Abstract
Tomato spotted wilt virus (TSWV), a member of the Tospovirus genus within the Bunyaviridae, is an economically important plant pathogen with a worldwide distribution. TSWV is transmitted to plants via thrips (Thysanoptera: Thripidae), which transmit the virus in a persistent propagative manner. The envelope glycoproteins, GN and GC, are critical for the infection of thrips, but they are not required for the initial infection of plants. Thus, it is assumed that the envelope glycoproteins play important roles in the entry of TSWV into the insect midgut, the first site of infection. To directly test the hypothesis that GN plays a role in TSWV acquisition by thrips, we expressed and purified a soluble, recombinant form of the GN protein (GN-S). The expression of GN-S allowed us to examine the function of GN in the absence of other viral proteins. We detected specific binding to thrips midguts when purified GN-S was fed to thrips in an in vivo binding assay. The TSWV nucleocapsid protein and human cytomegalovirus glycoprotein B did not bind to thrips midguts, indicating that the GN-S-thrips midgut interaction is specific. TSWV acquisition inhibition assays revealed that thrips that were concomitantly fed purified TSWV and GN-S had reduced amounts of virus in their midguts compared to thrips that were fed TSWV only. Our findings that GN-S binds to larval thrips guts and decreases TSWV acquisition provide evidence that GN may serve as a viral ligand that mediates the attachment of TSWV to receptors displayed on the epithelial cells of the thrips midgut.
Tomato spotted wilt virus (TSWV) is the prototypic member of the genus Tospovirus within the family Bunyaviridae. TSWV is a prominent plant pathogen with a worldwide distribution and a large host range (reviewed in reference 56). The virus infects 732 species of plants in 102 families (http://www.oznet.ksu.edu/tospovirus/hostlist.html), resulting in enormous annual monetary losses due to crop damage and pesticide applications (11, 18).
The family Bunyaviridae is made up of the Tospovirus, Hantavirus, Nairovirus, Phlebovirus, and Bunyavirus genera. TSWV, like all viruses in the family Bunyaviridae, has a tripartite, negative-strand RNA genome. All of these viruses encode a nucleocapsid (N) protein on a small (S) RNA segment, two membrane glycoproteins on a medium (M) RNA segment, and a large (L) protein on a large RNA segment. The glycoproteins are derived from a polyprotein that is proteolytically processed to yield the two glycoproteins (GPs). The GPs are designated GN and GC based on their positions relative to the amino and carboxy termini of the polyprotein. For most of the members of the Bunyaviridae studied, the GN protein has a Golgi retention sequence and the GC possesses an endoplasmic reticulum retention sequence (3, 28, 39, 57). When the TSWV glycoproteins are expressed together, they colocalize to the Golgi, the site of virion formation (27, 28).
TSWV is transmitted by at least seven species of thrips (Thysanoptera: Thripidae) in a persistent, replicative manner (64). Frankliniella occidentalis (Pergande), the Western flower thrips, is an efficient vector of TSWV and has a wide plant host range and a global distribution (34). Thrips acquire the virus as first or early second instar larvae, but adult thrips that acquire the virus are unable to transmit it (42, 62, 65). The insects ingest the virus, and the virus enters the midgut epithelial cells, where it replicates and spreads to surrounding muscle cells (12, 42, 62). Eventually, TSWV infects the salivary glands, enabling adult insects to transmit the virus for the duration of their lives (63, 68).
The hypothesis that TSWV acquisition involves a thrips midgut receptor(s) that binds the virus GPs is supported by several observations. First, the TSWV GPs are necessary for thrips acquisition but not for plant infection. Serial, mechanical inoculations of TSWV between plants lead to envelope-deficient mutants that have deletions and point mutations in the sequences encoding the GPs. These mutants are no longer transmissible by thrips, but they are not compromised in their ability to infect plants (41, 48). Second, anti-idiotypic antibodies that mimic the GPs specifically label the midgut, the expected location of the cellular receptor (5). Third, by analogy to other members of the Bunyaviridae, the GP-thrips receptor hypothesis is consistent with the role of GPs in the acquisition of bunyaviruses by arthropod vectors (37, 38, 58).
Several lines of evidence indicate that GN may serve as a viral attachment and/or entry protein. The RGD motif of GN is intriguing because this motif is known to interact with β-integrins on cell surfaces (47, 59). Several viruses have been shown to bind β-integrin receptors via RGD motifs in the context of their viral attachment proteins (2, 14, 50). Moreover, hantaviruses use integrins as receptors (15, 16). Research with La Crosse virus, another member of the Bunyaviridae, provides insight into the possible TSWV GN participation in virus entry. When La Crosse virions were subjected to a protease treatment, GC was cleaved but GN remained intact. The protease-treated virions exhibited increased binding to the insect vector midgut; however, they exhibited reduced binding to cultured mosquito and mammalian cells (37, 38). These results indicate that La Crosse GN may mediate attachment to insect midguts while GC may play a role in cell-to-cell spread in mammals and insects.
To determine the role(s) of GN in binding to thrips guts, we expressed and purified a soluble recombinant form of GN. Because GN is an integral membrane protein, we expressed the ectodomain of GN from a recombinant baculovirus in SF21 cells, thus creating a protein that was soluble in the absence of detergents (52). Soluble recombinant proteins are essential for functional studies with living organisms and cells in which membrane integrity is imperative for determinations of glycoprotein function. By expressing GN individually, we examined its role in virus binding and entry in the absence of other viral proteins. Here we report the first high-level expression and characterization of a soluble glycoprotein encoded by a member of the Tospovirus genus. We have characterized the truncated form of GN (GN-S) and found that it is soluble and recognized by monoclonal antibodies (MAbs) generated against wild-type GN. A comparison of TSWV GN and GN-S revealed that both proteins contain O-linked glycans and form dimers. We provide evidence that GN-S binds larval midguts and inhibits TSWV acquisition in a manner consistent with GN participation in virus binding and/or entry.
MATERIALS AND METHODS
Cells, insects, and virus.
Spodoptera frugiperda cells (SF21) were grown in IPL41 medium (Gibco-BRL) supplemented with 10% fetal calf serum (Gibco-BRL), 2.6 g of tryptose broth (Sigma)/liter, and 1% penicillin-streptomycin-amphotericin B (Gibco-BRL). A colony of F. occidentalis was maintained on green bean pods (Phaseolus vulgaris) as previously described (62). TSWV (isolate TSWV-L) was maintained by thrips transmission as described previously (62, 63), and the virus was mechanically transferred one time after thrips transmission, which did not affect the thrips transmissibility of the isolate. Infected leaves used for virus purification were harvested just prior to maximal symptom expression (at approximately 2 weeks postinoculation), and TSWV virions were isolated by the procedures of Gonsalves and Truijillo (19).
Sequence analysis.
The GN/GC open reading frame (ORF) encodes an 1,135-amino-acid polypeptide that is cleaved to generate the two glycoproteins. HMMTOP (60, 61), Tmpred (23), and PHDhtm (53, 54) were used to predict hydrophobic and transmembrane domains of GN. SignalP was used to identify a putative signal sequence and signal peptidase cleavage sites (44), Prosite was used to identify N-linked glycosylation sites and the lectin-like domain on the protein (13, 22), and NetOGlyc 2.0 (http://www.cbs.dtu.dk/services/NetOGlyc-2.0/) was used to predict O-linked N-acetylgalactosamine glycosylation sites.
Construction of a recombinant baculovirus encoding a soluble form of GN.
We PCR amplified the ectodomain of GN from pGF7, a plasmid containing the GN/GC ORF (1). Two transmembrane domains were consistently identified in the GN portion of the ORF by the prediction methods described above, and the GN-S construct was designed to exclude the putative signal sequence, the transmembrane domains, and the adjacent cytoplasmic tail. The forward primer used to generate the GN-S (amino acids 35 to 309) polypeptide started at nucleotide 109 of the ORF (5′ GTCATGAGCTCGGTAGAGATAATTCGTGGAGACCAT 3′), and the reverse primer started at nucleotide 946 (5′ ACTCAGCGGCCGCGGCTGTTTGTTTATAAATGCT 3′). The 5′ primer contained a recognition site for SacI, and the 3′ primer contained a recognition site for NotI (underlined). We used MasterAmp DNA polymerase (Epicentre) with PreMix 4 for PCRs. The PCR amplification protocol consisted of three cycles of denaturation at 94°C for 60 s, annealing at 50°C for 60 s, and extension at 72°C for 90 s. The next 40 cycles followed the same protocol except that the annealing temperature was increased to 55°C. The expected 0.9-kb product was cloned into the pBacgus-3 baculovirus transfer plasmid (Novagen, Madison, Wis.). The PCR product and the transfer plasmid were sequentially cut with NotI and SacI. The PCR product was ligated into the pBacgus-3 plasmid in frame with the GP64 signal sequence and a six-His tag and was transformed into Escherichia coli strain DH5α. The transformants were analyzed by diagnostic restriction digestion and DNA sequence analysis. The transfer plasmid DNA was prepared according to the manufacturer's instructions (Novagen). Baculovirus DNA (BacVector-1000; Novagen) and transfer plasmid DNA were cotransfected into SF21 cells. Cells containing recombinant viruses were visualized by staining with X-Gluc (5-bromo-4-chloro-3-indoyl-β-d-glucuronide). Recombinant viruses were subjected to three rounds of plaque purification, and high-titer virus stocks were made according to the manufacturer's instructions. Three recombinant viruses were screened for protein production by Western blot analysis using MAbs to GN (1) and the six-His tag (Invitrogen). To characterize the expression of GN-S, we harvested the cell pellets and supernatants of baculovirus-infected SF21 cells at 0, 24, 48, 72, and 96 h postinfection and analyzed the samples by Western blotting. For protein expression, SF21 cells were infected at a multiplicity of infection of 5 to 10, and the cell culture medium was harvested at 72 h postinfection.
Protein purification.
Protein purification was performed as described by Lopper and Compton (36), with a few modifications. The medium was harvested and the GN-S protein was purified from the cell-free supernatant. The medium was supplemented with a cocktail of protease inhibitors (2 μg each of antipain, aprotinin, chymostatin, leupeptin, and pepstatin/ml) and dialyzed against phosphate-buffered saline (PBS), pH 7.4. The resulting dialysate was incubated with nickel resin (Qiagen) by a batch procedure. After batch binding, the resin was poured into a column, and subsequent steps were performed according to a column procedure. The column was first washed with 2 bed volumes of a low-pH buffer (50 mM sodium phosphate, 10% glycerol, pH 6.0) and subsequently washed with 30 bed volumes of 10 mM imidazole (50 mM sodium phosphate, 0.5 M sodium chloride, 10% glycerol, pH 7.0) and 5 bed volumes of 50 mM imidazole. GN-S was eluted with 200 mM imidazole, dialyzed against PBS-10% glycerol, and stored in aliquots at −80°C.
SDS-PAGE, Western blots, and immunoprecipitations.
To monitor protein expression, glycosylation, and dimerization, we separated the proteins by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) in 10% polyacrylamide gels and analyzed them by Coomassie brilliant blue staining or Western blotting. For Western blot analysis, polyacrylamide gels were electrophoretically transferred to Hybond-C Extra membranes (Amersham) in transfer buffer (48 mM Tris, 39 mM glycine, 20% methanol, and 0.037% SDS). The membranes were blocked with 5% nonfat dry milk and then incubated with a GN MAb used at a 1:2,000 dilution (1, 5) or a six-His MAb (Clontech) diluted 1:7,500 in PBS-Tween 20 and 5% nonfat dry milk. Western blots were visualized with horseradish peroxidase-conjugated goat anti-mouse immunoglobulin G and ECLplus (Amersham).
To determine if the GN MAb recognized GN-S under native conditions, we performed immunoprecipitation by using a Seize X protein A IP kit (Pierce) according to the manufacturer's instructions. Briefly, anti-GN or -GC (500 μg) was incubated with immobilized protein A gel for 1 h and then covalently bound by the addition of disuccinimidyl suberate. Affinity-purified GN-S (0.02 mg) was incubated with the cross-linked antibody overnight at 4°C. The mixture was washed five times with BupH (0.14 M sodium chloride, 0.008 M sodium phosphate, 0.002 M potassium phosphate, and 0.01 M potassium chloride, pH 7.4) and then eluted with a low-pH elution buffer (Pierce). The fractions were analyzed by Western blotting.
Analysis of glycosylation.
For comparative analyses of GN and GN-S glycosylation, purified GN-S or TSWV virions were deglycosylated with enzymes to remove N-linked glycans (N-glycosidase F) and/or O-linked glycans (endo-α-N-acetylgalactosaminidase, α-2,3,6,8,9-neuraminidase, β-1,4-galactosidase, and β-N-acetylglucosaminidase). The proteins were denatured with 0.1% SDS and 50 mM β-mercaptoethanol (β-ME) at 100°C for 5 min. After heating, Triton X-100 was added to 0.75%, and then glycosidases were added. To assay for the addition of fucoses that were α-1,3-linked to N-acetylglucosamine, we incubated the proteins with N-glycosidase A (Calbiochem). Purified TSWV or GN-S was incubated with 1 U of N-glycosidase A in a solution containing 10 mM sodium acetate, 0.5 M sodium isothiocyanate, and 0.1 β-ME at pH 5.2. The proteins were incubated for a minimum of 3 h at 37°C. Protein deglycosylation was evaluated by observing protein mobility shifts when the proteins were analyzed by Western blotting.
Analysis of dimerization.
To determine if GN and GN-S exist as dimers, we added increasing concentrations of the reducing agent β-ME (concentrations ranged from 0 to 5%) to the gel loading buffer (62.5 mM Tris-HCl [pH 6.8], 2% SDS, 10% glycerol, 0.005% bromophenol blue). GN-S or freshly purified TSWV was mixed with the gel loading buffer and boiled for 5 min. Protein dimerization was analyzed by Western blotting.
In vivo binding assay.
An insect feeding assay was developed to determine if GN-S binds to thrips guts. First instar larval thrips were fed a solution of protein mixed with buffer TF (PBS, 10% glycerol, 0.01% Chicago sky blue, and 5 mg of bovine serum albumin [BSA]/ml) through a layer of Parafilm. Thrips were fed in cylindrical 25-mm-diameter containers similar to the method described by Hunter et al. (24). Immunolabeling treatments were as follows: (i) TF buffer alone, (ii) TF buffer and 0.1 nM GN-S, (iii) TF buffer and partially purified TSWV, (iv) TF buffer and 0.1 nM human cytomegalovirus (HCMV) gB protein tagged with a six-His tag (a soluble form of the gB viral attachment protein, expressed from a baculovirus and purified by the same method as GN-S), and (v) TF buffer and 0.2 nM TSWV nucleocapsid (N) protein tagged with a six-His tag (49). Thrips were allowed to feed for 2 h, and insects that ingested the feeding solution, as indicated by blue guts, were transferred to another feeding chamber containing a 7% sucrose solution. After 2 h, the midguts no longer contained visible amounts of the blue feeding solution, and these insect guts are hereafter referred to as cleared guts. Thrips were then dissected in insect physiological saline (150 mM NaCl, 2 mM NaHCO3, 2 mM MgCl2, 2 mM CaCl2, 2 mM KCl, and 20 mM C6H12O6) and fixed in 4% paraformaldehyde in 50 mM sodium phosphate buffer, pH 7.0, overnight at 4°C. The thrips were washed twice and permeabilized with 0.5% Triton X-100 for 30 min, after which the guts were blocked with 20% normal goat serum (NGS) in PBS. Insects that fed on purified virus were treated with GN MAb at a 1:20 dilution and then washed five times with PBS. MAb binding was detected with a fluorescein isothiocyanate-conjugated secondary antibody (1:100). Alternatively, thrips that were fed six-His-tagged proteins were labeled with Penta · His Alexa fluor 488 (Qiagen) diluted to 6 μg/ml in PBS-20% NGS. Actin was stained with Texas red phalloidin (Molecular Probes) to delineate cell boundaries and tissue types. The dissected insects were mounted in the Slow Fade Light reagent (Molecular Probes) and viewed with a Bio-Rad 1024 laser scanning confocal microscope. Images were collected by use of the same laser power and gain. The in vivo binding assay was repeated six times.
Inhibition of TSWV acquisition.
An assay was performed to determine if GN-S inhibits TSWV acquisition, with two types of experiments being performed. Experiment A treatments included buffer (n = 8), TSWV (n = 9), and TSWV and GN-S (n = 14). Experiment B treatments included buffer (n = 8), TSWV (n = 10), TSWV and GN-S (n = 12), and gB and TSWV (n = 16). The gB and TSWV treatment was included to test the specificity of TSWV acquisition inhibition by GN-S. Experiment A was conducted three times, and experiment B was conducted twice. For each treatment, a group of thrips were subjected to the assay and each gut served as a subsample of the group.
The feeding solutions contained 50 μl of sodium sulfite or TSWV in sodium sulfite, 10 μl (10 mg/ml) of BSA, 0.1% Chicago sky blue, 20 μl of 20% sucrose solution, and 50 μl of PBS-10% glycerol or 50 μl of GN (0.1 nM) or gB (0.1 nM) protein in PBS-10% glycerol. Thrips were fed, cleared, dissected, and fixed as described above for the in vivo binding assay. After being blocked, the guts were treated with a polyclonal antibody to the TSWV N protein at a 1:50 dilution in PBS-20% NGS for 2 h at room temperature (RT). The dissected insects were washed five times with PBS. Subsequently, the guts were incubated with Alexa fluor 647-conjugated anti-rabbit immunoglobulin G (Molecular Probes) diluted 1:50 in PBS-20% NGS for 1 h at RT. The guts were washed five times with PBS and then incubated with Texas red phalloidin (Molecular Probes) diluted 1:200 in PBS for 1 h at RT. The guts were washed six times with PBS and mounted in antifade solution (Molecular Probes). Images were collected with a Bio-Rad 1024 laser scanning confocal microscope. Images were collected by use of the same microscope settings (i.e., laser power and gain) for all treatments within each experimental repeat. Images collected were all of the same size and magnification and included the anterior region of the midgut and portions of the posterior midgut.
Image analyses were performed with Adobe Photoshop (v. 7.0) to quantify the amounts of virus in insect midguts. The average amount of fluorescence (intensity of fluorescent pixels/total number of pixels in the 512-by-512 pixel image) over the surface of the captured image was optically measured in the blue channel for each midgut, which represented a subsample within each treatment. The average fluorescence for each treatment was calculated. Each experimental repeat was considered a treatment replicate (i.e., there were three and two replicates for experiments A and B, respectively). With Minitab (v. 13.31) software, analysis of variance was performed on the average fluorescence to determine the treatment effects on virus acquisition separately for each experiment. Fisher's least significant differences were calculated to make pairwise comparisons between treatment means.
RESULTS
Sequence analysis.
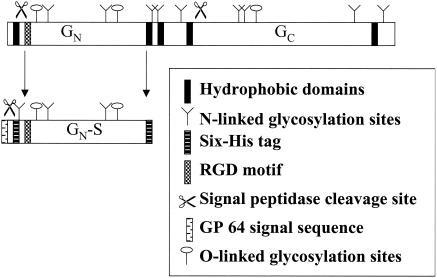
We used several protein sequence analysis tools to identify posttranslational modifications, hydrophobic domains, and motifs of the GN/GC polyprotein. The analyses revealed a putative signal sequence at the N terminus of GN, a signal peptidase cleavage site at amino acid 35, nine possible N-glycosylation sites in the polyprotein, and possible O-linked N-acetylgalactosamine glycosylation sites at amino acids 58 and 209. Another signal peptidase site is predicted to occur at amino acid 464, and if this site is cleaved by a signal peptidase, this would likely be the cleavage that generates the GN and GC proteins from the polyprotein. We also identified a region of GN from amino acids 132 to 231 that resembles a lectin-like domain. Schematics of the recombinant truncated GN-S protein and the GN/GC polyprotein are shown in Fig. 1.
FIG. 1.
Schematic of the TSWV glycoprotein ORF and of soluble, truncated GN (GN-S). The top figure represents the precursor polyprotein, with putative signal sequences, signal peptidase cleavages sites, N- and O-linked glycosylation sites, and transmembrane domains indicated. The bottom figure is a schematic of GN-S, from amino acids 35 to 309, expressed from a baculovirus. Note that the putative hydrophobic domains were removed and six-His tags were added. The figure is not drawn to scale.
The GN soluble polypeptide begins at amino acid 35 after the first hydrophobic domain and the putative signal peptidase cleavage site. To ensure the faithful translation and secretion of GN in the baculovirus system, we deleted the predicted GN signal sequence and replaced it with the signal sequence of the major baculovirus glycoprotein, GP64 (8). We also deleted the putative membrane-spanning domain and cytoplasmic tail so that the recombinant protein would be secreted. The expressed polypeptide continues to amino acid 309, where the predicted transmembrane region begins.
Expression and purification of GN-S.
To characterize the expression of GN-S, we performed a time course experiment and determined by Western blot analysis that maximal GN-S was expressed at approximately 72 h postinfection (data not shown). GN-S was secreted into the medium (Fig. 2, lane 2). The Coomassie blue-stained gel shows that the cell culture supernatant (Fig. 2, lane 3) was heavily stained due to the presence of 10% fetal bovine serum in the medium. GN-S was purified from the cell culture supernatant (Fig. 2, lanes 4 and 5) with a yield of approximately 5 mg/liter.
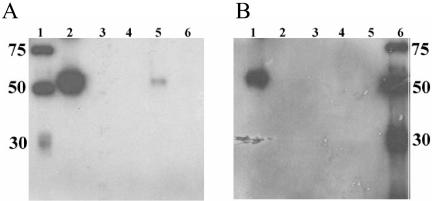
FIG. 2.
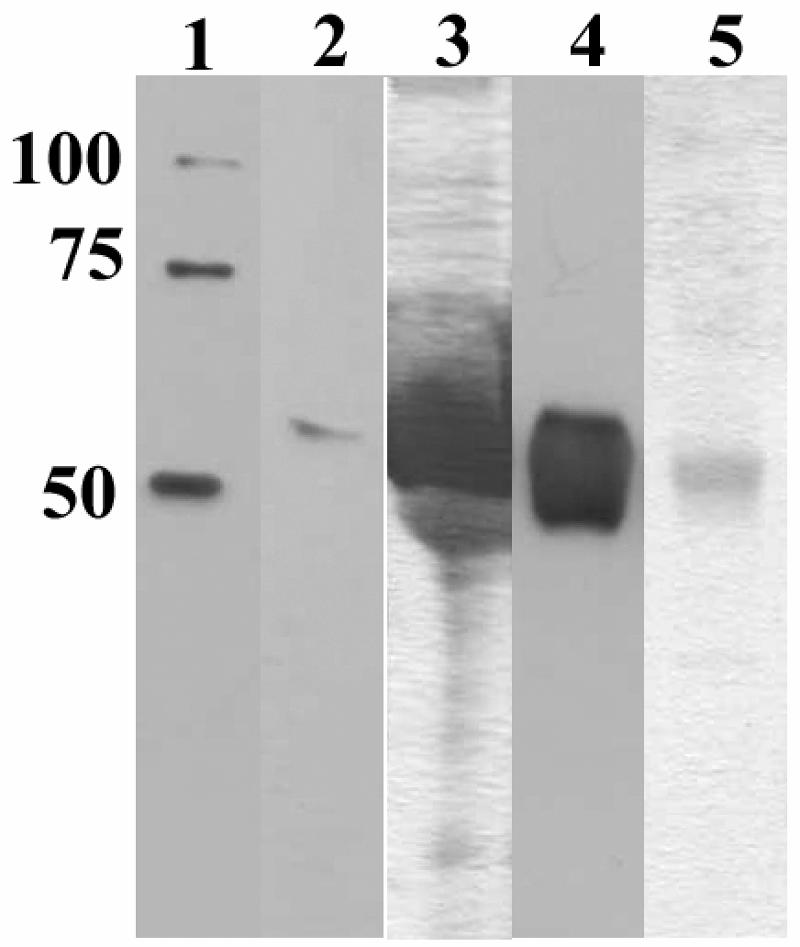
Purification of soluble GN (GN-S) by nickel affinity chromatography. Culture supernatants were harvested at 72 h postinfection, purified, and dialyzed against PBS. The samples were analyzed by SDS-PAGE, one gel was stained with Coomassie brilliant blue (lanes 3 and 5), and another gel was analyzed by Western blotting (lanes 1, 2, and 4). Equal volumes were added to each well. Lane 1, six-His-tag molecular weight marker; lanes 2 and 3, cell culture medium added to column; and lanes 4 and 5, GN-S protein eluate from the 200 mM imidazole wash.
GN MAb recognizes soluble GN-S.
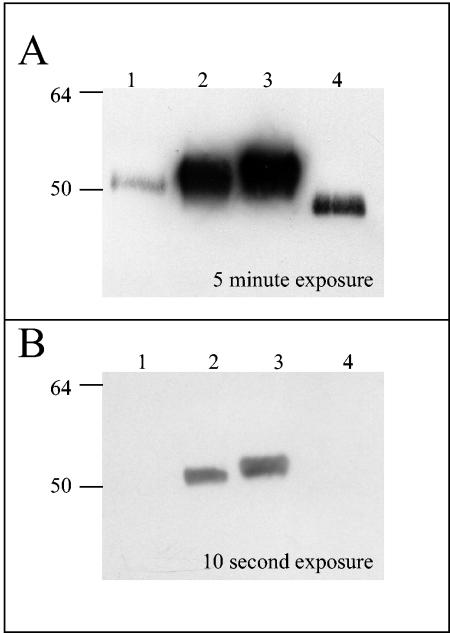
To ensure that a GN MAb generated against wild-type GN recognized GN-S, we attempted to immunoprecipitate GN-S with a GN MAb. GN-S bound to the cross-linked antibody and specifically eluted at a low pH (Fig. 3A, lane 5). As a control for nonspecific binding, anti-GC was also used, and no GN protein was eluted from this column (Fig. 3B, lanes 4 and 5). The GN MAb was able to bind GN-S under native and reducing (data not shown) conditions, suggesting that the GN epitope is a linear epitope and is accessible on the native protein. Furthermore, the GN MAb recognized the GN ectodomain and could be used to detect GN-S under nondenaturing conditions.
FIG. 3.
Immunoprecipitation of soluble GN (GN-S) with GN MAb. Antibodies were cross-linked to protein A gel, poured into a column, and incubated with GN-S. The columns were washed extensively, and the protein was eluted with a low-pH buffer. Fractions were analyzed by SDS-PAGE, and Western blots were probed with a six-His MAb. (A) GN-S incubated with GN MAb column. Lane 1, six-His marker; lane 2, GN-S added to the column; lanes 3 and 4, washes 1 and 5, respectively; lane 5, eluant 1; and lane 6, eluant 2. (B) GN-S incubated with GC MAb column. Lane 1, GN-S added to the column; lanes 2 and 3, washes 1 and 5, respectively; lane 4, eluant 1; lane 5, eluant 2; and lane 6, six-His molecular weight marker.
Analysis of GN glycosylation.
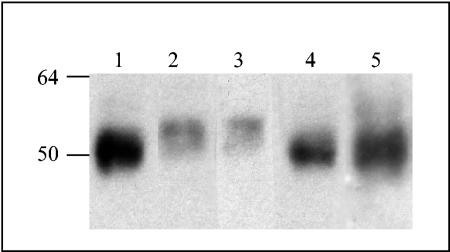
Enzymatic deglycosylation has been used as a tool to demonstrate the presence of carbohydrates on bunyavirus and tospovirus glycoproteins (43, 46). Because glycosylation can play an important role in protein stability and/or function, we compared GN and GN-S glycosylation. We incubated purified proteins and the virus with enzymes to specifically remove N- or O-linked glycans. Endoglycosidase F was used to remove N-linked glycans, and a cocktail of enzymes was used to remove O-linked glycans. Both GN and GN-S exhibited an increased mobility when glycans were removed. We consistently observed a small shift in protein mobility when the virus was incubated with O-glycosidases (Fig. 4, compare lanes 1 and 3), indicating that GN is modified by the addition of O-linked glycans. We observed no change in the mass (Fig. 4, compare lanes 2 and 3) when TSWV virion GN was incubated with enzymes to remove N-linked glycans. When the N- and O-glycosidases were used simultaneously (Fig. 4, lane 4), the shift in mobility was similar to the shift observed when just O-linked glycans were removed (Fig. 4, compare lanes 1 and 4). These results indicate that TSWV GN purified from infected plants is modified by the addition of O-linked glycans but not N-linked glycans. We did not detect fucose α-1,3-linked glycans on wild-type GN, as no additional shift was observed when N-glycosidase A was added (Fig. 4, lane 5) or when the protein was incubated with N-glycosidase A only (data not shown).
FIG. 4.
Analysis of wild-type GN glycosylation. Purified TSWV was incubated with glycosidases to remove oligosaccharides and then separated by SDS-PAGE. The proteins were transferred to a nitrocellulose membrane, and GN was detected with a GN MAb. GN was incubated with enzymes to remove the oligosaccharides. Lane 1, O-linked glycans; lane 2, N-linked glycans; lane 3, mock digestion, no glycosidases; lane 4, N- and O-linked glycans; and lane 5, endoglycosidase A and N- and O-linked glycans.
As observed with TSWV GN, the removal of O-linked glycans resulted in an increased mobility for GN-S, indicating that O-linked glycans comprise approximately 4 kDa of the molecular mass of GN-S (Fig. 5A, compare lanes 1 and 3). We found that GN-S was also N-glycosylated, and the removal of N-linked glycans from GN-S resulted in a 3-kDa shift in mobility (Fig. 5A and B, compare lanes 2 and 3). To ensure that the shift in mobility that we observed when GN-S was incubated with O-glycosidases was not due to the removal of N-linked glycans, we simultaneously subjected the proteins to both N- and O-glycosidases and observed that the shift in mobility was consistent with both the N- and O-linked glycans being removed (Fig. 5A, lane 4). Approximately 7 kDa of the molecular mass of GN-S was composed of N- and O-linked glycans, confirming that GN-S is N- and O-glycosylated.
FIG. 5.
Analysis of soluble GN (GN-S) glycosylation. Purified GN-S was incubated with glycosidases to remove oligosaccharides and then separated by SDS-PAGE. The proteins were transferred to nitrocellulose membranes, and GN-S was detected with a six-His MAb. (A) GN-S incubated with enzymes to remove oligosaccharides. Lane 1, O-linked glycans; lane 2, N-linked glycans; lane 3, mock digestion, no glycosidases; and lane 4, N- and O-linked glycans. (B) Short exposure of panel A showing the differences in size of GN-S incubated with enzymes to remove N-linked glycans (lane 2) and with no enzymes for a mock digestion (lane 3).
Analysis of dimerization.
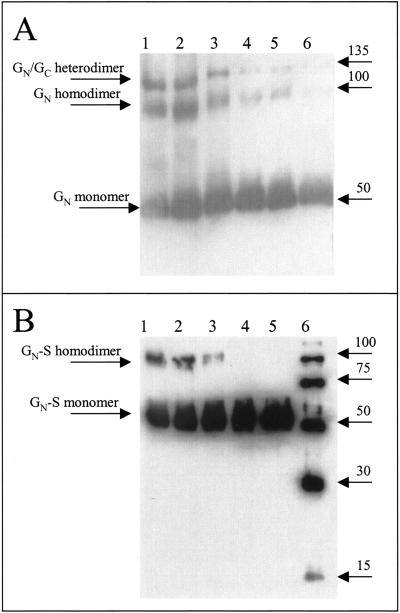
Viral envelope glycoproteins are generally found as oligomers such as dimers, trimers, and tetramers. In many instances, these higher-order structures are disulfide bond dependent. We analyzed GN and GN-S electrophoretic mobilities by SDS-PAGE under nonreducing and reducing conditions to determine if they formed oligomers. When TSWV purified from infected plants was subjected to SDS-PAGE with increasing amounts of β-ME and analyzed by Western blotting with a GN MAb, we found that GN existed as monomers, SDS-resistant homodimers, and heterodimers with GC (Fig. 6A, lanes 1 and 2). The 135-kDa band was confirmed to be GN-GC heterodimers by probing the blot with a GC MAb and observing the same band (data not shown). When the amount of β-ME in the sample loading buffer was increased, the disulfide bonds were reduced and the dimers resolved into monomers (Fig. 6A, lane 6). GN-S was also observed as a 50-kDa monomer and a 100-kDa dimer under nonreducing conditions (Fig. 6B, lanes 1 and 2). The addition of 0.1% β-ME to the sample loading buffer caused dimers of GN to partially shift to monomers (Fig. 6B, lane 3), and β-ME concentrations of 1% (Fig. 6B, lane 4) and 2.5% (Fig. 6B, lane 5) reduced all of the dimers to monomers. Because GN-S was still capable of forming dimers, we concluded that the domains involved in dimerization are located in the ectodomain.
FIG. 6.
Analysis of GN and GN-S dimerization. Increasing amounts of β-ME were added to TSWV purified from infected plants (GN) or to GN-S. Proteins were detected by Western blotting. (A) Purified TSWV detected with GN MAb. Lane 1, no β-ME and sample was not boiled; lane 2, no β-ME; lane 3, 0.1% β-ME; lane 4, 1.0% β-ME; lane 5, 2.5% β-ME; and lane 6, 5% β-ME. (B) Purified GN-S protein was detected with a six-His MAb. Lane 1, no β-ME and sample was not boiled; lane 2, no β-ME; lane 3, 0.1% β-ME; lane 4, 1.0% β-ME; lane 5, 2.5% β-ME; and lane 6, six-His-tagged molecular weight marker.
In vivo binding assay.
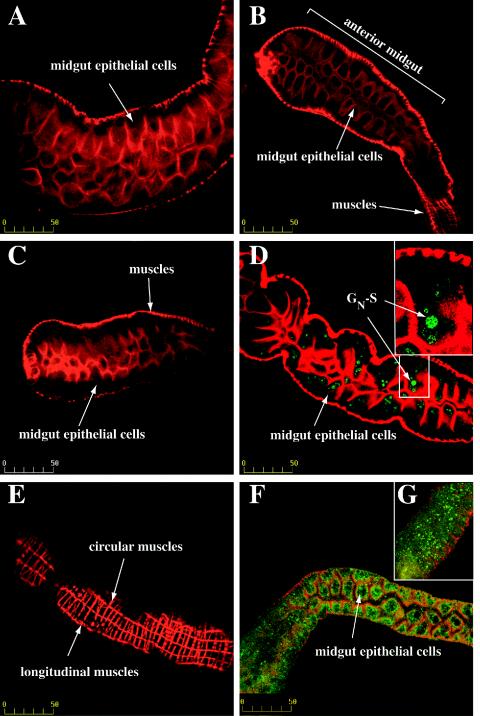
To examine the biological activity of GN-S, we assayed GN-S for the ability to bind to thrips guts. Thrips were fed purified protein and then cleared so that only proteins that were retained in the midgut were detected. We focused our study on the thrips midgut because it is the site of virus entry (63). The thrips midgut consists of a single layer of epithelial cells that is surrounded by longitudinal and circular muscle cells (40). Midgut muscle and epithelial cells had distinct labeling patterns with Texas red phalloidin, which allowed us to identify the tissues that we were optically sectioning in the gut. Thrips that were fed a BSA-buffer solution (Fig. 7A) did not become labeled with antibody. For an examination of the specificity of the GN-S gut interaction, thrips were fed a TSWV nucleocapsid (N) protein that was expressed in bacteria and purified via a six-His tag. The N protein did not label thrips guts (Fig. 7B). Another protein, glycoprotein B (gB) of HCMV, was fed to thrips to examine the specificity of protein-thrips midgut binding. The gB protein also failed to bind larval thrips midguts (Fig. 7C). GN-S (Fig. 7D) consistently bound to thrips midgut epithelial cells. We observed GN-S labeling (Fig. 7D) associated with the midgut epithelial cell layer and not with the muscle cells surrounding the midgut (Fig. 7E). Because GN-S bound to thrips guts and HCMV gB and TSWV N did not, we believe that there is a specific interaction between GN-S and a thrips midgut molecule.
FIG. 7.
In vivo binding assay. Larval thrips were fed BSA, TSWV N protein, HCMV glycoprotein gB, soluble GN (GN-S), or purified TSWV. After the feeding, thrips guts were cleared for 2 h in a 7% sucrose solution. Thrips were then dissected, fixed in 4% paraformaldehyde, and permeabilized. The guts were immunolabeled with a six-His MAb conjugated to Alexa fluor 488 (green), except for panels F and G, for which the samples were labeled with a GN MAb and a fluorescein isothiocyanate-conjugated goat anti-mouse antibody. Actin was stained with Texas red phalloidin (red). Staining was visualized by confocal microscopy. (A) Thrips fed BSA; (B) thrips fed six-His-tagged nucleocapsid (N) protein; (C) thrips fed purified, six-His-tagged HCMV gB protein; (D) thrips fed GN-S; (E) exterior of a gut from a thrips that was fed GN-S showing that labeling was associated with midgut epithelial cell layers and not with other tissues; (F) thrips fed purified TSWV; and (G) thrips fed purified GN-S. Bar, 50 μm.
For a comparison of GN-S binding and TSWV GN binding, thrips were fed the virus or GN-S and were immunolabeled with a GN MAb. The virus accumulated in the midgut epithelial cells of thrips that were fed TSWV (Fig. 7F), and a similar localization was observed for thrips that were fed GN-S (Fig. 7G). The in vivo binding experiment was repeated six times, and in all experiments we observed GN-S binding to thrips guts.
GN-S inhibition of TSWV acquisition by thrips.
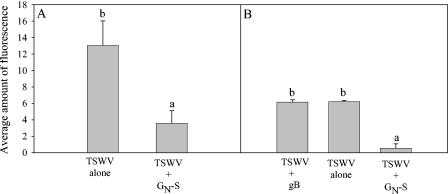
We further characterized the interaction between GN-S and thrips by testing GN-S for the ability to inhibit TSWV acquisition. The insects used for this experiment were from the same population and were 0 to 24 h old. All thrips chosen for experiments contained blue dye, indicating that they had fed on the virus solution. Acquisition was assessed by immunolabeling with an antibody to the TSWV N protein and was quantified by image analysis. Virus acquisition was reduced 4-fold (P = 0.009) and 12-fold (P = 0.003) by GN-S in experiments A and B, respectively (Fig. 8). In experiment B, HCMV gB, which did not bind to midguts in our in vivo binding assay (Fig. 7C), did not inhibit TSWV acquisition (Fig. 8B). These results indicate that GN-S inhibited TSWV acquisition and that another viral envelope protein, gB, did not inhibit TSWV acquisition by thrips.
FIG. 8.
Effect in vivo of purified, recombinant TSWV GN (GN-S) on thrips acquisition of TSWV in feeding experiments. Thrips were given 2-h acquisition access periods to BSA, TSWV alone, TSWV plus GN-S, and TSWV plus gB. All treatments contained the same concentrations of virus and/or buffers. Thrips were then allowed to feed on a sucrose solution to clear their guts. Acquisition was measured by immunolabeling with a TSWV nucleocapsid polyclonal antibody. The amount of immunolabeled TSWV was quantified by measuring the average amount of fluorescence (647 nm) in an optical section of a thrips gut, using Adobe Photoshop 7.0. Each bar represents a mean of three or two replicates for experiment, A or B, respectively. Bars headed by different letters are significantly different, with P values of <0.05. (A) Thrips were fed BSA in buffer, TSWV, and a combination of TSWV and GN-S. In a second set of experiments (B), thrips were also fed recombinant HCMV gB and TSWV, which served as another negative control. Thrips guts were imaged with a laser scanning confocal microscope.
DISCUSSION
Our findings that GN-S binds larval thrips guts and that it inhibits TSWV acquisition provide evidence that GN plays a role in virus binding and/or entry into the vector midgut. Recombinant soluble forms of viral envelope proteins have proven to be powerful tools for elucidating virus-host interactions (9, 32, 33, 66). By making a soluble form of the GN protein, we were able to use this protein in experiments that would not be feasible with wild-type purified GN because it is insoluble in the absence of detergents. Our data revealed that GN-S could bind the midgut epithelial cells of larval thrips without assistance from other TSWV proteins. The specificity of the GN-S-thrips interaction was supported by the failure of another structural TSWV protein, N, to bind thrips midgut epithelial cells. Furthermore, we demonstrated that the viral attachment protein, glycoprotein B, from another enveloped virus, HCMV, did not bind to thrips guts or inhibit TSWV acquisition.
Our observations regarding the tissue-specific localization and binding of GN to thrips gut tissue are supported by those of other studies with TSWV and animal-infecting members of the family Bunyaviridae. We found that both GN-S and TSWV are present in the midgut epithelia 2 to 4 h after feeding, which is consistent with studies that report the presence of TSWV in the anterior region of the midgut epithelia 16 h after acquisition access (12, 42, 62, 63). Immunolabeling experiments with anti-idiotypic GN and GC showed that anti-idiotypic GPs bound larval thrips guts (5). In support of a GN-vector interaction, researchers found that after the enzymatic removal of GC, and not GN, La Crosse virions exhibited an increased ability to bind mosquito midguts (37, 38). This finding highlights the importance of GN in virus binding to vector midguts. Furthermore, a sequence analysis of isolates of La Crosse virus with different passage histories revealed that the GN coding sequence is more stable than the GC coding sequence (7). The binding role of GN was further strengthened by the discovery of neutralizing antibodies to Hantaan virus GPs (29, 35).
Because GN-S bound to thrips guts and inhibited TSWV acquisition, it is likely that GN binding to the thrips midgut inhibited TSWV binding or entry. We consistently observed an inhibition of TSWV acquisition by GN-S, but there was variability in the levels of inhibition. This variation was likely attributable to differences between individual virus preparations. TSWV is a labile virus; therefore, it was necessary to purify a fresh batch of virus for each experiment. For experiment A, we observed higher acquisition levels for both TSWV alone and the TSWV and GN-S treatments, while for experiment B we observed lower acquisition levels for all treatments. The inhibition results with GN-S and TSWV are supported by the results of research with Rice ragged stunt virus, which is transmitted by rice brown planthoppers (20). In those experiments, the viral spike protein inhibited virus transmission and insects fed a nonstructural virus protein exhibited no transmission inhibition. These results support the finding that GN-S inhibited TSWV acquisition and the concept of disrupting the insect-mediated transmission of viruses via viral attachment proteins. The finding that GN-S can inhibit TSWV entry is the first step towards developing new control strategies for TSWV.
Our characterization of GN-S showed that the recombinant protein shares biochemical properties with GN even though the putative transmembrane domains, signal sequence, and cytoplasmic tail were removed and the remaining amino acids (35-309) were expressed with a six-His tag. Like virion GN, GN-S contains O-linked oligosaccharides and organizes into a homodimer. We also found that a MAb raised against virion GN recognized GN-S. The properties of GN-S compared to those of wild-type TSWV GN indicate that GN-S may serve as a surrogate for GN in experiments.
When the transmembrane domains were removed from the TSWV GN protein and the ectodomain was fused to the baculovirus GP64 signal sequence, GN-S was efficiently secreted from the cell. The GN proteins of several virus species within the family Bunyaviridae contain Golgi retention sequences (4, 17, 28), and the retention signals were mapped to the transmembrane domain and the cytosolic tail for Rift Valley fever virus (17) and the cytosolic tail for Uukuniemi virus (4). The TSWV GN Golgi localization signal has not been mapped, but by analogy with other members of the Bunyaviridae it likely resides in the transmembrane domain and/or cytosolic tail. Because these domains were removed from GN-S, the Golgi localization signal was likely removed, allowing the protein to be secreted, or the GP64 signal sequence negated any part of the Golgi retention motif that may have been maintained in the construct.
We found by enzymatic deglycosylation that GN-S and wild-type GN were modified by the addition of O-linked glycans. Sequence analysis results for our TSWV isolate predicted two sites on GN that may be O-glycosylated. O-linked glycosylation is a common form of posttranslational modification and may be involved in protein conformation (25), the stability of cell surface glycoproteins (31), and virus attachment to cell surfaces (45). Several virus glycoproteins have been shown to be O-glycosylated, including the human immunodeficiency virus type 1 envelope glycoprotein (6), the respiratory syncytial virus G protein (10), and equine herpesvirus type 1 gp300 (67). The GP ORF of Crimean-Congo hemorrhagic fever virus, another member of the Bunyaviridae, contains a variable mucin-like domain that is predicted to be extensively O-glycosylated (55), indicating that O-linked glycans may be an important modification of Bunyaviridae proteins. The findings of Naidu et al. (43), however, differ from ours to some extent. They did not detect O-linked glycans on an isolate of TSWV from Georgia. It is possible that our findings disagree because different TSWV isolates were examined in both studies and because TSWV isolates may be glycosylated differently. For example, a GP sequence reported by Kormelink et al. (30) contains eight sites that may be N-glycosylated, but a sequence reported by Adkins et al. (1) for a Hawaiian isolate of TSWV contains nine sites that may be N-glycosylated. Single amino acid changes could alter GP glycosylation and may explain the differences in our findings. Another possible explanation for the apparent differences in GN glycosylation may be due to the use of different methods for examining the glycosylation of GPs. We used enzymatic removal followed by SDS-PAGE to detect glycans, while Naidu et al. used lectin affinity blotting (43). Further studies of GN posttranslational modifications may elucidate the function(s) of glycans in protein folding, stability in the insect gut, or interactions with molecules on the thrips gut.
While both wild-type and recombinant GN contained O-linked glycans, only recombinant GN contained N-linked glycans. This difference may have been due to two events. First, during the construction of GN-S, we found upon sequencing that a new N-linked glycosylation site was added by the addition of the affinity purification tags. Second, the protein expression host may affect glycosylation (i.e., GN was isolated from TSWV-infected plants while GN-S was isolated from baculovirus-infected insect cells). In support of this hypothesis, Adkins et al. (1) found that a nontruncated GN protein expressed in a baculovirus was also N-glycosylated. This supports the claim that GN glycan modifications and/or site usage may vary in plants and insects.
We found that both wild-type GN and GN-S oligomerize and, more specifically, that both exist as monomers and dimers. As for GN-S, it was not surprising that the protein formed dimers because the GN ectodomain contains seven cysteines, and thus some of the amino acids expected to be involved in dimerization were retained in GN-S. We do not know which form of GN is involved in virus entry, but because GN and GN-S are capable of forming oligomers, this form of GN may interact with molecules on the surface of the thrips gut to mediate attachment and/or entry.
GPs encoded by other members of the Bunyaviridae have been shown to form oligomers. Uukuniemi virus GN maintains a pH-stable covalent homodimeric association (51). The GN protein of Sin Nombre virus was also found in monomeric and stable, SDS-resistant, multimeric forms, with the dimer being the only form present late in infection (57). Conversely, Punta Toro virus GN was found as a heterodimer with GC, but not as a homodimer (39). The ability of envelope glycoproteins to oligomerize seems to be conserved within the Bunyaviridae, indicating that this is an important part of the virus life cycle. Understanding the formation of higher-order oligomers may be important for determining how the GPs interact with molecules on the thrips gut to mediate acquisition or other virus processes such as assembly and replication.
We hypothesize that TSWV entry into the vector midgut entails a complex series of steps and that GN is involved in the virus accessing the midgut epithelia. Virus entry may begin with an initial docking step followed by binding to a cellular receptor. This binding may result in a GP becoming fusogenic and in a subsequent mixing of membrane bilayers, resulting in the release of virion contents. Our results suggest a role for GN in this process but do not preclude a role for GC. We found biochemical similarities between native GN and GN-S in their ability to form dimers, and we demonstrated that GN and GN-S are both modified by the addition of O-linked glycans. These biochemical similarities and functional data provide a basis for further studies to investigate the role of GN in virus binding and entry into thrips midgut cells by using GN-S. Our successful expression and characterization of GN-S provide a new understanding of TSWV GP biology. GN-S provides a significant new tool for delving deeper into the mechanisms of thrips-tospovirus interactions, which in time may help to elucidate the means of acquisition of other arthropod-transmitted viruses.
Acknowledgments
We thank Bruce Christensen and Teresa Compton for critical readings of the manuscript. HCMV gB was a gift from Teresa Compton, and the GN and GC monoclonal antibodies were a gift from John Sherwood. We also thank the Keck Biological Imaging Laboratory for the use of and assistance with the confocal microscope and Dorith Rotenberg for statistical consultations and a critical review of the manuscript.
This work was supported by United States Department of Agriculture grant 99-35303-8271 and by Hatch funds (WIS04316).
REFERENCES
- 1.Adkins, S., T. J. Choi, B. A. Israel, M. D. Bandla, K. E. Richmond, K. T. Schultz, J. L. Sherwood, and T. L. German. 1996. Baculovirus expression and processing of tomato spotted wilt tospovirus glycoproteins. Phytopathology 86:849-855. [Google Scholar]
- 2.Akula, S. M., N. P. Pramod, F. Z. Wang, and B. Chandran. 2002. Integrin α3β1 (CD49c/29) is a cellular receptor for Kaposi's sarcoma-associated herpesvirus (KSHV/JJV-8) entry into the target cells. Cell 108:407-419. [DOI] [PubMed] [Google Scholar]
- 3.Andersson, A. M., and R. F. Pettersson. 1998. Targeting of a short peptide derived from the cytoplasmic tail of the G1 membrane glycoprotein of Uukuniemi virus (Bunyaviridae) to the Golgi complex. J. Virol. 72:9585-9596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Andersson, A. M., L. Melin, A. Bean, and R. F. Pettersson. 1997. A retention signal necessary and sufficient for Golgi localization maps to the cytoplasmic tail of a Bunyaviridae (Uukuniemi virus) membrane glycoprotein. J. Virol. 71:4717-4727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bandla, M. D., L. R. Campbell, D. E. Ullman, and J. L. Sherwood. 1998. Interaction of tomato spotted wilt tospovirus (TSWV) glycoproteins with a thrips midgut protein, a potential cellular receptor for TSWV. Phytopathology 88:98-104. [DOI] [PubMed] [Google Scholar]
- 6.Bernstein, H. B., S. P. Tucker, E. Hunter, J. S. Schutzbach, and R. W. Compans. 1994. Human immunodeficiency virus type 1 envelope glycoprotein is modified by O-linked oligosaccharides. J. Virol. 68:463-468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Borucki, M. K., B. J. Kempf, C. D. Blair, and B. J. Beaty. 2001. The effect of mosquito passage on the La Crosse virus genotype. J. Gen. Virol. 82:2919-2926. [DOI] [PubMed] [Google Scholar]
- 8.Boublik, Y., P. Di Bonito, and I. M. Jones. 1995. Eukaryotic virus display: engineering the major surface glycoprotein of the Autographa californica nuclear polyhedrosis virus (AcNPV) for the presentation of foreign proteins on the virus surface. Biotechnology 13:1079-1084. [DOI] [PubMed] [Google Scholar]
- 9.Boyle, K. A., and T. Compton. 1998. Receptor-binding properties of a soluble form of human cytomegalovirus glycoprotein B. J. Virol. 72:1826-1833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Collins, P. L., and G. Mottet. 1992. Oligomerization and post-translational processing of glycoprotein G of human respiratory syncytial virus: altered O-glycosylation in the presence of brefeldin A. J. Gen. Virol. 73:849-863. [DOI] [PubMed] [Google Scholar]
- 11.Daughtrey, M. L., R. K. Jones, J. W. Moyer, M. E. Daub, and J. R. Baker. 1997. Tospoviruses strike the greenhouse industry: INSV has become a major pathogen on flower crops. Plant Dis. 81:1220-1230. [DOI] [PubMed] [Google Scholar]
- 12.de Assis Filho, F. M., R. A. Naidu, C. M. Deom, and J. L. Sherwood. 2002. Dynamics of tomato spotted wilt virus replication in the alimentary canal of two thrips species. Phytopathology 92:729-733. [DOI] [PubMed] [Google Scholar]
- 13.Faulquet, L., M. Pagni, N. Hulo, C. J. Sigrist, K. Hofmann, and A. Bairoch. 2002. The PROSITE database, its status in 2002. Nucleic Acids Res. 30:235-238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fox, G., N. R. Parry, P. V. Barnatt, B. McGinn, D. Rowlands, and F. Brown. 1989. The cell attachment sequence on foot-and-mouth disease virus includes the amino acid sequence RGD (arginine-glycine-aspartic acid). J. Gen. Virol. 70:625-637. [DOI] [PubMed] [Google Scholar]
- 15.Gavrilovskaya, I. N., E. J. Brown, M. H. Ginsberg, and E. R. Mackow. 1999. Cellular entry of hantaviruses which cause hemorrhagic fever with renal syndrome is mediated by β3 integrins. J. Virol. 73:3951-3959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gavrilovskaya, I. N., M. Shepley, R. Shaw, M. H. Ginsberg, and E. R. Mackow. 1998. β3 integrins mediate the cellular entry of hantaviruses that cause respiratory failure. Proc. Natl. Acad. Sci. USA 95:7074-7079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gerrard, S. R., and S. T. Nichol. 2002. Characterization of the Golgi retention motif of Rift Valley fever virus GN glycoprotein. J. Virol. 76:12200-12210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Goldbach, R. W., and D. Peters. 1994. Possible causes of the emergence of tospovirus diseases. Semin. Virol. 5:113-120. [Google Scholar]
- 19.Gonsalves, D., and E. E. Trujillo. 1986. Tomato spotted wilt virus in papaya and detection by ELISA. Plant Dis. 70:501-506. [Google Scholar]
- 20.Guoying, Z., L. Xiongbin, L. Huijuan, L. Juanli, C. Shengxiang, and G. Zuxun. 1999. Rice ragged stunt oryzavirus: role of the viral spike protein in transmission by the insect vector. Ann. Appl. Biol. 135:573-575. [Google Scholar]
- 21.Hanover, J. A., W. J. Lennarz, and J. D. Young. 1980. Synthesis of N- and O-linked glycopeptides in oviduct membrane preparations. J. Biol. Chem. 255:6713-6716. [PubMed] [Google Scholar]
- 22.Hofmann, K., P. Bucher, L. Falquet, and A. Bairoch. 1999. The PROSITE database, its status in 1999. Nucleic Acids Res. 27:215-219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hofmann, K., and W. Stoffel. 1993. TMbase—a database of membrane spanning protein segments. Biol. Chem. 374:166. [Google Scholar]
- 24.Hunter, W. B., H. T. Hsu, and R. H. Lawson. 1995. A novel method for tospovirus acquisition by thrips. Phytopathology 85:480-483. [Google Scholar]
- 25.Jentoft, N. 1990. Why are proteins O-glycosylated? Trends Biochem. Sci. 15:291-294. [DOI] [PubMed] [Google Scholar]
- 26.Kikkert, M., C. Meurs, F. van de Wetering, S. Dorfmuller, D. Peters, R. Kormelink, and R. Goldbach. 1998. Binding of tomato spotted wilt virus to a 94-kDa thrips protein. Phytopathology 88:63-69. [DOI] [PubMed] [Google Scholar]
- 27.Kikkert, M., J. van Lent, M. Storms, R. Bodegom, R. Kormelink, and R. Goldbach. 1999. Tomato spotted wilt virus particle morphogenesis in plant cells. J. Virol. 73:2288-2297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kikkert, M., A. Verschoor, R. Kormelink, P. Rottier, and R. Goldbach. 2001. Tomato spotted wilt virus glycoproteins exhibit trafficking and localization signals that are functional in mammalian cells. J. Virol. 75:1004-1012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Koch, J., M. Liang, I. Queitsch, A. A. Kraus, and E. K. F. Bautz. 2003. Human recombinant neutralizing antibodies against Hantaan virus G2 protein. Virology 308:64-73. [DOI] [PubMed] [Google Scholar]
- 30.Kormelink, R., P. de Haan, C. Meurs, D. Peters, and R. Goldbach. 1992. The nucleotide sequence of the M RNA segment of tomato spotted wilt virus, a bunyavirus with two ambisense RNA segments. J. Gen. Virol. 73:2795-2804. [DOI] [PubMed] [Google Scholar]
- 31.Krieger, M., P. Reddy, K. Kozarsky, D. Kingsley, L. Hobbie, and M. Penman. 1989. Analysis of the synthesis, intracellular sorting, and function of glycoproteins, using a mammalian cell mutant with reversible glycosylation defects. Methods Cell Biol. 32:57-84. [DOI] [PubMed] [Google Scholar]
- 32.Lasky, L. A., J. Groopman, C. Fennie, P. Benz, D. Capon, D. Dowbenko, G. Nakamura, W. Nunes, M. Renz, and P. Berman. 1986. Neutralization of the AIDS retrovirus by antibodies to a recombinant envelope glycoprotein. Science 233:209-212. [DOI] [PubMed] [Google Scholar]
- 33.Lasky, L. A., G. Nakamura, D. H. Smith, C. Fennie, C. Shimasaki, D. Patzer, P. Berman, T. Gregory, and D. J. Capon. 1987. Delineation of a region of the human immunodeficiency virus type I gp120 glycoprotein critical for interaction with the CD4 receptor. Cell 50:975-985. [DOI] [PubMed] [Google Scholar]
- 34.Lewis, T. 1997. Pest thrips in perspective, p. 13, 675-709. In T. Lewis (ed.), Thrips as crop pests. CAB International, Cambridge, United Kingdom.
- 35.Liang, M., M. Mahler, J. Koch, Y. Ji, L. Dexin, C. Schmaljohn, and E. K. F. Bautz. 2003. Generation of an HFRS patient-derived neutralizing recombinant antibody to Hantaan virus G1 protein and definition of the neutralizing domain. J. Med. Virol. 69:99-107. [DOI] [PubMed] [Google Scholar]
- 36.Lopper, M., and T. Compton. 2002. Disulfide bond formation of human cytomegalovirus glycoprotein B. J. Virol. 76:6073-6082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ludwig, G. V., B. M. Christensen, T. M. Yuill, and K. T. Schultz. 1989. Enzyme processing of La Crosse virus glycoprotein G1: a bunyavirus infection model. Virology 171:108-113. [DOI] [PubMed] [Google Scholar]
- 38.Ludwig, G. V., B. A. Israel, B. M. Christensen, T. M. Yuill, and K. T. Schultz. 1991. Role of La Crosse virus glycoproteins in attachment of virus to host cells. Virology 181:564-571. [DOI] [PubMed] [Google Scholar]
- 39.Matsuoka, Y., S.-Y. Chen, C. E. Holland, and R. W. Compans. 1996. Molecular determinants of Golgi retention in the Punta Toro virus G1 protein. Arch. Biochem. Biophys. 336:184-189. [DOI] [PubMed] [Google Scholar]
- 40.Moritz, G. 1997. Structure, growth and development, p. 15-63. In T. Lewis (ed.), Thrips as crop pests. CAB International, Cambridge, United Kingdom.
- 41.Nagata, T., A. K. Nagata-Inoue, M. Prins, R. Goldbach, and D. Peters. 2000. Impeded thrips transmission of defective tomato spotted wilt virus isolates. Phytopathology 90:454-459. [DOI] [PubMed] [Google Scholar]
- 42.Nagata, T., A. K. Nagata-Inoue, H. M. Smid, R. Goldbach, and D. Peters. 1999. Tissue tropism related to vector competence of Frankliniella occidentalis for tomato spotted wilt tospovirus. J. Gen. Virol. 80:507-515. [DOI] [PubMed] [Google Scholar]
- 43.Naidu, R. A., C. J. Ingle, C. M. Deom, and J. L. Sherwood. 2004. The two envelope membrane glycoproteins of Tomato spotted wilt virus show differences in lectin-binding properties and sensitivities to glycosidases. Virology 319:107-117. [DOI] [PubMed] [Google Scholar]
- 44.Nielsen, H., J. Engelbrecht, S. Brunak, and G. von Heijne. 1997. Identification of prokaryotic and eukaryotic signal peptides and prediction of their cleavage sites. Protein Eng. 10:1-6. [DOI] [PubMed] [Google Scholar]
- 45.Paulson, J. C. 1989. Glycoproteins: what are the sugars for? Trends Biochem. Sci. 14:272-275. [DOI] [PubMed] [Google Scholar]
- 46.Pekosz, A., C. Griot, N. Nathanson, and F. Gonzalez-Scarano. 1995. Tropism of bunyaviruses: evidence for a G1 glycoprotein-mediated entry pathway common to the California serogroup. Virology 214:339-348. [DOI] [PubMed] [Google Scholar]
- 47.Pierschbacher, M. D., and E. Ruoslahti. 1984. Cell attachment activity of fibronectin can be duplicated by small synthetic fragments of the molecule. Nature 309:30-33. [DOI] [PubMed] [Google Scholar]
- 48.Resende, O. R., P. de Haan, A. C. de Avila, E. W. Kitajima, R. Kormelink, R. Goldbach, and D. Peters. 1991. Generation of envelope defective interfering RNA mutants of tomato spotted wilt virus by mechanical passage. J. Gen. Virol. 72:2375-2383. [DOI] [PubMed] [Google Scholar]
- 49.Richmond, K. E., K. Chenault, J. L. Sherwood, and T. L. German. 1998. Characterization of the nucleic acid binding properties of tomato spotted wilt virus nucleocapsid protein. Virology 248:6-11. [DOI] [PubMed] [Google Scholar]
- 50.Roivainen, M., L. Piirainen, T. Hovi, I. Virtanen, T. Riikonen, J. Heino, and T. Hyypiä. 1994. Entry of coxsackievirus A9 into host cells: specific interactions with αvβ3 integrin, the vitronectin receptor. Virology 203:357-365. [DOI] [PubMed] [Google Scholar]
- 51.Rönkä, H., P. Hilden, C.-H. Bonsdorff, and E. von Kuismanen. 1995. Homodimeric association of the spike glycoproteins G1 and G2 of Uukuniemi virus. Virology 211:241-250. [DOI] [PubMed] [Google Scholar]
- 52.Rosenberg, I. M. 1996. Membrane and particulate-associated proteins, p. 135-152. In Protein analysis and purification. Birkhauser, Boston, Mass.
- 53.Rost, B. 1996. PHD: predicting one-dimensional protein structure by profile based neural networks. Methods Enzymol. 266:525-539. [DOI] [PubMed] [Google Scholar]
- 54.Rost, B., P. Fariselli, and R. Casadio. 1996. Topology prediction for helical transmembrane proteins at 86% accuracy. Protein Sci. 7:1704-1718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Sanchez, A. J., M. J. Vincent, and S. T. Nichol. 2002. Characterization of the glycoproteins of Crimean-Congo hemorrhagic fever virus. J. Virol. 76:7263-7275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Sherwood, J. L., T. L. German, A. E. Whitfield, J. W. Moyer, and D. E. Ullman. 2001. Tospoviruses, p 1034-1040. In O. C. Maloy and T. D. Murray (ed.), Encyclopedia of plant pathology. John Wiley & Sons, Inc., New York, N.Y.
- 57.Spiropolou, C. F., C. S. Goldsmith, T. R. Shoemaker, C. J. Peters, and R. W. Compans. 2003. Sin Nombre virus glycoprotein trafficking. Virology 308:48-63. [DOI] [PubMed] [Google Scholar]
- 58.Sundin, D. R., B. J. Beaty, N. Nathanson, and F. Gonzalez-Scarano. 1987. A G1 glycoprotein epitope of La Crosse virus: a determinant of infection of Aedes triseriatus. Science 235:591-593. [DOI] [PubMed] [Google Scholar]
- 59.Tamkun, J. W., D. W. DeSimone, D. Fonda, R. S. Patel, C. Buck, A. F. Horowitz, and R. O. Hynes. 1986. Structure of integrin, a glycoprotein involved in the transmembrane linkage between fibronectin and actin. Cell 46:271-282. [DOI] [PubMed] [Google Scholar]
- 60.Tusnády, G. E., and I. Simon. 1998. Principles governing amino acid composition of integral membrane proteins: applications to topology prediction. J. Mol. Biol. 283:489-506. [DOI] [PubMed] [Google Scholar]
- 61.Tusnády, G. E., and I. Simon. 2001. The HMMTOP transmembrane topology prediction server. Bioinformatics 17:849-850. [DOI] [PubMed] [Google Scholar]
- 62.Ullman, D. E., J. J. Cho, R. F. L. Mau, D. M. Westcot, and D. M. Custer. 1992. Midgut epithelial cells act as a barrier to tomato spotted wilt virus acquisition by adult Western flower thrips. Phytopathology 82:1333-1342. [Google Scholar]
- 63.Ullman, D. E., T. L. German, J. L. Sherwood, D. M. Westcot, and F. A. Cantone. 1993. Tospovirus replication in insect vector cells: immunocytochemical evidence that the nonstructural protein encoded by the S RNA of tomato spotted wilt tospovirus is present in thrips vector cells. Phytopathology 83:456-463. [Google Scholar]
- 64.Ullman, D. E., R. Meideros, L. R. Campbell, A. E. Whitfield, J. L. Sherwood, and T. L. German. 2002. Thrips as vectors of tospoviruses, p. 113-140. In R. T. Plumb and J. A. Callow (ed.), Advances in botanical research: plant virus vector interactions, vol. 36. Academic Press, London, United Kingdom. [Google Scholar]
- 65.van de Wetering, F., R. Goldbach, and D. Peters. 1996. Tomato spotted wilt tospovirus ingestion by first instar larvae of Frankliniella occidentalis is a prerequisite for transmission. Phytopathology 86:900-905. [Google Scholar]
- 66.Whitbeck, J. C., C. Peng, H. Lou, R. Xu, S. H. Willis, M. Ponce de Leon, T. Peng, A. V. Nicola, R. I. Montgomery, M. S. Warner, A. M. Soulika, L. A. Spruce, W. T. Moore, J. D. Lambris, P. G. Spear, G. H. Cohen, and R. J. Eisenberg. 1997. Glycoprotein D of herpes simplex virus (HSV) binds directly to HVEM, a member of the tumor necrosis factor receptor superfamily and a mediator of HSV entry. J. Virol. 71:6083-6093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Whittaker, G. R., L. A. Wheldon, L. E. Giles, J. M. Stocks, I. W. Halliburton, R. A. Killington, and D. M. Meredith. 1990. Characterization of the high Mr glycoprotein (gP300) of equine herpesvirus type 1 as a novel glycoprotein with extensive O-linked carbohydrate. J. Gen. Virol. 71:2407-2416. [DOI] [PubMed] [Google Scholar]
- 68.Wijkamp, I., J. van Lent, R. Kormelink, R. Goldbach, and D. Peters. 1993. Multiplication of tomato spotted wilt virus in its insect vector, Frankliniella occidentalis. J. Gen. Virol. 74:341-349. [DOI] [PubMed] [Google Scholar]