Abstract
Although inhibition of RNA interference (RNAi) by plant virus proteins has been shown to enhance viral replication and pathogenesis in plants, no viral gene product has as yet been shown to inhibit RNAi in vertebrate cells. Here, we present evidence demonstrating that the highly structured ∼160-nucleotide adenoviral VA1 noncoding RNA can inhibit RNAi at physiological levels of expression. VA1, which is expressed at very high levels in adenovirus-infected cells, potently inhibited RNAi induced by short hairpin RNAs (shRNAs) or human microRNA precursors but did not affect RNAi induced by artificial short interfering RNA duplexes. Inhibition appeared to be due both to inhibition of nuclear export of shRNA or premicro-RNA precursors, competition for the Exportin 5 nuclear export factor, and inhibition of Dicer function by direct binding of Dicer. Together, these data argue that adenovirus infection can result in inhibition of RNAi and identify VA1 RNA as the first viral gene product able to inhibit RNAi in human cells.
RNA interference (RNAi) is a mechanism of eukaryotic posttranscriptional gene silencing that relies on ∼21-nucleotide (nt) guide RNAs to confer gene specificity (9, 14, 26, 30). These short noncoding RNAs are incorporated into a large protein complex, the RNA-induced silencing complex (RISC), where they guide the RISC to complementary mRNA targets (13). RISC binding can lead to either mRNA degradation, in the case of a fully complementary mRNA target, or translation inhibition, in the case of mRNA targets that bear a partial mismatch (8, 16, 41).
Two similar but distinct types of guide RNA for RNAi, termed small interfering RNAs (siRNAs) and microRNAs (miRNAs), have been described. siRNAs are derived by the cytoplasmic processing of long double-stranded RNA (dsRNA) molecules into ∼21-bp duplex RNA intermediates by the Dicer enzyme, after which one strand of each duplex is incorporated into the RISC (3, 18). In contrast, miRNAs are encoded within the eukaryotic genome as part of a predicted RNA stem-loop structure, and more than 300 distinct miRNA genes have been identified in the genomes of humans and other eukaryotic species (2). miRNAs are processed from long, largely single-stranded RNA precursors termed pri-miRNAs by the initial excision of an ∼65-nt stem-loop RNA precursor, termed a pre-miRNA, by a nuclear RNA processing enzyme termed Drosha (19, 20, 40). The resultant pre-miRNA is then specifically recognized by a nuclear export factor termed Exportin 5 (Exp5) which transports the pre-miRNA to the cytoplasm (24, 39). There, the pre-miRNA is further processed by Dicer (15) to give an ∼21-bp RNA duplex intermediate, one strand of which is selectively incorporated into the RISC (29). Artificial short hairpin RNAs (shRNAs) (the structures of shRNAs are very similar to the structures of pre-miRNAs) have been widely used as precursors of siRNAs in cultured cells (5, 28). The evidence collected so far indicates that siRNAs and miRNAs program RISCs equivalently and that they can exert similar effects on mRNA expression, despite their somewhat different origins (8, 16, 41).
While miRNAs are expressed in a developmental stage- and/or tissue-specific manner and are believed to play a key role in the regulation of differentiation and development (2), the natural roles of siRNAs remain to be fully established, especially in vertebrate species. However, artificial siRNAs can be used to inhibit the expression of specific genes in vertebrate cells in culture or in vivo (10, 37), and siRNAs derived from dsRNAs generated during viral infection are believed to play a key role in antiviral defense in plant, and possibly also in animal, cells (6, 22).
The hypothesis that RNAi can form an important part of the innate response of plants to viral infection is strongly supported by the observation that many plant viruses encode proteins that inhibit RNAi and thereby greatly enhance viral replication and pathogenesis (6). In contrast, there is little evidence supporting a role for RNAi in antiviral defense in vertebrate cells, although several groups have reported that the replication of a range of human viruses is inhibited upon the introduction of virus-specific siRNAs into normally susceptible cells (11). However, if RNAi is indeed a major feature of the innate response of animal cells to viral infection, then one would predict that at least some animal viruses would also have developed gene products that can inhibit some aspect of the RNAi response. In the case of flock house virus (FHV), an RNA virus that can replicate in both plant and insect cells, the viral B2 protein has been shown to block the ability of both plant and insect cells to mount an FHV-specific RNAi response (21). As a result, the B2 protein is required for viral RNA accumulation in both drosophila and mosquito cells infected with FHV (22). Moreover, there is evidence that both the influenza virus NS1 protein and the E3L protein encoded by vaccinia virus can inhibit RNAi when expressed in drosophila S2 cells (22). However, no gene product encoded by an animal virus has as yet been shown to inhibit RNAi in vertebrate cells.
Adenoviruses are a family of dsDNA viruses that generate large amounts of dsRNA during their replication (25) and that therefore might be predicted to encode an inhibitor of RNAi. One attractive candidate inhibitor of RNAi is the adenovirus VA1 noncoding RNA, an ∼160-nt-long structured RNA that is expressed at extraordinarily high levels (∼108 molecules/cell) during adenoviral replication (27). VA1 is known to play a key role in blocking activation of protein kinase R (PKR) during the adenoviral replication cycle, presumably by viral dsRNAs. In the absence of VA1, activation of PKR induces phosphorylation of the translation factor eIF-2α and, hence, inhibition of viral mRNA translation (36). Therefore, while the major effect of VA1 is clearly to enhance viral mRNA translation, it has also been reported that VA1 can specifically increase the cytoplasmic stability and abundance of ribosome-bound mRNAs (34, 35). As RISCs are known to be closely associated with ribosomes (13), these reports raise the possibility that VA1 might also inhibit the ability of infected cells to mount an adenovirus mRNA-specific RNAi response.
Here, we report that adenovirus VA1 can indeed inhibit RNAi, even at expression levels significantly below those seen in virus-infected cells. VA1 RNA inhibits the biogenesis of both miRNAs and siRNAs by inhibiting nuclear export of the pre-miRNA and shRNA precursors for mature miRNAs and siRNAs and by inhibiting Dicer function by binding Dicer directly. Together, these data identify the first gene product encoded by a pathogenic human virus that is able to inhibit RNAi when expressed in human cells in culture.
MATERIALS AND METHODS
Plasmids and RNAs.
Plasmids pCMV-Luc-miR-30(P), pCMV-miR-30, pCMV-miR-21, pSUPER-miR-30, pSUPER-Luc (39), pSUPER (5), pKmyc and pKmyc-Exp5 (12) have been described. pRL is a Renilla luciferase reporter plasmid (Promega). pBS-VA1 was constructed by insertion of a 330-bp adenovirus type 5 (Ad5) VA1 gene fragment PCR amplified from pAdEasy-1 (Qbiogene) into the unique EcoRI and HindIII sites present in pBluescript KS+ (Stratagene). This VA1 gene fragment includes sequences located 72 bp 5′ of the transcription start site and 96 bp 3′ of the transcription termination site of VA1. Synthetic siRNAs and miRNAs were obtained from Dharmacon. shRNAs, U6 RNA, and VA1 RNA were produced in vitro using a T7 Megashortscript kit (Ambion). The first nucleotide of pre-miR-30 was changed to G in order to achieve efficient transcription by T7 polymerase, and the stem structure was restored by changing the complementary nucleotide to C. 32P-labeled pre-miR-30 RNA transcripts that were subjected to gel shift assay or in vitro Dicer cleavage were generated using a Riboprobe kit (Promega).
Cell culture and transfection.
Human 293T and HeLa cells were maintained as described previously (23) and transfected using either Fugene-6 (Roche) or Lipofectamine 2000 (Invitrogen). Both firefly luciferase and Renilla luciferase activity were assayed ∼48 h after transfection using a dual-luciferase reporter assay system (Promega). shRNAs, synthetic siRNAs, or miRNAs were cotransfected with the indicator constructs ∼20 h after transfection of pBS-VA1 or pBS. In certain experiments, poly(I-C) was added to the medium 2 h posttransfection to a final concentration of 100 μg/ml, and/or 2-aminopurine (2-AP) (Sigma) was added 4 h after transfection to a final concentration of 2.5 mM. Western blotting was performed as previously described (23). Anti-Exp5 rabbit antiserum (a gift of Ian Macara) and anti-Tap rabbit antiserum have been previously described (7, 12).
RNA isolation and analysis.
Total RNA was purified 18 h after infection with wild-type Ad5, 48 h after infection with E1-deficient Ad5, or 48 h after transfection, using Trizol reagent (Invitrogen). Wild-type Ad5 was a gift from Joseph Nevins, and E1-deficient Ad5 was provided by Andrea Amalfitano. Nuclear and cytoplasmic RNAs were isolated as previously described (39) using Trizol reagent (Invitrogen). Northern blotting was performed using ExpressHyb hybridization solution (BD Biosciences) as described previously (41). The probe sequence used for detection of VA1 was 5′-GATACCCTTGCGAATTTATCCACCAGACCACGGAA-3′.
In vitro assays.
For gel shift analysis, briefly, 3 μl of 10× Dicer buffer (Invitrogen), 0.2 μl of RNase inhibitor (Promega), 0.25 μl of yeast tRNA (10 mg/ml), 0.25 μl of Escherichia coli 16S and 23S rRNA (4 μg/μl; Roche), 1 μl of Dicer (Invitrogen), 32P-labeled VA1 RNA probe (∼10,000 cpm/10 ng), and diethyl pyrocarbonate-treated water were added to a final volume of 30 μl. Probe was preheated at 95°C for 3 min and then cooled to 37°C. The binding reaction was performed on ice for 20 to 30 min. The Dicer cleavage assay was performed at 37°C for 5 to 10 min using Dicer buffer (Invitrogen). Each reaction mixture contained ∼25 ng of 32P-labeled pre-miR30 and 0.5 U of Dicer.
RESULTS
Physiological levels of adenovirus VA1 RNA can inhibit miRNA biogenesis and function.
Zeng et al. have previously described (41) the indicator construct pCMV-Luc-miR-30(P), containing the firefly luciferase (Luc) gene (luc), transcribed under the control of a cytomegalovirus (CMV) immediate-early promoter, linked to eight 3′ untranslated region copies of a target sequence perfectly complementary to the human miR-30 miRNA. Cotransfection of pCMV-Luc-miR-30(P) with a miR-30 expression plasmid results in a marked drop in luciferase expression compared to cotransfection with a control vector expressing an irrelevant miRNA. This decrease is specific, as overexpression of miR-30 has no effect on the level of luciferase activity expressed from analogous indicator constructs bearing irrelevant target sites (41). It is interesting that because the introduced target sites are fully complementary to miR-30, miR-30 primarily inhibits luc expression by inducing cleavage of the luc mRNA within the introduced target sequences (41).
As noted above, miRNA biogenesis initiates with transcription of a long pri-miRNA transcript which is then processed by the nuclear enzyme Drosha to give the pre-miRNA intermediate, an RNA stem-loop of ∼65 nt bearing a 2-nt 3′ overhang (20, 40). The pre-miRNA is then exported to the cytoplasm by Exp5, a nuclear export factor that is also known to mediate nuclear export of VA1 RNA (12). Finally, in the cytoplasm, the pre-miRNA is processed by Dicer to give the duplex intermediate, and the miRNA strand is incorporated into the RISC (2).
It has previously been reported that the mature miR-30 miRNA can be overexpressed in human cells by entering this processing pathway at different steps (39, 41). The expression plasmid pCMV-miR-30 expresses miR-30 as part of a pri-miRNA precursor; therefore, miR-30 overexpression requires all the steps listed above for endogenous miRNAs. In contrast, the pSUPER-miR-30 expression plasmid expresses a 63-nt RNA, transcribed under the control of RNA polymerase III (Pol III), that is predicted to be identical to pre-miR-30 except that the 2-nt 3′ overhang is changed from 5′-GC-3′ to 5′-UU-3′ to accommodate the sequence requirements for transcription termination by Pol III (5). This miR-30 precursor still requires Exp5-dependent nuclear export and processing by Dicer (28, 39) but should no longer be dependent on Drosha function for miR-30 overexpression. Finally, we can also directly transfect cells with synthetic RNA molecules whose sequences are identical to those of the predicted pre-miR-30 or miR-30 duplex intermediates (39). Both pre-miR-30 and the miR-30 duplex, which is analogous in structure to an siRNA, are predicted to enter the cytoplasm directly, without any need for nuclear export by Exp5. In addition, at least the pre-miR-30 RNA precursor would still require processing by cytoplasmic Dicer to yield the miR-30 duplex intermediate.
To determine whether VA1 expression would alleviate inhibition mediated by miR-30, first we established conditions that permitted all four methods for miR-30 overexpression to inhibit luc expression in the linear range (data not shown). Moreover, because VA1 can enhance gene expression from transfected plasmids nonspecifically (17), presumably by inhibiting activation of PKR by low levels of dsRNAs that arise due to the presence of cryptic promoters, all experiments were internally controlled by cotransfection of a Renilla luciferase expression plasmid.
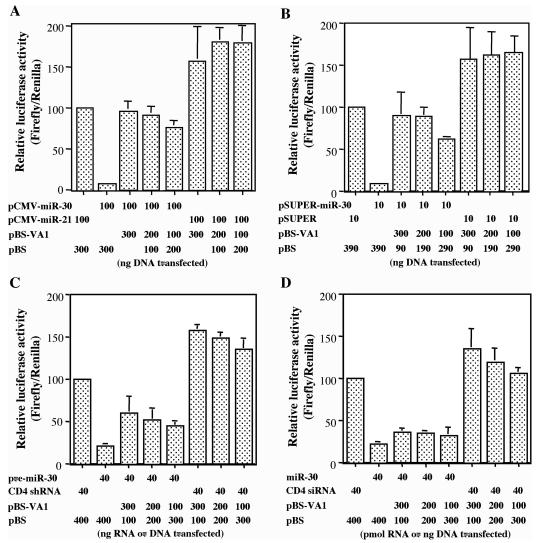
As shown in Fig. 1A and as previously reported (41), cotransfection of 293T cells with the pCMV-Luc-miR-30(P) indicator plasmid and pCMV-miR-30 results in profound inhibition of luc expression. However, cotransfection of a VA1 expression plasmid restored luc expression essentially back to the level seen in the absence of the pCMV-miR-30 effector plasmid. The VA1 expression plasmid used, pBS-VA1, consists of the VA1 gene, together with 168 bp of flanking sequences, inserted into the pBS vector backbone and therefore relies entirely on the cognate internal Pol III promoter and terminator for expression of VA1. Surprisingly, given that these data are given relative to a cotransfected Renilla luciferase internal control, cotransfection of the pBS-VA1 expression plasmid enhanced firefly luciferase expression by 1.5- to 2-fold relative to the internal control, even in the absence of the cotransfected pCMV-miR-30 expression vector. In Table 1, we corrected for this presumably nonspecific effect, and the data in Table 1 demonstrate that VA1 expression enhances firefly luciferase expression by 7- to 10-fold, depending on the level of VA1 expression analyzed, even when this nonspecific effect is taken into account.
FIG. 1.
Adenovirus VA1 RNA can rescue inhibition of gene expression caused by a pri-miRNA expression plasmid, a pre-miRNA expression plasmid, or a short hairpin pre-miRNA, but not by a synthetic miRNA duplex (A) 293T cells were cotransfected with pCMV-Luc-miR-30(P), the pRL internal control plasmid, and plasmids expressing pri-miR-30 (pCMV-miR-30) and/or VA1 (pBS-VA1). pCMV-miR21 and pBS served as negative controls. Data are shown normalized to the Renilla luciferase internal control and relative to 293T cells transfected with pCMV-Luc-miR-30(P) together with the pCMV-miR-21 control plasmid, which was set at 100. The experiments in panels B to D were set up like the experiment in panel A with the following differences. (B) pSUPER-miR-30, designed to directly express a pre-miR-30 RNA, was used. pSUPER served as the negative control. (C) In vitro-transcribed pre-miR-30 shRNA was transfected. A CD4-specific shRNA served as a negative control. (D) Synthetic miR-30 duplex RNA was used. A CD4-specific siRNA duplex served as a negative control. All data are the averages ± standard deviations (error bars) from three independent experiments.
TABLE 1.
Effect of VA1 on the level of firefly luciferase activity observed in cells transfected with pCMV-Luc-miR-30(P) and the indicated miR-30 expression vehicle
| Amt of pBS-VA1 (ng) | Fold increase in firefly luciferase activitya in cells transfected with:
|
|||
|---|---|---|---|---|
| pCMV-miR-30 | pSUPER-miR-30 | Synthetic pre-miR-30 | Synthetic miR-30 duplex | |
| 300 | 10.2 ± 3.5 | 6.2 ± 1.1 | 1.8 ± 0.1 | 1.2 ± 0.3 |
| 200 | 8.0 ± 1.1 | 6.0 ± 0.4 | 1.8 ± 0.4 | 1.3 ± 0.5 |
| 100 | 6.6 ± 1.1 | 4.1 ± 0.5 | 1.7 ± 0.3 | 1.4 ± 0.5 |
These data are derived from Fig. 1 and show the average fold increase in firefly luciferase expression relative to the Renilla luciferase internal control observed for the indicated level of pBS-VA1 used and corrected for the nonspecific increase seen in the absence of miR-30 expression. Values are averages ± standard deviations from three experiments.
The miR-30 expression vector utilized in Fig. 1A generates a Pol II transcript analogous to a pri-miRNA. In contrast, the pSUPER-miR-30 plasmid utilized in Fig. 1B generates a Pol III transcript closely similar to the pre-miR-30 RNA. Nevertheless, the effect of VA1 expression is very similar to that seen in Fig. 1A, i.e., VA1 blocks the inhibition of firefly luciferase expression caused by the encoded miR-30 miRNA (Fig. 1B and Table 1). In Fig. 1C, miR-30 was expressed by direct transfection of a synthetic pre-miR-30 RNA stem-loop. Again, we observed a marked and specific inhibition of firefly luciferase expression. However, in this case, the rescue caused by coexpression of VA1 was only partial, i.e., ∼2-fold instead of up to 10-fold (Table 1). Therefore, these data suggest that one target for VA1 is likely to be the Exp5-mediated nuclear export of the pre-miR-30 precursor encoded by both pCMV-miR-30 and pSUPER-miR-30.
Finally, direct transfection of cells with the predicted duplex intermediate for miR-30 (Fig. 1D) resulted in efficient inhibition of firefly luciferase expression from pCMV-Luc-miR-30(P), but in this case VA1 had no significant specific enhancing effect (Table 1). Therefore, these data suggest that Dicer function might also be a target for VA1, as the miR-30 duplex RNA is the only miR-30 precursor that does not require cytoplasmic processing by Dicer. In conclusion, these data show that miR-30 function is inhibited by expression of the adenovirus VA1 RNA. However, this inhibition is attenuated if miR-30 expression is independent of nuclear export, and this inhibition is lost entirely if the miR-30 precursor is fully processed. Therefore, these data suggest that VA1 may inhibit Exp5 and/or Dicer function but does not affect RISC function per se.
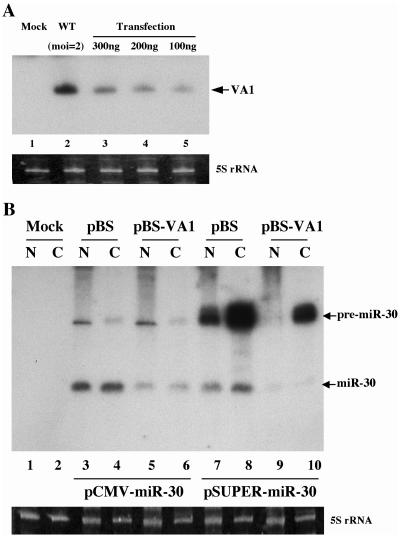
As noted above, VA1 RNA is expressed at an extraordinarily high level, ∼108 copies, in adenovirus-infected cells (27). Nevertheless, we wished to confirm that the level of VA1 present in cells transfected with the pBS-VA1 expression plasmid was equivalent to or lower than the level in infected cells. In Fig. 2A, we present the results of a Northern blot analysis comparing the level of VA1 RNA detected in 293T cells infected with wild-type adenovirus at a multiplicity of infection (MOI) of 2 with 293T cells transfected with the levels of pBS-VA1 used in Fig. 1. Even taking into account the fact that the transfection efficiency is less than 100% (we estimate ∼70% on the basis of transfections performed in parallel using a green fluorescent protein expression plasmid), these data nevertheless clearly reveal that the levels of VA1 expressed in pBS-VA1-transfected cells are lower than those in adenovirus-infected cells.
FIG. 2.
Northern blot analysis of VA1 expression in transfected or infected 293T cells and its effect on miR-30 production. (A) The level of VA1 expression in 293T cells infected with wild-type (WT) adenovirus at an MOI of 2 or mock infected or transfected with the indicated level of pBS-VA1 was determined by Northern blot analysis. The transfection efficiency was ∼70%. (B) Effect of VA1 on mature and pre-miR-30 RNA expression in 293T cells. 293T cells were cotransfected with pCMV-miR-30 or pSUPER-miR-30, together with pBS-VA1 or the pBS control plasmid, and the pre-miR-30 and mature miR-30 expression levels were determined by Northern blot analysis of nuclear (N) or cytoplasmic (C) RNA fractions. 5S rRNA was used as a loading control.
An important question is whether VA1 expression inhibits the production of mature miR-30 from the miR-30 expression plasmids pCMV-miR-30 and pSUPER-miR-30. Yi et al. demonstrated that inhibition of Exp5 expression by RNAi results in a drop in the cytoplasmic expression of both pre-miR-30 and miR-30 in cells transfected with pCMV-miR-30 but does not lead to nuclear accumulation of pre-miR-30 (39). This may reflect nuclear destabilization of the pre-miR-30 intermediate in the absence of Exp5. RNA analysis shows that VA1 expression also results in a marked decline in the accumulation of mature miR-30 not only in the cytoplasm but also in the nuclear fraction (Fig. 2B, lanes 3 to 6). The apparently nuclear miR-30 may be derived from RISCs associated with ribosomes bound to the outer membrane of the nucleus or might represent authentically nuclear miR-30. In contrast, VA1 did not affect the nuclear level of pre-miR-30 (Fig. 2B, compare lanes 3 and 5) and also had relatively little effect on cytoplasmic pre-miR-30 levels (compare lanes 4 and 6).
Analysis of cells transfected with pSUPER-miR-30 (Fig. 2B) showed very high levels of the primary transcript, whose sequence is predicted to be identical to that of pre-miR-30 except that it contains a U-rich 3′ overhang. In contrast, relatively modest levels of mature miR-30 were observed. The reason for the apparent inefficiency of processing of the pre-miRNA-like transcript encoded by pSUPER-miR-30 (Fig. 2B, lanes 7 and 8) compared to the analogous transcript encoded by pCMV-miR-30 (Fig. 2B, lanes 3 and 4), are unclear. However, it is possible that the cellular La protein, which is known to associate with the 3′ U tail present on Pol III transcripts (32, 38), interferes with the cytoplasmic processing of this shRNA. In any event, coexpression of VA1 clearly inhibited the production of mature miR-30 in cells expressing pSUPER-miR-30 (Fig. 2B, compare lanes 7 and 8 with lanes 9 and 10). Moreover, and unlike the situation in pCMV-miR-30-transfected cells, VA1 also appeared to reduce the steady-state level of the primary transcript from pSUPER-miR-30. However, the overall conclusion of this RNA analysis is that VA1 coexpression can inhibit the production of mature miR-30.
VA1 inhibition of miRNA function is not due to inhibition of PKR.
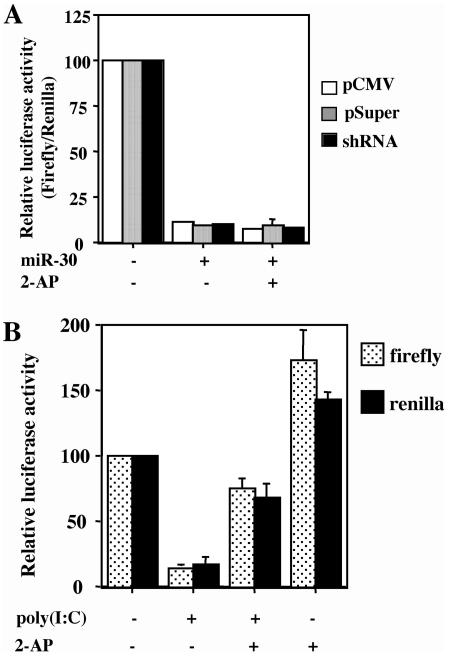
Although it has previously been documented that inhibition of luc expression from the pCMV-Luc-miR-30(P) expression plasmid by overexpressed miR-30 is specific both in terms of an absolute requirement for introduced miR-30 target sites and with reference to the Renilla luciferase internal control (39, 41), it has also been shown that some siRNAs can activate the interferon response and, possibly, the PKR enzyme (4, 33). To demonstrate that the inhibition of miR-30 function by VA1 is not due to the ability of VA1 to block PKR activation, we asked whether 2-AP, a potent inhibitor of PKR (17), would also inhibit miR-30 function. In fact, 2-AP had no specific effect on the expression of firefly luciferase in cells cotransfected with pCMV-Luc-miR-30(P) and either pCMV-miR-30 or pSUPER-miR-30 (Fig. 3A). However, 2-AP did have a nonspecific positive effect on the expression of both firefly luciferase and the Renilla luciferase internal control (Fig. 3B), consistent with earlier reports documenting a nonspecific enhancing effect of both 2-AP and VA1 on gene expression from transfected plasmids (17). In addition, 2-AP also largely rescued firefly and Renilla luciferase expression that had been inhibited by cotransfection of cells with the potent interferon inducer poly(I-C), as would be expected for an inhibitor of PKR (Fig. 3B). Therefore, we conclude that the effect of VA1 on miRNA function is independent of its ability to act as an inhibitor of PKR.
FIG. 3.
Effect of 2-AP on miR-30 function. (A) 2-AP does not rescue the inhibition of gene expression caused by pCMV-miR-30, pSUPER-miR-30, or the pre-miR-30 shRNA. This transfection experiment was performed as described in the legend to Fig. 1, except that 293T cells were treated with 2-AP (+) 4 h after certain transfections. (B) 2-AP relieves the nonspecific inhibition of gene expression caused by poly(I-C). The experiment in panel B is set up like the experiment in panel A, except that 293T cells were treated with poly(I-C) (+) 2 h after transfection. These data were not normalized to the Renilla luciferase internal control.
VA1 can inhibit siRNA function.
The data presented in Fig. 1 are from a single cell line, human 293T cells, and the function of a single miRNA, miR-30, is analyzed. While miRNAs and siRNAs have been shown to be functionally equivalent in human cells (8, 41), we wished to extend this analysis to a second human cell line and to a clearly artificial siRNA. The experiment shown in Fig. 4A was performed in HeLa cells and examined the effect of VA1 expression on RNAi directed against the firefly luciferase open reading frame using an siRNA encoded by the expression plasmid pSUPER-Luc (39) (Fig. 4A). As may be readily observed, the specific inhibition of firefly luciferase expression observed upon cotransfection of pSUPER-Luc was largely reversed by coexpression of VA1. Although VA1 again had an approximately twofold nonspecific enhancing effect in this assay, as seen in HeLa cells transfected with pCMV-Luc-miR-30(P) in the absence of pSUPER-Luc, the approximately eightfold enhancing effect observed in cells cotransfected with pSUPER-Luc is clearly more profound. Therefore, we conclude that the ability of VA1 to inhibit RNAi is not restricted to a particular cell line or siRNA.
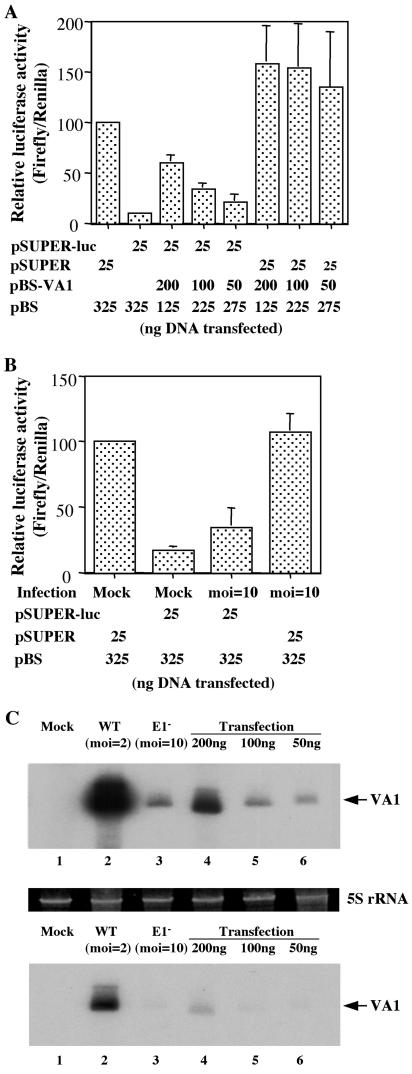
FIG. 4.
Effect of VA1 expression or adenovirus infection on RNAi. The experiment in panel A was set up like the experiment in Fig. 1B, except that this experiment was performed in HeLa cells using a pSUPER-based plasmid encoding a firefly luciferase-specific shRNA. The experiment in panel B was set up like the experiment in panel A, except that the HeLa cells were transfected immediately after infection with 10 infectious units of an E1A− E1B− adenovirus or mock infection. (C) Northern blot analysis of the level of VA1 expression detected in mock-infected HeLa cells (lane 1), HeLa cells infected with wild-type (WT) adenovirus at a MOI of 2 (lane 2) or an E1A− E1B− adenovirus mutant at a MOI of 10 (lane 3), or HeLa cells transfected with the indicated level of pBS-VA1 (lanes 4 to 6). The transfection efficiency in HeLa cells was ∼50%, so that the level of VA1 per transfected cell is approximately twofold higher than the levels indicated in lanes 4 to 6. The top and bottom gels are two different exposures of the same Northern blot analysis. 5S rRNA was used as a loading control.
An interesting question is whether inhibition of RNAi can be observed during adenovirus infection. Attempts to address this issue using wild-type adenovirus or a variety of adenovirus mutants were unsuccessful due to the rapid and severe cytopathic effect induced by adenovirus infection, which kills permissive cells ≤24 h after infection, while our RNAi assay requires 40 h for a readout. However, we have been able to test an adenovirus virus mutant with the early regulatory proteins E1A and E1B deleted (E1A− E1B− adenovirus). Expression of E1A and E1B is required for adenovirus to proceed into the late phase of the virus replication cycle, including amplification of the viral DNA genome; therefore, E1A− E1B− adenoviruses are effectively replication incompetent. Nevertheless, they can be grown to high titers in the 293 cell line, which constitutively expresses E1A and E1B.
As shown in Fig. 4B, infection of HeLa cells with an MOI of 10 of the E1A− E1B− adenovirus exerted a modest, approximately twofold enhancing effect on luc activity in the presence of pSUPER-Luc but had no nonspecific effect on the level of luc activity in the absence of pSUPER-Luc. The Renilla luciferase internal control was unaffected. Next, we asked whether the E1A− E1B− adenovirus mutant indeed expressed detectable levels of VA1 in infected HeLa cells. As shown in Fig. 4C, VA1 expression was detected but at levels that were much lower than seen in HeLa cells infected with wild-type adenovirus. Comparison of the level of VA1 RNA seen in HeLa cells infected with the E1A− E1B− adenovirus with the levels seen in pBS-VA1-transfected HeLa cells suggested that this low level of VA1 RNA expression explains the modest phenotype observed in Fig. 4B, as HeLa cells expressing a comparable level of VA1 after transfection (Fig. 4C) also exhibited only limited rescue of luciferase expression (Fig. 4A). Efforts to increase the level of VA1 expression by increasing the MOI of the E1A− E1B− adenovirus used resulted in cytopathic effects that rendered the experiment difficult to interpret. Nevertheless, these data are at least consistent with the hypothesis that VA1 RNA expressed during adenovirus infection can inhibit RNAi.
VA1 inhibition of RNAi is at least partly due to competition for Exp5.
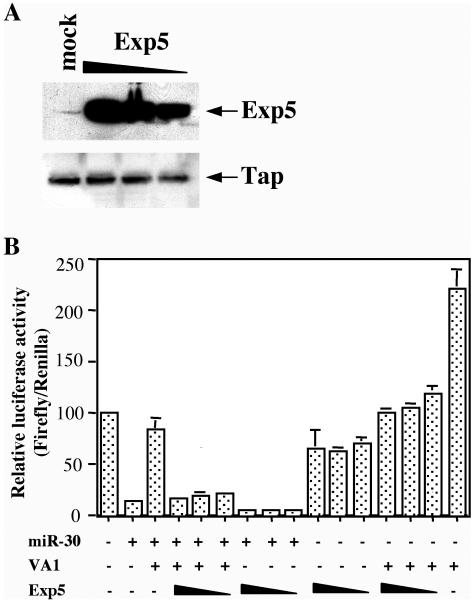
The data presented in Fig. 1 suggest that VA1 likely blocks miRNA function by inhibiting the nuclear export of miRNA and siRNA precursors and/or by inhibiting Dicer function. It has been previously demonstrated that VA1, pre-miRNAs, and artificial shRNAs all rely on Exp5 for their nuclear export (12, 24, 39). Therefore, given the extremely high level of expression of VA1 in adenovirus-infected cells, it is possible that VA1, which is known to bind Exp5 specifically (12), simply outcompetes pre-miRNAs and shRNAs for a limited pool of Exp5. Using a rabbit polyclonal antiserum specific for Exp5, we in fact observed very low levels of expression of Exp5 not only in 293T cells (Fig. 5A) but also in a variety of other cell lines (data not shown). However, transfection of 293T cells with an Exp5 expression plasmid resulted in a dramatic increase in Exp5 levels (Fig. 5A).
FIG. 5.
Overexpression of Exp5 partially relieves the inhibition of pre-miRNA function induced by VA1. (A) Western blot analysis of the level of Exp5 expression in transfected 293T cells. 293T cells were either mock transfected or transfected with 200, 150, or 100 ng of pKmyc-Exp5 (indicated by the thickness of the triangle above the gel). Exp5 and Tap, which serve as loading controls, were detected using rabbit polyclonal antisera. (B) This transfection experiment was performed as described in the legend to Fig. 1B, except that 200, 150, or 100 ng of pKmyc-Exp5 was included in certain transfections (indicated by the thickness of the triangle below the graph).
Next, we asked whether overexpression of Exp5 would rescue the ability of miR-30 expressed from pCMV-miR-30 to inhibit firefly luciferase expression from the pCMV-Luc-miR-30(P) indicator construct. In fact, as shown in Fig. 5B, Exp5 overexpression largely rescued this inhibition. Although these experiments were internally controlled by a Renilla luciferase expression plasmid, we also observed an approximately twofold reduction in firefly luciferase expression in the presence of miR-30 but absence of VA1 (Fig. 5B, compare column 2 with columns 7 to 9 [the leftmost column being column 1]) and in the presence of VA1 but absence of miR-30 (compare columns 13 to 15 with column 16 [the rightmost column]). This may suggest that some component of the inhibitory effect of Exp5 overexpression on firefly luciferase expression is nonspecific and that the rescue of miR-30 function is therefore not complete. Nevertheless, these data do clearly suggest that a major effect of VA1 is to competitively inhibit pre-miRNA and shRNA nuclear export and, hence, function.
VA1 RNA binds Dicer and inhibits Dicer function in vitro.
While inhibition of pre-miRNA and shRNA nuclear export clearly accounts for at least part of the ability of VA1 to inhibit miRNA and siRNA function, as documented in Fig. 1 and Fig. 5, the data presented in Fig. 1C suggest that VA1 may also act at the level of Dicer processing. One possibility is that VA1 acts as an RNA decoy that binds Dicer specifically and sequesters it away from its normal pre-miRNA and dsRNA substrates.
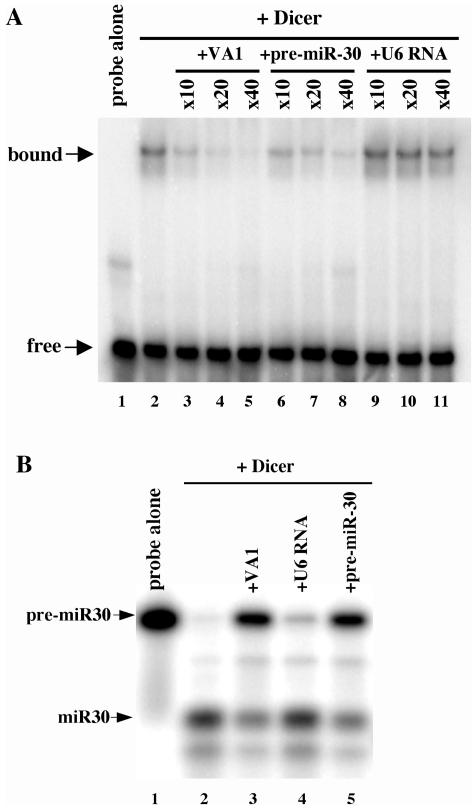
To test this hypothesis, we performed an RNA gel shift analysis with a 32P-labeled VA1 RNA probe to ask whether VA1 would be able to bind recombinant human Dicer specifically. As shown in Fig. 6A, we did indeed observe a shift in probe mobility in the presence of Dicer (lane 2) that was specifically competed by unlabeled VA1 RNA (lanes 3 to 5) but not by an irrelevant RNA competitor, the U6 small nuclear RNA (lanes 9 to 11). Importantly, VA1 binding by Dicer was also competed by pre-miR-30 RNA (lanes 6 to 8), thus suggesting that VA1 and pre-miRNAs may compete for specific binding by Dicer in vivo.
FIG. 6.
VA1 binds to Dicer and inhibits Dicer function in vitro. (A) Specific binding of VA1 by Dicer. A 32P-labeled VA1 RNA probe was incubated with recombinant Dicer in the presence (+) or absence of the indicated fold excess of various unlabeled competitor RNAs. Protein-RNA complexes were detected by nondenaturing gel electrophoresis and autoradiography. (B) Specific inhibition of Dicer cleavage activity by VA1. A 32P-labeled pre-miR-30 probe was incubated with recombinant Dicer in the presence or absence of an ∼40-fold excess of various unlabeled competitor RNAs. Pre-miR-30 and mature miR-30 were detected by 15% denaturing gel electrophoresis and autoradiography.
Next, we asked whether VA1 would interfere with in vitro Dicer processing of the pre-miR-30 intermediate. As shown in Fig. 6B, we observed efficient processing of a 32P-labeled pre-miR-30 RNA by Dicer to give products that migrate to the positions predicted for the strands of the mature miR-30 duplex. While the addition of excess U6 RNA to this in vitro reaction mixture had little or no inhibitory effect, an excess of either unlabeled VA1 RNA or pre-miR-30 RNA largely blocked processing of the 32P-labeled pre-miR-30 probe. Therefore, we conclude that VA1 has the ability to bind Dicer specificity and sequester it away from its normal physiological substrates when present in excess.
DISCUSSION
Given the known importance of RNAi as an innate antiviral defense mechanism in plants (6) and given that vertebrate cells express Dicer and are fully competent to perform RNAi (10, 28), it is reasonable to hypothesize that RNAi might also form part of the defense of vertebrate cells against viruses that produce dsRNAs during their life cycle. Consistent with this proposal, gene products encoded by influenza virus and vaccinia virus have been shown to inhibit RNAi in insect cells (22). However, no gene product derived from any human virus has as yet been shown to inhibit RNAi in human cells.
In considering which human viral gene product might function as an inhibitor of RNAi, we identified the adenovirus VA1 noncoding RNA as a possible candidate. Adenoviruses are known to produce significant levels of dsRNA during their replication cycle (25), thus potentially necessitating some form of inhibitor of RNAi. Moreover, the structure of VA1 RNA is quite similar to the structures of pre-miRNAs, endogenously encoded ∼65-nt precursors of miRNAs, and shRNAs, artificial ∼50-nt precursors of siRNAs. Yet, at ∼160 nt in length, it is both larger and has a more complex structure (27) than these short miRNA or siRNA stem-loop RNA precursors. Therefore, we hypothesized that VA1, which is expressed at extraordinarily high levels in adenovirus-infected cells (27), might function as some form of RNA decoy.
In this study, we demonstrate that VA1 does indeed potently inhibit RNAi induced by endogenously transcribed pre-miRNAs and shRNAs and weakly inhibits RNAi induced by in vitro-transcribed, transfected shRNAs. However, VA1 does not affect RNAi induced by artificial siRNA-miRNA duplexes (Fig. 1 and 4). Further analysis demonstrated that VA1 expression inhibited the production of mature miRNAs from pre-miRNA precursors (Fig. 2B). Finally, analysis of the step(s) in miRNA or siRNA biogenesis inhibited by VA1 RNA showed that not only did VA1 act as a potent inhibitor of pre-miRNA or shRNA nuclear export by competing for a limited pool of the cellular Exp5 nuclear export factor (Fig. 5) but it also bound Dicer specifically in vitro (Fig. 6A) and inhibited Dicer processing of a pre-miRNA when present in excess (Fig. 6B). Together these data identify adenovirus VA1 as the first viral gene product able to specifically block RNAi in human cells.
An important question is whether VA1 actually inhibits RNAi during the viral life cycle and whether this has any beneficial effect on virus replication. In this regard, it is important to note that the levels of VA1 that were found to effectively block RNAi in transfected cultures are significantly below the level of VA1 expressed in adenovirus-infected cells (Fig. 2A and 4C). Therefore, this inhibition is certainly predicted to occur during the adenovirus life cycle. However, we have found it difficult to demonstrate that inhibition of RNAi by adenovirus actually does occur during infection because the rapid cytopathic effect induced by wild-type and most mutant adenoviruses interferes with our assay for RNAi. Nevertheless, using a replication-incompetent adenovirus mutant that lacks both E1A and E1B function, we were able to detect a modest inhibition of RNAi in infected cells (Fig. 4B). While this weak effect likely results from the fact that this mutant produces only very low levels of VA1 (Fig. 4C), this result is at least consistent with the hypothesis that VA1 produced in adenovirus-infected cells can attenuate any RNAi response to viral infection.
While this hypothesis could at least in principle be further tested by direct analysis of VA1-deficient adenovirus mutants, this experiment is complicated by the well-established role of VA1 in inhibiting activation of PKR by the dsRNAs produced during adenoviral infection. While analysis of VA1-deficient adenovirus has previously identified an important role for VA1 in enhancing viral mRNA translation by blocking phosphorylation of eIF-2α by PKR (36), others have reported that VA1 can also induce an increase in the level of expression of specific target mRNA species (34, 35), which is at least consistent with inhibition of an RNAi response by VA1.
A final point of interest is that adenoviruses have been used by several groups as vectors for the delivery of shRNA expression cassettes that induce RNAi (1, 31). These adenovirus vectors are invariably defective, minimally lacking E1A and E1B, and the vestigial level of VA1 that is produced by these vectors (Fig. 4C) undoubtedly has only a modest inhibitory effect on the level of RNAi induced. Nevertheless, it seems possible that deletion of the VA1 gene from these adenovirus vectors might further enhance their effectiveness as mediators of RNAi.
Acknowledgments
We thank Ian Macara, Joseph Nevins, and Andrea Amalfitano for reagents used in this research.
REFERENCES
- 1.Arts, G.-J., E. Langemeijer, R. Tissingh, L. Ma, H. Pavliska, K. Dokic, R. Dooijes, E. Mešić, R. Clasen, F. Michiels, J. van der Schueren, M. Lambrecht, S. Herman, R. Brys, K. Thys, M. Hoffmann, P. Tomme, and H. van Es. 2003. Adenoviral vectors expressing siRNAs for discovery and validation of gene function. Genome Res. 13:2325-2332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bartel, D. P. 2004. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell 116:281-297. [DOI] [PubMed] [Google Scholar]
- 3.Bernstein, E., A. A. Caudy, S. M. Hammond, and G. J. Hannon. 2001. Role for a bidentate ribonuclease in the initiation step of RNA interference. Nature 409:295-296. [DOI] [PubMed] [Google Scholar]
- 4.Bridge, A. J., S. Pebernard, A. Ducraux, A.-L. Nicoulaz, and R. Iggo. 2003. Induction of an interferon response by RNAi vectors in mammalian cells. Nat. Genet. 34:263-264. [DOI] [PubMed] [Google Scholar]
- 5.Brummelkamp, T., R. Bernards, and R. Agami. 2002. A system for stable expression of short interfering RNAs in mammalian cells. Science 296:550-553. [DOI] [PubMed] [Google Scholar]
- 6.Carrington, J. C., K. D. Kasschau, and L. K. Johansen. 2001. Activation and suppression of RNA silencing by plant viruses. Virology 281:1-5. [DOI] [PubMed] [Google Scholar]
- 7.Coburn, G. A., H. L. Wiegand, Y. Kang, D. N. Ho, M. M. Georgiadis, and B. R. Cullen. 2001. Using viral species specificity to define a critical protein/RNA interaction surface. Genes Dev. 15:1194-1205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Doench, J. G., C. P. Petersen, and P. A. Sharp. 2003. siRNAs can function as miRNAs. Genes Dev. 17:438-442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dykxhoorn, D. M., C. D. Novina, and P. A. Sharp. 2003. Killing the messenger: short RNAs that silence gene expression. Nature Rev. 4:457-467. [DOI] [PubMed] [Google Scholar]
- 10.Elbashir, S. M., J. Harborth, W. Lendeckel, A. Yalcin, K. Weber, and T. Tuschl. 2001. Duplexes of 21-nucleotide RNAs mediate RNA interference in cultured mammalian cells. Nature 411:494-498. [DOI] [PubMed] [Google Scholar]
- 11.Gitlin, L., and R. Andino. 2003. Nucleic acid-based immune system: the antiviral potential of mammalian RNA silencing. J. Virol. 77:7159-7165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gwizdek, C., B. Ossarch-Nazari, A. M. Brownawell, A. Doglio, E. Bertrand, I. G. Macara, and C. Dargemont. 2003. Exportin-5 mediates nuclear export of minihelix-containing RNAs. J. Biol. Chem. 278:5505-5508. [DOI] [PubMed] [Google Scholar]
- 13.Hammond, S. M., E. Bernstein, D. Beach, and G. J. Hannon. 2000. An RNA-directed nuclease mediates post-transcriptional gene silencing in Drosophila cells. Nature 404:293-295. [DOI] [PubMed] [Google Scholar]
- 14.Hannon, G. J. 2002. RNA interference. Nature 418:244-251. [DOI] [PubMed] [Google Scholar]
- 15.Hutvágner, G., J. McLachlan, A. E. Pasquinelli, É. Bálint, T. Tuschl, and P. D. Zamore. 2001. A cellular function for the RNA-interference enzyme dicer in the maturation of the let-7 small temporal RNA. Science 293:834-838. [DOI] [PubMed] [Google Scholar]
- 16.Hutvágner, G., and P. D. Zamore. 2002. A microRNA in a multiple-turnover RNAi enzyme complex. Science 297:2056-2060. [DOI] [PubMed] [Google Scholar]
- 17.Kaufman, R. J., and P. Murtha. 1987. Translational control mediated by eucaryotic initiation factor-2 is restricted to specific mRNAs in transfected cells. Mol. Cell. Biol. 7:1568-1571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ketting, R. F., S. E. Fischer, E. Bernstein, T. Sijen, G. J. Hannon, and R. H. Plasterk. 2001. Dicer functions in RNA interference and in synthesis of small RNA involved in developmental timing in C. elegans. Genes Dev. 15:2654-2659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lee, Y., C. Ahn, J. Han, H. Choi, J. Kim, J. Yim, J. Lee, P. Provost, O. Rådmark, S. Kim, and V. N. Kim. 2003. The nuclear RNase III Drosha initiates microRNA processing. Nature 425:415-419. [DOI] [PubMed] [Google Scholar]
- 20.Lee, Y., K. Jeon, J.-T. Lee, S. Kim, and V. N. Kim. 2002. MicroRNA maturation: stepwise processing and subcellular localization. EMBO J. 21:4663-4670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Li, H., W. X. Li, and S. W. Ding. 2002. Induction and suppression of RNA silencing by an animal virus. Science 296:1319-1321. [DOI] [PubMed] [Google Scholar]
- 22.Li, W.-X., H. Li, R. Lu, F. Li, M. Dus, P. Atkinson, E. W. A. Brydon, K. L. Johnson, A. Garcia-Sastre, L. A. Ball, P. Palese, and S.-W. Ding. 2004. Interferon antagonist proteins of influenza and vaccinia viruses are suppressors of RNA silencing. Proc. Natl. Acad. Sci. USA 101:1350-1355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lu, S., and B. R. Cullen. 2003. Analysis of the stimulatory effect of splicing on mRNA production and utilization in mammalian cells. RNA 9:618-630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lund, E., S. Güttinger, A. Calado, J. E. Dahlberg, and U. Kutay. 2004. Nuclear export of microRNA precursors. Science 303:95-98. [DOI] [PubMed] [Google Scholar]
- 25.Maran, A., and M. B. Mathews. 1988. Characterization of the double-stranded RNA implicated in the inhibition of protein synthesis in cells infected with a mutant adenovirus defective for VA RNA1. Virology 164:106-113. [DOI] [PubMed] [Google Scholar]
- 26.Martinez, J., A. Patkaniowska, H. Urlaub, R. Lührmann, and T. Tuschl. 2002. Single-stranded antisense siRNAs guide target RNA cleavage in RNAi. Cell 110:563-574. [DOI] [PubMed] [Google Scholar]
- 27.Mathews, M. B., and T. Shenk. 1991. Adenovirus virus-associated RNA and translation control. J. Virol. 65:5657-5662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Paddison, P. J., A. A. Caudy, E. Bernstein, G. J. Hannon, and D. S. Conklin. 2002. Short hairpin RNAs (shRNAs) induce sequence-specific silencing in mammalian cells. Genes Dev. 16:948-958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Schwarz, D. S., G. Hutvágner, T. Du, Z. Xu, N. Aronin, and P. D. Zamore. 2003. Asymmetry in the assembly of the RNAi enzyme complex. Cell 115:199-208. [DOI] [PubMed] [Google Scholar]
- 30.Schwarz, D. S., G. Hutvágner, B. Haley, and P. D. Zamore. 2002. Evidence that siRNAs function as guides, not primers, in the Drosophila and human RNAi pathways. Mol. Cell 10:537-548. [DOI] [PubMed] [Google Scholar]
- 31.Shen, C., A. K. Buck, X. Liu, M. Winkler, and S. N. Reske. 2003. Gene silencing by adenovirus-delivered siRNA. FEBS Lett. 539:111-114. [DOI] [PubMed] [Google Scholar]
- 32.Simons, F. H., S. A. Rutjes, W. J. van Venrooij, and G. J. Pruijn. 1996. The interactions with Ro60 and La differentially affect nuclear export of hY1 RNA. RNA 2:264-273. [PMC free article] [PubMed] [Google Scholar]
- 33.Sledz, C. A., M. Holko, M. J. De Veer, R. H. Silverman, and B. R. Williams. 2003. Activation of the interferon system by short-interfering RNAs. Nat. Cell Biol. 5:834-839. [DOI] [PubMed] [Google Scholar]
- 34.Strijker, R., D. T. Fritz, and A. D. Levinson. 1989. Adenovirus VAI-RNA regulates gene expression by controlling stability of ribosome-bound RNAs. EMBO J. 8:2669-2675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Svensson, C., and G. Akusjarvi. 1990. A novel effect of adenovirus VA RNA1 on cytoplasmic mRNA abundance. Virology 174:613-617. [DOI] [PubMed] [Google Scholar]
- 36.Thimmappaya, B., C. Weinberger, R. J. Schneider, and T. Shenk. 1982. Adenovirus VAI RNA is required for efficient translation of viral mRNAs at late times after infection. Cell 31:543-551. [DOI] [PubMed] [Google Scholar]
- 37.Tiscornia, G., O. Singer, M. Ikawa, and I. M. Verma. 2003. A general method for gene knockdown in mice by using lentiviral vectors expressing small interfering RNA. Proc. Natl. Acad. Sci. USA 100:1844-1848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wolin, S. L., and T. Cedervall. 2002. The LA protein. Annu. Rev. Biochem. 71:375-403. [DOI] [PubMed] [Google Scholar]
- 39.Yi, R., Y. Qin, I. G. Macara, and B. R. Cullen. 2003. Exportin-5 mediates the nuclear export of pre-microRNAs and short hairpin RNAs. Genes Dev. 17:3011-3016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zeng, Y., and B. R. Cullen. 2003. Sequence requirements for micro RNA processing and function in human cells. RNA 9:112-123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zeng, Y., R. Yi, and B. R. Cullen. 2003. MicroRNAs and small interfering RNAs can inhibit mRNA expression by similar mechanisms. Proc. Natl. Acad. Sci. USA 100:9779-9784. [DOI] [PMC free article] [PubMed] [Google Scholar]