Abstract
The hemagglutinin-neuraminidase (HN) protein of Newcastle disease virus mediates attachment to sialic acid receptors, as well as cleavage of the same moiety. HN also interacts with the other viral glycoprotein, the fusion (F) protein, to promote membrane fusion. The ectodomain of the HN spike consists of a stalk and a terminal globular head. The most conserved part of the stalk consists of two heptad repeats separated by a nonhelical intervening region (residues 89 to 95). Several amino acid substitutions for a completely conserved proline residue in this region not only impair fusion and the HN-F interaction but also decrease neuraminidase activity in the globular domain, suggesting that the substitutions may alter HN structure. Substitutions for L94 also interfere with fusion and the HN-F interaction but have no significant effect on any other HN function. Amino acid substitutions at other positions in the intervening region also modulate only fusion. In all cases, diminished fusion correlates with a decreased ability of the mutated HN protein to interact with F at the cell surface. These findings indicate that the intervening region is critical to the role of HN in the promotion of fusion and may be directly involved in its interaction with the homologous F protein.
The Paramyxoviridae family of enveloped, negative-stranded RNA viruses includes human parainfluenza virus (hPIV) types 1 to 4, mumps and measles viruses, and the animal pathogens Sendai virus, simian virus 5, and Newcastle disease virus (NDV) (19). Virion and infected cell surfaces are characterized by two types of glycoproteins, which mediate early interactions with the target cell: the hemagglutinin-neuraminidase (HN) and the fusion (F) proteins (28, 29).
The HN glycoprotein mediates attachment to sialic acid-containing receptor(s) on target cell surfaces and, through its neuraminidase (NA) activity, performs the opposite activity of removing sialic acid from progeny virus particles to prevent viral self-aggregation (19). The HN protein is a type II homodimer that exists on the surface of virions and infected cells as a tetrameric spike. The ectodomain of the HN spike consists of a stalk that supports a terminal globular head in which receptor recognition, NA activity, and all of the known antigenic sites reside (23, 34). The monomers in each HN homodimer of some NDV strains, such as Australia-Victoria (NDV-AV), are linked by disulfide bonds involving the cysteines at position 123 in the stalk (30).
The X-ray crystallographic structure of the globular head (residues 124 to 570) of the HN dimer originally suggested that receptor recognition and NA activities are mediated by a single binding site that adopts two different conformations (3, 5). However, this was inconsistent with the ability to rescue the attachment activity of HN proteins carrying NA active site substitutions by treatment with exogenous neuraminidase (16). Instead, this result suggested that the lack of attachment activity of these mutated HN proteins was a secondary result of their lack of NA activity and that there may be another sialic acid binding site. Indeed, a second sialic acid binding site has recently been discovered at the membrane-distal end of the dimer interface in the NDV HN globular domain (40).
The paramyxovirus F protein is a type I homotrimer (2, 27). It is synthesized as a precursor protein, Fo, the activation of which requires proteolytic cleavage into the disulfide-linked polypeptides F1 and F2. The hydrophobic amino terminus of F1 is the fusion peptide, which is inserted into the target cell membrane, thereby disordering the lipid bilayer and preparing it for merger of the membranes (19).
In addition to its receptor binding activity, HN contributes to fusion in a virus-specific manner. When expressed alone or with heterologous HN proteins, the F proteins of most paramyxoviruses cannot mediate membrane fusion; they require the coexpression of the homologous HN protein (19). This specificity is consistent with the existence of an interaction between HN and F mediated by specific domains on the two proteins (13). Coimmunoprecipitation (co-IP) studies both with (21, 31) and without (7, 20, 39) cross-linking agents support the existence of an interaction between the attachment and fusion proteins at the cell surface.
Chimeric HN proteins with segments derived from heterologous paramyxoviruses have been evaluated to identify the F-interactive domain on the HN protein. These studies have shown that F specificity for several paramyxoviruses segregates with the stalk of HN (6, 8, 33, 35, 37). The simplest explanation for this finding is that a domain in the HN stalk interacts specifically with a complementary domain in F, though this has not yet been established.
Examination of the HN stalk identifies a partially conserved motif that could mediate the HN-F interaction and that has enough sequence heterogeneity to account for its virus specificity. This is a stretch of almost 40 residues, 74 to 110 in NDV HN, which includes two conserved heptad repeats (HR1 and HR2) (32). Analyses of HN proteins carrying substitutions for the heptadic residues in each heptad repeat demonstrate that they are important for fusion but not for receptor recognition activity (32). However, the significance of their effect on fusion is tempered somewhat by the observation that most of these mutated HN proteins also exhibit decreased NA activity in the globular domain (32, 36).
Between HR1 and HR2 is a seven-amino-acid intervening region, defined by residues 89 to 95 (36). In this region, two residues are highly conserved among paramyxovirus HN proteins: P93, which is completely conserved, and L94, which is conserved in type. We have previously shown that an alanine substitution for residue P93, but not residue L94, diminishes NA activity (36). Herein, we explore the role in fusion of these two residues and others in the intervening region. Amino acid substitutions for P93 and L94 severely impair fusion, but those for residue L94 do so without a significant effect on any other HN function. Substitutions resulting in a phenotype similar to those for L94 were identified at positions 89 and 90. The decreased fusion seen with each of these mutated HN proteins correlates with a diminished ability to interact with the F protein in a cell surface coimmunoprecipitation assay. These constitute the first amino acid substitutions in any region of the HN protein that modulate fusion and the HN-F interaction with no detectable effect on HN structure or its other functions.
MATERIALS AND METHODS
Recombinant plasmids and site-directed mutagenesis.
Construction of the NDV-AV HN and F recombinant pBluescript SK(+) (Stratagene Cloning Systems, La Jolla, Calif.) expression vectors has been described previously (24). Site-directed mutagenesis was performed as described previously (4), using primers obtained from Integrated DNA Technologies, Inc. (Coralville, Iowa). Identification of mutants was facilitated by screening for the presence of a unique restriction enzyme site introduced by the mutagenic primer. The presence of the desired mutation was verified by sequencing of double-stranded DNA using the Sequenase plasmid sequencing kit (United States Biochemical, Cleveland, Ohio), according to the protocol provided by the company. Multiple clones were characterized for each substitution.
Transient expression system and quantitation of cell surface expression.
Wild-type (wt) and mutated HN proteins were expressed in BHK-21 cells (American Type Culture Collection, Manassas, Va.), using the vaccinia virus-T7 RNA polymerase expression system (10). All experiments, except the NA assays, were done in 35-mm plates seeded a day earlier at 4 × 105 cells per well. Maintenance of cells, infection with recombinant vaccinia virus vTF7-3, and transfection were performed as described previously (24), using 1 μg of each plasmid for transfection. Cell surface expression was quantitated by flow cytometry with a mixture of monoclonal antibodies (MAbs) to at least five different antigenic sites in the HN globular domain (14, 15, 17, 18).
HN functional assays.
The hemadsorption (HAd) activities of HN proteins were determined by the abilities of the expressed proteins to adsorb guinea pig erythrocytes (Crane Laboratories, Syracuse, N.Y.). HN-expressing monolayers were incubated for 30 min at 4°C with a 2% suspension of erythrocytes in phosphate-buffered saline supplemented with 1% each of CaCl2 and MgCl2. After extensive washing, adsorbed erythrocytes were lysed in 50 mM NH4Cl, and the lysate was clarified by centrifugation. HAd activity was quantified by measuring the absorbance at 540 nm and subtracting the background absorbance obtained with cells expressing vector alone.
The NA activity of cell surface HN was determined as described previously (4, 24). NA assays were performed on 22.6-mm plates seeded a day earlier at 1.6 × 105 cells per well and transfected with 0.5 μg of DNA. Monolayers were incubated at 37°C for 20 min with 625 μg of neuraminlactose (Sigma Chemical Co., St. Louis, Mo.)/ml in 0.5 ml of 0.1 M sodium acetate (pH 6). NA activity was quantitated by measuring the absorbance at 590 nm. The background absorbance obtained with vector-expressing cells was subtracted, and the data were corrected for differences in cell surface expression.
The abilities of the mutated HN proteins to complement the F protein in the promotion of fusion were quantitated by using a content mixing assay (4).
Immunoprecipitation assay.
The immunoprecipitation protocol has been described previously (16). Briefly, at 22 h posttransfection, BHK cells were starved for 1 h at 37°C in medium lacking cysteine and methionine. Cells were then labeled with 1 ml of medium containing 100 μCi of Expre35S35S labeling mix (Perkin-Elmer Life and Analytical Sciences, Boston, Mass.) for 3 h at 37°C and chased for 90 min with medium. The cells were lysed, and HN was immunoprecipitated with the same mixture of anti-HN MAbs used for flow cytometry. Immune complexes were collected by using Ultralink-Immobilized Protein A Plus (Pierce, Rockford, Ill.) and then analyzed by sodium dodecyl sulfate-polyacrylamide gel electrophoresis.
Co-IP assay.
The ability of wt and mutated HN proteins to interact with F at the surface of transfected BHK cells was assayed at 16 h posttransfection using the co-IP assay described previously (20). This assay involves biotinylation of cell surface proteins and immunoprecipitation of both HN and F with only an anti-F MAb. The HN-F interaction is quantitated by comparing the amount of HN that coimmunoprecipitates with F in this assay to the total amount of HN present at the cell surface. The F protein is immunoprecipitated in greater amounts from fusing monolayers than from nonfusing monolayers, possibly due to aggregation of F during formation of the fusion pore. Thus, a cleavage site mutant form of F that does not fuse and that interacts efficiently with the HN protein was used (7).
RESULTS
Point mutations in the intervening region of the HN stalk.
Examination of an alignment of the heptad repeats in the stalk domains of several paramyxovirus HN proteins reveals that HR2, but not HR1, is highly conserved (Fig. 1). In addition, the alignment shows the conservation of the P-L(I,V) doublet in the intervening region between the repeats. In NDV-AV HN, the intervening region includes residues A89, L90, E91, S92, P93, L94, and A95. Initially, based on the conservation of P93 and L94, the role of these two residues in fusion was examined by the introduction of an individual alanine substitution at each position. Subsequently, additional amino acid substitutions were introduced at both positions, such that mutated HN proteins P93A, -L, and -S, as well as L94A, -G, -I, -P, and -R, were constructed. The remaining residues in the intervening region were also subjected to mutational analyses. They were initially changed to the corresponding amino acid in hPIV type 3 (hPIV3) HN, based on the assumption that these substitutions would be more likely to be tolerated. Subsequently, additional amino acid substitutions were introduced at each position. Thus, to investigate the role of the intervening region in HN, proteins carrying the following substitutions were prepared and characterized: A89I and -Q; L90A, -I, and -N; E91A and -Y; S92A, -L, and -R; P93A, -L, and -S; L94A, -G, -I, -P, and -R; and A95R and -S.
FIG. 1.
Comparison of the amino acid sequences in a conserved motif in the stalk of several paramyxovirus HN proteins. The conserved P-L (I,V) doublet in the intervening region is in the box. HR1 and HR2 are indicated. The sequences are as follows: NDV (22); hPIV3 (9); mumps virus (MuV) (38); simian virus 5 (SV5) (12); and Sendai virus (SV) (1).
Amino acid substitutions for P93 modulate fusion and the HN-F interaction but also result in a marked decrease in NA activity.
The abilities of the P93-mutated HN proteins to complement the homologous F protein in the promotion of fusion were quantitated with the content mixing assay (Fig. 2). All three of the P93-mutated HN proteins exhibit significantly reduced fusion promotion activity. HN proteins carrying P93A, P93L, and P93S substitutions promote fusion at only 19.6, 9.2, and 13.4% of wt HN activity, respectively, when coexpressed with the F protein. Figure 3 shows the extent of syncytium formation in monolayers of cells in which the weakly fusogenic proteins P93A-HN and P93L-HN are coexpressed with NDV F and stained for fusion. In contrast to vector-expressing cells, small syncytia can be seen in both monolayers. In each case, the syncytia are not only smaller but also fewer than those in cells expressing wt HN and F.
FIG. 2.
Cell surface expression and functional assays of the P93-mutated HN proteins. The cell surface expression (CSE) is determined by flow cytometry using a cocktail of at least five anti-HN MAbs specific for different antigenic sites on the globular domain of the protein. The HAd activity is determined by the ability of the expressed HN proteins to adsorb guinea pig erythrocytes. NA activity is determined by the ability of the expressed proteins to catalyze the release of sialic acid from neuraminlactose. NA data are corrected for differences in expression. The ability to complement wt NDV-AV F in the promotion of fusion is determined by the content mixing assay. For all four of these assays, the background level obtained with vector alone is subtracted. All data are expressed relative to the amount for the wt protein and represent the mean of at least four independent determinations.
FIG. 3.
Syncytium formation in monolayers coexpressing intervening region mutated HN proteins and the F protein. The extent of syncytium formation is shown in monolayers expressing wt F with the following: vector control, wt HN, P93A-HN, P93L-HN, L94A-HN, L94G-HN, L94P-HN, A89Q-HN, and L90N-HN. At 22 h posttransfection, the monolayers are fixed with methanol and stained with Giemsa. The arrows indicate small syncytia.
To determine whether reduced fusogenic activity correlates with diminished capacity to interact with the F protein, poorly fusogenic mutated HN proteins were assayed for their ability to be coimmunoprecipitated with the F protein by an anti-F MAb. Figure 4 shows the co-IP results for each of the P93-mutated HN proteins. Immunoprecipitation of HN and F from cells coexpressing the two proteins shows the maximum amounts of the two proteins that can be immunoprecipitated from the cell surface for each sample. Wt HN is coimmunoprecipitated efficiently with F (21.2% ± 3.1% of the total cell surface HN), especially considering that the HN-F complex may be transient and triggered by HN′s interaction with receptors. However, none of the P93-mutated HN proteins is coimmunoprecipitated with F in significant amounts. P93A-HN coimmunoprecipitates less than 2% (1.9% ± 0.9%) of the wt HN amount, and P93L-HN and P93S-HN are not detectable in coimmunoprecipitates. Thus, the weak fusogenic activity of P93-mutated proteins correlates with a loss of the ability to interact with the F protein, at least in amounts detectable by the co-IP assay.
FIG. 4.
Co-IP of NDV-AV HN and P93-mutated HNs with the homologous F protein. Equal numbers of cells were transfected as indicated. The cell surface proteins were biotinylated, and the cells were lysed by treatment with dodecylmaltoside. The lysate was split into two equal aliquots and immunoprecipitated with either a combination of an anti-F MAb and a cocktail of anti-HN MAbs (the first lane in each pair) or an anti-F MAb alone (the second lane in each pair). The percentage of the total amount of each P93-mutated HN protein at the cell surface that is coimmunoprecipitated with anti-F MAb is expressed relative to that of the wt HN protein. The data are means of three independent determinations. The percent wt fusion data shown are the results from the content mixing assay shown in Fig. 2. ND, none detected.
Figure 4 also shows critical controls for the experiment. The HN protein is not precipitated by the anti-F MAb in the absence of F, indicating that the co-IP occurs through its interaction with F. In addition, the protein that is coimmunoprecipitated with F is not present in cells that are not expressing HN. We have previously confirmed that the coimmunoprecipitated protein is authentic HN by sodium dodecyl sulfate-polyacrylamide gel electrophoresis under nonreducing conditions (20).
To determine the basis for the lack of fusion promotion activity by these mutated HN proteins, we have characterized their antigenic structure and function. The cell surface expression of the P93-mutated HN proteins was determined by flow cytometry with a cocktail of five different HN-specific MAbs. As shown in Fig. 2, the expression level of each of the P93-mutated HN proteins is comparable to the wt level. This is not surprising based on the ability to efficiently immunoprecipitate each of these proteins from the cell surface with the mixture of HN MAbs in the co-IP assay (Fig. 4). The mutated proteins are expressed at more than 90% of the wt level with the exception of P93L-HN, which is still expressed efficiently (72.1% of the wt level). In addition to flow cytometry, immunoprecipitation was used to confirm the expression levels of these proteins. All of the P93-mutated HN proteins were efficiently precipitated with the cocktail of HN-specific MAbs (data not shown). Since the HN MAbs used in both assays are conformation specific, this confirms that all of the P93-mutated proteins are efficiently expressed and strongly suggests that they are not misfolded.
The receptor recognition properties of the P93-mutated HN proteins were evaluated by assaying their ability to adsorb guinea pig erythrocytes at 4°C (Fig. 2). All of the P93-mutated HN proteins hemadsorb at a level comparable to that of wt HN, ranging from 84.0 to 108.3%. Even though P93L-mutated HN is expressed at 72.1% of the wt level, it is still able to hemadsorb at 88.7% of the wt level. Thus, the receptor recognition activity of HN is only minimally affected by substitutions for residue P93. This is consistent with our previous data for P93A-HN and P93L-HN (36). Moreover, the HAd activity of these mutated HN proteins is also similar to that of wt HN at 37°C, unlike another set of dimer interface mutants members of our group have previously described (4). Therefore, the fusion deficiency of these mutated proteins is not due to an effect on receptor recognition.
However, evaluation of the NA activities of the P93-mutated proteins gives a very different result. The NA activity of each of the P93-mutated HN proteins expressed at the cell surface was determined at 37°C (Fig. 2). All of the P93-mutated proteins show a drastic reduction in NA activity. HN carrying a P93A, P93L, or P93S substitution exhibits levels of NA that are 17.3, 4.0, and 34.7% of the wt amount, respectively. Since NA activity resides in the terminal globular domain of HN, an effect on this activity induced by a point mutation in the stalk raises the possibility that these mutations may alter the structure of the protein.
Amino acid substitutions for residue L94 modulate fusion and the HN-F interaction with no detectable effect on receptor recognition or NA activity.
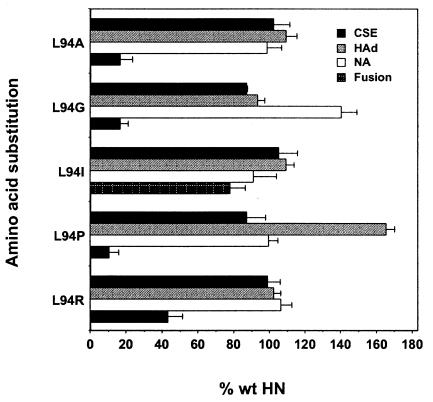
The abilities of the L94-mutated HN proteins to complement the homologous F protein in fusion promotion were quantitated with the content mixing assay (Fig. 5). Several different substitutions for residue L94 markedly reduce fusion. When L94 is replaced with alanine, glycine, or proline, the fusion promotion activity of HN is reduced to 10 to 17% of the wt level. An arginine substitution for L94 reduces fusion promotion to slightly less than 50% of the wt level. Only when L94 is replaced with isoleucine does fusion approach the wt level, consistent with the conservative nature of this substitution. The weak fusion promotion activities of L94A-HN and L94P-HN and the moderate activity of L94R-HN are visible in the stained monolayers coexpressing each of these mutated proteins with wt F (Fig. 3).
FIG. 5.
Cell surface expression and functional assays of the L94-mutated HN proteins. The assays are performed as described in the legend to Fig. 2. Data represent the means of at least four independent determinations.
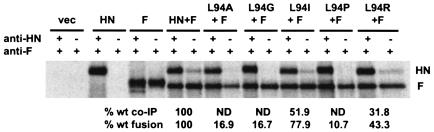
To determine whether decreased fusogenic activity correlates with a diminished ability of these L94-mutated proteins to interact with F, the amount of each mutated HN protein coimmunoprecipitated with F by an anti-F MAb was determined (Fig. 6). Wt HN is efficiently coimmunoprecipitated with F (23.7% ± 3.3% of the total cell surface HN). This is consistent with previous data, which showed that 32.6% ± 7.6% of the total amount of wt HN at the cell surface is coimmunoprecipitated with F (20). As seen with the P93-mutated proteins, no co-IP of HN carrying mutations L94A, L94G, or L94P can be detected, correlating with the extremely weak fusogenic activity of these proteins (Fig. 5). This is consistent with the co-IP assay requiring at least 20% of wt fusion to detect an interaction between HN and F. Furthermore, L94-mutated proteins that promote significant, though reduced, levels of fusion can be coimmunoprecipitated with antibody to the F protein. Thus, L94R-HN, which complements F almost 50% compared to wt HN, can be coimmunoprecipitated 31.8% ± 4.4% of the wt HN amount. Similarly, L94I-HN, which fuses approximately 80% as well as wt, can be coimmunoprecipitated the most efficiently of all the L94-mutated proteins (51.9% ± 5.1% of wt level).
FIG. 6.
Co-IP of NDV-AV HN and L94-mutated HNs with the homologous F protein. The experiment is performed and data are expressed as described in the legend to Fig. 4. The percentage of the total amount of each L94-mutated HN protein at the cell surface that is coimmunoprecipitated with anti-F MAb is expressed relative to that of the wt HN protein. The data are means of three independent determinations. The percent wt fusion data shown are the results from the content mixing assay shown in Fig. 5. ND, none detected.
The cell surface expression of the L94-mutated HN proteins was determined by flow cytometry. As shown in Fig. 5, the expression level of each of the L94-mutated HN proteins is comparable to the wt level. All of these mutated proteins are expressed at least 85% of wt HN. Immunoprecipitation studies also confirm the near-wt levels of expression for these mutated proteins, as well as their proper folding (data not shown).
The receptor recognition activities of the L94-mutated HN proteins were evaluated to determine if the observed effects on fusion and the HN-F interaction correlate with a deficiency in attachment. All L94-mutated HN proteins hemadsorb in amounts comparable to that for the wt protein, with the exception of L94P-HN, which shows a 65% increase in HAd over the wt level (Fig. 5). Interestingly, despite such significantly elevated HAd activity, this mutated protein still promotes fusion at only about 10% of the wt level.
HN proteins carrying substitutions for residue L94 exhibit wt levels of NA activity, with one exception, L94G-HN, which exhibits a 40% increase in NA activity. Thus, the fusion deficiency and failure to interact with F exhibited by L94A-HN, L94G-HN, and L94P-HN is not accompanied by a decrease in NA activity. This is in sharp contrast to the P93-mutated proteins, for which deficiencies in fusion and the HN-F interaction correlate with a deficiency in NA.
HN carrying an A89Q or L90N mutation is also deficient in fusion and the HN-F interaction but exhibits no significant effect on any other HN function.
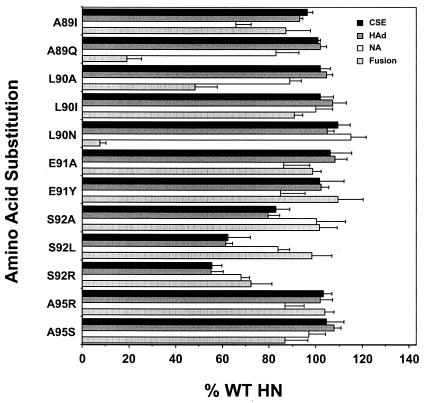
Based on the demonstrated specific effects on fusion and the HN-F interaction induced by the substitutions at residue L94, we performed a site-directed mutational analysis of the role in fusion of nonconserved residues in the intervening region by the introduction of substitutions A89I and -Q; L90A, -I, and -N; E91A and -Y; S92A, -L, and -R; and A95R and -S. The abilities of these mutated HN proteins to complement the homologous F protein in fusion promotion were determined (Fig. 7). Certain amino acid substitutions for residues A89 and L90 markedly reduce the fusion promotion activity of HN. A89Q-HN and L90N-HN promote only 19.1 and 7.7% of wt fusion, respectively. The extent of fusion promotion in monolayers coexpressing these mutated HN proteins with the F protein is shown in Fig. 3. A substitution of alanine for residue L90 also modulates fusion, but much less significantly, resulting in approximately 50% of wt fusion. Substitutions of isoleucine for residues A89 and L90 have no significant effect on fusion, resulting in 87.1 and 91.8% of wt fusion, respectively (Fig. 7). Similarly, all substitutions introduced for residues E91, S92, and A95 have no effect on fusion, with the exception of the S92R substitution, which reduces fusion to 72.2% of the wt level (Fig. 7).
FIG. 7.
Cell surface expression and the functional assays of HN proteins carrying substitutions for the remaining residues in the intervening region. Assays are performed as described in the legend to Fig. 2. The data represent the means of at least four independent determinations.
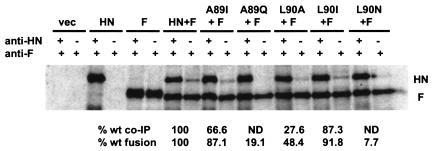
Figure 8 shows the co-IP results for each of the A89- and L90-mutated HN proteins. Wt HN is coimmunoprecipitated efficiently with F (22.3% ± 0.8% of the total cell surface HN). There is no detectable interaction of either A89Q-HN or L90N-HN with F in this assay, consistent with their less than 20% level of fusion. However, L90A-HN, which promotes 48.4% of wt fusion with F, can be coimmunoprecipitated with the anti-F MAb. In fact, 27.6% ± 1.3% of the total L90A-HN coexpressed with F at the cell surface can be coimmunoprecipitated with the anti-F MAb (Fig. 8). As positive controls, the highly fusogenic proteins A89I-HN and L90I-HN are also efficiently coimmunoprecipitated, 66.6% ± 6.7% and 87.3% ± 1.9% of the wt amount, respectively. Thus, the loss of fusogenic activity induced by substitutions for residues A89 and L90 correlates with a modulation of the abilities of these mutated HN proteins to interact with the F protein at the cell surface.
FIG. 8.
Co-IP of NDV-AV HN and A89- and L90-mutated HNs with the homologous F protein. The experiment was performed and data are expressed as described in the legend to Fig. 4. The percentage of the total amount of each A89- and L90-mutated HN protein at the cell surface that is coimmunoprecipitated with anti-F MAb is expressed relative to that of the wt HN protein. The data are means of two independent determinations. The percent wt fusion data shown are the results from the content mixing assay shown in Fig. 7. ND, none detected.
Again, we have explored the possibility that the fusion and F interaction defects in these mutated HN proteins are related to a modulation of their cell surface expression and/or functional properties (Fig. 7). The weakly fusogenic proteins, carrying A89Q, L90A, or L90N substitutions, are all expressed slightly more efficiently than wt HN. The receptor recognition and NA activities of these mutated proteins are also shown in Fig. 7. Similar to the findings obtained with the L94-mutated, poorly fusogenic proteins, HN carrying the A89Q, L90A, or L90N substitution exhibits near-wt receptor recognition and NA activities. HAd by each of these mutated proteins is slightly greater than the wt level, and NA activity ranges from 82.8 to 115.1% of the wt level. Thus, it is extremely unlikely that the fusion-related defects in these mutated proteins are the result of an alteration in either cell surface expression, receptor recognition, or NA activity. Both A89I-HN and L90I-HN exhibit wt levels of expression and HAd activity, yet A89I-HN has decreased NA activity.
Interestingly, all of the S92-mutated HN proteins exhibit a reduction in cell surface expression, especially the proteins carrying the S92L and S92R substitutions, which are expressed at only 62.6 and 55.4% of the wt HN level, respectively (Fig. 7). This lower level of expression may account for the slight deficiency in receptor recognition activity exhibited by these mutated proteins, which ranges from 55.2 to 79.4% of the wt level. These expression and binding defects may be responsible for the slight reduction in fusion (72.2% of wt level) exhibited by S92R-HN. However, S92L-HN still promotes 98.3% of wt fusion, despite slightly reduced expression. Among the S92-mutated proteins, only an arginine at this position results in a significant reduction in specific NA activity (68.1% of wt level).
The E91- and A95-mutated HN proteins are expressed at slightly more than the wt level, exhibit slightly greater-than-wt HAd activity, and have NA activities similar to that of the wt. These properties are all consistent with their near-wt levels of fusion (Fig. 7).
DISCUSSION
The virus-specific nature of the HN-F interaction suggests the existence of one or more domains on the NDV HN and F proteins that mediate a specific interaction between the two proteins. Analysis of the fusion-promoting activity of NDV-hPIV3 HN chimeras has demonstrated that the specificity of HN for its homologous F protein segregates with the stalk of HN (6, 8, 37). However, the actual determinants of the HN-F interaction on either protein have not been firmly established.
We previously identified two highly conserved amino acids, P93 and L94, in the intervening region between the two heptad repeats in the stalk of NDV HN (36). Here, we have investigated the effects of amino acid substitutions for these residues on fusion. The introduction of any of three different substitutions, alanine, leucine, or serine, for residue P93 results in a significant reduction in fusion promotion and the ability of HN to interact with the F protein at the cell surface. This phenotype is not due to changes in either expression or receptor recognition activity, both of which are comparable to those of wt HN. However, consistent with our earlier findings (36), substitutions for P93 do impair NA activity. The replacement of P93 may induce a subtle structural change in the HN stalk that is transmitted to its globular domain, thus affecting the NA active site.
The effect of substitutions for P93 on NA activity calls into question the basis for their effect on fusion. These substitutions could indirectly affect HN′s fusion promotion function by altering the transmission of a signal from the receptor recognition site in the globular domain to the F-specific region in the stalk, thereby abrogating the HN-F interaction. P93-mutated proteins have the same phenotype as proteins carrying substitutions at the heptadic positions, although the effect of the latter on the HN-F interaction was not determined (32, 36).
Our results for P93-mutated NDV HN are in agreement with the phenotype of a mutated protein carrying a serine substitution at the corresponding residue (P111) in hPIV3 HN. This substitution, initially identified in a NA-deficient hPIV3 variant virus (25), induces an F-triggering defect, despite wt receptor recognition activity and avidity for receptors (26). However, F insertion into the target membrane progressed to fusion more slowly with P111S-HN than with the wt HN protein. Based on this finding, it was proposed that this triggering-defective protein may have a diminished capacity to interact with F. It may be that the P111S-HN protein requires a longer period of contact with its cellular receptor in order to induce the conformational change in HN required for the conversion of F to its fusion-ready form. Perhaps P93-mutated NDV HN proteins also have a F-triggering defect, resulting in the weak fusion detected in the content mixing assay. Consistent with this, P93-mutated NDV HN proteins are unable to coimmunoprecipitate with F, suggesting that these substitutions interfere with their ability to interact with F.
A surprising result is the 65% increase in HAd activity over the wt level exhibited by L94P-HN. This is the only substitution introduced at this position or anywhere else in this domain that gives this phenotype. Evidently, the presence of the two prolines in succession in the stalk somehow stabilizes the receptor binding activity of HN, possibly by providing added rigidity to the peptide backbone.
Although substitutions for P93 appear to alter the structure of HN, some substitutions for the neighboring residue L94, as well as A89 and L90, modulate fusion and the HN-F interaction with no detectable effect on the other HN functions. When residue L94 is replaced with alanine, glycine, or proline, the mutated HN proteins display a significant reduction in fusion promotion activity while maintaining wt levels of NA and receptor recognition activities. These L94 substitutions also cause loss of the ability of HN to be coimmunoprecipitated by the anti-F MAb. Interestingly, L94R HN promotes a significant level of fusion, approximately 50% of the wt level. Consistent with this, we were able to detect an interaction between this protein and F. The L94I substitution has a minimal effect on fusion and the HN-F interaction, consistent with the conservative nature of this substitution. Thus, substitutions for L94 appear to directly affect fusion, since they have no significant effect on any other HN function. They may modulate fusion via a direct effect on the ability of HN to interact with the F protein. Apparently, the size of the residue at this position carries some importance to the HN-F interaction and fusion. A larger, though charged amino acid, such as arginine, is more functionally conservative than the smaller glycine or alanine.
Similarly, A89Q-HN and L90N-HN have the same phenotype as the weakly fusogenic L94-mutated proteins. They modulate fusion promotion and the HN-F interaction with no significant effect on NA or receptor recognition activity. Thus, these substitutions also appear to directly affect fusion promotion and the HN-F interaction.
Our data indicate that the co-IP assay can detect quantitative differences in the HN-F interaction at different levels of fusion. Mutated proteins that exhibit a significant level of fusion can be shown to interact with the F protein in the assay. For example, L94I-HN and L94R-HN, which fuse at 77.9 and 43.3% of the wt level, respectively, coimmunoprecipitate with an anti-F MAb at 51.9 and 31.8% of the wt level, respectively. Similarly, L90A-HN, which fuses at almost 50% of the wt level, is coimmunoprecipitated with an anti-F MAb at 27.6% of the wt amount. A89I-HN and L90I-HN coimmunoprecipitate 66.6 and 87.3% of the wt amount, consistent with their levels of fusion of approximately 90% of the wt level.
Thus, the co-IP assay can detect differences in the HN-F interaction that are consistent with differences in fusion promotion. However, the amount of co-IP of HN is not directly proportional to the extent of fusion. This is very likely a reflection of the limits of the co-IP assay. Of the P93-mutated HN proteins, an interaction can be detected, albeit at less than 2%, only with P93A-HN. This protein fuses at almost 20% of the wt level. An interaction between the other two P93-mutated HN proteins and F was not detected, presumably because these proteins promote fusion below 14% of the wt level. Similarly, L94A-HN, L94G-HN, and L94P-HN do not coimmunoprecipitate with F, consistent with their low fusion-promoting activities. Also, neither A89Q-HN nor L90N-HN, both of which fuse at less than 20% of the wt level, interacts with F in the co-IP assay. Thus, taken together, these data suggest that the limit of detection of the HN-F interaction coincides with a level of fusion of approximately 20% of the wt level. The one notable exception to this is I175E-HN, which is capable of interacting with F, despite a lack of receptor recognition activity (20). Thus, the co-IP assay has proven extremely useful in evaluating the relative effects of amino acid substitutions in both HN and F on the ability of the two proteins to interact with each other in the promotion of fusion.
Our results identify the intervening region in the NDV HN stalk as being critical to its ability to complement F in the promotion of fusion. These findings are consistent with NDV-hPIV3 HN chimera studies, which assign the F-specific region of NDV HN to its stalk (8). However, these findings are inconsistent with a recent peptide-based study, which asserts that residues 124 to 152 of HN directly mediate its interaction with the homologous F protein (11). It is suggested that this span of residues in the globular head of HN directly interacts with a heptad repeat domain in NDV F situated just outside the transmembrane. Point mutations in this domain were evaluated for their effect on HN function. While substitutions for residues I133 and L140 do result in a 70 to 80% reduction in fusion, they also impair NA activity by as much as 60% and attachment by as much as 50% (11). Thus, these mutated HN proteins are analogous to our P93-mutated HNs. The effect of the I133 and L140 substitutions on fusion may be a reflection of structural changes in HN rather than a direct effect on its interaction with F. Also, the effect of these two substitutions on the HN-F interaction was not tested. In addition, the peptide data contrast with the fact that several chimeras with NDV residues 124 to 152 intact not only fail to fuse with NDV F but fuse very efficiently with hPIV3 F (37).
In summary, the substitutions we have introduced at positions 89, 90, and 94 are the only ones for which a correlation has been demonstrated between fusion deficiency and decreased ability of HN to interact with F, with no other detectable negative effect on HN′s structure or function. But it still remains to be determined how these substitutions modulate the HN-F interaction. The intervening region may directly mediate an interaction with a complementary domain in the stalk region of the F protein. P93 may introduce a kink in the helix, so that the side chains of the surrounding amino acids can protrude away from the backbone in a bulge-like structure. Alternatively, we cannot strictly rule out the possibility that the domain does not make direct contact with F but may be critical to the conversion of HN to its F-interactive form.
Acknowledgments
We gratefully acknowledge Judith Alamares, Elizabeth Corey, Jianrong Li, Paul Mahon, Anne Mirza, Celia Schiffer, and Rajas Warke for critical reading of the manuscript and helpful discussions. We also thank Robert Lamb and Trudy Morrison for the NDV F and HN genes, respectively, and Bernard Moss for the recombinant vaccinia virus.
This work was made possible by grant AI-49268 from the National Institutes of Health.
REFERENCES
- 1.Blumberg, B., C. Giorgi, L. Roux, R. Raju, P. Dowling, A. Chollet, and D. Kolakofsky. 1985. Sequence determination of the Sendai virus HN gene and its comparison to the influenza virus glycoproteins. Cell 41:269-278. [DOI] [PubMed] [Google Scholar]
- 2.Chen, L., P. M. Colman, L. J. Cosgrove, M. C. Lawrence, L. J. Lawrence, P. A. Tulloch, and J. J. Gorman. 2001. Cloning, expression, and crystallization of the fusion protein of Newcastle disease virus. Virology 290:290-299. [DOI] [PubMed] [Google Scholar]
- 3.Connaris, H., T. Takimoto, R. Russell, S. Crennell, I. Moustafa, A. Portner, and G. Taylor. 2002. Probing the sialic acid binding site of the hemagglutinin-neuraminidase of Newcastle disease virus: identification of key amino acids involved in cell binding, catalysis, and fusion. J. Virol. 76:1816-1824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Corey, E. A., A. M. Mirza, E. Levandowsky, and R. M. Iorio. 2003. Fusion deficiency induced by mutations at the dimer interface in the Newcastle disease virus hemagglutinin-neuraminidase is due to a temperature-dependent defect in receptor binding. J. Virol. 77:6913-6922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Crennell, S., T. Takimoto, A. Portner, and G. Taylor. 2000. Crystal structure of the multifunctional paramyxovirus hemagglutinin-neuraminidase. Nat. Struct. Biol. 7:1068-1074. [DOI] [PubMed] [Google Scholar]
- 6.Deng, R., A. M. Mirza, P. J. Mahon, and R. M. Iorio. 1997. Functional chimeric HN glycoproteins derived from Newcastle disease virus and human parainfluenza virus-3. Arch. Virol. Suppl. 13:115-130. [DOI] [PubMed] [Google Scholar]
- 7.Deng, R., Z. Wang, P. J. Mahon, M. Marinello, A. M. Mirza, and R. M. Iorio. 1999. Mutations in the NDV HN protein that interfere with its ability to interact with the homologous F protein in the promotion of fusion. Virology 253:43-54. [DOI] [PubMed] [Google Scholar]
- 8.Deng, R., Z. Wang, A. M. Mirza, and R. M. Iorio. 1995. Localization of a domain on the paramyxovirus attachment protein required for the promotion of cellular fusion by its homologous fusion protein spike. Virology 209:457-469. [DOI] [PubMed] [Google Scholar]
- 9.Elango, N., J. E. Coligan, R. C. Jambou, and S. Venkatesan. 1986. Human parainfluenza type 3 virus hemagglutinin-neuraminidase glycoprotein: nucleotide sequence of mRNA and limited amino acid sequence of CNBr peptides of the purified protein. J. Virol. 57:481-489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fuerst, T. R., E. G. Niles, F. W. Studier, and B. Moss. 1986. Eucaryotic transient expression system based on recombinant vaccinia virus that synthesizes bacteriophage T7 RNA polymerase. Proc. Natl. Acad. Sci. USA 83:8122-8126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gravel, K., and T. G. Morrison. 2003. Interacting domains of the HN and F proteins of Newcastle disease virus. J. Virol. 77:11040-11049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hiebert, S. W., R. G. Paterson, and R. A. Lamb. 1985. Hemagglutinin-neuraminidase protein of simian virus 5: nucleotide sequence of the mRNA predicts an N-terminal anchor. J. Virol. 54:1-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hu, X., R. Ray, and R. W. Compans. 1992. Functional interactions between the fusion protein and hemagglutinin-neuraminidase of human parainfluenza viruses. J. Virol. 66:1528-1534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Iorio, R. M., J. B. Borgman, R. L. Glickman, and M. A. Bratt. 1986. Genetic variation within a neutralizing domain on the haemagglutinin-neuraminidase glycoprotein of Newcastle disease virus. J. Gen. Virol. 67:1393-1403. [DOI] [PubMed] [Google Scholar]
- 15.Iorio, R. M., and M. A. Bratt. 1983. Monoclonal antibodies to Newcastle disease virus: delineation of four epitopes on the HN glycoprotein. J. Virol. 48:440-450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Iorio, R. M., G. M. Field, J. M. Sauvron, A. M. Mirza, R. Deng, P. J. Mahon, and J. Langedijk. 2001. Structural and functional relationship between the receptor recognition and neuraminidase activities of the Newcastle disease virus hemagglutinin-neuraminidase protein: receptor recognition is dependent on neuraminidase activity. J. Virol. 75:1918-1927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Iorio, R. M., R. L. Glickman, A. M. Riel, J. P. Sheehan, and M. A. Bratt. 1989. Functional and neutralization profile of seven overlapping antigenic sites on the HN glycoprotein of Newcastle disease virus: monoclonal antibodies to some sites prevent viral attachment. Virus Res. 13:245-262. [DOI] [PubMed] [Google Scholar]
- 18.Iorio, R. M., R. J. Syddall, J. P. Sheehan, M. A. Bratt, R. L. Glickman, and A. M. Riel. 1991. Neutralization map of the HN glycoprotein of Newcastle disease virus: domains recognized by monoclonal antibodies that prevent receptor recognition. J. Virol. 65:4999-5006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lamb, R. A., and D. Kolakofsky. 2001. Paramyxoviridae: the viruses and their replication, p. 689-724. In D. M. Knipe and P. M. Howley (ed.), Fundamental virology, 4th ed. Lippincott Williams & Wilkins, Philadelphia, Pa.
- 20.Li, J., E. Quinlan, A. Mirza, and R. M. Iorio. 2004. Mutated form of the Newcastle disease virus hemagglutinin-neuraminidase interacts with the homologous fusion protein despite deficiencies in both receptor recognition and fusion promotion. J. Virol. 78:5299-5310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Malvoisin, E., and T. F. Wild. 1993. Measles virus glycoproteins: studies on the structure and interaction of the haemagglutinin and fusion proteins. J. Gen. Virol. 74:2365-2372. [DOI] [PubMed] [Google Scholar]
- 22.McGinnes, L. W., A. Wilde, and T. G. Morrison. 1987. Nucleotide sequence of the gene encoding the Newcastle disease virus hemagglutinin-neuraminidase protein and comparisons of paramyxovirus hemagglutinin-neuraminidase protein sequences. Virus Res. 7:187-202. [DOI] [PubMed] [Google Scholar]
- 23.Mirza, A. M., J. P. Sheehan, L. W. Hardy, R. L. Glickman, and R. M. Iorio. 1993. Structure and function of a membrane anchor-less form of the hemagglutinin-neuraminidase glycoprotein of Newcastle disease virus. J. Biol. Chem. 268:21425-21431. [PubMed] [Google Scholar]
- 24.Mirza, A. M., R. Deng, and R. M. Iorio. 1994. Site-directed mutagenesis of a conserved hexapeptide in the paramyxovirus hemagglutinin-neuraminidase glycoprotein: effects on antigenic structure and function. J. Virol. 68:5093-5099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Porotto, M., O. Greengard, N. Poltoratskaia, M. A. Horga, and A. Moscona. 2001. Human parainfluenza virus type 3 HN-receptor interaction: effect of 4-guanidino-Neu5Ac2en on a neuraminidase-deficient variant. J. Virol. 75:7481-7488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Porotto, M., M. Murrell, O. Greengard, and A. Moscona. 2003. Triggering of human parainfluenza virus 3 fusion protein (F) by the hemagglutinin-neuraminidase (HN) protein: an HN mutation diminishes the rate of F activation and fusion. J. Virol. 77:3647-3654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Russell, R., R. G. Paterson, and R. A. Lamb. 1994. Studies with cross-linking reagents on the oligomeric form of the paramyxovirus fusion protein. Virology 199:160-168. [DOI] [PubMed] [Google Scholar]
- 28.Scheid, A., and P. W. Choppin. 1973. Isolation and purification of the envelope proteins of Newcastle disease virus. J. Virol. 11:263-271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Scheid, A., and P. W. Choppin. 1974. Identification and biological activities of paramyxovirus glycoproteins. Activation of cell fusion, hemolysis and infectivity by proteolytic cleavage of an inactive precursor protein of Sendai virus. Virology 57:475-490. [DOI] [PubMed] [Google Scholar]
- 30.Sheehan, J. P., R. M. Iorio, R. J. Syddall, R. L. Glickman, and M. A. Bratt. 1987. Reducing agent-sensitive dimerization of the hemagglutinin-neuraminidase glycoprotein of Newcastle disease virus correlates with the presence of cysteine at residue 123. Virology 161:603-606. [DOI] [PubMed] [Google Scholar]
- 31.Stone-Hulslander, J., and T. G. Morrison. 1997. Detection of an interaction between the HN and F proteins in Newcastle disease virus-infected cells. J. Virol. 71:6287-6295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Stone-Hulslander, J., and T. G. Morrison. 1999. Mutational analysis of heptad repeats in the membrane-proximal region of Newcastle disease virus HN protein. J. Virol. 73:3630-3637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Tanabayashi, K., and R. W. Compans. 1996. Functional interaction of paramyxovirus glycoproteins: identification of a domain in Sendai virus HN which promotes cell fusion. J. Virol. 70:6112-6118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Thompson, S. D., W. G. Laver, K. B. Murti, and A. Portner. 1988. Isolation of a biologically active soluble form of the hemagglutinin-neuraminidase protein of Sendai virus. J. Virol. 62:4653-4660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Tsurodome, M., M. Kawano, T. Yuasa, N. Tabata, M. Nishio, H. Komada, and Y. Ito. 1995. Identification of regions on the hemagglutinin-neuraminidase protein of human parainfluenza virus type 2 important for promoting cell fusion. Virology 213:190-203. [DOI] [PubMed] [Google Scholar]
- 36.Wang, Z., and R. M. Iorio. 1999. Amino acid substitutions in a conserved region in the stalk of the Newcastle disease virus HN glycoprotein spike impair its neuraminidase activity in the globular domain. J. Gen. Virol. 80:749-753. [DOI] [PubMed] [Google Scholar]
- 37.Wang, Z., A. M. Mirza, J. Li, P. J. Mahon, and R. M. Iorio. 2004. An oligosaccharide at the C-terminus of the F-specific domain in the stalk of the human parainfluenza virus 3 hemagglutinin-neuraminidase modulates fusion. Virus Res. 99:177-185. [DOI] [PubMed] [Google Scholar]
- 38.Waxham, M. N., J. Aronowski, A. C. Server, J. A. Smith, J. S. Wolinsky, and H. M. Goodman. 1988. Sequence determination of the mumps virus HN gene. Virology 164:318-325. [DOI] [PubMed] [Google Scholar]
- 39.Yao, Q., X. Hu, and R. W. Compans. 1997. Association of the parainfluenza virus fusion and hemagglutinin-neuraminidase glycoproteins on cell surfaces. J. Virol. 71:650-656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zaitsev, V., M. von Itzstein, D. Groves, M. Kiefel, T. Takimoto, A. Portner, and G. Taylor. 2004. Second sialic acid binding site in Newcastle disease virus hemagglutinin-neuraminidase: implications for fusion. J. Virol. 78:3733-3741. [DOI] [PMC free article] [PubMed] [Google Scholar]