Abstract
SL3-3 murine leukemia virus is a potent inducer of T-lymphomas in mice. Using inbred NMRI mice, it was previously reported that a mutant of SL3-3 with all enhancer Runx (AML1/core) sites disrupted by 3-bp mutations (SL3-3dm) induces predominantly non-T-cell tumors with severely extended latency (S. Ethelberg, J. Lovmand, J. Schmidt, A. Luz, and F. S. Pedersen, J. Virol. 71:7273-7280, 1997). By use of three-color flow cytometry and molecular and histopathological analyses, we have now performed a detailed phenotypic characterization of SL3-3- and SL3-3dm-induced tumors in this mouse strain. All wild-type induced tumors had clonal T-cell receptor β rearrangements, and the vast majority were CD3+ CD4+ CD8− T-lymphomas. Such a consistent phenotypic pattern is unusual for murine leukemia virus-induced T-lymphomas. The mutant virus induced malignancies of four distinct hematopoietic lineages: myeloid, T lymphoid, B lymphoid, and erythroid. The most common disease was myeloid leukemia with maturation. Thus, mutation of all Runx motifs in the enhancer of SL3-3 severely impedes viral T-lymphomagenicity and thereby discloses a considerable and formerly unappreciated potential of this virus for myeloid leukemia induction. Proviral enhancers with complex structural alterations (deletions, insertions, and/or duplications) were found in most SL3-3dm-induced T-lymphoid tumors and immature myeloid leukemias but not in any cases of myeloid leukemia with maturation, mature B-lymphoma, or erythroleukemia. Altogether, our results indicate that the SL3-3dm enhancer in itself promotes induction of myeloid leukemia with maturation but that structural changes may arise in vivo and redirect viral disease specificity to induction of T-lymphoid or immature myeloid leukemias, which typically develop with moderately shorter latencies.
SL3-3 is a highly pathogenic ecotropic murine leukemia virus (MLV), which induces strictly T-lymphomas with mean latencies of usually 2 to 4 months dependent on the mouse strain (12, 19, 32, 50). The U3 transcriptional enhancer in the proviral long terminal repeat (LTR) is a critical determinant of tumor induction by this and other MLVs (6-8, 10, 12, 19, 31, 34, 41, 47, 50, 61). The SL3-3 enhancer contains 2.5 tandem copies of a 72-bp sequence with binding sites for several different cellular transcription factors, including two distinct Runx sites, here termed Runx sites I and II but also known as core and inverse core, respectively. The Runx sites are of major importance for T-lymphoma induction by SL3-3 (12, 19, 47). These motifs support high viral enhancer activity in T cells and may thereby ensure effective viral replication and potent activation of host genes at the integration site. Mutation of enhancer Runx sites in closely related T-lymphomagenic Moloney MLV (MoMLV) delays tumor induction and shifts disease specificity towards erythroleukemogenesis (61).
In vitro binding studies have shown that the Runx sites of SL3-3 and the corresponding enhancer sequences of other MLVs are specifically recognized by factors previously referred to as SEF1 (SL3-3 enhancer factor 1) (65, 66), S/A-CBF (SL3-3 and Akv core binding factor) (4, 36), CBFα2 (core binding factor α2) (77), or just CBF (core binding factor) (71) but now seen as members of the Runx (AML1/CBF/PEBP2) transcription factor family. Runx factors work as heterodimers of a DNA binding α subunit, encoded by three related genes, Runx1 to Runx3 (1, 33, 44, 52), and a non-DNA binding β subunit (CBFβ), encoded by Cbfb (35, 51, 70). Runx/CBFβ plays pivotal roles in normal developmental pathways and in various cancers (9, 35, 39, 44, 53, 62, 69). Runx1 to Runx3 are expressed throughout T-cell development, with gene- and stage-specific variation in expression and function (64, 67, 75). The SL3-3 enhancer is transiently activated by ectopic murine Runx1 (CBF2α-451), with most of the activity mediated through Runx site I and dependent on the binding of a heterologous factor to at least one of the neighboring c-Myb or Ets site motifs (76-78). Molecular interactions between nuclear proteins and MLV enhancer Runx sites are poorly described in vivo. Although Runx factors are the most likely candidates for regulating MLV T-lymphomagenicity through these motifs (34), they may, at least in some cell types, be recognized by other proteins (36, 40, 63).
SL3-3 pathogenicity is severely impaired by the mutation of Runx site I and even further inhibited if Runx site II is also mutated (12, 19). A 1-nucleotide substitution in Runx site I, converting it to the equivalent enhancer Runx site sequence of weakly pathogenic Akv MLV, increases the latency of viral tumor induction (47). Tumors induced by attenuated Runx site mutant viruses often accumulate altered proviral enhancer structures in vivo, containing reversions or suppressor mutations of the mutated Runx sites or more complex second-site changes. Such altered enhancers appear to be selected for increased pathogenicity during the tumorigenic process, as verified in some cases by molecular cloning and reintroduction into mice (11-13, 41, 42, 47).
We previously reported that an SL3-3 virus with all enhancer Runx sites mutated by 3-bp substitutions that abolish Runx (SEF1) binding in vitro (65, 66) is virtually nontumorigenic in random-bred NMRI mice (19) and induces tumors only after severely extended latency in more-susceptible inbred NMRI mice (12). Most SL3-3 double mutant (SL3-3dm)-induced tumors were nonthymic and lacked clonal T-cell receptor β (TCRβ) rearrangements by Southern blotting analysis, indicating that they were not of T-cell type and, as such, differed from tumors induced by SL3-3 (12). Preliminary histopathology suggested that some of the SL3-3dm-induced tumors may derive from B-cells; however, the tumor phenotype(s) were not fully characterized (12). SL3-3 pathogenicity is reduced to a similar extent by mutation of the c-Myb site in the enhancer, but the few tumors analyzed so far provides little evidence for a possible change in viral disease specificity (50). Indeed, SL3-3dm is the only mutant of SL3-3 for which a significant shift in cell type specificity of viral tumor induction has been clearly indicated in the literature.
To better characterize the phenotype(s) of these tumors, we have examined here a new and larger panel of SL3-3dm-induced tumors from inbred NMRI mice by molecular analyses, flow cytometric immunophenotyping, and histopathological examination. SL3-3-induced lymphomas of the same mouse strain were analyzed in parallel, revealing a strikingly homogeneous phenotypic pattern. In contrast, the mutant virus induced malignancies of four hematopoietic lineages (myeloid, T lymphoid, B lymphoid, and erythroid), with a preference for myeloid leukemias. Proviral enhancer structures with complex in vivo-derived alterations were detected with high frequency in SL3-3dm-induced T-lymphoid tumors and immature myeloid leukemias but never in cases of myeloid leukemia with maturation, histiocyte-associated diffuse large B-cell lymphoma, erythroleukemia, or wild-type-induced T-cell tumors. This strongly indicates that structural enhancer alterations arise during the prolonged process of tumor induction by SL3-3dm and that such newly generated structures may determine the cell type specificity of viral transformation.
MATERIALS AND METHODS
Virus stocks.
The generation of SL3-3 and SL3-3dm viruses has been described previously (19). Virus titers were determined by end point dilution on NIH 3T3 murine fibroblast cells (60).
Pathogenicity experiments and tissue preparations.
Newborn inbred NMRI mice, without endogenous ecotropic MLVs (30), were injected intraperitoneally with ca. 106 infectious virus particles (0.1 ml), and control mice were injected with 0.1 ml of complete medium. The mice were observed for tumor development 5 days per week. Moribund animals were sacrificed by cervical dislocation and examined for gross pathological changes including enlargement of thymus, spleen, liver, and mesenteric and peripheral lymph nodes, as previously described in detail (59). Lymphomatous tissues were dissected, and samples were stored frozen and/or fixed in 4% paraformaldehyde for further analysis. In addition, spleen, thymus, and lymph node tissue samples were minced in cold RPMI medium (Gibco BRL, Invitrogen Corporation) by the use of surgical scissors to prepare single-cell suspensions for flow cytometry analysis. Single cells were pelleted by centrifugation and incubated on ice for 5 min with lysis buffer (0.16 M NH4Cl, 1 mM NaHCO3, 0.12 mM EDTA) to hemolyze erythrocytes and washed twice in cold (4°C) phosphate-buffered saline containing 2% fetal calf serum and 2 mM NaN3 (Flow-I buffer). Viable cells were enumerated by trypan blue exclusion.
Three-color flow cytometry.
For each assay, 106 cells from the spleen, thymus, or lymph node were suspended in 50 μl of Flow-I buffer and preincubated for 5 min on ice with 1 μg of rat anti-mouse CD16/CD32 (Fcγ III/II receptor) monoclonal antibody (Mouse Fc Block; BD Pharmingen) to block unspecific antibody binding to Fcγ receptors. For staining of surface antigens, cells were incubated on ice for 30 min with a mixture of three different antibodies (0.5 μg of each). In all cases, appropriate isotype controls were made in parallel. The cells were washed twice in cold Flow-I buffer and resuspended in cold phosphate-buffered saline containing 2% fetal calf serum, 2 mM NaN3, and 1% formaldehyde. Nine different fluorochrome-conjugated rat anti-mouse monoclonal antibodies were used, all obtained from BD Pharmingen: CD11b-fluorescein isothiocyanate (FITC) (clone M1/70), CD3-phycoerythrin (PE) (clone 17A2), B220-peridinin chlorophyll α protein (PerCP) (clone RA3-6B2), CD4-FITC (clone GK1.5), CD8a-PerCP-CY5.5 (clone 53-6.7), immunoglobulin D (IgD)-FITC (clone 11-26c.2a), IgM-PE (clone R6-60.2), CD43-FITC (clone S7), and CD138-PE (clone 281-2). Isotype controls were rat IgG2a (clone R35-95) and rat IgG2b (clone A95-1), light chain monoclonal Igs conjugated with FITC, PE, PerCP, or PerCP-CY5.5 (BD Pharmingen).
The antibodies were used in four combinations: CD3-B220-CD11b, CD3-CD4-CD8a, B220-IgM-IgD, and B220-CD43-CD138. CD3 is a pan-T marker (15), B220 is a pan-B marker (21), and CD11b (Mac-1) is highly expressed on myeloid cells (granulocytes, monocytes, and macrophages) and NK cells (27, 68). Immature T-lymphoid cells are CD4− CD8− or CD4+ CD8+ while mature T cells express either CD4 or CD8 (15). Anti-CD8a recognizes the α and α′ chains of the CD8 coreceptor. Antibodies to B220-IgM-IgD allow identification of immature and mature B cells before isotype switching (56). Plasma cells do not express any of these markers but are positive for CD138 (syndecan 1), as are a substantial proportion of early B cells in the bone marrow (58). CD43 is expressed prior to the small pre-B-cell stage (72) on plasma cells (18) but not on resting conventional B cells (72). A variety of other hematopoietic cells are CD43 positive, including T cells (thymocytes, cytotoxic or suppressor T cells, and most T-helper cells), granulocytes, monocytes, macrophages, hematopoietic stem cells, and myeloid and lymphoid progenitors (18, 25, 45, 46, 72).
Surface staining was analyzed on a FACSCalibur flow cytometer (BD Bioscience, San Jose, Calif.) equipped with a single 15-mW argon laser (488 nm) for excitation and standard emission optics. Data were recorded for 50,000 events of each sample. Forward and side scatters were used to measure cell size and granularity to gate out cell debris. Data analysis, including automatic software compensation based on single-color stainings of cells from control mice, was performed by using WinList 3D software (version 5.0; Verity Software House, Inc.). Flow cytometry data are presented in two-dimensional dot plots showing representative results of 10,000 events. The quadrants of all plots were defined on the basis of appropriate isotype controls measured in parallel to determine background fluorescence levels.
Histopathological examination and immunohistochemical analysis.
Paraformaldehyde- and formalin-fixed paraffin-embedded tissues from lymph nodes, thymus, and spleen were analyzed. Three- to five-micrometer-thick sections were cut and stained with hematoxylin and eosin (H&E) and with chloroacetate esterase when indicated. All cases were classified according to the Bethesda proposals for classification of lymphoid and nonlymphoid hematopoietic neoplasms in mice (26, 48). In this proposal, myeloid leukemias without maturation, by definition, are not monocytic, and 90% of the blasts are immature. Myeloid leukemia with maturation, by definition, shows differentiation to neutrophils or granulocytes and lacks a monocytic component. Leukemias with a monocytic and neutrophilic component are termed myelomonocytic leukemias while leukemias with an exclusively monocytic component are called monocytic leukemias. Immunohistochemistry was performed on an automated immunostainer (Ventana Medical System Inc., Tucson, Ariz.) according to the company's protocols, with slight modifications. After deparaffinization and rehydration, the slides were placed in a microwave pressure cooker in 0.01 M citrate buffer, pH 6.0, containing 0.1% Tween 20 and heated in a microwave oven at maximum power for 30 min. After cooling in Tris-buffered saline, the sections were incubated with 3% goat serum for 20 min. The antibody panel used included CD3 (Dako, Hamburg, Germany), B220 (Pharmingen), TdT (Dako), and myeloperoxidase (Dako). Appropriate positive controls were used to confirm the adequacy of the staining.
Southern blotting and hybridization probes.
Genomic DNA was isolated from frozen tumor tissues by using the DNeasy tissue kit (QIAGEN) according to the instructions provided by the manufacturer. Twenty micrograms of each tumor DNA was cleaved with HindIII or EcoRI, and the products were separated by electrophoresis into 0.8% agarose gels. DNA fragments were transferred to nylon membranes (Zeta-Probe; Bio-Rad) by alkaline blotting (0.4 M NaOH), and membranes were preincubated twice with (pre)hybridization buffer (0.25 M Na2HPO4 [pH 7.2], 7% sodium dodecyl sulfate) for 1 h at 65°C. 32P-labeled random-priming probes were added to the hybridization buffer, and hybridizations were performed overnight (ca. 16 h) at 65°C. Hybridization patterns were analyzed by autoradiography or phosphorimaging (Molecular Imager FX [Bio-Rad]; Quantity One software). The TCRβ-specific probes (J1 and J2), the Ig heavy and kappa light chain-specific probes [IgH-J11 and Ig(κ)], and the ecotropic virus-specific probe (env) were as previously described (37). The TCRδ-specific probes, TCRδ-J1 (corresponding to nucleotides 363100 to 363599, GenBank accession no. AE008686) and TCRδ-J2 (nucleotides 369900 to 370499, GenBank accession no. AE008686) were generated by PCR with primers 5′-GTTGTCTATTTAAAAAGTAGGATTTGGGGG-3′ plus 5′-TTGGAAGTCCCACATTCTCGTTAATTCTTC-3′ and 5′-GATGCTGAATTCCTAACATGCTTCTGTGTA-3′ plus 5′-CAGACCTTCTCCTCTTATGAGCACTCATAT-3′, respectively. TCRδ-J1 was used with EcoRI-digested genomic DNA, and TCRδ-J2 was used with HindIII-digested genomic DNA.
PCR amplification of proviral DNA and sequencing analysis.
PCR was performed on genomic tumor DNA with primers 440 (5′-TTCATAAGGCTTAGCCAGCTAACTGCAG-3′) and 1112 (5′-GATGCCGGCACACACACACACACTCTCCC-3′), located in the 5′ end of the U3 region and in the 5′ untranslated region, respectively (Fig. 1A). Endogenous sequences from control DNA of noninjected inbred NMRI mice were not amplified by these primers. Each tumor DNA sample typically gave 2 to 5 amplification products, which were separated by gel electrophoresis. Single bands were cut out of the gel, purified by the GFX PCR DNA and gel band purification kit (Amersham Pharmacia Biotech), and sequenced with primers 440 and P2-rev (5′-ATACACGGGTACCCGGGCGACTCAGTCT-3′, located in R) with the DYEnamic ET terminator cycle sequencing kit (Amersham Pharmacia Biotech).
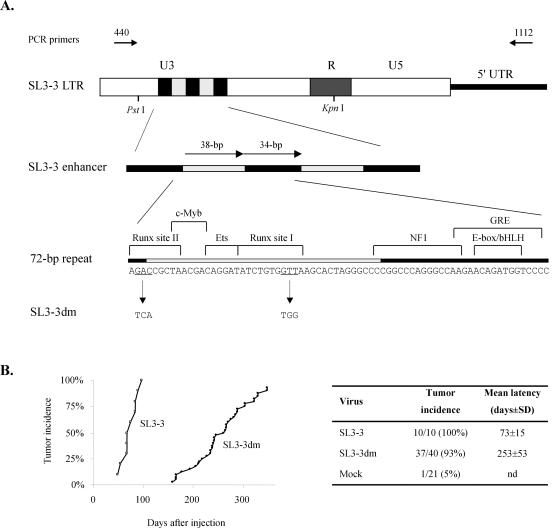
FIG. 1.
(A) LTR structures of SL3-3 and the Runx site double mutant virus SL3-3dm. The 38- and 34-bp repeat elements of the SL3-3 proviral enhancer in the U3 region are represented by grey and black boxes, respectively. The exact nucleotide sequences are given below the diagram, and known binding site sequences for cellular transcription factors are indicated. The 3-bp substitutions present in both copies of Runx sites I and II of the mutant virus are shown at the bottom. Locations of PCR primers used for amplification of genomic tumor DNA and some restriction sites are also presented. 5′ UTR, 5′ untranslated region; GRE, glucocorticoid response element; NF1, nuclear factor 1. (B) Mortality curves show the cumulative incidence of tumor development versus the number of days after infection of newborn inbred NMRI mice. Statistics are given to the right. nd, not determined.
RESULTS
SL3-3-induced lymphomas of inbred NMRI mice share the surface phenotype of mature helper T cells.
Wild-type SL3-3 (Fig. 1A) was injected into 10 newborn mice, which all developed lymphomas after a mean latency of about 2 months (Fig. 1B). The mice presented with enlarged thymuses and in most cases also enlarged spleens and lymph nodes. We looked at multiple diseased organs from nine animals. By Southern blotting analysis, clonal TCRβ but not Ig(κ) light chain gene rearrangements were found in all investigated tumors (Table 1), consistent with T-cell lymphoma. The tumors contained up to 5 clonal ecotropic provirus integrations with an average of 2 to 3, as detected with an ecotropic env gene-specific hybridization probe (data not shown). These results verify previously reported findings (12, 38).
TABLE 1.
Characterization of wild-type SL3-3-induced lymphomas in inbred NMRI mice
| Mouse no. | DNA rearrangements ina:
|
Lymphoma immunophenotype inb:
|
||||
|---|---|---|---|---|---|---|
| TCRβ | Ig(κ) | Tissue | Thymus | Mesenteric lymph node | Spleen | |
| 511-1 | 2/2 | 0/2 | t, m | CD4+ | CD4+ | CD4+ |
| 511-2 | NDc | ND | ND | ND | ND | ND |
| 511-3 | 2/2 | 0/2 | t, m | CD4+ | CD4+ | CD4+ |
| 511-4 | 2/2 | 0/2 | t, m | CD4+ | CD4+ | CD4+ |
| 511-5 | 2/2 | 0/2 | t, m | CD4+ | CD4+ | CD4+ |
| 511-6 | 2/2 | 0/2 | t, m | CD4+ | CD4+ | CD4+ |
| 511-7 | 2/2 | 0/2 | t, m | CD4+ CD8+ | CD4+ | CD4+ |
| 511-8 | 2/2 | 0/2 | t, s | CD4+ CD8+ | ND | CD4+ CD8−/dim+ |
| 511-9 | 2/2 | 0/2 | t, m | CD4+ | CD4+ | CD4+ |
| 511-10 | 2/2 | 0/2 | t, s | CD4+ | ND | CD4+ |
Detected by Southern blot analysis of HindIII-digested genomic tumor DNAs with the TCRβ-specific probes J1 and J2 or the Ig(κ)gene-specific probe, respectively (number of tumor samples with rearrangement/number of tumor samples analyzed). Specific tumor tissues analyzed from each mouse are indicated m, mesenteric lymph node; s, spleen; t, thymus.
Surface expression of CD4 and CD8 in lymphomas of different anatomical sites, as determined by flow cytometry analysis. All lymphomas were CD3+ (data not shown).
ND, not determined.
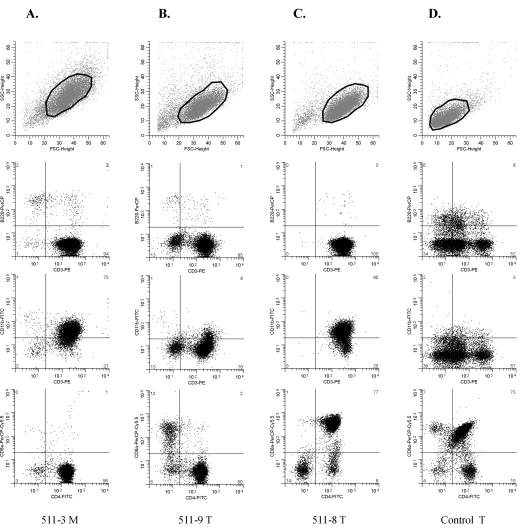
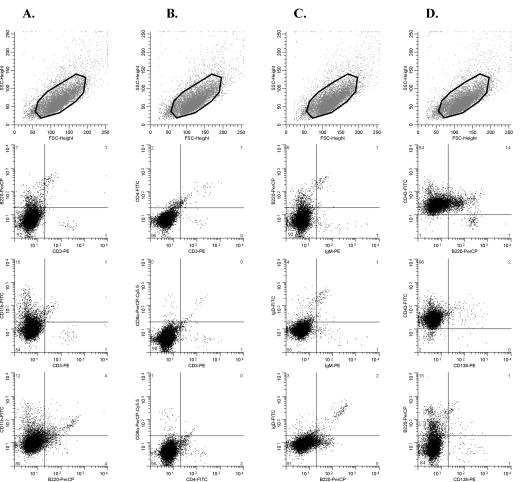
For further characterization, tumor cells were stained with antibodies to CD3, B220, and CD11b to distinguish T, B, and myeloid cells and with T-cell-specific antibodies to CD3, CD4, and CD8a and analyzed by three-color flow cytometry. In all cases, the great majority of cells (typically ≥85%) were CD3+ and B220− (Fig. 2A to C and data not shown), in agreement with the molecular diagnosis of T-cell lymphoma. Several tumors showed elevated expression of CD11b, with 20 to 85% of cells CD11b+, which consistently coexpressed CD3 (Fig. 2A and C and data not shown). CD3+ CD11b+ cells were barely detected in the thymuses, spleens, or mesenteric lymph nodes of healthy control mice analyzed in parallel (Fig. 2D and data not shown). Concomitant expression of markers of different lineages by tumors cells, termed lineage infidelity, has been observed also in other MLV-induced murine leukemia models (54, 57) and in some human hematopoietic malignancies (24).
FIG. 2.
Characterization of SL3-3-induced tumors by three-color flow cytometry. Dot plots of data obtained from three different tumor samples (A to C) and the thymus of an age-matched noninjected control mouse (D) stained with antibodies to B220-CD3-CD11b (middle panels) and CD3-CD4-CD8a (bottom) are shown. The region used for gating of the cells, as defined by forward (FSC) and side (SSC) scatter properties, is shown in each case (top). Mouse numbers and tissue types are shown below each column. M, mesenteric lymph node; T, thymus. The percentages of cells within each quadrant are indicated in the outer corners.
In 7 of 9 mice, all tumors analyzed for expression of CD4 and CD8 showed positivity for CD4 only (Fig. 2A and B; Table 1). One mouse (no. 511-8) had a CD4+ CD8+ thymic tumor (Fig. 2C) and a CD4+ CD8−/dim+ spleen tumor (Table 1). As judged from Southern hybridization patterns, the two diseased organs examined from each of these 8 mice harbored the same clonal ecotropic provirus integrations (data not shown), indicating that the tumors derive from the same transformation event. Mouse no. 511-7 had T-cell lymphomas of CD4+ and CD4+ CD8+ types (Table 1) showing different patterns of ecotropic virus integrations (data not shown) and, hence, seemed to represent distinct clones. Altogether, our results showed that the great majority of tumors induced by SL3-3 in inbred NMRI mice share the CD4+ phenotype of mature helper T cells, whereas a few present with the CD4+ CD8+ phenotype of immature T cells. The tumors contained one major cell population (usually ≥80%) with these CD4 and CD8 expression patterns.
SL3-3dm induces hematopoietic malignancies of four distinct lineages.
To characterize SL3-3dm-induced hematopoietic disease, a new pathogenicity experiment was initiated. Thirty-seven of 40 inbred NMRI mice injected with SL3-3dm (Fig. 1A) developed tumors with a mean latency of 8 months while 3 mice remained healthy throughout the 1-year observation period (Fig. 1B). Most of the affected mice presented with enlarged spleens and lymph nodes, but only a minority showed thymic enlargement, as also observed in a previous work with a smaller cohort (12). Nine different antibodies were used for three-color flow cytometry analysis of 68 tumor samples from 33 mice. TCRβ, Ig heavy, and Ig(κ) light chain gene rearrangements were analyzed by Southern blotting, and an ecotropic env gene-specific hybridization probe was used to detect clonal provirus integrations. Finally, more than 50 tumor samples from 28 mice were examined histopathologically. The results are summarized in Tables 2 and 3.
TABLE 2.
Characterization of SL3-3dm-induced tumors by flow cytometry and molecular analysis
| Tumor phenotypea | No. of miceb | No. of tumorsc | Location of tumor tissued
|
Ecotropic env positivee | DNA rearrangements inf:
|
||||
|---|---|---|---|---|---|---|---|---|---|
| S | LN | T | TCRβ | Ig(κ) | 1gH | ||||
| CD3+ | 13 | 26 | 4 | 16 | 6 | 17/21 | 10/21g | 3/21g | 6/21g |
| CD3+ CD138+ | 1 | 3 | 1 | 1 | 1 | 2/2 | 2/2 | 0/2 | 2/2 |
| CD3− CD4+ | 1 | 2 | 1 | 1 | 0 | 2/2 | 2/2 | 0/2 | 1/1 |
| CD11b+ | 14 | 23 | 14 | 9 | 0 | 17/18 | 0/18 | 0/18 | 0/19 |
| CD43+ only | 7 | 12 | 5 | 6 | 1 | 10/12 | 0/12 | 0/12 | 0/12 |
| CD138+ CD43+ | 1 | 1 | 1 | 0 | 0 | 1/1 | NDh | ND | 0/1 |
| Heterogeneous | 1 | 1 | 1 | 0 | 0 | 1/1 | 0/1 | 0/1 | 0/1 |
Immunophenotype determined by three-color flow cytometry with antibodies to CD3, B220, CD11b, CD4, CD8a, IgM, IgD, CD43, and CD138.
Number of mice, some with tumors of more than one type.
Number of analyzed tumors of each immunophenotype.
S, spleen; LN, lymph node (mesenteric or cervical); T, thymus.
Number of tumor samples with clonal ecotropic provirus integration(s)/number of tumor samples analyzed (as detected by Southern blot hybridization).
As determined by Southern blot analysis of genomic tumor DNA using TCRβ (J1 and J2)-, Ig(κ)-, and IgH-specific hybridization probes (number of tumor samples with rearrangement/number of tumor samples analyzed).
See the text for further description of different tumor types in the CD3+ subgroup.
ND, not determined.
TABLE 3.
Incidence, latency, and molecular characterization of SL3-3dm-induced tumors of different types
| Tumor phenotypea | No. of miceb | Mean tumor latency (days) ± SD | No. of clonal ecotropic integrationsc
|
No. of second-site altered proviral enhancer structuresd | |
|---|---|---|---|---|---|
| Range | Avg | ||||
| CD3+, CD3+ CD138+, or CD3− CD4+ | 15 | 266 ± 63 | 0-3 | 1-2 | |
| T-cell lymphomas [TCRβ r, IgH g/r, and Ig(κ) g]e | 7 | 240 ± 72 | 0-3 | 1-2 | 11/13 (5/7) |
| T-cell lymphoblastic lymphoma | 4 | 189 ± 32 | 1-2 | 1-2 | 10/10 (4/4) |
| Small T-cell lymphoma | 2 | 290 ± 19 | 0-3 | 1-2 | 0/2 (0/2) |
| No histologic diagnosis | 1 | 349 | 1 | 1 | 1/1 (1/1) |
| Other CD3+ typesf | 9 | 285 ± 43 | 0-3 | 1-2 | 4/17 (2/9) |
| Mature T-cell lymphoma, not otherwise specified | 3 | 246 ± 16 | 0-2 | 1-2 | 0/7 (0/3) |
| Small T-cell lymphoma | 2 | 339 ± 14 | 0-2 | 1-2 | 4/4 (2/2) |
| Diffuse large B-cell lymphoma, histiocyte-associated | 2 | 313 ± 14 | 2 | 2 | 0/3 (0/2) |
| No histologic diagnosis | 2 | 287 ± 60 | 2-3 | 2-3 | 0/2 (0/1) |
| CD11b+ (Myeloid leukemia with maturation) | 14 | 267 ± 49 | 0-4 | 1-2 | 0/22 (0/14) |
| CD43+ only | 7 | 258 ± 41 | 0-3 | 1-2 | 7/9 (5/7) |
| Erythroid leukemia | 2 | 292 ± 16 | 0-2 | 1-2 | 0/2 (0/2) |
| Myeloid leukemia without maturation | 3 | 232 ± 47 | 0-2 | 1 | 5/5 (3/3) |
| Pre-T-cell lymphoblastic lymphoma | 1 | 288 | 2 | 2 | 1/1 (1/1) |
| No histologic diagnosis | 1 | 242 | 3 | 3 | 1/1 (1/1) |
| CD138+ CD43+ | 1 | 177 | 1 | 1 | 0/1 (0/1) |
| Heterogeneous | 1 | 260 | 1 | 1 | 0/1 (0/1) |
| Total, all types | 37 | 253 ± 53 | 0-4 | 1-2 | |
As determined by three-color flow cytometry and molecular and histopathological analyses.
A few mice had tumors of two different types and, hence, appear twice in the table.
Number of clonal ecotropic provirus integrations in tumors of each phenotypic group, as detected by Southern blot hybridization.
Number of tumors with complex proviral enhancer structures/number of tumors analyzed (number of mice with complex enhancer structures detected/number of mice analyzed), determined by PCR amplification and sequencing of proviral enhancer sequences from genomic tumor DNAs. See Fig. 8 for a schematic presentation of enhancer structures with second-site alterations.
T-cell lymphoma subgroup of tumors with TCRβ clonally rearranged (r), IgH in germline configuration or clonally rearranged (g/r), and Ig(κ) in germline configuration (g), as determined by Southern blot analyses.
CD3+ type tumors not included in the T-cell lymphoma subgroup. Based on histopathological evidence; these cases included mature T-cell lymphomas (not otherwise specified), small T-cell lymphomas without detectable TCRβ rearrangements, and histiocyte-associated diffuse large B-cell lymphomas.
We found that SL3-3dm induced multiple hematopoietic malignancies in inbred NMRI mice, including myeloid leukemia, T-cell lymphoma, B-cell lymphoma, and erythroid leukemia. Of 33 analyzed mice, 11 presented with only lymphoma, 15 presented with only leukemia, and 4 presented with both leukemia and lymphoma at different anatomical sites. Conclusive diagnoses were not reached for 3 mice. Based on flow cytometry data, the mutant-induced tumors were initially divided into three overall phenotypic groups: CD3+ (lymphoid, Fig. 3), CD11b+ (myeloid, Fig. 4), and CD43+ only (Fig. 5). Minor variations in surface expression patterns were observed in each group. Also, a few tumors could not be sorted by these criteria (see below).
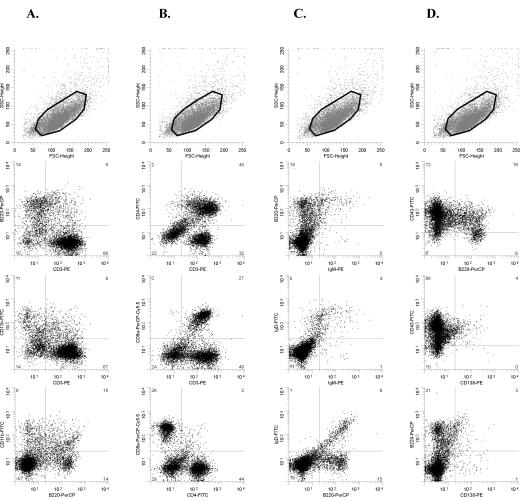
FIG. 3.
Flow cytometry analysis of an SL3-3dm-induced mesenteric lymph node tumor (mouse no. 498-6) with the CD3+ immunophenotype after staining with antibodies to CD3-B220-CD11b (A), CD3-CD4-CD8a (B), B220-IgM-IgD (C), and B220-CD43-CD138 (D). The gated population is indicated in the top panel in each column. The tumor contains a large population of CD3+ T cells that do not coexpress CD11b (A) and show heterogeneous expression of CD4 and CD8 (B). Cells positive for B-cell surface markers B220, IgM, IgD, and CD138 are relatively rare (A, C, and D). Most cells are CD43+ (D). This tumor had TCRβ but not Ig(κ) clonally rearranged (data not shown), and the mouse was histologically diagnosed as having a small T-cell lymphoma. FSC, forward scatter; SSC, side scatter.
FIG. 4.
Flow cytometry data from an SL3-3dm-induced tumor with the CD11b+ phenotype. Dot plots show results after staining with fluorochrome-conjugated antibodies to CD3-B220-CD11b (A), CD3-CD4-CD8a (B), B220-IgM-IgD (C), and B220-CD43-CD138 (D) of cells from the spleen tumor of a mouse (no. 498-2) diagnosed with myeloid leukemia with maturation. The region used for gating is also indicated (top panel of each column). FSC, forward scatter; SSC, side scatter.
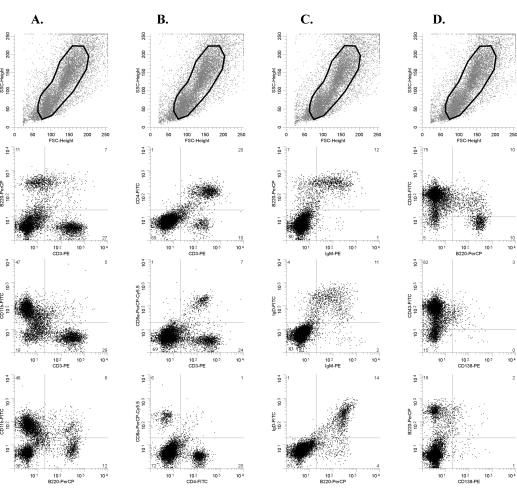
FIG. 5.
Flow cytometry analysis of an SL3-3dm-induced CD43+ only type mesenteric lymph node tumor (mouse no. 499-25). Dot plots show results obtained after staining with antibodies to CD3-B220-CD11b (A), CD3-CD4-CD8a (B), B220-IgM-IgD (C), and B220-CD43-CD138 (D). The gating region is shown in each case (top panel of each column). The vast majority of cells are CD43+ (D) and negative for all other markers investigated (A to D). The case shown is a myeloid leukemia without maturation, as determined by histopathology. FSC, forward scatter; SSC, side scatter.
CD3+ subgroup (T- and B-cell lymphomas).
About one-third (13 of 33) of the mice analyzed by flow cytometry showed a CD3+ phenotype (in most cases, 70 to 90% of the cells), whereas B220- and CD11b-expressing cells were rare (Fig. 3), consistent with a T-cell lymphoma. Nevertheless, only 6 of 13 mice had detectable TCRβ rearrangements by Southern analysis. Five of 6 mice had tumors with TCRβ rearrangements that seemed mono- or oligoclonal and clearly lacked Ig(κ) rearrangements, although occasionally IgH was rearranged along with TCRβ (4 of 8 tumors investigated from 5 mice) (Fig. 6A; Table 2). This strongly indicates a T-cell origin of the tumors. Histopathologically, two of these mice were diagnosed as having T-cell lymphoblastic lymphomas (Fig. 7A), another two were diagnosed as having small T-cell lymphomas (Fig. 7B-C), and one mouse was not analyzed.
FIG. 6.
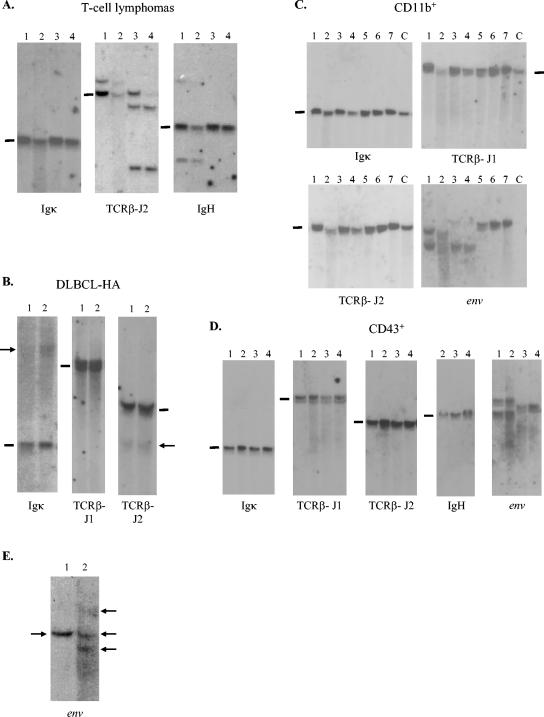
Southern blot analysis of genomic DNA extracted from SL3-3dm-induced tumors of distinct types. (A) DNA from SL3-3dm-induced T-cell tumors cleaved with HindIII and hybridized with the Ig(κ), TCRβ-J2, and IgH probes. Results are shown for 4 tumors from two different mice histologically diagnosed as having T-cell lymphoblastic lymphomas. Lanes: 1 and 2, spleen and lymph node tumors of mouse no. 499-8; 3 and 4, spleen and lymph node tumors of mouse no. 499-12. Black bars indicate the positions of the unrearranged genomic bands. (B) Hybridization with Ig(κ), TCRβ-J1, and TCRβ-J2 of HindIII-digested DNA from spleen (lane 1) and thymic (lane 2) tumors of an SL3-3dm-infected mouse (no. 499-30) diagnosed with histiocyte-associated DLBCL. Arrows mark weak rearranged bands. (C) HindIII-digested DNA from SL3-3dm-induced tumors of the CD11b+ immunophenotype hybridized with probes as indicated. Results are presented for 7 tumors from three different mice diagnosed with myeloid leukemia with maturation. Lanes: 1 and 2, spleen and lymph node tumors of mouse no. 499-10; 3 and 4, spleen and lymph node tumors of mouse no. 499-16; 5 to 7, spleen, cervical, and mesenteric lymph node tumors of mouse no. 498-5. Results are also shown for a negative control (lane C, DNA from the spleen of a noninjected inbred NMRI mouse). (D) DNA from SL3-3dm-induced CD43+ only type tumors cleaved with HindIII and analyzed with Ig(κ), J1, J2, and env hybridization probes or cleaved with EcoRI and hybridized with the IgH probe. Results are displayed for tumors from two mice diagnosed with erythroleukemia (lanes 1 and 2, spleen and mesenteric lymph node tumors of mouse no. 499-24) and myeloid leukemia without maturation (lanes 3 and 4, spleen and thymic tumors of mouse no. 499-25). (E) Hybridization with the ecotropic env gene-specific probe of HindIII-cleaved DNA from two tumors of different types (lane 1, spleen, myeloid leukemia with maturation; lane 2, mesenteric lymph node, DLBCL-HA) found in the same mouse (no. 499-33). Arrows mark the detected bands.
FIG. 7.
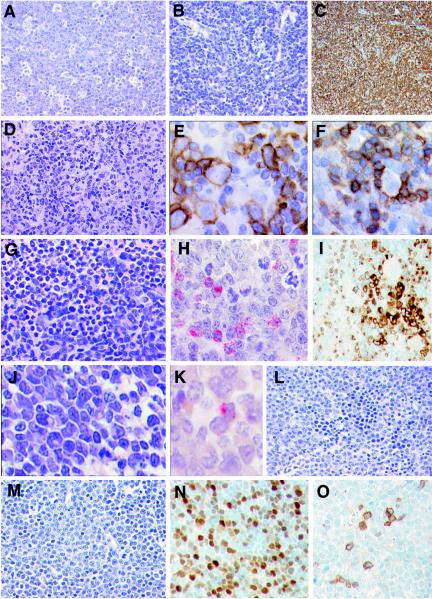
Histopathology of SL3-3dm-induced tumors. (A) T-cell lymphoblastic lymphoma. A diffuse infiltrate of medium-sized cells with blastic chromatin, central nucleolus, and inconspicuous cytoplasm is shown. Note the presence of numerous tingible body macrophages, conferring a starry-sky pattern (H&E staining). Magnification, ×360. (B and C) Small T-cell lymphoma. (B) A diffuse infiltrate of small- to medium-sized cells with minimal atypia is shown (H&E staining) Magnification, ×360. (C) Anti-CD3 reveals that the neoplastic infiltrate is of T-cell lineage (immunohistochemistry). Magnification, ×200. (D to F) Diffuse large B-cell lymphoma, histiocyte-associated. (D) A pleomorphic population of large lymphoid cells intermingled with histiocytes is shown (H&E staining). Magnification, ×360. (E) Anti-B220 highlights the large neoplastic Bcells, some of which are in mitosis (immunohistochemistry) Magnification, ×520. (F) Anti-CD3 reveals the small, reactive T cells surrounding a large pleomorphic B cell (immunohistochemistry). Magnification, ×520. (G to I) Myeloid leukemia with maturation. (G) The spleen is diffusely infiltrated with myeloid cells in all stages of maturation (H&E staining). Magnification, ×360. (H) Chloroacetate esterase reveals the presence of myeloblasts, promyelocytes, and myelocytes (cytochemistry). Magnification, ×520. (I) Myeloperoxidase staining shows the presence of groups of blasts surrounded by mature neutrophils (immunohistochemistry). Magnification, ×360. (J and K) Myeloid leukemia without maturation. (J) Diffuse infiltrate of blastic cells, some of which have a prominent Golgi area (H&E staining). Magnification, ×520. (K) Chloroacetate esterase revealed that at least 3% of the blast populations have granules in the cytoplasm (cytochemistry). Magnification, ×520. (L) Erythroleukemia. Note the presence of blastic cells intermingled with normoblasts with some dyserythropoietic features (H&E staining). Magnification, ×360. (M to O) Pre-T-cell lymphoblastic lymphoma. (M) A diffuse infiltrate of medium-sized cells with blastic chromatin, central nucleolus, and inconspicuous cytoplasm is shown. Several mitotic figures are present (H&E staining). Magnification, ×360. (N) The tumor cells show nuclear positivity for TdT (immunohistochemistry). Magnification, ×360. (O) Only a minority of the tumor cells show cytoplasmic positivity for CD3, indicating the early stage of differentiation of this lymphoblastic lymphoma (immunohistochemistry). Magnification, ×360.
Rearranged TCRβ bands of relatively weak intensity were detected together with both Ig(κ) (3 of 3 tumors) and IgH (2 of 2 tumors) rearrangements (Fig. 6B; Table 2) in tumors from 2 of 6 mice. The spleen tumor from one mouse was histologically diagnosed as a diffuse large B-cell lymphoma rich in histiocytes (DLBCL-HA). It contained numerous small reactive T cells, whereas the neoplastic B cells were rare (Fig. 7D to F). We note that this particular mouse also had a small T-cell lymphoma (described above) with TCRβ and IgH, but not Ig(κ), clonally rearranged and located at a different site (mesenteric lymph node), and it was, hence, diagnosed as having a mixed tumor (T and B lymphoid). The second mouse with both TCRβ, Ig(κ), and IgH clonally rearranged was not examined by histology.
In 7 of 13 mice, no clonal TCRβ or Ig rearrangements were identified, although many (10 of 15 tumors) contained clonally integrated ecotropic provirus(es) (Table 2). Following histopathological examination, 2 mice were diagnosed as having small T-cell lymphomas, whereas in 3 mice, the T-cell proliferation did not fit into any of the categories of the Bethesda classification system (26, 48) and were diagnosed as mature T-cell lymphoma, not otherwise specified. The absence of detectable TCRβ rearrangements in these 3 mice, and the lack of clonal ecotropic provirus integrations, suggest that these cases correspond to polyclonal T-cell proliferations and may be reactive. TCRδ rearrangements were also absent (Southern analysis; data not shown), excluding a clonal gamma/delta T-cell lymphoma type. In addition, one mouse in this group was diagnosed as having histiocyte-associated DLBCL. The rarity of neoplastic B cells may explain the absence of Ig rearrangements by Southern analysis. Thus, CD3+-rich tumors without TCRβ rearrangements were shown by histology to include T- as well as B-cell lymphomas and possibly reactive T-cell proliferations. It is of note that the antibody panel used here for flow cytometry analysis did not discriminate neoplastic versus reactive T-cell proliferations. Histology was not available for the last mouse.
Two mice developed T-cell lymphomas of distinct immunophenotypes. One mouse had a large population of CD3− CD4+ cells, and the other contained CD3+ cells that coexpressed CD138 (data not shown). Identification of clonal TCRβ and IgH, but not Ig(κ), gene rearrangements revealed the T-cell origin of these tumors (Table 2). Morphologically, both mice were diagnosed with T-cell lymphoblastic lymphomas.
Expression of CD4 and CD8 varied considerably in the SL3-3dm-induced T-cell tumors. Some contained a mixture of two T-cell populations (CD3+ CD4+ and CD3+ CD8+) (Fig. 3 and data not shown), whereas others had only one major population but they were of distinct types (CD3+ CD4− CD8−, CD3+ CD4+ CD8+, CD3+ CD4+, and CD3− CD4+; data not shown). The phenotypic heterogeneity is in contrast to the consistent CD4 and CD8 expression pattern of T-cell lymphomas induced by SL3-3 (Fig. 2; Table 1). Also, CD3+ cells of mutant-induced T-cell tumors did not coexpress CD11b in any of the cases analyzed (Fig. 3A and data not shown). It is notable that SL3-3dm-induced T-cell lymphoblastic lymphomas developed with considerably shorter mean latency than other tumors of the CD3+ immunophenotypic group (Table 3). We also note that in the previous work with inbred NMRI, only 1 of 10 SL3-3dm-induced tumors had clonal TCRβ rearrangements by Southern analysis (12) compared to 14 of 56 tumors analyzed in this study (Table 2), but the seeming difference can probably be ascribed to distinct sample sizes.
CD11b+ subgroup (myeloid leukemia).
About one-half (14 of 33) of the examined mice displayed signs of myeloid disease by flow cytometry analysis. Twenty-three tumors analyzed from these mice showed highly similar surface expression of the nine markers investigated (Fig. 4 and data not shown). The affected lymphoid organs contained an abnormally large population of CD11b+ cells (40 to 80%) together with distinct populations of CD3+ T cells and B220+ B cells (Fig. 4A). The T and B cells appeared normal based on coexpression patterns of CD4 and CD8 (Fig. 4B), IgM and IgD (Fig. 4C), and CD43 and CD138 (Fig. 4D). By molecular analysis, almost all tumors contained clonal ecotropic provirus integrations, but none had TCRβ, IgH, or Ig(κ) genes clonally rearranged (Fig. 6C; Table 2), consistent with the diagnosis of myeloid leukemia. Tumor tissues from the 14 mice were examined histopathologically (Fig. 7G to I and data not shown). In all cases, a diagnosis of myeloid leukemia with maturation was made. Monocytic and myelomonocytic leukemias were not seen. Almost one-third of the mice with mature myeloid leukemia also presented with lymphoma, as described above in detail. Thus, three mice had mixed mature myeloid and T-lymphoid tumors (small T-cell lymphoma or mature T-cell lymphoma, not otherwise specified), and one mouse had a mixed mature myeloid and B-lymphoid (DLBCL-HA) tumor.
CD43+ only subgroup (immature myeloid leukemia, erythroleukemia, and pre-T-cell lymphoblastic lymphoma).
Seven of 33 mice analyzed by flow cytometry presented with tumors that contained a large population of CD43+ cells (in most cases, >80%) and were negative for all other hematopoietic lineage markers investigated (Fig. 5). Most tumors had clonal ecotropic provirus integrations, but all cases were negative for TCRβ, IgH, and Ig(κ) rearrangements (Fig. 6D; Table 2). By histopathological examination, 3 mice had myeloid leukemia without maturation, i.e., immature myeloid leukemia (Fig. 7J to K). At least 3% of the blasts were positive for chloroacetate esterase (Fig. 7K). Two other mice showed clear morphological evidence of erythroid leukemia (Fig. 7L). The last mouse had a pre-T-cell lymphoblastic lymphoma (Fig. 7M), positive for TdT (Fig. 7N) but negative for TCRβ rearrangements (Table 2). Approximately 10% of the tumor cells showed cytoplasmic positivity for CD3 (Fig. 7O), revealing the early T-lineage differentiation stage of this lymphoma.
Minor tumor types.
Tumors from two mice were incompatible with the phenotypic subgroups described above. One mouse had a clonal CD138+ CD43+ B220− tumor without detectable IgH rearrangements, despite surface expression of B-lineage-specific CD138 (Table 2 and data not shown). The spleen tumor of another mouse contained clonal ecotropic proviral integrations, lacked TCRβ and Ig rearrangements (Table 2), and had no major abnormal cell population identified by flow cytometry analysis with the nine antibodies (data not shown). It was therefore regarded as heterogeneous. These minor cases were not further analyzed.
Ecotropic provirus integrations in SL3-3dm-induced tumors.
Southern hybridizations with the env gene-specific probe did not reveal any significant differences with respect to numbers and distribution of clonal ecotropic provirus integrations between tumors of the different phenotypic groups (Table 3). Tumors of SL3-3dm-infected mice with the same phenotype generally also contained the same ecotropic provirus integrations (Fig. 6C and D), indicating that they derive from the same transformation event. In two mice with mixed tumors (mature myeloid plus T lymphoid) located at distinct sites, both tumors were negative for ecotropic provirus integrations and lacked clonal Ig and TCRβ rearrangements, and their potential clonal relatedness was not evaluated. However, in another two cases, lymph node tumors (lymphomas) of mice with mixed tumors (mature myeloid plus T lymphoid and mature myeloid plus B lymphoid) contained one to two additional clonal ecotropic virus integration(s) compared to the myeloid spleen tumor (Fig. 6E and data not shown). The common bands may mirror infiltrating cells from one tumor to the other; however, neither histopathology nor flow cytometry analysis of these cases showed significant evidence thereof. Hence, it is possible that phenotypically distinct end-stage tumors of these mice originate from the same hematopoietic (progenitor) cell.
Proviral enhancer structures with complex alterations are commonly found in some types of SL3-3dm-induced tumors.
Secondary mutations are frequently present in the enhancer of proviruses found in tumors induced by SL3-3 with mutated Runx sites and have in some cases been shown to partially rescue viral T lymphomagenicity (11-13, 41, 42, 47). We therefore analyzed proviral enhancer structures in genomic DNA from SL3-3dm-induced tumors by bulk PCR amplification of a 5′ LTR fragment that contains the viral U3 enhancer with a primer located upstream of the enhancer repeats and a primer located in the 5′ untranslated region, respectively (Fig. 1A). The amplification products were purified, and nucleotide sequences were determined from a position 90-nucleotides upstream of the enhancer repeats to the 3′ end of the U3 region.
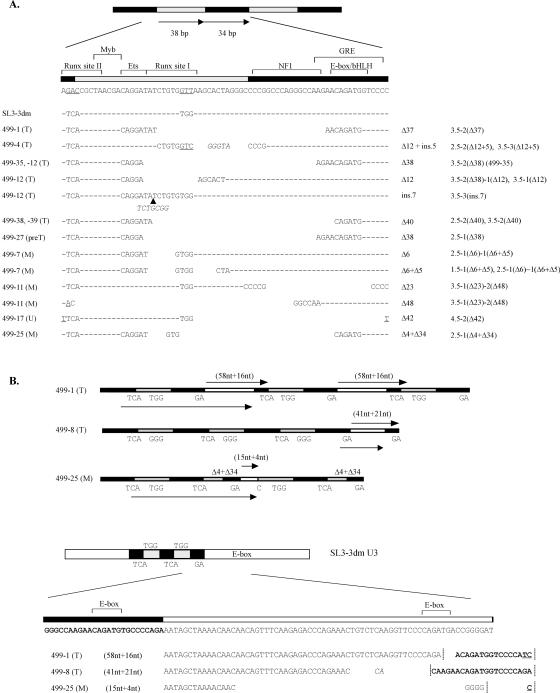
In several SL3-3dm-induced tumors, only simple fluctuations in numbers of 72-bp repeat copies were detected (loss of 1 or gain of 1 to 2), and the 3-bp Runx site mutations were retained. Variations in repeat copy numbers were also the only changes in proviral enhancers amplified from SL3-3-induced T-cell lymphomas of this study (data not shown), in agreement with previous reports (11, 12, 47, 50). Provirus enhancer structures with complex alterations were found in tumors from 12 different mice infected with SL3-3dm. The altered structures contained short deletions and/or insertions within the 72-bp repeat elements (Fig. 8A). In tumors from three of the mice, enhancer structures were identified with alternative repeats generated by duplication of U3 sequence fragments containing various parts of the 72-bp repeat elements and U3 sequences immediately downstream thereof (Fig. 8B). In one of these (no. 499-8), the TGG mutation in Runx site I had partially reverted to GGG, as also observed in an enhancer structure from an SL3-3dm-induced T-cell tumor of a former study (11). The same enhancer structure was typically found in more than one tumor of each mouse and amplified from several independent PCRs. Furthermore, by Southern blotting analysis of genomic tumor DNA digested with PstI and KpnI, cutting at both ends of the LTR (Fig. 1A), bands corresponding in length to the different enhancer structures amplified by PCR were detected with an SL3-3 repeat-specific hybridization probe (data not shown). We can therefore exclude that the complex structures are PCR artifacts. The original SL3-3dm enhancer, with or without copy number variation, was found together with complex altered structures only in a few of these tumors. However, not all PCR-amplified bands were sequenced, and additional proviral forms may exist in the tumors.
FIG. 8.
Structures of altered proviral enhancers found in CD3+ and CD43+ only tumors of mice infected with SL3-3dm. The number of the mouse in which each structure was found is given to the left together with an indication of the tumor type. T, T-cell lymphoma; preT, pre-T-cell lymphoblastic lymphoma; M, myeloid leukemia without maturation; U, CD43+ only, no histologic diagnosis. (A) Nucleotide sequences are shown for 72-bp repeat elements with in vivo-derived deletions (Δ) or insertions (ins.). Point mutations are underlined; insertions are in italics. Repeat organizations of proviral enhancer structures with the given alterations are indicated to the right [e.g., 3.5-2(Δ37) consists of 3.5 copies of the 72-bp repeat element, two of which have the 37-bp deletion shown]. Complete sequences of all enhancer structures detected in SL3-3dm-induced tumors are available upon request. (B) Schematic presentation of enhancer structures with alternative sequence duplications found in three different mice infected with SL3-3dm. The duplicated sequence is indicated by an arrow below each structure, and the alternative repeat element generated thereby is marked by arrow(s) above. Nucleotide sequences of the alternative repeats are shown at the bottom, and vertical lines indicate the borders between repeat elements.
Although different and complex, many of the alterations seemed to follow a common pattern. The short deletions typically included the NF1 binding site and, to various extents, the flanking sequences, often including the mutated Runx site I sequence (Fig. 8A). The observed changes were highly similar to proviral enhancer alterations reported previously in tumors induced by SL3-3 Runx site mutants (11, 12), strongly indicating that they were selected for increased pathogenicity during the tumorigenic process in vivo. Interestingly, proviral enhancers with complex structural changes were found only in some SL3-3dm-induced tumor types (Fig. 8; Table 3). Altered enhancers were frequently detected in T-cell lymphomas, pre-T-cell lymphoblastic lymphoma, and in cases of myeloid leukemia without maturation, which indicates that the SL3-3dm enhancer is suboptimal for induction of these disease types. Proviral enhancer structures with complex changes were not amplified from any of the tumors diagnosed as myeloid leukemia with maturation, DLBCL-HA, or erythroid leukemia (Table 3). The only changes found in these tumors were simple fluctuations in 72-bp repeat copy numbers, and the introduced Runx site mutations also had not reverted, indicating that the SL3-3dm enhancer is sufficient for induction these tumor types.
DISCUSSION
Two markedly different MLV-induced disease models were characterized in this study: a fast and homogeneous T-lymphoma model induced by wild-type SL3-3 MLV in inbred NMRI mice and a highly variegated model of myeloid, T-lymphoid, B-lymphoid, and erythroid tumors induced with prolonged latency in the same mouse strain by a mutant of SL3-3 (SL3-3dm) with disrupted Runx binding sites in the proviral enhancer.
Our analyses showed that the vast majority of SL3-3-induced T lymphomas displayed the CD3+ CD4+ surface phenotype of mature helper T cells while tumors with features of more immature T cells, CD3+ CD4+ CD8+, were rare. The consistent phenotypic pattern and marked predominance of tumor cells with surface expression similar to mature T cells is atypical for MLV-induced T-lymphoma models. Most other cases investigated, including SL3-3-induced T lymphomas of AKR mice (22, 23) and tumors induced by closely related T-lymphomagenic MoMLV in NIH/Swiss mice (54) and rats (29), exhibit pronounced variation with respect to expression of CD4 and CD8.
We also found that obstruction of rapid T-cell lymphoma induction by mutation of all Runx binding sites in the enhancer of SL3-3 allowed this otherwise strictly T-lymphomagenic MLV to induce a multitude of hematopoietic malignancies in inbred NMRI mice. The most common disease was myeloid leukemia, followed by T-lymphoid tumors, a few cases of mature B-cell lymphoma, and erythroid leukemia, in addition to some minor types not fully characterized. Such a broad disease induction potential is unusual for replication-competent MLVs without oncogenes, although SRS19-6 and Cas-Br-M (or Cas-Br-E) induce tumors of four distinct hematopoietic lineages (5, 14). The phenotypic heterogeneity suggests that SL3-3dm infects and transforms several different cell types or targets an early hematopoietic progenitor, but our results do not resolve this question.
This is the first demonstration of a significant potential for myeloid leukemia induction by SL3-3. However, the capacity to induce myeloid disease has been exposed for other MLVs that normally induce almost exclusively lymphoid or erythroid leukemias, also after prevention of the more rapid disease by manipulation of virus-host interactions (73). Exchanging most of the U3 region of T-lymphomagenic MoMLV with that of 4070A MLV causes the recombinant virus (MOL4070LTR) to induce both myeloid and T-lymphoid tumors (74) with incidences similar to SL3-3dm. However, MOL4070LTR induces predominantly myelomonocytic leukemia in FVB and BALB/c mice (74), a disease type not seen here for SL3-3dm in inbred NMRI mice. A few MLVs induce myeloid malignancies with significant incidence (28, 73). SRS19-6 and Graffi MLV induce granulocytic or myeloblastic leukemias (2, 5, 28, 49, 57, 73), compatible with diagnoses of myeloid leukemia with or without maturation (26), whereas Cas-Br-E-induced non-T-, non-B-cell leukemia is composed of very immature cells (3, 55) and may represent an even less differentiated disease than the SL3-3dm-induced CD43+ only type of myeloid leukemia without maturation. Moreover, expression of endogenous ecotropic MLVs is causally associated with myeloid leukemia development in BXH-2 mice (predominantly granulocytic with mild to complete maturation) and AKXD-23 mice (primarily myelomonocytic) (2, 28, 49, 57, 73).
Proviral enhancer structures isolated from SL3-3dm-induced myeloid leukemia with maturation, mature B-cell lymphoma, and erythroleukemia showed variation only in repeat copy numbers, indicating that the SL3-3dm enhancer in itself is sufficient for induction of these malignancies. In contrast, the SL3-3dm enhancer appeared to be suboptimal for induction of T-lymphoid tumors and the CD43+ only type of immature myeloid leukemias, since complex structural changes were detected in proviral enhancer structures from most of these tumors. Although the overall patterns of second-site changes were similar in T-lymphoid versus immature myeloid tumors, some differences attracted further attention.
Provirus enhancers with short insertions were identified in two T-cell lymphomas induced by SL3-3dm, in one case (no. 499-12) generating a sequence (ATCTGCGGTCT) with a perfect match (underlined) to the Runx consensus sequence PyGPyGGTPy (43) and strikingly similar to the equivalent motif of T-lymphomagenic Soule MLV (ATCTGCGGTCA). The Soule Runx site sequence supports T-cell lymphoma induction effectively in the context of MoMLV (34) and is a frequent in vivo-derived suppressor mutation of attenuated T lymphomagenesis by the SAA mutant of SL3-3, which has Runx site I converted to that of weakly pathogenic Akv MLV (42, 47). In the other case (no. 499-4), the mutated Runx site I was replaced by a consensus binding motif (ATCTGTGGTCGG) identical to the Runx site sequence of Akv (and SAA) except for two 3′ flanking guanosines. This exact sequence has been identified also as a second-site mutation in prelymphomatous thymus tissue of an SAA-infected mouse (42), and its generation in two distinct experimental systems by different molecular mechanisms indicates a biological function.
Sequence alterations at the mutated Runx site I were detected in proviral enhancers from two immature myeloid leukemias (no. 499-7, TGTGGAA and TGTGGCT; no. 499-25, TGTGCAG) but did not match the Runx consensus sequence (43), indicating that the pathogenic effects of these structures, most probably, are not mediated by Runx factors. Likewise, enhancer structures isolated from the remaining majority of SL3-3dm-induced T-lymphoid tumors are more likely to promote viral T-cell lymphoma induction independently of Runx. The altered proviral enhancers found in T-lineage tumors typically had short deletions affecting the NF1 site and, to various extents, the glucocorticoid response element and mutated Runx site I. It was previously shown that this type of alterations can partially restore viral enhancer activity and to some degree rescue the pathogenicity of SL3-3 Runx site mutants in T cells without recovery of Runx responsiveness (11, 12). All SL3-3dm-induced T-cell lymphomas, in which in vivo-derived LTR changes were detected, showed amplification of an enhancer fragment containing the basic helix-loop-helix (bHLH) site, the mutated Runx site II, the c-Myb site, and an Ets core motif (GGAA/T), in all cases combined with other alterations. Deletions in enhancer structures isolated from immature myeloid leukemias also involved the mutated Runx site I and/or the NF1 site, but flanking c-Myb, Ets, and bHLH binding site sequences were not consistently preserved. This indicates that arrays of bHLH, c-Myb, and Ets (core) sites promote viral enhancer activity and oncogenicity specifically in T cells together with other changes such as deletion of inhibitory NF1 sites, and in rare cases, possible reconstitution of a functional Runx site.
The c-Myb motif is critical for SL3-3 enhancer activity in T cells and for viral T lymphomagenicity (50, 78), consistent with the strong selection observed for this site during T-lymphoma induction by SL3-3dm. Enhancer Ets sites play a minor role for SL3-3 pathogenicity (50), but their significance may increase upon mutation of all Runx sites, as supported by our finding of the same Ets core site sequence (GGAA) in proviral enhancer structures from different tumors. Also, the preservation of bHLH motifs in proviral enhancer structures isolated from different SL3-3dm-induced T-lymphoid tumors suggests a significant contribution to viral T-cell lymphoma induction, as reported for the equivalent GRE/bHLH sites in MoMLV (61) but yet unexamined for SL3-3. Notably, bHLH site sequences were multiplied in the proviral LTR structures isolated from two T-cell lymphomas with downstream U3 sequence stretches copied into the SL3-3dm repeat region but not in the one case of immature myeloid leukemia with enhancer changes of similar type. The U3 region immediately 3′ to the enhancer repeats in SL3-3 may contain additional genetic determinants of viral disease specificity, as shown for MoMLV (20) and could be manifested by sequence amplification in the context of weakly pathogenic SL3-3dm.
The preference of SL3-3dm for induction of myeloid leukemia with maturation and the absence of proviral enhancer structures with complex changes in these tumors indicate that Runx factors are of little importance for induction of this disease type by SL3-3. This is similar to the suggested minor role of these sites for erythroleukemia induction by mutants of MoMLV but in striking contrast to the critical role of enhancer Runx sites for T-cell lymphoma induction by SL3-3 and MoMLV, as shown here and reported previously by various groups (12, 19, 34, 47, 61). Both SRS19-6 and GV1.4 Graffi MLV have a single-copy enhancer with a nonconsensus Runx motif (43). Effective induction of myeloid leukemia therefore seems to not require strong Runx factor binding sites in the proviral enhancer. However, a high-affinity Runx site also does not rule out the ability to induce myeloid disease with high incidence, as evident for Cas-Br-E (3, 14, 55) sharing the enhancer Runx site sequence of MoMLV (16). Hence, the function of Runx sites in MLV tumorigenesis may depend on the exact sequence as well as the context in which these sites are located. The role of Runx site binding factors for induction of the CD43+ only type of immature myeloid leukemia is less clear. Our repeated finding of altered proviral enhancers in these tumors may reflect a need for compensatory mutations, and we thereby propose a significant role of enhancer Runx sites for development of this disease type, which is not revealed in the context of the wild-type virus.
Despite some variation in the distribution of different tumor types, the broad disease pattern induced by SL3-3dm in inbred NMRI resembles that of SRS19-6 MLV in NIH/Swiss mice (5), which is also influenced by LTR rearrangements in vivo (17). A short fragment, including putative Ets and Runx but not NF1 and bHLH binding sites of the single-copy SRS19-6 enhancer, is frequently multiplied in proviral enhancers isolated from T-cell lymphomas but not from other tumor types induced by this virus (17). The Ets-Runx site arrays seem to support viral enhancer activity most effectively in T cells and to thereby promote T-cell lymphoma induction by SRS19-6 (17). C-Myb, Ets, and bHLH sites may have a similar function in the SL3-3dm enhancer to compensate for the introduced Runx site mutations. Disease induction by SL3-3dm is relatively slow compared to SRS19-6 (5), which seems to have evolved several different means to ensure potent induction of multiple tumor types, involving viral genetic determinants both within and outside of the enhancer region (17). SL3-3dm, on the other hand, is a severely impeded mutant of T-lymphomagenic SL3-3 not specialized in the same way for induction of multiple disease types.
In summary, we have shown that mutation of all Runx binding sites in the enhancer of SL3-3 impairs rapid T-lymphomagenesis and in turn exposes a significant potential of this virus for induction of other hematopoietic malignancies. In itself, SL3-3dm seemed to primarily induce myeloid leukemia with maturation and to less frequently induce other tumor types such as diffuse large B-cell lymphoma and erythroleukemia. Occasionally, proviral enhancer structures with complex second-site changes arose in vivo, which appeared to promote induction of yet other disease types. Such altered enhancer structures were not detected in T-cell lymphomas induced by wild-type SL3-3, altogether suggesting that the Runx site mutations in SL3-3dm facilitate in vivo molecular evolution of viral enhancer structures with altered cell type specificity, most probably as a consequence of high selection pressures for enhancer sequences with increased pathogenicity during the greatly prolonged latency period of tumor induction by the mutated virus. Finally, we emphasize that the Runx site mutations unmasked a considerable and formerly unrevealed potential of SL3-3 for the induction of myeloid leukemia. This finding may distinguish SL3-3 MLV from another strongly T-lymphomagenic virus, MoMLV, which becomes erythroleukemogenic when all enhancer Runx sites are mutated (61).
Acknowledgments
We thank Alexander Schmitz and Søren Warming for help with flow cytometry analysis. The technical assistance of Lone Højgaard, Angelika Appold, Katrin Reindl, Jaqueline Müller, Nadine Kink, and Elenore Samson is gratefully acknowledged.
This work was supported by the Danish Cancer Society, the Novo Nordic Foundation, the Karen Elise Jensen Foundation, the John and Birte Meyer Foundation, and the Danish Natural Sciences and Medical Research Councils.
REFERENCES
- 1.Bae, S. C., Y. Yamaguchi-Iwai, E. Ogawa, M. Maruyama, M. Inuzuka, H. Kagoshima, K. Shigesada, M. Satake, and Y. Ito. 1993. Isolation of PEBP2 alpha B cDNA representing the mouse homolog of human acute myeloid leukemia gene, AML1. Oncogene 8:809-814. [PubMed] [Google Scholar]
- 2.Bedigian, H. G., D. A. Johnson, N. A. Jenkins, N. G. Copeland, and R. Evans. 1984. Spontaneous and induced leukemias of myeloid origin in recombinant inbred BXH mice. J. Virol. 51:586-594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bergeron, D., J. Houde, L. Poliquin, B. Barbeau, and E. Rassart. 1993. Expression and DNA rearrangement of proto-oncogenes in Cas-Br-E-induced non-T-, non-B-cell leukemias. Leukemia 7:954-962. [PubMed] [Google Scholar]
- 4.Boral, A. L., S. A. Okenquist, and J. Lenz. 1989. Identification of the SL3-3 virus enhancer core as a T-lymphoma cell-specific element. J. Virol. 63:76-84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bundy, L. M., M. Ru, B.-F. Zheng, L. Cheng, P. K. Pattengale, J. L. Portis, and H. Fan. 1995. Biological characterization and molecular cloning of murine c-type retrovirus derived from the TSZ complex from mainland China. Virology 212:367-382. [DOI] [PubMed] [Google Scholar]
- 6.Celander, D., and W. A. Haseltine. 1984. Tissue-specific transcription preference as a determinant of cell tropism and leukaemogenic potential of murine retroviruses. Nature 312:159-162. [DOI] [PubMed] [Google Scholar]
- 7.Chatis, P. A., C. A. Holland, J. W. Hartley, W. P. Rowe, and N. Hopkins. 1983. Role for the 3′ end of the genome in determining disease specificity of Friend and Moloney murine leukemia viruses. Proc. Natl. Acad. Sci. USA 80:4408-4411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chatis, P. A., C. A. Holland, J. E. Silver, T. N. Frederickson, N. Hopkins, and J. W. Hartley. 1984. A 3′ end fragment encompassing the transcriptional enhancers of nondefective Friend virus confers erythroleukemogenicity on Moloney leukemia virus. J. Virol. 52:248-254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Coffman, J. A. 2003. Runx transcription factors and the developmental balance between cell proliferation and differentiation. Cell Biol. Int. 27:315-324. [DOI] [PubMed] [Google Scholar]
- 10.DesGroseillers, L., and P. Jolicoeur. 1984. The tandem direct repeats within the long terminal repeat of murine leukemia viruses are the primary determinant of their leukemogenic potential. J. Virol. 52:945-952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ethelberg, S., B. Hallberg, J. Lovmand, J. Schmidt, A. Luz, T. Grundström, and F. S. Pedersen. 1997. Second-site proviral enhancer alterations in lymphomas induced by enhancer mutants of SL3-3 murine leukemia virus: negative effect of nuclear factor 1 binding site. J. Virol. 71:1196-1206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ethelberg, S., J. Lovmand, J. Schmidt, A. Luz, and F. S. Pedersen. 1997. Increased lymphomagenicity and restored disease specificity of AML1 site (core) mutant SL3-3 murine leukemia virus by a second-site enhancer variant evolved in vivo. J. Virol. 71:7273-7280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ethelberg, S., A. B. Sørensen, J. Schmidt, A. Luz, and F. S. Pedersen. 1997. An SL3-3 murine leukemia virus enhancer variant more pathogenic than the wild type obtained by assisted molecular evolution in vivo. J. Virol. 71:9796-9799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fredrickson, T. N., W. Y. Langdon, P. M. Hoffman, J. W. Hartley, and H. C. Morse III. 1984. Histologic and cell surface antigen studies of hematopoietic tumors induced by Cas-Br-M murine leukemia virus. J. Natl. Cancer Inst. 72:447-454. [PubMed] [Google Scholar]
- 15.Goldrath, A. W., and M. J. Bevan. 1999. Selecting and maintaining a diverse T-cell repertoire. Nature 402:255-262. [DOI] [PubMed] [Google Scholar]
- 16.Golemis, E. A., N. A. Speck, and N. Hopkins. 1990. Alignment of U3 region sequences of mammalian type C viruses: identification of highly conserved motifs and implications for enhancer design. J. Virol. 64:534-542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Granger, S. W., L. Bundy, and H. Fan. 1999. Tandemerization of a subregion of the enhancer sequences from SRS 19-6 murine leukemia virus associated with T-lymphoid but not other leukemias. J. Virol. 73:7175-7184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gulley, M. L., L. C. Ogata, J. A. Thorson, M. O. Dailey, and J. D. Kemp. 1988. Identification of a murine pan-T cell antigen which is also expressed during the terminal phases of B cell differentiation. J. Immunol. 140:3751-3757. [PubMed] [Google Scholar]
- 19.Hallberg, B., J. Schmidt, A. Luz, F. S. Pedersen, and T. Grundström. 1991. SL3-3 enhancer factor 1 transcriptional activators are required for tumor formation by SL3-3 murine leukemia virus. J. Virol. 65:4177-4181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hanecak, R., P. K. Pattengale, and H. Fan. 1991. Deletion of a GC-rich region flanking the enhancer element within the long terminal repeat sequences alters the disease specificity of Moloney murine leukemia virus. J. Virol. 65:5357-5363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hardy, R. R., C. E. Carmack, S. A. Shinton, J. D. Kemp, and K. Hayakawa. 1991. Resolution and characterization of pro-B and pre-pro-B cell stages in normal mouse bone marrow. J. Exp. Med. 173:1213-1225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hays, E. F., and G. Bristol. 1992. Observations on lymphomagenesis and lymphoma in AKR mice. A description of prelymphoma changes in the thymus and phenotypic diversity of lymphomas induced by SL3-3 virus. Thymus 19:219-234. [PubMed] [Google Scholar]
- 23.Hays, E. F., G. C. Bristol, S. McDougall, J. L. Klotz, and M. Kronenberg. 1989. Development of lymphoma in the thymus of AKR mice treated with the lymphomagenic virus SL 3-3. Cancer Res. 49:4225-4230. [PubMed] [Google Scholar]
- 24.Jaffe, E. S., N. L. Harris, H. Stein, and J. W. Vardiman (ed.). 2001. Pathology and genetics of tumours of hematopoietic and lymphoid tissues. IARC Press, Lyon, France.
- 25.Jones, A. T., B. Federsppiel, L. G. Ellies, M. J. Williams, R. Burgener, V. Duronio, C. A. Smith, F. Takei, and H. J. Ziltener. 1994. Characterization of the activation-associated isoform of CD43 on murine T lymphocytes. J. Immunol. 153:3426-3439. [PubMed] [Google Scholar]
- 26.Kogan, S. C., J. M. Ward, M. R. Anver, J. J. Berman, C. Brayton, R. D. Cardiff, J. S. Carter, S. de Coronado, J. R. Downing, T. N. Fredrickson, D. C. Haines, A. W. Harris, N. L. Harris, H. Hiai, E. S. Jaffe, I. C. MacLennan, P. P. Pandolfi, P. K. Pattengale, A. S. Perkins, R. M. Simpson, M. S. Tuttle, J. F. Wong, and H. C. Morse III. 2002. Bethesda proposals for classification of nonlymphoid hematopoietic neoplasms in mice. Blood 100:238-245. [DOI] [PubMed] [Google Scholar]
- 27.Lagasse, E., and I. L. Weissman. 1996. Flow cytometric identification of murine neutrophils and monocytes. J. Immunol. Methods 197:139-150. [DOI] [PubMed] [Google Scholar]
- 28.Largaespada, D. A. 2000. Genetic heterogeneity in acute myeloid leukemia: maximizing information flow from MuLV mutagenesis studies. Leukemia 14:1174-1184. [DOI] [PubMed] [Google Scholar]
- 29.Lazo, P. A., A. J. Klein-Szanto, and P. N. Tsichlis. 1990. T-cell lymphoma lines derived from rat thymomas induced by Moloney murine leukemia virus: phenotypic diversity and its implications. J. Virol. 64:3948-3959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Leib-Mösch, C., J. Schmidt, M. Etzerodt, F. S. Pedersen, R. Hehlmann, and V. Erfle. 1986. Oncogenic retrovirus from spontaneous murine osteomas. II. Molecular cloning and genomic characterization. Virology 150:96-105. [DOI] [PubMed] [Google Scholar]
- 31.Lenz, J., D. Celander, R. L. Crowther, R. Patarca, D. W. Perkins, and W. A. Haseltine. 1984. Determination of the leukaemogenicity of a murine retrovirus by sequences within the long terminal repeat. Nature 308:467-470. [DOI] [PubMed] [Google Scholar]
- 32.Lenz, J., R. Crowther, S. Klimenko, and W. Haseltine. 1982. Molecular cloning of a highly leukemogenic, ecotropic retrovirus from an AKR mouse. J. Virol. 43:943-951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Levanon, D., V. Negreanu, Y. Bernstein, I. Bar-Am, L. Avivi, and Y. Groner. 1994. AML1, AML2, and AML3, the human members of the runt domain gene-family: cDNA structure, expression, and chromosomal localization. Genomics 23:425-432. [DOI] [PubMed] [Google Scholar]
- 34.Lewis, A. F., T. Stacy, W. R. Green, L. Taddesse-Heath, J. W. Hartley, and N. A. Speck. 1999. Core-binding factor influences the disease specificity of Moloney murine leukemia virus. J. Virol. 73:5535-5547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Liu, P., S. A. Tarle, A. Hajra, D. F. Claxton, P. Marlton, M. Freedman, M. J. Siciliano, and F. S. Collins. 1993. Fusion between transcription factor CBF beta/PEBP2 beta and a myosin heavy chain in acute myeloid leukemia. Science 261:1041-1044. [DOI] [PubMed] [Google Scholar]
- 36.LoSardo, J. E., A. L. Boral, and J. Lenz. 1990. Relative importance of elements within the SL3-3 virus enhancer for T-cell specificity. J. Virol. 64:1756-1763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lovmand, J., A. B. Sørensen, J. Schmidt, M. Østergaard, A. Luz, and F. S. Pedersen. 1998. B-Cell lymphoma induction by Akv murine leukemia viruses harboring one or both copies of the tandem repeat in the U3 enhancer. J. Virol. 72:5745-5756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lund, A. H., J. Schmidt, A. Luz, A. B. Sørensen, M. Duch, and F. S. Pedersen. 1999. Replication and pathogenicity of primer binding site mutants of SL3-3 murine leukemia viruses. J. Virol. 73:6117-6122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lund, A. H., and M. van Lohuizen. 2002. RUNX: a trilogy of cancer genes. Cancer Cell 1:213-215. [DOI] [PubMed] [Google Scholar]
- 40.Manley, N. R., M. O'Connell, W. Sun, N. A. Speck, and N. Hopkins. 1993. Two factors that bind to highly conserved sequences in mammalian type C retroviral enhancers. J. Virol. 67:1967-1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Martiney, M. J., L. S. Levy, and J. Lenz. 1999. Suppressor mutations within the core binding factor (CBF/AML1) binding site of a T-cell lymphomagenic retrovirus. J. Virol. 73:2143-2152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Martiney, M. J., K. Rulli, R. Beaty, L. S. Levy, and J. Lenz. 1999. Selection of reversions and suppressors of a mutation in the CBF binding site of a lymphomagenic retrovirus. J. Virol. 73:7599-7606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Melnikova, I. N., B. E. Crute, S. Wang, and N. A. Speck. 1993. Sequence specificity of the core-binding factor. J. Virol. 67:2408-2411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Miyoshi, H., K. Shimizu, T. Kozu, N. Maseki, Y. Kaneko, and M. Ohki. 1991. t(8;21) breakpoints on chromosome 21 in acute myeloid leukemia are clustered within a limited region of a single gene, AML1. Proc. Natl. Acad. Sci. USA 88:10431-10434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Moore, T., M. Bennett, and V. Kumar. 1995. Transplantable NK cell progenitors in murine bone marrow. J. Immunol. 154:1653-1663. [PubMed] [Google Scholar]
- 46.Moore, T., S. Huang, L. W. Terstappen, M. Bennett, and V. Kumar. 1994. Expression of CD43 on murine and human pluripotent hematopoietic stem cells. J. Immunol. 153:4978-4987. [PubMed] [Google Scholar]
- 47.Morrison, H. L., B. Soni, and J. Lenz. 1995. Long terminal repeat enhancer core sequences in proviruses adjacent to c-myc in T-cell lymphomas induced by a murine retrovirus. J. Virol. 69:446-455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Morse, H. C., III, M. R. Anver, T. N. Fredrickson, D. C. Haines, A. W. Harris, N. L. Harris, E. S. Jaffe, S. C. Kogan, I. C. MacLennan, P. K. Pattengale, and J. M. Ward. 2002. Bethesda proposals for classification of lymphoid neoplasms in mice. Blood 100:246-258. [DOI] [PubMed] [Google Scholar]
- 49.Mucenski, M. L., B. A. Taylor, N. A. Jenkins, and N. G. Copeland. 1986. AKXD recombinant inbred strains: models for studying the molecular genetic basis of murine lymphomas. Mol. Cell. Biol. 6:4236-4243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Nieves, A., L. S. Levy, and J. Lenz. 1997. Importance of a c-Myb binding site for lymphomagenesis by the retrovirus SL3-3. J. Virol. 71:1213-1219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Ogawa, E., M. Inuzuka, M. Maruyama, M. Satake, M. Naito-Fujimoto, Y. Ito, and K. Shigesada. 1993. Molecular cloning and characterization of PEBP2 beta, the heterodimeric partner of a novel Drosophila runt-related DNA binding protein PEBP2 alpha. Virology 194:314-331. [DOI] [PubMed] [Google Scholar]
- 52.Ogawa, E., M. Maruyama, H. Kagoshima, M. Inuzuka, J. Lu, M. Satake, K. Shigesada, and Y. Ito. 1993. PEBP2/PEA2 represents a family of transcription factors homologous to the products of the Drosophila runt gene and the human AML1 gene. Proc. Natl. Acad. Sci. USA 90:6859-6863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Okuda, T., J. van Deursen, S. W. Hiebert, G. Grosveld, and J. R. Downing. 1996. AML1, the target of multiple chromosomal translocations in human leukemia, is essential for normal fetal liver hematopoiesis. Cell 84:321-330. [DOI] [PubMed] [Google Scholar]
- 54.Ott, D. E., J. Keller, K. Sill, and A. Rein. 1992. Phenotypes of murine leukemia virus-induced tumors: influence of 3′ viral coding sequences. J. Virol. 66:6107-6116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Rassart, E., J. Houde, C. Denicourt, M. Ru, C. Barat, E. Edouard, L. Poliquin, and D. Bergeron. 1996. Molecular analysis and characterization of two myeloid leukemia inducing murine retroviruses. Curr. Top. Microbiol. Immunol. 211:201-210. [DOI] [PubMed] [Google Scholar]
- 56.Rolink, A. G., E. ten Boekel, T. Yamagami, R. Ceredig, J. Andersson, and F. Melchers. 1999. B cell development in the mouse from early progenitors to mature B cells. Immunol. Lett. 68:89-93. [DOI] [PubMed] [Google Scholar]
- 57.Ru, M., C. Shustik, and E. Rassart. 1993. Graffi murine leukemia virus: molecular cloning and characterization of the myeloid leukemia-inducing agent. J. Virol. 67:4722-4731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Sanderson, R. D., P. Lalor, and M. Bernfield. 1989. B lymphocytes express and lose syndecan at specific stages of differentiation. Cell Regul. 1:27-35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Schmidt, J., V. Erfle, F. S. Pedersen, H. Rohmer, H. Schetters, K. H. Marquart, and A. Luz. 1984. Oncogenic retrovirus from spontaneous murine osteomas. I. Isolation and biological characterization. J. Gen. Virol. 65:2237-2248. [DOI] [PubMed] [Google Scholar]
- 60.Schmidt, J., A. Luz, and V. Erfle. 1988. Endogenous murine leukemia viruses: frequency of radiation-activation and novel pathogenic effects of viral isolates. Leuk. Res. 12:393-403. [DOI] [PubMed] [Google Scholar]
- 61.Speck, N. A., B. Renjifo, E. Golemis, T. N. Fredrickson, J. W. Hartley, and N. Hopkins. 1990. Mutation of the core or adjacent LVb elements of the Moloney murine leukemia virus enhancer alters disease specificity. Genes Dev. 4:233-242. [DOI] [PubMed] [Google Scholar]
- 62.Speck, N. A., T. Stacy, Q. Wang, T. North, T. L. Gu, J. Miller, M. Binder, and M. Marin-Padilla. 1999. Core-binding factor: a central player in hematopoiesis and leukemia. Cancer Res. 59:1789s-1793s. [PubMed] [Google Scholar]
- 63.Sun, W., M. O'Connell, and N. A. Speck. 1993. Characterization of a protein that binds multiple sequences in mammalian type C retrovirus enhancers. J. Virol. 67:1976-1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Taniuchi, I., M. Osato, T. Egawa, M. J. Sunshine, S. C. Bae, T. Komori, Y. Ito, and D. R. Littman. 2002. Differential requirements for Runx proteins in CD4 repression and epigenetic silencing during T lymphocyte development. Cell 111:621-633. [DOI] [PubMed] [Google Scholar]
- 65.Thornell, A., B. Hallberg, and T. Grundström. 1991. Binding of SL3-3 enhancer factor 1 transcriptional activators to viral and chromosomal enhancer sequences. J. Virol. 65:42-50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Thornell, A., B. Hallberg, and T. Grundström. 1988. Differential protein binding in lymphocytes to a sequence in the enhancer of the mouse retrovirus SL3-3. Mol. Cell. Biol. 8:1625-1637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Vaillant, F., K. Blyth, L. Andrew, J. C. Neil, and E. R. Cameron. 2002. Enforced expression of Runx2 perturbs T cell development at a stage coincident with beta-selection. J. Immunol. 169:2866-2874. [DOI] [PubMed] [Google Scholar]
- 68.Voura, E. B., F. Billia, N. N. Iscove, and R. G. Hawley. 1997. Expression mapping of adhesion receptor genes during differentiation of individual hematopoietic precursors. Exp. Hematol. 25:1172-1179. [PubMed] [Google Scholar]
- 69.Wang, Q., T. Stacy, J. D. Miller, A. F. Lewis, T. L. Gu, X. Huang, J. H. Bushweller, J. C. Bories, F. W. Alt, G. Ryan, P. P. Liu, A. Wynshaw-Boris, M. Binder, M. Marin-Padilla, A. H. Sharpe, and N. A. Speck. 1996. The CBFbeta subunit is essential for CBFalpha2 (AML1) function in vivo. Cell 87:697-708. [DOI] [PubMed] [Google Scholar]
- 70.Wang, S., Q. Wang, B. E. Crute, I. N. Melnikova, S. R. Keller, and N. A. Speck. 1993. Cloning and characterization of subunits of the T-cell receptor and murine leukemia virus enhancer core-binding factor. Mol. Cell. Biol. 13:3324-3339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Wang, S. W., and N. A. Speck. 1992. Purification of core-binding factor, a protein that binds the conserved core site in murine leukemia virus enhancers. Mol. Cell. Biol. 12:89-102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Wells, S. M., A. B. Kantor, and A. M. Stall. 1994. CD43 (S7) expression identifies peripheral B cell subsets. J. Immunol. 153:5503-5515. [PubMed] [Google Scholar]
- 73.Wolff, L. 1997. Contribution of oncogenes and tumor suppressor genes to myeloid leukemia. Biochim. Biophys. Acta 1332:F67-F104. [DOI] [PubMed] [Google Scholar]
- 74.Wolff, L., R. Koller, X. Hu, and M. R. Anver. 2003. A Moloney murine leukemia virus-based retrovirus with 4070A long terminal repeat sequences induces a high incidence of myeloid as well as lymphoid neoplasms. J. Virol. 77:4965-4971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Woolf, E., C. Xiao, O. Fainaru, J. Lotem, D. Rosen, V. Negreanu, Y. Bernstein, D. Goldenberg, O. Brenner, G. Berke, D. Levanon, and Y. Groner. 2003. Runx3 and Runx1 are required for CD8 T cell development during thymopoiesis. Proc. Natl. Acad. Sci. USA 100:7731-7736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Zaiman, A. L., and J. Lenz. 1996. Transcriptional activation of a retrovirus enhancer by CBF (AML1) requires a second factor: evidence for cooperativity with c-Myb. J. Virol. 70:5618-5629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Zaiman, A. L., A. F. Lewis, B. E. Crute, N. A. Speck, and J. Lenz. 1995. Transcriptional activity of core binding factor-alpha (AML1) and beta subunits on murine leukemia virus enhancer cores. J. Virol. 69:2898-2906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Zaiman, A. L., A. Nieves, and J. Lenz. 1998. CBF, Myb, and Ets binding sites are important for activity of the core I element of the murine retrovirus SL3-3 in T lymphocytes. J. Virol. 72:3129-3137. [DOI] [PMC free article] [PubMed] [Google Scholar]