruses. For the WT, A-3, A-5, T-2, T-5, and A-2-5 viruses, there was a dramatic increase in the proportions of 2-LTR circle junctions containing a consensus sequence in the IN− background. However, there was not a comparable increase in the percentage of consensus 2-LTR circle junctions for the T-2-5 double mutant. The percentage of consensus 2-LTR circle junctions was 6% in the presence of integration and 3% in the absence of integration. The IN+ virus had short (1 bp) insertions in 56% of the 2-LTR circle junctions. This number increased to 82% in the absence of IN. When the titer of the T-2-5 virus was measured, it was 10% that of the wild type. The T-2-5 mutation leads to miscleavage either at the time that the PPT is generated or when it is removed. Either of these events would increase the proportion of 2-LTR circle junctions containing a one-base insertion. The proportion of 2-LTR circle junctions containing these short insertions increased when the T-2-5 mutation was present in the IN− background, indicating that the viral IN can successfully use linear DNAs with one extra nucleotide. This indicates that the 3′-end-processing activity of HIV-1 IN that normally removes two nucleotides from the 3′ end of the linear viral DNA can also process substrates that are one nucleotide longer than normal. In vitro assays have shown that purified HIV integrase can use U3 ends containing an extra G as a substrate for the end-processing reaction (3). Previously, it was shown that the RU5 ends of linear viral DNAs in preintegration complexes do not all conform to a consensus. It was proposed that the IN processing reaction, which removes a dinucleotide from each end of the linear viral DNA, may serve to produce defined ends from a heterogeneous population. The data that we have obtained suggest that IN can use an aberrant U3 end, which is consistent with a role for the end-processing function of IN in generating uniform ends from a heterogeneous population of linear DNAs (22).
The differences in the proportions of consensus 2-LTR circle junctions in the presence and absence of integration indicate that the efficiency of integration of the linear viral DNA in infected cells is high. When the wild-type PPT was present in the vector, the proportion of consensus 2-LTR circle junctions in the integration-competent vector was 56%. In the IN− background, the proportion of 2-LTR circle junctions containing a consensus sequence was 95%. This represents a ninefold enrichment in aberrant ends when integration occurs. This increase in the proportions of aberrant ends correlates with an integration efficiency of approximately 90%, which is similar to the integration efficiency measured by Buckman et al. by quantitative PCR (2).
We were initially surprised by the increase in the proportion of 2-LTR circle junctions containing a tRNA insertion when the C-1 and C-4 mutations were present in the PPT. These mutations did not significantly affect the proportion of 2-LTR circle junctions containing PPT insertions. However, the presence of the tRNA sequences can be explained as follows. If these mutations delayed the cleavage of the PPT and if there was a loss of RT activity from the virion core, then there might not have been sufficient RNase H activity to completely remove the tRNA. Alternatively, it is possible that these mutations in the PPT affected the assembly or structure of the virion core in such a way that the processing (removal) of the tRNA was also affected.
When G-tract mutants were analyzed to determine which classes of aberrant 2-LTR circle junction sequence they gave rise to, we found that the A-2 mutant caused a statistically significant increase in the proportion of 2-LTR circle junctions containing simple PPT insertions. These PPT segments may have been retained through the following different mechanisms. As has already been discussed, if the PPT was processed slowly, the RNase H activity in the viral core may have been low when the PPT should have been removed. RT may have generated a short PPT by miscleavage at the 3′ end of the PPT. If RT used this primer to initiate plus-strand DNA synthesis, then the removal of the primer from the RNA-DNA junction would have left behind part of the PPT sequence because DNA synthesis started in the PPT. Alternatively, RT may have generated a correct PPT but failed to remove all of the RNA nucleotides after plus-strand DNA synthesis was initiated.
Several in vitro approaches have been used to probe the PPT structure and to monitor the RNase H cleavages that generate and remove the PPT. Our results are similar but not identical to those obtained from in vitro reactions using model substrates that mimic the PPT. The results of Pullen et al. indicated that G-to-C transversions at the 3rd and 5th positions of the PPT had the largest effects on the specificity of RNase H cleavage (27). We found that positions 2 and 5 are the most important and that G-to-T transversions have the most dramatic effect in vivo. Despite the effects of the PPT mutations, some cleavages still occur at the PPT-U3 junction in our in vivo assays. Mutations at several other positions in the G tract had subtle effects on the specificity of RNase H cleavage. Kvaratskhelia et al. recently showed that G-to-C mutations alter the structure of an RNA-DNA duplex containing the PPT when the duplex is free in solution (18) When pyrrolo-deoxycytosine was used to examine the G tract of the HIV-1 PPT, a weakening of the intrastrand base pairing at position −2 was observed (5). The authors concluded that the interaction of Q475 and the −2 base pair may play an important role in PPT recognition. Taken together, the results of these different approaches are consistent with our observations.
Mutations at several positions in the PPT have subtle effects on PPT cleavage in vitro and in vivo, which suggests that the entire sequence of the PPT is involved, either directly or indirectly, in its interactions with HIV-1 RT. For this reason, individual mutations rarely disrupt the structure of the entire PPT enough to dramatically affect the specificity of RNase H cleavage. None of the mutations at position 6 in the G tract (the nucleotide immediately upstream of the RNase H cleavage site) had a dramatic effect on RNase H cleavage. Double mutations upstream in the PPT (in the A tract, which is 5′ of the G tract), as well as previously characterized mutations (some of which were complex in nature) further upstream, had larger effects, indicating that the mutations within the PPT have a larger effect on the interaction of the PPT and RNase H than do mutations at the actual site of cleavage.
Acknowledgments
We thank Louise Finch and colleagues and the SAIC-Frederick Laboratory of Molecular Technology for technical support. We also thank Hilda Marusiodis for assistance with manuscript preparation.
The research described in this publication was funded in part with federal funds from the National Cancer Institute, National Institutes of Health, under contract no. NO1-CO-12400, and by the National Cancer Institute.
The content of this publication does not necessarily reflect the views or policies of the U.S. Department of Health and Human Services, nor does the mention of trade names, commercial products, or organizations imply endorsement by the U.S. Government.
REFERENCES
- 1.Bartz, S. R., and M. A. Vodicka. 1997. Production of high-titer human immunodeficiency virus type 1 pseudotyped with vesicular stomatitis virus glycoprotein. Methods 12:337-342. [DOI] [PubMed] [Google Scholar]
- 2.Buckman, J. S., W. J. Bosche, and R. J. Gorelick. 2003. Human immunodeficiency virus type 1 nucleocapsid Zn2+ fingers are required for efficient reverse transcription, initial integration processes, and protection of newly synthesized viral DNA. J. Virol. 77:1469-1480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bushman, F. D., and R. Craigie. 1991. Activities of human immunodeficiency virus (HIV) integration protein in vitro: specific cleavage and integration of HIV DNA. Proc. Natl. Acad. Sci. USA 88:1339-1343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Coffin, J. M., S. H. Hughes, and H. E. Varmus (ed.). 1997. Retroviruses. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y. [PubMed]
- 5.Dash, C., J. W. Rausch, and S. F. Le Grice. 2004. Using pyrrolo-deoxycytosine to probe RNA/DNA hybrids containing the human immunodeficiency virus type-1 3′ polypurine tract. Nucleic Acids Res. 32:1539-1547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Di Marzo Veronese, F., T. D. Copeland, A. L. DeVico, R. Rahman, S. Oroszlan, R. C. Gallo, and M. G. Sarngadharan. 1986. Characterization of highly immunogenic p66/p55 as the reverse transcriptase of HTLVIII/LAV. Science 231:1289-1291. [DOI] [PubMed] [Google Scholar]
- 7.Fedoroff, O. Y., M. Salazar, and B. R. Reid. 1993. Structure of a DNA:RNA hybrid duplex: why RNase H does not cleave pure RNA. J. Mol. Biol. 233:509-523. [DOI] [PubMed] [Google Scholar]
- 8.Furfine, E. S., and J. E. Reardon. 1991. Human immunodeficiency virus reverse transcriptase ribonuclease H: specificity of tRNA-lys-primer excision. Biochemistry 30:7041-7046. [DOI] [PubMed] [Google Scholar]
- 9.Furfine, E. S., and J. E. Reardon. 1991. Reverse-transcriptase-RNaseH from human immunodeficiency virus: relationship of the DNA polymerase and RNA hydrolysis activities. J. Biol. Chem. 266:406-412. [PubMed] [Google Scholar]
- 10.Gopalakrishnan, V., J. A. Peliska, and S. J. Benkovic. 1992. Human immunodeficiency virus type 1 reverse transcriptase: spatial and temporal relationship between the polymerase and RNase H activities. Proc. Natl. Acad. Sci. USA 89:10763-10767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Han, G. W., M. L. Kopka, D. Cascio, K. Grzeskowiak, and R. E. Dickerson. 1997. Structure of a DNA analog of the primer for HIV-1 RT second strand synthesis. J. Mol. Biol. 269:811-826. [DOI] [PubMed] [Google Scholar]
- 12.Horton, N. C., and B. C. Finzel. 1996. The structure of an RNA/DNA hybrid: a substrate of the ribonuclease activity of HIV-1 reverse transcriptase. J. Mol. Biol. 264:521-533. [DOI] [PubMed] [Google Scholar]
- 13.Jacobo-Molina, A., J. Ding, R. G. Nanni, A. D. Clark, Jr., X. Lu, C. Tantillo, R. L. Williams, G. Kamer, A. L. Ferris, P. Clark, A. Hizi, S. H. Hughes, and E. Arnold. 1993. Crystal structure of human immunodeficiency virus type 1 reverse transcriptase complexed with double-stranded DNA at 3.0Å resolution shows bent DNA. Proc. Natl. Acad. Sci. USA 90:6320-6324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Julias, J. G., A. L. Ferris, P. L. Boyer, and S. H. Hughes. 2001. Replication of phenotypically mixed human immunodeficiency virus type 1 virions containing catalytically active and catalytically inactive reverse transcriptase. J. Virol. 75:6537-6546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Julias, J. G., M. J. McWilliams, S. Sarafianos, E. Arnold, and S. H. Hughes. 2002. Mutations in the RNase H domain of HIV-1 reverse transcriptase affect the initiation of DNA synthesis and the specificity of RNase H cleavage in vivo. Proc. Natl. Acad. Sci. USA 99:9515-9520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kohlstaedt, L. A., J. Wang, J. M. Friedman, P. A. Rice, and T. A. Steitz. 1992. Crystal structure at 3.5 Å resolution of HIV-1 reverse transcriptase complexed with an inhibitor. Science 256:1783-1790. [DOI] [PubMed] [Google Scholar]
- 17.Kopka, M. L., L. Lavelle, G. W. Han, H. L. Ng, and R. E. Dickerson. 2003. An unusual sugar conformation in the structure of an RNA/DNA decamer of the polypurine tract may affect recognition by RNase H. J. Mol. Biol. 334:653-665. [DOI] [PubMed] [Google Scholar]
- 18.Kvaratskhelia, M., S. R. Budihas, and S. F. Le Grice. 2002. Pre-existing distortions in nucleic acid structure aid polypurine tract selection by HIV-1 reverse transcriptase. J. Biol. Chem. 277:16689-16696. [DOI] [PubMed] [Google Scholar]
- 19.Le Grice, S. F. J., T. Naas, B. Wohlgensinger, and O. Schatz. 1991. Subunit selective mutagenesis indicates minimal polymerase activity in heterodimer-associated p51 HIV-1 reverse transcriptase. EMBO J. 10:3905-3911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Li, L., J. M. Olvera, K. E. Yoder, R. S. Mitchell, S. L. Butler, M. Lieber, S. L. Martin, and F. D. Bushman. 2001. Role of the non-homologous DNA end joining pathway in the early steps of retroviral infection. EMBO J. 20:3273-3281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.McWilliams, M. J., J. G. Julias, S. G. Sarafianos, W. G. Alvord, E. Arnold, and S. H. Hughes. 2003. Mutations in the 5′ end of the human immunodeficiency virus type 1 polypurine tract affect RNase H cleavage specificity and virus titer. J. Virol. 77:11150-11157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Miller, M. D., C. M. Farnet, and F. D. Bushman. 1997. Human immunodeficiency virus type 1 preintegration complexes: studies of organization and composition. J. Virol. 71:5382-5390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Post, K., J. Guo, E. Kalman, T. Uchida, R. J. Crouch, and J. G. Levin. 1993. A large deletion in the connection subdomain of murine leukemia virus reverse transcriptase or replacement of the RNase H domain with Escherichia coli RNase H results in altered polymerase and RNase H activities. Biochemistry 32:5508-5517. [DOI] [PubMed] [Google Scholar]
- 24.Powell, M. D., and J. G. Levin. 1996. Sequence and structural determinants required for priming of plus-strand DNA synthesis by the human immunodeficiency virus type 1 polypurine tract. J. Virol. 70:5288-5296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Pullen, K. A., and J. J. Champoux. 1990. Plus-strand origin for human immunodeficiency virus type 1: implications for integration. J. Virol. 64:6274-6277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Pullen, K. A., L. K. Ishimoto, and J. J. Champoux. 1992. Incomplete removal of the RNA primer for minus-strand DNA synthesis by human immunodeficiency virus type 1 reverse transcriptase. J. Virol. 66:367-373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Pullen, K. A., A. J. Rattray, and J. J. Champoux. 1993. The sequence features important for plus strand priming by human immunodeficiency virus type 1 reverse transcriptase. J. Biol. Chem. 268:6221-6227. [PubMed] [Google Scholar]
- 28.Rattray, A. J., and J. J. Champoux. 1989. Plus-strand priming by Moloney murine leukemia virus. The sequence features important for cleavage by RNase H. J. Mol. Biol. 208:445-456. [DOI] [PubMed] [Google Scholar]
- 29.Rausch, J. W., D. Lener, J. T. Miller, J. G. Julias, S. H. Hughes, and S. F. Le Grice. 2002. Altering the RNase H primer grip of human immunodeficiency virus reverse transcriptase modifies cleavage specificity. Biochemistry 41:4856-4865. [DOI] [PubMed] [Google Scholar]
- 30.Repaske, R., J. W. Hartley, M. F. Kavlick, R. R. O'Neill, and J. B. Austin. 1989. Inhibition of RNase H activity and viral replication by single mutations in the 3′ region of Moloney murine leukemia virus reverse transcriptase. J. Virol. 63:1460-1464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sarafianos, S. G., K. Das, C. Tantillo, A. D. Clark, Jr., J. Ding, J. Whitcomb, M. Gait, P. L. Boyer, S. H. Hughes, and E. Arnold. 2001. Crystal structure of HIV-1 reverse transcriptase in complex with a polypurine tract RNA:DNA. EMBO J. 20:1449-1461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Schatz, O., F. V. Cromme, T. Naas, D. Lindemann, J. Mous, and S. F. J. Le Grice. 1990. Inactivation of the RNase H domain of HIV-1 reverse transcriptase blocks viral infectivity, p. 293-404. In T. S. Papas (ed.), Gene regulation and AIDS. Portfolio, Houston, Tex.
- 33.Smith, J. S., and M. Roth. 1992. Specificity of human immunodeficiency virus-1 reverse transcriptase-associated ribonuclease H in removal of the minus-strand primer, tRNA(Lys3). J. Biol. Chem. 267:15071-15079. [PubMed] [Google Scholar]
- 34.Telesnitsky, A., S. W. Blain, and S. P. Goff. 1992. Defects in Moloney murine leukemia virus replication caused by a reverse transcriptase mutation modeled on the structure of Escherichia coli ribonuclease H. J. Virol. 66:615-622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Tisdale, M., T. Schultze, B. A. Larder, and K. Moelling. 1991. Mutations within the RNase H domain of HIV-1 reverse transcriptase abolish viral infectivity. J. Gen. Virol. 72:59-66. [DOI] [PubMed] [Google Scholar]
- 36.Whitcomb, J. M., R. Kumar, and S. H. Hughes. 1990. Sequence of the circle junction of human immunodeficiency virus type 1: implications for reverse transcription and integration. J. Virol. 64:4903-4906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Whitcomb, J. M., and S. H. Hughes. 1992. Retroviral reverse transcription and integration: progress and problems. Annu. Rev. Cell Biol. 8:275-306. [DOI] [PubMed] [Google Scholar]
- 38.Wohrl, B. M., and K. Moelling. 1990. Interaction of HIV-1 ribonuclease H with polypurine tract containing RNA-DNA hybrids. Biochemistry 29:10141-10147. [DOI] [PubMed] [Google Scholar]
- 39.Yee, J. K., T. Friedmann, and J. C. Burns. 1994. Generation of high-titer pseudotyped retroviral vectors with very broad host range. Methods Cell Biol. 43:99-112. [DOI] [PubMed] [Google Scholar]