Abstract
Final envelopment of the cytoplasmic herpes simplex virus type 1 (HSV-1) nucleocapsid is thought to occur by budding into trans-Golgi network (TGN)-derived membranes. The highly membrane-associated proteins UL20p and glycoprotein K (gK) are required for cytoplasmic envelopment at the TGN and virion transport from the TGN to extracellular spaces. Furthermore, the UL20 protein is required for intracellular transport and cell surface expression of gK. Independently expressed gK or UL20p via transient expression in Vero cells failed to be transported from the endoplasmic reticulum (ER). Similarly, infection of Vero cells with either gK-null or UL20-null viruses resulted in ER entrapment of UL20p or gK, respectively. In HSV-1 wild-type virus infections and to a lesser extent in transient gK and UL20p coexpression experiments, both gK and UL20p localized to the Golgi apparatus. In wild-type, but not UL20-null, viral infections, gK was readily detected on cell surfaces. In contrast, transiently coexpressed gK and UL20p predominantly localized to the TGN and were not readily detected on cell surfaces. However, TGN-localized gK and UL20p originated from endocytosed gK and UL20p expressed at cell surfaces. Retention of UL20p to the ER through the addition of an ER retention motif forced total ER retention of gK, indicating that transport of gK is absolutely dependent on UL20p transport. In all experiments, gK and UL20p colocalized at intracellular sites, including the ER, Golgi, and TGN. These results are consistent with the hypothesis that gK and UL20p directly interact and that this interaction facilitates their TGN localization, an important prerequisite for cytoplasmic virion envelopment and egress.
It is widely accepted that herpesvirus egress entails a two-stage envelopment process. Initially, capsids assemble within the nuclei and virions acquire an initial envelope by budding into the perinuclear spaces. Subsequently, these enveloped virions lose their envelope by fusion with the outer nuclear lamellae. Within the cytoplasm, tegument proteins associate with the viral nucleocapsid and final envelopment occurs by budding of cytoplasmic capsids into specific trans-Golgi network (TGN)-associated membranes. Mature virions subsequently traffic to cell surfaces, presumably following the cellular secretory pathway (31, 54, 65). Although several lines of evidence support this model of TGN assembly and egress (8, 30, 54, 76), the specific viral and cellular mechanisms responsible for the targeted trafficking of viral envelope glycoproteins to intracellular sites of virion envelopment are only recently being elucidated.
Herpes simplex viruses (HSVs) specify at least 11 virally encoded glycoproteins, as well as several nonglycosylated membrane-associated proteins, that are pivotal in membrane fusion processes during a productive viral infection. Mutations that cause extensive virus-induced cell-to-cell fusion have been mapped to at least four regions of the viral genome: the UL20 gene (1, 50, 53), the UL24 gene (38, 64), the UL27 gene encoding glycoprotein B (gB) (9, 57), and the UL53 gene coding for gK (3, 16, 34, 58, 59, 63). Of these four membrane-associated proteins, UL20p and gK are essential for the intracellular transport of virions to extracellular spaces (1, 25, 29, 35, 39, 55).
The UL20 and UL53(gK) genes encode multipass transmembrane proteins of 222 and 338 amino acids, respectively, that are conserved in all alphaherpesviruses (16, 50, 60). Although both proteins have multiple sites where posttranslational modification can occur, only gK is posttranslationally modified by N-linked carbohydrate addition (16, 34, 60). Viruses that specify deletions in either of these proteins are unable to translocate from the cytoplasm to extracellular spaces and accumulate enveloped virions within TGN-like cytoplasmic vesicles (1, 24, 25, 29, 35, 39, 53). Moreover, both transport of gK to cell surfaces and gK-mediated cell-to-cell fusion were abolished in UL20-null virus-infected cells (25). Taken together, these results suggest a functional interdependence between gK and UL20 for intracellular protein trafficking, virus egress, and virus-mediated cell-to-cell fusion (19, 23, 25, 29, 53).
Recent observations that both UL20-null and gK-null virions accumulated within TGN-like cytoplasmic vesicles (24, 25, 39) prompted the detailed examination of the cellular localization and interdependent transport of gK and UL20p. In this report, we show that UL20p and gK are mutually dependent for post-endoplasmic reticulum (ER) transport and specifically colocalize to the TGN after endocytosis from cell surfaces. These findings suggest that TGN localization of gK and UL20p may be required for cytoplasmic virion envelopment and subsequent virus trafficking from the TGN to extracellular spaces.
MATERIALS AND METHODS
Cells and viruses.
African green monkey kidney (Vero) cells were obtained from the American Type Culture Collection (Rockville, Md.). The UL53-gK V5 epitope-containing virus, gKDIV5, was propagated in Vero cells as described previously (23, 25, 26). The corresponding gK domain I V5-tagged UL20-null virus, ΔUL20/gKDIV5, was described previously (25) and was propagated on UL20-null-complementing G5 cells (18). The gK-null virus, ΔgK, was described previously (39) and was propagated on the gK-null-complementing cell line VK302 (35). For isolation and replication of the ΔUL20/ΔgK virus, the UL20-null/gK-null-complementing cell line, CV-1(c20cgK), was constructed in a CV-1 cell background through FLP recombinase target recombination essentially as described previously (53).
Plasmid construction.
The untagged UL20 plasmid, pUL20, and the pgKDIV5 plasmid, which specifies a V5 epitope tag within domain I of gK, were described previously (23). The pUL20amFLAG expression plasmid was generated by PCR amplification of the UL20 gene with the 5′-primer-coding sequence specifying an in-frame insertion of the FLAG epitope (MDYKDDDDK) immediately prior to the start codon of the HSV type 1 (HSV-1) UL20 gene. The resultant PCR product was inserted into the eukaryotic expression TOPO cloning vector pEF6 (Invitrogen, Carlsbad, Calif.), tested for orientation, and sequenced to confirm that the amino-terminal FLAG epitope fusion was present without intervening mutations in UL20. From this initial amino-terminal FLAG-tagged construct, the ER retention (pUL20KKSL) and ER retention control (pUL20KKSLAL) plasmids were generated by modifying the 3′ primers to contain the additional motifs inserted at the carboxyl terminus of UL20p. The pUL20DIVFLAG expression plasmid, which specifies UL20p with the FLAG epitope (DYKDDDDK) inserted within domain IV of UL20p, was generated by splice overlap extension PCR utilizing primers that contained the coding sequence for the FLAG epitope and were specific for the insertion region, in a manner that was essentially as described previously for domain-specific epitope tagging of gK (23, 24, 27).
UL20/gK double-null and ΔgK/UL20amFLAG recombinant virus construction.
The UL20/UL53(gK) double-null recombinant virus, ΔgK/ΔUL20, was generated essentially as described previously for the isolation of the UL20-null virus ΔUL20/gKDIV5 (25), with the exception that the UL20 gene was deleted from a gK-null virus background. Briefly, gK-null/UL20-null-complementing CV-1(c20cgK) cells were transfected with plasmid pΔ20-EGFP (25) and subsequently infected with the gK-null virus, ΔgK (Fig. 1) (39). Thirty-six hours postinfection (hpi), virus stocks were isolated and replated onto CV-1(c20cgK) cells for virus plaque isolation. Potential recombinant viruses were initially selected by the presence of green fluorescence in isolated viral plaques. Viral plaque isolates were picked, plaque purified at least seven times, and tested by diagnostic PCR and DNA sequencing for contaminating background virus, deletion of the UL20 gene, and the absence of the UL53(gK) gene. The ΔgK/UL20amFLAG virus, which specifies a 3xFLAG epitope tag at the amino terminus of UL20p in a gK-null viral background, was generated by a similar methodology. Plasmid pF20-AM3xFLAG contains the coding sequence for a 3xFLAG epitope inserted at the amino terminal of the UL20 gene, as well as upstream and downstream HSV genomic flanking regions, to facilitate homologous recombination with viral genomes (Fig. 1). gK-null-complementing VK302 cells were transfected with pF20-AM3xFLAG and subsequently infected with the ΔgK/ΔUL20 double-null virus. Recombinant virus isolates that had rescued the lack of the UL20 gene with the UL20am3xFLAG gene (Fig. 1) were detected by their growth on VK302 cells, as well as the absence of green fluorescence. Viral plaque isolates were picked, plaque purified at least seven times, and tested by diagnostic PCR, DNA sequencing, and immunofluorescence using anti-FLAG for the presence of the UL20am3xFLAG fusion, as well as the absence of the UL53(gK) gene.
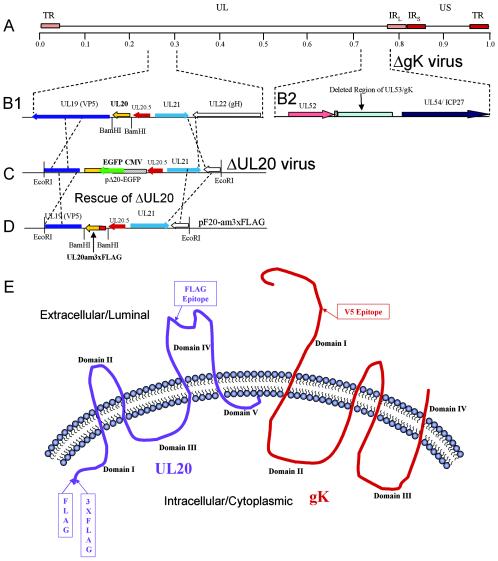
FIG. 1.
Schematic of gK-null and UL20-null recombinant viruses expressing epitope-tagged UL20p and gK, respectively. (A) The top line represents the prototypic arrangement of the HSV-1 genome with the unique long (UL) and unique short (US) regions flanked by the terminal repeat (TR) and internal repeat (IR) regions. (B) An expanded genomic region between map units 0.25 and 0.3 containing the UL19, UL20, UL20.5, UL21, and UL22 genes (panel 1) or the region between map units 0.7 and 0.8 containing the UL52, UL53, and UL54 open reading frames (panel 2). (C) The recombination plasmid, pΔ20-EGFP, which contains UL20 flanking sequences for recombination with the viral genome, was used through homologous recombination with the ΔgK viral genome to generate a ΔUL20/ΔgK double-null virus. (D) The recombination plasmid pF20-am3xFLAG, which encodes a 3xFLAG epitope-tagged UL20 gene and flanking sequences for recombination, was used to rescue the UL20 deletion and transfer the 3xFLAG epitope into the virus. (E) Schematic depicting the experimentally determined membrane topology of gK and the predicted membrane topology of UL20p, as well as the relative sites of insertion of each of the epitope tags.
Confocal microscopy.
Cell monolayers grown on coverslips in six-well plates were infected with the indicated virus at a multiplicity of infection of 10. Twelve hours postinfection, infected cell monolayers were prepared for confocal microscopy essentially as described previously (23, 25, 26). Alternatively, cell monolayers were transfected with the indicated UL20 and/or gK plasmid combinations by using Superfect reagent (QIAGEN) according to the manufacturer's instructions and prepared for confocal microscopy approximately 30 h posttransfection.
For cell surface biotinylation, prior to fixation cells were washed with Tris-buffered saline with Ca and Mg (TBS-Ca/Mg) and incubated for 15 min at room temperature in EZ-Link sulfo-NHS-LC biotin cell-impermeable biotinylation reagent (Pierce Chemical), which reacts with primary amines on cell surface proteins (25, 26). Cells were washed with TBS and fixed with electron microscopy-grade 3% paraformaldehyde (Electron Microscopy Sciences, Fort Washington, Pa.) for 15 min, washed twice with phosphate-buffered saline-50 mM glycine, and permeabilized with 1.0% Triton X-100. Monolayers were subsequently blocked for 1 h with 7% normal goat serum and 7% bovine serum albumin in TBS (TBS blocking buffer) before incubation for 2 h with either anti-V5 (Invitrogen), for recognition of gK, or anti-FLAG (Sigma Chemical), for recognition of UL20p, diluted 1:500 in TBS blocking buffer. Alternatively, simultaneous detection of gK and UL20p in cotransfected cells was accomplished by concurrent incubation with murine anti-V5 and rabbit anti-FLAG (Sigma Chemical) diluted 1:500 in TBS blocking buffer. Cells were then washed extensively and incubated for 30 min with Alexa Fluor 594-conjugated anti-immunoglobulin G diluted 1:500 in TBS blocking buffer. After incubation, excess antibody was removed by washing five times with TBS. For cell surface labeling, biotinylated cells were reacted with 1:1,000-diluted Alexa Fluor 647-conjugated streptavidin for 20 min. For Golgi and ER organelle labeling, cell monolayers were incubated with a 1:750 dilution of Alexa Fluor 488-conjugated lectins GSII and concanavalin A, respectively (12, 26, 46). TGN were identified with a donkey anti-TGN46 primary antibody and an Alexa Fluor 488-conjugated sheep anti-donkey secondary antibody (52). Specific immunofluorescence was examined using a Leica TCS SP2 laser scanning confocal microscope (Leica Microsystems, Exton, Pa.) fitted with a CS APO 63× Leica objective (1.4 numerical aperture). Individual optical sections in the z axis, averaged six times, were collected at the indicated zoom in series in the different channels at 1,024- by 1,024-pixel resolution as described previously (23, 25, 26). Images were compiled and rendered with Adobe Photoshop. Image analysis and subcellular colocalization fluorograms were generated and analyzed using the Leica confocal microscopy software package and were modified from protocols described previously (17). Positive and negative image masks of significant colocalization of two fluorophores were generated from the image fluorogram data sets by defining the specific regions of interest with a bounding box on the fluorogram. To determine an approximate percentage of subcellular colocalization, pixel enumeration and intensity statistics within the Leica software package were applied to a series of individual optical sections. The average percentage of pixels colocalized within a given organelle marker image mask (i.e., Golgi, TGN) relative to the total average pixel counts yielded an approximation of the percentage of protein localized to that organelle at that time point. In order to determine the approximate stoichiometric ratio of UL20p to gK, the TGN was defined as an end point of transport of the two proteins and an image mask was set according to the αTGN46 subcellular marker. Image statistics across a series of individual optical sections were used to determine the percentage of UL20p within the TGN mask relative to the percentage of gK outside of the subcellular image mask. This quantity was compared to the percentage of gK within the subcellular mask relative to the percentage of gK outside the TGN image mask in order to approximate the ratio of the two proteins.
UL20p/gK cell surface internalization assay.
Internalization assays were modified from similar assays performed previously (6, 66, 67). Briefly, Vero cells were transfected with pUL20amFLAG, pgKDIV5, pUL20DIVFLAG, or a combination with either pgKDIV5/pUL20amFLAG or pgKDIV5/pUL20DIVFLAG. Twenty hours posttransfection, cells were incubated under live conditions for 6 h at 37°C with either mouse anti-FLAG, mouse anti-V5, or a combination of mouse anti-V5 and rabbit anti-FLAG antibodies. Cells were extensively washed, fixed with paraformaldehyde, and processed for confocal microscopy as described above, with the exception that the internalized antibodies served as the primary antibody in all assays.
RESULTS
Insertion of in-frame epitope tags to facilitate detection of gK and UL20p.
The detection of highly hydrophobic, membrane-associated proteins, such as UL20p and gK, has been difficult mainly due to the lack of specific immunological reagents. Previously, our investigators analyzed the cellular localization and processing of gK through the insertion of a V5 antigenic epitope tag within the amino-terminal extracellular domain of gK (23, 25) (Fig. 1E). A similar methodology was utilized to facilitate detection of UL20p. Specifically, the FLAG epitope (DYKDDDDK) was inserted either at the amino terminus of UL20p, which is predicted to lie intracellularly, or within the predicted extracellular UL20p domain IV (53) (Fig. 1E) as described in Materials and Methods.
To facilitate the isolation of recombinant viruses carrying modifications or mutations in either gK- or UL20p-null genetic backgrounds, the UL53(gK)/UL20 double-null virus, ΔgK/ΔUL20, was isolated by insertional replacement of the UL20 gene with a CMV immediate-early promoter-enhanced green fluorescent protein (EGFP) gene cassette in the ΔgK (gK-null) KOS genetic background (39) (Fig. 1B and C). Subsequently, a gK-null recombinant virus that specified the amino-terminal 3xFLAG epitope (MDYKDHDGDYKDHDIDYKDDDDK)-tagged UL20p, designated here as ΔgK/UL20amFLAG, was isolated by rescuing the UL20 gene within the UL53(gK)/UL20 double-null virus (Fig. 1C and D). The generated plasmids and recombinant viruses were utilized to localize gK and UL20p relative to cellular markers demarcating ER, Golgi, TGN, and plasma membranes (cell surface) (Table 1).
TABLE 1.
Antibody markers for delineation of cellular organelles
| Cellular organelle | Antibody or marker |
|---|---|
| ER | Concanavalin A |
| Golgi | Lectin GSII |
| TGN | αTGN46 (sheep) |
| Plasma membranes | Live cell surface biotinylation |
| Early endosomes | αEEA-1 |
In the absence of UL20p, gK fails to be transported past the ER.
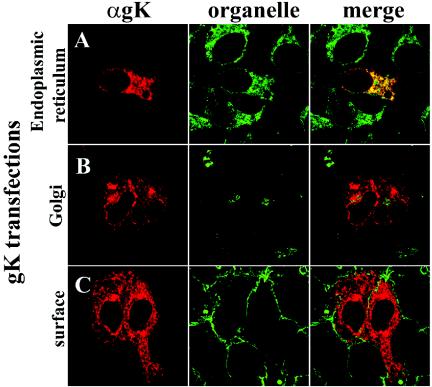
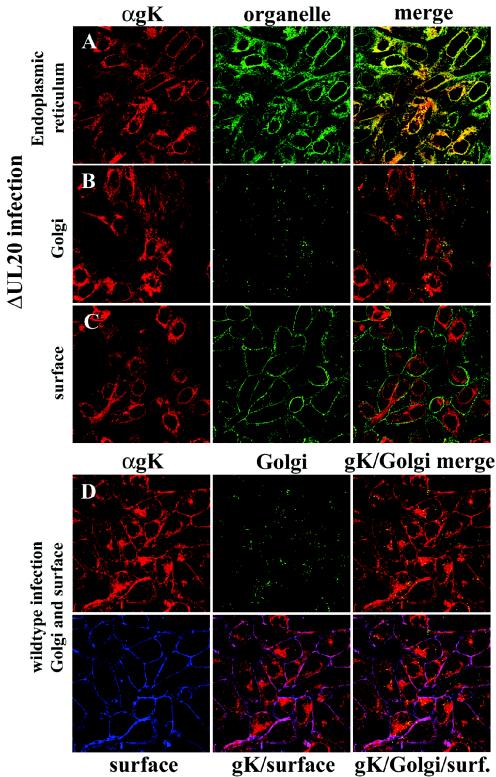
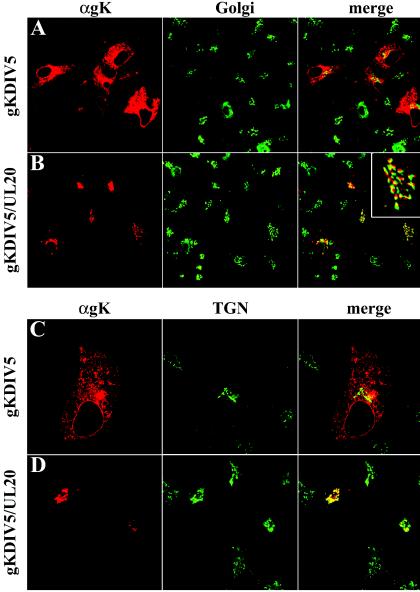
Transient expression of gK in Vero cells resulted in accumulation of gK in the ER (Fig. 2A), while no gK was detected in either Golgi or plasma membranes (Fig. 2B and C). Similarly, in the context of viral infections, gK was exclusively detected within the ER of UL20-null-infected Vero cells (Fig. 3A) and was absent from Golgi and plasma membranes (Fig. 3B and C). In contrast, gK colocalized with both Golgi and cell surface markers (Fig. 3D) in Vero cells infected with wild-type virus (UL20+). Therefore, gK requires at least UL20p for transportation past the ER to Golgi, TGN, and cell surfaces.
FIG. 2.
Intracellular localization of transiently expressed gK. Cells transfected with pgKDIV5 were fixed at 25 h posttransfection and stained with anti-V5 antibodies for gK (red) (A, B, and C) or specific organelle markers (green) that recognized the ER (A), Golgi (B), or plasma membranes (C). Magnification, ×63; zoom, ×4.
FIG. 3.
Digital images of confocal micrographs showing ΔUL20/gKDIV5 (A, B, and C) or gKDIV5/UL20amFLAG (D) virus-infected Vero cells. Infected cells were fixed at 12 hpi and stained with anti-V5 antibodies for gK (red) or specific organelle markers (green and blue) that specifically recognized ER (A), Golgi (green) (B and D), or plasma membranes (green in panel C, blue in panel D). Magnification, ×63; zoom, ×2.
Interdependence of gK and UL20p for post-ER-to-TGN intracellular transport and localization.
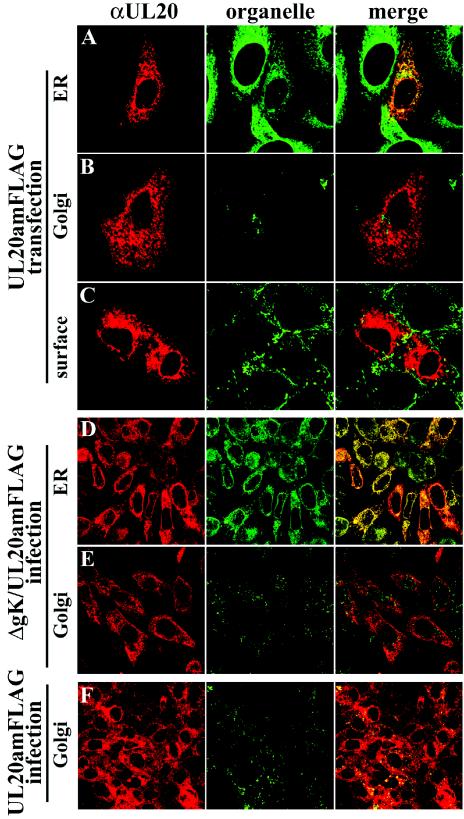
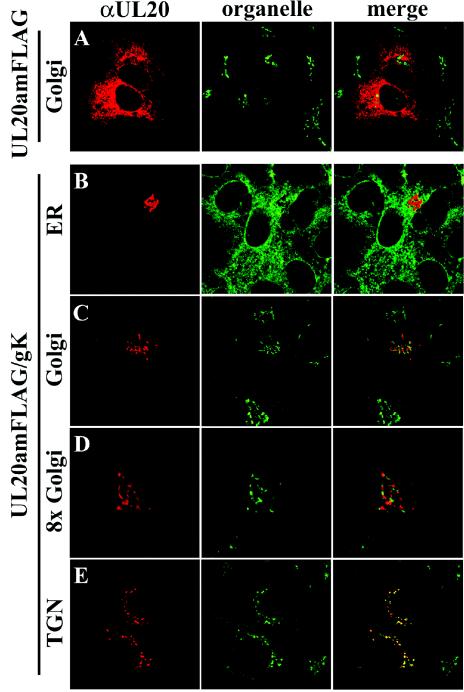
The inability of gK to be transported post-ER in the absence of UL20p suggests that UL20p may exhibit a similar defect in the absence of gK. Therefore, the cellular localization of transiently expressed UL20p and UL20p in a gK-null virus background was examined. As described previously, in a wild-type HSV-1 infection, UL20p was found throughout the cytoplasm and within the Golgi (Fig. 4F) (72). However, UL20p localized exclusively within the ER (Fig. 4D) and was absent from Golgi membranes (Fig. 4E) in gK-null virus infections. Similarly, transiently expressed UL20p was found within the ER (Fig. 4A) but was not detected in Golgi (Fig. 4B) and plasma membranes (Fig. 4C). Therefore, it appears that both UL20p and gK are mutually dependent for cellular transport and localization.
FIG. 4.
Intracellular localization of UL20p in transfected (A to C), ΔgK/UL20amFLAG (D and E), or gKDIV5/UL20amFLAG (F) virus-infected cells. Cells were fixed at either 24 h posttransfection (A to C) or 12 hpi (D to F) and stained with anti-FLAG antibodies for UL20p (red) or specific organelle markers (green) that specifically recognized ER (A and D), Golgi (B, E, and F), or plasma membranes (C). Magnification, ×63; zoom, ×4 (A to C) or ×2 (D to F).
The interdependence of gK and UL20p for intracellular transport and localization was further assessed by transient coexpression of gK and UL20p and simultaneous detection of Golgi and TGN compartments. As shown earlier, in the absence of UL20p, gK was localized exclusively to reticular-like compartments and was absent from the Golgi and TGN (Fig. 5A and C). Similarly, in the absence of gK, UL20p produced a localization pattern which resembled gK localization and was not detected in either Golgi (Fig. 6A) or TGN (data not shown). In contrast, coexpression of gK and UL20p significantly altered the distribution pattern of both gK and UL20p. Both proteins were detected in multilayered curvilinear compartments reminiscent of Golgi/TGN intracellular sites (Fig. 5B and D and 6B to E). Although some gK and UL20p localized to Golgi (Fig. 5B and inset; Fig. 6C and D), the majority of UL20p and gK were localized within compartments immediately adjacent to the Golgi apparatus as evidenced by the distinct and nonoverlapping fluorescent staining of gK and UL20p relative to that of the Golgi-specific marker (Fig. 5B and inset; Fig. 6C and D). Colocalization analysis indicated that approximately 5% of either gK or UL20p colocalized with Golgi-specific markers, whereas approximately 95% was localized in compartments immediately adjacent to the Golgi. Therefore, we examined the distribution of gK and UL20p relative to that of the TGN marker, TGN46. In transient-coexpression experiments, both gK and UL20p specifically localized within the TGN (Fig. 5D and 6E), the organelle where final virion envelopment is thought to occur.
FIG. 5.
Digital images of confocal micrographs depicting gK localization in either gK-transfected (A and C) or UL20p- and gK-cotransfected (B and D) Vero cells. Transfected cells were fixed at 24 h posttransfection and stained with anti-V5 antibodies for gK (red) or specific organelle markers (green) that specifically recognized either Golgi (A and B) or TGN (C and D). Magnification, ×63; zoom, ×4.
FIG. 6.
Digital images of confocal micrographs depicting UL20p localization in either UL20p-transfected (A) or UL20p- and gK-cotransfected (B to E) Vero cells. Transfected cells were fixed at 24 h posttransfection and stained with anti-FLAG antibodies for UL20p (red) or organelle markers (green) that specifically recognized ER (B), Golgi (A, C, and D), or TGN (E). Magnification, ×63; zoom, ×4 (A, B, C, and E) or ×8 (D).
Subcellular colocalization of UL20p and gK.
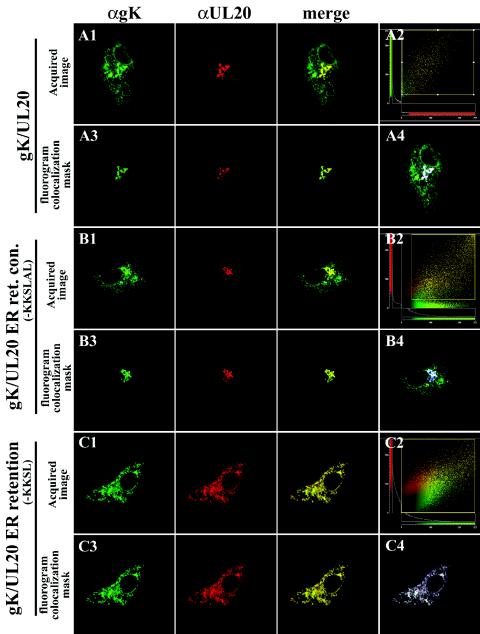
Simultaneous detection of gK and UL20p indicated that the two proteins colocalized within TGN-like structures (Fig. 7A, panels 1 and 3). Quantitative image assessment of gK and UL20p colocalization was determined by local image correlation confocal microscopy (17). In this method, a two-dimensional pixel fluorogram generated from the confocal image section was used to analyze colocalization of two fluorophores within a specific subcellular domain or organelle. Regions of specific intracellular areas, such as the TGN, exhibiting significant colocalization of gK and UL20p were defined by masking a portion of the fluorogram where significant colocalization occurred (Fig. 7A, panel 2). Corresponding images of the significant colocalization masks were generated to highlight specific subcellular areas of colocalization (Fig. 7A, panel 3). In addition, a subtractive blue-masked image was generated to further demonstrate the specific subcellular domains where colocalization occurred (Fig. 7A, panel 4). The generated masked images indicated that gK and UL20p specifically colocalized to TGN-like compartments (Fig. 7A, panels 3 and 4). In these experiments, gK was also visualized within reticulate-like intracellular sites that did not colocalize with UL20p (Fig. 7A, panels 1 and 4). However, in experiments where UL20p was overexpressed in relation to gK, the inverse pattern was observed, in which UL20p exhibited reticular-like and TGN localization patterns, whereas gK exhibited only TGN-like localization (data not shown). Taken together, these results suggest a stoichiometric relationship between gK and UL20p for intracellular transport and localization to TGN compartments. This conclusion is further supported by examination of pixel and intensity counts in site-specific colocalization masks of images, which indicated a probable 1:1 ratio for levels of the two proteins within the subcellular TGN-like regions.
FIG.7.
Colocalization of gK and UL20p in transient-coexpression experiments with or without UL20p containing an ER retention motif. Vero cells coexpressing gK (green) and either UL20amFLAG (A), the ER retention control motif protein UL20p(KKSLAL) (B), or the ER retention motif protein UL20p(KKSL) (C) were fixed at 24 h posttransfection, and the subcellular distributions of gK (green) and UL20p (red) were determined. Fluorograms, which depict pixel intensity and distribution, are shown for each image subset, as well as the fluorogram regions that showed significant colocalization between gK and UL20p (panel 2 in A, B, and C). Image correlation masks where significant specific colocalization occurred are shown for each image subset (panel 3 in A to C). In addition, a negative image correlation mask is shown with a blue-masked image depicting the specific subcellular region of significant colocalization (panel 4 in A to C). Magnification, ×63; zoom, ×4.
Intracellular transport of UL20p is required for post-ER transport of gK to TGN compartments.
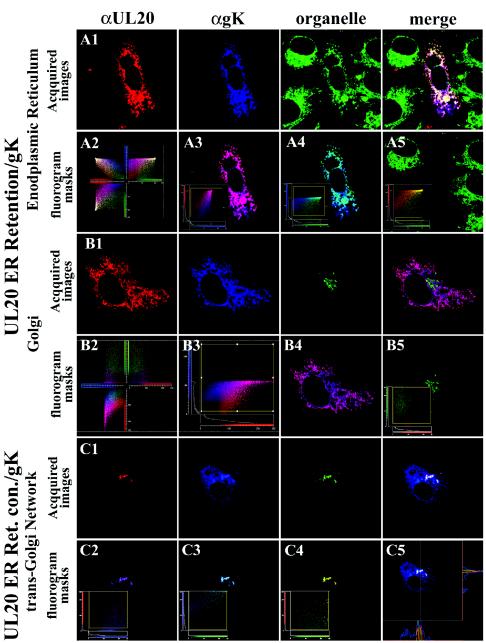
To determine whether transport of UL20p was coordinately required for transport of gK, UL20p was retained within the ER through the addition of either an ER retention motif (KKSL) or a control motif (KKSLAL) at the carboxyl terminus of UL20p. Insertion of the KKXX motif acts as a retrieval signal, transporting membrane proteins from a post-ER sorting compartment back to the ER. The addition of two amino acids to the KKXX motif (KKSLAL) overrides the retention signal, enabling proteins to traffic out of the ER to Golgi and cell surfaces (8, 36, 37, 75). As expected, coexpression of gK with the UL20p(KKSL) ER-retained protein caused drastic modifications in the intracellular transport and localization of both gK and UL20p (compare Fig. 7C and B, panels 1, and 8A and B with Fig. 8C). Specifically, both UL20p and gK were colocalized and visualized in a reticular-like staining pattern (Fig. 7C, panels 2 to 4). In addition, UL20p(KKSL) and gK were shown to localize exclusively within the ER, as evidenced by colocalization with an ER-specific marker (Fig. 8A, panels 1 to 4). The specific localization of UL20p(KKSL) and gK within the ER was further confirmed by defining the fluorogram region which lacked significant colocalization (Fig. 8A, panel 4 inset). The corresponding masked image was devoid of ER-, UL20p(KKSL)-, and gK-specific costaining for the cell of interest (Fig. 8A, panel 4), whereas the ER was readily visualized for the adjacent cells that showed no UL20p- or gK-specific expression (Fig. 8A, panel 4). Neither UL20p(KKSL) nor gK exhibited any colocalization with the Golgi (Fig. 8B, panels 1 and 2) or TGN (data not shown). Application of a subcellular image colocalization mask for UL20p(KKSL) and Golgi (Fig. 8B, panel 5 inset) indicated that UL20p(KKSL) was not transported to the Golgi (Fig. 8B, panel 5). In contrast, transient coexpression of gK with the UL20p(KKSLAL) control protein exhibited cellular localization and transport of gK and UL20p indistinguishable to that produced with the wild-type UL20p protein (compare Fig. 7B, panel 1, and 8C, panel 1, with Fig. 7A, panel 1). UL20p(KKSLAL) and gK were found colocalized within TGN-like compartments (Fig. 7B, panels 2 to 4, and 8C, panels 1 and 2) and were also found to specifically colocalize with the TGN marker αTGN46 (Fig. 8C, panels 3 and 4). Fluorohistogram sectional analysis also indicated that small percentages of UL20p(KKSLAL) and gK were also present within subcellular Golgi-like compartments that were immediately adjacent but distinguishable from the TGN, as indicated by the absence of TGN46 specific recognition but the presence of both gK and UL20p(KKSLAL) (Fig. 8C, panel 5). These results indicate that a functional association between gK and UL20p must form at the ER, and its formation is necessary for subsequent cotransport and localization of both gK and UL20p to Golgi and TGN membranes.
FIG.8.
Requirement for UL20p transport from ER to TGN for specific colocalization of gK, UL20p, and TGN markers. Vero cells coexpressing gK (green) and either the ER retention motif protein UL20p(KKSL) (A and B) or the ER retention control motif protein UL20p(KKSLAL) (C) were fixed at 24 h posttransfection, and the subcellular distributions of gK (blue) and UL20p (red) were determined relative to subcellular organelle markers (green) that specifically recognized ER (A), Golgi (B), or TGN (C). Three-dimensional fluorograms that depicted pixel intensity, distribution, and colocalization are shown for UL20p(KKSL) image subsets (panel 2 in A and B). Two-dimensional fluorograms and local image correlation masks between two fluorophores in each subset are shown for gK and UL20p(KKSL) (A, panel 3, and B, panels 3 and 4); gK and ER (A, panel 4); UL20p(KKSL) and Golgi (B, panel 5); UL20p(KKSLAL) and gK (C, panel 2); UL20p(KKSLAL) and TGN (C, panel 4); gK and TGN (C, panel 3). In addition, the region where no significant colocalization of gK, UL20p(KKSL), and ER occurred was masked (A, panel 5 inset), and as expected the relative image of this masked generated depicts no ER, gK, or UL20p(KKSL) proteins present. A sectional fluorohistogram for UL20p(KKSLAL), gK, and TGN colocalization is presented (C, panel 5) to show that all three markers specifically colocalized within most subcellular regions, except a few regions where only gK and UL20p(KKSLAL) colocalized outside of the TGN, most likely within Golgi membranes. Magnification, ×63; zoom, ×4.
UL20p and gK endocytose from cell surfaces and localize to TGN compartments.
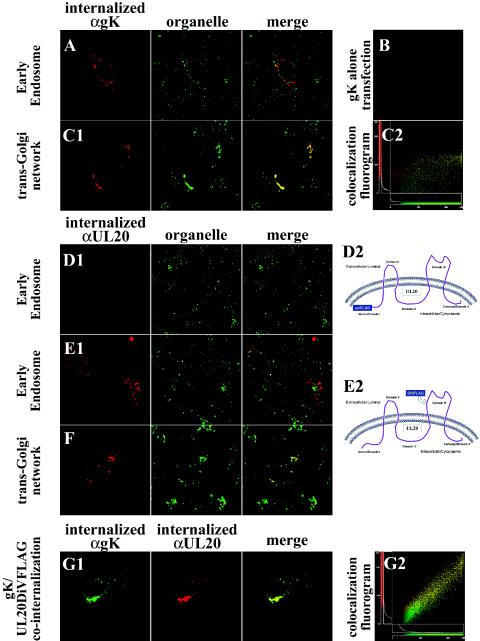
In virus-infected cells, gK was readily detected on infected plasma membranes (Fig. 3D). However, in transient-coexpression experiments, UL20p and gK were predominantly localized in the TGN and were not detected within plasma membranes. Both UL20p and gK contain cytoplasmic motifs that have been implicated in the recycling of membrane proteins from plasma membranes to the TGN (4, 13, 14, 51, 56, 66, 68-71). Specifically, the amino terminus of UL20p, which is predicted to lie intracellularly, contains both an acidic cluster as well as tyrosine-based (YXXΦ) internalization motifs (53). Furthermore, gK contains a YXXΦ motif within domain II, which has been shown to lie intracellularly (23, 24). To determine whether cell surface-expressed gK recycled to the TGN, Vero cells that coexpressed gK and UL20p were reacted with anti-V5 antibody under live conditions (see Materials and Methods). The fate of the internalized V5-tagged gK was assessed at different times postlabeling. By 4 h postlabeling, gK was internalized to and colocalized with TGN compartments (Fig. 9C, panels 1 and 2), while only minimal amounts of gK were detected within early endosomes (Fig. 9A). As expected, independent expression of gK did not produce any gK-specific labeling (Fig. 9B), since in the absence of UL20p gK fails to be transported past the ER to cell surfaces. In addition, this experiment showed that Vero cells did not internalize anti-V5 antibody nonspecifically.
FIG.9.
Cell surface-expressed gK and UL20p endocytose to TGN. Antibodies bound to cell surface-expressed and internalized gK (red in A to C; green in G) or UL20p (red) (D to G) were detected at 24 h posttransfection relative to organelle markers that specifically recognize early endosomes (green in A, D, and E) or TGN (green in C and F). Fluorograms of image subsets depicting gK and TGN colocalization (C, panel 2) or gK and UL20p colocalization (G, panel 2) are shown. Schematics show the predicted membrane topology of UL20p and the relative sites of FLAG epitope tag insertion within either the intracellular domain I (D2) or extracellular domain IV (E2). Magnification, ×63; zoom, ×4.
A similar series of experiments was performed to assess UL20p internalization from plasma membranes. UL20p tagged at its amino terminus with a FLAG tag did not react with anti-FLAG antibody in the internalization assay (Fig. 9D, panel 1). However, this result was anticipated, since the amino terminus of UL20p is predicted to lie intracellularly, rendering this domain inaccessible to the anti-FLAG antibody (Fig. 9D, panel 2). Therefore, the FLAG epitope was also inserted within UL20p domain IV, which was predicted to lie extracellularly (Fig. 9E, panel 2). Coexpression of UL20DIVFLAG with gK resulted in UL20p internalization from plasma membranes to TGN compartments (Fig. 9E, panel 1, and F), in a similar fashion to that of gK.
In the previous experiments, the intracellular localization of gK and UL20p was individually assessed with respect to intracellular organelle markers. To assess whether gK and UL20p were simultaneously cointernalized from plasma membranes, gK and UL20p were reacted with anti-V5 and anti-FLAG antibodies, respectively. Both gK and UL20p exhibited identical cellular distributions following internalization (Fig. 9G, panel 1), as also evidenced by the relevant colocalization fluorogram (Fig. 9G, panel 2). These experiments revealed that both gK and UL20p are transported to cell surfaces and subsequently are cointernalized to specific TGN compartments.
DISCUSSION
UL20p and gK have been shown to function in cytoplasmic virion envelopment at TGN-like membranes, as well as in virus-induced cell fusion. Collectively, the similarities between gK and UL20p suggest a functional relationship between gK and UL20p. To investigate the interrelationship between gK and UL20p, a series of experiments were performed to track the intracellular transport and localization of gK and UL20p in transient-coexpression and virus infection experiments. The salient features of the obtained results were the following: (i) neither gK nor UL20p is transported from the ER when expressed alone, while coexpression of gK and UL20p results in efficient export from the ER; (ii) both gK and UL20p localize to TGN compartments; (iii) in infected cells, but not transfected cells, gK and UL20p can be readily detected by cell surface labeling; (iv) forced retention of UL20p to the ER results in ER retention of gK; (v) gK and UL20p localize to the TGN after internalization from plasma membranes; (vi) gK and UL20p assume a tight colocalization pattern within subcellular organelles, including the TGN; and (vii) the amino terminus (domain I) of UL20p lies intracellularly, while UL20p domain IV lies extracellularly. Therefore, UL20p appears to assume a membrane topology that is a topological mirror image of gK.
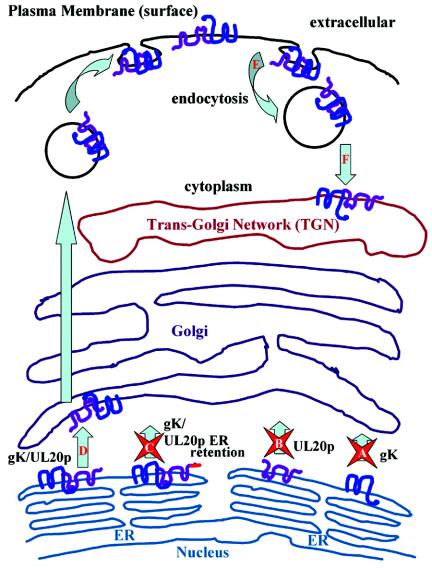
Transient-coexpression experiments revealed an absolute interdependence between gK and UL20p for intracellular transport past the ER, which was exemplified by the retention of gK or UL20p in the ER in the absence of their heterologous partner. Likewise, a mutual dependence for transport was observed in the context of gK- or UL20-null viral infections, in which the corresponding heterologous protein remained localized to the ER and failed to traffic to the Golgi, TGN, or cell surfaces (Fig. 10). Moreover, coexpression of gK and UL20p resulted in specific trafficking and colocalization to the TGN. These findings are in agreement with the secondary envelopment defects observed in gK- and UL20p-null viruses. Acquisition of the final viral envelope (secondary envelopment) is thought to occur by cytoplasmic capsids budding into TGN-derived membranes. In this regard, lack of gK or UL20p within TGN membranes may lead to accumulation of capsids into the cytoplasm and a failure of enveloped virions to be transported from TGN-like compartments.
FIG. 10.
Diagrammatic summary of gK and UL20p transport from the ER to TGN and cell surfaces. (A and B) Neither gK nor UL20p is capable of transport out of the ER in the absence of its heterologous partner. (C) Moreover, retention of UL20p to the ER via a carboxyl terminal ER retention motif retained both gK and UL20p within ER membranes. (D) Coexpression of gK and UL20p resulted in transport of both UL20p and gK to post-ER compartments, including Golgi, TGN, and cell surfaces. (E and F) UL20p and gK colocalized within TGN following endocytosis from cell surfaces (E) and specific transport to TGN (F).
UL20p and gK have been detected throughout the cytoplasm and Golgi apparatus of infected cells and in purified virions (1, 23, 27, 41, 72). In virus-infected cells, both gK and UL20p were readily detected on infected cell surfaces. In contrast, transiently coexpressed gK and UL20p were not detected on cell surfaces because they were rapidly internalized to TGN-like intracellular organelles (Fig. 10). Both gK and UL20p contain amino acid motifs located within their cytoplasmic domains, which are known to facilitate endocytosis to TGN. These motifs include the YXXΦ motif located within gK domain II (24) and within UL20p domain I (amino terminus) (53), as well as a cluster of acidic amino acids located in UL20p domain I (53). Alteration of the critical tyrosine residue in the YXXΦ motifs within either gK or UL20p was shown to inhibit cytoplasmic virion morphogenesis, egress, and cell-to-cell spread (24, 53). Similarly, mutagenesis of certain acidic amino acid clusters at the amino terminus of UL20p reduced infectious virus production and cytoplasmic virion envelopment (53). Therefore, internalization of either gK or UL20p to TGN may be required for cytoplasmic envelopment by budding of capsids into the TGN. There are several possible explanations for the apparent differences in cell surface localization of gK and UL20p between transient coexpression and viral infection. Virus infection may alter endocytic membrane trafficking mechanisms, effectively decreasing the rate of endocytosis of cell surface-expressed viral membrane proteins. Alternatively, other viral proteins may be involved in increasing retention of gK and UL20p on cell surfaces via direct or indirect interactions. The fact that both gK and UL20p contain syncytial mutations (1, 16, 53, 58, 59, 63) suggests that they may be required at cell surfaces to modulate virus-induced cell-to-cell fusion. Thus, HSV-1 may have evolved mechanisms for balancing levels of gK and UL20p within the TGN and plasma membranes required for efficient cytoplasmic envelopment and virus-induced cell-to-cell fusion, respectively.
Recently, gK was shown to assume a tetramembrane-spanning topology that oriented both amino and carboxyl termini in extracellular spaces, while two other gK domains were located in the cytoplasm (23). UL20p is also predicted to span membranes four times; however, unlike gK, UL20p is predicted to orient its amino and carboxyl termini within the cytoplasm, while two other internal domains are predicted to be located extracellularly (53). In this study, this predicted UL20p topology was partially confirmed by investigating the cell surface expression of UL20p derivatives tagged with FLAG epitopes inserted within either domain I or IV (Fig. 1). In agreement with the predicted UL20p membrane topology, the potential extracellular domain IV was readily accessible to anti-FLAG antibodies, while domain I was not accessible in live cell surface labeling internalization assays. Thus, UL20p seems to assume a membrane orientation that is a topological mirror image of gK. This corresponding topological orientation will be important in future studies that aim to determine possible direct or indirect interactions between domains of gK and UL20p.
HSV-1 codes for a number of membrane proteins that require direct interaction with a partner protein to facilitate their intracellular transport and function. Examples of such protein-protein interactions include the following: (i) the gB complex, which consists of two disulfide bond-linked gB monomers that are posttranslationally proteolytically cleaved into two subunits (10, 11, 32, 45, 73); (ii) the essential gH/gL complex, as coexpression of HSV-1 gL is required for the proper folding, cell surface expression, and virion incorporation of gH (33, 42, 43, 61, 62); (iii) the noncovalently linked gE/gI heterodimeric complex that functions in direct cell-to-cell spread in certain cell types and neuronal systems (2, 20, 74); and (iv) the disulfide-linked gM/UL49.5 (gN) complex, in which PRV gM is required for virion incorporation and transport of gN (40). These heterodimeric protein complexes involve direct interactions between the partner proteins. Therefore, it is possible that gK and UL20p directly interact and that this interaction is necessary for their intracellular transport, localization, and functions. This conclusion is supported by the quantitative subcellular image correlation of fluorescent pixels. This analysis shows that the actual pixel counts within a specific subcellular site of TGN compartments suggest a 1:1 stoichiometric relationship between these two proteins. In this regard, our group's recent observations that overexpression of gK severely inhibited virion morphogenesis and intracellular trafficking of gK by collapsing Golgi into the ER (26) may be due to the lack of sufficient amounts of UL20p to transport gK from the ER to cell surfaces and TGN. The accumulation of gK within the ER, due to its inability to be transported to subsequent compartments, could mediate a cellular ER stress response and subsequent Golgi collapse (15, 47). Additional biochemical experiments are needed to more accurately define gK/UL20p stoichiometric relationships and test whether these two proteins directly interact. Alternatively, gK and UL20p may not necessarily interact but may be mutually required for independent transport to TGN compartments. However, this latter possibility is improbable, since ER retention of UL20p by addition of an ER retention motif to its carboxyl terminus resulted in ER retention of gK.
The observed interdependence of gK and UL20p for intracellular transport and localization implies that direct or indirect interactions between gK and UL20p may be required for their role in both cytoplasmic virion envelopment and virus-induced cell-to-cell fusion. With regard to virion morphogenesis, UL20 and gK may function to tether tegument proteins to TGN budding sites, as it has also been suggested for UL11, gM, gE, and VP22 (5, 7, 21, 22, 28, 44, 48, 49). Although both gK and UL20p contain syncytial mutations, it is unclear whether they directly participate in virus-induced membrane fusion or function to regulate other fusogenic viral proteins, such as gB. The inability of syncytial mutations in gB to cause membrane fusion in the absence of either gK (J. M. Melancon, T. P. Foster, and K. G. Kousoulas, unpublished data) or UL20p (25, 53) suggests that both UL20p and gK exert regulatory functions on gB-mediated membrane fusion. Recently our investigators showed that specific UL20 mutations can selectively inhibit either gB- or gK-mediated virus-induced cell fusion, while certain other UL20 mutations inhibited both gB- and gK-mediated cell fusion (25, 53). Collectively, these results suggest that a UL20p/gK interactive complex may regulate gB-mediated cell fusion through either direct or indirect protein-protein interactions.
Acknowledgments
This work was supported by National Institute of Allergy and Infectious Diseases grant AI43000 to K.G.K. We acknowledge support by the LSU School of Veterinary Medicine to BIOMMED.
REFERENCES
- 1.Baines, J. D., P. L. Ward, G. Campadelli-Fiume, and B. Roizman. 1991. The UL20 gene of herpes simplex virus 1 encodes a function necessary for viral egress. J. Virol. 65:6414-6424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Basu, S., G. Dubin, M. Basu, V. Nguyen, and H. M. Friedman. 1995. Characterization of regions of herpes simplex virus type 1 glycoprotein E involved in binding the Fc domain of monomeric IgG and in forming a complex with glycoprotein I. J. Immunol. 154:260-267. [PubMed] [Google Scholar]
- 3.Bond, V. C., and S. Person. 1984. Fine structure physical map locations of alterations that affect cell fusion in herpes simplex virus type 1. Virology 132:368-376. [DOI] [PubMed] [Google Scholar]
- 4.Bonifacino, J. S., M. S. Marks, H. Ohno, and T. Kirchhausen. 1996. Mechanisms of signal-mediated protein sorting in the endocytic and secretory pathways. Proc. Assoc. Am. Physicians 108:285-295. [PubMed] [Google Scholar]
- 5.Brack, A. R., B. G. Klupp, H. Granzow, R. Tirabassi, L. W. Enquist, and T. C. Mettenleiter. 2000. Role of the cytoplasmic tail of pseudorabies virus glycoprotein E in virion formation. J. Virol. 74:4004-4016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Brideau, A. D., M. G. Eldridge, and L. W. Enquist. 2000. Directional transneuronal infection by pseudorabies virus is dependent on an acidic internalization motif in the Us9 cytoplasmic tail. J. Virol. 74:4549-4561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Brignati, M. J., J. S. Loomis, J. W. Wills, and R. J. Courtney. 2003. Membrane association of VP22, a herpes simplex virus type 1 tegument protein. J. Virol. 77:4888-4898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Browne, H., S. Bell, T. Minson, and D. W. Wilson. 1996. An endoplasmic reticulum-retained herpes simplex virus glycoprotein H is absent from secreted virions: evidence for reenvelopment during egress. J. Virol. 70:4311-4316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bzik, D. J., B. A. Fox, N. A. DeLuca, and S. Person. 1984. Nucleotide sequence of a region of the herpes simplex virus type 1 gB glycoprotein gene: mutations affecting rate of virus entry and cell fusion. Virology 137:185-190. [DOI] [PubMed] [Google Scholar]
- 10.Cai, W. Z., S. Person, C. DebRoy, and B. H. Gu. 1988. Functional regions and structural features of the gB glycoprotein of herpes simplex virus type 1. An analysis of linker insertion mutants. J. Mol. Biol. 201:575-588. [DOI] [PubMed] [Google Scholar]
- 11.Claesson-Welsh, L., and P. G. Spear. 1986. Oligomerization of herpes simplex virus glycoprotein B. J. Virol. 60:803-806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cottin, V., A. A. Van Linden, and D. W. Riches. 2001. Phosphorylation of the tumor necrosis factor receptor CD120a (p55) recruits Bcl-2 and protects against apoptosis. J. Biol. Chem. 276:17252-17260. [DOI] [PubMed] [Google Scholar]
- 13.Crump, C. M., C. H. Hung, L. Thomas, L. Wan, and G. Thomas. 2003. Role of PACS-1 in trafficking of human cytomegalovirus glycoprotein B and virus production. J. Virol. 77:11105-11113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Crump, C. M., Y. Xiang, L. Thomas, F. Gu, C. Austin, S. A. Tooze, and G. Thomas. 2001. PACS-1 binding to adaptors is required for acidic cluster motif-mediated protein traffic. EMBO J. 20:2191-2201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dascher, C., and W. E. Balch. 1994. Dominant inhibitory mutants of ARF1 block endoplasmic reticulum to Golgi transport and trigger disassembly of the Golgi apparatus. J. Biol. Chem. 269:1437-1448. [PubMed] [Google Scholar]
- 16.Debroy, C., N. Pederson, and S. Person. 1985. Nucleotide sequence of a herpes simplex virus type 1 gene that causes cell fusion. Virology 145:36-48. [DOI] [PubMed] [Google Scholar]
- 17.Demandlox, D., and J. Davoust. 1997. Multicolour analysis and local image correlation in confocal microscopy. J. Microsc. 185:21-36. [Google Scholar]
- 18.Desai, P., N. A. DeLuca, J. C. Glorioso, and S. Person. 1993. Mutations in herpes simplex virus type 1 genes encoding VP5 and VP23 abrogate capsid formation and cleavage of replicated DNA. J. Virol. 67:1357-1364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Dietz, P., B. G. Klupp, W. Fuchs, B. Kollner, E. Weiland, and T. C. Mettenleiter. 2000. Pseudorabies virus glycoprotein K requires the UL20 gene product for processing. J. Virol. 74:5083-5090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Dingwell, K. S., and D. C. Johnson. 1998. The herpes simplex virus gE-gI complex facilitates cell-to-cell spread and binds to components of cell junctions. J. Virol. 72:8933-8942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Elliott, G., G. Mouzakitis, and P. O'Hare. 1995. VP16 interacts via its activation domain with VP22, a tegument protein of herpes simplex virus, and is relocated to a novel macromolecular assembly in coexpressing cells. J. Virol. 69:7932-7941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Farnsworth, A., K. Goldsmith, and D. C. Johnson. 2003. Herpes simplex virus glycoproteins gD and gE/gI serve essential but redundant functions during acquisition of the virion envelope in the cytoplasm. J. Virol. 77:8481-8494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Foster, T. P., X. Alvarez, and K. G. Kousoulas. 2003. Plasma membrane topology of syncytial domains of herpes simplex virus type 1 glycoprotein K (gK): the UL20 protein enables cell surface localization of gK but not gK-mediated cell-to-cell fusion. J. Virol. 77:499-510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Foster, T. P., and K. G. Kousoulas. 1999. Genetic analysis of the role of herpes simplex virus type 1 glycoprotein K in infectious virus production and egress. J. Virol. 73:8457-8468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Foster, T. P., J. M. Melancon, J. D. Baines, and K. G. Kousoulas. 2004. The herpes simplex virus type 1 UL20 protein modulates membrane fusion events during cytoplasmic virion morphogenesis and virus-induced cell fusion. J. Virol. 78:5347-5357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Foster, T. P., G. V. Rybachuk, X. Alvarez, O. Borkhsenious, and K. G. Kousoulas. Overexpression of gK in gK-transformed cells collapses the Golgi apparatus into the endoplasmic reticulum inhibiting virion egress, glycoprotein transport, and virus-induced cell fusion. Virology 317:237-252. [DOI] [PubMed]
- 27.Foster, T. P., G. V. Rybachuk, and K. G. Kousoulas. 2001. Glycoprotein K specified by herpes simplex virus type 1 is expressed on virions as a Golgi complex-dependent glycosylated species and functions in virion entry. J. Virol. 75:12431-12438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Fuchs, W., B. G. Klupp, H. Granzow, C. Hengartner, A. Brack, A. Mundt, L. W. Enquist, and T. C. Mettenleiter. 2002. Physical interaction between envelope glycoproteins E and M of pseudorabies virus and the major tegument protein UL49. J. Virol. 76:8208-8217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Fuchs, W., B. G. Klupp, H. Granzow, and T. C. Mettenleiter. 1997. The UL20 gene product of pseudorabies virus functions in virus egress. J. Virol. 71:5639-5646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Granzow, H., B. G. Klupp, W. Fuchs, J. Veits, N. Osterrieder, and T. C. Mettenleiter. 2001. Egress of alphaherpesviruses: comparative ultrastructural study. J. Virol. 75:3675-3684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Harley, C. A., A. Dasgupta, and D. W. Wilson. 2001. Characterization of herpes simplex virus-containing organelles by subcellular fractionation: role for organelle acidification in assembly of infectious particles. J. Virol. 75:1236-1251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Highlander, S. L., W. F. Goins, S. Person, T. C. Holland, M. Levine, and J. C. Glorioso. 1991. Oligomer formation of the gB glycoprotein of herpes simplex virus type 1. J. Virol. 65:4275-4283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hutchinson, L., H. Browne, V. Wargent, N. Davis-Poynter, S. Primorac, K. Goldsmith, A. C. Minson, and D. C. Johnson. 1992. A novel herpes simplex virus glycoprotein, gL, forms a complex with glycoprotein H (gH) and affects normal folding and surface expression of gH. J. Virol. 66:2240-2250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hutchinson, L., K. Goldsmith, D. Snoddy, H. Ghosh, F. L. Graham, and D. C. Johnson. 1992. Identification and characterization of a novel herpes simplex virus glycoprotein, gK, involved in cell fusion. J. Virol. 66:5603-5609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hutchinson, L., and D. C. Johnson. 1995. Herpes simplex virus glycoprotein K promotes egress of virus particles. J. Virol. 69:5401-5413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Jackson, M. R., T. Nilsson, and P. A. Peterson. 1990. Identification of a consensus motif for retention of transmembrane proteins in the endoplasmic reticulum. EMBO J. 9:3153-3162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Jackson, M. R., T. Nilsson, and P. A. Peterson. 1993. Retrieval of transmembrane proteins to the endoplasmic reticulum. J. Cell Biol. 121:317-333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Jacobson, J. G., S. H. Chen, W. J. Cook, M. F. Kramer, and D. M. Coen. 1998. Importance of the herpes simplex virus UL24 gene for productive ganglionic infection in mice. Virology 242:161-169. [DOI] [PubMed] [Google Scholar]
- 39.Jayachandra, S., A. Baghian, and K. G. Kousoulas. 1997. Herpes simplex virus type 1 glycoprotein K is not essential for infectious virus production in actively replicating cells but is required for efficient envelopment and translocation of infectious virions from the cytoplasm to the extracellular space. J. Virol. 71:5012-5024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Jons, A., J. M. Dijkstra, and T. C. Mettenleiter. 1998. Glycoproteins M and N of pseudorabies virus form a disulfide-linked complex. J. Virol. 72:550-557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Klupp, B. G., J. Baumeister, P. Dietz, H. Granzow, and T. C. Mettenleiter. 1998. Pseudorabies virus glycoprotein gK is a virion structural component involved in virus release but is not required for entry. J. Virol. 72:1949-1958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Klupp, B. G., J. Baumeister, A. Karger, N. Visser, and T. C. Mettenleiter. 1994. Identification and characterization of a novel structural glycoprotein in pseudorabies virus, gL. J. Virol. 68:3868-3878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Klupp, B. G., W. Fuchs, E. Weiland, and T. C. Mettenleiter. 1997. Pseudorabies virus glycoprotein L is necessary for virus infectivity but dispensable for virion localization of glycoprotein H. J. Virol. 71:7687-7695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kopp, M., H. Granzow, W. Fuchs, B. Klupp, and T. C. Mettenleiter. 2004. Simultaneous deletion of pseudorabies virus tegument protein UL11 and glycoprotein M severely impairs secondary envelopment. J. Virol. 78:3024-3034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Laquerre, S., S. Person, and J. C. Glorioso. 1996. Glycoprotein B of herpes simplex virus type 1 oligomerizes through the intermolecular interaction of a 28-amino-acid domain. J. Virol. 70:1640-1650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Lee, B. S., X. Alvarez, S. Ishido, A. A. Lackner, and J. U. Jung. 2000. Inhibition of intracellular transport of B cell antigen receptor complexes by Kaposi's sarcoma-associated herpesvirus K1. J. Exp. Med. 192:11-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Lee, T. H., and A. D. Linstedt. 1999. Osmotically induced cell volume changes alter anterograde and retrograde transport, Golgi structure, and COPI dissociation. Mol. Biol. Cell 10:1445-1462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Loomis, J. S., J. B. Bowzard, R. J. Courtney, and J. W. Wills. 2001. Intracellular trafficking of the UL11 tegument protein of herpes simplex virus type 1. J. Virol. 75:12209-12219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Loomis, J. S., R. J. Courtney, and J. W. Wills. 2003. Binding partners for the UL11 tegument protein of herpes simplex virus type 1. J. Virol. 77:11417-11424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.MacLean, C. A., S. Efstathiou, M. L. Elliott, F. E. Jamieson, and D. J. McGeoch. 1991. Investigation of herpes simplex virus type 1 genes encoding multiply inserted membrane proteins. J. Gen. Virol. 72:897-906. [DOI] [PubMed] [Google Scholar]
- 51.Marks, M. S., L. Woodruff, H. Ohno, and J. S. Bonifacino. 1996. Protein targeting by tyrosine- and di-leucine-based signals: evidence for distinct saturable components. J. Cell Biol. 135:341-354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.McMillan, T. N., and D. C. Johnson. 2001. Cytoplasmic domain of herpes simplex virus gE causes accumulation in the trans-Golgi network, a site of virus envelopment and sorting of virions to cell junctions. J. Virol. 75:1928-1940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Melancon, J. M., T. P. Foster, and K. G. Kousoulas. 2004. Genetic analysis of the herpes simplex virus type 1 UL20 protein domains involved in cytoplasmic virion envelopment and virus-induced cell fusion. J. Virol. 78:7329-7343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Mettenleiter, T. C. 2002. Herpesvirus assembly and egress. J. Virol. 76:1537-1547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Mo, C., J. Suen, M. Sommer, and A. Arvin. 1999. Characterization of varicella-zoster virus glycoprotein K (open reading frame 5) and its role in virus growth. J. Virol. 73:4197-4207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Mukherjee, S., R. N. Ghosh, and F. R. Maxfield. 1997. Endocytosis. Physiol. Rev. 77:759-803. [DOI] [PubMed] [Google Scholar]
- 57.Pellett, P. E., K. G. Kousoulas, L. Pereira, and B. Roizman. 1985. Anatomy of the herpes simplex virus 1 strain F glycoprotein B gene: primary sequence and predicted protein structure of the wild type and of monoclonal antibody-resistant mutants. J. Virol. 53:243-253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Pogue-Geile, K. L., G. T. Lee, S. K. Shapira, and P. G. Spear. 1984. Fine mapping of mutations in the fusion-inducing MP strain of herpes simplex virus type 1. Virology 136:100-109. [DOI] [PubMed] [Google Scholar]
- 59.Pogue-Geile, K. L., and P. G. Spear. 1987. The single base pair substitution responsible for the syn phenotype of herpes simplex virus type 1, strain MP. Virology 157:67-74. [DOI] [PubMed] [Google Scholar]
- 60.Ramaswamy, R., and T. C. Holland. 1992. In vitro characterization of the HSV-1 UL53 gene product. Virology 186:579-587. [DOI] [PubMed] [Google Scholar]
- 61.Roberts, S. R., M. Ponce de Leon, G. H. Cohen, and R. J. Eisenberg. 1991. Analysis of the intracellular maturation of the herpes simplex virus type 1 glycoprotein gH in infected and transfected cells. Virology 184:609-624. [DOI] [PubMed] [Google Scholar]
- 62.Roop, C., L. Hutchinson, and D. C. Johnson. 1993. A mutant herpes simplex virus type 1 unable to express glycoprotein L cannot enter cells, and its particles lack glycoprotein H. J. Virol. 67:2285-2297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Ryechan, W. T., L. S. Morse, D. M. Knipe, and B. Roizman. 1979. Molecular genetics of herpes simplex virus. II. Mapping of the major viral glycoproteins and of the genetic loci specifying the social behavior of infected cells. J. Virol. 29:677-697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Sanders, P. G., N. M. Wilkie, and A. J. Davison. 1982. Thymidine kinase deletion mutants of herpes simplex virus type 1. J. Gen. Virol. 63:277-295. [DOI] [PubMed] [Google Scholar]
- 65.Skepper, J. N., A. Whiteley, H. Browne, and A. Minson. 2001. Herpes simplex virus nucleocapsids mature to progeny virions by an envelopment→deenvelopment→reenvelopment pathway. J. Virol. 75:5697-5702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Tirabassi, R. S., and L. W. Enquist. 1999. Mutation of the YXXL endocytosis motif in the cytoplasmic tail of pseudorabies virus gE. J. Virol. 73:2717-2728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Tirabassi, R. S., and L. W. Enquist. 1998. Role of envelope protein gE endocytosis in the pseudorabies virus life cycle. J. Virol. 72:4571-4579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Trowbridge, I. S. 1991. Endocytosis and signals for internalization. Curr. Opin. Cell Biol. 3:634-641. [DOI] [PubMed] [Google Scholar]
- 69.Trowbridge, I. S., J. F. Collawn, and C. R. Hopkins. 1993. Signal-dependent membrane protein trafficking in the endocytic pathway. Annu. Rev. Cell Biol. 9:129-161. [DOI] [PubMed] [Google Scholar]
- 70.Tugizov, S., E. Maidji, J. Xiao, and L. Pereira. 1999. An acidic cluster in the cytosolic domain of human cytomegalovirus glycoprotein B is a signal for endocytosis from the plasma membrane. J. Virol. 73:8677-8688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Voorhees, P., E. Deignan, E. van Donselaar, J. Humphrey, M. S. Marks, P. J. Peters, and J. S. Bonifacino. 1995. An acidic sequence within the cytoplasmic domain of furin functions as a determinant of trans-Golgi network localization and internalization from the cell surface. EMBO J. 14:4961-4975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Ward, P. L., G. Campadelli-Fiume, E. Avitabile, and B. Roizman. 1994. Localization and putative function of the UL20 membrane protein in cells infected with herpes simplex virus 1. J. Virol. 68:7406-7417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Whealy, M. E., A. K. Robbins, and L. W. Enquist. 1990. The export pathway of the pseudorabies virus gB homolog gII involves oligomer formation in the endoplasmic reticulum and protease processing in the Golgi apparatus. J. Virol. 64:1946-1955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Whitbeck, J. C., A. C. Knapp, L. W. Enquist, W. C. Lawrence, and L. J. Bello. 1996. Synthesis, processing, and oligomerization of bovine herpesvirus 1 gE and gI membrane proteins. J. Virol. 70:7878-7884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Whiteley, A., B. Bruun, T. Minson, and H. Browne. 1999. Effects of targeting herpes simplex virus type 1 gD to the endoplasmic reticulum and trans-Golgi network. J. Virol. 73:9515-9520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Zhu, Z., M. D. Gershon, Y. Hao, R. T. Ambron, C. A. Gabel, and A. A. Gershon. 1995. Envelopment of varicella-zoster virus: targeting of viral glycoproteins to the trans-Golgi network. J. Virol. 69:7951-7959. [DOI] [PMC free article] [PubMed] [Google Scholar]