Abstract
The hepatitis C virus (HCV) RNA-dependent RNA polymerase (RdRp), represented by nonstructural protein 5B (NS5B), belongs to a class of integral membrane proteins termed tail-anchored proteins. Its membrane association is mediated by the C-terminal 21 amino acid residues, which are dispensable for RdRp activity in vitro. For this study, we investigated the role of this domain, termed the insertion sequence, in HCV RNA replication in cells. Based on a structural model and the amino acid conservation among different HCV isolates, we designed a panel of insertion sequence mutants and analyzed their membrane association and RNA replication. Subgenomic replicons with a duplication of an essential cis-acting replication element overlapping the sequence that encodes the C-terminal domain of NS5B were used to unequivocally distinguish RNA versus protein effects of these mutations. Our results demonstrate that the membrane association of the RdRp is essential for HCV RNA replication. Interestingly, certain amino acid substitutions within the insertion sequence abolished RNA replication without affecting membrane association, indicating that the C-terminal domain of NS5B has functions beyond serving as a membrane anchor and that it may be involved in critical intramembrane protein-protein interactions. These results have implications for the functional architecture of the HCV replication complex and provide new insights into the expanding spectrum of tail-anchored proteins.
Hepatitis C virus (HCV) infection is a major cause of chronic hepatitis, liver cirrhosis, and hepatocellular carcinoma worldwide (29). Similar to all positive-strand RNA viruses investigated thus far (reviewed in reference 2), HCV forms a membrane-associated replication complex composed of viral proteins, replicating RNA, and altered cellular membranes (9, 13). Determinants for membrane association of the HCV nonstructural proteins have been mapped (reviewed in reference 8), and a specific membrane alteration, designated the membranous web, was recently identified as the site of RNA replication in Huh-7 cells harboring subgenomic HCV replicons (13).
Members of our laboratories have recently shown that the HCV RNA-dependent RNA polymerase (RdRp), represented by nonstructural protein 5B (NS5B), belongs to a relatively small class of membrane proteins termed tail-anchored proteins (16, 32). Characteristic features of these proteins include (i) posttranslational membrane targeting via a hydrophobic C-terminal insertion sequence (which in the case of NS5B was mapped to the C-terminal 21 amino acid residues), (ii) integral membrane association, and (iii) a cytosolic orientation of the functional protein domain (reviewed in references 3 and 34). The prototype of this class of proteins is cytochrome b5. Other examples include the soluble N-ethylmaleimide-sensitive factor attachment protein receptor (SNARE) proteins and Bcl-2. NS5B represents the first polymerase within the family of tail-anchored proteins.
Interestingly, the deletion of its hydrophobic C-terminal domain increases the solubility of recombinant NS5B expressed in Escherichia coli without compromising the in vitro RdRp activity (10, 36). As a consequence, C-terminally truncated forms of NS5B were used for biochemical characterization and crystal structure determination (1, 4, 20) of this essential viral enzyme as well as for the development and validation of specific inhibitors (6).
The aim of this study was to investigate the role of the NS5B membrane insertion sequence in HCV RNA replication in cells. To this end, we designed a panel of insertion sequence mutants and analyzed their membrane association and RNA replication. Our results demonstrate that membrane association of the RdRp is essential for HCV RNA replication in cells. More importantly, we identified a mutant that could not replicate despite having a fully preserved membrane association. This observation indicates that the NS5B insertion sequence has functions beyond serving as a membrane anchor and that it may be involved in specific intramembrane protein-protein interactions that are essential for the formation of the HCV replication complex.
MATERIALS AND METHODS
Plasmids.
Plasmids pCMVNS5Bcon, pCMVNS5BconΔC12, and -ΔC21 were described previously (32). A fragment carrying the R568/570A mutations (Fig. 1) was amplified from pBRTM/HCV1-3011con (17) (kindly provided by Charles M. Rice, The Rockefeller University, New York, N.Y.) by a PCR using primers NS5B139fwd (27) and R568/570A (Table 1). The amplification product was digested with EcoRI and PstI and ligated with the 1,124-bp PstI-AvrII and 4,899-bp AvrII-EcoRI fragments of pCMVNS5Bcon to yield pCMVNS5Bcon-R568/570A. For the construction of pCMVNS5Bcon-LVL and pCMVNS5Bcon-R591A, the 1,750-bp BamHI-PstI fragment of pCMVNS5Bcon was ligated together with the annealed oligonucleotides LVLfwd and LVLrev or R591Afwd and R591Arev, respectively (Table 1), into the BamHI-XbaI sites of pcDNA3.1 (Invitrogen, San Diego, Calif.). All constructs allow both mammalian cell expression from a cytomegalovirus (CMV) promoter and in vitro transcription from a T7 RNA polymerase promoter.
FIG. 1.
NS5B insertion sequence mutants. NS5B amino acid positions are indicated at the top (17) (GenBank accession number AF009606). Fully conserved, conserved, and similar residues found among 269 HCV isolates are symbolized by asterisks, colons, and dots, respectively (32). #, minimal transmembrane segment, as deduced from various prediction methods (32); +, positively charged Arg residues. Mutated amino acid residues are underlined.
TABLE 1.
Oligonucleotide sequences
| Oligonucleotide | Sequence (5′-3′)a | Restriction enzyme site(s) |
|---|---|---|
| R568/570A | GTAGATGCCTACCCCTGCAGCGAGCAGGAGTAGGCAAAACCAGAACCAGGCGGGCGCGGCATGAGACACGCTGTG | PstI |
| LVLfwd | CTGGTACTCATCTACCTCCTCCCCAACCGATAAT | PstI, XbaI |
| LVLrev | CTAGATTATCGGTTGGGGAGGAGGTAGATGAGTACCAGTGCA | XbaI, PstI |
| R591Afwd | GGGGTAGGCATCTACCTCCTCCCCAACGCTTAAT | PstI, XbaI |
| R591Arev | CTAGATTAAGCGTTGGGGAGGAGGTAGATGCCTACCCCTGCA | XbaI, PstI |
| S UGA/Xbal | CCAACCGATGATCTAGACGGGGAGCTAAAC | |
| A UGA/Xbal | GTTTAGCTCCCCGTCTAGATCATCGGTTGG | XbaI |
| A1bΔC12 | CGGTCTAGAGTGTTTAGCTCCCCGTTCATCGGTTGGGGAGTAGATAGATGCCTACCCCTACTCAAAGTAGGAG | XbaI |
| S1bR568/570A | CCTGTCTCGTGCCGCACCCGCCTGGTTCATGTG | |
| A1bR568/570A | CACATGAACCAGGCGGGTGCGGCACGAGACAGG | |
| A1bLVL | CGGTCTAGAGTGTTTAGCTCCCCGTTCATCGGTTGGGGAGTAGATAGATGAGTACCAGTACAGAAAGTAG | XbaI |
| A1bR591A | CGGTCTAGAGTGTTTAGCTCCCCGTTCATGCGTTGGGGAGTAG | XbaI |
Mutagenic nucleotides are double underlined. Restriction enzyme recognition sites are underlined, and stop codons are shown in bold. The Pstl sites in LVLfwd and LVLrev were preserved only in the overhanging nucleotides due to the introduced mutations.
All replicon constructs used for this study harbored two cell culture-adaptive mutations in NS3 (E1202G and T1280I) and one in NS5A (2202ΔS) (21), a combination designated “JT” (the numbers refer to the amino acid sequence of the HCV Con1 polyprotein [GenBank accession number AJ238799]). An XbaI restriction site directly following the UGA stop codon of the HCV polyprotein was inserted into pFKI341PINeo/NS3-3′/JT (originally designated pFKnt341-sp-PI-neoEI3420-9605 [11]) by a PCR using primers S UGA/Xba and A UGA/Xba (Table 1) and NcoI and SpeI restriction sites in NS5B and the vector, yielding pFKI341PINeo/NS3-3′/JT/Xba (16). The replicon construct pFKPI-JT/Xba, harboring a gene encoding firefly luciferase instead of neomycin phosphotransferase, was generated by the exchange of an XhoI-SpeI fragment in PI-JT (pFKI341PILuc/NS3-3′/E1202G/T1280I/2202ΔS [21]) with the same fragment from pFKI341PINeo/NS3-3′/JT/Xba. Mutations of the NS5B insertion sequence were introduced into the replicon by replacement of the 386-bp NcoI-XbaI fragment of pFKPI-JT/Xba with the NcoI-XbaI fragment of pCMVNS5Bcon, -R568/570A, -LVL, -R591A, -ΔC12, or -ΔC21, yielding pFKPI-JT/1b-1a and its respective mutants.
A 4,877-bp SfiI fragment obtained from plasmid pFKI341PILuc/NS3-3′/JT was replaced in pFKPI-ET/dv1-3 and pFK PI-ET/mut1-3/dv1-3 (12) to generate the replicon constructs pFKPI-JT/dv1-3 and pFKPI-JT/mut1-3/dv1-3, respectively. The mutations R568/570A, LVL, R591A, and ΔC12 were introduced into PCR products by use of the S1bR568/570A primer and primers A1bR568/570A, A1bLVL, A1bR591A, and A1bΔC12, respectively (Table 1), and then were subcloned into plasmid pBSK8499-9605/dv1-3 (12) by use of an NcoI restriction site in NS5B and a newly generated XbaI site at position 9391 of the HCV Con1 genome (12). Replicon plasmids pFKPI-JT/dv1-3/R568/570A, -LVL, -R591A, and -ΔC12 were generated by the exchange of a SfiI-SpeI fragment in pFKPI-JT/dv1-3 with the same fragment taken from pBSK8499-9605/dv1-3/R568/570A, -LVL, -R591A, or -ΔC12, respectively.
Replication assays.
In vitro transcription, transfection of Huh-7 human hepatoma cells by RNA electroporation, and RNA replication assays with luciferase reporter gene replicons as well as selectable HCV replicons were performed as previously described (18, 21). Besides naïve Huh-7 cells, a cured replicon cell clone, designated Huh-7/lunet (12), was used for some transient replication assays, as indicated in the figure legends.
Antibodies.
The monoclonal antibodies (MAbs) 5B-3B1 and 5B-12B7 against NS5B were described previously (27).
Confocal laser scanning microscopy.
Indirect immunofluorescence staining was performed as described previously (28). Coverslips were mounted in SlowFade (Molecular Probes, Eugene, Oreg.) and examined with a Zeiss LSM 510 laser scanning system. Images were processed with Zeiss Image 3.1.0.99 and Adobe Photoshop 7.0 software.
Membrane flotation.
Membrane flotation assays were performed essentially as described previously (25). In brief, 2 × 107 transiently transfected U-2 OS cells were subjected to Dounce homogenization in a hypotonic buffer (10 mM Tris-HCl [pH 7.5], 2 mM MgCl2), followed by centrifugation at 1,000 × g for 5 min to pellet nuclei, unlysed cells, and large debris. Nycodenz (Axis Shield, Oslo, Norway) was added to postnuclear supernatants to a final concentration of 37.5% (wt/vol) in phosphate-buffered saline. A 400-μl lysate was placed at the bottom of a 1.5-ml thick-walled ultracentrifugation tube and overlaid with 900 μl of 35% and 100 μl of 5% Nycodenz in phosphate-buffered saline. Equilibrium centrifugation was carried out at 100,000 × g for 20 h at 4°C in a Beckman TLS 55 rotor. Subsequently, gradients were separated into upper (low density) and lower (high density) fractions, analyzed by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE), and subjected to immunoblotting as described previously (28).
In vitro transcription-translation (IVTT).
The TNT T7 coupled reticulocyte lysate system (Promega, Madison, Wis.) was used essentially according to the manufacturer's recommendations. When indicated, 1.5 μl of canine pancreatic microsomes (kindly provided by Martin Spiess, University of Basel, Basel, Switzerland) was added. Membrane sedimentation analyses were performed as previously described (32). Subsequently, pellet and supernatant fractions were analyzed by SDS-PAGE followed by autoradiography. Gels were scanned on a Fuji BAS1000 phosphorimager and analyzed with Fuji MacBAS v. 2.4 software.
RESULTS
Subcellular localization of NS5B insertion sequence mutants.
The design of the NS5B insertion sequence mutants (Fig. 1) was based on a structural model of this domain (16) and the conservation of amino acid residues observed among 269 different HCV isolates (32). From a structural point of view, all mutations were expected to preserve the overall α-helical fold of the membrane insertion domain. The absolutely conserved Arg568 and Arg570 residues preceding the transmembrane domain were replaced with Ala, yielding construct R568/570A. Mutation of the Gly residues at positions 582 and 584 in the center of the α-helix into Leu residues resulted in construct LVL. The replacement of the absolutely conserved Arg591 residue with Ala yielded construct R591A. Finally, constructs ΔC12 and ΔC21 were obtained by deletion of the C-terminal 12 or 21 aa residues of NS5B, respectively (32).
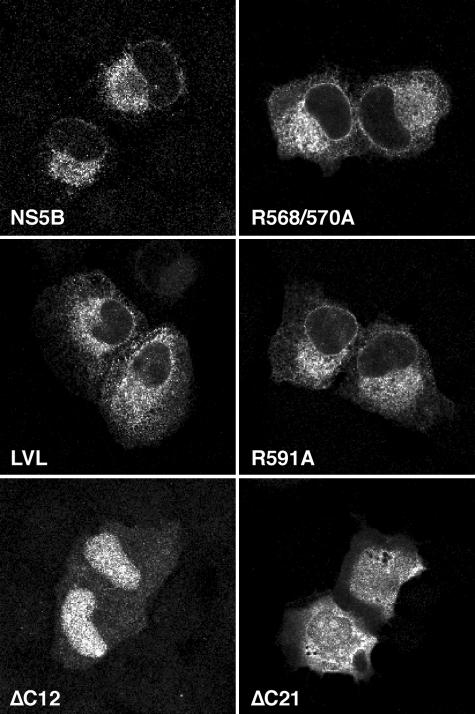
The subcellular localization of these mutants was analyzed by confocal laser scanning microscopy, as shown in Fig. 2. Wild-type NS5B was found to have a reticular staining pattern which surrounded the nucleus, extended through the cytoplasm, and included the nuclear membrane. No plasma membrane or nuclear staining was observed. This typical staining pattern reflects the ER localization of NS5B when it is expressed alone (32). Identical staining patterns were observed for the R568/570A, LVL, and R591A mutants. In contrast, constructs ΔC12 and ΔC21 showed diffuse staining with an accumulation of the C-terminally truncated proteins in nuclei and nucleoli, as previously reported (32). The ΔC21 construct showed the same diffuse staining pattern with nucleolar accumulation when it was expressed in the context of the HCV polyprotein, indicating that the C-terminal insertion sequence is essential for the proper subcellular localization of NS5B (R. Gosert and D. Moradpour, unpublished data). Taken together, these results demonstrate that the R568/570A, LVL, and R591A mutants retained their typical ER localization and apparent membrane association.
FIG. 2.
Subcellular localization of NS5B insertion sequence mutants. U-2 OS cells were transiently transfected with pCMVNS5Bcon, -R568/570A, -LVL, -R591A, -ΔC12, and -ΔC21, as indicated. At 36 h posttransfection, the cells were subjected to immunofluorescence staining with MAb 5B-12B7 against NS5B. The slides were analyzed by confocal laser scanning microscopy as described in Materials and Methods.
Membrane association of NS5B insertion sequence mutants.
Flotation analyses were performed to confirm the membrane association of the R568/570A, LVL, and R591A mutants. U-2 OS cells transfected with the corresponding expression constructs were lysed in a hypotonic buffer, and the proteins were analyzed by equilibrium centrifugation in a Nycodenz gradient. Under these conditions, membrane proteins float to the upper, low-density fractions, as shown in Fig. 3 for wild-type NS5B as well as the R591A, LVL, and R568/570A mutants, confirming the unimpaired membrane association of these mutants. In contrast, the ΔC12 (Fig. 3) and ΔC21 (data not illustrated) mutants remained in the high-density fraction.
FIG. 3.
Membrane association of NS5B insertion sequence mutants. Hypotonic lysates of U2-OS cells transiently transfected with pCMVNS5Bcon, -R568/570A, -LVL, -R591A, and -ΔC12 were analyzed by membrane flotation as described in Materials and Methods. Low-density (LD) and high-density (HD) fractions were collected, separated by SDS-12% PAGE, and analyzed by immunoblotting with MAb 5B-3B1 against NS5B.
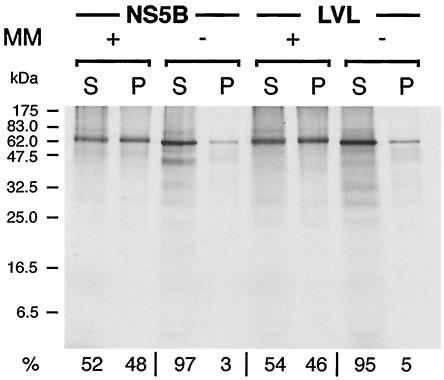
Tail-anchored proteins associate with membranes by a posttranslational mechanism. IVTT and membrane sedimentation analyses were performed to investigate whether the same was true for the NS5B mutants. The constructs were translated by use of a coupled rabbit reticulocyte lysate system. Subsequently, puromycin was added to the reaction mixture to stop translation, and the mixtures were incubated with microsomal membranes isolated from the canine pancreas. Finally, membrane association was analyzed by sedimentation analysis. In the case of wild-type NS5B, the majority of the protein remained in the supernatant fraction in the absence of microsomal membranes (Fig. 4, left side). In contrast, 52% of the protein sedimented into the membrane-containing pellet fraction when the IVTT reaction was posttranslationally incubated with microsomal membranes. Comparable results were obtained for the LVL (Fig. 4, right side), R568/570A, and R591A (data not illustrated) mutants, indicating that the posttranslational membrane association of these mutants also occurs very efficiently in vitro. In addition, the differential extraction of membranes with 1 M NaCl, 100 mM sodium carbonate (pH 11.5), and 4 M urea revealed that the R568/570A, LVL, and R591A mutants, like wild-type NS5B (32), behaved as integral membrane proteins (data not shown).
FIG. 4.
Posttranslational membrane association of NS5B and the LVL mutant in vitro. IVTT reactions of pCMVNS5Bcon and -LVL were performed in the absence of microsomal membranes. After 1 h, translation was blocked by puromycin, followed by the addition of microsomal membranes (MM) to an aliquot of the reactions and incubation for one additional hour. Subsequently, membrane sedimentation analyses were performed as described in Materials and Methods. Supernatant (S) and pellet (P) fractions were analyzed by SDS-12% PAGE. Relative amounts of [35S]methionine-labeled translation products, as quantified by phosphorimaging, are given at the bottom.
Taken together, these results demonstrate that the R568/570A, LVL, and R591A mutants have a membrane association that is indistinguishable from that of wild-type NS5B and thus fulfill the criteria of tail-anchored proteins. In contrast, deletion of the C-terminal 12 aa residues was sufficient to abolish the membrane association of NS5B.
Replication of NS5B insertion sequence mutants.
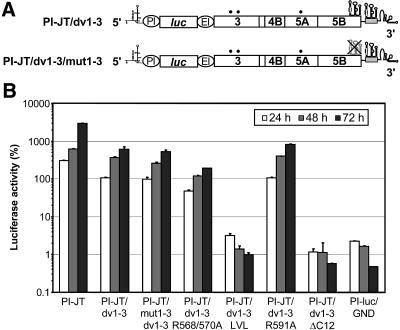
To analyze the effect of mutations in the NS5B membrane insertion sequence on HCV RNA replication, we introduced mutations into a reporter replicon carrying two cell culture-adaptive changes in NS3 (E1202G and T1280I) and one in NS5A (2202ΔS) (21) (Fig. 5A). In this construct, translation of the firefly luciferase gene is directed by the internal ribosome entry site (IRES) of poliovirus, whereas the IRES of encephalomyocarditis virus governs translation of the HCV nonstructural proteins. RNA replication was analyzed by measuring the luciferase activity at various time points after transfection into Huh-7 cells. The parental replicon and a replication-deficient RNA (designated GND) were used as positive and negative controls, respectively.
FIG. 5.
Replication efficiencies of NS5B insertion sequence mutants. (A) Structures of replicons PI-JT/Xba and PI-JT/1b-1a. The XbaI site introduced after the stop codon of the HCV open reading frame and an NcoI site present at the same position in the 1a and 1b isolates are indicated. The sequence corresponding to the genotype 1a isolate is shaded in gray. The adaptive mutations E1202G, T1280I, and 2202ΔS are indicated by black dots. PI, poliovirus IRES; luc, firefly luciferase; EI, encephalomyocarditis virus IRES. (B) Different replicon RNAs, as specified at the bottom, were transfected into Huh-7 cells by electroporation. Luciferase activities were determined for the cell lysates and are given as percentages of the relative light units at 24, 48, and 72 h in comparison to those 4 h after transfection (100%). Replicon PI-luc/GND, which harbors an inactivating mutation in the GDD motif of NS5B, was used as a negative control.
In a first set of experiments, a fragment encoding the C-terminal one-quarter of NS5B (aa 467 to 591) of genotype 1a from construct pCMVNS5Bcon, -R567/570A, -LVL, -R591A, -ΔC12, or -ΔC21 was introduced into a genotype 1b replicon with an XbaI site engineered directly after the stop codon of NS5B (PI-JT/Xba) (Fig. 5A). Thus, these constructs represent genotype 1b-1a chimeras with the N-terminal three-quarters of NS5B derived from the genotype 1b Con1 isolate (22) and the C-terminal one-quarter derived from the genotype 1a HCV H consensus clone (17). Interestingly, the chimeric construct containing the wild-type NS5B fragment from genotype 1a (PI-JT/1b-1a) (Fig. 5A) showed a replication efficiency comparable to that of the PI-JT/Xba replicon harboring only genotype 1b NS5B (Fig. 5B). As shown in Fig. 5B, the R591A mutant replicated as efficiently as the wild type while the R568/570A, LVL, ΔC12, and ΔC21 mutants did not show signals above the background of this assay, as defined with the GND construct. These results were confirmed for the R568/570A and LVL mutants by colony formation analyses of selectable subgenomic replicons, which did not yield any G418-resistant colonies (data not shown).
It was recently reported that the sequence encoding the C-terminal domain of NS5B contains a cis-acting replication element (CRE) that is essential for HCV RNA replication (37). An essential stem-loop, designated SL3.2, was identified within a larger cruciform RNA element, designated SL3. In view of these observations, it was important to distinguish whether the mutations in the NS5B insertion sequence affected protein or overlapping RNA functions. Therefore, a second set of experiments was performed with replicon constructs carrying a duplication of SL3 in the variable region of the 3′ nontranslated region (3′ NTR) (dv1-3) (Fig. 6A). A detailed characterization of this and related constructs is reported elsewhere (12). In this case, mutations were generated in a pure Con1 genotype 1b background with the same set of adaptive changes as that described above, and ΔC12 was created by the introduction of a stop codon after NS5B aa 579. As shown in Fig. 6B, the dv1-3 replicon, containing two functional SL3 elements, replicated nearly as efficiently as the parental PI-JT replicon. The replication efficiency was fully preserved in the mut1-3/dv1-3 replicon, although the SL3 element in NS5B was destroyed in this replicon by multiple silent mutations interfering with the predicted stem-loop structures (Fig. 6). In the absence of the SL3 duplication, this combination of silent mutations results in a complete loss of replication competence (12). Therefore, the first, nonfunctional SL3 element was efficiently rescued by the engineered second SL3 in the mut1-3/dv1-3 replicon, demonstrating that the duplication of the SL3 element in dv1-3 is capable of compensating for possible adverse effects of the insertion sequence mutations on the function of the CRE. The R591A mutant replicated with the same efficiency as the wild type, confirming the results obtained with the first set of experiments. Similarly, the LVL and ΔC12 mutants were completely replication defective. However, when analyzed in the context of the SL3 duplication replicons, the R568/570 mutant replicated in an only slightly impaired fashion compared to the reference construct dv1-3. Therefore, the replication defect of this mutant observed in the first set of experiments could be attributed primarily to an RNA effect as opposed to a protein effect.
FIG. 6.
Discrimination of protein versus RNA effects of mutations in the NS5B insertion sequence. (A) Schematic structures of replicons with a duplication of SL3 in the variable region of the 3′ NTR. The gray box indicates the duplication of part of the 3′ coding region of NS5B that carries the predicted stem-loop structures. Replicon PI-JT/dv1-3 contained two identical intact copies of the SL3 sequence and was used as a parental construct to introduce the NS5B insertion sequence mutations. The predicted SL3 RNA secondary structure in the coding region of NS5B was disrupted by multiple silent mutations in replicon PI-JT/mut1-3/dv1-3. For further details, see the legend to Fig. 5A. (B) Replicons containing two intact copies of SL3 and the indicated mutations in the NS5B insertion sequence were transfected into Huh-7/lunet cells by electroporation. The percentage of luciferase activity at a given time point is relative to the relative light units determined 4 h after transfection, which was set to 100%. Replicon PI-luc/GND, which contains an inactivating mutation in the GDD motif of NS5B, was used as a negative control.
Taken together, these results unequivocally demonstrate that deletion of the C-terminal 12 aa residues, which results in a loss of the membrane association of NS5B, abolishes RNA replication. Replacement of the positively charged Arg residues flanking the NS5B insertion sequence at both ends was tolerated under the conditions of the assays employed. Interestingly, the LVL mutant, which displayed a fully preserved membrane association phenotype, was replication defective, indicating that another function of the tail anchor was affected.
DISCUSSION
In this report, we showed that the C-terminal insertion sequence is essential not only for membrane association of the RdRp, but also for HCV RNA replication. Although not directly addressed experimentally, the most likely explanation is that membrane association as such is required for RNA replication. Deletion of the C-terminal 12 aa (ΔC12) resulted in the loss of the membrane association of NS5B and abolished RNA replication. The duplication of an essential CRE that was recently identified in the sequence encoding the C-terminal domain of NS5B (12, 37) did not rescue RNA replication in the ΔC12 mutant. This demonstrates that the replication defect of this mutant was due to a loss of protein function and not to an effect on overlapping RNA functions. While this paper was in preparation, a report by Lee et al. (19) described replication defects of NS5B mutants with a deletion of the insertion sequence or an introduction of two stop codons at aa 571. However, it was not experimentally validated whether these mutations affected the function of the overlapping CRE.
The R591A and R568/570A mutations did not affect or only moderately affected RNA replication. The replication defect of the R568/570A mutant observed in our first set of experiments could be rescued by the insertion of an intact copy of SL3 into the variable region of the 3′ NTR. Thus, the defect initially observed for this mutant could be attributed primarily to an inactivation of the overlapping RNA element. This was in good agreement with the findings of You et al. (37), who mapped the essential CRE, designated SL3.2, to the sequence coding for NS5B aa 555 to 571. Indeed, an analysis of 404 HCV nucleotide sequences reported in GenBank revealed that of the six possible codons for Arg, only two each, CGA and CGG or CGC and CGT, are used for Arg568 and Arg570, respectively (F. Penin, unpublished data). This very limited usage of specific codons clearly indicates that conservation of the nucleotide sequence is critical for RNA replication. In contrast, conservation of the codon for Arg591 is not critical, as reflected by the use of all six possible codons among different HCV isolates. In this case, the unimpaired replication of R591A may reflect functions or interactions that are dispensable in the replicon system or a subtle aspect of membrane association that is not detectable by our assays.
Interestingly, there was a striking discordance between the membrane association and RNA replication of the LVL mutant. This mutant displayed a subcellular localization and membrane association that were indistinguishable from those of wild-type NS5B. However, its RNA replication was completely abolished. It was previously shown that the absolutely conserved GVG motif in the center of the NS5B insertion sequence could theoretically form a flexible hinge within the α-helix but that it is not essential for translocation of the tail anchor across the phospholipid bilayer (16). Our assays did not permit us to detect subtle alterations in the kinetics of membrane integration. However, the fact that the RNA replication of this mutant was completely abolished suggests that the GVG motif may play another role. As illustrated in a recently reported structural model (16) (Fig. 7), the absence of side chains for the Gly residues within the GVG motif yields “holes” along the transmembrane α-helix. These holes were filled up by the bulky Leu side chains in the LVL mutant (Fig. 7, compare NS5B and LVL). Gly residues are frequently involved in transmembrane helix-helix interactions (33), during which the holes formed by Gly residues are filled by large hydrophobic residues (particularly Phe or Leu) present in the transmembrane segments of interaction partners, yielding a knob-into-hole hydrophobic interaction pattern (24). Thus, our current working model is that the GVG motif may play a role in a critical protein-protein interaction within the lipid bilayer that was blocked by the replacement of Gly with Leu. In principle, both the homo-oligomerization of multiple RdRp molecules, as described for the 3D polymerase of poliovirus (14, 23), and interactions with other viral or cellular components of the HCV replication complex can be envisioned.
FIG. 7.
Membrane association and RNA replication of NS5B insertion sequence mutants. A molecular model of the NS5B segment from aa 567 to 591 was constructed by using aa 5 to 30 of bacteriorhodopsin as a template (PDB entry 1BHB) and the Swiss Model server facilities (http://www.expasy.ch/swissmod/) as previously described (16). Models for mutants were constructed with the SwissPDB Viewer program, using the above model as a template. Space-filling representations are shown. Arg, Cys, and Gly residues are shown in blue, yellow, and orange, respectively. Leu residues in the LVL mutant are shown in red. The Cys residues are shown to facilitate comparisons between the various models. The models were manually positioned in the phospholipid bilayer that was built by using the coordinates of phospholipids reported in PDB entry 1BCC (38). The polar heads and the aliphatic tails of the phospholipids are light gray and very light gray, respectively. The figures were generated with Rasmol 2.7 (31).
HCV NS5B is one of the few RdRps with an experimentally determined transmembrane domain (16). Indeed, to our knowledge the only other RdRp with a similarly characterized membrane association is flock house virus protein A (25, 26). The incorporation of the RdRp into membrane-bound replication complexes of other positive-strand RNA viruses occurs by interactions with other viral proteins. For example, the membrane association of the poliovirus 3D polymerase is mediated by an interaction with the protein precursor 3AB (15, 23, 35). Similarly, mouse hepatitis virus RdRp is not membrane associated per se, but is incorporated into membrane-bound replication complexes via interactions with as yet unidentified viral or virus-induced factors (5). Finally, brome mosaic virus RNA polymerase-like protein 2a depends on multifunctional replicase protein 1a for recruitment to the ER (7, 30). In this context, we have recently found that interactions among the cytosolic domains of HCV nonstructural proteins are not sufficiently strong to rescue the ΔC21 mutant to membranes when expressed in the context of the HCV polyprotein (R. Gosert and D. Moradpour, unpublished data). Thus, the C-terminal membrane insertion sequence of HCV NS5B represents an essential and unique element and therefore an attractive target for antiviral intervention.
In conclusion, we have shown that the membrane association of the RdRp is essential for HCV RNA replication in cells. More importantly, the C-terminal membrane insertion sequence, which is dispensable for RdRp activity in vitro, represents an essential element that may be involved in critical intramembrane protein-protein interactions within the HCV replication complex. Such interactions may be exploited as novel antiviral targets.
Acknowledgments
This work was supported by grants Mo 799/1-3 and SFB 638/Teilprojekt A5 from the Deutsche Forschungsgemeinschaft, QLK2-CT1999-00356 and QLK2-CT2002-01329 from the European Commission, and 01 KI 9951 from the Bundesministerium für Bildung und Forschung, by the Bristol-Myers Squibb Foundation, and by the French Centre National de la Recherche Scientifique.
We gratefully acknowledge Sandra Hoffmann for excellent technical assistance, Shihyun You, Charles M. Rice, Denise Egger, and Kurt Bienz for helpful discussions, Charles M. Rice for pBRTM/HCV1-3011con, and Martin Spiess for microsomal membranes.
Footnotes
This work is dedicated to Madeleine and Morad Moradpour with affection and gratitude.
REFERENCES
- 1.Ago, H., T. Adachi, A. Yoshida, M. Yamamoto, N. Habuka, K. Yatsunami, and M. Miyano. 1999. Crystal structure of the RNA-dependent RNA polymerase of hepatitis C virus. Struct. Fold. Des. 7:1417-1426. [DOI] [PubMed] [Google Scholar]
- 2.Ahlquist, P., A. O. Noueiry, W. M. Lee, D. B. Kushner, and B. T. Dye. 2003. Host factors in positive-strand RNA virus genome replication. J. Virol. 77:8181-8186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Borgese, N., S. Colombo, and E. Pedrazzini. 2003. The tale of tail-anchored proteins: coming from the cytosol and looking for a membrane. J. Cell Biol. 161:1013-1019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bressanelli, S., L. Tomei, A. Roussel, I. Incitti, R. L. Vitale, M. Mathieu, R. De Francesco, and F. A. Rey. 1999. Crystal structure of the RNA-dependent RNA polymerase of hepatitis C virus. Proc. Natl. Acad. Sci. USA 96:13034-13039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Brockway, S. M., C. T. Clay, X. T. Lu, and M. R. Denison. 2003. Characterization of the expression, intracellular localization, and replication complex association of the putative mouse hepatitis virus RNA-dependent RNA polymerase. J. Virol. 77:10515-10527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Carroll, S. S., J. E. Tomassini, M. Bosserman, K. Getty, M. W. Stahlhut, A. B. Eldrup, B. Bhat, D. Hall, A. L. Simcoe, R. LaFemina, C. A. Rutkowski, B. Wolanski, Z. Yang, G. Migliaccio, R. De Francesco, L. C. Kuo, M. MacCoss, and D. B. Olsen. 2003. Inhibition of hepatitis C virus RNA replication by 2′-modified nucleoside analogs. J. Biol. Chem. 278:11979-11984. [DOI] [PubMed] [Google Scholar]
- 7.Chen, J., and P. Ahlquist. 2000. Brome mosaic virus polymerase-like protein 2a is directed to the endoplasmic reticulum by helicase-like viral protein 1a. J. Virol. 74:4310-4318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dubuisson, J., F. Penin, and D. Moradpour. 2002. Interaction of hepatitis C virus proteins with host cell membranes and lipids. Trends Cell Biol. 12:517-523. [DOI] [PubMed] [Google Scholar]
- 9.Egger, D., B. Wölk, R. Gosert, L. Bianchi, H. E. Blum, D. Moradpour, and K. Bienz. 2002. Expression of hepatitis C virus proteins induces distinct membrane alterations including a candidate viral replication complex. J. Virol. 76:5974-5984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ferrari, E., J. Wright-Minogue, J. W. Fang, B. M. Baroudy, J. Y. Lau, and Z. Hong. 1999. Characterization of soluble hepatitis C virus RNA-dependent RNA polymerase expressed in Escherichia coli. J. Virol. 73:1649-1654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Friebe, P., V. Lohmann, N. Krieger, and R. Bartenschlager. 2001. Sequences in the 5′ nontranslated region of hepatitis C virus required for RNA replication. J. Virol. 75:12047-12057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Friebe, P., J. Boudet, J.-P. Simorre, and R. Bartenschlager. A kissing loop interaction in the 3′ end of the hepatitis C virus genome essential for RNA replication. J. Virol., in press. [DOI] [PMC free article] [PubMed]
- 13.Gosert, R., D. Egger, V. Lohmann, R. Bartenschlager, H. E. Blum, K. Bienz, and D. Moradpour. 2003. Identification of the hepatitis C virus RNA replication complex in Huh-7 cells harboring subgenomic replicons. J. Virol. 77:5487-5492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hobson, S. D., E. S. Rosenblum, O. C. Richards, K. Richmond, K. Kirkegaard, and S. C. Schultz. 2001. Oligomeric structures of poliovirus polymerase are important for function. EMBO J. 20:1153-1163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hope, D. A., S. E. Diamond, and K. Kirkegaard. 1997. Genetic dissection of interaction between poliovirus 3D polymerase and viral protein 3AB. J. Virol. 71:9490-9498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ivashkina, N., B. Wölk, V. Lohmann, R. Bartenschlager, H. E. Blum, F. Penin, and D. Moradpour. 2002. The hepatitis C virus RNA-dependent RNA polymerase membrane insertion sequence is a transmembrane segment. J. Virol. 76:13088-13093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kolykhalov, A. A., E. V. Agapov, K. J. Blight, K. Mihalik, S. M. Feinstone, and C. M. Rice. 1997. Transmission of hepatitis C by intrahepatic inoculation with transcribed RNA. Science 277:570-574. [DOI] [PubMed] [Google Scholar]
- 18.Krieger, N., V. Lohmann, and R. Bartenschlager. 2001. Enhancement of hepatitis C virus RNA replication by cell culture-adaptive mutations. J. Virol. 75:4614-4624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lee, K. J., J. Choi, J. H. Ou, and M. M. Lai. 2004. The C-terminal transmembrane domain of hepatitis C virus (HCV) RNA polymerase is essential for HCV replication in vivo. J. Virol. 78:3797-3802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lesburg, C. A., M. B. Cable, E. Ferrari, Z. Hong, A. F. Mannarino, and P. C. Weber. 1999. Crystal structure of the RNA-dependent RNA polymerase from hepatitis C virus reveals a fully encircled active site. Nat. Struct. Biol. 6:937-943. [DOI] [PubMed] [Google Scholar]
- 21.Lohmann, V., S. Hoffmann, U. Herian, F. Penin, and R. Bartenschlager. 2003. Viral and cellular determinants of hepatitis C virus RNA replication in cell culture. J. Virol. 77:3007-3019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lohmann, V., F. Körner, J.-O. Koch, U. Herian, L. Theilmann, and R. Bartenschlager. 1999. Replication of subgenomic hepatitis C virus RNAs in a hepatoma cell line. Science 285:110-113. [DOI] [PubMed] [Google Scholar]
- 23.Lyle, J. M., E. Bullitt, K. Bienz, and K. Kirkegaard. 2002. Visualization and functional analysis of RNA-dependent RNA polymerase lattices. Science 296:2218-2222. [DOI] [PubMed] [Google Scholar]
- 24.MacKenzie, K. R., J. H. Prestegard, and D. M. Engelman. 1997. A transmembrane helix dimer: structure and implications. Science 276:131-133. [DOI] [PubMed] [Google Scholar]
- 25.Miller, D. J., and P. Ahlquist. 2002. Flock house virus RNA polymerase is a transmembrane protein with amino-terminal sequences sufficient for mitochondrial localization and membrane insertion. J. Virol. 76:9856-9867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Miller, D. J., M. D. Schwartz, B. T. Dye, and P. Ahlquist. 2003. Engineered retargeting of viral RNA replication complexes to an alternative intracellular membrane. J. Virol. 77:12193-12202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Moradpour, D., E. Bieck, T. Hügle, W. Wels, J. Z. Wu, Z. Hong, H. E. Blum, and R. Bartenschlager. 2002. Functional properties of a monoclonal antibody inhibiting the hepatitis C virus RNA-dependent RNA polymerase. J. Biol. Chem. 277:593-601. [DOI] [PubMed] [Google Scholar]
- 28.Moradpour, D., P. Kary, C. M. Rice, and H. E. Blum. 1998. Continuous human cell lines inducibly expressing hepatitis C virus structural and nonstructural proteins. Hepatology 28:192-201. [DOI] [PubMed] [Google Scholar]
- 29.National Institutes of Health. 2002. National Institutes of Health Consensus Development Conference statement: management of hepatitis C: 2002. Hepatology 36(Suppl. 1):S2-S20. [Google Scholar]
- 30.Restrepo-Hartwig, M., and P. Ahlquist. 1999. Brome mosaic virus RNA replication proteins 1a and 2a colocalize and 1a independently localizes on the yeast endoplasmic reticulum. J. Virol. 73:10303-10309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sayle, R. A., and E. J. Milner-White. 1995. RASMOL: biomolecular graphics for all. Trends Biochem. Sci. 20:374. [DOI] [PubMed] [Google Scholar]
- 32.Schmidt-Mende, J., E. Bieck, T. Hügle, F. Penin, C. M. Rice, H. E. Blum, and D. Moradpour. 2001. Determinants for membrane association of the hepatitis C virus RNA-dependent RNA polymerase. J. Biol. Chem. 276:44052-44063. [DOI] [PubMed] [Google Scholar]
- 33.Senes, A., M. Gerstein, and D. M. Engelman. 2000. Statistical analysis of amino acid patterns in transmembrane helices: the GxxxG motif occurs frequently and in association with beta-branched residues at neighboring positions. J. Mol. Biol. 296:921-936. [DOI] [PubMed] [Google Scholar]
- 34.Wattenberg, B., and T. Lithgow. 2001. Targeting of C-terminal (tail)-anchored proteins: understanding how cytoplasmic activities are anchored to intracellular membranes. Traffic 2:66-71. [DOI] [PubMed] [Google Scholar]
- 35.Xiang, W. K., A. Cuconati, D. Hope, K. Kirkegaard, and E. Wimmer. 1998. Complete protein linkage map of poliovirus P3 proteins: interaction of polymerase 3D(pol) with VPg and with genetic variants of 3AB. J. Virol. 72:6732-6741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Yamashita, T., S. Kaneko, Y. Shirota, W. Qin, T. Nomura, K. Kobayashi, and S. Murakami. 1998. RNA-dependent RNA polymerase activity of the soluble recombinant hepatitis C virus NS5B protein truncated at the C-terminal region. J. Biol. Chem. 273:15479-15486. [DOI] [PubMed] [Google Scholar]
- 37.You, S., D. D. Stump, A. D. Branch, and C. M. Rice. 2004. A cis-acting replication element in the sequence encoding the NS5B RNA-dependent RNA polymerase is required for hepatitis C virus RNA replication. J. Virol. 78:1352-1366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zhang, Z., L. Huang, V. M. Shulmeister, Y. I. Chi, K. K. Kim, L. W. Hung, A. R. Crofts, E. A. Berry, and S. H. Kim. 1998. Electron transfer by domain movement in cytochrome bc1. Nature 392:677-684. [DOI] [PubMed] [Google Scholar]