Abstract
The initiation of reverse transcription and nucleocapsid assembly in hepatitis B virus (HBV) depends on the specific recognition of an RNA signal (the packaging signal, ɛ) on the pregenomic RNA (pgRNA) by the viral reverse transcriptase (RT). RT-ɛ interaction in the duck hepatitis B virus (DHBV) was recently shown to require the molecular chaperone complex, the heat shock protein 90 (Hsp90). However, the requirement for RT-ɛ interaction in the human HBV has remained unknown due to the inability to obtain a purified RT protein active in specific ɛ binding. We now report that Hsp90 is also required for HBV RT-ɛ interaction. Inhibition of Hsp90 led to diminished HBV pgRNA packaging into nucleocapsids in cells, which depends on RT-ɛ interaction. Furthermore, using truncated HBV RT proteins purified from bacteria and five purified Hsp90 chaperone factors, we have developed an in vitro RT-ɛ binding assay. Our results demonstrate that Hsp90, in a dynamic process that was dependent on ATP hydrolysis, facilitated RT-ɛ interaction in HBV, as in DHBV. Specific ɛ binding required sequences from both the amino-terminal terminal protein and the carboxy-terminal RT domain. Only the cognate HBV ɛ, but not the DHBV ɛ, could bind the HBV RT proteins. Furthermore, the internal bulge, but not the apical loop, of ɛ was required for RT binding. The establishment of a defined in vitro reconstitution system has now paved the way for future biochemical and structural studies to elucidate the mechanisms of RT-ɛ interaction and chaperone activation.
Hepatitis B virus (HBV) infection is a major global public health problem with over 300 million chronically infected patients worldwide (34). Patients with chronic HBV infection carry a great risk of developing severe liver diseases, including cirrhosis and liver cancer, which result in a million mortalities annually (4, 10). HBV is a member of the Hepadnaviridae family, a group of small hepatotropic DNA viruses that also includes related animal viruses, such as the duck hepatitis B virus (DHBV) and the woodchuck hepatitis virus. All hepadnaviruses carry a small (ca. 3.2 kb), relaxed circular, partially double-stranded DNA genome and replicate this DNA genome through an RNA intermediate, the pregenomic RNA (pgRNA), by reverse transcription (52). The reverse transcription pathway employed by hepadnaviruses is similar to, yet distinct from, that used by retroviruses (for reviews, see references 49 and 50).
All hepadnaviruses encode a novel, multifunctional reverse transcriptase (RT). Like its retroviral counterparts, the hepadnavirus RT catalyzes RNA- and DNA-dependent DNA polymerization and has an intrinsic RNase H activity (12, 43, 56). Reflecting this functional conservation, the central catalytic RT domain and carboxy (C)-terminal RNase H domain of the hepadnavirus RT are homologous to the corresponding domains of retroviral RTs. However, all hepadnavirus RTs share an amino (N)-terminal domain, called the terminal protein (TP) (2, 12, 43). The TP domain is absent from retroviral RTs. Sequence database searches indicate that the TP does not share significant homology to any other known proteins. It is separated from the RT domain by a nonessential and nonconserved spacer (tether) region. The unique TP domain is used as a protein primer to initiate reverse transcription catalyzed by the conserved RT domain (59, 64, 66). In addition to this dual role as a primer and a polymerase in viral DNA synthesis, the RT is essential also for the packaging of the pgRNA into viral nucleocapsids (1, 12, 19), the locale of reverse transcription. By contrast, retroviruses can package their RNA genome independently of the RT (14).
Critical to both protein-primed initiation of reverse transcription and assembly of replication-competent nucleocapsids is the ability of the RT to specifically recognize, and form a ribonucleoprotein (RNP) complex with, a short RNA signal called ɛ, located at the 5′ end of the pgRNA (41, 60). Initially identified as the RNA packaging signal (27) that directs the specific encapsidation of the pgRNA into nucleocapsids, ɛ was subsequently found also to be the origin of reverse transcription (36, 55, 58). Binding of the RT to ɛ thus triggers two critical early steps in hepadnavirus replication, the initiation of reverse transcription via protein priming (15, 32, 36, 58, 60) and nucleocapsid assembly, leading to the selective incorporation of both the RT and pgRNA by the core protein (3, 41).
Biochemical studies on the hepadnavirus RT have been hampered by difficulties in obtaining an active recombinant protein due to problems of low expression, instability, and insolubility (for a review, see reference 21). However, progress has been made recently in deciphering the requirement of RT-ɛ interaction by using the DHBV model system. In 1992, Wang and colleagues succeeded in expressing a DHBV RT active in specific ɛ binding and protein priming by using the rabbit reticulocyte lysate in vitro translation system (59, 60). More recently, purified recombinant DHBV RT proteins were obtained by using the bacterial expression system and truncated mini-RT constructs that proved to be more readily expressed (20, 24, 61-63). Studies using these in vitro systems, as well as cell cultures replicating the virus, have demonstrated that DHBV RT-ɛ interaction requires sequences from both the TP and RT domains of the RT protein (41, 48, 60) and both sequence and structural determinants of the ɛ RNA (6, 8, 41, 60). Furthermore, it has been shown that the DHBV RT requires the assistance of a cellular molecular chaperone complex, consisting of the heat shock protein 90 (Hsp90) and several cofactors, in order to establish and maintain an ɛ-binding competent state (22, 25). By using purified DHBV mini-RT proteins and Hsp90 chaperone components, a defined in vitro biochemical reconstitution system has recently been developed, leading to the identification of the minimal set of host factors that are sufficient to activate the DHBV RT in vitro (20, 24, 61).
Genetic studies in transfected cells have confirmed that as in DHBV, the interaction between the HBV RT and its ɛ RNA is essential for the initiation of reverse transcription and the assembly of replication-competent nucleocapsids containing the pgRNA and the RT (1, 3, 15, 36, 44). However, biochemical studies on the HBV RT, and on the crucial interaction between the RT and the ɛ RNA in particular, have lagged behind. In contrast to DHBV, in vitro assays to measure specific RT-ɛ interaction in HBV have remained unavailable and specific RNP formation between the HBV RT and its ɛ RNA has not yet been demonstrated in vitro. Consequently, the sequence and structural determinants of the HBV RT and ɛ important for RNP formation are not yet defined. So far, the only in vitro system for studying the HBV RT function is the recombinant baculovirus-insect cell system developed by Lanford et al. (32, 33) and Urban et al. (57). The HBV RT partially purified from this system is active in protein priming in vitro. However, it seems that less than 0.1% of the purified HBV RT is active in protein priming; in contrast, 10 to 30% of DHBV RT is active following in vitro translation (41, 59, 60) or reconstitution with Hsp90 (20, 24). The very low specific activity in in vitro protein priming of the purified HBV RT, which apparently can occur independently of the ɛ RNA (33, 57), is consistent with the suggestion that host cell factors may be required for HBV RT-ɛ interaction and, thus, for efficient, ɛ-dependent protein priming, but the nature of these host factors remains obscure.
We report here in vivo and in vitro evidence that supports a critical role of Hsp90 for the RT-ɛ interaction in HBV, as in DHBV. Guided by our recent efforts in the DHBV system, we have succeeded in purifying truncated HBV RT proteins, analogous to the mini-DHBV RT proteins, using the bacterial expression system. Furthermore, we have developed a defined biochemical reconstitution system using the purified HBV RT and host chaperone proteins, which, for the first time, demonstrates a specific HBV RT-ɛ interaction in vitro and will greatly facilitate efforts to elucidate the viral and cellular requirements for specific RNP formation in HBV.
MATERIALS AND METHODS
Plasmids.
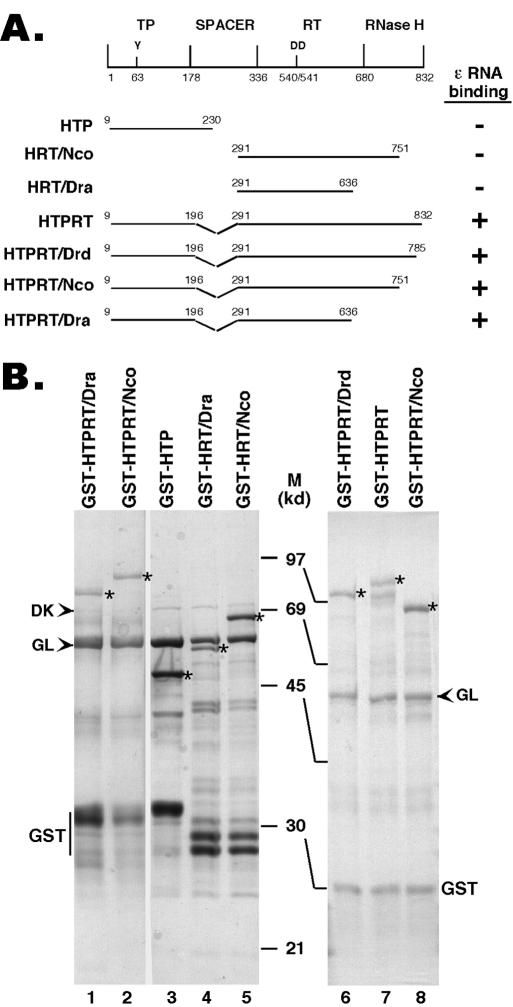
The HBV (ayw subtype) RT coding sequences were derived from pCMVHBV (15), kindly provided by Christoph Seeger. The starting and ending amino acid positions of the different segments of the RT that were expressed are as indicated in Fig. 1. Deletions and truncations were generated by using appropriate restriction enzyme digestions, with the C-terminal truncations denoted by the appropriate restriction sites. DNA coding sequences for these fragments were subcloned into pGEX-KT (17) so that the glutathione S-transferase (GST) was fused in frame to the N terminus of the RT segments in the resulting fusion proteins. A GST-tagged, truncated DHBV mini-RT construct, GST-MiniRT2, was described before (20).
FIG. 1.
Expression of truncated HBV RT proteins and domains. (A) Schematic diagram of the HBV RT proteins and domains expressed. Shown on the top is the domain structure of the HBV RT, with the primer tyrosine (Y63) and the double aspartate (D540/D541) in the RT active site denoted. The ends of the truncations and deletions are indicated. The truncated HBV RT proteins and domains were expressed as GST fusion proteins in bacteria, purified by using glutathione resins, resolved by SDS-PAGE, and detected by Coomassie blue staining (B). The intact fusion proteins are indicated by asterisks. The two bacterial chaperone proteins, DnaK (DK) and GroEL (GL), copurifying with the RT proteins, are indicated by arrowheads. The major degradation product, GST, is also indicated. M, molecular mass in kDa; HTP, HBV terminal protein.
Antibodies and reagents.
The monoclonal antibody (MAb) against Hsp90 (clone 3G3) was purchased from Affinity Bioreagents. The MAbs against DnaK and GroEL were purchased from Stressgen. Polyclonal rabbit anti-HBV core antibody was from DAKO. The MAb against p23 (clone JJ3) has been described previously (26). Geldanamycin (GA) was obtained from the Drug Synthesis and Chemistry Branch of the National Cancer Institute. Two GA derivatives, 17-allylamino-17-demethoxygeldanamycin (17-AAG; MSC 330507) and 17-DMAG (MSC 707545), were obtained from the National Cancer Institute. Novabiocin and radicicol were purchased from Sigma. GA, 17-AAG, 17-DMAG, and radicicol were dissolved in dimethyl sulfoxide as concentrated stocks, and novabiocin was dissolved in distilled H2O.
Cell culture and drug treatment.
The inducible HBV-replicating cell line, HepAD38, harboring a stable HBV integrant under the control of a tetracycline (TET)-regulated promoter (TET-off) (30), was maintained in Dulbecco's modified Eagle medium-nutrient mix F-12 medium supplemented with 10% fetal bovine serum and TET (1 μg/ml). To induce HBV expression and replication, the TET was removed from the culture medium. At the same time, various inhibitors of Hsp90 were added. Two days later, the cells were harvested for the analysis of viral replication. Preliminary experiments showed that induction for less than 2 days resulted in only weak HBV expression and replication, while induction for longer than 2 days led to the appearance of cytotoxicity in cultures treated with the highest concentrations of the Hsp90 inhibitors. The 2-day induction was therefore chosen to minimize drug toxicity while allowing significant HBV induction in the control cells.
Extraction of core DNA and Southern and Western blot analyses.
HepAD38 cells were lysed, and replicative viral DNA intermediates were purified from cytoplasmic core particles and analyzed by Southern blot analysis, as described previously (22, 39). HBV core protein levels were measured by Western blot analysis of cytoplasmic extracts resolved by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE), by using the anti-HBV core antibody and chemiluminescence detection as described previously (61).
RNA packaging assay by native agarose gel electrophoresis of nucleocapsids.
Encapsidated pgRNA in cytoplasmic nucleocapsid particles obtained by polyethylene glycol precipitation (42) was detected by resolving the particles on native agarose gels followed by Southern blot analysis using a 32P-labeled anti-sense HBV riboprobe, as described previously (61, 65). The amount of assembled capsid particles was determined by subsequent reprobing of the same membrane with the anti-HBV core antibody (61).
Bacterial expression and purification of RT fusion proteins.
GST-tagged HBV and DHBV RT fusion proteins were expressed in Escherichia coli and purified by using the glutathione affinity resin as described previously (20), except that BL21 CodonPlus-RIL cells (Stratagene) were used.
Purification of Hsp90.
Human Hsp90β was expressed in Sf9 cells and purified as described previously (16). Cell lysates were fractionated by DEAE-cellulose column chromatography followed by chromatography on heparin-agarose, Mono Q, and Superdex 200 columns. The preparation was greater than 99% pure as assessed by densitometry of SDS-PAGE gels.
Purification of Hsp70.
Human Hsp70 was expressed in Sf9 cells and purified as described previously for avian Hsp70 (46). Cell lysates were fractionated by DEAE-cellulose and ATP-agarose column chromatography. Hsp70 was precipitated by using ammonium sulfate (75% saturation), and the redissolved Hsp70 was fractionated by 16/60 Superdex 200 fast protein liquid chromatography (FPLC). The monomer peak of Hsp70 only was used. The preparation was approximately 97% pure, as assessed by densitometry of SDS-PAGE gels.
Purification of Hop.
Human Hop (p60) expressed in bacteria was prepared essentially as described previously (47). Bacterial lysates were fractionated by DEAE-cellulose and hydroxylapatite column chromatography. Additional purification was achieved by fractionating the pool from hydroxylapatite on a Mono Q 10/10 column (Pharmacia). The preparation was approximately 94% pure as assessed by densitometry of SDS-PAGE gels.
Purification of Ydj1, Hdj1, and Hdj2.
Ydj1 was expressed in bacteria and purified as described previously (29). Bacterial lysates were fractionated by DEAE-cellulose column chromatography, followed by chromatography on Mono Q FPLC and Superdex FPLC. The preparation was >90% pure as assessed by densitometry of SDS-PAGE gels. Hdj2 was purified similarly to Ydj1, as described previously (18). Hdj1 was expressed in bacteria, and the lysate was prepared by sonication in 20 mM Tris, 1 mM EDTA, and 10 mM monothioglycerol (pH 7.5). The cleared lysate was run through a DEAE column where Hdj1 appeared in the flowthrough fractions. These fractions were loaded on a 10-ml column of hydroxylapatite equilibrated in 10 mM potassium phosphate, 2 mM dithiothreitol, and 1 mM EDTA (pH 7). Hdj1 was eluted with a gradient of 10 mM to 500 mM potassium phosphate. Finally, the pooled fractions were concentrated and fractionated on a Superdex 200 FPLC column.
Purification of p23.
The bacterial expression and purification of human p23 have been described previously (51). The soluble fraction of bacterial lysate was fractionated by DEAE-cellulose column chromatography followed by phenyl-Sepharose (hp 1660) FPLC. The preparation was greater than 99% pure as assessed by densitometry of SDS-PAGE gels.
RNA binding.
The HBV and DHBV ɛ RNAs were made by in vitro transcription with a MegaScript kit (Ambion) as described previously (20). To make labeled RNA, [α-32P]UTP was included in the transcription reaction. The wild-type or “long” HBV ɛ RNA corresponded to that described before (23, 27). The “short” HBV ɛ RNA had 6 bp deleted from the bottom of the ɛ lower stem. In other deletion mutants, either the 6-nucleotide internal bulge (dB) or the apical loop (dL) was deleted. Approximately 20 ng (unless otherwise indicated) of purified GST RT fusion proteins was incubated, in a total volume of 10 μl, with approximately 20 ng of 32P-labeled RNA, in TMNK (20 mM Tris [pH 7.5], 2 mM MgCl2, 15 mM NaCl, 20 mM KCl, and 4 mM dithiothreitol), an RNA binding buffer, plus an RNase inhibitor (RNaseIN; Ambion), a protease inhibitor cocktail (Roche), and 6 μg of yeast tRNA. Hsp90, Hsp70, Hsp40 (Hdj1, Hdj2, or Ydj1), Hop, and p23 were added at the indicated amounts per reaction to stimulate RT-ɛ binding. In addition, an ATP regenerating system consisting of 5 mM ATP, 10 mM creatine phosphate, and 50 μg of creatine phosphokinase/ml (20), or the nonhydrolyzable ATP analog ATP-γ-S, was added as indicated. After incubation for 2 h at 30°C, the reactions were resolved on 5% native polyacrylamide gels. Autoradiography of the dried gels was then used to detect the labeled RNA and RNA-protein complexes.
RESULTS
Inhibition of HBV DNA replication and RNA packaging by Hsp90 inhibitors.
It was recently shown that the cellular chaperone Hsp90 is required for the RT-ɛ interaction in DHBV, including viral DNA replication and pgRNA packaging in transfected cells (20, 22, 24, 25, 62, 63). As in vitro systems to measure HBV RT-ɛ interaction were not available, we initially decided to test if inhibitors of Hsp90 could block RT functions in cells, as measured by HBV DNA replication and, more specifically, pgRNA packaging. For this purpose, we used an inducible HBV replicating cell line, HepAD38 (30), where HBV replication can be rapidly induced upon the removal of tetracycline. When Hsp90 inhibitors were added to the culture medium during the 2-day period of induction, we found that HBV DNA synthesis was dramatically inhibited (Fig. 2). To ensure the specificity of Hsp90 inhibition, we employed several different classes of inhibitors, which are structurally distinct and are known to bind to the different regions of Hsp90 (37, 45). Thus, GA and its two derivatives, 17-AAG and 17-DMAG, as well as the structurally distinct radicicol, all bind to the N-terminal ATP binding domain of Hsp90, whereas novabiocin, which is distinct structurally from both GA and radicicol, binds to the C-terminal domain of Hsp90. Although it is possible that each of these different agents may have a target(s) other than Hsp90, the fact that they all inhibited HBV replication strongly indicated that Hsp90 function was required for HBV DNA synthesis. Under conditions in which HBV DNA synthesis was strongly inhibited, the levels of cell viability or viral core proteins were not affected by the Hsp90 inhibitors, indicating that the inhibition of viral DNA synthesis was not a result of any potential pleiotropic effect of inhibiting Hsp90 function on these cells. Indeed, these results were essentially the same as what we have seen with DHBV (22, 25) (J. Hu, unpublished results).
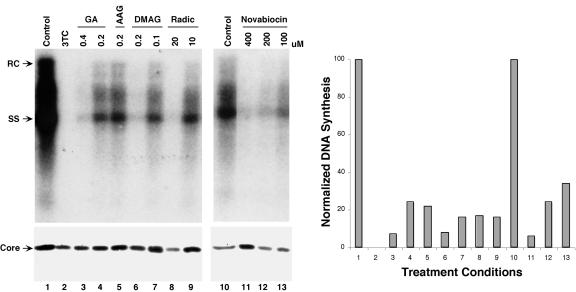
FIG. 2.
Inhibition of HBV DNA synthesis by Hsp90 inhibitors. The levels of HBV DNA synthesis were measured by Southern blot analysis of viral replicative intermediates extracted from HepAD38 cells following treatment with the various inhibitors as indicated (left panel, top). The levels of HBV core protein in cellular extracts prepared from the same cells were measured by Western blot analysis using an anti-HBV core antibody (left panel, bottom). RC, relaxed circular DNA; SS, single stranded DNA; Radic, radicicol; AAG, 17-AAG; DMAG, 17-DMAG. Levels of HBV DNA were normalized to the core protein levels at each treatment condition and are expressed as a percentage of the control untreated culture (right panel). The numbers on the right and left panels each represent the same treatment conditions.
If Hsp90 was required for HBV RT-ɛ interaction, as in the case of DHBV, the inhibition of HBV DNA synthesis we observed above was most likely due to the block of pgRNA packaging, since RT-ɛ interaction is known to be required for pgRNA packaging into nucleocapsids in both HBV and DHBV (3, 41). To test this prediction, we measured HBV pgRNA packaging directly by using the native agarose gel electrophoresis method originally developed by Yu and Summers (65). Although both the pgRNA and plus-strand DNA could in principle be detected by this procedure, the signals detected under our selected conditions were predominantly due to the presence of the pgRNA packaged in the nucleocapsids, with only a minor contribution by the plus DNA. This was evidenced by the fact that little or no decrease was observed in the packaging assay signal detected in the 3TC-treated sample (Fig. 3, lane 2), despite the complete block of viral DNA synthesis (no minus- or plus-strand DNA), as shown by Southern blot analysis of viral DNA extracted from the nucleocapsids (Fig. 2, lane 2). Since 3TC is known to block HBV DNA synthesis but not pgRNA packaging (25, 48), this result indicated that the packaging signal we detected was mostly due to the presence of the pgRNA. Otherwise, if the plus strand DNA had contributed significantly to the packaging signal detected, a dramatic reduction in the packaging signal in the 3TC-treated sample would have occurred. In addition, control experiments demonstrated that the detection of the pgRNA signal associated with the nucleocapsids in the packaging assay was dependent on the RT (data not shown), as expected from the known requirement of the RT for pgRNA packaging. Thus, this assay could be used as a valid surrogate for HBV pgRNA packaging under our experimental conditions. As shown in Fig. 3, the different Hsp90 inhibitors indeed inhibited HBV pgRNA packaging, without affecting the assembly of capsids per se. The extent of inhibition of pgRNA packaging correlated well with that of DNA synthesis, with the different inhibitors and at different concentrations (Fig. 2 and 3), thus indicating that the suppressive effect of the Hsp90 inhibitors on HBV DNA synthesis was mostly, if not exclusively, due to their inhibition of HBV pgRNA packaging. When the levels of viral DNA synthesis (Fig. 2) were normalized to those of pgRNA packaging (Fig. 3), the results indicated that the amount of viral DNA synthesis per pgRNA packaged was not significantly affected by the Hsp90 inhibitors. These results thus suggested that inhibition of Hsp90 did not inhibit HBV DNA synthesis per se; i.e., they did not inhibit the catalysis by the HBV RT. This again agreed well with the results that were obtained previously with the DHBV system, where Hsp90 was shown to be required for DHBV RT-ɛ interaction but not for DHBV RT catalysis once the RT-ɛ binding has occurred (20, 22, 25).
FIG. 3.
Inhibition of HBV RNA packaging by Hsp90 inhibitors. The levels of HBV RNA packaging were measured by native agarose gel analysis of cytoplasmic nucleocapsid particles extracted from HepAD38 cells following treatment with the various inhibitors as indicated (left). HBV RNA and plus-strand HBV DNA (+DNA) present in the resolved nucleocapsids were detected by Southern blot analysis (top) by using an anti-sense HBV riboprobe following their transfer to a nitrocellulose membrane, and the capsid protein was detected by subsequent Western blot analysis (bottom) of the same membrane with an anti-HBV core antibody. Radic, radicicol; AAG, 17-AAG; DMAG, 17-DMAG. Levels of HBV RNA and DNA were normalized to the capsid protein levels at each treatment condition and are expressed as percentages of the control untreated culture (right panel). The numbers on the right and left panels each represent the same treatment conditions.
Reconstitution of HBV RT-ɛ interaction in vitro by using purified RT and Hsp90 chaperone proteins.
Although the results described above obtained in cell cultures were consistent with a critical role of Hsp90 in HBV RT-ɛ interaction, it was conceivable that Hsp90 was required for pgRNA packaging through a mechanism independent of RT-ɛ interaction. To test directly whether Hsp90 was indeed required for HBV RT-ɛ interaction, an in vitro assay to measure directly HBV RT-ɛ interaction was required. As discussed earlier, biochemical studies on the interaction between the HBV RT and ɛ RNA have been hampered by the lack of appropriate in vitro systems. As a first step towards overcoming this difficulty, we have begun to construct mini-HBV RT proteins, guided by the results we have obtained recently with the DHBV mini-RT proteins (20, 24, 63). So far, we have been able to construct four HBV mini-RT proteins that contained sequences from both the TP and RT domains but harbored truncations from the N- and C-termini and an internal deletion in the spacer region, analogous to the DHBV mini-RT proteins we have made before. These HBV mini-RT proteins, like their DHBV counterparts, could be stably expressed as GST fusion proteins in bacteria (Fig. 1). In fact, a construct containing almost the entire TP domain (with only eight residues deleted from the N terminus) and the entire RT-RNase H domain, but with the spacer deletion, could be readily expressed, in contrast to the full-length RT, which was expressed only at very low levels (data not shown). In addition, we also expressed the TP and RT domains separately as GST fusion proteins.
The truncated HBV RT proteins and domains can be readily purified by using an affinity purification approach similar to that used for the DHBV RT (20). Interestingly, as observed with the DHBV mini-RT proteins (7, 20), the purified HBV mini-RT proteins and the individual TP and RT domains were associated with two bacterial chaperones, GroEL and DnaK (Fig. 1), as identified by Western blot analysis using specific antibodies against these proteins (20). These results suggested that the HBV mini-RT proteins might behave similarly to the DHBV counterparts in terms of their stability and chaperone association (both likely reflections of their similar folding properties).
Using these recombinant HBV RT proteins and domains, we have begun to characterize their activities in protein priming and ɛ RNA binding. It was recently shown that five components of the Hsp90 chaperone complex, i.e., Hsp90, Hsp70, Hop, Hsp40, and p23, can strongly stimulate both the ɛ binding and protein priming activities of the purified DHBV mini-RT proteins (24). Therefore, we have tried to measure the ɛ binding and protein priming activities of the purified HBV RT proteins, both with and without chaperone reconstitution. So far, none of the purified HBV RT proteins showed any detectable protein priming activity in vitro (see Discussion). However, upon reconstitution with the same five components of the Hsp90 complex as those used in the DHBV RT reconstitution, the HBV RT proteins containing both the TP and RT domains showed an efficient and specific (see also below) ɛ binding activity (Fig. 4 and 5). No or very little ɛ binding activity was detected by using any of the HBV RT proteins in the absence of chaperone reconstitution. Neither the TP nor the RT domain alone (Fig. 5, lanes 1 to 3) displayed any ɛ binding activity, indicating that both the TP and RT domains of the HBV RT were required for ɛ binding, as has been shown for the DHBV RT (24, 41, 48, 60). On the other hand, deletion of the entire C-terminal RNase H domain did not affect ɛ binding (Fig. 1 and 5), indicating that the RNase H domain was dispensable for ɛ binding.
FIG. 4.
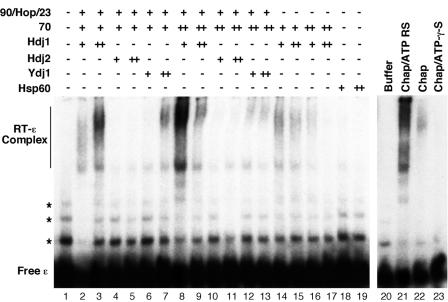
Reconstitution of HBV RT-ɛ interaction as detected by a gel-mobility shift assay. Purified GST-HTPRT-Drd (20 ng) was incubated with 32P-labeled HBV ɛ RNA, in the presence of the indicated chaperone (Chap) factors. The amount of chaperone factors used per reaction was as follows: Hsp90 (90), 360 ng; Hop, 375 ng; p23 (23), 100 ng; Hsp70 (70), 1 μg (+) or 3 μg (++); Hdj1 (or Hdj2 or Ydj1), 200 ng (+) or 600 ng (++); and Hsp60, 170 ng (+) or 510 ng (++). An ATP regenerating system was added in all reactions except in lane 22, where it was omitted, and lane 23, where it was replaced by the nonhydrolyzable ATP analog ATP-γ-S. In lanes 21 to 23, all five chaperone factors were added: Hsp90, 360 ng; Hop, 375 ng; p23, 100 ng; Hsp70, 3 μg; and Hdj1, 200 ng. After a 2-h incubation at 30°C, the reactions were resolved on a native polyacrylamide gel, which was then dried and subjected to autoradiography. The labeled free ɛ RNA and the RT-ɛ complex are indicated. The minor bands (marked by asterisks) above the marked free probe species represented minor structural isoforms of the free RNA, which were not always detectable (see also Fig. 5 to 7).
FIG. 5.
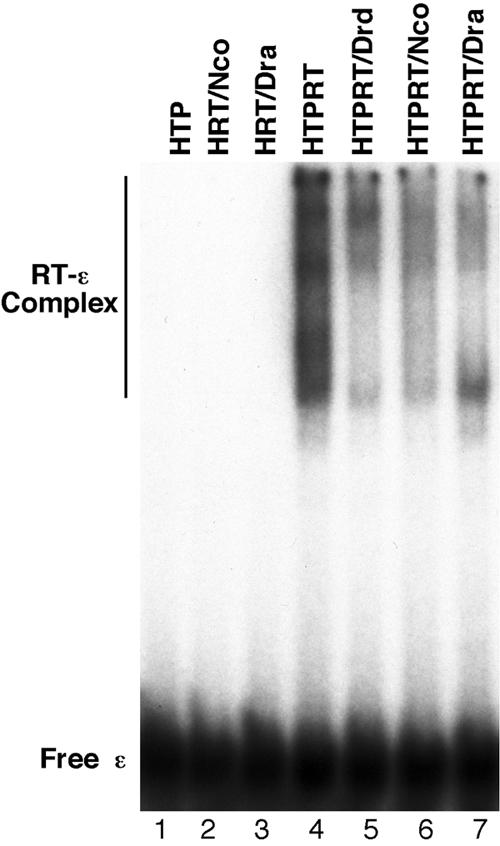
Reconstitution of HBV RT-ɛ interaction using different mini-HBV RT proteins and domains. HTP, HBV terminal protein. GST-HTP (lane 1), HRT/Nco (lane 2), HRT/Dra (lane 3), HTPRT (lane 4), HTPRT/Drd (lane 5), HTPRT/Nco (lane 6), and HTPRT/Dra (lane 7) were purified from bacteria (see Fig. 1) and incubated with 32P-labeled HBV ɛ in the presence of Hsp90 (360 ng), Hsp70 (3 μg), Hop (275 ng), Hdj1 (200 ng), and p23 (100 ng). Approximately 20 ng of all RT proteins or domains were used (except HTPRT, for which 40 ng was used). All reactions contained the ATP regenerating system. The labeled ɛ RNA and the RT-ɛ complex are indicated.
Optimal reconstitution of RT-ɛ binding required careful titration of each of the five chaperone proteins and the RT (Fig. 4 and data not shown). With approximately 20 to 40 ng of the RT in the binding reaction, the optimal amounts of the different chaperone factors were found to be as follows: 360 ng for Hsp90, 375 ng for Hop, 3 μg for Hsp70, 200 ng for Hdj1, and 100 ng for p23 (Fig. 4, lane 8). These amounts were similar but not identical to the optima found for the reconstitution of the mini-DHBV RT in vitro (24). As expected from the known energy requirement of the Hsp90 chaperone system, reconstitution of HBV RT-ɛ binding with the chaperone proteins required ATP hydrolysis (Fig. 4), as in the case of DHBV (24). Under the optimal conditions, approximately 30% of the HBV RT proteins became active in ɛ binding, based on the total amount of RT used in the reaction and that which was in the RT-ɛ complex and assuming a 1:1 stoichiometry of RT to ɛ in the complex. This level of specific activity was similar to that of the currently available mini-DHBV RT proteins (24). Two broad species of RT-ɛ complex seemed to be resolved, but their nature remains to be defined. An interesting possibility is that they may represent different RT and/or ɛ conformational states. Alternatively, partial degradation of the RT proteins could lead to mobility heterogeneity of the RT-ɛ complex although RT proteins with different C-terminal truncations all seemed to produce the same two broad RNP species (Fig. 4 and 5), arguing against such an explanation.
Interestingly, the different Hsp40 isoforms showed differential activity in the reconstitution assay. The human Hdj1 was found to be the most active, the yeast homolog Ydj1 showed less activity, and the human Hdj2 was essentially inactive. This differential activity mirrored the activities in the DHBV RT reconstitution assay (5, 24) (Hu, unpublished). Although optimal reconstitution of HBV RT-ɛ binding required all five chaperone proteins, Hsp70 and Hsp40 (Hdj1 and, to a lesser extent, Ydj1) were able to weakly activate the HBV RT (Fig. 4), as has been shown for the reconstitution of the DHBV RT (5, 24). A recent report claimed that Hsp60, which is a mitochondrial chaperone protein and not part of the Hsp90 chaperone system (11), could modestly stimulate the HBV protein priming activity in vitro (38). We therefore attempted to reconstitute RT-ɛ binding with this chaperone protein but were unable to demonstrate any stimulating effect of Hsp60 on HBV RT-ɛ interaction (Fig. 4, lanes 18 and 19). Although the particular Hsp60 preparation used was not known to be specifically tested for chaperone activity with a different substrate, it was shown to be an active ATPase by the supplier (Stressgen).
HBV RT-ɛ interaction in vitro was specific and depended on the internal bulge, but not the apical loop, of the ɛ RNA.
Competition experiments demonstrated that the binding of the HBV RT to the HBV ɛ RNA was specific. Thus, HBV RT-ɛ interaction could be efficiently competed by the unlabeled HBV ɛ, but not by the DHBV ɛ or tRNA (Fig. 6A). In addition, a large (300-fold) molar excess of tRNA was present in all binding reactions (see Materials and Methods). As expected, the HBV RT did not bind to the labeled DHBV ɛ RNA, which was bound by its cognate DHBV RT (Fig. 6B) (20, 24, 41, 60).
FIG. 6.
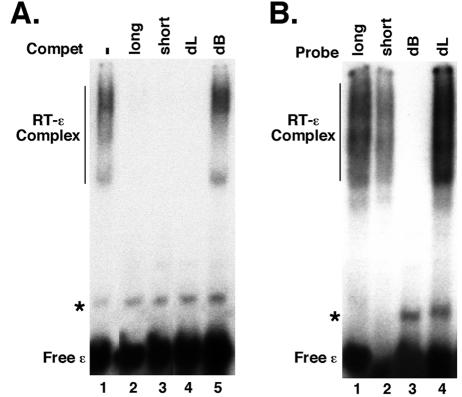
HBV or DHBV RT bound only its cognate ɛ RNA in vitro. (A) GST-tagged HTPRT/Drd (20 ng) was incubated with 32P-labeled HBV ɛ RNA, in the presence of five chaperone factors: Hsp90 (360 ng), Hsp70 (3 μg), Hop (275 ng), Hdj1 (200 ng), and p23 (100 ng). As indicated, the following competitors were added: unlabeled HBV ɛ (lane 3; 100-fold excess), DHBV ɛ (lane 4; 100-fold excess), and yeast tRNA (lane 2; 200-fold excess). In addition, all reactions contained a 300-fold molar excess of tRNA over the labeled probe. (B) The GST-tagged HBV RT (HTPRT/Drd) (lanes 2 and 5) or DHBV RT (MiniRT2) (lanes 3 and 6) was incubated with the 32-P labeled HBV (lanes 1 to 3) or DHBV ɛ RNA (lanes 4 to 6), in the presence of the same five Hsp90 chaperone factors as in panel A. Lanes 1 and 4 show reactions where no RT proteins were added. The labeled ɛ RNA and the RT-ɛ complex are indicated. The minor band (marked by an asterisk) represented a minor structural isoform of the free RNA.
To further confirm the specificity of the HBV RT-ɛ interaction in vitro and to begin to identify the RNA determinants recognized by the RT, we have so far tested three HBV ɛ RNA variants using our in vitro binding assay. The first ɛ variant (short) had the lower half of the lower stem of the ɛ stem loop deleted. The analogous ɛ variant in the DHBV ɛ RNA has been shown to be fully functional in both RT binding and protein priming in vitro (20, 24). We thus expected the short HBV ɛ to be active in RT binding, which was indeed shown to be the case (Fig. 7). Thus, the unlabeled short ɛ competed efficiently for RT binding by the labeled wild-type (long) ɛ (Fig. 7A, lane 3). Also, the labeled short ɛ was bound by the RT (Fig. 7B, lane 2). The other two ɛ variants had a deletion of either the internal bulge (dB) or the apical loop (dL) of the ɛ stem loop structure (27). Both the bulge and loop have been shown to be required for pgRNA packaging in vivo for both HBV and DHBV (40, 41) and for DHBV protein priming in vitro (41; Hu, unpublished). However, certain loop mutations in the DHBV ɛ have been shown to be functional in RT binding but defective in protein priming (6, 8, 41) and pgRNA packaging (41). We were therefore interested in determining if either the ɛ bulge or loop was required for RT binding in our in vitro assay. As shown clearly in Fig. 7, our results indicated that the internal bulge was essential for RT binding in vitro, whereas the apical loop was completely dispensable, as tested both by their ability to compete with the wild type ɛ for RT binding (Fig. 7A) and by their ability to bind RT directly (Fig. 7B).
FIG. 7.
The internal bulge, but not the apical loop, of ɛ was required for RT binding. (A) GST-HTPRT/Drd (20 ng) was incubated with 32P-labeled HBV ɛ RNA, in the presence of five chaperone factors: Hsp90 (360 ng), Hsp70 (3 μg), Hop (275 ng), Hdj1 (200 ng), and p23 (100 ng). As indicated, the following competitors (in 100-fold excess) were added: unlabeled HBV ɛ (wild-type or long) (lane 2), a truncated HBV ɛ variant with a shorter lower stem (short) (lane 3), an HBV ɛ variant with its apical loop deleted (dL) (lane 4), or with its internal bulge deleted (dB) (lane 5). (B) The wild type and three different HBV ɛ variants were 32P-labeled and used in the RT binding assay. The labeled ɛ RNA and the RT-ɛ complex are indicated. The minor band (marked by an asterisk in each panel) represented a minor structural isoform of the free RNA.
DISCUSSION
The molecular chaperone complex, Hsp90, has been identified as an essential host factor required for the interaction between the RT and the ɛ RNA in the DHBV system (20, 22, 24, 25, 61, 63). Here, we provide evidence that the Hsp90 complex is also required for the same protein-RNA interaction in the HBV system. This possibility was initially suggested by the suppressive effect of a variety of different pharmacological Hsp90 inhibitors on HBV pgRNA packaging in transfected cells. Furthermore, in vitro reconstitution experiments demonstrated that Hsp90, together with four cochaperones, Hsp70, Hop, Hsp40, and p23, was able to stimulate the specific ɛ binding activity of purified HBV RT proteins.
Guided by our recent success in expressing truncated mini-DHBV RT proteins (20, 24, 63), we have succeeded in expressing and purifying analogous mini-HBV RT proteins using the bacterial GST fusion system. The purified HBV RT proteins showed little or no ɛ binding activity. However, as we have demonstrated previously with the DHBV system, five purified factors from the Hsp90 chaperone system could strongly simulate their ɛ binding activity. The requirements for reconstituting HBV RT-ɛ interaction were found to be similar to those for DHBV (20, 24). Both systems required the same five chaperone factors and ATP hydrolysis. Specific ɛ binding by both the HBV (this report) and DHBV (24, 41, 48, 60) RT required their respective TP and RT domains but not the C-terminal RNase H domain, although detailed mapping and site directed mutagenesis, as have been done for the DHBV RT (24, 41, 48, 60), remain to be performed for the HBV RT.
We and others have noticed that the different isoforms of Hsp40 show differential activity in RT reconstitution. Thus, the human type II Hsp40, Hdj1, was the most active, among the three Hsp40 proteins tested, in stimulating DHBV and HBV RT activity in vitro (5, 24) (this report; Hu, unpublished). The yeast type I Hsp40, Ydj1, was shown to be active in both HBV (this report) and DHBV (24) RT reconstitution albeit less active than Hdj1. Another group reported that Ydj1 was inactive in DHBV RT reconstitution, although it was not certain if the particular Ydj1 preparation used was biologically active (5). Surprisingly, both we and others found that the human type I Hsp40, Hdj2, was completely inactive in either DHBV (5) (Hu, unpublished) or HBV (this report) RT reconstitution. It is important to emphasize that all of the chaperone proteins we used for HBV and DHBV RT reconstitution, including the different Hsp40 isoforms, had been shown to be functional in a different substrate (the progesterone receptor) system (29). The type I and II Hsp40 proteins show structural and functional similarities as well as differences (13, 28). Within a given type, the Hsp40 proteins show additional structural similarities that are not shared across the different types. However, the exact substrates and functional specificities of the different Hsp40 proteins remain to be clearly defined. Our results with both the DHBV and HBV RT reconstitutions suggest that Hsp40 proteins from the different types, i.e., Hdj1 (type II) and Ydj1 (type I), can share the same substrates and those from the same type, i.e., Hdj2 and Ydj1 (both type I), may not necessarily share the same substrate specificity, although much more work needs to be done to clearly define the substrate and functional specificity of the different Hsp40 isoforms. Moreover, it is not yet clear which Hsp40 protein(s) is truly responsible for RT activation in the cell.
Strikingly, although we could efficiently reconstitute the HBV RT-ɛ interaction, no protein priming activity could be detected with any of the HBV RT proteins expressed so far. A mutational analysis of the HBV RT sequence required for in vitro protein priming, using the baculovirus-insect cell system, showed that most of the sequences from the C-terminal region, including most of the RNase H domain, are required for HBV protein priming (31), even though the corresponding sequences are dispensable for protein priming by the DHBV RT (54, 62, 63). Specifically, those results indicated that the TP domain sequences from 20 to 175 and RT sequences from 300 to 775 are required for protein priming activity in vitro (31). These sequences were contained in two of the four mini-RT proteins we constructed here. Since none of the mini-HBV RT proteins showed any protein priming activity in vitro, the lack of protein priming activity was unlikely due to the absence of essential RT sequences, at least for the two longer RT proteins tested.
The lack of HBV RT protein priming activity despite efficient ɛ binding activity is consistent with earlier observations made with the DHBV that mere physical binding of the ɛ RNA to the RT is not sufficient for protein priming; rather, a functional protein-RNA interaction is required (6, 8, 41). It has been suggested that both the RT and the ɛ RNA undergo some conformational changes upon binding and these conformational changes are, in turn, required for the RT to gain catalytic competence or the ɛ RNA to serve as a functional template to initiate protein priming (6, 53, 54). It is thus possible that in our present reconstitution system, the putative conformational change of either the RT or the ɛ RNA could not occur and the physical binding of the ɛ to the RT did not proceed to the functional interaction required for protein priming. This, in turn, could be due to the lack of additional cellular factors required to establish this functional RT-ɛ interaction.
The lack of protein priming activity of the HBV RT contrasts sharply with the efficient protein priming activity obtained with similar DHBV mini-RT proteins using the same Hsp90 reconstitution procedure. These results suggest that whatever additional cellular factor required for HBV protein priming is apparently not essential for the DHBV RT. Interestingly, it has been reported that certain DHBV ɛ RNA mutants, particularly at the apical loop of the ɛ stem-loop structure, allow wild-type levels of RT binding but show severe defects in protein priming or pgRNA packaging (6, 8, 41). These results led to the suggestion that cellular factors, perhaps by binding to the apical loop of the ɛ RNA, may be required for protein priming and pgRNA packaging. However, recent reconstitution experiments using purified components clearly showed that host factors other than the Hsp90 chaperone complex are not essential for DHBV RT-ɛ interaction or protein priming (5, 24, 63). As there is no evidence to suggest that components of the Hsp90 chaperone binds to the DHBV ɛ RNA, these results indicate that the putative ɛ binding cellular factor, whatever it may be, is unlikely to be essential for ɛ-dependent protein priming by the DHBV RT. In fact, severely truncated DHBV mini-RT proteins that are active in ɛ binding and protein priming without the help of any host factors were made recently (63). However, these results do not exclude the possibility that such a putative ɛ binding factor, or some other cellular factor, can be critical for ɛ-dependent protein priming by the HBV RT. Indeed, a differential requirement of protein priming by the DHBV versus HBV RT has been suggested by the long-standing observation that the HBV RT expressed in the rabbit reticulocyte lysate in vitro translation system showed no detectable protein priming activity, despite the fact that this system was the first to demonstrate the ɛ-dependent protein priming activity of the DHBV RT (59). The lack of protein priming in our in vitro system, together with the results that the ɛ internal bulge, but not the apical loop, was required for HBV RT binding in vitro, are also consistent with the idea that a host cell factor, which is missing in our reconstitution system, may be required for HBV protein priming by binding the ɛ apical loop.
A critical role for Hsp90 in HBV as well as DHBV RT-ɛ interaction in vivo is supported by the very similar suppressive effect of a diverse array of Hsp90 inhibitors on HBV as well as DHBV pgRNA packaging in cells, which is known to require RT-ɛ interaction, without affecting either core protein expression or assembly (references 22 and 25 and this report). Recently, it has been suggested that Hsp90 may also be required for the function of the HBV X protein signaling, which, in turn, may be required for HBV assembly and replication (9). However, since the same results were obtained in the DHBV system, which does not encode an X protein important for viral replication (35), and since our in vitro reconstitution assays clearly showed that Hsp90 could stimulate HBV RT-ɛ interaction, we believe that most, if not all, of the effect of Hsp90 inhibition on HBV pgRNA packaging (and DNA synthesis) we observed here was mediated through its inhibitory effect on the RT-ɛ interaction. In addition, components of the Hsp90 complex, including Hsp90 and p23, have been found to be associated with purified DHBV nucleocapsids (25) (Hu, unpublished). Similarly, both Hsp90 and Hsp70 can be found in purified HBV nucleocapsids (D. Nguyen and J. Hu, unpublished results).
The establishment of a defined reconstitution system for HBV RT-ɛ interaction will allow further detailed analyses of the viral and cellular requirements for HBV RT-ɛ interaction. Furthermore, building on this system, efforts are under way to establish an in vitro HBV RT protein priming system that is dependent on RT-ɛ interaction. The development of these systems should facilitate biochemical and structural studies to dissect the molecular mechanisms of RT-ɛ interaction and protein priming in HBV and help to develop novel antiviral agents targeting these critical steps in HBV replication for the treatment of chronic HBV infections.
Acknowledgments
We thank Joslynn Jordan for technical assistance and Christoph Seeger for providing the pCMVHBV plasmid.
This work was supported by Public Health Service grant R01 AI43453 (to J.H.).
REFERENCES
- 1.Bartenschlager, R., M. Junker-Niepmann, and H. Schaller. 1990. The P gene product of hepatitis B virus is required as a structural component for genomic RNA encapsidation. J. Virol. 64:5324-5332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bartenschlager, R., and H. Schaller. 1988. The amino-terminal domain of the hepadnaviral P-gene encodes the terminal protein (genome-linked protein) believed to prime reverse transcription. EMBO J. 7:4185-4192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bartenschlager, R., and H. Schaller. 1992. Hepadnaviral assembly is initiated by polymerase binding to the encapsidation signal in the viral RNA genome. EMBO J. 11:3413-3420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Beasley, R. P., C. C. Lin, L. Y. Hwang, and C. S. Chien. 1981. Hepatocellular carcinoma and hepatitis B virus. Lancet 2:1129-1133. [DOI] [PubMed] [Google Scholar]
- 5.Beck, J., and M. Nassal. 2003. Efficient Hsp90-independent in vitro activation by Hsc70 and Hsp40 of duck hepatitis B virus reverse transcriptase, an assumed Hsp90 client protein. J. Biol. Chem. 278:36128-36138. [DOI] [PubMed] [Google Scholar]
- 6.Beck, J., and M. Nassal. 1998. Formation of a functional hepatitis B virus replication initiation complex involves a major structural alteration in the RNA template. Mol. Cell. Biol. 18:6265-6272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Beck, J., and M. Nassal. 2001. Reconstitution of a functional duck hepatitis B virus replication initiation complex from separate reverse transcriptase domains expressed in Escherichia coli. J. Virol. 75:7410-7419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Beck, J., and M. Nassal. 1997. Sequence- and structure-specific determinants in the interaction between the RNA encapsidation signal and reverse transcriptase of avian hepatitis B viruses. J. Virol. 71:4971-4980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bouchard, M. J., R. J. Puro, L. Wang, and R. J. Schneider. 2003. Activation and inhibition of cellular calcium and tyrosine kinase signaling pathways identify targets of the HBx protein involved in hepatitis B virus replication. J. Virol. 77:7713-7719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Buendia, M. A. 1992. Hepatitis B viruses and hepatocellular carcinoma. Adv. Cancer Res. 59:167-226. [DOI] [PubMed] [Google Scholar]
- 11.Bukau, B., and A. Horwich. 1998. The Hsp70 and Hsp60 chaperone machines. Cell 92:351-366. [DOI] [PubMed] [Google Scholar]
- 12.Chang, L. J., R. C. Hirsch, D. Ganem, and H. E. Varmus. 1990. Effects of insertional and point mutations on the functions of the duck hepatitis B virus polymerase. J. Virol. 64:5553-5558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cheetham, M. E., and A. J. Caplan. 1998. Structure, function and evolution of DnaJ: conservation and adaptation of chaperone function. Cell Stress Chaperones. 3:28-36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Coffin, J. M. 1996. Retroviridae: the viruses and their replication, p. 1810-1814. In B. N. Fields, D. M. Knipe, and P. M. Howley (ed.), Fields Virology, 3rd ed., vol. 2. Lippincott-Raven Publishers, Philadelphia, Pa. [Google Scholar]
- 15.Fallows, D. A., and S. P. Goff. 1995. Mutations in the ɛ sequences of human hepatitis B virus affect both RNA encapsidation and reverse transcription. J. Virol. 69:3067-3073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Grenert, J. P., W. P. Sullivan, P. Fadden, T. A. J. Haystead, J. Clark, E. Mimnaugh, H. Krutzsch, H.-J. Ochel, T. W. Schulte, E. Sausville, L. M. Neckers, and D. O. Toft. 1997. The amino-terminal domain of heat shock protein 90 (hsp90) that binds geldanamycin is an ATP/ADP switch domain that regulates hsp90 conformation. J. Biol. Chem. 272:23843-23850. [DOI] [PubMed] [Google Scholar]
- 17.Hakes, D. J., and J. E. Dixon. 1992. New vectors for high level expression of recombinant proteins in bacteria. Anal. Biochem. 202:293-298. [DOI] [PubMed] [Google Scholar]
- 18.Hernandez, M. P., A. Chadli, and D. O. Toft. 2002. HSP40 binding is the first step in the HSP90 chaperoning pathway for the progesterone receptor. J. Biol. Chem. 277:11873-11881. [DOI] [PubMed] [Google Scholar]
- 19.Hirsch, R. C., J. E. Lavine, L. J. Chang, H. E. Varmus, and D. Ganem. 1990. Polymerase gene products of hepatitis B viruses are required for genomic RNA packaging as well as for reverse transcription. Nature 344:552-555. [DOI] [PubMed] [Google Scholar]
- 20.Hu, J., and D. Anselmo. 2000. In vitro reconstitution of a functional duck hepatitis B virus reverse transcriptase: posttranslational activation by Hsp90. J. Virol. 74:11447-11455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hu, J., and C. Seeger. 1996. Expression and characterization of hepadnavirus reverse transcriptases. Methods Enzymol. 275:195-208. [DOI] [PubMed] [Google Scholar]
- 22.Hu, J., and C. Seeger. 1996. Hsp90 is required for the activity of a hepatitis B virus reverse transcriptase. Proc. Natl. Acad. Sci. USA 93:1060-1064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hu, J., and C. Seeger. 1997. RNA signals that control DNA replication in hepadnaviruses. Semin. Virol. 8:205-211. [Google Scholar]
- 24.Hu, J., D. Toft, D. Anselmo, and X. Wang. 2002. In vitro reconstitution of functional hepadnavirus reverse transcriptase with cellular chaperone proteins. J. Virol. 76:269-279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hu, J., D. O. Toft, and C. Seeger. 1997. Hepadnavirus assembly and reverse transcription require a multi-component chaperone complex which is incorporated into nucleocapsids. EMBO J. 16:59-68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Johnson, J. L., T. G. Beito, C. J. Krco, and D. O. Toft. 1994. Characterization of a novel 23-kilodalton protein of unactive progesterone receptor complexes. Mol. Cell. Biol. 14:1956-1963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Junker-Niepmann, M., R. Bartenschlager, and H. Schaller. 1990. A short cis-acting sequence is required for hepatitis B virus pregenome encapsidation and sufficient for packaging of foreign RNA. EMBO J. 9:3389-3396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kelley, W. L. 1998. The J-domain family and the recruitment of chaperone power. Trends Biochem. Sci. 23:222-227. [DOI] [PubMed] [Google Scholar]
- 29.Kosano, H., B. Stensgard, M. C. Charlesworth, N. McMahon, and D. Toft. 1998. The assembly of progesterone receptor-hsp90 complexes using purified proteins. J. Biol. Chem. 273:32973-32979. [DOI] [PubMed] [Google Scholar]
- 30.Ladner, S. K., M. J. Otto, C. S. Barker, K. Zaifert, G. H. Wang, J. T. Guo, C. Seeger, and R. W. King. 1997. Inducible expression of human hepatitis B virus (HBV) in stably transfected hepatoblastoma cells: a novel system for screening potential inhibitors of HBV replication. Antimicrob. Agents Chemother. 41:1715-1720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lanford, R. E., Y. H. Kim, H. Lee, L. Notvall, and B. Beames. 1999. Mapping of the hepatitis B virus reverse transcriptase TP and RT domains by transcomplementation for nucleotide priming and by protein-protein interaction. J. Virol. 73:1885-1893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lanford, R. E., L. Notvall, and B. Beames. 1995. Nucleotide priming and reverse transcriptase activity of hepatitis B virus polymerase expressed in insect cells. J. Virol. 69:4431-4439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lanford, R. E., L. Notvall, H. Lee, and B. Beames. 1997. Transcomplementation of nucleotide priming and reverse transcription between independently expressed TP and RT domains of the hepatitis B virus reverse transcriptase. J. Virol. 71:2996-3004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lee, W. 1997. Hepatitis B virus infection. N. Engl. J. Med. 337:1733-1745. [DOI] [PubMed] [Google Scholar]
- 35.Meier, P., C. A. Scougall, H. Will, C. J. Burrell, and A. R. Jilbert. 2003. A duck hepatitis B virus strain with a knockout mutation in the putative X ORF shows similar infectivity and in vivo growth characteristics to wild-type virus. Virology 317:291-298. [DOI] [PubMed] [Google Scholar]
- 36.Nassal, M., and A. Rieger. 1996. A bulged region of the hepatitis B virus RNA encapsidation signal contains the replication origin for discontinuous first-strand DNA synthesis. J. Virol. 70:2764-2773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Neckers, L. 2002. Hsp90 inhibitors as novel cancer chemotherapeutic agents. Trends Mol. Med. 8:S55-61. [DOI] [PubMed] [Google Scholar]
- 38.Park, S. G., and G. Jung. 2001. Human hepatitis B virus polymerase interacts with the molecular chaperonin Hsp60. J. Virol. 75:6962-6968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Perlman, D., and J. Hu. 2003. Duck hepatitis B virus virion secretion requires a double-stranded DNA genome. J. Virol. 77:2287-2294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Pollack, J. R., and D. Ganem. 1993. An RNA stem-loop structure directs hepatitis B virus genomic RNA encapsidation. J. Virol. 67:3254-3263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Pollack, J. R., and D. Ganem. 1994. Site-specific RNA binding by a hepatitis B virus reverse transcriptase initiates two distinct reactions: RNA packaging and DNA synthesis. J. Virol. 68:5579-5587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Pugh, J. C., K. Yaginuma, K. Koike, and J. Summers. 1988. Duck hepatitis B virus (DHBV) particles produced by transient expression of DHBV DNA in a human hepatoma cell line are infectious in vitro. J. Virol. 62:3513-3616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Radziwill, G., W. Tucker, and H. Schaller. 1990. Mutational analysis of the hepatitis B virus P gene product: domain structure and RNase H activity. J. Virol. 64:613-620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Rieger, A., and M. Nassal. 1996. Specific hepatitis B virus minus-strand DNA synthesis requires only the 5′ encapsidation signal and the 3′-proximal direct repeat DR1. J. Virol. 70:585-589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Scheibel, T., and J. Buchner. 1998. The Hsp90 complex—a super-chaperone machine as a novel drug target. Biochem. Pharmacol. 56:675-682. [DOI] [PubMed] [Google Scholar]
- 46.Schumacher, R. J., W. J. Hansen, B. C. Freeman, E. Alnemri, G. Litwack, and D. O. Toft. 1996. Cooperative action of Hsp70, Hsp90, and DnaJ proteins in protein renaturation. Biochemistry 35:14889-14898. [DOI] [PubMed] [Google Scholar]
- 47.Schumacher, R. J., R. Hurst, W. P. Sullivan, N. J. McMahon, D. O. Toft, and R. L. Matts. 1994. ATP-dependent chaperoning activity of reticulocyte lysate. J. Biol. Chem. 269:9493-9499. [PubMed] [Google Scholar]
- 48.Seeger, C., E. H. Leber, L. K. Wiens, and J. Hu. 1996. Mutagenesis of a hepatitis B virus reverse transcriptase yields temperature-sensitive virus. Virology 222:430-439. [DOI] [PubMed] [Google Scholar]
- 49.Seeger, C., and W. S. Mason. 2000. Hepatitis B virus biology. Microbiol. Mol. Biol. Rev. 64:51-68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Seeger, C., and W. S. Mason. 1996. Replication of the hepatitis virus genome, p. 815-831. In M. L. DePamphilis (ed.), DNA replication in eukaryotic cells. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 51.Sullivan, W., B. Stensgard, G. Caucutt, B. Bartha, N. McMahon, E. Alnemri, G. Litwack, and D. Toft. 1997. Nucleotides and two functional states of hsp90. J. Biol. Chem. 272:8007-8012. [DOI] [PubMed] [Google Scholar]
- 52.Summers, J., and W. S. Mason. 1982. Replication of the genome of a hepatitis B-like virus by reverse transcription of an RNA intermediate. Cell 29:403-415. [DOI] [PubMed] [Google Scholar]
- 53.Tavis, J. E., and D. Ganem. 1996. Evidence for activation of the hepatitis B virus polymerase by binding of its RNA template. J. Virol. 70:5741-5750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Tavis, J. E., B. Massey, and Y. Gong. 1998. The duck hepatitis B virus polymerase is activated by its RNA packaging signal, ɛ. J. Virol. 72:5789-5796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Tavis, J. E., S. Perri, and D. Ganem. 1994. Hepadnavirus reverse transcription initiates within the stem-loop of the RNA packaging signal and employs a novel strand transfer. J. Virol. 68:3536-3543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Toh, H., H. Hayashida, and T. Miyata. 1983. Sequence homology between retroviral reverse transcriptase and putative polymerases of hepatitis B virus and cauliflower mosaic virus. Nature 305:827-829. [DOI] [PubMed] [Google Scholar]
- 57.Urban, M., D. J. McMillan, G. Canning, A. Newell, E. Brown, J. S. Mills, and R. Jupp. 1998. In vitro activity of hepatitis B virus polymerase: requirement for distinct metal ions and the viral epsilon stem-loop. J. Gen. Virol. 79:1121-1131. [DOI] [PubMed] [Google Scholar]
- 58.Wang, G. H., and C. Seeger. 1993. Novel mechanism for reverse transcription in hepatitis B viruses. J. Virol. 67:6507-6512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Wang, G. H., and C. Seeger. 1992. The reverse transcriptase of hepatitis B virus acts as a protein primer for viral DNA synthesis. Cell 71:663-670. [DOI] [PubMed] [Google Scholar]
- 60.Wang, G. H., F. Zoulim, E. H. Leber, J. Kitson, and C. Seeger. 1994. Role of RNA in enzymatic activity of the reverse transcriptase of hepatitis B viruses. J. Virol. 68:8437-8442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Wang, X., N. Grammatikakis, and J. Hu. 2002. Role of p50/CDC37 in hepadnavirus assembly and replication. J. Biol. Chem. 277:24361-24367. [DOI] [PubMed] [Google Scholar]
- 62.Wang, X., and J. Hu. 2002. Distinct requirement for two stages of protein-primed initiation of reverse transcription in hepadnaviruses. J. Virol. 76:5857-5865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Wang, X., X. Qian, H.-C. Guo, and J. Hu. 2003. Hsp90-independent activation of truncated hepadnavirus reverse transcriptase. J. Virol. 77:4471-4480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Weber, M., V. Bronsema, H. Bartos, A. Bosserhoff, R. Bartenschlager, and H. Schaller. 1994. Hepadnavirus P protein utilizes a tyrosine residue in the TP domain to prime reverse transcription. J. Virol. 68:2994-2999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Yu, M., and J. Summers. 1994. Multiple functions of capsid protein phosphorylation in duck hepatitis B virus replication. J. Virol. 68:4341-4348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Zoulim, F., and C. Seeger. 1994. Reverse transcription in hepatitis B viruses is primed by a tyrosine residue of the polymerase. J. Virol. 68:6-13. [DOI] [PMC free article] [PubMed] [Google Scholar]