Abstract
Integrase has been implicated in human immunodeficiency virus type 1 (HIV-1) nuclear import. Integrase analyses, however, can be complicated by the pleiotropic nature of mutations: whereas class I mutants are integration defective, class II mutants display additional assembly and/or reverse transcription defects. We previously determined that HIV-1V165A, originally reported as defective for nuclear import, was a class II mutant. Here we analyzed mutants containing changes in other putative nuclear localization signals, including 186KRK188/211KELQKQITK219 and Cys-130. Previous work established HIV-1K186Q, HIV-1Q214L/Q216L, and HIV-1C130G as replication defective, but phenotypic classification was unclear and nuclear import in nondividing cells was not addressed. Consistent with previous reports, most of the bipartite mutants studied here were replication defective. These mutants as well as HIV-1V165A synthesized reduced cDNA levels, but a normal fraction of mutant cDNA localized to dividing and nondividing cell nuclei. Somewhat surprisingly, recombinant class II mutant proteins were catalytically active, and class II Vpr-integrase fusion proteins efficiently complemented class I mutant virus. Since a class I Vpr-integrase mutant efficiently complemented class II mutant viruses under conditions in which class II Vpr-integrases failed to function, we conclude that classes I and II define two distinct complementation groups and suggest that class II mutants are primarily defective at a postnuclear entry step of HIV-1 replication. HIV-1C130G was also defective for reverse transcription, but Vpr-integraseC130G did not efficiently complement class I mutant HIV-1. Since HIV-1C130A grew like the wild type, we conclude that Cys-130 is not essential for replication and speculate that perturbation of integrase structure contributed to the pleiotropic HIV-1C130G phenotype.
Retroviral replication requires the integration of the viral cDNA made by reverse transcription into a host cell chromosome. After entering a susceptible target cell, reverse transcriptase converts genomic RNA into linear double-stranded cDNA with a copy of the viral long terminal repeat (LTR) at each end. Integration is mediated by the viral integrase protein acting on the ends of the linear cDNA (reviewed in reference 14). Replication-defective integrase mutants of human immunodeficiency virus type 1 (HIV-1) can be grouped into one of two phenotypic classes. Class I mutants are specifically blocked at the integration step and are typified by changes in the Asp and Glu residues that constitute the enzyme active site. In contrast, class II mutants are pleiotropic, as additional defects at the postintegration step of viral particle assembly and/or at the preintegration step of reverse transcription are observed (reviewed in reference 20).
In addition to catalyzing the DNA breakage and joining steps essential for integration, integrase has been implicated in reverse transcription (33, 63, 68) and in the nuclear import of HIV-1 preintegration complexes (PICs) (reviewed in reference 53). When expressed on its own in the absence of other HIV-1 proteins, integrase localizes to cell nuclei and is therefore karyophilic (6, 12, 16, 17, 43, 47, 52, 54, 58). Despite numerous viral genetic and integrase localization analyses, the mechanism of integrase import and the role of integrase in PIC nuclear import have not been fully elucidated. Numerous human proteins interact with HIV-1 integrase, and some of these, including importin α (31), importin 7 (28), and lens epithelium-derived growth factor (LEDGF/p75) (46, 47), have been implicated in integrase and PIC nuclear import (reviewed in references 53 and 59).
One phenotype that is predicted for HIV-1 nuclear localization mutants is reduced infectivity and cDNA import specifically in nondividing compared to dividing cell types (reviewed in references 53 and 56). Since the integrase mutant HIV-1Y143G displayed a selective two- to fourfold defect in monocyte-derived macrophages (MDMs) compared to cycling cells, Tyr-143 contributed to PIC nuclear import in this classic nondividing cell model (58). However, the results of other mutagenic studies have likely been confounded by the pleiotropic nature of class II integrase mutant viruses (6, 18, 43). For example, a bipartite nuclear localization signal (NLS) comprised of NLS-proximal (NLSP) (186KRK188) and NLS-distal (NLSD) (211KELQKQITK219) sequences was originally described as being important for integrase-importin α interactions in vitro (31), yet HIV-1 mutated in either NLSP or NLSD residues was replication defective in both cycling and nondividing cells (9, 51, 58).
Whereas the NLSP mutant HIV-1K186Q (9, 51) and NLSD mutant HIV-1Q214L/Q216L (51) supported wild-type levels of cDNA synthesis and nuclear import in dividing cells, a separate study reported that HIV-1K186Q as well as a deletion of NLSP (HIV-1ΔKRK) caused defects in reverse transcription (58). The effects of NLSP and NLSD changes on reverse transcription and nuclear import in nondividing cells have not been reported. Although the K186Q and Q214L/Q216L changes altered the nuclear accumulation of Flag-tagged integrase in cells (51), K186Q and ΔKRK mutants were efficiently transported into cell nuclei as green fluorescent protein-integrase fusion proteins (58). Due to these conflicting reports on the phenotypes of NLSP and NLSD mutant viruses and the roles of these sequences in integrase localization, we analyzed previously described as well as novel NLSP and NLSD mutants in assays for virus replication, particle release, reverse transcription, and nuclear import. The effects of the K186Q and Q214L/Q216L changes on integrase localization were analyzed with a recently described codon-optimized expression system (43).
Residues in addition to Tyr-143 and those that constitute NLSP and NLSD have been implicated in integrase-PIC nuclear import. Among these is a noncanonical NLS comprised of Val-165 and Arg-166 (6) that was subsequently assessed as unimportant for viral import (18), as Flag-tagged INV165A localized to cell nuclei and HIV-1V165A and HIV-1R166A were classified as class II integrase mutant viruses (43). However, a recent report has resurrected the notion that this region of integrase comprises a functional NLS (3). Integrase residue Cys-130 has also been implicated in HIV-1 nuclear import through affecting integrase multimerization more than functioning as an NLS per se (52). Cys-130 resides in the integrase catalytic core domain (CCD) which harbors two additional Cys residues at positions 56 and 65. To investigate the potential role(s) of CCD cysteines in HIV-1 replication and nuclear localization, HIV-1C56A, HIV-1C65A, and HIV-1C130A were constructed. In total, 10 new NLSP, NLSD, and Cys mutant viruses were analyzed alongside previously described HIV-1K186Q (9, 51, 58), HIV-1Q214L/Q216L (9, 51), HIV-1C130G (52), and HIV-1V165A (6, 18, 43).
Integrase comprises three functional domains, the N-terminal domain, CCD, and C-terminal domain (reviewed in reference 14). This three-domain structure was in large part inferred from the results of functional complementation assays that showed that two different catalytically inactive integrase mutant proteins restored in vitro integration activity only when the proteins were altered in different domains (22, 61). Functional complementation can also occur in cells as integrase mutant infectivity defects were overcome by incorporating wild-type integrase into assembling virus particles as fusions to the virion accessory protein Vpr (29, 64). Certain changes like V165A and R199A that by themselves rendered HIV-1 replication defective nonetheless functionally complemented class I integrase active-site mutant viruses when assayed as Vpr-integrase fusion proteins (6, 29).
Since both the active site and Val-165 reside in the CCD (Fig. 1A), functional complementation implied that Val-165 plays a noncatalytic role in HIV-1 infection (6, 18). Here we extended this observation by showing that Vpr fusions to CCD NLSP mutants or C-terminal domain NLSD mutants efficiently restored class I mutant infectivity under conditions in which they failed to efficiently complement class II integrase mutant function. Since a CCD class I Vpr-integrase mutant fusion protein efficiently complemented class II mutant viruses but not class I HIV-1, these results demonstrate that class I and II are two distinct complementation groups and that this grouping extends beyond the domain boundaries that were defined by studying catalytically defective integrase mutant proteins in vitro (22, 61). Consistent with these observations, recombinant integrase proteins derived from complementing class II mutant viruses displayed activity in in vitro integration assays. Based on these results and the behavior of viral and integrase mutants in nuclear import assays, we conclude that NLSP, NLSD, Val-165, and Cys-130 residues do not play specific roles in HIV-1 nuclear import and suggest instead that altered nuclear trafficking contributes to the pleiotropic class II mutant phenotype.
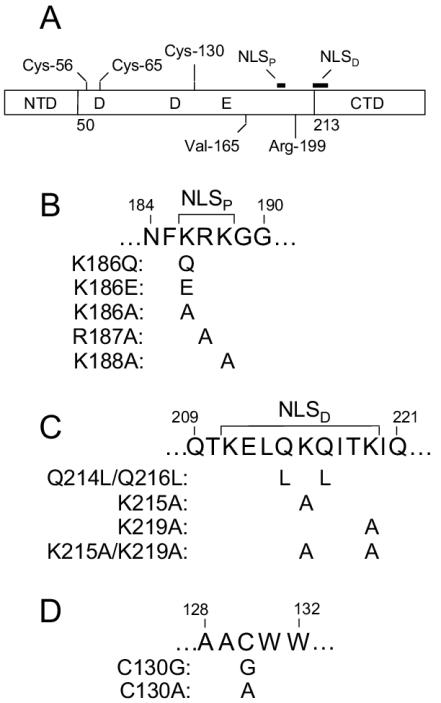
FIG. 1.
Integrase domain structure and mutant viruses. (A) Schematic diagram of integrase, highlighting the three functional domains and residues and sequences pertinent to this study. The locations of active-site residues Asp-64, Asp-116, and Glu-152 in the central CCD are indicated. Residues and regions that were targeted to generate novel mutant viruses are indicated above the protein. The numbers below the protein indicate the first residue of the catalytic core and C-terminal domains (26). NTD, N-terminal domain; CTD, C-terminal domain. (B) NLSP mutants. NLSP (186KRK188) (31) is indicated above the integrase coding sequence, and individual mutant changes are indicated underneath the corresponding wild-type amino acid residue. (C) NLSD (211KELQKQITK219) (31) and NLSD mutants. (D) Cys-130 mutants.
MATERIALS AND METHODS
Plasmids.
Integrase from the NL4-3 strain of HIV-1 was expressed in Escherichia coli with pINSD.His (43) or pKBIN6H (47). Flag-tagged HIV-1 integrase was expressed in HeLa cells with a previously described codon-optimized vector (43). Mutagenesis was performed with QuikChange mutagenesis as recommended by the manufacturer (Stratagene, La Jolla, Calif.). Viral integrase changes were introduced into pUCWTpol (43) and 1.8-kb AgeI-PflMI pol fragments were swapped for the corresponding fragments in pNL43/XmaI (7) and pNLXΔenv (43) to generate full-length and envelope (Env)-deleted molecular clones of pNL4-3, respectively. Integrase mutant plasmids pNL43(F185K) (35), pNLX(N/N), pNL43(1-212), (50), pNLX(V165A), and pNL43(R166A) (43) were described previously.
Plasmid pNLX(Vpr-) encoding nonfunctional Vpr protein (30) was constructed by digesting pNL43/XmaI with EcoRI, filling in with Klenow fragment, and religation. Plasmid pNLX.Luc was built by amplifying the gene for firefly luciferase (Luc) from pGL3-Basic (Promega, Madison, Wis.) with ClaI- and XhoI-tagged primers, digestion with ClaI and XhoI, and ligation to ClaI- and XhoI-digested pNLXΔenvCAT (44). The 1.6-kb PflMI-NheI fragment from pNLX(Vpr-) was swapped for the corresponding fragment in pNLX.Luc to create pNLX.Luc(R−). The 1.8-kb AgeI-PflMI fragments of various integrase mutant constructs were then switched for the corresponding fragment in pNLX.Luc(R−).
The Env expression vector encoding vesicular stomatitis virus G (VSV-G) glycoprotein was described previously (66). An expression vector carrying the dualtropic HIV-189.6 Env was made by swapping the KpnI-BamHI fragment from p89.6 (13) for the corresponding fragment in pNLXE7 (44). The Vpr-integrase expression vector pRL2P-Vpr-IN (64) (a generous gift from J. C. Kappes, University of Alabama at Birmingham) was altered with QuikChange mutagenesis. Targeted regions of plasmid DNAs were analyzed by sequencing to confirm desired mutations and rule out unwanted secondary changes.
Cells.
293T and HeLa cells were grown in Dulbecco's modified Eagle's medium supplemented to contain 10% fetal calf serum, 100 IU of penicillin per ml, and 100 μg of streptomycin per ml. Jurkat and C8166 T cells were grown in RPMI 1640 fortified with 10% fetal calf serum, 100 IU of penicillin per ml, and 100 μg of streptomycin per ml (RPMI). MDMs were derived from HIV-1-seronegative donor blood by absorbance onto plastic as previously described (50). Cell cycle status was determined by staining ethanol-fixed cells with propidium iodide and quantification by flow cytometry with ModFit LT version 3.0 (Verity Software House Inc., Topsham, Maine).
Nuclear localization of integrase.
Flag-tagged integrase in transiently transfected HeLa cells was detected by indirect immunostaining as previously described (43). Chromosomal DNA was visualized by staining with 4′,6′-diamidino-2-phenylindole as previously described (43).
Viruses and infections.
Virus stocks were generated in either 293T or HeLa cells by transfection with calcium phosphate, and cell-free virus titers were determined with an exogenous 32P-based reverse transcriptase assay as previously described (24, 50). Infections for virus replication assays were performed as previously described (50).
For real-time quantitative PCR (RQ-PCR) assays, 293T cells were cotransfected with a viral vector and an Env expression plasmid with FuGene 6 (Roche Molecular Biochemicals, Indianapolis, Ind.). The resulting cell-free supernatants were treated with DNase for 1 h at 37°C to degrade the bulk of residual plasmid DNA left over from transfection essentially as previously described (11, 43) with the exception that 40 U of Turbo DNase (Ambion, Austin, Tex.) per ml was used in most applications. Jurkat cells (106) were infected with 106 reverse transcriptase cpm of HIV-189.6 Env-containing virus by spinoculation as previously described (44, 45). For VSV-G-pseudotyped virus, Jurkat cells (4 × 106) were infected with 2.5 × 106 reverse transcriptase cpm by spinoculation, and MDMs were infected by incubation at 37°C for 2 h. Viruses for Southern blotting were derived from 293T cells as previously described (11) except that FuGene 6 was used as the transfection reagent instead of calcium phosphate.
RQ-PCR assays.
DNA from infected cells was harvested with the DNeasy tissue kit as recommended by the manufacturer (Qiagen, Valencia, Calif.), and 10 μl was amplified by PCR (30-μl reaction volume) as previously described (44, 45). Two different HIV-1 amplicons, late reverse transcription products and two-LTR-containing circles, were normalized to total cellular DNA with either spectrophotometry (44) or endogenous retrovirus 3 (43, 45) as previously described. Virus particles lacking Env were used in parallel infection and RQ-PCR assays to assess residual levels of plasmid DNA that remained after transfection and DNase treatment, and these values were subtracted from values obtained with matched Env-positive infections.
Southern blot analysis.
C8166 cells were infected and Southern blotting was performed essentially as previously described (7, 11) with the exception that DNA was extracted from infected cells with the Hirt protocol (34, 39). Hirt supernatant DNA digested with BsgI (6) and DpnI was recovered following extraction with phenol-chloroform and precipitation with ethanol. Viral DNA signals were quantified by PhosphorImager with ImageQuant v1.11 (Amersham Biosciences, Inc., Piscataway, N.J.).
Purification of recombinant integrase protein and in vitro integration assay.
N-terminally hexahistidine-tagged integrase expressed from pINSD.His was purified from inclusion bodies and refolded as previously described (43). The His tag was removed by cleavage with thrombin as previously described (43). C-terminally tagged integrase expressed from pKBIN6H was purified from soluble extracts of Escherichia coli as previously described (12).
Oligonucleotide substrate DNA representing the 21 bp of the HIV-1 U5 end was radiolabeled and annealed as previously described (43). Integration reactions (16 μl) contained 25 mM morpholinepropanesulfonic acid (pH 7.2), 0.1 mg of bovine serum albumin per ml, 10 mM β-mercaptoethanol, 10% glycerol, 25 nM labeled DNA substrate, 20 mM NaCl, 7.5 mM MnCl2, and 0.4 μM integrase. Reactions were terminated after 60 min at 37°C and electrophoresed through denaturing polyacrylamide sequencing gels, and the results were quantified by PhosphorImager.
Vpr-integrase complementation.
293T cells in six-well plates were cotransfected with 2 μg of pNLX.Luc(R−), 1 μg of pRL2P-Vpr-IN, and 0.067 μg of pNLXE7 by calcium phosphate, and the resulting cell-free viruses were titered by a 32P-labeled reverse transcriptase assay. Baseline integrase mutant infectivities in the absence of added Vpr-integrase were conducted by substituting pcDNA3 (Invitrogen) for pRL2P-Vpr-IN. Jurkat cells (2 × 106 in 0.5 ml) were infected with 5 × 105 reverse transcriptase cpm for 17 h at 37°C, at which time culture volumes were expanded by the addition of 5 ml of RPMI. Two days postinfection, cells were washed with phosphate-buffered saline (Mediatech, Herndon, Va.) and lysed with 75 μl of passive lysis buffer (Promega Corp.). After a freeze-thaw cycle, lysates were clarified by centrifugation at 18,730 × g for 15 min at 4oC. Supernatants (20 μl) were tested in duplicate for luciferase activity (Luciferase Assay System; Promega, Corp.) with an EG&G Berthold Microplate LB 96V luminometer and a Microlite 1 flat-bottomed microtiter plate (Thermo Labsystems, Franklin, Mass.). Luciferase activity was normalized to total protein concentration as determined by the Bio-Rad protein assay (Bio-Rad Laboratories, Hercules, Calif.). Background values derived from infections with Env-minus HIV-1NLX.Luc(R−) were subtracted from Env-positive wild-type and mutant integrase activities.
RESULTS
Mutagenesis strategy.
Although previous studies reported that NLSP mutant HIV-1K186Q (9, 51, 58), NLSD mutant HIV-1Q214L/Q216L (9, 51), and HIV-1C130G (52) were replication defective, phenotypic details of these putative NLS mutant viruses in cycling and nondividing target cells were unclear. To obtain a comprehensive picture of the roles of NLSP and NLSD in HIV-1 replication and nuclear import, seven novel mutant viruses were constructed. Whereas K186E, K186A, R187A, and K188A were designed to alter NLSP (Fig. 1B), K215A, K219A, and K215A/K219A targeted NLSD (Fig. 1C). HIV-1 integrase contains six Cys residues, two in the N-terminal domain that help coordinate zinc, three in the CCD, and one in the C-terminal domain (for a review of integrase domain structure, see references 14 and 26). Whereas zinc finger mutant HIV-1C43L was noninfectious (48), C-terminal domain mutant HIV-1C280S grew like the wild type (4). To obtain a comprehensive view of the role of each CCD cysteine in HIV-1 replication, Ala was substituted for Cys-56, Cys-65, and Cys-130 (Fig. 1A and 1D). Novel integrase changes as well as previously described K186Q (9, 51, 58), Q214L/Q216L (9, 51), and C130G (52) changes were introduced into the NL4-3 strain of HIV-1 (1).
Replication phenotypes of NLSP, NLSD, and Cys mutant viruses.
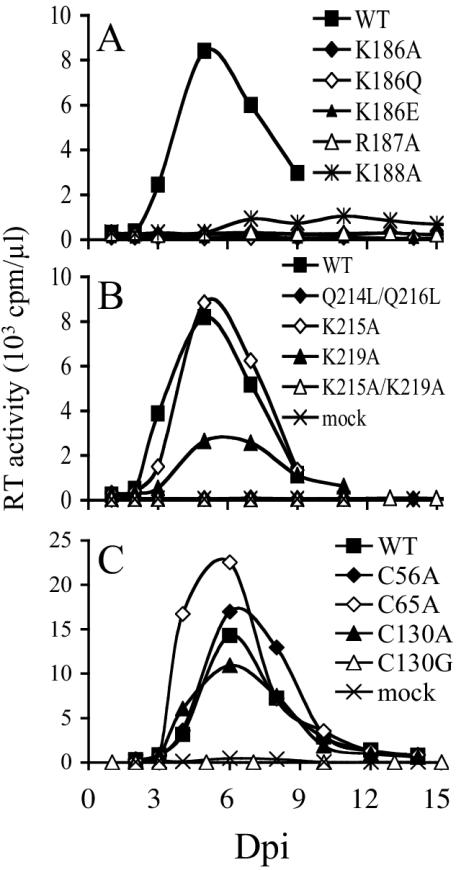
In repeat experiments, Jurkat T cells inoculated with 106 reverse transcriptase cpm (an approximate multiplicity of infection of 0.04) (44, 50) supported peak levels of HIV-1NL4-3 replication 5 to 6 days postinfection (Fig. 2). In agreement with previous reports (9, 51, 58), the K186Q change rendered HIV-1NL4-3 replication defective (Fig. 2A). Integrase mutants HIV-1K186A, HIV-1K186E, and HIV-1R187A were also replication defective. In contrast, HIV-1K188A supported a low but reproducible level of virus spread (Fig. 2A and data not shown).
FIG. 2.
Wild-type (WT) and integrase mutant replication kinetics in Jurkat T cells. (A) Supernatants of cells infected with the indicated viruses were analyzed for reverse transcriptase (RT) activity at the indicated time points. (B) Replication kinetics of NLSD mutant viruses. (C) Reverse transcriptase profiles of Cys mutant viruses. Similar integrase mutant profiles were observed in independent sets of infections. Extended observation (2 months) of cells infected with HIV-1K186Q, HIV-1K186A, HIV-1K186E, HIV-1R187A, HIV-1Q214L/Q216L, HIV-1K215A/K219A, or HIV-1C130G failed to reveal any evidence of virus replication. Dpi, days postinfection.
Akin to the previous reports (9, 51), we also found HIV-1Q214L/Q216L replication defective (Fig. 2B). Since NLSD mutant HIV-1G189R/K211N was infectious (58) and Los Alamos sequence alignments indicated that NLSD residues Lys-215 and Lys-219 were more highly conserved among HIV-1 isolates than Lys-211 (38), Lys-215 and Lys-219 were targeted in this study. Whereas HIV-1K215A grew like HIV-1NL4-3, HIV-1K219A was modestly perturbed in its ability to replicate, as HIV-1K219A-infected cultures yielded approximately threefold fewer total reverse transcriptase cpm than wild-type-infected cells in repeat experiments (Fig. 2B). In contrast, HIV-1K215A/K219A was replication defective.
Consistent with the previous report (52), HIV-1C130G was replication defective (Fig. 2C). Somewhat surprisingly, HIV-1C130A replicated with wild-type kinetics. HIV-1C56A and HIV-1C65A also yielded the wild-type replication profile in Jurkat T cells (Fig. 2C).
Integrase mutants and HIV-1 release.
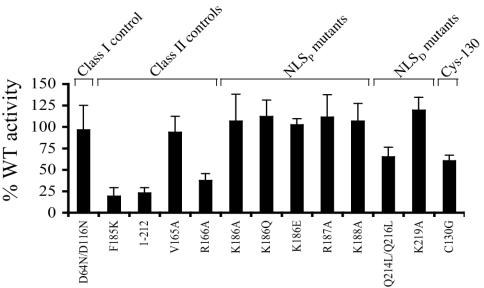
Having determined that a subset of NLSP, NLSD, and Cys-130 mutant viruses were replication defective, we next investigated the step(s) in the HIV-1 life cycle where these viruses were blocked. Defective particle assembly and release is one of the phenotypes associated with pleiotropic class II mutants (20). Particle release was assayed by transfecting CD4− cells and measuring HIV-1 in the resulting cell-free supernatants. This assay is well suited for quantifying the effects of integrase mutations on virus release because (i) it bypasses the requirement for integration as a prerequisite for gene expression and (ii) receptor-minus cells afford comparison of replication-defective and replication-competent strains. As controls we included wild-type HIV-1NL4-3, active-site class I mutant HIV-1D64N/D116N (50), assembly-defective class II mutants HIV-1F185K (35) and HIV-11-212 (50), and class II mutants HIV-1V165A and HIV-1R166A which we and others previously determined were released from 293T and COS-7 cells at the wild-type level (6, 43, 62). We note that previous analyses of virus-expressing cells revealed that wild-type and assembly-defective class II integrase mutants supported similar levels of HIV-1 gene expression (2, 8, 24, 25).
Whereas the class I HIV-1D64N/D116N mutant released from HeLa cells at the wild-type level, class II assembly-defective controls HIV-1F185K and HIV-11-212 were released at 20% to 25% of the wild-type level (Fig. 3). Consistent with previous reports (6, 43), HIV-1V165A was released as efficiently as the wild type. In contrast, HIV-1R166A release was reduced approximately threefold from that of the wild type (Fig. 3). This result agrees with previous findings that a class II integrase deletion mutant was released more efficiently from 293T and COS-7 cells than from HeLa cells (8). Each of the replication-defective NLSP mutants released with wild-type efficiency. In contrast, HIV-1C130G and HIV-1Q214L/Q216L release was reduced to 60% to 70% of the wild-type level (Fig. 3). In separate experiments, HIV-1K215A and HIV-1K215A/K219A behaved like HIV-1K219A and thus did not reveal any release defect in this assay (data not shown).
FIG. 3.
Integrase mutant virus release from cells. Supernatants of HeLa cells transfected with the indicated viruses were analyzed for reverse transcriptase activity. Results are averages of duplicate reverse transcriptase assays and a minimum of four independent transfections. Virus release was expressed as a percentage of wild-type reverse transcriptase activity. Radioimmunoprecipitation analyses of cell extracts revealed wild-type levels of gene expression for HIV-1F185K (25), HIV-11-212, HIV-1Q214L/Q216L, and HIV-1R166A (data not shown).
Reverse transcription defect of HIV-1V165A.
In addition to particle release, integrase mutations can affect reverse transcription (20). Reverse transcription is a multistep process requiring two template switches to generate full-length duplex cDNA (reviewed in reference 57), and PCR primers can be designed to quantify specific steps in the sequential process (36, 67). We previously devised an RQ-PCR assay (43, 44) with LTR- and gag-specific primers to quantify levels of full-length and nearly full-length late reverse transcription products. A second set of primers was designed to amplify two-LTR circles, a ligation product formed by host-mediated nonhomologous DNA end joining (42). Since two-LTR circles form in the nuclei of HIV-1-infected cells, the two-LTR/late reverse transcription ratio can be used as an indicator of HIV-1 nuclear import (reviewed in reference 56).
Previous results established that class II integrase mutant viruses were defective for two-LTR circle formation (2, 6, 24, 25, 40, 43). Although we previously proposed that defective nuclear import might be a phenotype common to all class II mutant viruses (43), other results indicated that reduced levels of reverse transcription might in large part account for the observed reductions in two-LTR circle formation (40). Based on these observations as well as other discrepancies, for example, whether HIV-1K186Q synthesized wild-type (9, 51) or diminished (58) cDNA levels, we reevaluated our RQ-PCR assay to ensure that it detected bona fide levels of wild-type and integrase mutant reverse transcription.
Since class II integrase mutants are replication defective, virus stocks for RQ-PCR assays must be generated by transfection. Supernatants derived from transiently transfected cells contain residual plasmid DNA, and it is therefore important to treat such viruses with a DNA-degrading enzyme such as DNase I prior to infection. We originally determined by Southern blotting (11) and RQ-PCR (44) that 2 U of RQ1 DNase I (Promega Corp.) per ml reduced plasmid DNA signals to background levels.
To investigate whether residual plasmid might in some instances survive this treatment, single-round Env-positive and Env-negative HIV-1NLX.Luc was treated with various concentrations of DNase I prior to infection, and cell lysates prepared at 3 h, 6 h, and 24 h postinfection were analyzed by RQ-PCR as previously described (44). Although Env-positive virus treated with 2 U of DNase I per ml yielded more HIV-1 DNA than the Env-negative control, the Env-negative signal was significantly above the background level of detection at 3 h postinfection (Fig. S1 in the supplemental material). Whereas the Env-positive signal increased from 3 to 6 h postinfection, the Env-negative signal decreased during this time frame and approached the detection limit of the assay by 24 h postinfection (Fig. S1). By increasing the concentration of DNase I in the pretreatment, the levels of DNA detected after mock infection were reduced to near background levels without affecting the level of Env-dependent reverse transcription (Fig. S1). These results confirmed our suspicion that plasmid DNA in viral stocks could in some instances survive DNase I treatment. Based on these results, we shifted to treating viruses for RQ-PCR assays with relatively high concentrations of DNase I. The results of additional preliminary experiments revealed that Turbo DNase reproducibly yielded lower levels of RQ-PCR signals after mock infection than did RQ1 DNase I (data not shown).
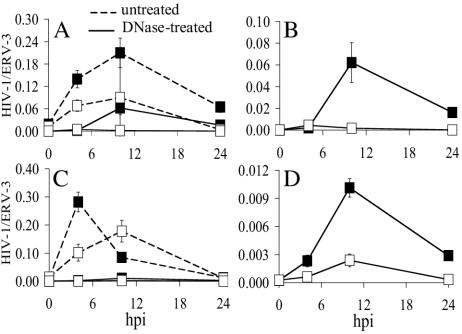
The kinetics of HIV-1NLX.Luc and HIV-1V165A.Luc reverse transcription were compared with and without Turbo DNase pretreatment. Untreated Env-positive and Env-negative HIV-1NLX.Luc yielded surprisingly similar RQ-PCR profiles (Fig. 4A, dashed lines). Whereas similarly low levels of DNA were detected at time zero, both signals rose until 10 h postinfection and subsided thereafter (Fig. 4A). DNase treatment collapsed the Env-negative profile nearly to background levels, revealing the kinetics of HIV-1NLX.Luc reverse transcription under these conditions (Fig. 4A and B). Similar to the wild type, untreated HIV-1V165A.Luc yielded robust RQ-PCR signals independent of Env function (Fig. 4C, dashed lines). In this case, DNase significantly reduced both Env-positive and Env-negative signals (Fig. 4C). Rescaling unmasked the level of integrase mutant reverse transcription which, after correcting for the residual Env-negative signal, revealed approximately eightfold defective cDNA synthesis for HIV-1V165A.Luc (compare Fig. 4D to 4B).
FIG. 4.
Analysis of HIV-1V165A reverse transcription by RQ-PCR. (A) Jurkat cells were infected with untreated (dashed line) or DNase-treated (solid line) Env-positive (▪) or Env-negative (□) HIV-1NLX.luc. Cellular DNA prepared at the indicated times was evaluated for HIV-1 by RQ-PCR as previously described (43, 45). (B) Same as in panel A except that DNase-treated samples were regraphed to highlight the level of HIV-1NLX.Luc reverse transcription (▪) over that of residual plasmid DNA (□). (C and D) Same as in panels A and B except that integrase mutant V165A was analyzed. Error bars represent variations between duplicate RQ-PCR assays. ERV-3, endogenous retrovirus 3; hpi, hours postinfection.
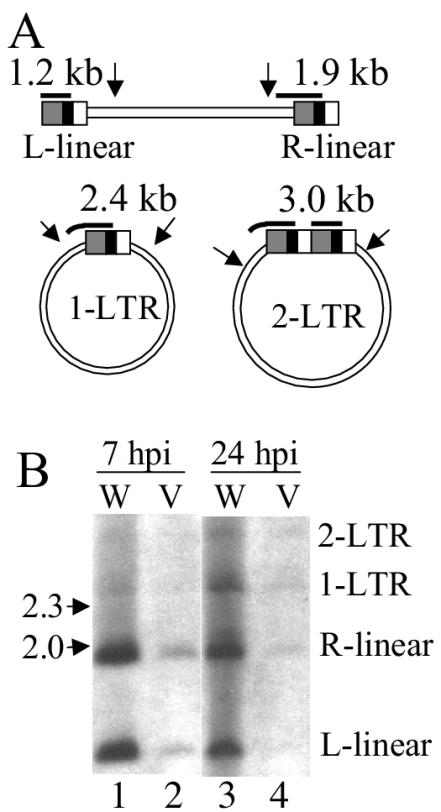
We note that the class II mutation reduced the quantity of late reverse transcription products without detectably altering the kinetics of cDNA synthesis or the stability of completed reverse transcripts (Fig. 4B and D). HIV-1NL4-3 and HIV-1V165A reverse transcription was also analyzed by Southern blotting. Hirt supernatant DNAs were treated with BsgI to yield 1.2-kb and 1.9-kb products representing the left and right ends of linear HIV-1 cDNA, respectively (Fig. 5A). HIV-1V165A yielded about fivefold less cDNA than HIV-1NL4-3 at 7 h postinfection (Fig. 5B, lanes 1 and 2). Since Southern blotting confirmed a reverse transcription defect for HIV-1V165A, we concluded that the modified RQ-PCR assay (Fig. 4) accurately determined levels of integrase mutant reverse transcription.
FIG. 5.
Southern blot analysis of HIV-1NL4-3 and HIV-1V165A DNA. (A) Major forms of unintegrated HIV-1 cDNA in infected cells. Arrows denote the approximate locations of relevant BsgI sites. Bold horizontal lines represent the nef/U3 minus-strand-specific probe used for blotting (7, 11), with the predicted size of each hybridizing fragment indicated above the probe. (B) Wild-type (W) and HIV-1V165A (V) DNA synthesis. The migration positions of DNA standards are indicated to the left of the blot (in kilobases), and the identities of HIV-1 DNA species are indicated on the right. hpi, h postinfection.
Reverse transcription and nuclear import profiles of integrase mutant viruses.
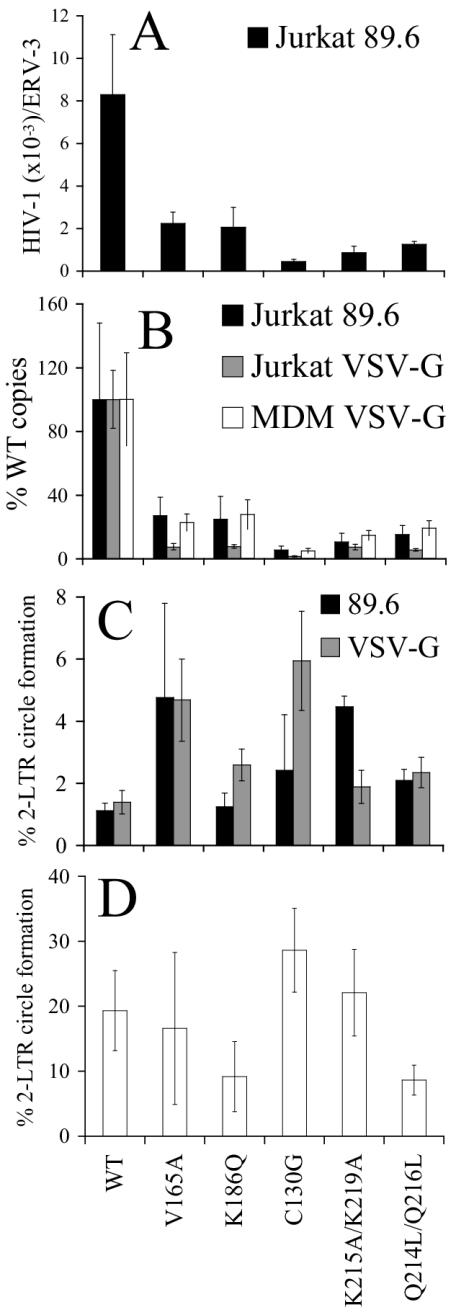
We next analyzed the cDNA synthesis and nuclear import profiles of a subset of replication-defective integrase mutant viruses in cycling and nondividing cells. Since each NLSP mutant was released from HeLa cells at the wild-type level (Fig. 3), HIV-1K186Q was chosen as a representative mutant. As the replication-defective HIV-1Q214L/Q216L and HIV-1K215A/K219A NLSD mutants displayed slightly different release profiles, both of these were selected for study. HIV-1V165A and HIV-1C130G were also analyzed.
Infectivity and reverse transcription profiles were initially tested in cycling T cells. As anticipated from the results of spreading infection assays (Fig. 2), the single-round infectivities of these replication-defective integrase mutants were reduced to near the detection limit of the luciferase assay (approximate 400- to 10,000-fold reductions compared to HIV-1NLX.Luc[R−]; Table 1). Since wild-type and integrase mutant reverse transcription levels peak approximately 7 to 8 h postinfection (10, 11, 37) (Fig. 4), late reverse transcription values were determined at 7 h postinfection. HIV-1V165A.Luc and HIV-1K186Q.Luc synthesized about fourfold less cDNA than HIV-1NLX.Luc (Fig. 6A). HIV-1Q214L/Q216L.Luc and HIV-1K215A/K219A.Luc yielded similar low levels of reverse transcripts, while HIV-1C130G.Luc yielded even lower cDNA levels, which amounted to approximately 10% of the wild-type level in repeat experiments (Fig. 6A and data not shown).
TABLE 1.
Complementation of mutant viruses with Vpr-integrase fusionsa
| Integrase mutant | % of wild-type activityb | Luciferase activity of Vpr-integrase fusions
|
|||||||
|---|---|---|---|---|---|---|---|---|---|
| Wild-type activityc | Complementation relative to Vpr-INWTd
|
||||||||
| V165A | C130G | K186Q | K215A/K219A | Q214L/Q216L | Q62K | R199A | |||
| V165A | 0.00 (0.01) | 26.5 (3.0) | − | − | +/− | +/− | +/− | ++e | + |
| C130G | 0.02 (0.04) | 71.3 (28.2) | − | − | +/− | +/− | +/− | +/− | +/− |
| K186Q | 0.23 (0.28) | 31.4 (8.4) | − | − | +/− | +/− | +/− | ++ | + |
| K215A/K219A | 0.02 (0.02) | 27.9 (4.3) | − | − | +/− | +/− | +/− | ++ | + |
| Q214L/Q216L | 0.11 (0.05) | 36.1 (12.0) | − | − | + | + | +/− | ++ | + |
| D64N/D116N | 0.07 (0.03) | 18.2 (9.7) | ++ | +/− | ++ | ++ | ++ | − | ++ |
| R199A | 0.39 (0.04) | 37.5 (18.5) | − | − | +/− | + | +/− | + | + |
Values represent averages of a minimum of two infections.
Luciferase activity of the indicated mutant virus relative to HIV-1NLX.Luc(R−), with standard deviations in parentheses.
Activity of the indicated mutant viruses complemented with Vpr-INWT relative to WT HIV-1NLX.Luc(R−), with the standard deviations in parentheses.
++, ≥50% of Vpr-INWT complementation; +, 10 to 50%; +/−, 1 to 10%; −, <1%.
Whereas Vpr-INQ62K displayed 51% of Vpr-INWT complementation activity, the class I active-site Vpr-IND116A mutant protein (29) displayed about 4% activity. Thus, we did not observe the same pattern of efficient reciprocal complementation between active-site and V165A mutant viruses and Vpr-integrase proteins as noted in reference 6.
FIG. 6.
DNA synthesis and nuclear import profiles of integrase mutant viruses. (A) Jurkat cells were infected with the indicated viruses, and DNA prepared 7 h postinfection was analyzed for total HIV-1 DNA by RQ-PCR. (B) Same as in panel A except VSV-G pseudotypes were used to infect Jurkat cells (grey bars) or MDMs (white bars). Integrase mutant DNA levels were normalized to the wild-type (WT) level, which was set to 100%. DNA levels from panel A are included for comparison. (C) Percentage of total HIV-1 DNA converted to two-LTR circles, determined by dividing the two-LTR circle value obtained at 24 h postinfection by the total DNA level from 7 h postinfection. Black bars, Jurkat cells infected via HIV-189.6 Env; grey bars, infections mediated via VSV-G. (D) Percentage of total HIV-1 DNA converted to two-LTR circles in MDMs infected by the indicated VSV-G-pseudotypes. Values were determined as in panel C except that two-LTR circle levels peaked 48 h postinfection in MDMs. Cell cycle analysis revealed that the MDM cultures used in these experiments comprised approximately 2% cells in S phase. The error bars in panels A through D represent variation between duplicate RQ-PCR assays. Similar results were determined in duplicate sets of infections. VSV-G-pseudotyped HIV-1K188A, HIV-1K215A, and HIV-1K219A yielded two-LTR/total cDNA fractions in MDMs that were greater than or equal to the wild-type value (data not shown). ERV-3, endogenous retrovirus 3.
The fraction of wild-type and integrase mutant cDNA converted into two-LTR circles at 24 h postinfection is shown in Fig. 6C. Although each integrase mutant formed a lower absolute number of DNA circles, mutant two-LTR/late reverse transcription ratios were equal to or greater than the wild-type value (Fig. 6C). Thus, none of the class II integrase mutants was defective for nuclear import under these infection conditions. Southern blotting confirmed that a normal fraction of HIV-1V165A cDNA was converted to LTR circles at 24 h postinfection (Fig. 5B).
Terminally differentiated MDMs present a paradigm for active HIV-1 nuclear import (53, 56). Thus, for the purpose of this work, it was important to address integrase mutant nuclear import in this cell type. Preliminary experiments, however, revealed that HIV-1NLX.Luc pseudotyped with the dual-tropic HIV-189.6 Env failed to yield detectable levels of two-LTR circles in MDMs derived from most blood donors (data not shown). Furthermore, class II integrase mutant viruses yielded less two-LTR circle DNA than wild-type virus under the favorable condition of dividing cell infection (2, 24, 25, 40, 43) (Fig. 5B, 6B, and 6C). For these reasons, the VSV-G Env glycoprotein was used to enhance levels of HIV-1 entry. Class II integrase mutant reverse transcription (Fig. 6B) and infectivity (data not shown) defects were observed in Jurkat T cells following VSV-G-mediated entry. Similar to HIV-1 Env-mediated infections, the percentages of integrase mutant cDNAs converted into two-LTR circles were at or above the wild-type level following VSV-G-mediated entry (Fig. 6C).
The reverse transcription profiles of integrase mutant viruses in MDMs mirrored their profiles in Jurkat T cells (Fig. 6B). The frequencies of integrase mutant two-LTR circle formation in MDMs were again quite similar to that of HIV-1NLX.Luc, with HIV-1Q214L/Q216L.Luc and HIV-1K186Q.Luc revealing minor defects (approximately twofold) in repeat experiments (Fig. 6D and data not shown). However, since the partially defective NLSP mutant HIV-1K188A.Luc (data not shown) and replication-defective NLSD mutant HIV-1K215A/K219A.Luc (Fig. 6D) supported the wild-type frequency of two-LTR circle formation in MDMs, we conclude that neither NLSP, NLSD, Cys-130, nor Val-165 plays a critical role in HIV-1 nuclear import in cycling cells or in primary MDMs (Fig. 6).
Cellular localization of integrase mutant proteins.
The results of the previous section discounted significant roles for NLSP, NLSD, Val-165, and Cys-130 in PIC nuclear import and instead highlighted diminished levels of cDNA synthesis as the primary reason for reduced levels of nuclear HIV-1. We next tested the roles of NLSP and NLSD in the nuclear import of integrase protein. This interest stemmed from inconsistencies in the literature; whereas one study reported that Flag-tagged NLSP and NLSD mutants mislocalized and were dispersed throughout transfected cells (51), a separate study reported wild-type localization properties for green fluorescent protein-integrase fusion proteins (58).
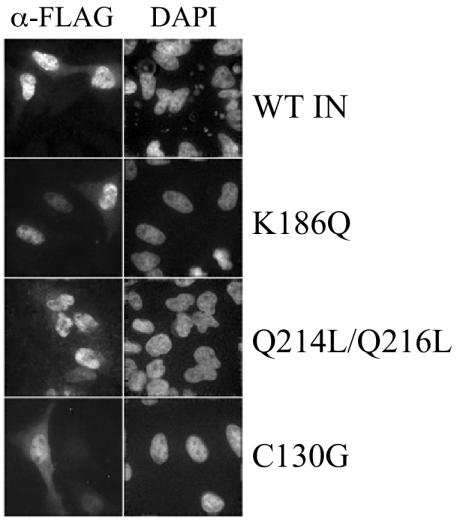
The effects of K186Q and Q214L/Q216L changes on integrase localization were assayed in the context of a recently developed codon-optimized Flag-tagged expression system (17, 43). As expected, wild-type integrase efficiently accumulated in cell nuclei (Fig. 7). The integrase mutants INK186Q and INQ214/Q216L also localized to cell nuclei under these experimental conditions (Fig. 7). Since the C130G change was also reported to alter the cellular distribution of Flag-tagged integrase (52), we also tested this mutant in our expression system. Although INC130G was predominantly nuclear, it displayed substantially more cytoplasmic staining than the wild-type, NLSP, or NLSD mutant proteins (Fig. 7). Our results indicate that neither K186Q, Q214L/Q216L, nor C130G dramatically altered the nuclear accumulation of Flag-tagged integrase, although, similar to the previous report (52), INC130G in part localized to the cell cytoplasm.
FIG. 7.
Distribution of Flag-tagged wild-type (WT) and mutant integrase (IN) proteins in HeLa cells. Cells transfected with codon-optimized Flag-tagged expression vectors were fixed and mounted in 4′,6′-diamidino-2-phenylindole (DAPI)-containing medium to visualize nuclear DNA. Integrase was visualized by indirect immunofluorescence microscopy with the anti-Flag M2 antibody.
Analyses of mutant catalytic activities. (i) In vitro integration.
Our results highlighted that replication-defective integrase mutants HIV-1V165A, HIV-1K186Q, HIV-1Q214L/Q216L, HIV-1K215A/K219A, and HIV-1C130G were 4- to 10-fold defective in their ability to synthesize HIV-1 cDNA. Since single-round mutant infectivities were reduced approximately 400- to 10,000-fold from the wild-type level, we reasoned that the primary replication defect(s) of these viruses was at a step downstream of reverse transcription and nuclear localization. We reasoned that the defect was likely to be at integration because (i) mutant virus assembly and release were comparable to those of the wild type (Fig. 3), (ii) mutant virus was transported into the nucleus with nearly wild-type efficiency, as judged from two-LTR/late reverse transcription ratios (Fig. 6C and D), and (iii) the viruses were altered in integrase. To further dissect the replication blocks of these viruses, the catalytic activities of the mutant integrase proteins were analyzed.
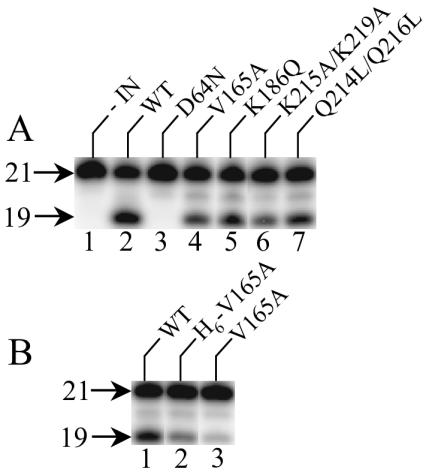
In cells, integrase catalyzes two distinct activities: 3′ processing of newly synthesized reverse transcripts, which is followed by the transfer of the nicked viral ends to the 5′ phosphates of a double-stranded cut in chromosomal DNA at the site of integration (reviewed in reference 14). Recombinant integrase proteins were expressed as fusions to a noncleavable C-terminal hexahistidine tag to enable purification by nickel nitrilotriacetate chromatography from soluble extracts of E. coli (12, 47). To assess catalytic activity, purified proteins were reacted with an oligonucleotide substrate that modeled the U5 end of HIV-1 cDNA. Wild-type integrase converted about 43% of the substrate to the nicked 3′-processing product (Fig. 8A, lane 2). The IND64N active-site mutant protein was predictably inactive (23) under these reaction conditions (<0.2% of substrate converted to product; Fig. 8A, lane 3). In stark contrast, the integrases derived from replication-defective HIV-1V165A, HIV-1K186Q, HIV-1Q214L/Q216L, and HIV-1K215A/K219A were catalytically active (Fig. 8A, lanes 4 to 7). Of note, INV165A displayed about 60% of wild-type integrase activity under these conditions (Fig. 8A, lane 4, and data not shown).
FIG. 8.
Catalytic activity of class II integrase mutant proteins. (A) Activities of C-terminally His6-tagged proteins. Integrase (IN) was omitted from the reaction in lane 1. (B) Effect of His6 tag on the 3′-processing activity of INV165A. Whereas the protein in lane 2 retained the tag, the tag was removed from the protein in lane 3. The tag was also removed from wild-type integrase (lane 1). 21, migration position of the 21-base labeled strand in substrate DNA; 19, position of cleaved DNA product.
Using similar reaction conditions, we noted previously that INV165A displayed only about 3% of wild-type integrase activity (43). INV165A was previously solubilized with guanidine-HCl, purified in denatured form, and refolded, and the N-terminal His6 tag was removed by protease cleavage. To see if the His6 tag and/or purification scheme affected INV165A activity, the activity of refolded and tagged protein was compared to that of refolded and thrombin-cleaved INV165A. The tagged protein supported about 45% of wild-type integrase activity (Fig. 8B, compare lane 2 to lane 1). Removing the tag reduced INV165A activity approximately threefold, to about 14% of wild-type integrase activity (Fig. 8B, lanes 1 and 3). Based on this, we conclude that although the His6 tag can influence the activity of recombinant integrase protein derived from replication-defective class II integrase mutant viruses, these enzymes posses significantly more in vitro integration activity than active-site mutant proteins derived from replication-defective class I mutant viruses (Fig. 8).
(ii) Vpr-integrase trans-complementation.
Vpr-integrase complementation was next used to investigate the infectivity defects and catalytic activities of class I and class II integrase mutants. The infectivity of integration-defective HIV-1 can be restored by incorporating wild-type integrase into assembling particles as a Vpr-integrase fusion protein (29, 64). In addition, certain replication-incompetent integrase mutants were shown to complement class I integrase mutant virus when expressed as Vpr-integrase fusions. Three different noninfectious CCD mutants were previously shown to restore function to class I integrase mutants as Vpr-integrase fusions: V165A (6), L172A/K173A (55), and R199A (29).
Based on these observations, the following changes were incorporated into Vpr-integrase alongside V165A and R199A controls: C130G, K186Q, K215A/K219A, and Q214L/Q216L. The Q62K change, which perturbs integrase-cDNA binding (11, 27, 32) and causes an integration-specific defect (11; R. Lu, A. Limón, H. Ghory, and A. Engelman, submitted for publication), was engineered as a CCD class I Vpr-integrase mutant protein, and HIV-1D64N/D116N (50) was used as a CCD class I mutant virus. Functional Vpr was deleted from the single-round luciferase-expressing viruses used in these assays to enhance trans-incorporation of Vpr-integrase during particle assembly. Table 1 reports the levels at which Vpr-wild-type integrase fusions complemented each integrase mutant virus as a percentage of wild-type HIV-1NLX.Luc(R−) infectivity. The level at which each Vpr-integrase mutant protein functioned was then expressed as the percentage of Vpr-wild-type integrase activity.
Consistent with previous reports, Vpr-INR199A (29) and Vpr-INV165A (6) efficiently complemented HIV-1D64N/D116N.Luc(R−) (Table 1). Vpr-INK186Q, Vpr-INK215A/K219A, and Vpr-INQ214L/Q216L behaved like Vpr-INR199A and Vpr-INV165A in that efficient complementation of HIV-1D64N/D116N.Luc(R−) was observed (Table 1). In contrast, the class I mutant Vpr-INQ62K protein failed to complement HIV-1D64N/D116N.Luc(R−). The inability of Vpr-INQ62K to complement HIV-1D64N/D116N.Luc(R-) is consistent with the results of in vitro assays that demonstrated that different catalytically inactive CCD integrase mutants failed to complement each other (61). Thus, these results define Vpr-INQ62K and HIV-1D64N/D116N.Luc(R−) as belonging to the same complementation group and Vpr-INV165A, Vpr-INR199A, Vpr-INK186Q, Vpr-INK215A/K219A, and Vpr-INQ214L/Q216L as belonging to a different group. Of note, V165A, R199A, and K186Q are CCD changes and K215A/K219A and Q214L/Q216L are C-terminal domain changes (Fig. 1A). We also note that Vpr-INC130G displayed an intermediately low level of activity (about 3% of that of Vpr-wild-type integrase) when tested in trans to HIV-1D64N/D116N.Luc(R−) (Table 1).
Each of the class II Vpr-integrase fusions was next assayed in trans to its respective virus as well as the other class II mutant viruses. Unexpectedly, none of the class II Vpr-integrase mutants complemented a class II mutant virus as well as they complemented HIV-1D64N/D116N.Luc(R−). This was especially evident with Vpr-INV165A (Table 1). Although Vpr-INK186Q, Vpr-INK215A/K219A, and Vpr-INQ214L/Q216L displayed partial activities with class II mutant viruses, we note that each of these Vpr-integrase mutants displayed similar low levels of activity when tested against their cognate mutant viruses (Table 1). The ability of a particular Vpr-integrase mutant protein to functionally complement its own otherwise defective virus has been reported previously (55). Whereas Vpr-INR199A also displayed this phenotype (Table 1), Vpr-INQ62K did not (data not shown). These results suggest that HIV-1R199A is a class II mutant virus, which is consistent with previous findings that strains carrying this change (i) failed to yield detectable levels of two-LTR circles (62) and (ii) were approximately 1,000-fold less infectious than active-site class I mutants in the multinuclear activation of galactosidase indicator assay (62), a single-round infectivity assay that is particularly sensitive to transient levels of class I mutant gene expression (50; reviewed in reference 20). In contrast to the results obtained with class II Vpr-integrase mutants, Vpr-INQ62K efficiently complemented class II mutant viruses HIV-1V165A.Luc(R−), HIV-1K186Q.Luc(R−), HIV-1K215A/K219A.Luc(R−), and HIV-1Q214L/Q216L.Luc(R−) (Table 1).
DISCUSSION
Integrase plays a key role during the early phase of retroviral replication by catalyzing the integration of the cDNA made by reverse transcription into a cell chromosome (14). However, mutations in integrase can cause a range of pleiotropic phenotypes. We previously defined two classes of replication-defective HIV-1 integrase mutant viruses (20). Whereas class I mutants are specifically blocked at the integration step, the mutations in class II viruses are pleiotropic, as they display defects at the relatively late step of particle assembly and/or at the early preintegration step of reverse transcription. Whereas class I mutants are typified by changes in the integrase active site, integrase deletion mutants as well as point mutants with mutations in the N-terminal zinc finger domain are typical class II mutant viruses (2, 20, 24, 40, 48).
The results presented here clarify that replication-defective class II integrase mutant viruses can encode catalytically active integrase. This interpretation was based on the in vitro integration activities of purified recombinant proteins (Fig. 8) and efficient complementation of class I integrase mutant HIV-1D64N/D116N.Luc(R−) with class II Vpr-integrase mutant proteins (Table 1). Of note, a subset of these proteins, including Vpr-INV165A, Vpr-INK186Q, and Vpr-INR199A, carried alterations of CCD residues (Fig. 1A). These results highlight that class II Vpr-integrase mutant proteins can functionally complement class I integrase mutant viruses even when the mutations reside in the same functional domain. This unanticipated finding confirms and extends the results of Fletcher et al. (29), Bouyac-Bertoia et al. (6), and Priet et al. (55), who reported similar phenomena with Vpr-INR199A, Vpr-INV165A, and Vpr-INL172A/K173A, respectively, in trans to active-site mutant viruses.
Although we previously reported that PICs purified from HIV-1V165A-infected cells failed to support detectable levels of in vitro integration activity, recombinants INV165A (Fig. 8), INK186Q (Fig. 8) (60), INR199S (60), INR199C (41), and INL172A/K173A (55) supported 15 to 100% of wild-type integrase activity. Additional experiments will be required to determine if other replication-defective class II integrase mutant viruses that encode functional integrase proteins and Vpr-integrase fusions (Table 1) yield active or defective PICs.
Integrase and PIC nuclear import.
HIV-1V165A was originally reported as defective for nuclear import (6). The results reported here highlight that although fewer HIV-1V165A two-LTR circles formed in cell nuclei, this was due to an overall decrease in the level of reverse transcription rather than a defect in nuclear import (Fig. 5 and 6). We had previously proposed that defective nuclear import was a phenotype common to many replication-defective integrase mutant viruses (43). In light of our current results, we now believe that this interpretation was premature and that it was likely due to residual levels of plasmid DNA that resisted DNase I digestion (supplemental Fig. S1). Our experience highlights that caution should be exercised when using sensitive RQ-PCR assays to quantify levels of retroviral reverse transcription, especially when transient transfection is used to generate virus stocks that may be defective for reverse transcription (Fig. 4).
When expressed in human cells, HIV-1 integrase localizes to cell nuclei (6, 12, 16, 17, 43, 47, 52, 54, 58). These observations, coupled with prior results that NLSP change K186Q and NLSD change Q214L/Q216L reduced the affinity of integrase for importin α in vitro, led to suggestions that integrase plays a role in PIC nuclear import (reviewed in references 53 and 56). Although the NLSP and NLSD mutant viruses were replication defective, they failed to reveal evidence of altered nuclear localization in dividing cells (9, 51, 58). Since active nuclear import of the HIV-1 PIC might play a particularly important role in nondividing cells (53, 56), reverse transcription and two-LTR-circle formation were analyzed in MDMs. Similar to the results observed in dividing T cells, VSV-G-pseudotyped HIV-1V165A, HIV-1K186Q, HIV-1C130G, HIV-1K215A/K219A, and HIV-1Q214L/Q216L formed fewer two-LTR circles than HIV-1NL4-3 in MDMs, but this was primarily due to lowered levels of reverse transcription rather than defective nuclear import (Fig. 6). These results confirm those of Tsurutani et al. (58), who reported that NLSP mutants HIV-1K186Q and HIV-1ΔKRK were defective for reverse transcription, and extend those findings to include the physiologically relevant MDM cell type. Considered alongside the results that NLSP and NLSD mutant proteins effectively localize to cell nuclei (Fig. 7) (58), we conclude that the basic amino acid residues of the NLSP and NLSD do not play a discernible role in integrase or HIV-1 nuclear import. Analyses of peptide-based nuclear import in semipermeabilized cells similarly discounted functional NLS roles for NLSP and NLSD (15).
Cys-130 and HIV-1 replication.
Our results confirmed the previous finding that HIV-1C130G was replication defective (52). Because of this, we were somewhat surprised to discover HIV-1C130A grew like wild-type HIV-1NL4-3 (Fig. 2C). These results demonstrate that Cys-130 is not essential for HIV-1 replication and highlight that HIV-1C130G was replication defective due to a negative impact of the Cys-to-Gly substitution compared to the loss of a positive role for Cys at this position. We note that Cys-130 constitutes part of the third alpha helix in the three-dimensional structure of the CCD (19). Since Gly is notorious for destabilizing alpha helices (5, 49), we speculate that the C130G change negatively impacted overall integrase structure and that this in large part contributed to the observed replication defect.
Consistent with this interpretation, Vpr-INC130G displayed a low level of activity (about 3%) when tested in trans to HIV-1D64N/D116N.Luc(R−) (Table 1), the C130G change caused partial mislocalization (Fig. 7) (52) and reduced the stability of Flag-tagged integrase in human cells (52), and the C130S change grossly affected integrase stability in insect cells (4). We note that a recent study reported replication-defective phenotypes for HIV-1 strains carrying the C130S change and that analogous mutant proteins displayed levels of in vitro DNA strand transfer activity that were significantly reduced from that of the wild type (68). In contrast to the differences observed with strains carrying different amino acid substitutions at Cys-130, strains mutated at Cys-56 or Cys-65 with Ala (Fig. 2C) or Ser (68) displayed wild-type replication profiles.
Class II integrase mutant viruses and postnuclear entry interactions.
Replication-defective class I integrase mutant viruses are typified by changes in the integrase active site, and thus these viruses encode integrase proteins that lack detectable levels of in vitro activity (20). Paradoxically, class I integrase mutants can support relatively high levels of activity under certain single-round infection conditions such as the multinuclear activation of galactosidase indicator assay, which is likely due to transient expression from unintegrated DNA templates (20, 50, 65).
A variety of integrase changes, including large deletions, cause the class II phenotype (20). In this case the mutant integrase protein will lack detectable levels of catalytic activity. Yet the results presented here extend those of Bouyac-Bertoia et al. (6) and suggest that in contrast to integrases encoded by replication-defective class I mutant viruses, class II mutants can support substantial levels of integrase activity in vitro (Fig. 8) and in the context of HIV-1 infection (Table 1). Reciprocal complementation assays employing class I and class II mutant viruses and Vpr-integrases defined the class I and class II phenotypes as genetically distinct: each class of Vpr-integrase mutant protein (i) only efficiently complemented the other class of mutant virus and (ii) either failed to complement or only partially complemented its own virus class (Table 1). Since CCD class II Vpr-integrase mutant proteins efficiently complemented CCD class I mutant HIV-1D64N/D116N.Luc(R−) and C-terminal domain class II Vpr-INK215A/K219A and Vpr-INQ214L/Q216L proteins did not efficiently complement CCD class II mutant HIV-1V165A.Luc(R−), the pattern of functional class I-class II complementation extends beyond the traditional boundaries of integrase functional domains (Fig. 1A and Table 1). Thus, class I and class II mutant viruses are defective for what would appear to be completely different reasons.
Considering that some class II mutant viruses can encode functional integrase protein, our findings pose the question of the basis of the class II mutant phenotype. Due to the pleiotropic nature of the phenotype, many different factors may contribute. However, in certain cases, for example, HIV-1V165A and HIV-1K186Q, viral late events appeared to proceed normally (Fig. 3). In these cases, we therefore propose that the virus mutation perturbs an interaction between integrase and a host cell or viral factor(s) that occurs after virus entry but before locating a chromosomal target for integration and that this interaction(s) is necessary to ensure a productive infectious route. A similar postnuclear entry, preintegration defect was previously proposed for HIV-1V165A (18).
Because the common class II phenotype is reduced levels of cDNA synthesis and because integrase and reverse transcriptase interact with each other in vitro (33, 63, 68), it seems possible that an aberrant integrase-reverse transcriptase interaction could contribute to the class II phenotype. Alternatively, a variety of host cell factors have been shown to interact with integrase, and a subset of these have been proposed to help PICs find their chromosomal targets for integration (reviewed in references 21 and 59). INL172A/K173A is defective for interaction with uracil DNA glycosidase (UNG2) (55), and INV165A is defective for interaction with LEDGF/p75 (59). Replication-defective HIV-1L172A/K173A supported wild-type levels of reverse transcription, but the levels of two-LTR circles were not reported (55). Thus, it is currently unclear if HIV-1L172A/K173A is primarily a class I or class II mutant virus. Despite this, it seems possible that the inability to interact with UNG2 and/or LEDGF/p75 might contribute to the class II integrase mutant phenotype. Determining the type(s) of protein-protein interactions that is perturbed during class II integrase mutant viral infection is an active area of research that might lead to novel ways to block HIV-1 integration without necessarily inhibiting the enzymatic activity of integrase protein.
Supplementary Material
Acknowledgments
We thank D. Gabuzda and J. Kappes for generous gifts of plasmid DNA and N. Vandegraaff for comments on the manuscript.
This work was supported by an HHMI Predoctoral Fellowship (E.D.), the Claudia Adams Barr Fund (P.A.S.), and NIH grants AI52014 (A.E.) and AI28691 (Dana-Farber Cancer Institute Center for AIDS Research).
Footnotes
Supplemental material for this article may be found at http://jvi.asm.org.
REFERENCES
- 1.Adachi, A., H. E. Gendelman, S. Koenig, T. Folks, R. Willey, A. Rabson, and M. A. Martin. 1986. Production of acquired immunodeficiency syndrome-associated retrovirus in human and nonhuman cells transfected with an infectious molecular clone. J. Virol. 59:284-291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ansari-Lari, M. A., L. A. Donehower, and R. Gibbs. 1995. Analysis of human immunodeficiency virus type 1 integrase mutants. Virology 211:332-335. [DOI] [PubMed] [Google Scholar]
- 3.Armon-Omer, A., A. Graessmann, and A. Loyter. 2004. A synthetic peptide bearing the HIV-1 integrase 161-173 amino acid residues mediates active nuclear import and binding to importin alpha: characterization of a functional nuclear localization signal. J. Mol. Biol. 336:1117-1128. [DOI] [PubMed] [Google Scholar]
- 4.Bischerour, J., H. Leh, E. Deprez, J.-C. Brochon, and J.-F. Mouscadet. 2003. Disulfide-linked integrase oligomers involving C280 residues are formed in vitro and in vivo but are not essential for human immunodeficiency virus replication. J. Virol. 77:135-141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Blaber, M., X. J. Zhang, and B. W. Matthews. 1993. Structural basis of amino acid alpha helix propensity. Science 260:1637-1640. [DOI] [PubMed] [Google Scholar]
- 6.Bouyac-Bertoia, M., J. D. Dvorin, R. A. M. Fouchier, Y. Jenkins, B. E. Meyer, L. I. Wu, M. Emerman, and M. H. Malim. 2001. HIV-1 infection requires a functional integrase NLS. Mol. Cell 7:1025-1035. [DOI] [PubMed] [Google Scholar]
- 7.Brown, H. E. V., H. Chen, and A. Engelman. 1999. Structure-based mutagenesis of the human immunodeficiency virus type 1 DNA attachment site: effects on integration and cDNA synthesis. J. Virol. 73:9011-9020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bukovsky, A., and H. Göttlinger. 1996. Lack of integrase can markedly affect human immunodeficiency virus type 1 particle production in the presence of an active viral protease. J. Virol. 70:6820-6825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cannon, P. M., E. D. Byles, S. M. Kingsman, and A. J. Kingsman. 1996. Conserved sequences in the carboxyl terminus of integrase that are essential for human immunodeficiency virus type 1 replication. J. Virol. 70:651-657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chen, H., and A. Engelman. 2001. Asymmetric processing of human immunodeficiency virus type 1 cDNA in vivo: implications for functional end coupling during the chemical steps of DNA transposition. Mol. Cell. Biol. 21:6758-6767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chen, H., S.-Q. Wei, and A. Engelman. 1999. Multiple integrase functions are required to form the native structure of the human immunodeficiency virus type I intasome. J. Biol. Chem. 274:17358-17364. [DOI] [PubMed] [Google Scholar]
- 12.Cherepanov, P., G. Maertens, P. Proost, B. Devreese, J. Van Beeumen, Y. Engelborghs, E. De Clercq, and Z. Debyser. 2003. HIV-1 integrase forms stable tetramers and associates with LEDGF/p75 protein in human cells. J. Biol. Chem. 278:372-381. [DOI] [PubMed] [Google Scholar]
- 13.Collman, R., J. W. Balliet, S. A. Gregory, H. Friedman, D. L. Kolson, N. Nathanson, and A. Srinivasan. 1992. An infectious molecular clone of an unusual macrophage-tropic and highly cytopathic strain of human immunodeficiency virus type 1. J. Virol. 66:7517-7521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Craigie, R. 2002. Retroviral DNA integration, p. 613-630. In N. L. Craig, R. Craigie, M. Gellert, and A. M. Lambowitz (ed.), Mobile DNA II. ASM Press, Washington, D.C.
- 15.Depienne, C., A. Mousnier, H. Leh, E. Le Rouzic, D. Dormont, S. Benichou, and C. Dargemont. 2001. Characterization of the nuclear import pathway for HIV-1 integrase. J. Biol. Chem. 276:18102-18107. [DOI] [PubMed] [Google Scholar]
- 16.Depienne, C., P. Roques, C. Creminon, L. Fritsch, R. Casseron, D. Dormont, C. Dargemont, and S. Benichou. 2000. Cellular distribution and karyophilic properties of matrix, integrase, and Vpr proteins from the human and simian immunodeficiency viruses. Exp. Cell Res. 260:387-395. [DOI] [PubMed] [Google Scholar]
- 17.Devroe, E., A. Engelman, and P. A. Silver. 2003. Intracellular transport of human immunodeficiency virus type 1 integrase. J. Cell Sci. 116:4401-4408. [DOI] [PubMed] [Google Scholar]
- 18.Dvorin, J. D., P. Bell, G. G. Maul, M. Yamashita, M. Emerman, and M. H. Malim. 2002. Reassessment of the roles of integrase and the central DNA flap in human immunodeficiency virus type 1 nuclear import. J. Virol. 76:12087-12096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Dyda, F., A. B. Hickman, T. M. Jenkins, A. Engelman, R. Craigie, and D. R. Davies. 1994. Crystal structure of the catalytic domain of HIV-1 integrase: similarity to other polynucleotidyl transferases. Science 266:1981-1986. [DOI] [PubMed] [Google Scholar]
- 20.Engelman, A. 1999. In vivo analysis of retroviral integrase structure and function. Adv. Virus Res. 52:411-426. [DOI] [PubMed] [Google Scholar]
- 21.Engelman, A. 2003. The roles of cellular factors in retroviral integration. Curr. Top. Microbiol. Immunol. 281:209-238. [DOI] [PubMed] [Google Scholar]
- 22.Engelman, A., F. D. Bushman, and R. Craigie. 1993. Identification of discrete functional domains of HIV-1 integrase and their organization within an active multimeric complex. EMBO J. 12:3269-3275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Engelman, A., and R. Craigie. 1992. Identification of conserved amino acid residues critical for human immunodeficiency virus type 1 integrase function in vitro. J. Virol. 66:6361-6369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Engelman, A., G. Englund, J. M. Orenstein, M. A. Martin, and R. Craigie. 1995. Multiple effects of mutations in human immunodeficiency virus type 1 integrase on viral replication. J. Virol. 69:2729-2736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Engelman, A., Y. Liu, H. Chen, M. Farzan, and F. Dyda. 1997. Structure-based mutagenesis of the catalytic domain of human immunodeficiency virus type 1 integrase. J. Virol. 71:3507-3514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Esposito, D., and R. Craigie. 1999. HIV integrase structure and function. Adv. Virus Res. 52:319-333. [DOI] [PubMed] [Google Scholar]
- 27.Esposito, D., and R. Craigie. 1998. Sequence specificity of viral end DNA binding by HIV-1 integrase reveals critical regions for protein-DNA interaction. EMBO J. 17:5832-5843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Fassati, A., D. Gorlich, I. Harrison, L. Zaytseva, and J. M. Mingot. 2003. Nuclear import of HIV-1 intracellular reverse transcription complexes is mediated by importin 7. EMBO J. 22:3675-3685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Fletcher, T. M., III, M. A. Soares, S. McPhearson, H. Hui, M. Wiskerchen, M. A. Muesing, G. M. Shaw, A. D. Leavitt, J. D. Boeke, and B. H. Hahn. 1997. Complementation of integrase function in HIV-1 virions. EMBO J. 16:5123-5138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Freed, E. O., G. Englund, and M. A. Martin. 1995. Role of the basic domain of human immunodeficiency virus type 1 matrix in macrophage infection. J. Virol. 69:3949-3954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Gallay, P., T. Hope, D. Chin, and D. Trono. 1997. HIV-1 infection of nondividing cells through the recognition of integrase by the importin/karyopherin pathway. Proc. Natl. Acad. Sci. USA 94:9825-9830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Gerton, J. L., S. Ohgi, M. Olsen, J. DeRisi, and P. O. Brown. 1998. Effects of mutations in residues near the active site of human immunodeficiency virus type 1 integrase on specific enzyme-substrate interactions. J. Virol. 72:5046-5055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hehl, E. A., P. Joshi, G. V. Kalpana, and V. R. Prasad. 2004. Interaction between human immunodeficiency virus type 1 reverse transcriptase and integrase proteins. J. Virol. 78:5056-5067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hirt, B. 1967. Selective extraction of polyoma DNA from infected mouse cell cultures. J. Mol. Biol. 26:365-369. [DOI] [PubMed] [Google Scholar]
- 35.Jenkins, T. M., A. Engelman, R. Ghirlando, and R. Craigie. 1996. A soluble active mutant of HIV-1 integrase: involvement of both the core and the C-terminal domains in multimerization. J. Biol. Chem. 271:7712-7718. [DOI] [PubMed] [Google Scholar]
- 36.Julias, J. G., A. L. Ferris, P. L. Boyer, and S. H. Hughes. 2001. Replication of phenotypically mixed human immunodeficiency virus type 1 virions containing catalytically active and catalytically inactive reverse transcriptase. J. Virol. 75:6537-6546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kim, S. Y., R. Byrn, J. Groopman, and D. Baltimore. 1989. Temporal aspects of DNA and RNA synthesis during human immunodeficiency virus infection: evidence for differential gene expression. J. Virol. 63:3708-3713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Korber, B., C. Kuiken, B. Foley, B. Hahn, F. McCutchan, J. W. Mellors, and J. Sodroski. 1998. Hum. retroviruses and AIDS 1998: a compilation and analysis of nucleic acid and amino acid sequences. Theoretical Biology and Biophysics Group T-10, Los Alamos National Laboratory, Los Alamos, N.Mex.
- 39.Kumar, R., N. Vandegraaff, L. Mundy, C. J. Burrell, and P. Li. 2002. Evaluation of PCR-based methods for the quantitation of integrated HIV-1 DNA. J. Virol. Methods 105:233-246. [DOI] [PubMed] [Google Scholar]
- 40.Leavitt, A. D., G. Robles, N. Alesandro, and H. E. Varmus. 1996. Hum. immunodeficiency virus type 1 integrase mutants retain in vitro integrase activity yet fail to integrate viral DNA efficiently during infection. J. Virol. 70:721-728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Leavitt, A. D., L. Shiue, and H. E. Varmus. 1993. Site-directed mutagenesis of HIV-1 integrase demonstrates differential effects on integrase functions in vitro. J. Biol. Chem. 268:2113-2119. [PubMed] [Google Scholar]
- 42.Li, L., J. M. Olvera, K. E. Yoder, R. S. Mitchell, S. L. Butler, M. Lieber, S. L. Martin, and F. D. Bushman. 2001. Role of the non-homologous DNA end joining pathway in the early steps of retroviral infection. EMBO J. 20:3272-3281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Limón, A., E. Devroe, R. Lu, H. Z. Ghory, P. A. Silver, and A. Engelman. 2002. Nuclear localization of human immunodeficiency virus type 1 preintegration complexes (PICs): V165A and R166A are pleiotropic integrase mutants primarily defective for integration, not PIC nuclear import. J. Virol. 76:10598-10607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Limón, A., N. Nakajima, R. Lu, H. Z. Ghory, and A. Engelman. 2002. Wild-type levels of nuclear localization and human immunodeficiency virus type 1 replication in the absence of the central DNA flap. J. Virol. 76:12078-12086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Lu, R., N. Nakajima, W. Hofmann, M. Benkirane, K.-T. Jeang, J. Sodroski, and A. Engelman. 2004. Simian virus 40-based replication of catalytically inactive human immunodeficiency virus type 1 integrase mutants in nonpermissive T cells and monocyte-derived macrophages. J. Virol. 78:658-668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Maertens, G., P. Cherepanov, Z. Debyser, Y. Engelborghs, and A. Engelman. 2004. Identification and characterization of a functional nuclear localization signal in the HIV-1 integrase interactor LEDGF/p75. J. Biol. Chem. 279:33421-33429. [DOI] [PubMed] [Google Scholar]
- 47.Maertens, G., P. Cherepanov, W. Pluymers, K. Busschots, E. De Clercq, Z. Debyser, and Y. Engelborghs. 2003. LEDGF/p75 is essential for nuclear and chromosomal targeting of HIV-1 integrase in human cells. J. Biol. Chem. 278:33528-33539. [DOI] [PubMed] [Google Scholar]
- 48.Masuda, T., V. Planelles, P. Krogstad, and I. S. Chen. 1995. Genetic analysis of human immunodeficiency virus type 1 integrase and the U3 att site: unusual phenotype of mutants in the zinc finger-like domain. J. Virol. 69:6687-6696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Myers, J. K., C. N. Pace, and J. M. Scholtz. 1997. A direct comparison of helix propensity in proteins and peptides. Proc. Natl. Acad. Sci. USA 94:2833-2837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Nakajima, N., R. Lu, and A. Engelman. 2001. Hum. immunodeficiency virus type 1 replication in the absence of integrase-mediated DNA recombination: definition of permissive and nonpermissive T-cell lines. J. Virol. 75:7944-7955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Petit, C., O. Schwartz, and F. Mammano. 2000. The karyophilic properties of human immunodeficiency virus type 1 integrase are not required for nuclear import of proviral DNA. J. Virol. 74:7119-7126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Petit, C., O. Schwartz, and F. Mammano. 1999. Oligomerization within virions and subcellular localization of human immunodeficiency virus type 1 integrase. J. Virol. 73:5079-5088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Piller, S. C., L. Caly, and D. A. Jans. 2003. Nuclear import of the pre-integration complex (PIC): the Achilles heel of HIV? Curr. Drug Targets 4:409-429. [DOI] [PubMed] [Google Scholar]
- 54.Pluymers, W., P. Cherepanov, D. Schols, E. De Clercq, and Z. Debyser. 1999. Nuclear localization of human immunodeficiency virus type 1 integrase expressed as a fusion protein with green fluorescent protein. Virology 258:327-332. [DOI] [PubMed] [Google Scholar]
- 55.Priet, S., J.-M. Navarro, G. Querat, and J. Sire. 2003. Reversion of the lethal phenotype of an HIV-1 integrase mutant virus by overexpression of the same integrase mutant protein. J. Biol. Chem. 278:20724-20730. [DOI] [PubMed] [Google Scholar]
- 56.Stevenson, M. 2002. Molecular biology of lentivirus-mediated gene transfer. Curr. Top. Microbiol. Immunol. 261:1-30. [DOI] [PubMed] [Google Scholar]
- 57.Telesnitsky, A., and S. P. Goff. 1997. Reverse transcriptase and the generation of retroviral DNA, p. 121-160. In J. M. Coffin, S. H. Hughes, and H. E. Varmus (ed.), Retroviruses. Cold Spring Harbor Laboratory Press, Plainview, N.Y. [PubMed]
- 58.Tsurutani, N., M. Kubo, Y. Maeda, T. Ohashi, N. Yamamoto, M. Kannagi, and T. Masuda. 2000. Identification of critical amino acid residues in human immunodeficiency virus type 1 IN required for efficient proviral DNA formation at steps prior to integration in dividing and nondividing cells. J. Virol. 74:4795-4806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Turlure, F., E. Devroe, P. A. Silver, and A. Engelman. 2004. Hum. cell proteins and human immunodeficiency virus DNA integration. Front. Biosci. 9:3187-3208. [DOI] [PubMed] [Google Scholar]
- 60.van Gent, D. C., A. A. M. O. Groeneger, and R. H. A. Plasterk. 1992. Mutational analysis of the integrase protein of human immunodeficiency virus type 2. Proc. Natl. Acad. Sci. USA 89:9598-9602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.van Gent, D. C., C. Vink, A. A. M. O. Groeneger, and R. H. A. Plasterk. 1993. Complementation between HIV integrase proteins mutated in different domains. EMBO J. 12:3261-3267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Wiskerchen, M., and M. A. Muesing. 1995. Human immunodeficiency virus type 1 integrase: effects of mutations on viral ability to integrate, direct viral gene expression from unintegrated viral DNA templates, and sustain viral propagation in primary cells. J. Virol. 69:376-386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Wu, X., H. Liu, H. Xiao, J. A. Conway, E. Hehl, G. V. Kalpana, V. Prasad, and J. C. Kappes. 1999. Hum. immunodeficiency virus type 1 integrase protein promotes reverse transcription through specific interactions with the nucleoprotein reverse transcription complex. J. Virol. 73:2126-2135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Wu, X., H. Liu, H. Xiao, J. A. Conway, E. Hunter, and J. C. Kappes. 1997. Functional RT and IN incorporated into HIV-1 particles independently of the Gag/Pol precursor protein. EMBO J. 16:5113-5122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Wu, Y., and J. W. Marsh. 2003. Early transcription from nonintegrated DNA in human immunodeficiency virus infection. J. Virol. 77:10376-10382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Yee, J., A. Miyanohara, P. LaPorte, K. Bouic, J. C. Burns, and T. Friedmann. 1994. A general method for the generation of high-titer, pantropic retroviral vectors: highly efficient infection of primary hepatocytes. Proc. Natl. Acad. Sci. USA 91:9564-9568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Zack, J. A., S. J. Arrigo, S. R. Weitsman, A. S. Go, A. Haislip, and I. S. Chen. 1990. HIV-1 entry into quiescent primary lymphocytes: molecular analysis reveals a labile, latent viral structure. Cell 61:213-222. [DOI] [PubMed] [Google Scholar]
- 68.Zhu, K., C. Dobard, and S. A. Chow. 2004. Requirement for integrase during reverse transcription of human immunodeficiency virus type 1 and the effect of cysteine mutations of integrase on its interactions with reverse transcriptase. J. Virol. 78:5045-5055. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.