Abstract
Broadly neutralizing monoclonal antibodies (MAbs) are potentially important tools in human immunodeficiency virus type 1 (HIV-1) vaccine design. A few rare MAbs have been intensively studied, but we still have a limited appreciation of their neutralization breadth. Using a pseudovirus assay, we evaluated MAbs from clade B-infected donors and a clade B HIV+ plasma against 93 viruses from diverse backgrounds. Anti-gp120 MAbs exhibited greater activity against clade B than non-B viruses, whereas anti-gp41 MAbs exhibited broad interclade activity. Unexpectedly, MAb 4E10 (directed against the C terminus of the gp41 ectodomain) neutralized all 90 viruses with moderate potency. MAb 2F5 (directed against an epitope adjacent to that of 4E10) neutralized 67% of isolates, but none from clade C. Anti-gp120 MAb b12 (directed against an epitope overlapping the CD4 binding site) neutralized 50% of viruses, including some from almost every clade. 2G12 (directed against a high-mannose epitope on gp120) neutralized 41% of the viruses, but none from clades C or E. MAbs to the gp120 V3 loop, including 447-52D, neutralized a subset of clade B viruses (up to 45%) but infrequently neutralized other clades (≤7%). MAbs b6 (directed against the CD4 binding site) and X5 (directed against a CD4-induced epitope of gp120) neutralized only sensitive primary clade B viruses. The HIV+ plasma neutralized 70% of the viruses, including some from all major clades. Further analysis revealed five neutralizing immunotypes that were somewhat associated with clades. As well as the significance for vaccine design, our data have implications for passive-immunization studies in countries where clade C viruses are common, given that only MAbs b12 and 4E10 were effective against viruses from this clade.
Neutralizing antibodies (NAbs) against viral envelope proteins (Env) provide the first line of adaptive immune defense against human immunodeficiency virus type 1 (HIV-1) exposure by blocking the infection of susceptible cells (64, 89, 94, 108). The efficacy of vaccines against several viruses has been attributed to their ability to elicit NAbs (21, 150). However, despite enormous efforts, for HIV-1 there has been limited progress toward an effective immunogen (21, 84, 90).
HIV-1 is among the most genetically diverse viral pathogens described to date. There are three main branches of the HIV-1 phylogenetic tree, the M (main), N (new), and O (outlier) groups. Group M viruses are the most widespread, accounting for >99% of global infections. This group is presently divided into nine distinct genetic subtypes, or clades (A through K), originally based on short sequences mostly in the Env gene (80, 122) but more recently based on full-length sequences. Env is the most variable HIV-1 gene, with up to 35% sequence diversity between clades, 20% sequence diversity within clades, and up to 10% sequence diversity in a single infected person (61, 130). Clade B is dominant in Europe, the Americas, and Australia (60). Clade C is common in southern Africa, China, and India and presently infects more people worldwide than any other clade (80). Clades A and D are prominent in central and eastern Africa. However, many viruses are difficult to classify into clades due to the common intermixing of cocirculating viruses that leads to interclade recombinants (52, 82). Some recombinant forms have in fact given rise to important epidemic lineages, called circulating recombinant forms (CRFs). The two most common of these are CRF01 (AE), discovered in Thailand, which was initially classified as clade E, though later it was found to be clade E only in Env and clade A in other parts of the genome, and CRF02, an AG recombinant form common in Western Africa (122). Globally, clades A through D and the CRF01 AE and CRF02 AG recombinants account for >90% of global infections.
Although clades provide a useful means to categorize HIV-1 based on genetic relationships, the relevance of clades in distinguishing virus neutralization sensitivities remains unproven. There is so far no consistent evidence that HIV+ sera preferentially neutralize autologous viruses more effectively than they do viruses from other clades (6, 10, 43, 58, 59, 71, 75, 90, 93, 102, 145), although some studies have suggested stronger intraclade neutralizing responses for clades C (17) and AE (CRF01) (75). The overall difficulty of determining cladeassociated neutralizing immunotype groups may be because sequence differences that determine genetic clades do not influence neutralization epitopes (71) or that limited sampling and high background noise activity complicate cross-clade neutralization analyses (92, 101, 152).
Distinct grouping of primary isolates into neutralization immunotypes may be feasible using a few rare broadly neutralizing human monoclonal antibodies (MAbs) that have been isolated from HIV+ clade B-infected human donors (35, 108). These MAbs neutralize many primary HIV-1 isolates, including some from different clades, indicating that certain elements of Env structure are well conserved (20, 22, 56, 57, 98, 109, 138, 139). Four relatively conserved epitopes have been defined by a set of five neutralizing human MAbs. Two MAbs recognize epitopes located on the gp120 surface unit of the Env spike: MAb b12 is directed against an epitope overlapping the CD4 binding site (5, 19, 22, 121), and MAb 2G12 recognizes a unique epitope in a carbohydrate-rich region on the outer domain of gp120 (14, 23, 62, 126, 129, 139). Three MAbs recognize epitopes located on the membrane-proximal external region of the gp41 transmembrane protein: MAb 2F5 has been mapped to a region overlapping the conserved sequence ELDKWA (98, 106, 114). MAbs 4E10 and Z13 recognize an epitope involving the sequence NWF(D/N)IT located adjacent and carboxy terminal to the 2F5 epitope (14, 153). Passive-transfer studies in macaques have shown that these MAbs can provide protection against HIV-1 challenge (4, 73, 74, 79, 107). The very existence of these MAbs provides hope that a vaccine able to elicit broadly neutralizing antibodies may be possible in the future. Unfortunately, all attempts to elicit antibodies with specificities similar to those of these MAbs have so far failed (7, 27, 37-39, 69, 78, 96, 119, 132, 140). Since neutralizing antibodies against these epitopes do not appear to constitute a significant fraction of the plasma response against Env generated during HIV-1 infection or by vaccination, their epitopes appear to be poorly immunogenic (18). Typically, immune sera from vaccine trials to date at best neutralize only the matched vaccine strain and a few neutralization-sensitive primary isolates (30, 32, 111, 141).
Some other anti-Env MAbs are of significant interest. These include MAbs against the gp120 V3 loop (9, 28, 46, 144). The V3 loop is highly immunogenic and was once considered to be the principal neutralizing determinant of HIV-1. However, it has since been shown that many primary HIV-1 isolates resist neutralization by sequestering the V3 loop, rendering it inaccessible to MAbs (13). The V3 loop is highly variable and, barring some notable exceptions, tends to induce type-specific antibodies with limited breadth (9, 28, 47, 49, 50). Despite its sequence variation, the V3 loop retains certain conformational features that may preserve its ability to interact with chemokine receptors during infection (31, 53, 131, 137, 147). MAb 447-52D is an unusual V3 loop-specific MAb in that it recognizes the well-conserved GPGR motif at the tip of the V3 loop of clade B isolates. This specificity lends it greater breadth than most V3 MAbs described to date (47, 133).
Another group of MAbs recognize discontinuous epitopes that overlap the conserved coreceptor binding site of gp120 and that are preferentially exposed by gp120 binding to CD4. This group of MAbs is represented by MAbs X5 and 17b (66). As whole immunoglobulin Gs (IgGs), these MAbs have very limited neutralizing activity against primary isolates. However, Fab and single-chain Fv fragments neutralize primary isolates more effectively (66). Thus, their activity against primary viruses appears to be limited by steric constraints that are less significant for smaller IgG fragments.
Many studies have evaluated the neutralization activities of the above-mentioned MAbs (33-36, 45, 48, 49, 72, 95, 114, 115, 138). However, these all made only a limited assessment of cross-clade neutralization: at most, only three or four isolates of each non-B clade were analyzed. Here, we have analyzed the neutralization activities of b12; b6 (a MAb overlapping a CD4 binding site); 2G12; 2F5; 4E10; X5; 447-52D; 58.2 (a V3 loop MAb); a five-MAb cocktail of b12, 2G12, 447-52D, 2F5, and 4E10; and a broadly cross-reactive HIV+ donor plasma against a total of 93 viruses, including 61 non-clade B isolates. Depending on availability, we included viruses from as many different parts of the world as possible, so that neutralizing activity could be collated on a geographic basis as well as on a genetic basis. To make this large study manageable, we used a sensitive and highly reproducible single-round pseudovirus infectivity assay. We found that MAbs b12, 2G12, 4E10, and 2F5; the five-MAb cocktail; and the HIV+ plasma exhibited various degrees of cross-clade neutralization. Neutralization by the V3 loop-specific MAbs was mostly restricted to clade B viruses, and MAbs b6 and X5 neutralized only sensitive clade B viruses.
MATERIALS AND METHODS
MAbs and HIV+ human plasma.
Seven anti-Env MAbs were derived from clade B HIV-infected donors. These included the anti-gp120 MAbs b12 (19, 22) and b6 (5, 19, 121), both directed against an epitope overlapping the CD4 binding site; 2G12, directed against a unique epitope in a carbohydrate-rich region on the outer domain of gp120 (14, 23, 62, 126, 129, 139); IgG X5 (66, 95), which recognizes an epitope on gp120 whose exposure is increased when gp120 complexes with CD4; and 447-52D, which was selected with a clade B-derived peptide and recognizes the GPXR motif at the tip of the V3 loop (28, 45, 47-50, 133). 58.2 is a mouse MAb generated against a clade B Env immunogen and is directed against the V3 loop HIGPGRAF motif (55). Two anti-gp41 MAbs were 2F5 (14, 29, 62, 97, 98, 106, 114, 115) and 4E10 (14, 134, 153), directed against epitopes located on the membrane-proximal external region of the gp41 transmembrane protein. 2F5 recognizes the conserved sequence ELDKWA (residues 667 to 672, based on the numbering of residues in HXBc2 to facilitate comparison with structural information published on this Env protein) (64, 98, 106), and 4E10 recognizes an epitope involving the sequence NWF(D/N)IT (residues 671 to 676) motif located carboxy terminal to the 2F5 epitope (134, 153). Twenty-seven HIV+ human plasmas from HIV+ clade B-infected donors were obtained from NABI Biopharmaceuticals (Rockville, Md.). These plasmas were available in large volumes, which is an important criterion for their use as assay controls. All plasmas were initially tested in neutralization assays against 10 viruses. Plasma N16 exhibited the most potent and cross-reactive neutralization and was selected for further use. There is no clinical information available about the donor of this plasma.
Viruses.
Pseudoviruses capable of a single round of infection were produced by cotransfecting HEK293 cells with a subgenomic plasmid, pHIVlucΔU3, that incorporates a firefly luciferase indicator gene and a second plasmid, pCXAS, that expresses HIV-1 Env libraries or clones. Recombinant viruses pseudotyped with Env proteins were harvested 48 h posttransfection. Env gp160 DNAs used for cloning into pCXAS were derived either from shotgun cloning of viral quasispecies by reverse transcription-PCR of HIV genomic RNA from HIV+ donor plasma or from primary virus that had been cultured in primary peripheral blood mononuclear cells (PBMCs), as described previously (120). Others were from well-established molecular clones. An amphotropic control murine leukemia virus (MuLV) Env plasmid was also used to make pseudovirus to control for nonspecific neutralization.
Uncloned plasma-derived quasispecies were all designated VLGC, with the following codes that reflect their clades (origins, where known, are given in parentheses): ABEVLGC1 (Belgium), AxVLGC2 (unknown); AGxVLGC1 and AGxVLGC2 (both unknown, but CRF_02 recombinants like these are common in Northern Africa); BBRVLGC1, BBRVLGC2, and BBRVLGC3 (Brazil); BGPTVLGC3 (Portugal); BFITVLGC1 and BFITVLGC2 (Italy); BFBRVLGC3 and FBRVLGC4 (a mosaic Env that appears to be mostly clade F with traces of clade B sequence) (Brazil); GxxVLGC1 (unknown); GPTVLGC2 (Portugal); JGEVLGC1 (Germany); and JxxVLGC2 (unknown).
For consistency, we used a systematic and standardized eight-character nomenclature to define viruses by their Env proteins by clade, country of origin, year isolated, number, and a “c” to indicate a molecular clone. This also assisted mathematical analysis. Previous names, where known, are given in parentheses, and references, if available, are cited. Quasispecies derived from primary virus cultures were as follows. Clade A viruses were ARW93029 (93RW029), ARW92024 (92RW024), ARW92009 (92RW009), ARW92026 (92RW026), ARW92008 (92RW008), ARW92021 (92RW021), ARW92020 (92RW020) (Rwanda) (41), AUG92031 (92UG031), AUG92037 (92UG037), AUG93077 (93UG077), and AUG94103 (94UG103) (Uganda) (40, 41). Clade AC recombinants were ACKE94105 (94KE105) (Kenya) and ACUG93086 (93UG086) (Uganda) (40, 41).
Clade B viruses were BUSxx692 (692), BUSx1168 (1168), BUSx1196 (1196), BUSx5768 (5768; previously P27), and BUSx6101 (6101; previously P15) (United States) (16); BUSxPAVO (PAVO) and BUSTORNO (TORNO) (Italy) (16); BTTxx515 (QH0515), BTTxx692 (QH0692), and BTTZ4589 (QZ4589) (Trinidad) (26); BHT92593 (92HT593) and BHT92594c (92HT594) (Haiti) (40); BBR92020 (92BR020), BBR92021 (92BR021), and BBR92028 (92BR028) (Brazil) (41); BUSxxME1 (ME1) and BUSxME46 (ME46) (United States) (24); BUS92712 (92US712) and BUS93073 (93US073) (United States) (40); BTH93305 (93TH305) (Thailand; unpublished; previously thought to be clade E); BUSxxBaL (BaL) (United States) (54); BFRxBXO8 (BX08) (France) (87); BUSxxx44c (clone 44; derived from a patient designated AC10 [D. Montefiori, unpublished data]), BUSxJRFLc (JR-FL), and BUSJRCSFc (JR-CSFc; a clone related to the published JR-CSF sequence) (United States) (103); and BUSSF162c (United States) (25). Molecular clones of 3 T-cell line-adapted viruses were BFRxIIIBc (HXB2c; a clone related to the published HXB2 sequence) (France) (117), BFRxNL43c (NL4-3) (France) (125), and BUSxxxMNc (MN) (United States) (105).
Clade C viruses were CIN93101 (93IN101) (85), CIN93905 (93IN905; also known as 301905) (70), CIN93999 (93IN999; 301999) (70), 21068 (70), CIN11246 (94IN11246-3; also known as 11246-3) (70), and CIN98022 (98IN022) (India) (123); CMW93959 (93MW959; 301959), and CMW93960 (93MW960, 301960) (Malawi) (40); CBR98004 (98BR004) (Brazil) (123); CZA97012 (97ZA012) (South Africa) (123); and CCN98006 (98CN006) and CCN98009 (98CN009) (China) (123).
Clade D viruses were DUG93082 (93UG082), DUG94118 (94UG118), DUG94114 (94UG114), DUG92021 (92UG021), DUG92001 (92UG001), DUG92035 (92UG035), DUG93070 (93UG070), DUG92046 (92UG046), DUG92005 (92UG005), DUG92038 (92UG038), and DUG92024 (92UG024) (Uganda) (40, 41).
Clade AE viruses (CRF01_AE recombinants comprised of almost full-length clade E Env with small fragments of clade A sequence at the N terminus of gp120 and the C terminus of gp41) were ETH92001 (92TH001), ETH92005 (92TH005), ETH92006 (92TH006), ETH92019 (92TH019), ETH92021 (92TH021), ETH92022 (92TH022), ETH92024 (92TH024) (3), ETHCMU02 (CMU02) (81), and CMU06 (Thailand) (116).
Clade F viruses were FBR93020 (93BR020) and FBR93029 (93BR029) (Brazil) (40). BFBR93019 (93BR019) is a BF hybrid (Brazil) (40).
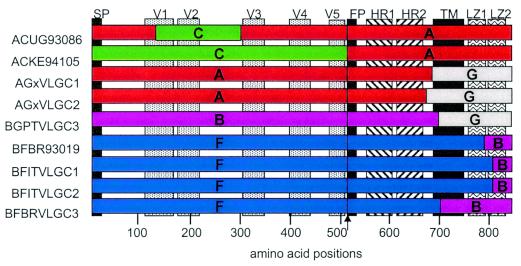
Some of the Env sequences are from quasispecies and are therefore of uncertain length in the variable domains. The Env sequence from the ARW92021 Env quasispecies was so heterogeneous that it could not be determined with certainty. Accordingly, this sequence was not submitted to the database. A depiction of novel interclade recombinant Env genes in this panel is shown in Fig. 1, to give an indication of the sites of recombination between Envs from different clades that form these hybrids. Many of these recombinants have breakpoints downstream of the gp41 transmembrane domain, so that the entire ectodomain is from a single clade (viruses AGxVLGC1, AGxVLGC2, BGPTVLGC3, BFITVLGC1, BFITVLGC2, BFBRVLGC3, and BFBR93019). In contrast, the AC hybrids ACUG93086 and ACKE94105 have mixed gp120/gp41 ecotodomains.
FIG. 1.
Depiction of the genetic composition of recombinant viruses. Along the top, from left to right, on the gp160 sequence, we annotate the signal peptide (SP), the gp120 variable loops, the fusion peptide (FP) of gp41, the helical regions HR1 and HR2, the transmembrane domain (TM), and the leucine zipper domains in the cytoplasmic tail (LZ1 and LZ2). Amino acid positions are depicted along the bottom. Env fragments from different clades are represented in color. The approximate recombination junctions were gleaned from sequence information from each of these viruses submitted to the Los Alamos database.
Live primary-virus isolates provided by the National Institutes of Health AIDS Reference Reagent Program were grown in PBMCs, as described previously (154). The 25 viruses that matched viruses in our pseudovirus panel were ARW92021, DUG92035, BUS91056, BBR92020, BBR92021, BBR92028, BHT92593, BHT92594c, BUS92712, BUSxxBaL, BUSJRCSF, BUSxJRFL, BUSxxxME1, BUSxxME46, BTTQZ458, BUSSF162, BFBR92019, CIN93101, CMW93959, CMW93960, CIN11246, DUG92021, DUG92038, DUG92046, and FBR93020.
Neutralization assays. (i) Single-round pseudovirus neutralization assay.
A recombinant-virus assay involving a single round of virus infection was used to measure neutralization (110, 120). Recombinant luciferase pseudoviruses were incubated for 1 h at 37°C with 10 serial fourfold dilutions of MAbs or heat-inactivated plasma, usually starting from 50 μg/ml (MAbs) or a 1:20 dilution (plasma). In a variant protocol, virus was incubated with antibody for 18 h before the mixture was added to target cells. U87 cells expressing CD4 plus the CCR5 and CXCR4 coreceptors were inoculated with virus-antibody (Ab) dilutions in the absence of added cations. Virus stocks were screened to ensure that they were functional and yielded a high luciferase reporter light signal in target cell lysates. Input virus used in each experiment was not standardized because prior experiments (not shown) had demonstrated that the input virus dose does not affect the neutralizing 50% inhibitory concentrations (IC50s) of antibodies. Virus infectivity was determined 72 h postinoculation by measuring the amount of luciferase activity expressed in infected cells. Neutralizing activity is reported as the concentration or dilution of each MAb or plasma required to confer 50% (IC50) or 90% (IC90) inhibition of infection (percent inhibition = {1 − [luciferase + Ab/luciferase − Ab]} × 100). To eliminate nonspecific neutralization, the criterion for genuine neutralization is that the titer must be at least 2.5-fold higher against HIV-1 than it is against the amphotropic control MuLV. Due to the large size of this study, each individual virus-Ab combination was in general tested only once. To ensure that the results were reproducible, the control viruses JR-CSF (R5-tropic) and NL4-3 (X4-tropic) were run at least six times in all assays. The reproducibility of the assay within and between runs was assessed by looking at these controls and was found to be within 2.5-fold.
(ii) PBMC neutralization assay.
Traditional neutralization assays were performed using primary virus grown in PBMCs, as described previously (153). Each experiment was performed in triplicate. Briefly, using 96-well round-bottom tissue culture plates, 50 μl of MAb at 100 μg/ml and 50 μl of primary-isolate virus (100 50% tissue culture infective doses) were coincubated for 1 h at 37°C, and then 100 μl of phytohemagglutinin-activated target PBMCs (105/ml) was added. Neutralization was evaluated at a single MAb concentration (50 μg/ml, according to the MAb concentration in the virus-MAb mixture before being added to the plate). After an overnight incubation, the cells were washed twice with tissue culture medium. On day 4, 100 μl of the medium was replaced with fresh tissue culture medium. On day 7, the supernatants were collected and treated with 1% (vol/vol) Empigen (Calbiochem). Neutralization was measured as a reduction in p24 production in the supernatants. p24 was assayed using an in-house enzyme-linked immunosorbent assay, as described previously (88). Production of p24 in the MAb-containing cultures was compared to p24 production in cultures without MAb run in the same assay. Neutralization was recorded in assays that resulted in a 10-fold reduction in the mean p24 content. Each virus-MAb combination was assayed at least twice to ensure consistency. The PBMCs used came from a pool of three anonymous healthy donors and were constant throughout all neutralization assays.
Two-dimensional hierarchical clustering of viruses and MAbs based on neutralization susceptibility profiles.
Viruses were clustered according to their IC50 neutralization susceptibility profiles so that those with shared susceptibilities were grouped together. The MAbs were also arranged according to their abilities to neutralize the panel of viruses. Dendrograms to show the clustering patterns for viruses and for MAbs were then derived. The most common clades in the study were assigned colors, indicated on the ends of dendrogram branches. Dendrograms were generated using methods described by Hastie et al. (51) and implemented using the hierarchical clustering tool and the Mcquitty clustering algorithm in R (version 1.8.1, 2003; The R Foundation for Statistical Computation) (54a). The actual neutralizing scores were changed to 0, 1, 2, 3, and 4, encoding undetectable to highly sensitive neutralization, and the vectors of the scores were used as the basis for clustering.
Nucleotide sequence accession numbers.
Sequence data for each of the Envs have been submitted to GenBank (accession numbers AY669697 to AY669786).
RESULTS
Comparison of neutralization by pseudovirus and traditional PBMC assays.
To verify that the pseudovirus assay yields meaningful neutralization data, we initially compared b12, 2G12, X5, 2F5, and 4E10 activities against 25 viruses in pseudovirus and traditional PBMC neutralization assays (Fig. 2). In the PBMC assay, neutralization was scored as positive if >90% neutralization was observed at a MAb concentration of 50 μg/ml. In the pseudovirus assay, the MAb concentration that gave 90% neutralization was determined, and the data were coded so that the warmer the color, the more potent the neutralization. We arbitrarily took 50 μg/ml as the neutralization cutoff.
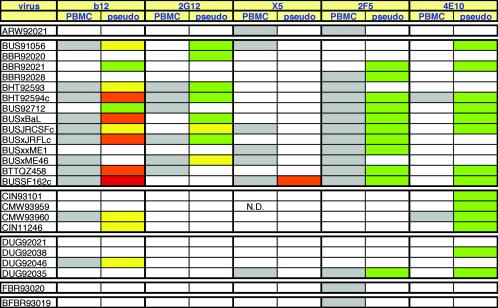
FIG. 2.
Comparison of neutralization activities of MAbs b12, 2G12, X5, 2F5, and 4E10 in pseudovirus and PBMC neutralization assays against 25 viruses. For the PBMC assay, an IC90 of <50 μg/ml is depicted by a gray square, and an IC90 of >50 μg/ml is shown as a white square. In the pseudovirus assay, IC90 data are shown. To assist comprehension, neutralization in the pseudovirus assay has been color coded so that the warmer the color, the more potent the neutralization: a white box indicates an IC90 of >50 μg/ml, a green box indicates 50 μg/ml > IC90 > 10 μg/ml, a yellow box indicates 10 μg/ml > IC90 > 1 μg/ml, an orange box indicates 1 μg/ml > IC90 > 0.1 μg/ml, and a red box indicates an IC90 of <0.1 μg/ml. N.D., not done.
Overall, we observed fairly concordant patterns in the two assays: 73% of the 125 assay reactions compared were either positive in both assays or negative in both assays, and 27% of the reactions were discordant. Comparing the neutralizations in PBMC and pseudovirus assays purely on the basis of the presence or absence of any detectable neutralization, we observed the following concordance: b12, 84%; 2G12, 76%; X5, 72%; 2F5, 76%; and 4E10, 52%. Therefore, for b12, 2F5, and 2G12, the concordance was 75 to 85%, but for X5 it was somewhat lower and for 4E10 it was considerably lower. Clearly, then, much of the discordance can be attributed to 4E10, which weakly neutralized 14 of 25 viruses in the pseudovirus assay, yet only 2 of these viruses were neutralized in the PBMC assay (IC90 < 50 μg/ml). It appears that the pseudovirus assay is more sensitive than the PBMC assay for detecting 4E10 activity. When 4E10 is removed from the overall MAb comparison, the data concordance of the remaining reactions increases from 73 to 78%. In contrast, MAb X5 neutralization is more easily detected in the PBMC assay. This might be explained by differing levels of CCR5 coreceptor expression on the target cells, which could affect the window of opportunity for X5 to neutralize virus after it has engaged CD4. Experiments to evaluate the discordant X5 results are ongoing. Since X5 does not neutralize particularly effectively in either assay, giving many double-negative results, this discordance does not have as large an impact on the overall concordance as does 4E10. Similarly, 2G12 shows 43% discordance in neutralization against clade B viruses, but this is compensated for by the 100% double-negative results against the non-B isolates in the panel. For the remaining two MAbs, b12 and 2F5, the sensitivities of the two neutralization assays appear to be quite similar.
Effect of virus-MAb incubation time on neutralization.
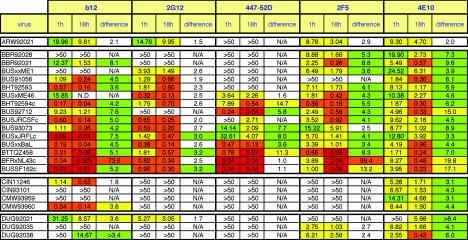
To determine the most appropriate pseudovirus neutralization assay protocol for our analysis, we initially determined the effect of the virus-MAb incubation time on IC50 titers. Comparing 1- and 18-h incubations with a subset of mostly clade B viruses (Fig. 3), we observed a mean 6.2-fold increase in neutralization sensitivity in the 18-h format. However, there was significant variability in the factor increase that appeared to be highly virus and MAb dependent. Some virus-MAb mixtures resulted in dramatically increased neutralization in the 18-h format. For example, the increase in BFRxNL4-3c neutralization was 73.6-fold with b12 and 99.4-fold with 2F5 but only 2.5-fold with 2G12. In general, 2G12 showed a disproportionately smaller increase in neutralization (mean, 2.5-fold increase) compared to the other MAbs. The dramatically increased neutralization with 18 h of incubation was not restricted to neutralization-sensitive viruses like BFRxNL4-3c. For example, BUS92712 was 15-fold better neutralized by 4E10 in the 18-h format. Since the discrepancies are neither entirely virus nor MAb specific, it is difficult to be sure what underlies this phenomenon. It is possible that the discrepancies are related to MAb-induced virus shedding of gp120 that might progressively increase with longer MAb-virus incubation times (88, 112). We decided to use the 1-h format for subsequent analysis both to avoid this caveat and to generate data that can be compared more easily to previously published data, where a 1-h virus-MAb incubation phase is the norm.
FIG. 3.
Effect of MAb-virus incubation time in pseudovirus neutralization assays. Viruses were incubated with MAb for either 1 or 18 h as indicated, and the IC50s (μg/ml) were determined. The neutralization titers are color coded as in Fig. 2. The differences in IC50, expressed in ratio form, are also color coded, so that those with bigger discrepancies are shown in warmer colors and vice versa. N/A, not applicable; N.D., not done.
Cross-clade neutralization analysis. (i) Overview.
We next sought to comprehensively assay a collection of antibodies against a large panel of viruses. In light of the difficulties of growing large numbers of primary viruses, we took advantage of the relatively convenient pseudovirus assay. We chose a genetically and geographically diverse panel of viruses, including those for which live virus stocks are available, to enable a comparison of results to those from traditional PMBC-based neutralization assays (as discussed above and shown in Fig. 2). Our panel included some viruses from early in the pandemic in an attempt to provide data that could be compared with data already published for these viruses. We also included some contemporary and recently transmitted viruses that are typically available as uncloned quasispecies. Overall, we emphasize that, while the 93 viruses were selected to represent the broadest possible cross section of Env diversity from those available at ViroLogic, our panel should not be interpreted to accurately represent the percentages of each type of virus circulating globally. For example, compared to global estimates, our panel has a relatively high emphasis on clade B viruses and relatively little emphasis on clade C isolates. In addition, since we are concerned about selecting naturally occurring Env proteins, we did not limit our analysis to viruses of “pure” clade pedigree but instead selected several recombinant viruses in an attempt to factor in the relatively common populations of interclade recombinant viruses.
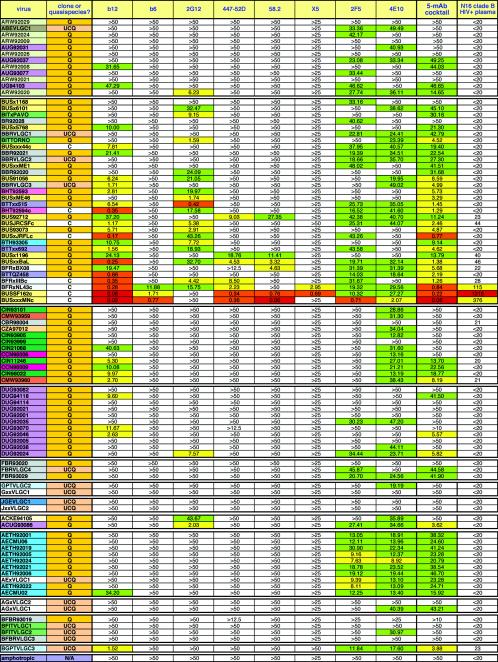
The collection of antibodies we tested included five known neutralizing MAbs (b12, 2G12, 2F5, 4E10, and 447-52D), three control MAbs known to have modest neutralizing activities (58.2, X5, and b6), an equimolar five-MAb cocktail (b12, 2G12, 2F5, 4E10, and 447-52D), and a plasma sample (N16) from a clade B HIV-1-infected donor. The neutralizing IC50s (and dilutions, in the case of the N16 plasma) using a 1-h virus-Ab incubation phase are presented in Fig. 4. The Env-expressing plasmids encoded either molecular clones (C) or Env quasispecies cloned by reverse transcription-PCR from virus in HIV+ donor plasma (uncultured quasispecies [UCQ]) or from virus that had been cultured in primary lymphocytes (quasispecies [Q]).
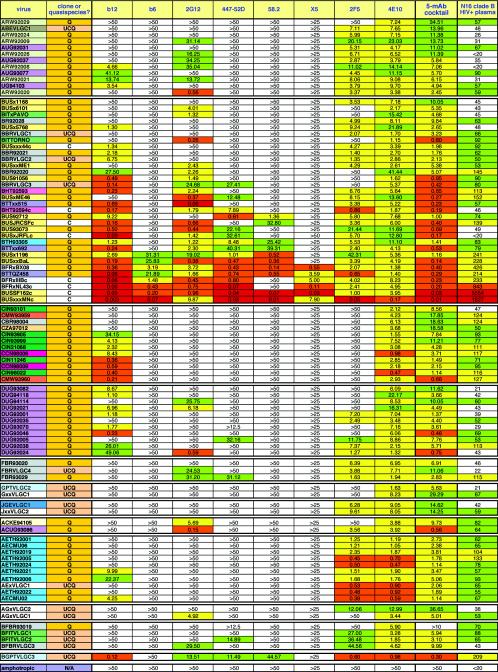
FIG. 4.
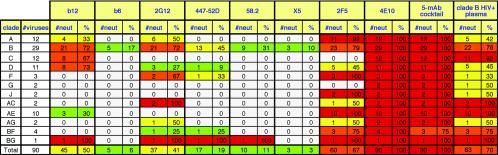
IC50 titers in the pseudovirus neutralization assay, using eight MAbs, the five-MAb cocktail, and the clade B HIV+ plasma against 93 HIV-1 isolates. The MAbs are organized horizontally, and the viruses are organized from top to bottom. In the leftmost columns, viruses have been grouped into clades A through J and are also color coded to reflect their country of origin where known. The viruses are also categorized as molecular clones (C), cultured quasispecies (Q), or uncloned quasispecies made directly from plasma samples (UCQ). The MAb neutralization titers have been color coded in exactly the same way as in Fig. 2. For the plasma, the color codes are as follows: a white box indicates an IC50 of <1:50 dilution, a green box indicates 1:50 < IC50 < 1:100, a yellow box indicates 1:100 < IC50 < 1:500, an orange box indicates 1:500 < IC50 < 1:1,000, and a red box indicates an IC50 of >1:1,000. Based on overall neutralizing potency, the viruses in each clade have been organized from the most resistant at the top to the most sensitive at the bottom.
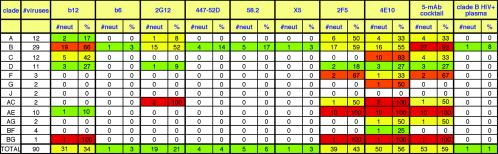
Figure 5 summarizes the IC50 data, showing the total number of viruses neutralized by each MAb with an IC50 below 50 μg/ml, regardless of potency. 4E10 neutralized all the viruses; 2F5, b12, and 2G12 neutralized 41 to 67%, and the V3 loop MAbs neutralized <20% of the total. At IC90 (Fig. 6 and 7), 4E10 still neutralized 56% of the viruses, a larger proportion of isolates than any other MAb. 2F5 also retained neutralizing activity against a large proportion of the isolates. The neutralizing activities of b12, 2G12, and 2F5 outside clade B were diminished, so that most of the b12 and 2G12 activity was now concentrated in clade B. V3 loop MAbs were only weakly effective at IC90. 2F5 and 4E10 still neutralized all AE recombinant viruses but were no longer the most broadly neutralizing MAbs against clade B viruses; the most broadly neutralizing MAb was now b12. At IC95, we observed that 4E10 still neutralized several more isolates than any other MAb, but its potency was very low (data not shown).
FIG. 5.
Summary of the total number of primary viruses neutralized with an IC50, of <50 μg/ml. We excluded the data from T-cell line-adapted isolates BUSxxxMNc, BFRxNL43c, and BFRxIIIBc. The data have been color coded as follows: a white box indicates that no viruses were neutralized, a green box indicates that 1 to 30% of viruses were neutralized, a yellow box indicates that 31 to 60% of viruses were neutralized, an orange box indicates that 61 to 90% of viruses were neutralized, and a red box indicates that >90% of viruses were neutralized.
FIG. 6.
IC90 titers against the panel of 93 viruses. The data are organized as in Fig. 4.
FIG. 7.
Summary of the total number of primary viruses neutralized with an IC90 of <50 μg/ml, regardless of potency. The data are organized as in Fig. 5.
Considering that all the MAbs were isolated from clade B-infected persons, it is interesting to compare the neutralization of clade B to that of non-clade B viruses. Each MAb neutralized (IC50, <50 μg/ml) clade B and non-B isolates, respectively, as follows: b12, 72 vs. 39%; b6, 17 vs. 0%; 2G12, 72 vs. 26%; 447-52D, 45 vs. 7%; 58.2, 31 vs. 2%; X5, 10 vs. 0%; 2F5, 79 vs. 61%; and 4E10, 100 vs. 100%. From this, we can infer that gp120 MAbs neutralize a lower percentage of non-clade B viruses than they do clade B viruses but that b12 and 2G12 are relatively cross-clade neutralizing. Gp41 MAbs are, however, significantly more cross-reactive outside clade B than the gp120 MAbs. The mean neutralization titers of each MAb against clade B and non-B viruses were also calculated from Fig. 4 as follows (values are in micrograms per milliliter): b12, 2.65 and 10.22; b6, 16.48 and not applicable; 2G12, 3.46 and 17.06; 447-52D, 11.90 and 22.42; 58.2, 11.11 and 44.57; X5, 1.39 and not applicable; 2F5, 6.26 and 7.33; and 4E10, 7.37 and 5.40. Again, it is apparent that each gp120 MAb neutralizes clade B viruses more effectively (with a range of differences from 1.88- to 4.93-fold). As mentioned above, this may be because some clade B viruses in our panel were derived from viruses that have been more extensively passaged in culture and are therefore inherently more neutralization sensitive. However, if this is the case, it does not appear to have significantly affected the activities of the gp41 MAbs. X5 has an apparently very high mean titer against clade B viruses, but this is artificially enhanced by the fact that the MAb neutralizes only a few very sensitive isolates and has no detectable activity against more resistant isolates.
(ii) Conformational masking of Env epitopes.
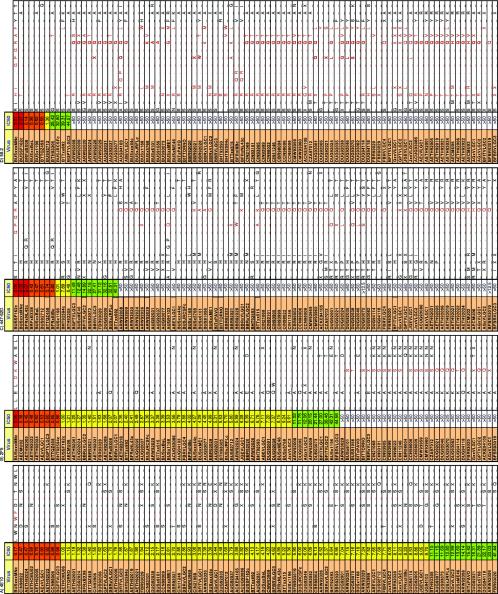
In Fig. 8, we present sequence data relevant to the neutralization of MAbs directed to linear epitopes (4E10, 2F5, 447-52D, and 58.2). This helps define the minimal epitope for recognition by these MAbs. It is clear that, while substitutions in the conserved core epitopes can predict neutralization resistance, the presence of the correct core sequence cannot predict neutralization. Instead, distal regions of Env may influence neutralization potency. For example, the presence of the correct V3 sequence motifs resulted in 447-52D neutralization of 20 of 32 (62%) GPGR-containing viruses. Neither 58.2 nor 447-52D neutralizes BUSTORNO, BBRVLGC2, BFBRVLGC1, or BFBRVLGC3, despite the presence of HIGPGRAF at the tip of the V3 loop. This is probably because the V3 loop is frequently sequestered in neutralization-resistant isolates (13, 146). Conformational masking is not limited to MAbs directed to linear epitopes; those that recognize discontinuous epitopes, like the CD4 binding site and CD4-induced epitopes, have very well-conserved epitopes that are frequently subject to steric occlusion. A more detailed analysis of the relationship of Env sequence polymorphisms to MAb neutralization, particularly for those that recognize conformational epitopes, will be presented elsewhere.
FIG. 8.
Mapping the sequence requirements for neutralization by MAbs to linear epitopes. Env sequences from each virus are depicted to show the epitopes of gp41 MAbs 4E10 (amino acids 667 to 677) (A) and 2F5 (amino acids 658 to 667) (B) and V3 MAbs 447-52D (C) and 58.2 (amino acids 306 to 320 are shown) (D). For each MAb, to determine which amino acid substitutions are permissible, viruses that were most potently neutralized were organized at the top of each column and those that were not neutralized were placed at the bottom. Those residues that appear to be important for MAb binding are highlighted in red. A dash indicates consistency with the sequence at the top of each figure. A dot indicates a deletion. An X represents a residue where there is variation at an amino acid position. Definitive sequencing of ARW92021 was not possible due to extensive sequence heterogeneity in the quasispecies.
(iii) Analysis of the neutralizing activity of each MAb. (a) 4E10.
The most remarkable finding from our analysis is that MAb 4E10 neutralized all 93 viruses with IC50s <50 μg/ml in the pseudovirus assay (Fig. 4). This is consistent with, and significantly expands, a recent study in which 4E10 neutralized an entire panel of clade B viruses in a pseudovirus neutralization assay (72). The neutralization observed is specific, as confirmed by the lack of nonspecific activity against the amphotropic MuLV control. Although panreactive, 4E10 is only modestly potent; rather than neutralizing a few viruses very potently, 4E10 consistently neutralizes viruses with an even potency. For example, it does not neutralize sensitive clade B isolates as potently as some of the other MAbs (the average IC50 of 4E10 against clade B viruses is 7.4 μg/ml; for b12, it is 2.6 μg/ml; for 2G12, it is 3.5 μg/ml; for 2F5, it is 6.3 μg/ml). The sequence data (Fig. 8A) indicate the minimum requirements for 4E10 recognition. Previously, 4E10 was mapped to an epitope in gp41, NWFDIT (Env amino acid positions 671 to 676), with some sequence promiscuity at positions 671, 674, and 676 (134, 153). Given the present data, we can now further define the WFXI motif as the minimum core requirement for 4E10 neutralization. This motif is preserved in 2,828 of 2,894 (98%) M group HIV-1 sequences that span this portion of gp41 presently available in the Los Alamos HIV database (http://www.hiv.lanl.gov) and is highly conserved in all pure-clade and recombinant forms. A variant with an F→L substitution at position 673 is the most common variant; it is predominant in the O (outlier) virus group and was found in 31 of 2,894 (1%) M group sequences. However, the precise sequence of the 4E10 epitope in each virus is not the only factor that determines the strength of neutralization. For example, while BUSxxxMNc is neutralized with an IC50 of 0.17 μg/ml, BUS93073, bearing an identical 4E10 epitope sequence, is neutralized with an IC50 of 11.7 μg/ml (i.e., >67-fold weaker neutralization). This suggests that expression of the 4E10 epitope is dependent on structural context.
(b) 2F5.
MAb 2F5 is also broadly neutralizing, although not to the extent of 4E10. Our data confirm previous studies that showed that 2F5 can neutralize viruses from clades A, B, D, and E (35, 36, 114, 134, 138). However, 2F5 had no activity against any of the clade C viruses tested. Scant activity was found against clade D viruses. Sequence information (Fig. 8B), together with previous peptide-mapping studies, defines the minimal requirement for 2F5 recognition as DKW (Env positions 664 to 666 in gp41). Many viruses resistant to 2F5 neutralization had a substitution in the DKW motif. For example, most clade C viruses had DSW in place of the DKW motif. The lack of clade C virus neutralization in our panel does not rule out the possibility that 2F5 may occasionally neutralize viruses from this clade. For example, it was previously shown that 2F5 neutralized a Zambian isolate, ZAM20 (138). However, ZAM20 has an unusual 2F5 epitope for a clade C virus (ALDKWN). Similarly, 2F5 was able to neutralize the AC hybrid ACUG93086. This is consistent with the entire gp41 segment of this hybrid originating from a clade A isolate (2F5 epitope sequence, ALDKWA). In the Los Alamos HIV database, the motif DKW is preserved in only 54 of 212 (25%) clade C gp41 sequences sampled from 19 nations. The 2F5-reactive motif, DKW, is enriched in certain regional clade C samples; for example, it was found in 15 of 17 (88%) clade C sequences from Burundi, 6 of 6 (100%) sequences from Brazil, and 8 of 10 (80%) sequences from Ethiopia, while it is very rare in other countries (for example, it was found in only 1 of 71 [1%] clade C sequences from Botswana). It is very likely that there is a geographic lineage association with 2F5 susceptibility.
2F5 neutralized only 5 of 11 (45%) clade D viruses, while the motif DKW is present in 64 of 91 (70%) clade D virus gp41 sequences available in the HIV sequence database. The clade D viruses available for this study were all derived from Ugandan samples, and those that were neutralized by 2F5 (5 of 11) all exhibited the DKW motif, whereas the remaining 6 exhibited a DQW motif. In the sequences from the database, 41 of 67 (61%) Ugandan samples carried DKW, while outside of Uganda, almost all available clade D viruses—22 of 23 (96%) clade D gp41 sequences from nine nations—carried the DKW motif and so had the potential to react with 2F5. Thus, our relatively low clade D susceptibility may be a reflection of sampling from Uganda; clade D viruses from other regions are likely to be more susceptible. Overall, as with clade C viruses, there may be a geographic lineage association with the observed 2F5 susceptibility among clade D viruses.
As with all the MAbs to linear epitopes, the presence of the target sequence did not always predict 2F5 neutralization: the exceptions here were the Brazilian isolates BFBR93019, BFR93020, and CBR98004; the Rwandan isolate ARW93029; and BITxPAVO. The DKW epitope motif may be conformationally masked in these viruses, so that neutralization was not detectable at <50 μg/ml. Overall, widely different neutralization potencies against viruses with the same 2F5 epitope sequence were observed (Fig. 8).
(c) b12.
b12 was also broadly and potently neutralizing (Fig. 4 and 5) but not quite as broadly neutralizing as 2F5, and the distribution of neutralized viruses was clearly quite different. b12's activity was distributed more evenly across clades. Whereas b12 neutralized a fraction of viruses from almost every clade and geographic location, consistent with previous findings (138), 2F5 neutralized most viruses from some clades but was very poor against others. b12 was also significantly more potent than 2F5 and 4E10 against many virus isolates, especially those from clade B (as noted above).
(d) b6.
b6 is a prototype “nonneutralizing” CD4bs-specific MAb derived from the same HIV+ human donor as b12 (5, 19, 121). Inclusion of this MAb provides a benchmark for the activity of “garden variety” CD4bs MAbs. In the single-round assay, b6 did not neutralize any isolate that was not neutralized by b12, and it was never as potent. Here, we found that b6 neutralizes only a small number of highly neutralizationsensitive clade B primary viruses and T-cell line-adapted virus strains (BUSx1196, BUSxxBaL, BFRxBX08, BTTQZ458, BUSSF162, BFRxIIIBc, BFRxNL43c, and BUSxxxMNc). It is likely that steric restrictions limit the access of this MAb to its epitope on the vast majority of primary viruses (13, 63, 121, 127).
(e) 2G12.
MAb 2G12 neutralized a proportion of isolates in our panel similar to that neutralized by b12 but with a pattern that was more restricted by clade (Fig. 4 and 5). It had no activity against clade C or E isolates in our panel. This is consistent with, and expands, previous findings (17, 35, 134, 138). 2G12 is thought to recognize glycans at residues 332 and 392, with some dependence on glycans at positions 295, 339, and 386 (23, 126, 129). The lack of clade C neutralization may in some cases be due to a loss of the glycan at position 295 at the C-terminal base of the V3 loop. In our group of 12 clade C viruses, the only one that retains this site is CBR98004 (8%); in the Los Alamos database, a somewhat higher fraction (474 of 2,042 [24%]) of C clade viruses retain this glycosylation site. The lack of clade C neutralization may also be due to the loss of the N-linked glycan at position 339 in viruses CIN98022, CZA97012, and CBR98004 (25% of our viruses). In the database (and in our group of clade C viruses), the glycan at position 339 is fairly well conserved among clade C Env sequences (664 of 891 [75%]), comparable to its level of retention in most other clades (149). Since CBR98004 retains the glycan at position 295, it is likely that the loss of one at position 339 leads to loss of 2G12 reactivity. Overall, the lack of a glycan at position 295 appears to be most important in explaining the lack of clade C neutralization. In contrast to its lack of neutralization of pure clade C viruses, 2G12 does neutralize the clade AC hybrids ACKE94105 and ACUG93086 in which the glycosylation sites at 295 and 339 are retained.
2G12 does not neutralize CRF01 clade AE viruses, perhaps due to an N-to-E mutation at position 332 at the C-terminal side of the V3 loop that abolishes a critical glycan in the epitope. The lack of clade AE neutralization may also be partly explained by the presence of an additional disulfide bond in the V4 region (129). Overall, with very few exceptions, it appears that when the residues important for 2G12 are present, neutralization is predictable. This may be because the 2G12 epitope is fully exposed on the outer domain of gp120 and not subject to conformational masking as MAbs to other epitopes appear to be.
(f) 447-52D.
The V3 loop MAb 447-52D neutralizes 45% of the clade B viruses tested at IC50 <50 μg/ml (Fig. 4 and 5) but only 14% at IC90 <50 μg/ml (Fig. 6 and 7). It is particularly effective against the globally most sensitive viruses (35, 91).
MAb 447-52D has been reported to occasionally neutralize some viruses from non-B clades, including clades A, D, and F (45, 48, 49, 101). However, in our panel, neutralization outside clade B was rare (4 of 61 [∼7%]). In one case (isolate BFITVLGC2), 447-52D neutralized when no other anti-gp120 MAbs neutralized. Alignment of the V3 loop sequences confirms that the 447-52D core epitope is GPGR (Fig. 8C) (28, 45). While the dominant V3 sequence motif in clade B viruses is GPGR (appearing in 1,272 of 1,911 clade B V3 sequences [67%]), it appears in only 140 of 1,753 (8%) non-clade B M group viruses, where the dominant V3 sequence motif is GPGQ (42, 43). Therefore, 447-52D activity against non-B viruses depends on the unusual presence of a GPGR V3 sequence (45, 49).
BUSJRCSFc virus was found to be resistant to 447-52D here, but in other studies it was sensitive to this MAb (47). This might be explained by two amino acid differences (503 R/A and 674 G/D) in distal locations between the BUSJRCSFc clone used here and the previously published sequence. However, it is not immediately clear how such mutations in the gp120 C5 domain and the C terminus of the gp41 ectodomain might influence neutralization by V3 loop MAbs. As an alternative explanation, it is possible that the particular BUSJRCSFc virus stock used in previous studies underwent serial PBMC passages that rendered it more sensitive to V3 MAb neutralization (8, 113).
(g) 58.2.
Like 447-52D, MAb 58.2 neutralized a significant proportion of clade B primary isolates but did not neutralize any viruses outside clade B (Fig. 4 and 5). It is narrower in breadth than 447-52D but not remarkably so. V3 loop sequencing suggested that its minimal epitope is (H/T)IGPGR(A/T)(F/L) (Fig. 8D).
(h) X5.
X5 is a MAb whose epitope on gp120 is preferentially exposed by engagement of CD4; in other words, its epitope is CD4 induced (CD4i). Like b6, X5 neutralized only highly sensitive clade B isolates, as described recently (66). The CD4i epitope therefore appears to be accessible in neutralization-sensitive isolates, but it appears to be sterically occluded in most primary isolates. However, smaller fragments of CD4i MAbs are known to be able to neutralize primary isolates by overcoming the steric restriction (66). Unlike the V3 loop, the CD4i epitope is well conserved, since it overlaps the coreceptor binding site. X5 was able to neutralize 2 of 25 non-B isolates in the PBMC assay (Fig. 2). It was not able to neutralize these viruses in the pseudovirus assay, perhaps due to differential coreceptor expression, as discussed above.
(i) The five-MAb cocktail.
It has been questioned whether MAbs to different epitope clusters can synergize in achieving greater neutralization potency or breadth. We found that a cocktail of five MAbs (b12, 2G12, 447-52D, 2F5, and 4E10) at an equimolar ratio neutralized all viruses with an IC50 <50 μg/ml. This was not surprising, considering that the cocktail includes 4E10, which neutralizes every virus. The IC50 of the IgG cocktail in each case was similar to the average IC50 of its component MAbs (adjusting for the fact that in the five-MAb cocktail, any one MAb concentration is reduced to 20%), indicating a lack of obvious synergy. The IC90 data (Fig. 6 and 7) allow examination of the activity of the cocktail at a level of stringency where 4E10 is no longer panneutralizing. Here again, however, there is no evidence for strong synergistic neutralization, nor was there a significant improvement in breadth. Although the effects of the cocktail appear to be near additive, our analysis is not sophisticated enough to rule out the possibility of subtle synergistic effects.
(j) N16 HIV+ plasma.
Plasma from an HIV-1-positive donor infected with a clade B virus exhibited weak neutralization of a significant fraction of viruses from all clades (IC50s were generally below 1:100 plasma dilution). As reported previously by other groups (6, 10, 43, 58, 59, 71, 75, 90, 93, 102, 145), there is a great breadth of response in interclade (non-B) reactions. However, there was a modest preference for clade B neutralization: excluding the sensitive viruses HXB2r, MN, SF162, and NL4-3, 22 of 28 (79%) clade B viruses were neutralized, while 40 of 61 (66%) non-clade B viruses were neutralized. The neutralization potency of the plasma within clade B was also statistically higher. If we exclude the four sensitive clade B isolates, using a conservative nonparametric Wilcoxon test, the clade B data have a distribution of IC50s that is higher, with a median IC50 titer of 72 and an interquartile range of 50 to 120 compared to the non-clade B median IC50 of 60 with an interquartile range of 43 to 89 (P = 0.042). Therefore, there is an indication of somewhat higher intraclade neutralization.
Neutralization of viruses bearing interclade recombinant Envs.
Our virus panel included several viruses with interclade recombinant Envs (Fig. 1). We found that the neutralization profiles of these viruses were largely predictable from the compositions of the hybrid Envs. The BF and BG recombinants were entirely composed of Env ectodomains of clade B, the junction with another Env clade arising either in the gp41 transmembrane domain or the cytoplasmic tail (Fig. 1). Accordingly, their neutralization profiles were similar to those of clade B viruses. Similarly, the two AG recombinants behaved like clade A viruses, with the possible exception that AGxVLGC1 is not susceptible to 2F5. Since the breakpoint between clade A and G Env proteins is close to this epitope, clade G sequence could explain this 2F5 resistance, a point that could not be verified in the sequence due to some heterogeneity in the DKW epitope (Fig. 8B). The AC recombinants ACUG93086 and ACKE94105 are true Env hybrids and appear to have unique neutralization profiles. Unlike the pure clade C viruses, as mentioned above, they are sensitive to 2G12. Also, despite having an entirely clade A gp41, ACKE94105 still resists 2F5, due to a mutation in its core epitope (Fig. 8). It is difficult to comment on the behavior of AE recombinants in the absence of any pure clade E viruses to compare. However, these viruses do not behave like clade A viruses, in keeping with the fact that most of their Env sequence derives from clade E parents.
Two-dimensional hierarchical clustering of viruses and antibodies based on IC50 neutralization data.
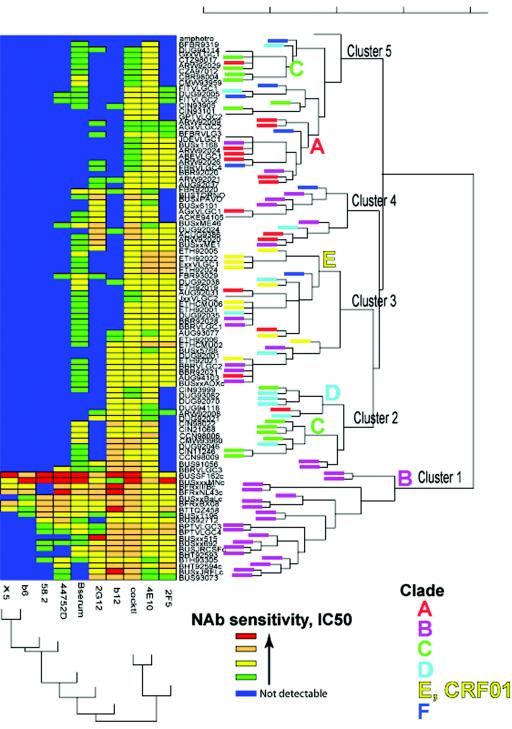
Dendrograms were constructed (Fig. 9) in which viruses were clustered according to their profiles of neutralization susceptibility (IC50 data) to the panel of antibodies, and antibodies were clustered according to their patterns of reactivity with the panel of viruses, using a two-dimensional clustering strategy (51). To our knowledge, this represents the first attempt to classify viruses based on their levels of neutralization sensitivity to MAbs. The viruses segregated into relatively clear clusters, 1 to 5 (Fig. 3). The most common clades in this study, A to F, were assigned colors, indicated on the end of each branch in the dendrogram. When there is no colored box on the end of a branch, this is because the virus was either from one of the clades not represented by many samples in this study or was a recombinant.
FIG. 9.
Two-dimensional hierarchical clustering of viruses and MAbs based on IC50 neutralization profiles. Viruses were clustered according to their neutralization profiles along the vertical axis. Simultaneously, the MAbs were arranged according to their abilities to neutralize the panel of viruses. Dendrogram patterns are shown to the right (for viruses) and bottom (for MAbs). The same color scheme for neutralization data points as in Fig. 2 was used. The most common clades in this study, A to F, were assigned a color, indicated on the end of the branches in the dendrograms. No color was assigned for viruses from clades that were not represented by many samples in this study or for interclade recombinants.
The dendrogram revealed that MAb neutralization patterns are somewhat associated with clades, although imperfectly. Statistical analysis confirmed that the clade association with IC50 neutralization clusters is highly significant (using a chi-square test, P = 10−15). This test was used to test the null hypothesis that viruses were randomly distributed among the different clusters defined on the basis of neutralization susceptibility. Only clade B is found in cluster 1, the cluster that defines the group of viruses that are most susceptible to neutralization. This may be in part because clade B infections gave rise to all the antibodies under investigation. Considering the fundamental tenets of immune reactivity, it might be expected that these MAbs would be most reactive with clade B viruses. The presence of clade B Envs in cluster I may also be partly attributed to the fact that some of the clade B isolates are, in general, particularly neutralization sensitive, perhaps because some of them have been cultured more extensively in vitro or selected and cloned, a process that tends to render viruses more neutralization sensitive.
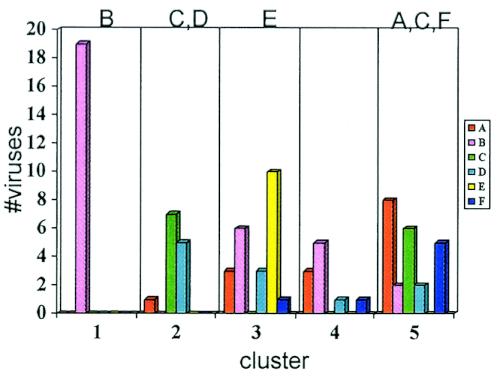
It could be argued that the association between clades and neutralization susceptibility profiles is solely driven by clade B viruses in cluster 1, especially since clusters 2 to 5 contain viruses from more than one clade. Therefore, we performed a chi-square test on the distribution of non-clade B viruses in clusters 2 to 5. Here, the association remained highly significant (P = 0.00002). In Fig. 10, the clades and neutralizing clusters have been plotted histogramatically using the information from Fig. 9. It can be seen more clearly that there is an association of clade with cluster. Cluster 2 is formed mostly from clade C and D viruses and is characterized by resistance to 2G12 and 2F5. All CRF01 AE viruses (10 of 10) are found in cluster 3, with some contributions from A and B viruses. Resistance to 2G12 but sensitivity to 2F5 and the clade B plasma characterize this cluster. Cluster 4 contains viruses from several clades. Cluster 5 includes clade A, C, and F viruses, and five of seven of the clade F viruses are in this cluster. Cluster 5 generally shows resistance to b12 and 2G12. The only geographic trend readily apparent in the clustering data was that no clade B viruses from Brazil appeared in the highly reactive cluster 1; four of five of the Brazilian B viruses were located in cluster 3 and were distinguishable from other B viruses in being quantitatively less susceptible to 2F5, 4E10, and b12 and resistant to 2G12.
FIG. 10.
Neutralization patterns are associated with clades. The total number of viruses of each clade in each neutralization immunotype cluster are shown.
Clustering based on IC90 neutralization profiles (data not shown) was less informative than the IC50 profile, in part because there were so many reactions that had undetectable neutralization. Still, some patterns are clearly evident: clade C viruses tended to be sensitive to 4E10, and clade E (CRF01) tended to be sensitive to both 4E10 and 2F5. Many clade A, D, and F viruses could not be neutralized by any MAbs, even at the highest concentration tested. Among these three clades, those that could be neutralized by one or two of the MAbs showed sporadic, clade-independent sensitivity to particular MAbs.
DISCUSSION
Comparison of the PBMC and pseudovirus neutralization assays.
We observed reasonable agreement between the pseudovirus and PBMC neutralization assays reported here (Fig. 2) and elsewhere (17, 48, 49, 134, 153). Our results lend support to the pseudovirus assay as a useful and relevant approach for measuring neutralization. In an unpublished multilaboratory blinded comparison of the neutralization activities of several MAbs and HIV+ sera in various neutralization assays (D. Montefiori, personal communication), the pseudovirus assay also showed good concordance with PBMC neutralization assays, although there were some differences in assay sensitivities. We found the greatest discrepancies between the two assays for the MAbs 4E10 and X5. As discussed below, it is possible that these inconsistencies are related to different virus infection kinetics with the target cells used in each assay, as reported by Reeves et al. (118), or perhaps to differences in the nature of the viruses used.
(i) Discrepancy in 4E10 neutralization.
A significant observation was that 4E10 neutralized all the viruses in our panel in the pseudovirus assay (Fig. 4). However, in PBMC assays here (Fig. 2) and elsewhere, 4E10 did not neutralize all viruses (134, 153). To explain this discrepancy, it could be argued that the PBMC assay is not sensitive enough to detect 4E10 neutralization, especially given that PBMC assay neutralization is usually reported at IC90. Alternatively, it could be argued that the ability of 4E10 to neutralize after CD4 and CCR5 binding (postattachment) (11) may make it particularly sensitive to the receptor expression levels or the nature of the target cell types. Notably, MAb 2F5, which has also been suggested to be capable of some virus neutralization after CD4 and CCR5 engagement, neutralized equivalently in both neutralization formats. Overall, the basis of the differential activity of 4E10 in these neutralization assays remains unresolved. The panneutralizing activity of 4E10 in the pseudovirus assay suggests that it recognizes a very highly conserved site on the viral Env protein that may be essential for virus replication; this is consistent with the preservation of the core epitope for 4E10 in 98% of the sequences in the HIV database (however, 4E10 does not neutralize SIVmac239 and SIVmac316 [J. Binley, unpublished observations]). For this reason, it would be of interest to determine if escape from 4E10 neutralization is possible, and if so, how this affects the resistant virus.
(ii) Discrepancy in X5 neutralization.
In contrast to 4E10, MAb X5 exhibited neutralization in the PBMC assay but not in the pseudovirus assay (Fig. 2). X5 recognizes Env that has engaged CD4 but not coreceptor (66). Differing coreceptor densities on the target cells could affect the window of time during which this epitope is available. X5 neutralization in the pseudovirus assay was limited, perhaps because of the relatively high levels of coreceptor on the U87 target cells (C. J. Petropoulos and T. Wrin, preliminary data) that increase infection kinetics and decrease the opportunity for X5 neutralization.
(iii) Other discrepancies.
Other discrepancies occurred in which there was neutralization in the pseudovirus assay but not in the PBMC assay (Fig. 2). These might be explained by differences in Env sequence in the pseudovirion and PBMC-derived virus populations, which might be related to the passage history of the viruses and Envs used. It is known that prolonged PBMC culturing renders primary viruses progressively more neutralization sensitive (8, 113). The Env on the surfaces of the pseudovirions may be derived from a molecular or biological clone, low- or high-passage PBMC stock, or circulating virus that may differ somewhat from the PBMC-derived live-virus stock to which it is compared in the other neutralization assay format. It is also possible to explain some discrepancies by the fact that some viruses have relatively rapid replication kinetics in the PBMC assay, and in these cases, the nonneutralized virus subpopulation may rebound significantly by the time of harvesting on day 7.
There are also some differences between our PBMC assay results and PBMC assay results reported previously. In an earlier report, 4E10 neutralized isolate ARW92021 potently (134), as it did in the pseudovirus assay here. However, we did not observe neutralization in our PBMC assays (Fig. 2). This may be due to differences in the assay sensitivity or passage history.
It has been reported that b12 can, at certain concentrations, lead to enhancement of virus infection (usually <40%) (104). However, we did not observe any infectivity enhancement in pseudovirus neutralization assays. Although enhancement might arise with particular clade A Envs (104), it is more likely to be a phenomenon associated with the conditions of neutralization assays, for example, the receptor density on the target cells.
Comparison of neutralization against gp120 and gp41 epitopes.
Our results highlight a dichotomy in the behavior of the anti-gp120 and anti-gp41 MAbs. Compared to the anti-gp120 MAbs, the anti-gp41 MAbs neutralize more evenly, with relatively greater breadth but with a somewhat lower potency. The greater breadth of the gp41 MAbs might be anticipated, given the conservation of their epitopes. Their relatively modest potencies, even against neutralization-sensitive clade B viruses, may originate from their recognition of structures that have limited accessibility on native Env trimers (153) with possibly somewhat better exposure at a postattachment stage of virus entry (1, 11). The limited availability of the gp41 epitopes may be responsible for a reduced MAb on rate, decreased affinity, and lower neutralization potency. Considering the impressive breadth of neutralization, it would be of interest to try to increase the neutralization potency of 4E10 by recombinant methods to increase its affinity.
Neutralization-resistant isolates.
A previous study identified isolates resistant to neutralization by 2G12, 2F5, b12, and HIV-1 patient sera (109). Here, we also identified several resistant isolates. In the PBMC assay, 7 of the 25 viruses were not neutralized by any of the NAbs (Fig. 2). Therefore, even the broadest of the NAbs are unable to neutralize all isolates in a PBMC assay. Five of the seven resistant isolates were non-clade B. The only two clade B isolates that were resistant to neutralization (IC90) by all MAbs in the PBMC assay were BBR92020 and BBR92021. In the case of BBR92020, the presence of a substitution (boldface) adjacent to the 2F5 epitope, ELDKWDS, may contribute to its resistance.
Assay recommendations.
One of the most significant issues relating to neutralization is assay standardization, especially when evaluating immune sera from vaccine trials. This is important so that data from different studies can be compared in a meaningful fashion. Variations in protocol may lead to different neutralization titers (35, 128), as seen here in comparing the pseudovirus and PBMC assays. Here, the use of a CD4+ CCR5+ CXCR4+ cell line and pseudoviruses gave qualitative results reasonably similar to those of the PBMC assay (they differed in about one-third of the tests performed), but the pseudovirus assay offers greater assay consistency and convenience, allowing an accurate measurement of titer and greater scope for comparative analyses.
Ultimately, the most important factor to consider when assessing neutralization in vitro is its relevance to protection in vivo. Virus challenge studies in animal models indicate that protection may require much higher concentrations of antibody than that required to achieve, for example, an IC90 in a PBMC assay (44, 73, 74, 79, 107). While the pseudovirus assay has the sensitivity to allow MAb comparisons, great care needs to be taken not to read too much into the absolute titers until we can correlate specific pseudovirus assay titers with passive protection in relevant animal challenge models.
In some assays, “neutralization” may result from a nonspecific cytotoxic effect of the antibody sample on the target cells. False-positive neutralization can be a significant problem when assessing serum or plasma neutralization at high concentrations. The use of the amphotropic MuLV as a control in the pseudotyped neutralization assay reported here helps to eliminate this problem. Any neutralization against this control virus is really a direct measure of toxicity that should be taken into account when considering neutralization against HIV-1 isolates. There is no convenient equivalent in the PBMC assay, making the pseudovirus assay particularly valuable in this regard.
It has been the accepted norm recently to report IC90 neutralization titers in PBMC neutralization assays (134, 153). This is because the high degree of experimental scatter associated with PBMC assays requires a more stringent cutoff point. However, in the pseudovirus assay, IC50s may be more sensible, because the neutralization curves are more reproducible. Since the IC50 is taken at the inflection of a sigmoid curve, it can be estimated with much greater confidence than the IC90, which approaches an asymptote and is therefore more prone to error. In addition, to detect subtler levels of potency and comparisons of cross-reactivity, the IC50 provides more comprehensive information (fewer responses below detection level). This point is illustrated by the fact that the clade associations with reactivity patterns are clear using the IC50 data (Fig. 4, 5, 9, and 10) but are hard to distinguish using the IC90 data, as negative reactions tend to dominate outside of clade B (Fig. 6 and 7).
Differences in neutralization assay sensitivities due to variations in reagents and protocols have historically made it very difficult to compare the activities of MAbs and sera described by different groups (77, 128). For example, we compared the pseudovirus assay using two protocols, with either a 1- or an 18-h virus-antibody incubation. We found that neutralization in the 18-h format was generally ∼6-fold more sensitive (Fig. 3). While high sensitivity is useful up to a point, the increase in sensitivity is not always predictable, and there are sometimes much larger discrepancies between 1- and 18-h formats than would normally be expected. We suggest the continued use of the traditional 1-h virus-Ab incubation.
An important step toward standardizing neutralization assays is to use common positive controls so that neutralization can be measured relative to a known standard. This provides a measure of assay sensitivity in context with previously published data. Possible MAbs useful for this purpose include the broadly reactive MAbs b12, 2G12, 2F5, and 4E10, and perhaps an equimolar MAb cocktail.
In the present analysis, we were forced to make decisions about how to categorize neutralization potencies and where to make the neutralization cutoff. Our chosen cutoff of 50 μg/ml was picked arbitrarily and is not to be taken as an absolute but rather as a guide. However, MAb concentrations higher than this are probably not relevant in natural infection or immunization, in which the total polyclonal anti-Env Ab concentrations are typically ∼100 to 1,000 μg/ml (12). Overall, the sensitivity and reproducibility of the pseudovirus neutralization assay (120) makes it particularly attractive, and the use of carefully standardized reagents and protocols eliminates inaccuracies and adds greater confidence in the validity of the results.
Synergy.
We determined if a five-MAb cocktail offered greater neutralization breadth or potency. Several groups have investigated whether MAbs against different epitope clusters achieve greater neutralization potency in combination than would be predicted by simple additive effects. Synergy was observed in some scenarios (2, 15, 65, 67, 68, 76, 83, 86, 135, 136, 143, 148, 154) but has not been universal (76, 142). A review of the literature reveals several complexities and contradictions. Some of the present authors previously observed weak synergy using cocktails of three or more MAbs against primary isolates but not T-cell line-adapted isolates (68, 154). Others observed synergistic effects on T-cell line-adapted isolates but not primary isolates (142). One study reported synergy only when MAbs were used at high concentrations (77). In other cases, synergy was manifest when unequal concentrations of MAbs were tested (154). Given the algorithms required to measure synergy and the relative inaccuracies incumbent with PBMC-based assays, a definitive study of neutralization synergy is still lacking. Our data indicated no strong synergy: neutralization by the five-MAb cocktail in each case was largely determined by the most active of the five MAbs. Our analysis of the equimolar five-MAb cocktail was designed only to detect strong synergy similar to that observed previously between CD4-IgG2 and the T-20 peptide (100) but was not sophisticated enough to detect weak synergy. Overall, we can conclude that if synergy among these MAbs does exist, it is likely to be quite limited.
Our MAb cocktail analysis was also intended to determine whether neutralization breadth could be improved. However, since 4E10 neutralizes every virus for which there is an IC50, it was not surprising that the cocktail also neutralized all the viruses. At IC90, although 4E10 is no longer panneutralizing, the situation was a little different (Fig. 6 and 7): the cocktail neutralized only four more viruses than 4E10 alone. It was previously reported (57) that a cocktail of b12, 2G12, 2F5, and 4E10 broadly neutralizes clade A, B, C, and D viruses. However, our results, consistent with other observations (17), indicate that of the MAbs tested, only b12 and 4E10 are capable of neutralizing the majority of clade C viruses, with little or no contribution from MAbs 2G12 and 2F5. This has important implications for passive-immunization studies of mother-child transmission in Africa (124, 148), since the inclusion of 2F5 and 2G12 in MAb cocktails against these viruses may be largely ineffective.
Neutralizing immunotypes and the importance of clade for vaccines.
Here, we grouped viruses into immunotypes based on their susceptibilities to a panel of neutralizing MAbs and one HIV+ plasma. We identified five distinct immunotype clusters that were highly significantly, albeit imperfectly, associated with clades. This was unexpected and perhaps controversial, considering that several previous studies have found little evidence of an association of HIV-1 immunotypes with genetic clades (6, 10, 43, 58, 59, 71, 90, 93, 102, 145). This raises the question of why we found something different here. The basis for this discrepancy is probably related to the exact kind of immunotype being studied, the number of viral isolates and clades being compared, and the reliability of the assay(s) being used. Immunotyping analyses using HIV+ sera have in general not found clade-specific neutralization patterns (6, 10, 43, 58, 59, 71, 75, 90, 93, 102, 145). However, practical limitations on the number of samples, the polyclonal nature of sera, the typically low serum neutralizing titers, background noise and limited sampling, and the difficulty of controlling for false-positive neutralization data have meant that it has been difficult to conduct a definitive analysis. Previous studies describing immunotyping with MAbs were mostly based on binding and not on neutralization (99, 101, 151). Interestingly, one study of immunotypes based on binding (rather than neutralization) of a panel of MAbs to the V3 loop, C5 region, or gp41 immunodominant loop, all known to be either weakly or nonneutralizing, resulted in three immunotype clusters (99).
The present analysis differs from previous studies in several respects. We defined viral immunotypes with neutralizing MAbs (and one serum). The use of MAbs rather than polyclonal sera for immunotyping probably increased the likelihood of finding distinct patterns. In addition, our analysis was empowered with more data than any other and with a highly reproducible and reliable assay. We also controlled false positives using the MuLV pseudovirus. This is an especially important consideration when using sera or plasma that can sometimes give false positives due to cytotoxicity. With these considerations in mind, the description of clade-specific immunotypes can be reconciled with those of previous studies.
What impact, if any, do the newly defined immunotypes have on the debate over whether clades are an important consideration for design of vaccine candidates? Certainly, a vaccine able to elicit antibodies similar to b12, 2G12, 2F5, 447-52D, and 4E10 would offer significant cross-clade neutralization crucial for a successful vaccine. All these antibodies arose following immunization (through infection) with clade B Env. However, the current study does reveal gaps in the cross-clade neutralization coverage. Neither 2G12 nor 2F5 neutralized the clade C viruses studied. 2G12 did not neutralize the AE recombinant viruses. The V3 MAbs that neutralized sensitive clade B viruses were dependent on a GPGR sequence at the crown of the loop and did not neutralize non-clade B viruses expressing a GPGQ sequence. Therefore, in seeking immunogens that will elicit neutralizing antibodies with the greatest breadth against global HIV-1, it would seem valuable to study neutralizing responses in non-clade B-infected individuals. In particular, broadly neutralizing MAbs derived from such individuals would be useful reagents for further exploration of neutralization serotypes and to assist in vaccine design.
Acknowledgments
We thank the IAVI Neutralizing Antibody Consortium and the Pendleton Trust for financial support. D.R.B. acknowledges NIH grant AI33292. M.B.Z. is grateful for support from a fellowship from the Elizabeth Glaser Pediatric AIDS Foundation. S.Z.-P. acknowledges grants HL 59725 and the NYU CFAR Immunology Core support by AI 27742.
We are grateful to James Rusche of Repligen for providing MAb 58.2 and James Theiler of Los Alamos National Laboratory for statistical advice. We thank Lon Trinh, Randi Spears, and Eric Lam for technical support.
REFERENCES
- 1.Abrahamyan, L. G., R. M. Markosyan, J. P. Moore, F. S. Cohen, and G. B. Melikyan. 2003. Human immunodeficiency virus type 1 Env with an intersubunit disulfide bond engages coreceptors but requires bond reduction after engagement to induce fusion. J. Virol. 77:5829-5836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Allaway, G. P., A. M. Ryder, G. A. Beaudry, and P. J. Maddon. 1993. Synergistic inhibition of HIV-1 envelope-mediated cell fusion by CD4-based molecules in combination with antibodies to gp120 or gp41. AIDS Res. Hum. Retrovir. 9:581-587. [DOI] [PubMed] [Google Scholar]
- 3.Anonymous.1994. HIV type 1 variation in World Health Organization-sponsored vaccine evaluation sites: genetic screening, sequence analysis, and preliminary biological characterization of selected viral strains. AIDS Res. Hum. Retrovir. 10:1327-1343. [DOI] [PubMed] [Google Scholar]
- 4.Baba, T. W., V. Liska, R. Hofmann-Lehmann, J. Vlasak, W. Xu, S. Ayehunie, L. A. Cavacini, M. R. Posner, H. Katinger, G. Stiegler, B. J. Bernacky, T. A. Rizvi, R. Schmidt, L. R. Hill, M. E. Keeling, Y. Lu, J. E. Wright, T. C. Chou, and R. M. Ruprecht. 2000. Human neutralizing monoclonal antibodies of the IgG1 subtype protect against mucosal simian-human immunodeficiency virus infection. Nat. Med. 6:200-206. [DOI] [PubMed] [Google Scholar]
- 5.Barbas, C. F. D., E. Bjorling, F. Chiodi, N. Dunlop, D. Cababa, T. M. Jones, S. L. Zebedee, M. A. Persson, P. L. Nara, E. Norrby, et al. 1992. Recombinant human Fab fragments neutralize human type 1 immunodeficiency virus in vitro. Proc. Natl. Acad. Sci. USA 89:9339-9343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Barin, F., S. Brunet, D. Brand, C. Moog, R. Peyre, F. Damond, P. Charneau, and F. Barre-Sinoussi. 2004. Interclade neutralization and enhancement of human immunodeficiency virus type 1 identified by an assay using HeLa cells expressing both CD4 receptor and CXCR4/CCR5 coreceptors. J. Infect. Dis. 189:322-327. [DOI] [PubMed] [Google Scholar]
- 7.Barnett, S. W., S. Lu, I. Srivastava, S. Cherpelis, A. Gettie, J. Blanchard, S. Wang, I. Mboudjeka, L. Leung, Y. Lian, A. Fong, C. Buckner, A. Ly, S. Hilt, J. Ulmer, C. T. Wild, J. R. Mascola, and L. Stamatatos. 2001. The ability of an oligomeric human immunodeficiency virus type 1 (HIV-1) envelope antigen to elicit neutralizing antibodies against primary HIV-1 isolates is improved following partial deletion of the second hypervariable region. J. Virol. 75:5526-5540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Beaumont, T., E. Quakkelaar, A. van Neunen, R. Pantophlet, and H. Schiutemaker. 2004. Increased sensitivity to CD4 binding site directed neutralization following in vitro propagation on primary lymphocytes of a neutralization-resistant human immunodeficiency virus IIIB strain isolated from an accidentally infected laboratory worker. J. Virol. 78:5651-5657 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Beddows, S., S. Louisirirotchanakul, R. Cheingsong-Popov, P. J. Easterbrook, P. Simmonds, and J. Weber. 1998. Neutralization of primary and T-cell line adapted isolates of human immunodeficiency virus type 1: role of V3-specific antibodies. J. Gen. Virol. 79:77-82. [DOI] [PubMed] [Google Scholar]
- 10.Beirnaert, E., P. Nyambi, B. Willems, L. Heyndrickx, R. Colebunders, W. Janssens, and G. van der Groen. 2000. Identification and characterization of sera from HIV-infected individuals with broad cross-neutralizing activity against group M (env clade A-H) and group O primary HIV-1 isolates. J. Med. Virol. 62:14-24. [PubMed] [Google Scholar]
- 11.Binley, J. M., C. S. Cayanan, C. Wiley, N. Schulke, W. C. Olson, and D. R. Burton. 2003. Redox-triggered infection by disulfide-shackled human immunodeficiency virus type 1 pseudovirions. J. Virol. 77:5678-5684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Binley, J. M., P. J. Klasse, Y. Cao, I. Jones, M. Markowitz, D. D. Ho, and J. P. Moore. 1997. Differential regulation of the antibody responses to Gag and Env proteins of human immunodeficiency virus type 1. J. Virol. 71:2799-2809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bou-Habib, D. C., G. Roderiquez, T. Oravecz, P. W. Berman, P. Lusso, and M. A. Norcross. 1994. Cryptic nature of envelope V3 region epitopes protects primary monocytotropic human immunodeficiency virus type 1 from antibody neutralization. J. Virol. 68:6006-6013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Buchacher, A., R. Predl, K. Strutzenberger, W. Steinfellner, A. Trkola, M. Purtscher, G. Gruber, C. Tauer, F. Steindl, A. Jungbauer, et al. 1994. Generation of human monoclonal antibodies against HIV-1 proteins; electrofusion and Epstein-Barr virus transformation for peripheral blood lymphocyte immortalization. AIDS Res. Hum. Retrovir. 10:359-369. [DOI] [PubMed] [Google Scholar]
- 15.Buchbinder, A., S. Zolla-Pazner, S. Karwowska, M. K. Gorny, and S. T. Burda. 1992. Synergy between human monoclonal antibodies to HIV extends their effective biologic activity against homologous and divergent strains. AIDS Res. Hum. Retrovir. 8:1395. [DOI] [PubMed] [Google Scholar]
- 16.Bures, R., A. Gaitan, T. Zhu, C. Graziosi, K. M. McGrath, J. Tartaglia, P. Caudrelier, R. El Habib, M. Klein, A. Lazzarin, D. M. Stablein, M. Deers, L. Corey, M. L. Greenberg, D. H. Schwartz, and D. C. Montefiori. 2000. Immunization with recombinant canarypox vectors expressing membrane-anchored glycoprotein 120 followed by glycoprotein 160 boosting fails to generate antibodies that neutralize R5 primary isolates of human immunodeficiency virus type 1. AIDS Res. Hum. Retrovir. 16:2019-2035. [DOI] [PubMed] [Google Scholar]
- 17.Bures, R., L. Morris, C. Williamson, G. Ramjee, M. Deers, S. A. Fiscus, S. Abdool-Karim, and D. C. Montefiori. 2002. Regional clustering of shared neutralization determinants on primary isolates of clade C human immunodeficiency virus type 1 from South Africa. J. Virol. 76:2233-2244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Burton, D. 1997. A vaccine for HIV type 1: the antibody perspective. Proc. Natl. Acad. Sci. USA 94:10018-10023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Burton, D., C. F. Barbas III, M. Persson, S. Koenig, R. Chanock, and R. Lerner. 1991. A large array of human monoclonal antibodies to type 1 human immunodeficiency virus from combinatorial libraries of asymptomatic seropositive individuals. Proc. Natl. Acad. Sci. USA 88:10134-10137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Burton, D., and D. Montefiori. 1997. The antibody response in HIV-1 infection. AIDS 11(Suppl. A):S87-S98. [PubMed] [Google Scholar]
- 21.Burton, D. R. 2002. Antibodies, viruses and vaccines. Nat. Rev. Immunol. 2:706-713. [DOI] [PubMed] [Google Scholar]
- 22.Burton, D. R., J. Pyati, R. Koduri, S. J. Sharp, G. B. Thornton, P. W. Parren, L. S. Sawyer, R. M. Hendry, N. Dunlop, P. L. Nara, et al. 1994. Efficient neutralization of primary isolates of HIV-1 by a recombinant human monoclonal antibody. Science 266:1024-1027. [DOI] [PubMed] [Google Scholar]
- 23.Calarese, D. A., C. N. Scanlan, M. B. Zwick, S. Deechongkit, Y. Mimura, R. Kunert, P. Zhu, M. R. Wormald, R. L. Stanfield, K. H. Roux, J. W. Kelly, P. M. Rudd, R. A. Dwek, H. Katinger, D. R. Burton, and I. A. Wilson. 2003. Antibody domain exchange is an immunological solution to carbohydrate cluster recognition. Science 300:2065-2071. [DOI] [PubMed] [Google Scholar]
- 24.Chen, M., M. K. Singh, R. Balachandran, and P. Gupta. 1997. Isolation and characterization of two divergent infectious molecular clones of HIV type 1 longitudinally obtained from a seropositive patient by a progressive amplification procedure. AIDS Res. Hum. Retrovir. 13:743-750. [DOI] [PubMed] [Google Scholar]
- 25.Cheng-Mayer, C., M. Quiroga, J. W. Tung, D. Dina, and J. A. Levy. 1990. Viral determinants of human immunodeficiency virus type 1 T-cell or macrophage tropism, cytopathogenicity, and CD4 antigen modulation. J. Virol. 64:4390-4398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Cleghorn, F. R., N. Jack, J. K. Carr, J. Edwards, B. Mahabir, A. Sill, C. B. McDanal, S. M. Connolly, D. Goodman, R. Q. Bennetts, T. R. O'Brien, K. J. Weinhold, C. Bartholomew, W. A. Blattner, and M. L. Greenberg. 2000. A distinctive clade B HIV type 1 is heterosexually transmitted in Trinidad and Tobago. Proc. Natl. Acad. Sci. USA 97:10532-10537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Coeffier, E., J. M. Clement, V. Cussac, N. Khodaei-Boorane, M. Jehanno, M. Rojas, A. Dridi, M. Latour, R. El Habib, F. Barre-Sinoussi, M. Hofnung, and C. Leclerc. 2000. Antigenicity and immunogenicity of the HIV-1 gp41 epitope ELDKWA inserted into permissive sites of the MalE protein. Vaccine 19:684-693. [DOI] [PubMed] [Google Scholar]
- 28.Conley, A. J., M. K. Gorny, J. A. Kessler II, L. J. Boots, M. Ossorio-Castro, S. Koenig, D. W. Lineberger, E. A. Emini, C. Williams, and S. Zolla-Pazner. 1994. Neutralization of primary human immunodeficiency virus type 1 isolates by the broadly reactive anti-V3 monoclonal antibody, 447-52D. J. Virol. 68:6994-7000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Conley, A. J., J. A. Kessler II, L. J. Boots, J. S. Tung, B. A. Arnold, P. M. Keller, A. R. Shaw, and E. A. Emini. 1994. Neutralization of divergent human immunodeficiency virus type 1 variants and primary isolates by IAM-41-2F5, an anti-gp41 human monoclonal antibody. Proc. Natl. Acad. Sci. USA 91:3348-3352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Connor, R. I., B. T. Korber, B. S. Graham, B. H. Hahn, D. D. Ho, B. D. Walker, A. U. Neumann, S. H. Vermund, J. Mestecky, S. Jackson, E. Fenamore, Y. Cao, F. Gao, S. Kalams, K. J. Kunstman, D. McDonald, N. McWilliams, A. Trkola, J. P. Moore, and S. M. Wolinsky. 1998. Immunological and virological analyses of persons infected by human immunodeficiency virus type 1 while participating in trials of recombinant gp120 subunit vaccines. J. Virol. 72:1552-1576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Cormier, E. G., and T. Dragic. 2002. The crown and stem of the V3 loop play distinct roles in human immunodeficiency virus type 1 envelope glycoprotein interactions with the CCR5 coreceptor. J. Virol. 76:8953-8957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Donners, H., T. Vermoesen, B. Willems, D. Davis, and G. van der Groen. 2003. The first generation of candidate HIV-1 vaccines can induce antibodies able to neutralize primary isolates in assays with extended incubation phases. Vaccine 22:104-111. [DOI] [PubMed] [Google Scholar]
- 33.D'Souza, M. P., P. Durda, C. V. Hanson, G. Milman, et al. 1991. Evaluation of monoclonal antibodies to HIV-1 by neutralization and serological assays: an international collaboration. AIDS 5:1061-1070. [DOI] [PubMed] [Google Scholar]
- 34.D'Souza, M. P., K. A. Kent, C. Thiriart, C. Collignon, and G. Milman. 1993. International collaboration comparing neutralization and binding assays for monoclonal antibodies to simian immunodeficiency virus. AIDS Res. Hum. Retrovir. 9:415-422. [DOI] [PubMed] [Google Scholar]
- 35.D'Souza, M. P., D. Livnat, J. A. Bradac, S. H. Bridges, et al. 1997. Evaluation of monoclonal antibodies to human immunodeficiency virus type 1 primary isolates by neutralization assays: performance criteria for selecting candidate antibodies for clinical trials. J. Infect. Dis. 175:1056-1062. [DOI] [PubMed] [Google Scholar]
- 36.D'Souza, M. P., G. Milman, J. A. Bradac, D. McPhee, C. V. Hanson, R. M. Hendry, et al. 1995. Neutralization of primary HIV-1 isolates by anti-envelope monoclonal antibodies. AIDS 9:867-874. [DOI] [PubMed] [Google Scholar]
- 37.Earl, P. L., C. C. Broder, D. Long, S. A. Lee, J. Peterson, S. Chakrabarti, R. W. Doms, and B. Moss. 1994. Native oligomeric human immunodeficiency virus type 1 envelope glycoprotein elicits diverse monoclonal antibody reactivities. J. Virol. 68:3015-3026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Earl, P. L., W. Sugiura, D. C. Montefiori, C. C. Broder, S. A. Lee, C. Wild, J. Lifson, and B. Moss. 2001. Immunogenicity and protective efficacy of oligomeric human immunodeficiency virus type 1 gp140. J. Virol. 75:645-653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Eckhart, L., W. Raffelsberger, B. Ferko, A. Klima, M. Purtscher, H. Katinger, and F. Ruker. 1996. Immunogenic presentation of a conserved gp41 epitope of human immunodeficiency virus type 1 on recombinant surface antigen of hepatitis B virus. J. Gen. Virol. 77:2001-2008. [DOI] [PubMed] [Google Scholar]
- 40.Gao, F., S. G. Morrison, D. L. Robertson, C. L. Thornton, S. Craig, G. Karlsson, J. Sodroski, M. Morgado, B. Galvao-Castro, H. von Briesen, et al. 1996. Molecular cloning and analysis of functional envelope genes from human immunodeficiency virus type 1 sequence subtypes A through G. J. Virol. 70:1651-1667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Gao, F., L. Yue, S. Craig, C. L. Thornton, D. L. Robertson, F. E. McCutchan, J. A. Bradac, P. M. Sharp, B. H. Hahn, et al. 1994. Genetic variation of HIV type 1 in four World Health Organization-sponsored vaccine evaluation sites: generation of functional envelope (glycoprotein 160) clones representative of sequence subtypes A, B, C, and E. AIDS Res. Hum. Retrovir. 10:1359-1368. [DOI] [PubMed] [Google Scholar]
- 42.Gaschen, B., B. Korber, and D. T. Foley. 1999. Global variation in the HIV-1 V3 region, p. 594-602. In C. Kuiken, B. Foley, B. H. Hahn, P. Marx, F. McCutchan, J. W. Mellors, J. Mullins, S. Wolinsky, and B. Korber (ed.), Human retroviruses and AIDS 1999. Los Alamos National Laboratory, Los Alamos, N. Mex.
- 43.Gaschen, B., J. Taylor, K. Yusim, B. Foley, F. Gao, D. Lang, V. Novitsky, B. Haynes, B. H. Hahn, T. Bhattacharya, and B. Korber. 2002. Diversity considerations in HIV-1 vaccine selection. Science 296:2354-2360. [DOI] [PubMed] [Google Scholar]
- 44.Gauduin, M. C., P. W. Parren, R. Weir, C. F. Barbas, D. R. Burton, and R. A. Koup. 1997. Passive immunization with a human monoclonal antibody protects hu-PBL− SCID mice against challenge by primary isolates of HIV-1. Nat. Med. 3:1389-1393. [DOI] [PubMed] [Google Scholar]
- 45.Gorny, M. K., A. J. Conley, S. Karwowska, A. Buchbinder, J. Y. Xu, E. A. Emini, S. Koenig, and S. Zolla-Pazner. 1992. Neutralization of diverse human immunodeficiency virus type 1 variants by an anti-V3 human monoclonal antibody. J. Virol. 66:7538-7542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Gorny, M. K., J. P. Moore, A. J. Conley, S. Karwowska, J. Sodroski, C. Williams, S. Burda, L. J. Boots, and S. Zolla-Pazner. 1994. Human anti-V2 monoclonal antibody that neutralizes primary but not laboratory isolates of human immunodeficiency virus type 1. J. Virol. 68:8312-8320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Gorny, M. K., K. Revesz, C. Williams, B. Volsky, M. K. Louder, C. A. Anyangwe, C. Krachmarov, S. C. Kayman, A. Pinter, A. Nadas, P. N. Nyambi, J. R. Mascola, and S. Zolla-Pazner. 2004. The V3 loop is accessible on the surface of most human immunodeficiency virus type 1 primary isolates and serves as a neutralization epitope. J. Virol. 78:2394-2404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Gorny, M. K., T. C. VanCott, C. Hioe, Z. R. Israel, N. L. Michael, A. J. Conley, C. Williams, J. A. Kessler II, P. Chigurupati, S. Burda, and S. Zolla-Pazner. 1997. Human monoclonal antibodies to the V3 loop of HIV-1 with intra- and interclade cross-reactivity. J. Immunol. 159:5114-5122. [PubMed] [Google Scholar]
- 49.Gorny, M. K., C. Williams, B. Volsky, K. Revesz, S. Cohen, V. R. Polonis, W. J. Honnen, S. C. Kayman, C. Krachmarov, A. Pinter, and S. Zolla-Pazner. 2002. Human monoclonal antibodies specific for conformation-sensitive epitopes of V3 neutralize human immunodeficiency virus type 1 primary isolates from various clades. J. Virol. 76:9035-9045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Gorny, M. K., J. Y. Xu, S. Karwowska, A. Buchbinder, and S. Zolla-Pazner. 1993. Repertoire of neutralizing human monoclonal antibodies specific for the V3 domain of HIV-1 gp120. J. Immunol. 150:635-643. [PubMed] [Google Scholar]
- 51.Hastie, T., R. Tibshirani, and J. Friedman. 2001. The elements of statistical learning, data mining inference and prediction, p. 453-480. Springer-Verlag, New York, N.Y.
- 52.Heyndrickx, L., W. Janssens, L. Zekeng, R. Musonda, S. Anagonou, G. Van der Auwera, S. Coppens, K. Vereecken, K. De Witte, R. Van Rampelbergh, M. Kahindo, L. Morison, F. E. McCutchan, J. K. Carr, J. Albert, M. Essex, J. Goudsmit, B. Asjo, M. Salminen, A. Buve, G. van Der Groen, et al. 2000. Simplified strategy for detection of recombinant human immunodeficiency virus type 1 group M isolates by gag/env heteroduplex mobility assay. J. Virol. 74:363-370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Hill, C. M., H. Deng, D. Unutmaz, V. N. Kewalramani, L. Bastiani, M. K. Gorny, S. Zolla-Pazner, and D. R. Littman. 1997. Envelope glycoproteins from human immunodeficiency virus types 1 and 2 and simian immunodeficiency virus can use human CCR5 as a coreceptor for viral entry and make direct CD4-dependent interactions with this chemokine receptor. J. Virol. 71:6296-6304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Hwang, S. S., T. J. Boyle, H. K. Lyerly, and B. R. Cullen. 1992. Identification of envelope V3 loop as the major determinant of CD4 neutralization sensitivity of HIV-1. Science 257:535-537. [DOI] [PubMed] [Google Scholar]
- 54a.Ihaka, R., and R. Gentleman. 1996. R: a language for data analysis and graphics. J. Comput. Graph. Stat. 5:299-314. [Google Scholar]
- 55.Jellis, C. L., T. J. Cradick, P. Rennert, P. Salinas, J. Boyd, T. Amirault, and G. S. Gray. 1993. Defining critical residues in the epitope for a HIV-neutralizing monoclonal antibody using phage display and peptide array technologies. Gene 137:63-68. [DOI] [PubMed] [Google Scholar]
- 56.Kessler, J. A., II, P. M. McKenna, E. A. Emini, C. P. Chan, M. D. Patel, S. K. Gupta, G. E. Mark III, C. F. Barbas III, D. R. Burton, and A. J. Conley. 1997. Recombinant human monoclonal antibody IgG1b12 neutralizes diverse human immunodeficiency virus type 1 primary isolates. AIDS Res. Hum. Retrovir. 13:575-582. [DOI] [PubMed] [Google Scholar]
- 57.Kitabwalla, M., F. Ferrantelli, T. Wang, A. Chalmers, H. Katinger, G. Stiegler, L. A. Cavacini, T. C. Chou, and R. M. Ruprecht. 2003. Primary African HIV clade A and D isolates: effective cross-clade neutralization with a quadruple combination of human monoclonal antibodies raised against clade B. AIDS Res. Hum. Retrovir. 19:125-131. [DOI] [PubMed] [Google Scholar]
- 58.Kostrikis, L. G., Y. Cao, H. Ngai, J. P. Moore, and D. D. Ho. 1996. Quantitative analysis of serum neutralization of human immunodeficiency virus type 1 from subtypes A, B, C, D, E, F, and I: lack of direct correlation between neutralization serotypes and genetic subtypes and evidence for prevalent serum-dependent infectivity enhancement. J. Virol. 70:445-458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Kostrikis, L. G., Z. H. Michalopoulou, Y. Cao, J. P. Moore, and D. D. Ho. 1996. Determining neutralization serotypes of HIV type 1 by neural networks. AIDS Res. Hum. Retrovir. 12:1667-1669. [DOI] [PubMed] [Google Scholar]
- 60.Kuiken, C., R. Thakallapalli, A. Esklid, and A. de Ronde. 2000. Genetic analysis reveals epidemiologic patterns in the spread of human immunodeficiency virus. Am. J. Epidemiol. 152:814-822. [DOI] [PubMed] [Google Scholar]
- 61.Kuiken, C. L., V. V. Lukashov, E. Baan, J. Dekker, J. A. Leunissen, and J. Goudsmit. 1996. Evidence for limited within-person evolution of the V3 domain of the HIV-1 envelope in the Amsterdam population. AIDS 10:31-37. [DOI] [PubMed] [Google Scholar]
- 62.Kunert, R., F. Ruker, and H. Katinger. 1998. Molecular characterization of five neutralizing anti-HIV type 1 antibodies: identification of nonconventional D segments in the human monoclonal antibodies 2G12 and 2F5. AIDS Res. Hum. Retrovir. 14:1115-1128. [DOI] [PubMed] [Google Scholar]
- 63.Kwong, P. D., M. L. Doyle, D. J. Casper, C. Cicala, S. A. Leavitt, S. Majeed, T. D. Steenbeke, M. Venturi, I. Chaiken, M. Fung, H. Katinger, P. W. Parren, J. Robinson, D. Van Ryk, L. Wang, D. R. Burton, E. Freire, R. Wyatt, J. Sodroski, W. A. Hendrickson, and J. Arthos. 2002. HIV-1 evades antibody-mediated neutralization through conformational masking of receptor-binding sites. Nature 420:678-682. [DOI] [PubMed] [Google Scholar]
- 64.Kwong, P. D., R. Wyatt, J. Robinson, R. W. Sweet, J. Sodroski, and W. A. Hendrickson. 1998. Structure of an HIV gp120 envelope glycoprotein in complex with the CD4 receptor and a neutralizing human antibody. Nature 393:648-659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Laal, S., S. Burda, M. K. Gorny, S. Karwowska, A. Buchbinder, and S. Zolla-Pazner. 1994. Synergistic neutralization of human immunodeficiency virus type 1 by combinations of human monoclonal antibodies. J. Virol. 68:4001-4008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Labrijn, A. F., P. Poignard, A. Raja, M. B. Zwick, K. Delgado, M. Franti, J. Binley, V. Vivona, C. Grundner, C. C. Huang, M. Venturi, C. J. Petropoulos, T. Wrin, D. S. Dimitrov, J. Robinson, P. D. Kwong, R. T. Wyatt, J. Sodroski, and D. R. Burton. 2003. Access of antibody molecules to the conserved coreceptor binding site on glycoprotein gp120 is sterically restricted on primary human immunodeficiency virus type 1. J. Virol. 77:10557-10565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Li, A., T. W. Baba, J. Sodroski, S. Zolla-Pazner, M. K. Gorny, J. Robinson, M. R. Posner, H. Katinger, C. F. Barbas III, D. R. Burton, T. C. Chou, and R. M. Ruprecht. 1997. Synergistic neutralization of a chimeric SIV/HIV type 1 virus with combinations of human anti-HIV type 1 envelope monoclonal antibodies or hyperimmune globulins. AIDS Res. Hum. Retrovir. 13:647-656. [DOI] [PubMed] [Google Scholar]
- 68.Li, A., H. Katinger, M. R. Posner, L. Cavacini, S. Zolla-Pazner, M. K. Gorny, J. Sodroski, T. C. Chou, T. W. Baba, and R. M. Ruprecht. 1998. Synergistic neutralization of simian-human immunodeficiency virus SHIV-vpu+ by triple and quadruple combinations of human monoclonal antibodies and high-titer anti-human immunodeficiency virus type 1 immunoglobulins. J. Virol. 72:3235-3240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Liang, X., S. Munshi, J. Shendure, G. Mark III, M. E. Davies, D. C. Freed, D. C. Montefiori, and J. W. Shiver. 1999. Epitope insertion into variable loops of HIV-1 gp120 as a potential means to improve immunogenicity of viral envelope protein. Vaccine 17:2862-2872. [DOI] [PubMed] [Google Scholar]
- 70.Lole, K. S., R. C. Bollinger, R. S. Paranjape, D. Gadkari, S. S. Kulkarni, N. G. Novak, R. Ingersoll, H. W. Sheppard, and S. C. Ray. 1999. Full-length human immunodeficiency virus type 1 genomes from subtype C-infected seroconverters in India, with evidence of intersubtype recombination. J. Virol. 73:152-160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Louisirirotchanakul, S., S. Beddows, R. Cheingsong-Popov, N. Shaffer, T. D. Mastro, P. Auewarakul, S. Likanonsakul, C. Wasi, and J. Weber. 1998. Characterization of sera from subjects infected with HIV-1 subtypes B and E in Thailand by antibody binding and neutralization. J. Acquir. Immune Defic. Syndr. Hum. Retrovirol. 19:315-320. [DOI] [PubMed] [Google Scholar]
- 72.Markowitz, M., T. Wrin, G. Stiegler, B. Vcelar, A. Hurley, C. Hogan, S. Vasan, H. Katinger, C. J. Petropoulos, and J. Galovich. 2003. Neutralization profiles by monoclonal antibodies 2F5, 2G12 and 4E10 of plasma-derived HIV-1 from 44 newly infected individuals. AIDS Vaccine 2003, abstract 212. Foundation for AIDS Research and Development, Alexandria,Va. http://www.aidsvaccine2003.org/2003/(Poster session 27).
- 73.Mascola, J. R. 2002. Passive transfer studies to elucidate the role of antibody-mediated protection against HIV-1. Vaccine 20:1922-1925. [DOI] [PubMed] [Google Scholar]
- 74.Mascola, J. R., M. G. Lewis, G. Stiegler, D. Harris, T. C. VanCott, D. Hayes, M. K. Louder, C. R. Brown, C. V. Sapan, S. S. Frankel, Y. Lu, M. L. Robb, H. Katinger, and D. L. Birx. 1999. Protection of macaques against pathogenic imian/uman immunodeficiency virus 89.6PD by passive transfer of neutralizing antibodies. J. Virol. 73:4009-4018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Mascola, J. R., M. K. Louder, S. R. Surman, T. C. Vancott, X. F. Yu, J. Bradac, K. R. Porter, K. E. Nelson, M. Girard, J. G. McNeil, F. E. McCutchan, D. L. Birx, and D. S. Burke. 1996. Human immunodeficiency virus type 1 neutralizing antibody serotyping using serum pools and an infectivity reduction assay. AIDS Res. Hum. Retrovir. 12:1319-1328. [DOI] [PubMed] [Google Scholar]
- 76.Mascola, J. R., M. K. Louder, T. C. VanCott, C. V. Sapan, J. S. Lambert, L. R. Muenz, B. Bunow, D. L. Birx, and M. L. Robb. 1997. Potent and synergistic neutralization of human immunodeficiency virus (HIV) type 1 primary isolates by hyperimmune anti-HIV immunoglobulin combined with monoclonal antibodies 2F5 and 2G12. J. Virol. 71:7198-7206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Mascola, J. R., M. K. Louder, C. Winter, R. Prabhakara, S. C. De Rosa, D. C. Douek, B. J. Hill, D. Gabuzda, and M. Roederer. 2002. Human immunodeficiency virus type 1 neutralization measured by flow cytometric quantitation of single-round infection of primary human T cells. J. Virol. 76:4810-4821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Mascola, J. R., S. W. Snyder, O. S. Weislow, S. M. Belay, R. B. Belshe, D. H. Schwartz, M. L. Clements, R. Dolin, B. S. Graham, G. J. Gorse, M. C. Keefer, M. J. McElrath, M. C. Walker, K. F. Wagner, J. G. McNeil, F. E. McCutchan, D. S. Burke, et al. 1996. Immunization with envelope subunit vaccine products elicits neutralizing antibodies against laboratory-adapted but not primary isolates of human immunodeficiency virus type 1. J. Infect. Dis. 173:340-348. [DOI] [PubMed] [Google Scholar]
- 79.Mascola, J. R., G. Stiegler, T. C. VanCott, H. Katinger, C. B. Carpenter, C. E. Hanson, H. Beary, D. Hayes, S. S. Frankel, D. L. Birx, and M. G. Lewis. 2000. Protection of macaques against vaginal transmission of a pathogenic HIV-1/SIV chimeric virus by passive infusion of neutralizing antibodies. Nat. Med. 6:207-210. [DOI] [PubMed] [Google Scholar]
- 80.McCutchan, F. E. 2000. Understanding the genetic diversity of HIV-1. AIDS 14(Suppl. 3):S31-S44. [PubMed] [Google Scholar]
- 81.McCutchan, F. E., A. W. Artenstein, E. Sanders-Buell, M. O. Salminen, J. K. Carr, J. R. Mascola, X. F. Yu, K. E. Nelson, C. Khamboonruang, D. Schmitt, M. P. Kieny, J. G. McNeil, and D. S. Burke. 1996. Diversity of the envelope glycoprotein among human immunodeficiency virus type 1 isolates of clade E from Asia and Africa. J. Virol. 70:3331-3338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.McCutchan, F. E., J. K. Carr, M. Bajani, E. Sanders-Buell, T. O. Harry, T. C. Stoeckli, K. E. Robbins, W. Gashau, A. Nasidi, W. Janssens, and M. L. Kalish. 1999. Subtype G and multiple forms of A/G intersubtype recombinant human immunodeficiency virus type 1 in Nigeria. Virology 254:226-234. [DOI] [PubMed] [Google Scholar]
- 83.McKeating, J. A., J. Cordell, C. J. Dean, and P. Balfe. 1992. Synergistic interaction between ligands binding to the CD4 binding site and V3 domain of human immunodeficiency virus type I gp120. Virology 191:732-742. [DOI] [PubMed] [Google Scholar]
- 84.McMichael, A. J., and T. Hanke. 2003. HIV vaccines 1983-2003. Nat. Med. 9:874-880. [DOI] [PubMed] [Google Scholar]
- 85.Mochizuki, N., N. Otsuka, K. Matsuo, T. Shiino, A. Kojima, T. Kurata, K. Sakai, N. Yamamoto, S. Isomura, T. N. Dhole, Y. Takebe, M. Matsuda, and M. Tatsumi. 1999. An infectious DNA clone of HIV type 1 subtype C. AIDS Res. Hum. Retrovir. 15:1321-1324. [DOI] [PubMed] [Google Scholar]
- 86.Montefiori, D. C., B. S. Graham, J. Zhou, R. A. Bucco, D. H. Schwartz, L. A. Cavacini, M. R. Posner, et al. 1993. V3-specific neutralizing antibodies in sera from HIV-1 gp160-immunized volunteers block virus fusion and act synergistically with human monoclonal antibody to the conformation-dependent CD4 binding site of gp120. J. Clin. Investig. 92:840-847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Moog, C., H. J. Fleury, I. Pellegrin, A. Kirn, and A. M. Aubertin. 1997. Autologous and heterologous neutralizing antibody responses following initial seroconversion in human immunodeficiency virus type 1-infected individuals. J. Virol. 71:3734-3741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Moore, J., J. McKeating, R. Weiss, and Q. Sattentau. 1990. Dissociation of gp120 from HIV-1 virions induced by soluble CD4. Science 250:1139-1142. [DOI] [PubMed] [Google Scholar]
- 89.Moore, J., Q. Sattentau, R. Wyatt, and J. Sodroski. 1994. Probing the structure of the human immunodeficiency virus surface glycoprotein gp120 with a panel of monoclonal antibodies. J. Virol. 68:469-484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Moore, J. P., Y. Cao, J. Leu, L. Qin, B. Korber, and D. D. Ho. 1996. Inter- and intraclade neutralization of human immunodeficiency virus type 1: genetic clades do not correspond to neutralization serotypes but partially correspond to gp120 antigenic serotypes. J. Virol. 70:427-444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Moore, J. P., Y. Cao, L. Qing, Q. J. Sattentau, J. Pyati, R. Koduri, J. Robinson, C. F. Barbas III, D. R. Burton, and D. D. Ho. 1995. Primary isolates of human immunodeficiency virus type 1 are relatively resistant to neutralization by monoclonal antibodies to gp120, and their neutralization is not predicted by studies with monomeric gp120. J. Virol. 69:101-109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Moore, J. P., F. E. McCutchan, S. W. Poon, J. Mascola, J. Liu, Y. Cao, and D. D. Ho. 1994. Exploration of antigenic variation in gp120 from clades A through F of human immunodeficiency virus type 1 by using monoclonal antibodies. J. Virol. 68:8350-8364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Moore, J. P., P. W. Parren, and D. R. Burton. 2001. Genetic subtypes, humoral immunity, and human immunodeficiency virus type 1 vaccine development. J. Virol. 75:5721-5729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Moore, J. P., and J. Sodroski. 1996. Antibody cross-competition analysis of the human immunodeficiency virus type 1 gp120 exterior envelope glycoprotein. J. Virol. 70:1863-1872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Moulard, M., S. K. Phogat, Y. Shu, A. F. Labrijn, X. Xiao, J. M. Binley, M. Y. Zhang, I. A. Sidorov, C. C. Broder, J. Robinson, P. W. Parren, D. R. Burton, and D. S. Dimitrov. 2002. Broadly cross-reactive HIV-1-neutralizing human monoclonal Fab selected for binding to gp120-CD4-CCR5 complexes. Proc. Natl. Acad. Sci. USA 99:6913-6918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Muster, T., B. Ferko, A. Klima, M. Purtscher, A. Trkola, P. Schulz, A. Grassauer, O. G. Engelhardt, A. Garcia-Sastre, P. Palese, et al. 1995. Mucosal model of immunization against human immunodeficiency virus type 1 with a chimeric influenza virus. J. Virol. 69:6678-6686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Muster, T., R. Guinea, A. Trkola, M. Purtscher, A. Klima, F. Steindl, P. Palese, and H. Katinger. 1994. Cross-neutralizing activity against divergent human immunodeficiency virus type 1 isolates induced by the gp41 sequence ELDKWAS. J. Virol. 68:4031-4034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Muster, T., F. Steindl, M. Purtscher, A. Trkola, A. Klima, G. Himmler, F. Ruker, and H. Katinger. 1993. A conserved neutralizing epitope on gp41 of human immunodeficiency virus type 1. J. Virol. 67:6642-6647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Nadas, A., P. Zhong, S. Burda, L. Zekeng, M. Urbanski, M. K. Gorny, S. Zolla-Pazner, and P. Nyambi. 2004. Defining human immunodeficiency virus (HIV) type 1 immunotypes with six human monoclonal antibodies. AIDS Res. Hum. Retrovir. 20:55-65. [DOI] [PubMed] [Google Scholar]
- 100.Nagashima, K. A., D. A. Thompson, S. I. Rosenfield, P. J. Maddon, T. Dragic, and W. C. Olson. 2001. Human immunodeficiency virus type 1 entry inhibitors PRO 542 and T-20 are potently synergistic in blocking virus-cell and cell-cell fusion. J. Infect. Dis. 183:1121-1125. [DOI] [PubMed] [Google Scholar]
- 101.Nyambi, P. N., A. Nadas, H. A. Mbah, S. Burda, C. Williams, M. K. Gorny, and S. Zolla-Pazner. 2000. Immunoreactivity of intact virions of human immunodeficiency virus type 1 (HIV-1) reveals the existence of fewer HIV-1 immunotypes than genotypes. J. Virol. 74:10670-10680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Nyambi, P. N., J. Nkengasong, P. Lewi, K. Andries, W. Janssens, K. Fransen, L. Heyndrickx, P. Piot, and G. van der Groen. 1996. Multivariate analysis of human immunodeficiency virus type 1 neutralization data. J. Virol. 70:6235-6243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.O'Brien, W. A., Y. Koyanagi, A. Namazie, J. Q. Zhao, A. Diagne, K. Idler, J. A. Zack, and I. S. Chen. 1990. HIV-1 tropism for mononuclear phagocytes can be determined by regions of gp120 outside the CD4-binding domain. Nature 348:69-73. [DOI] [PubMed] [Google Scholar]
- 104.Painter, S. L., R. Biek, D. C. Holley, and M. Poss. 2003. Envelope variants from women recently infected with clade A human immunodeficiency virus type 1 confer distinct phenotypes that are discerned by competition and neutralization experiments. J. Virol. 77:8448-8461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Park, E. J., L. K. Vujcic, R. Anand, T. S. Theodore, and G. V. Quinnan, Jr. 1998. Mutations in both gp120 and gp41 are responsible for the broad neutralization resistance of variant human immunodeficiency virus type 1 MN to antibodies directed at V3 and non-V3 epitopes. J. Virol. 72:7099-7107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Parker, C. E., L. J. Deterding, C. Hager-Braun, J. M. Binley, N. Schulke, H. Katinger, J. P. Moore, and K. B. Tomer. 2001. Fine definition of the epitope on the gp41 glycoprotein of human immunodeficiency virus type 1 for the neutralizing monoclonal antibody 2F5. J. Virol. 75:10906-10911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Parren, P. W., P. A. Marx, A. J. Hessell, A. Luckay, J. Harouse, C. Cheng-Mayer, J. P. Moore, and D. R. Burton. 2001. Antibody protects macaques against vaginal challenge with a pathogenic R5 simian/human immunodeficiency virus at serum levels giving complete neutralization in vitro. J. Virol. 75:8340-8347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Parren, P. W., J. P. Moore, D. R. Burton, and Q. J. Sattentau. 1999. The neutralizing antibody response to HIV-1: viral evasion and escape from humoral immunity. AIDS 13:S137-S162. [PubMed] [Google Scholar]
- 109.Parren, P. W. H. I., M. Wang, A. Trkola, J. M. Binley, M. Purtscher, H. Katinger, J. P. Moore, and D. R. Burton. 1998. Antibody neutralization-resistant primary isolates of human immunodefiency virus type 1. J. Virol. 72:10270-10274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Petropoulos, C. J., N. T. Parkin, K. L. Limoli, Y. S. Lie, T. Wrin, W. Huang, H. Tian, D. Smith, G. A. Winslow, D. J. Capon, and J. M. Whitcomb. 2000. A novel phenotypic drug susceptibility assay for human immunodeficiency virus type 1. Antimicrob. Agents Chemother. 44:920-928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Pitisuttithum, P., S. Nitayaphan, P. Thongcharoen, C. Khamboonruang, J. Kim, M. de Souza, T. Chuenchitra, R. P. Garner, D. Thapinta, V. Polonis, S. Ratto-Kim, P. Chanbancherd, J. Chiu, D. L. Birx, A. M. Duliege, J. G. McNeil, and A. E. Brown. 2003. Safety and immunogenicity of combinations of recombinant subtype E and B human immunodeficiency virus type 1 envelope glycoprotein 120 vaccines in healthy Thai adults. J. Infect. Dis. 188:219-227. [DOI] [PubMed] [Google Scholar]
- 112.Poignard, P., T. Fouts, D. Naniche, J. Moore, and Q. Sattentau. 1996. Neutralizing antibodies to human immunodeficiency virus type-1 gp120 induce envelope glycoprotein subunit dissociation. J. Exp. Med. 183:473-484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Pugach, P., S. Kuhmann, J. Taylor, A. J. Marozsan, A. Snyder, T. Ketas, S. Wolinsky, B. Korber, and J. P. Moore. 2004. The prolonged culture of human immunodeficiency virus type 1 in primary lymphocytes increases its sensitivity to neutralization by soluble CD4. Virology 321:8-22. [DOI] [PubMed] [Google Scholar]
- 114.Purtscher, M., A. Trkola, A. Grassauer, P. M. Schulz, A. Klima, S. Dopper, G. Gruber, A. Buchacher, T. Muster, and H. Katinger. 1996. Restricted antigenic variability of the epitope recognized by the neutralizing gp41 antibody 2F5. AIDS 10:587-593. [DOI] [PubMed] [Google Scholar]
- 115.Purtscher, M., A. Trkola, G. Gruber, A. Buchacher, R. Predl, F. Steindl, C. Tauer, R. Berger, N. Barrett, A. Jungbauer, et al. 1994. A broadly neutralizing human monoclonal antibody against gp41 of human immunodeficiency virus type 1. AIDS Res. Hum. Retrovir. 10:1651-1658. [DOI] [PubMed] [Google Scholar]
- 116.Quinones-Mateu, M. E., S. C. Ball, A. J. Marozsan, V. S. Torre, J. L. Albright, G. Vanham, G. van Der Groen, R. L. Colebunders, and E. J. Arts. 2000. A dual infection-competition assay shows a correlation between ex vivo human immunodeficiency virus type 1 fitness and disease progression. J. Virol. 74:9222-9233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Ratner, L., W. Haseltine, R. Patarca, K. J. Livak, B. Starcich, S. F. Josephs, E. R. Doran, J. A. Rafalski, E. A. Whitehorn, K. Baumeister, et al. 1985. Complete nucleotide sequence of the AIDS virus, HTLV-III. Nature 313:277-284. [DOI] [PubMed] [Google Scholar]
- 118.Reeves, J. D., S. A. Gallo, N. Ahmad, J. L. Miamidian, P. E. Harvey, M. Sharron, S. Pohlmann, J. N. Sfakianos, C. A. Derdeyn, R. Blumenthal, E. Hunter, and R. W. Doms. 2002. Sensitivity of HIV-1 to entry inhibitors correlates with envelope/coreceptor affinity, receptor density, and fusion kinetics. Proc. Natl. Acad. Sci. USA 99:16249-16254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Richardson, T. M., Jr., B. L. Stryjewski, C. C. Broder, J. A. Hoxie, J. R. Mascola, P. L. Earl, and R. W. Doms. 1996. Humoral response to oligomeric human immunodeficiency virus type 1 envelope protein. J. Virol. 70:753-762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Richman, D. D., T. Wrin, S. J. Little, and C. J. Petropoulos. 2003. Rapid evolution of the neutralizing antibody response to HIV type 1 infection. Proc. Natl. Acad. Sci. USA 100:4144-4149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Roben, P., J. P. Moore, M. Thali, J. Sodroski, C. F. Barbas III, and D. R. Burton. 1994. Recognition properties of a panel of human recombinant Fab fragments to the CD4 binding site of gp120 that show differing abilities to neutralize human immunodeficiency virus type 1. J. Virol. 68:4821-4828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Robertson, D. L., J. P. Anderson, J. A. Bradac, J. K. Carr, B. Foley, R. K. Funkhouser, F. Gao, B. H. Hahn, M. L. Kalish, C. Kuiken, G. H. Learn, T. Leitner, F. McCutchan, S. Osmanov, M. Peeters, D. Pieniazek, M. Salminen, P. M. Sharp, S. Wolinsky, and B. Korber. 2000. HIV-1 nomenclature proposal. Science 288:55-56. [DOI] [PubMed] [Google Scholar]
- 123.Rodenburg, C. M., Y. Li, S. A. Trask, Y. Chen, J. Decker, D. L. Robertson, M. L. Kalish, G. M. Shaw, S. Allen, B. H. Hahn, and F. Gao. 2001. Near full-length clones and reference sequences for subtype C isolates of HIV type 1 from three different continents. AIDS Res. Hum. Retrovir. 17:161-168. [DOI] [PubMed] [Google Scholar]
- 124.Ruprecht, R. M., F. Ferrantelli, M. Kitabwalla, W. Xu, and H. M. McClure. 2003. Antibody protection: passive immunization of neonates against oral AIDS virus challenge. Vaccine 21:3370-3373. [DOI] [PubMed] [Google Scholar]
- 125.Salminen, M. O., C. Koch, E. Sanders-Buell, P. K. Ehrenberg, N. L. Michael, J. K. Carr, D. S. Burke, and F. E. McCutchan. 1995. Recovery of virtually full-length HIV-1 provirus of diverse subtypes from primary virus cultures using the polymerase chain reaction. Virology 213:80-86. [DOI] [PubMed] [Google Scholar]
- 126.Sanders, R. W., M. Venturi, L. Schiffner, R. Kalyanaraman, H. Katinger, K. O. Lloyd, P. D. Kwong, and J. P. Moore. 2002. The mannose-dependent epitope for neutralizing antibody 2G12 on human immunodeficiency virus type 1 glycoprotein gp120. J. Virol. 76:7293-7305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Saphire, E. O., P. W. Parren, R. Pantophlet, M. B. Zwick, G. M. Morris, P. M. Rudd, R. A. Dwek, R. L. Stanfield, D. R. Burton, and I. A. Wilson. 2001. Crystal structure of a neutralizing human IgG against HIV-1: a template for vaccine design. Science 293:1155-1159. [DOI] [PubMed] [Google Scholar]
- 128.Sawyer, L. S., M. T. Wrin, L. Crawford-Miksza, B. Potts, Y. Wu, P. A. Weber, R. D. Alfonso, and C. V. Hanson. 1994. Neutralization sensitivity of human immunodeficiency virus type 1 is determined in part by the cell in which the virus is propagated. J. Virol. 68:1342-1349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Scanlan, C. N., R. Pantophlet, M. R. Wormald, E. O. Saphire, R. Stanfield, I. A. Wilson, H. Katinger, R. A. Dwek, P. M. Rudd, and D. R. Burton. 2002. The broadly neutralizing anti-HIV-1 antibody 2G12 recognizes a cluster of α1→2 mannose residues on the outer face of gp120. J. Virol. 76:7306-7321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Shankarappa, R., J. B. Margolick, S. J. Gange, A. G. Rodrigo, D. Upchurch, H. Farzadegan, P. Gupta, C. R. Rinaldo, G. H. Learn, X. He, X. L. Huang, and J. I. Mullins. 1999. Consistent viral evolutionary changes associated with the progression of human immunodeficiency virus type 1 infection. J. Virol. 73:10489-10502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Sharon, M., N. Kessler, R. Levy, S. Zolla-Pazner, M. Gorlach, and J. Anglister. 2003. Alternative conformations of HIV-1 V3 loops mimic beta hairpins in chemokines, suggesting a mechanism for coreceptor selectivity. Structure 11:225-236. [DOI] [PubMed] [Google Scholar]
- 132.Srivastava, I. K., L. Stamatatos, E. Kan, M. Vajdy, Y. Lian, S. Hilt, L. Martin, C. Vita, P. Zhu, K. H. Roux, L. Vojtech, C. D. Montefiori, J. Donnelly, J. B. Ulmer, and S. W. Barnett. 2003. Purification, characterization, and immunogenicity of a soluble trimeric envelope protein containing a partial deletion of the V2 loop derived from SF162, an R5-tropic human immunodeficiency virus type 1 isolate. J. Virol. 77:11244-11259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Stanfield, R. L., M. K. Gorny, C. Williams, S. Zolla-Pazner, and I. A. Wilson. 2004. Structural rationale for the broad neutralization of HIV-1 by human monoclonal antibody 447-52D. Structure 12:193-204. [DOI] [PubMed] [Google Scholar]
- 134.Stiegler, G., R. Kunert, M. Purtscher, S. Wolbank, R. Voglauer, F. Steindl, and H. Katinger. 2001. A potent cross-clade neutralizing human monoclonal antibody against a novel epitope on gp41 of human immunodeficiency virus type 1. AIDS Res. Hum. Retrovir. 17:1757-1765. [DOI] [PubMed] [Google Scholar]
- 135.Thali, M., C. Furman, B. Wahren, M. Posner, D. D. Ho, J. Robinson, and J. Sodroski. 1992. Cooperativity of neutralizing antibodies directed against the V3 and CD4 binding regions of the human immunodeficiency virus gp120 envelope glycoprotein. J. Acquir. Immune Defic. Syndr. 5:591-599. [PubMed] [Google Scholar]
- 136.Tilley, S. A., W. J. Honnen, M. E. Racho, T. C. Chou, and A. Pinter. 1992. Synergistic neutralization of HIV-1 by human monoclonal antibodies against the V3 loop and the CD4-binding site of gp120. AIDS Res. Hum. Retrovir. 8:461-467. [DOI] [PubMed] [Google Scholar]
- 137.Trkola, A., T. Dragic, J. Arthos, J. M. Binley, W. C. Olson, G. P. Allaway, C. Cheng-Mayer, J. Robinson, P. J. Maddon, and J. P. Moore. 1996. CD4-dependent, antibody-sensitive interactions between HIV-1 and its co-receptor CCR-5. Nature 384:184-187. [DOI] [PubMed] [Google Scholar]
- 138.Trkola, A., A. B. Pomales, H. Yuan, B. Korber, P. J. Maddon, G. P. Allaway, H. Katinger, C. F. Barbas III, D. R. Burton, and D. D. Ho. 1995. Cross-clade neutralization of primary isolates of human immunodeficiency virus type 1 by human monoclonal antibodies and tetrameric CD4-IgG. J. Virol. 69:6609-6617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Trkola, A., M. Purtscher, T. Muster, C. Ballaun, A. Buchacher, N. Sullivan, K. Srinivasan, J. Sodroski, J. P. Moore, and H. Katinger. 1996. Human monoclonal antibody 2G12 defines a distinctive neutralization epitope on the gp120 glycoprotein of human immunodeficiency virus type 1. J. Virol. 70:1100-1108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.VanCott, T. C., S. C. Veit, V. Kalyanaraman, P. Earl, and D. L. Birx. 1995. Characterization of a soluble, oligomeric HIV-1 gp160 protein as a potential immunogen. J. Immunol. Methods 183:103-117. [DOI] [PubMed] [Google Scholar]
- 141.Verrier, F., S. Burda, R. Belshe, A. M. Duliege, J. L. Excler, M. Klein, and S. Zolla-Pazner. 2000. A human immunodeficiency virus prime-boost immunization regimen in humans induces antibodies that show interclade cross-reactivity and neutralize several X4-, R5-, and dualtropic clade B and C primary isolates. J. Virol. 74:10025-10033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Verrier, F., A. Nadas, M. K. Gorny, and S. Zolla-Pazner. 2001. Additive effects characterize the interaction of antibodies involved in neutralization of the primary dualtropic human immunodeficiency virus type 1 isolate 89.6. J. Virol. 75:9177-9186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Vijh-Warrier, S., A. Pinter, W. J. Honnen, and S. A. Tilley. 1996. Synergistic neutralization of human immunodeficiency virus type 1 by a chimpanzee monoclonal antibody against the V2 domain of gp120 in combination with monoclonal antibodies against the V3 loop and the CD4-binding site. J. Virol. 70:4466-4473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Warrier, S. V., A. Pinter, W. J. Honnen, M. Girard, E. Muchmore, and S. A. Tilley. 1994. A novel, glycan-dependent epitope in the V2 domain of human immunodeficiency virus type 1 gp120 is recognized by a highly potent, neutralizing chimpanzee monoclonal antibody. J. Virol. 68:4636-4642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Weber, J., E. M. Fenyo, S. Beddows, P. Kaleebu, A. Bjorndal, et al. 1996. Neutralization serotypes of human immunodeficiency virus type 1 field isolates are not predicted by genetic subtype. J. Virol. 70:7827-7832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Wei, X., J. M. Decker, S. Wang, H. Hui, J. C. Kappes, X. Wu, J. F. Salazar-Gonzalez, M. G. Salazar, J. M. Kilby, M. S. Saag, N. L. Komarova, M. A. Nowak, B. H. Hahn, P. D. Kwong, and G. M. Shaw. 2003. Antibody neutralization and escape by HIV-1. Nature 422:307-312. [DOI] [PubMed] [Google Scholar]
- 147.Wu, L., N. P. Gerard, R. Wyatt, H. Choe, C. Parolin, N. Ruffing, A. Borsetti, A. A. Cardoso, E. Desjardin, W. Newman, C. Gerard, and J. Sodroski. 1996. CD4-induced interaction of primary HIV-1 gp120 glycoproteins with the chemokine receptor CCR-5. Nature 384:179-183. [DOI] [PubMed] [Google Scholar]
- 148.Xu, W., B. A. Smith-Franklin, P. L. Li, C. Wood, J. He, Q. Du, G. J. Bhat, C. Kankasa, H. Katinger, L. A. Cavacini, M. R. Posner, D. R. Burton, T. C. Chou, and R. M. Ruprecht. 2001. Potent neutralization of primary human immunodeficiency virus clade C isolates with a synergistic combination of human monoclonal antibodies raised against clade B. J. Hum. Virol. 4:55-61. [PubMed] [Google Scholar]
- 149.Zhang, M. Y., B. Gaschen, W. Blay, B. Foley, N. Haigwood, C. Kuiken, and B. Korber. Tracking global patterns of N-linked glycosylation site variation in highly variable viral glycoproteins: HIV, SIV and HCV envelopes and influenza haemagglutinin. Glycobiology, in press. [DOI] [PubMed]
- 150.Zinkernagel, R. M., A. LaMarre, A. Ciurea, L. Hunziker, A. F. Ochsenbein, K. D. McCoy, T. Fehr, M. F. Bachmann, U. Kalinke, and H. Hengartner. 2001. Neutralizing antiviral antibody responses. Adv. Immunol. 79:1-53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Zolla-Pazner, S., M. K. Gorny, P. N. Nyambi, T. C. VanCott, and A. Nadas. 1999. Immunotyping of human immunodeficiency virus type 1 (HIV): an approach to immunologic classification of HIV. J. Virol. 73:4042-4051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Zolla-Pazner, S., J. O'Leary, S. Burda, M. K. Gorny, M. Kim, J. Mascola, and F. McCutchan. 1995. Serotyping of primary human immunodeficiency virus type 1 isolates from diverse geographic locations by flow cytometry. J. Virol. 69:3807-3815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Zwick, M. B., A. F. Labrijn, M. Wang, C. Spenlehauer, E. O. Saphire, J. M. Binley, J. P. Moore, G. Stiegler, H. Katinger, D. R. Burton, and P. W. Parren. 2001. Broadly neutralizing antibodies targeted to the membrane-proximal external region of human immunodeficiency virus type 1 glycoprotein gp41. J. Virol. 75:10892-10905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Zwick, M. B., M. Wang, P. Poignard, G. Stiegler, H. Katinger, D. R. Burton, and P. W. Parren. 2001. Neutralization synergy of human immunodeficiency virus type 1 primary isolates by cocktails of broadly neutralizing antibodies. J. Virol. 75:12198-12208. [DOI] [PMC free article] [PubMed] [Google Scholar]