Abstract
The epitope determinants of chimpanzee Fab antibody 1A5, which have been shown to be broadly reactive to flaviviruses and efficient for cross-neutralization of dengue virus type 1 and type 2 (DENV-1 and DENV-2), were studied by analysis of DENV-2 antigenic variants. Sequence analysis showed that one antigenic variant contained a Gly-to-Val substitution at position 106 within the flavivirus-conserved fusion peptide loop of the envelope protein (E), and another variant contained a His-to-Gln substitution at position 317 in E. Substitution of Gly106Val in DENV-2 E reduced the binding affinity of Fab 1A5 by approximately 80-fold, whereas substitution of His317Gln had little or no effect on antibody binding compared to the parental virus. Treatment of DENV-2 with β-mercaptoethanol abolished binding of Fab 1A5, indicating that disulfide bridges were required for the structural integrity of the Fab 1A5 epitope. Binding of Fab 1A5 to DENV-2 was competed by an oligopeptide containing the fusion peptide sequence as shown by competition enzyme-linked immunosorbent assay. Both DENV-2 antigenic variants were shown to be attenuated, or at least similar to the parental virus, when evaluated for growth in cultured cells or for neurovirulence in mice. Fab 1A5 inhibited low pH-induced membrane fusion of mosquito C6/36 cells infected with DENV-1 or DENV-2, as detected by reduced syncytium formation. Both substitutions in DENV-2 E lowered the pH threshold for membrane fusion, as measured in a fusion-from-within assay. In the three-dimensional structure of E, Gly106 in domain II and His317 in domain III of the opposite E monomer were spatially close. From the locations of these amino acids, Fab 1A5 appears to recognize a novel epitope that has not been mapped before with a flavivirus monoclonal antibody.
The four dengue virus serotypes (DENV-1 to DENV-4) constitute the dengue virus complex within the Flavivirus genus of the Flaviviridae. Dengue outbreaks and epidemics continue to pose a public health problem in most tropical and subtropical countries. Dengue illnesses range from mild dengue fever to severe dengue, characterized by dengue hemorrhagic fever and dengue shock syndrome, which has a high mortality rate. According to one estimate, approximately 50 to 100 million dengue virus infections and up to 250,000 dengue hemorrhagic fever cases occur every year worldwide (12, 34). Despite six decades of research, a safe and effective dengue vaccine has not been developed, nor is a specific, short-term preventive measure available. Currently, prevention of dengue is carried out by mosquito vector control, which is rather ineffective. Several other arthropod-borne flaviviruses are also important human pathogens, including the yellow fever virus (YFV), tick-borne encephalitis virus (TBEV), Japanese encephalitis virus (JEV), and West Nile virus (WNV), which has recently emerged in North America (23, 25). Vaccines against all of these viruses except WNV have been developed.
The flavivirus genome contains a positive-strand RNA with one open reading frame coding for a polyprotein. The polyprotein is processed to produce the three structural proteins, i.e., the capsid (C), precursor membrane (prM), and envelope (E) proteins, plus seven nonstructural proteins, designated NS1, NS2A, NS2B, NS3, NS4A, NS4B, and NS5. The E protein is responsible for viral attachment to the putative cell surface receptor(s), fusion with the endosomal membranes upon entry, and mediating protective immune responses in the infected host. Mouse monoclonal antibodies (MAbs) against the E proteins of most major flaviviruses have been identified (19, 42). Studies using these MAbs have allowed identification of flavivirus group-, complex-, and type-specific epitopes on the flavivirus E proteins. With few exceptions, neutralizing MAbs are flavivirus type or subtype specific, consistent with the flavivirus classification determined with polyclonal sera (8).
The three-dimensional (3-D) structure of the flat homodimeric E glycoprotein that is organized in a direction parallel to the viral membrane has been determined for TBEV (40) and DENV-2 (32). The E subunit, approximately 500 amino acids in length, is folded into three structurally distinct domains, termed domains I, II, and III. Domain I organizes the entire E structure and contains a flavivirus-conserved glycosylated asparagine. Domain II is folded into an elongated structure containing at its distal end the fusion peptide sequence, commonly called the fusion loop, which is conserved among the flaviviruses. The outward glycan unit in domain I protrudes to cover the fusion loop of the other subunit. There is an extensive interface contact between domain II and each of the three domains of the neighboring subunit. Domain III is an immunoglobulin-like region and lies at the end of the subunit. The dimeric E structure realigns to become trimeric when triggered by lowering the pH, while the three domains remain intact structurally (7, 33). During the transition, the fusion loop becomes exposed and reoriented outward, making it available for membrane contact.
Antigenic determinants of flavivirus cross-reactive antibodies have been mapped to domain II, whereas determinants of subtype- and type-specific antibodies have been assigned to domains I and III (19, 27, 42, 43). Most epitopes of neutralizing antibodies have been placed on the outer surface of the E glycoprotein, consistent with their accessibility to antibody binding. Mutations present in variant viruses that have escaped neutralization by antibodies blocking virus adsorption to Vero cells have been assigned to the lateral side of E in domain III (10). Similarly, the mutations of antigenic variants that affect mouse neurovirulence have been mapped to this domain (9, 21, 22). These findings suggest that the sequence in domain III may mediate viral attachment to the receptor on susceptible cells.
The antigenic model of flavivirus E proteins established thus far from studies with the large repertoire of mouse MAbs has provided much information about serological specificities and functional activities (19, 42). The question remains whether these antigenic epitopes are mouse specific or whether in fact they represent immuno-dominant sites on E recognized by the immune systems of other host species as well. Unfortunately, there is a lack of flavivirus MAbs from other host species, especially higher primates or humans.
We have recently turned to the identification of chimpanzee Fab fragments by repertoire cloning and construction of full-length humanized immunoglobulin G (IgG) antibodies in an effort to develop a passive immunization strategy for prevention of dengue virus infection. Our investigators have described a DENV-4-specific chimpanzee Fab fragment and a derived full-length humanized IgG antibody highly efficient for neutralization of DENV-4 (31). Our group has also identified chimpanzee Fab fragments, including 1A5, that exhibit a broad cross-reactivity to members of the flavivirus group and cross-neutralize DENV-1 and DENV-2 efficiently (11). The present study describes mapping the epitope determinants of Fab 1A5 by analysis of DENV-2 antigenic variants. A determinant critically involved in Fab 1A5 antibody binding and neutralization maps to Gly106 within the flavivirus conserved fusion loop in domain II of E. Another determinant affecting antibody neutralization, but not antibody binding, maps to His317 in domain III of the neighboring E monomer. Amino acid substitutions in these DENV-2 variants lower the pH threshold for membrane fusion of the infected cells. From the locations of these amino acids in the 3-D structure, the Fab 1A5 antibody appears to recognize a novel epitope on E that has not been mapped before.
MATERIALS AND METHODS
Dengue viruses and cultured cells.
Simian Vero cells and mosquito C6/36 cells were grown in minimum essential medium (MEM) plus 10% fetal bovine serum (FBS), 2 mM l-glutamine, 0.05 mg of gentamicin/ml, and 2.5 U of amphotericin B (Fungizone)/ml. Mouse-adapted DENV-2 New Guinea B (NGB) and New Guinea C (NGC) strains were used for selection of antigenic variants. DENV-2 NGC was provided by K. Eckels (6), and DENV-2 NGB at mouse passage 11 was provided by W. Schlesinger. Stocks of the dengue viruses were prepared from infected C6/36 cells grown in VP-SFM medium (Invitrogen). The titers of these viruses were approximately 107 PFU/ml and were determined on Vero cell monolayers.
Antibodies.
Chimpanzee Fab 1A5 was identified by panning of a phage library using DENV-2 as described previously (11). Polyhistidine-tagged Fab 1A5, expressed in Escherichia coli, was affinity purified using TALON affinity resin (Clontech). The concentration of Fab was determined colorimetrically using the BCA protein assay kit (Pierce). Hyperimmune mouse ascites fluid (HMAF) raised against DENV-2 and DENV-4 was purchased from the American Type Culture Collection. Mouse MAb 3H5, specific to DENV-2, was kindly provided by R. Putnak (20).
Plaque reduction neutralization test (PRNT).
Approximately 50 PFU of DENV-2, or other viruses to be tested, were mixed with Fab 1A5 serially diluted in 250 μl of MEM. The mixture was incubated at 37°C for 1 h prior to use for infection of Vero cells or C6/36 cells in duplicate wells of a 24-well plate. Infected Vero cells were added with a medium overlay containing 1% gum tragacanth (Sigma) and incubated at 37°C for 3 days. Infected C6/36 cells were overlaid with medium containing 0.8% methyl cellulose and incubated at 32°C for 5 days. Foci of infected cells were visualized by immuno-staining, using HMAF and anti-mouse IgG peroxidase (Pierce). The Fab titer in micrograms per milliliter that produced 50% reduction of foci (PRNT50) was calculated from at least three experiments.
Selection of DENV-2 antigenic variants.
Affinity-purified Fab 1A5 was used for selection of antigenic variants from mouse-passaged DENV-2 NGB and DENV-2 NGC, both of which had been previously sequenced in the C-prM-E region (6; I. Tokimatsu, unpublished observations). Parental DENV-2 NGB or DENV-2 NGC, approximately 107 PFU, was mixed with Fab 1A5 at 80 μg/ml (equivalent to 100 PRNT50 titers) in MEM and incubated at 37°C for 1 h. The mixture was added to the Vero cell monolayer in a 35-mm culture plate for adsorption at 37°C for 1 h. The monolayer was rinsed once with phosphate-buffered saline (PBS), 3 ml of MEM containing 2% FBS plus 5 μg of Fab 1A5/ml was added, and then the cells were incubated at 37°C for 7 days. Progeny virus in the culture medium was collected for neutralization with Fab 1A5, followed by infection of Vero cells again. The neutralization cycle was repeated, and the Fab 1A5-resistant phenotype of progeny virus was monitored. Fab 1A5-resistant variants were isolated by plaque-to-plaque purification three times on Vero cells prior to amplification in C6/36 cells in the absence of the antibody.
Sequence analysis of antigenic variants.
Genomic RNA of each antigenic variant following amplification in C6/36 cells was extracted using TRIzol solution (Life Technologies). Reverse transcription of RNA with the primer AGTCTTGTTACTGAGCGGATTCC at nucleotide positions 2587 to 2565 in DENV-2 NS1 was carried out using the Superscript kit (Life Technologies). Amplification of C-prM-E DNA with appropriate primers by PCR was performed with AmpliTaq DNA polymerase (Perkin-Elmer). The DNA product was sequenced using primers spanning the DNA segment with an ABI sequencer (Perkin-Elmer, Applied Biosystems). The sequences of 8 to 10 plaque-purified isolates from each variant were analyzed. Sequence assembly was performed using the Vector NTI suite (InforMax). Structural modeling of the mutant E protein was performed using SwissModel and the crystal coordinates of DENV-2 (1OAN.pdb) as the template (13, 32). Swiss-Pdb Viewer was used for graphical development.
Construction of DENV-2/DENV-4 chimeras.
Construction of chimeric cDNA containing the C-prM-E sequence of parental DENV-2 NGB, DENV-2 NGC, or their antigenic variants on the DENV-4 background was as described elsewhere (5). Briefly, parental or variant DENV-2 C-prM-E DNA was generated by reverse transcription of virion RNA and PCR amplification. The DNA product was digested with BglII and XhoI and then cloned into plasmid p5′-2, replacing the corresponding DENV-4 sequence. The ClaI-XhoI fragment of p5′-2 DNA containing the DENV-2 C-prM-E sequence was then used to replace the corresponding fragment of full-length DENV-4 DNA, generating full-length chimeric DENV-2/DENV-4 DNA. Confluent C6/36 cells were transfected with the RNA transcripts of the chimeric DENV-2/DENV-4 DNA construct as described previously (5, 24). Three weeks after transfection, the culture medium had a titer greater than 106 PFU/ml as determined by focus assay on C6/36 cells. The C-prM-E DNA segment of progeny virus was prepared for sequence verification.
Construction of DENV-4 variants.
Two silent mutations, A to C at nucleotide 378 and C to T at nucleotide 381 near the fusion loop encoding sequence in E, were first introduced to create a unique AgeI site in full-length DENV-4 DNA (24). Site-directed mutagenesis by PCR was performed using a forward primer, GTTTGACAGCTTATCATCGATAAGC, corresponding to nucleotides 8 to 32 of pBR322, and a reverse primer containing the AgeI cleavage sequence and the following nucleotide substitution(s) in E: G to C at nucleotide 310 and G to A at nucleotide 311 for generating the Gly104His substitution; G to T at nucleotide 317 for the Gly106Val substitution; and G to C at nucleotide 321 for the Leu107Phe substitution. The PCR products, digested with ClaI and AgeI, were each cloned into full-length DENV-4 DNA. RNA transcription and transfection of C6/36 cells and recovery of virus were performed as described above.
Polyacrylamide gel electrophoresis and Western blotting.
Dengue virus was mixed with an equal volume of 2× sample buffer (2% sodium dodecyl sulfate, 20% glycerol, 20 mM Tris-HCl, 0.02% bromophenol blue) with or without 0.5% β-mercaptoethanol. The virus mixture was boiled for 10 min prior to loading for separation by polyacrylamide gel electrophoresis. The protein gel was blot transferred onto a nitrocellulose membrane electrophoretically. The protein blot was treated with 5% skim milk and reacted with Fab 1A5 or MAb 3H5 for 1 h. The blot was then washed with Tris-buffered saline containing 0.05% Tween 20 three times and reacted with goat anti-human IgG or anti-mouse IgG peroxidase (Pierce) at room temperature for 1 h. The protein blot was developed with Sigma Fast 3,3′-diaminobenzidine (Sigma-Aldrich).
Antibody binding affinity assay.
An enzyme-linked immunosorbent assay (ELISA) was performed to determine the binding affinity of Fab 1A5 to parental DENV-2 and its antigenic variants (26, 35, 38). Briefly, MAb 3H5-coated wells of a microtiter plate were blocked with 3% bovine serum albumin (BSA), and then each virus was added to separate wells. Following incubation at 37°C for 1 h, affinity-purified Fab 1A5 in serial dilution was added and the plate was incubated at 37°C for 1 h. Fab 1A5 that bound to DENV-2 on the microtiter plate was detected using goat anti-human IgG-alkaline phosphatase (Sigma). The apparent affinity constant, termed the ELISA Kd was calculated for the Fab 1A5 concentration (in nanomolar) that produced 50% of maximum binding.
Binding of Fab 1A5 to oligopeptides.
Three oligopeptides were analyzed: control peptide 1, GAMHSALAGATEVD; control peptide 2, WWWQTFDAR (48); and fusion peptide, DRGWGNGSGLFGKGG. The control peptides contained sequences unrelated to the fusion sequence, and the fusion peptide contained the entire fusion sequence with a Ser substitution for Cys. In a direct binding assay, a 96-well microtiter plate was coated with each of the oligopeptides at 5 μg/well in 0.1 M carbonate buffer, pH 9.6. After washing with PBS containing 0.05% Tween 20 and then blocking with PBS containing 3% BSA, Fab 1A5 in PBS containing 1% BSA was added. Fab 1A5 that bound to the oligopeptides was detected using goat anti-human IgG-alkaline phosphatase (Sigma). The competition binding assay was performed essentially as described elsewhere (48). Briefly, purified Fab 1A5 at 0.05 μg/ml was preincubated with each of the oligopeptides in serial dilution at 37°C for 2 h. The reaction mixture was added to the wells of a microtiter plate coated with 25 μl of DENV-2 at 105 PFU/ml in PBS plus 1% BSA. Fab 1A5 bound to DENV-2 was detected as described above.
Plaque morphology and growth analysis.
Vero cells in a six-well plate were infected with parental DENV-2 NGB, DENV-2 NGC, or an antigenic variant and overlaid with medium containing 1% gum tragacanth. After incubation at 37°C for 5 days, viral plaques were visualized by immuno-staining. The diameters of 20 plaques from each virus were measured on a digital image using Adobe Photoshop. For growth analysis, confluent monolayers of Vero cells or C6/36 cells in a 24-well plate were infected with each C6/36 cell-amplified virus at a multiplicity of infection (MOI) of 0.01 in duplicate. Infected Vero cells were incubated at 37°C and C6/36 cells at 32°C, and the culture medium was collected daily for 7 days. The virus sample was clarified by centrifugation, and the titer was determined by focus assay on Vero cells.
Mouse neurovirulence.
Neurovirulence of parental DENV-2 NGB and its antigenic variants was evaluated in outbred Swiss mice. Three-day-old suckling mice, in groups of 8 to 11, were inoculated by the intracranial (i.c.) route with 100, 10, or 1 PFU of each virus in 20 μl of MEM containing 0.25% human serum albumin. Inoculated mice were observed for symptoms of encephalitis, including ruffled hair, hunched back, paralysis, and death. Paralyzed, moribund mice were euthanized and scored during the 4-week observation period. Student's t test was used to compare the 50% lethal dose (LD50) in PFU between the parental DENV-2 and its antigenic variants.
Fusion activity assay.
Fusion-from-within (FFWI) assays were performed for the DENV-2 parent and its antigenic variants as described previously (14, 39). C6/36 cell monolayers in a 24-well plate were infected with each virus at 0.2 MOI in MEM plus 10% FBS, buffered with 10 mM 4-(2-hydroxyethyl)piperazine-1-ethanesulfonic acid (HEPES) at pH 7.7, and incubated at 32°C. Four to five days after infection, the infected cell monolayer was rinsed once with PBS, and fusion medium (MEM plus 20 mM HEPES for pH 7.0 to 7.8, or 20 mM 2-morpholinoethanesulonic acid for pH 5.4 to 6.6) was added before incubation at 40°C for 2 h. The infected cells were stained using the Diff-Quik stain set (Dade Behring) and examined for syncytium formation microscopically. The fusion index, defined as (1 − [number of cells/number of nuclei]), was calculated by counting 300 nuclei for each virus in at least five microscopic fields. The percentage of infected cells was determined by immunofluorescence assay with HMAF. Fusion inhibition by Fab 1A5 was performed as described elsewhere (16). In brief, DENV-1- or DENV-2-infected C6/36 cells were incubated with Fab 1A5 at 37°C for 1 h prior to exposure to the low-pH medium. Infected cells were also incubated with MAb 3H5 in parallel as the control.
RESULTS
Selection of DENV-2 antigenic variants using Fab 1A5.
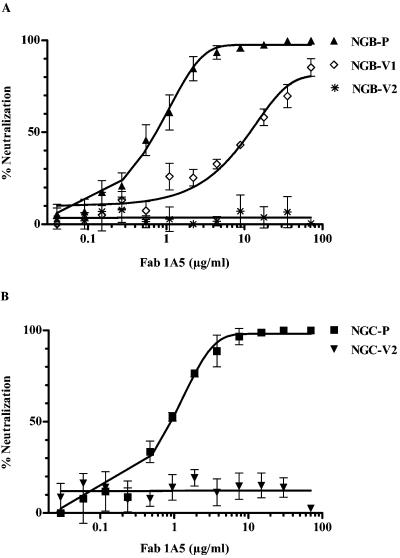
Mouse-adapted, neurovirulent DENV-2 NGB and DENV-2 NGC were used for selection of antigenic variants resistant to Fab 1A5 by neutralization in vitro. One DENV-2 NGB antigenic variant, designated as NGB-V1, was isolated after eight cycles of neutralization and Vero cell passage. The PRNT50 titer of NGB-V1 was 12.0 μg/ml, while that of parental DENV-2 NGB was 0.74 μg/ml (Fig. 1A). A second antigenic variant, designated NGB-V2, was isolated after 11 rounds of neutralization. NGB-V2 was completely resistant to neutralization by Fab 1A5 (>70 μg/ml). In parallel, selection of DENV-2 NGC variants of Fab 1A5 was also performed to provide additional information. This effort yielded one antigenic variant, termed NGC-V2. The PRNT50 titer of NGC-V2 was >70 μg/ml, while that of parental DENV-2 NGC was 0.89 μg/ml (Fig. 1B).
FIG. 1.
Neutralization of DENV-2 parental viruses and their variants using Fab 1A5. (A) Results for NGB-P (DENV-2 NGB parent), DENV-2 variant NGB-V1, and DENV2 variant NGB-V2. (B) Results for NGC-P (DENV-2 NGC parent) and DENV-2 variant NGC-V2. PRNT was performed using approximately 50 PFU of each virus for incubation with serially diluted Fab 1A5 at 37°C for 1 h. The reaction mixture was used to infect Vero cells. Foci of infected cells were detected by immuno-staining.
Sequence analysis of DENV-2 antigenic variants.
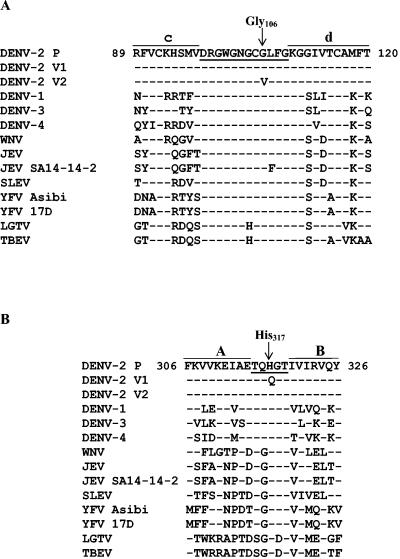
To map the Fab 1A5 epitope, the C-prM-E genes of antigenic variants NGB-V1, NGB-V2, and NGC-V2 and the parental viruses were sequenced. Variant NGB-V1 contained five nucleotide mutations in E, compared to the sequence of parental DENV-2 NGB (Table 1). Only the mutation at nucleotide 951 resulted in an amino acid substitution, Gln for His, at position 317 in E, whereas other nucleotide changes were silent mutations. The E sequence of variant NGB-V2 contained two nucleotide changes: a silent mutation of C to T at nucleotide 222, which was also present in NGB-V1, and a G-to-T mutation at position 317 that resulted in substitution of Val for Gly at position 106. Nucleotide changes were not found in the C-prM genes of variant NGB-V1 or NGB-V2. Variant NGC-V2 contained only a G-to-T change at nucleotide 317 in E that resulted in substitution of Val for Gly at position 106, identical to that found in NGB-V2. Figure 2 shows alignment of the flavivirus E sequences surrounding Gly106 (panel A) and His317 (panel B). Gly106 is located within the 12-amino-acid fusion peptide sequence (positions 98 to 109) that is nearly conserved among the arthropod-borne flaviviruses. His317 in E is also conserved among flaviviruses, although the surrounding sequences vary. In the 3-D structure, Gly106 is located in the cd loop at the tip of domain II and His317 is located between β-sheets A and B in domain III (Fig. 3A and B). Despite their locations in different domains, Gly106 and His317 of the opposite E monomer are spatially close, approximately 16 Å apart, as calculated with the SwissModel program (13).
TABLE 1.
Nucleotide and amino acid changes in the E proteins of antigenic variants compared to their parental viruses
| Variant | Nucleotide change | Amino acid change | Domain |
|---|---|---|---|
| NGB-V1a | 222C → T | No | |
| 402T → C | No | ||
| 468A → G | No | ||
| 526A → G | No | ||
| 951T → A | 317His → Gln | III | |
| 222C → T | No | ||
| NGB-V2a | 222C → T | No | |
| 317G → T | 106Gly → Val | II | |
| NGC-V2b | 317G → T | 106Gly → Val | II |
No amino acid changes were found in the C-PreM region.
A substitution of Ala for Thr at position 280, the last amino acid of prM, was found.
FIG. 2.
Alignment of amino acid sequences among flaviviruses. (A) Sequences surrounding Val106 found in DENV-2 variants NGB-V2 and NGC-V2. The fusion sequence (loop) between the c and d β-strands is underlined. (B) Sequences surrounding Gln317 present in DENV-2 variant NGB-V1. The sequence between the A and B β-strands is underlined. Flavivirus sequences were obtained from the following references: DENV-1 (30); DENV-2 (18); DENV-3 (37); DENV-4 (52); WNV (25, 50); St. Louis encephalitis virus (SLEV) (49); JEV JaOAr S982 (47); JEV SA14-14-2 (36); YFV 17D (41); YFV Asibi (17); LGTV (29); TBEV (28).
FIG. 3.
Localization of Fab 1A5 epitope determinants in 3-D structure of DENV-2 E. (A) Positions of Gly106 and His317 (both in light blue) as viewed from the top of the dimeric E structure with domain I (red), domain II (yellow), and domain III (blue), based on the published coordinates (32). (B) Expanded area of the insert in panel A.
Neutralization of DENV-2/DENV-4 chimeras by Fab 1A5.
Sequence analysis of antigenic variants indicated that Fab 1A5 appeared to recognize a novel epitope involving two closely spaced amino acids in different domains and from two interacting homodimers of DENV-2 E. The antigenic variants containing these mutations differed from the parent viruses in their Fab 1A5 neutralization titer. To provide additional evidence, we constructed DENV-2/DENV-4 chimeras composed of the parental DENV-2 NGB C-prM-E sequence or the variant C-prM-E sequence specifying the His317-Gln or Gly106-Val substitution present in NGB-V1 and NGB-V2, respectively, on the DENV-4 genetic background. As predicted, Fab 1A5 neutralized the chimeric DENV-2 (NGB-P)/DENV-4 at a PRNT50 titer of 0.64 μg/ml, similar to that measured for parental DENV-2 NGB (data not shown). Substitution of Gly106Val or His317Gln in DENV-2 E of these chimeras conferred resistance to neutralization by Fab 1A5. The chimera containing Gly106Val had a PRNT50 titer of >70 μg/ml and the chimera containing His317Gln had a PRNT50 titer of 31.7 μg/ml, similar to that measured for NGB-V2 and NGB-V1, respectively.
Binding affinity of Fab 1A5 to antigenic variants.
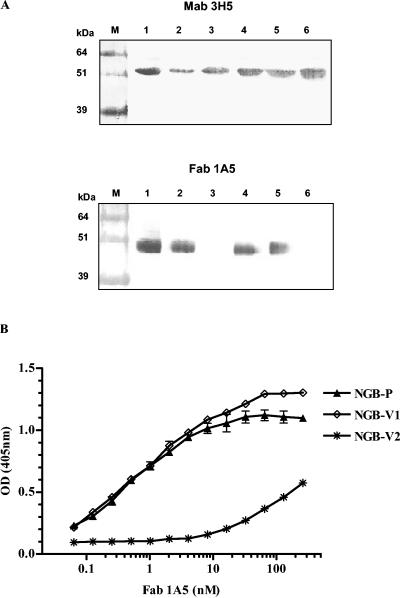
To gain an insight into the neutralizing mechanism, the Fab 1A5 binding activities of the DENV-2 NGB parent virus and its variants were first analyzed by Western blotting. MAb 3H5, which had been shown to recognize an epitope at or near positions 383 to 385 of DENV-2 E (20), was used for comparison. MAb 3H5 reacted to the DENV-2 NGB parent, variant NGB-V1, NGB-V2, and each of the chimeras similarly. Under the same conditions, Fab 1A5 reacted with the DENV-2 NGB parent and variant NGB-V1, but not with variant NGB-V2 (Fig. 4A, top panel). Similarly, binding of Fab 1A5 to the DENV-2 NGB-V1/DENV-4 chimera, but not to the DENV-2 NGB-V2/DEN4 chimera, was observed (Fig. 4A, bottom panel).
FIG. 4.
Reactivity of Fab 1A5 to DENV-2 NGB parent and its antigenic variants. (A) Top: binding of control MAb 3H5 (which does not bind to the fusion peptide) to various viruses by Western blot analysis. Lanes: 1, DENV-2, NGB parent; 2, DENV-2 NGB-V1; 3, DENV-2 NGB-V2; 4, NGB-parent/DENV-4 chimera; 5, NGB V1/DENV-4 chimera; 6, NGB-V2/DENV-4 chimera. (Bottom) Binding of Fab 1A5 to the viruses listed above by Western blot analysis. Boiled dengue virus samples in the absence of β-mercaptoethanol were separated on sodium dodecyl sulfate-polyacrylamide gels by electrophoresis for Western blot analysis. Note that the electrophoretic mobility of the DENV-2 E bands that reacted with MAb 3H5 and with Fab 1A5 varied on the gel blot, presumably reflecting E protein species glycosylated differently. (B) Binding of Fab 1A5 to the DENV-2 NGB parent and its antigenic variants by ELISA.
An ELISA was performed to semiquantify the binding affinity of Fab 1A5 for DENV-2 NGB and its two variants (Fig. 4B and Table 2). The apparent binding affinity ELISA Kd of Fab 1A5 for highly resistant variant NGB-V2 was the lowest among the three viruses. Thus, Gly106 represented a major determinant of the Fab 1A5 epitope on the DENV-2 E. On the other hand, the binding affinity of Fab 1A5 for variant NGB-V1 was not appreciably reduced compared to that for the DENV-2 NGB parent. It is possible that His317 represented a minor determinant of the Fab 1A5 epitope and affected Fab 1A5 neutralization through a steric effect.
TABLE 2.
Apparent binding affinities of Fab 1A5 for parental DENV-2 NGB and its variants
| DENV-2 | ELISA Kd (nM) | Affinity reduction (fold) |
|---|---|---|
| NGB-Pa | 0.47 ± 0.18 | |
| NGB-V1 | 0.75 ± 0.31 | 1.60 |
| NGB-V2 | 37.75 ± 1.11 | 80.32 |
NGB-P, parental DENV-2 NGB.
Disulfide bridge dependency of the Fab 1A5 epitope.
In the DENV-2 E sequence, Fab 1A5 epitope determinant Gly106 is followed by Cys105, which forms a disulfide bridge with Cys74. It was of interest to provide data in support of the requirements of this and other disulfide bridges for functional integrity of the Fab 1A5 epitope. Treatment of DENV-2 NGB with β-mercaptoethanol abolished binding of Fab 1A5, as determined by Western blot analysis. MAb 3H5, which recognizes a conformational epitope on DENV-2 E, also failed to bind DENV-2 NGB that was similarly treated (data not shown).
Reactivity of Fab 1A5 to an oligopeptide containing the fusion peptide sequence.
Two separate assays were performed to detect the reactivity of Fab 1A5 with oligopeptides bearing the fusion peptide sequence or unrelated sequences. Binding of Fab 1A5 to each of these oligopeptides (see Materials and Methods) immobilized on wells of a microtiter plate was not detected (data not shown). Competition binding was then performed in which Fab 1A5 was allowed to bind the individual oligopeptides in solution prior to testing for binding to DENV-2. The result in Fig. 5 indicates that binding of Fab 1A5 to DENV-2 was competed by the fusion peptide sequence at the 50% inhibitory concentration of 0.17 mM, whereas each of the two control peptides containing unrelated sequences failed to compete, or only poorly. The concentration of Fab 1A5 used in the inhibition assay was as low as 1.04 nM. One interpretation of this result is that the oligopeptide in solution was able to assume the conformation that is required for binding to Fab 1A5, but rather inefficiently.
FIG. 5.
Inhibition of Fab 1A5 binding to DENV-2 by a fusion peptide. In the binding competition assay, Fab 1A5 was mixed with serial dilutions of an oligopeptide containing the entire fusion peptide sequence (cd loop peptide) or a control peptide with an unrelated sequence (see Materials and Methods). The mixtures were tested for binding to an ELISA plate coated with DENV-2.
Growth analysis of DENV2 NGB antigenic variants.
Four days after infection of Vero cells, parental DENV-2 NGB, DENV-2 NGC, and variant NGBV-1 containing a His317Gln substitution produced plaques similar in size, averaging 1.2 ± 0.2, 1.3 ± 0.1, and 1.1 ± 0.2 mm, respectively (means ± standard errors). Under the same conditions, variant NGB-V2 and NGC-V2 containing the Gly106Val substitution produced plaques of 0.4 ± 0.1 and 0.6 ± 0.1 mm, respectively, appreciably smaller than their parental virus. The growth kinetics of variant NGB-V1 and its parental virus were similar in C6/36 cells and in Vero cells (Fig. 6A and B). On the other hand, variant NGB-V2 consistently yielded a titer 10-fold lower than its parental virus in C6/36 cells and in Vero cells during the log-phase period, i.e., at 3, 4, and 5 days after infection. Similarly, Gly106Val substitution reduced replication of DENV-2/DENV-4 chimeras in C6/36 and Vero cells (Fig. 6C and D). The chimera containing His317Gln replicated to a level that was comparable to that of NGB-V1 in C6/36 cells. For reasons not understood, the chimeras containing His317Gln failed to replicate in Vero cells. Thus, Fab 1A5 selected antigenic variants that were attenuated or, at least, similar to the parental virus for growth in mammalian or insect cells.
FIG. 6.
Growth analysis of antigenic variants, chimeras, and the DENV-2 NGB parent in cultured cells. The DENV-2 NGB parent and its antigenic variants were analyzed for growth in C6/36 cells (A) and in Vero cells (B). Chimeras that contained C-prM-E of the parental NGB, variant NGB V-1, or NGB V-2 on the DENV-4 background were similarly analyzed for growth in C6/36 cells (C) and in Vero cells (D). Cells were infected with each virus at an MOI of 0.01, and the culture medium was collected daily for titer determination by focus assay on Vero cells.
Mouse neurovirulence of DENV-2 antigenic variants.
Mouse neurovirulence of the DENV-2 NGB antigenic variants was evaluated by i.c. inoculation of 3-day-old outbred Swiss mice. Mice infected with the DENV-2 NGB parent developed symptoms of encephalitis and eventually succumbed to infection (Tokimatsu, unpublished). Table 3 shows that the LD50 of variant NGB-V1 was 8.9 PFU, not significantly different from the LD50 of 4.5 PFU calculated for the parental virus. The LD50 of variant NGB-V2 at 16.4 PFU was significantly lower than that of the parental virus, indicating that the variant containing the Gly106Val substitution was attenuated.
TABLE 3.
Neurovirulence of parental DENV-2 NGB and its variants following i.c. inoculation in suckling Swiss mice
| Virus | Mortality (%)b of mice after i.c. inoculation at PFU of:
|
Mean LD50 ± SE (PFU)c | ||
|---|---|---|---|---|
| 100 | 10 | 1 | ||
| NGB-Pa | 20/20 (100) | 19/21 (90.5) | 3/10 (30) | 4.52 ± 0.07 |
| NGB-V1 | 19/20 (95) | 14/20 (70) | 2/10 (20) | 8.9 ± 3.6* |
| NGB-V2 | 18/18 (100) | 9/19 (47.4) | 2/10 (20) | 16.4 ± 0.28** |
NGB-P, parental DENV-2 NGB.
The mortality rates at 100 and 10 PFU are based on the cumulative results of two experiments. Values are the number dead per number inoculated.
*, P = 0.23; **, P = 0.0065.
Fusion activity of DENV-2 antigenic variants.
Since the mutation site of variant NGB-V2 was mapped within the flavivirus conserved fusion peptide loop, the attenuating phenotype of the variant might be associated with alteration of membrane fusion. Initially, the fusion activities of the DENV-2 NGB parent and its variants were examined on infected C6/36 cells. Syncytium formation of the cell monolayer was evident 2 days after infection with parental DENV-2 NGB. At 4 to 5 days after infection, cells of the entire monolayer formed syncytia and the cytopathic effect was extensive. In contrast, formation of syncytium was not observed on cells infected with either NGB-V1 or NGB-V2 under the same conditions, and the cytopathic effect was not seen until 7 days of infection. Reduced fusion of C6/36 cells infected with the DENV-2/DENV-4 chimeras containing the amino acid substitution present in NGB-V1 or NBG-V2 was also evident, compared to cells infected with the chimera containing the parental sequence (data not shown).
We also studied fusion of C6/36 cells infected with DENV-2 NGB and its antigenic variants at various pHs in the FFWI assay. Little or no fusion was observed at pH 7.0, 7.4, and 7.8. At pH 6.8, approximately 84% of the cells infected with parental DENV-2 formed syncytia. In contrast, 37% of cells infected with variant NGB-V1 and 46% of cells infected with variant NGB-V2 formed syncytia. Figure 7 shows the fusion activity in terms of the index for the DENV-2 NGB parent and its variants determined at various pHs. Accordingly, the DENV-2 NGB parent had a pH threshold for 50% maximum fusion activity (fusion index = 0.5) at pH 6.77, for variant NGB-V1 it was at pH 6.55, and for variant NGB-V2 it was at pH 6.41.
FIG. 7.
Fusion activity of DENV-2 NGB parent or its variants. FFWI assay was performed on C6/36 cells infected with each of the viruses at an MOI of 0.2 for 4 to 5 days at 32°C. The fusion activities of infected cells in the fusion medium at various pHs were detected by syncytium formation. The fusion index was calculated to determine the pH threshold for each virus.
Neutralizing activity of Fab 1A5 against DENV-4 mutants containing a Gly106Val or Leu107Phe substitution in the fusion loop.
Alignment of the flavivirus fusion sequences indicated JEV SA 14-14-2 contains a substitution of Phe for Leu at position 107 and Langat virus (LGTV) contains a His substitution for the Gly at position 104 (Fig. 2). The neutralizing activity of Fab 1A5 against JEV SA 14-14-2 and LGTV was the lowest among the flaviviruses tested (11). We questioned if substitution of Leu107Phe or Gly104His contributed to the resistance of these viruses to Fab 1A5 neutralization. The question of whether Gly106 represented a determinant of the Fab 1A5 epitope on DENV-4 E was also raised.
To address the above questions in aggregate, full-length DENV-4 DNA was used to construct mutants containing various substitutions in the fusion peptide for analysis. DENV-4 mutants containing either Gly106Val or Leu107Phe were successfully constructed; however, a DENV-4 mutant containing the Gly104His substitution was apparently not viable. Figure 8A shows the binding of Fab 1A5 to the DENV-4 parent and mutants containing a Leu107Phe or Gly106Val substitution. Fab 1A5 for the DENV-4 mutant containing Gly106Val had a binding affinity (ELISA Kd) of >40 nM, significantly reduced compared to that of the DENV-4 parent (ELISA Kd = 0.65 nM; P < 0.0001). Similarly, substitution of Leu107Phe in DENV-4 lowered the binding affinity of Fab 1A5 to an ELISA Kd of 3.07 ± 0.27 nM (P < 0.001). Figure 8B presents Fab 1A5 neutralization of the DENV-4 parent and mutants. The PRNT50 titers of the parental DENV-4, mutant Gly106Val, and mutant Leu107Phe were 4.3, >50, and approximately 50 μg/ml, respectively. The neutralizing titer of Fab 1A5 against each of the DENV-4 mutants was greatly reduced compared to that against DENV-4. These observations suggest that both Gly106 and Leu107 are Fab 1A5 epitope determinants on DENV-4 E.
FIG. 8.
Binding activity of Fab 1A5 to DENV-4 parent and DENV-4 mutants containing a substitution of Gly106Val or Leu107Phe in the fusion loop (A) and neutralizing activity of Fab 1A5 against these viruses (B). The binding activity of Fab 1A5 to the DENV-4 parent and its derived mutants was determined by ELISA. PRNT was performed to determine the neutralizing activity. OD, optical density.
DISCUSSION
Epitope analysis of chimpanzee Fab 1A5 was performed to gain an insight into the binding site on the E protein responsible for the flavivirus cross-reactivity and the mechanism for cross-neutralization of dengue viruses and other flaviviruses. Our current understanding of the flavivirus E antigenic structure and functional activities has been obtained from studies using mouse MAbs against these viruses. The present study represents the first analysis of any primate-derived MAbs against an arthropod-borne flavivirus.
Analysis of mutations in the C-prM-E region of DENV-2 antigenic variants established that Fab 1A5 recognized a novel epitope on the DENV-2 E protein that is defined by two closely spaced amino acids from the two neighboring E subunits of the homodimer. Gly106 at the tip of the flavivirus conserved fusion loop in domain II represents a major determinant of the Fab 1A5 epitope on DENV-2 E. DENV-2 variants and chimeras containing a Val substitution at this position failed to bind Fab 1A5, and these viruses were completely resistant to Fab 1A5. Substitution of Val for Gly106 is a nonconserved amino acid change, resulting in an increase of the mean hydrophobicity in the Cys-Gly-Leu tripeptide segment (positions 105 to 107) from 1.69 for parental DENV-2 to 2.69 for the variant.
Evidence suggests that His317 in domain III from the adjacent E monomer represents another determinant of the Fab 1A5 epitope. First, the DENV-2 antigenic variant and the DENV-2/DENV-4 chimera containing the His317Glu substitution were partially resistant to neutralization by Fab 1A5. Second, in the 3-D structure both Gly106 and His317 were spatially close, calculated to be 16.2 Å apart, well within the distance for antibody binding. Substitution of His317Gln in DENV-2 E did not significantly alter the hydrophobicity of this region and had little, if any, effect on the binding of Fab 1A5l. Thus, the mechanism for the involvement of this amino acid in Fab 1A5 neutralization is more complicated.
Mutations in flavivirus or other virus escape mutants have been localized outside the antibody binding sites (4, 21, 51). Such mutations might affect epitopes for antibody neutralization indirectly. Thus, His317 might play a lesser role than did Gly106 in the interactions with Fab 1A5. It can be assumed that the existence of the Fab 1A5 epitope requires interdomain interactions occurring either before or following the conformational shift from dimeric to trimeric E. MAbs that react with an epitope located in the interface between two adjacent subunits have been described for the influenza virus neuraminidase (44).
There is ample evidence indicating that the flavivirus fusion loop directly participates in the low-pH-induced membrane fusion believed to take place following viral entry. The low pH-triggered membrane fusion involves the dissociation of the flavivirus E dimer and subsequent formation of the E trimer (2, 15, 45, 46). Recent structural studies indicate that such a conformational switch allows the fusion loop in the interface between domains I and III of the dimeric E to become exposed, realigned, and made available for fusion interaction with the endosomal membrane (7, 33). Using a liposomal membrane model, it has been shown that recombinant subviral particles of TBEV prM and E containing a substitution of Asp or Phe for Leu107 in the fusion peptide sequence are defective for membrane fusion (1). It has been reported that several MAbs recognizing the domain A epitope (domain II) lost and regained their reactivities to TBEV upon lowering the pH (15). It is possible that these MAbs recognize a site in E that involves membrane fusion. DENV-2 MAbs capable of blocking membrane fusion of DENV-2-infected cells have also been shown to react with a tryptic peptide fragment consisting of the first 120 amino acids of E, although the epitope determinants of these antibodies have not been precisely mapped (42).
The present study with MAb Fab 1A5 has provided additional data supporting the involvement of the fusion loop in membrane fusion. First, a major epitope determinant of Fab 1A5 mapped to Gly106 within the fusion peptide loop. Second, Fab 1A5 inhibited membrane fusion, as demonstrated by reduced syncytium formation of DENV-2- and DENV-1-infected C6/36 cells. Third, the DENV-2 antigenic variants of Fab 1A5 that contained a Gly106Val substitution lowered the pH threshold for fusion of infected C6/36 cells. Binding of Fab 1A5 to DENV-2 and DENV-1 E probably inhibits the low-pH-induced conformational change of E, rendering the subsequent fusion step defective following viral entry.
Structural analysis of TBEV E indicates that His323 (His317 in DENV-2) also plays a role in the pH-induced transition of the E homodimer to become a homotrimer (7). The flavivirus conserved His323 of domain III forms a hydrogen bond with another flavivirus conserved residue in domain I to stabilize the interdomain contact between two E monomers. These investigators further suggested that protonation of this His residue serves to facilitate dissociation of the E homodimers at a low pH. In trimer E, domain III packs the interface of domain I and domain II. His323 also participates in the interaction. Our result showing that the DENV-2 variant NGB-V1 containing a His317Gln substitution has a lower fusion threshold for membrane fusion than the parental DENV-2 provides evidence for the involvement of His317 in membrane fusion.
Conservation of the fusion peptide sequence among flaviviruses suggests that the fusion peptide plays a central role in viral replication, and variants containing mutations in this region would be at an evolutionary disadvantage. Examination of the sequences in this region among flaviviruses indicates that JEV SA14-14-2 contains a Leu107Phe substitution along with many other substitutions, compared to the sequence of the parental JEV SA14 strain (13, 47). JEV strain SA14-14-2 is attenuated and has been used widely as a live vaccine in China. The contributions of Leu107Phe substitution alone and together with several others to the attenuating phenotype of JEV SA14-14-2 were studied in detail by analysis of the mouse neurovirulence phenotypes of mutants constructed to contain reversions to the wild-type sequence (3). The virulent parental JEV SA14 strain has a fusion peptide sequence identical to that of the dengue viruses and WNV. Importantly, other biosafety level 3 flaviviruses of medical importance, including SLEV and YFV, also have fusion peptides that share this exact sequence (Fig. 2). It will be interesting to determine if these viruses are also neutralized by Fab 1A5. However, it is clear that mutations in or near the flavivirus fusion peptide are not the sole determinants of attenuation, since the relatively attenuated LGTV and the virulent TBEV have identical sequences in the two regions studied here (28, 29), as do the attenuated YFV 17D strain and its virulent parent Asibi virus (17, 41).
Compared to the parental virus, the DENV-2 antigenic variant containing Gly106Val replicated less favorably in cultured simian Vero and mosquito cells in vitro and exhibited reduced neurovirulence in mice. Naturally occurring dengue virus variants, if they should ever emerge in the presence of the Fab 1A5 antibody, would be expected to be attenuated or at least similar to the parental virus. In this regard, Fab 1A5 or its humanized antibody appears to have an added safety feature for use in passive immunization against dengue virus and other flaviviruses.
Acknowledgments
We thank Kim Taylor and her staff for providing animal care, Walter Schlesinger for providing DENV-2 NGB, Kenneth Eckels for providing DENV-2 NGC, Robert Putnak for providing MAb 3H5, Ruhe Men for helpful discussions, and Lynn Rasmussen for nucleotide sequencing.
REFERENCES
- 1.Allison, S. L., J. Schalich, K. Stiasny, C. W. Mandl, and F. X. Heinz. 2001. Mutational evidence for an internal fusion peptide in flavivirus envelope protein E. J. Virol. 75:4268-4275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Allison, S. L., J. Schalich, K. Stiasny, C. W. Mandl, C. Kunz, and F. X. Heinz. 1995. Oligomeric rearrangement of tick-borne encephalitis virus envelope proteins induced by an acidic pH. J. Virol. 69:695-700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Arroyo, J., F. Guirakhoo, S. Fenner, Z. X. Zhang, T. P. Monath, and T. J. Chambers. 2001. Molecular basis for attenuation of neurovirulence of a yellow fever virus/Japnese encephalitis virus chimera vaccine (ChimeriVax-JE). J. Virol. 75:934-942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Blondel, B., R. Crainic, O. Fichot, G. Dufraisse, A. Candrea, D. Diamond, M. Girard, and F. Horaud. 1986. Mutations conferring resistance to neutralization with monoclonal antibodies in type 1 poliovirus can be located outside or inside the antibody-binding site. J. Virol. 57:81-90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bray, M., and C. J. Lai. 1991. Construction of intertypic chimeric dengue viruses by substitution of structural protein genes. Proc. Natl. Acad. Sci. USA 88:10342-10346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bray, M., R. Men, I. Tokimatsu, and C. J. Lai. 1998. Genetic determinants responsible for acquisition of dengue type 2 virus mouse neurovirulence. J. Virol. 72:1647-1651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bressanelli, S., K. Stiasny, S. L. Allison, E. A. Stura, S. Duquerroy, J. Lescar, F. X. Heinz, and F. A. Rey. 2004. Structure of a flavivirus envelope glycoprotein in its low-pH-induced membrane fusion conformation. EMBO J. 23:728-738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Calisher, C. H., N. Karabatsos, J. M. Dalrymple, R. E. Shope, J. S. Porterfield, E. G. Westaway, and W. E. Brandt. 1989. Antigenic relationships between flaviviruses as determined by cross-neutralization tests with polyclonal antisera. J. Gen. Virol. 70:37-43. [DOI] [PubMed] [Google Scholar]
- 9.Cecilia, D., and E. A. Gould. 1991. Nucleotide changes responsible for loss of neuroinvasiveness in Japanese encephalitis virus neutralization-resistant mutants. Virology 181:70-77. [DOI] [PubMed] [Google Scholar]
- 10.Crill, W. D., and J. T. Roehrig. 2001. Monoclonal antibodies that bind to domain III of dengue virus E glycoprotein are the most efficient blockers of virus adsorption to Vero cells. J. Virol. 75:7769-7773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Goncalvez, A. P., R. Men, C. Wernly, R. H. Purcell, and C.-J. Lai. 2004. Chimpanzee Fab fragments and a derived humanized immunoglobulin G1 antibody that efficiently cross-neutralize dengue type 1 and 2 viruses. J. Virol. 78:12910-12918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gubler, D. J. 1998. Dengue and dengue hemorrhagic fever. Clin. Microbiol. Rev. 11:480-496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Guex, N., and M. C. Peitsch. 1997. SWISS-MODEL and the Swiss-PdbViewer: an environment for comparative protein modeling. Electrophoresis 18:2714-2723. [DOI] [PubMed] [Google Scholar]
- 14.Guirakhoo, F., R. A. Bolin, and J. T. Roehrig. 1992. The Murray Valley encephalitis virus prM protein confers acid resistance to virus particles and alters the expression of epitopes within the R2 domain of E glycoprotein. Virology 191:921-931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Guirakhoo, F., F. X. Heinz, and C. Kunz. 1989. Epitope model of tick-borne encephalitis virus envelope glycoprotein E: analysis of structural properties, role of carbohydrate side chain, and conformational changes occurring at acidic pH. Virology 169:90-99. [DOI] [PubMed] [Google Scholar]
- 16.Guirakhoo, F., F. X. Heinz, C. W. Mandl, H. Holzmann, and C. Kunz. 1991. Fusion activity of flaviviruses: comparison of mature and immature (prM-containing) tick-borne encephalitis virus. J. Gen. Virol. 72:1323-1329. [DOI] [PubMed] [Google Scholar]
- 17.Hahn, C. S., J. M. Dalrymple, J. H. Strauss, and C. M. Rice. 1987. Comparison of the virulent Asibi strain of yellow fever virus with the 17D vaccine strain derived from it. Proc. Natl. Acad. Sci. USA 84:2019-2023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hahn, Y. S., R. Galler, T. Hunkapillar, J. M. Dalrymple, J. H. Strauss, and E. G. Struaa. 1988. Nucleotide sequence of dengue 2 RNA and comparison of the encoded proteins with those of other flaviviruses. Virology 162:167-180. [DOI] [PubMed] [Google Scholar]
- 19.Heinz, F. X. 1986. Epitope mapping of flavivirus glycoprotein. Adv. Virus Res. 31:103-168. [DOI] [PubMed] [Google Scholar]
- 20.Hiramatsu, K., M. Tadano, R. Men, and C. J. Lai. 1996. Mutational analysis of a neutralization epitope on the dengue type 2 virus (DEN2) envelope protein: monoclonal antibody resistant DEN2/DEN4 chimeras exhibit reduced mouse neurovirulence. Virology 224:437-445. [DOI] [PubMed] [Google Scholar]
- 21.Holzmann, H., K. Stiasny, M. Ecker, C. Kunz, and F. X. Heinz. 1997. Characterization of monoclonal antibody-escape mutants of tick-borne encephalitis virus with reduced neuroinvasiveness in mice. J. Gen. Virol. 78:31-37. [DOI] [PubMed] [Google Scholar]
- 22.Jiang, W. R., A. Lowe, S. Higgs, H. Reid, and E. A. Gould. 1993. Single amino acid codon changes detected in louping ill virus antibody-resistant mutants with reduced neurovirulence. J. Gen. Virol. 74:931-935. [DOI] [PubMed] [Google Scholar]
- 23.Komar, N. 2003. West Nile virus: epidemiology and ecology in North America. Adv. Virus Res. 61:185-234. [DOI] [PubMed] [Google Scholar]
- 24.Lai, C. J., B. T. Zhao, H. Hori, and M. Bray. 1991. Infectious RNA transcribed from stably cloned full-length cDNA of dengue type 4 virus. Proc. Natl. Acad. Sci. USA 88:5139-5143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lanciotti, R. S., J. T. Roehrig, V. Deubel, J. Smith, M. Parker, K. Steele, B. Crise, K. E. Volpe, M. E. Crabtree, J. H. Scherret, R. A. Hall, J. S. MacKenzie, C. B. Cropp, B. Panigrahy, E. Ostlund, B. Schmitt, M. Malkinson, C. Banet, J. Weissman, N. Komar, H. M. Savage, W. Stone, T. McNamara, and D. J. Gubler. 1999. Origin of the West Nile virus responsible for an outbreak of encephalitis in the northeastern United States. Science 286:2333-2337. [DOI] [PubMed] [Google Scholar]
- 26.Lin, C.-W. and S.-C. Wu. 2003. A functional epitope determinant on domain III of the Japanese encephalitis virus envelope protein interacted with neutralizing antibody combining sites. J. Virol. 77:2600-2606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mandl, C. W., F. Guirakhoo, H. Holzmann, F. X. Heinz, and C. Kunz. 1989. Antigenic structure of the flavivirus envelope protein E at the molecular level, using tick-borne encephalitis virus as a model. J. Virol. 63:564-571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mandl, C. W., F. X. Heinz, and C. Kunz. 1988. Sequence of the structural proteins of tick-borne encephalitis virus (western subtype) and comparative analysis with other flaviviruses. Virology 166:197-205. [DOI] [PubMed] [Google Scholar]
- 29.Mandl, C. W., L. Iacono-Connors, G. Wallner, H. Holzmann, C. Kunz, and F. X. Heinz. 1991. Sequence of the genes encoding the structural proteins of the low-virulence tick-borne flavivirus Langat TP21 and Yelantsev. Virology 185:891-895. [DOI] [PubMed] [Google Scholar]
- 30.Mason, P. W., P. C. McAda, T. L. Mason, and M. J. Fournier. 1987. Sequence of the dengue-1 virus genome in the region encoding the three structural proteins and the major nonstructural protein NS1. Virology 161:262-267. [DOI] [PubMed] [Google Scholar]
- 31.Men, R., T. Yamashiro, A. P. Goncalvez, C. Wernly, D. J. Schofield, S. U. Emerson, R. H. Purcell, and C.-J. Lai. 2004. Identification of chimpanzee Fab fragments by repertoire cloning and production of a full-length humanized immunoglobulin G1 antibody that is highly efficient for neutralization of dengue type 4 virus. J. Virol. 78:4665-4674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Modis, Y., S. Ogata, D. Clements, and S. C. Harrison. 2003. A ligand-binding pocket in the dengue virus envelope glycoprotein. Proc. Natl. Acad. Sci. USA 100:6986-6991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Modis, Y., S. Ogata, D. Clements, and S. C. Harrison. 2004. Structure of the dengue virus envelope protein after membrane fusion. Nature 427:313-319. [DOI] [PubMed] [Google Scholar]
- 34.Monath, T. P. 1994. Dengue: the risk to developed and developing countries. Proc. Natl. Acad. Sci. USA 91:2395-2400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Moore, J. P., Y. Cao, L. Qing, Q. J. Sattentau, J. Pyati, R. Koduri, J. Robinson, C. F. Barbas III, D. R. Burton, and D. D. Ho. 1995. Primary isolates of human immunodeficiency virus type I are relatively resistant to neutralization by monoclonal antibodies to gp120, and their neutralization is not predicted by studies with monmeric gp120. J. Virol. 69:101-109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Nitayaphan, S., J. A. Grant, G. J. Chang, and D. W. Trent. 1990. Nucleotide sequence of the virulent SA-14 strain of Japanese encephalitis virus and its attenuated vaccine derivative, SA-14-14-2. Virology 177:541-552. [DOI] [PubMed] [Google Scholar]
- 37.Osatomi, K., and H. Sumiyoshi. 1990. Complete nucleotide sequence of dengue type 3 genome RNA. Virology 176:643-647. [DOI] [PubMed] [Google Scholar]
- 38.Raffai, R., K. H. Weisgraber, R. MacKenzie, B. Rupp, E. Rassart, T. Hirama, T. L. Innerarity, and R. Milne. 2000. Binding of an antibody mimetic of the human low density lipoprotein receptor to apolipoprotein E is governed through electrostatic forces. Studies using site-directed mutagenesis and molecular modeling. J. Biol. Chem. 275:7109-7116. [DOI] [PubMed] [Google Scholar]
- 39.Randolph, V. B., and V. Stollar. 1990. Low pH-induced cell fusion in flavivirus-infected Aedes albopictus cell cultures. J. Gen. Virol. 71:1845-1850. [DOI] [PubMed] [Google Scholar]
- 40.Rey, F. A., F. X. Heinz, C. Mandl, C. Kunz, and S. C. Harrison. 1995. The envelope glycoprotein from tick-borne encephalitis virus at 2 Å resolution. Nature 375:291-298. [DOI] [PubMed] [Google Scholar]
- 41.Rice, C. M., E. M. Lenches, S. R. Eddy, S. J. Shin, R. L. Sheets, and J. H. Strauss. 1985. Nucleotide sequence of yellow fever virus: implications for flavivirus gene expression and evolution. Science 229:726-733. [DOI] [PubMed] [Google Scholar]
- 42.Roehrig, J. T. 2003. Antigenic structure of flavivirus proteins. Adv. Virus Res. 59:141-175. [DOI] [PubMed] [Google Scholar]
- 43.Roehrig, J. T., R. A. Bolin, and R. G. Kelly. 1998. Monoclonal antibody mapping of the envelope glycoprotein of the dengue 2 virus, Jamaica. Virology 246:317-328. [DOI] [PubMed] [Google Scholar]
- 44.Saito, T., G. Taylor, W. G. Laver, Y. Kawaoka, and R. G. Webster. 1994. Antigenicity of the N8 influenza A virus neuraminidase: existence of an epitope at the subunit interface of the neuraminidase. J. Virol. 68:1790-1796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Stiasny, K., S. L. Allison, J. Schalich, and F. X. Heinz. 2002. Membrane interactions of the tick-borne encephalitis virus fusion protein E at low pH. J. Virol. 76:3784-3790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Stiasny, K., S. Bressanelli, J. Lepault, F. A. Rey, and F. X. Heinz. 2004. Characterization of a membrane-associated trimeric low-pH-induced form of the class II viral fusion protein E from tick-borne encephalitis virus and its crystalization. J. Virol. 78:3178-3183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Sumiyoshi, H., C. Mori, I. Fuke, K. Morita, S. Kuhara, J. Kondou, Y. Kikuchi, H. Nagamatsu, and A. Igarashi. 1987. Complete nucleotide sequences of the Japanese encephalitis virus genome RNA. Virology 161:497-510. [DOI] [PubMed] [Google Scholar]
- 48.Thullier, P., C. Demangel, H. Bedouelle, F. Megret, A. Jouan, V. Deubel, J. C. Mazie, and P. Lafaye. 2001. Mapping of a dengue virus neutralizing epitope critical for the infectivity of all serotypes: insight into the neutralization mechanism. J. Gen. Virol. 82:1885-1982. [DOI] [PubMed] [Google Scholar]
- 49.Trent, D. W., R. M. Kinney, B. J. B. Johnson, A. V. Vorndam, J. A. Grant, V. Deubel, C. M. Rice, and C. Hahn. 1987. Partial nucleotide sequence of St. Louis encephalitis virus RNA: structural proteins, NS1, ns2a, and ns2b. Virology 156:293-304. [DOI] [PubMed] [Google Scholar]
- 50.Wengler, G., E. Castle, U. Leidner, T. Nowak, and G. Wengler. 1985. Sequence analysis of the membrane protein V3 of the flavivirus West Nile virus and its gene. Virology 147:264-274. [DOI] [PubMed] [Google Scholar]
- 51.Wilson, C., M. S. Reitz, Jr., K. Aldrich, P. J. Klasse, J. Blomberg, R. C. Gallo, and M. Robert-Guroff. 1990. The site of an immune-selected point mutation in the transmembrane protein of human immunodeficiency virus type 1 does not constitute the neutralization epitope. J. Virol. 64:3240-3248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Zhao, B., E. Mackow, A. Buckler-White, L. Markoff, R. M. Chanock, C.-J. Lai, and Y. Makino. 1986. Cloning full-length dengue type 4 virus DNA sequences: analysis of genes coding for structural proteins. Virology 155:77-88. [DOI] [PubMed] [Google Scholar]