Abstract
Hepatitis C virus (HCV) has evolved complex strategies to evade host immune responses and establish chronic infection. The only treatment available for HCV infections, alpha interferon (IFN-α), is effective in a limited percentage of patients. The mechanisms by which IFN-α interferes with the HCV life cycle and the reasons for limited effectiveness of IFN-α therapy have not yet been fully elucidated. Using a cell-based HCV replication system and specific kinase inhibitors, we examined the role played by various signaling pathways in the IFN-α-mediated HCV clearance. We reported that conventional protein kinase C (cPKC) activity is important for the effectiveness of IFN-α treatment. In cells treated with a cPKC-specific inhibitor, IFN-α failed to induce an efficient HCV RNA degradation. The lack of cPKC activity leads to a broad reduction of IFN-α-stimulated gene expression due to a significant impairment of STAT1 and STAT3 tyrosine phosphorylation. Thus, modulation of cPKC function by either host or viral factors could influence the positive outcome of IFN-α-mediated antiviral therapies.
The interferon (IFN) system is the first line of defense against viral infection in mammals (15). Transcriptional activation of type I IFN (IFN-α and IFN-β) genes is mainly triggered by viral double-stranded RNA present in infected cells (39, 40). Most of the viruses have evolved molecular mechanisms to evade the IFN-mediated cellular response (22). One of the most significant examples is the hepatitis C virus (HCV) which, in a high percentage of cases (70 to 80% of infected individuals), escapes the host defenses and establishes a chronic infection (12, 23). Pathological consequences of HCV infection differ significantly from individuals, varying from asymptomatic state to liver fibrosis, cirrhosis, and hepatocellular carcinoma (12, 23).
Alpha interferon (IFN-α), alone or in combination with ribavirin, represents the only treatment available for HCV infections, with a moderate percentage of virus eradication (30 to 40% of the patients) (32). The mechanisms by which IFN-α interferes with HCV replication have not yet been elucidated, nor are the reasons for limited effectiveness of IFN-α therapy known (33). It has been hypothesized that a difference in HCV genomic sequence may affect the structure and function of the viral RNA and proteins, which in turn can alter the host's response. For example, specific nucleotide sequence mutations within a region known as the interferon sensitivity-determining region of the NS5A protein have been associated with clinical resistance to IFN-α treatment (10, 54).
Type I IFNs induce the transcription of a large number of genes (IFN-stimulated genes [ISGs]) (14, 42). Specific antiviral activities have been described for some of these genes, including the double-stranded RNA-activated protein kinase (PKR), which inhibits protein synthesis by phosphorylating the α subunit of the translation initiation factor eIF-2 and disrupting the critical delivery of methionyl-tRNA to the 40S ribosome (53). Similarly, ISG56 has been identified as a suppressor of translation by binding and inhibiting the translation initiation factor eIF3 (20). Another IFN-α-inducible gene with antiviral activity is the 2′-5′ oligoadenylate synthase (2′-5′OAS). 2′-5′OAS enzymatic products allow RNase L activation which, besides RNA degradation, can lead to translational suppression by the cleavage of the 28S rRNA (41). Finally, the GTPase protein MxA has a well-described antiviral activity, probably due to a direct interference on either viral nucleocapsid transport or viral RNA synthesis (19).
IFN-α and IFN-β bind to a common receptor expressed on the surface of target cells (14, 42). Receptor engagement leads to the activation of Jak1 and Tyk2 protein kinases, which in turn catalyze phosphorylation events leading to the heterodimerization and nuclear translocation of the signal transducer and activator of transcription (STAT) proteins STAT1 and STAT2 (42). In the nucleus, STAT1/2 heterocomplex associates with p48/IRF-9 to form the ISG factor 3 (ISGF3) which binds to the IFN-α-stimulated response element present in the ISG promoter (14). Sequence motifs within the IFN-α-stimulated response element also serve as target sites for interferon regulatory factors, whose actions contribute to define the overall spectrum and duration of ISG expression (43). STAT3 has also been shown to respond to type I IFN receptor signaling and has been proposed to link IFNs with cell growth regulation and the phosphatidylinositol 3-kinase (PI3K) pathway (34, 55).
Although the Jak-STAT pathway is essential for triggering the IFN-α response, a variety of other signaling cascades are stimulated following IFN-α/β receptor activation (6, 36), such as IRS/PI3K (34, 48), c-Cbl/Crk/Rap1 (1, 45), Rac1/p38 mitogen-activated protein kinase (MAPK) (13, 46), p42/p44 MAPK (7), protein kinase C (PKC) (35, 47), and mTOR (25). While various evidences of the contribution of these pathways to the IFN-α response have been reported, more effort is required to better understand their functions in IFN-α-mediated antiviral activity. In particular, uncovering the role played by different pathways in the IFN-α response could be relevant to elucidate cellular functions whose activities influence the outcome of the anti-HCV therapy.
An important improvement for HCV studies has been the recent establishment of a cell culture system able to sustain viral replication (2, 26). In this system, a subgenomic HCV RNA (replicon) in which structural proteins have been replaced by a selection marker has allowed the isolation of adapted HCV genomes able to replicate in HuH7 hepatoma cells (3, 26). Importantly, IFN-α treatment leads to the clearance of the HCV replicon, making this system suitable for dissecting the IFN-α-regulated antiviral response (3, 12, 18).
The aim of this work was to elucidate the role of different signaling pathways during the IFN-α-mediated HCV clearance. We report that conventional PKC (cPKC) activity is involved in the HCV RNA clearance. Furthermore, our results suggest that inhibition of cPKC activity impairs STAT activation and IFN-α-induced gene expression.
MATERIALS AND METHODS
Cells and reagents.
HuH7, Hep3B, and HepG2 cells were purchased from the American Type Culture Collection and grown in Dulbecco's modified Eagle's medium (Sigma-Aldrich) supplemented with 10% fetal bovine serum (Sigma-Aldrich), 100 U of penicillin/ml, 100 μg of streptomycin/ml, and 2 mM l-glutamine in the presence of 0.5 mg of G418 (Invitrogen)/ml. Rep60 is a HuH7 cell line harboring an HCV replicon obtained by the transfection of HuH7 cells with the HCV replicon RNA synthesized from pHCVneo17.wt (28, 29), a template for T7 transcription of RNA identical in sequence to the replicon I377neo/NS3-3′/wt (26). In vitro transcription, transfection, and selection were carried out as previously described (28, 29). HCV replicon present in Rep60 cells carries the previously described adaptative mutation A2199T (29), as determined by sequencing. Recombinant human IFN-α was purchased from PeproTech. All kinase inhibitors added to the culture medium 60 min before IFN-α stimulation and the concentrations used are as follows: the pan-PKC inhibitor GF109203X at 5 μM, the PKCδ-specific inhibitor rottlerin at 1 μM, the cPKC inhibitor Gö6976 at 200 nM, the p38 MAPK inhibitor SB202190 at 10 μM, the PI3K inhibitor LY294002 at 10 μM, the MEK1/2 inhibitor U0126 at 10 μM, and the TOR kinase inhibitor rapamycin at 100 nM. Stock solutions were prepared in dimethyl sulfoxide at a 1,000× concentration with respect to the working concentrations indicated above; dimethyl sulfoxide alone was added to control cells. All inhibitors were purchased from Calbiochem.
RNA isolation, RT-PCR, and Northern blot analysis.
Total RNA was isolated from cells by using TRIzol reagent (Invitrogen) according to the manufacturer's instructions. cDNA was synthesized by using a reverse transcription (RT) system (Promega) with random primers (random hexamers), and PCR amplification was performed with Taq DNA polymerase (Promega) using the following primers: HCV NS4A (sense), 5′-GCA CCT GGG TGC TGG TAG GCG GAG TCC-3′; HCV NS4A (antisense), 5′-CAC TCT TCC ATC TCA TCG AAC TCC CG-3′; β-actin (sense), 5′-ATG GAT GAC GAT ATC GCT GCG-3′; β-actin (antisense), 5′-ATC TTC ATG AGG TAG TCT GTC AGG-3′. The PCR conditions used were as follows: 28 cycles of 94°C for 30s, 65°C for 30s, and 72°C for 1 min for HCV primers and for 20 cycles of 94°C for 30s, 65°C for 30s, and 72°C for 1 min for β-actin.
For Northern blot analysis, 10 μg of total RNA was separated by electrophoresis in a 0.8% denaturing agarose gel, transferred to nitrocellulose membranes (PROTRAN; Schleicher & Schuell), and fixed by UV cross-linking (Stratagene). Hybridization was carried out with [32P]dCTP-labeled cDNA probes in a solution containing 50% formamide, 5× SSC (1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate), 5 mM phosphate buffer (pH 6.8), 5× Denhardt solution (0.1% Ficoll 400, 0.1% polyvinylpyrrolidone, 0.1% bovine serum albumin), 0.5% sodium dodecyl sulfate (SDS), and salmon sperm DNA (100 μg/ml) for 18 h at 42°C. Autoradiography and quantification were performed with a Typhoon 9200 PhosphorImager (Amersham Bioscience). The cDNA probes were generated by RT-PCR using the following primers: HCV NS4B (sense), 5′-CTC ACA CCT CCC TTA CAT CGA ACA G-3′; HCV NS4B (antisense), 5′-CAT GGC GTG GAG CAG TCC TCG TTG-3′; β-actin (sense), 5′-ATG GAT GAC GAT ATC GCT GCG-3′; β-actin (antisense), 5′-ATC TTC ATG AGG TAG TCT GTC AGG-3′; 2′-5′OAS (sense), 5′-AAA GTG CCG GTA AAA GTC AT-3′; 2′-5′OAS (antisense), 5′-CTG TAG TGC AAG GGT TCT CA-3′; MxA (sense), 5′-CTG TGG CCA TAC TGC CAG GA-3′; MxA (antisense), 5′-ACT CCT GAC AGT GCC TCC AA-3′; ISG54 (sense), 5′-AGA AAT CAA GGG AGA AAG AA-3′; ISG54 (antisense), 5′-AAG GTG ACT AAG CAA ATG GT-3′; ISG60 (sense), 5′-GAT CTC GCT GAG TTC CTG GAG AC-3′; ISG60 (antisense), 5′-AGC TCT CTG GGA CTG GAG CTG AC-3′; PKR (sense), 5′-GGT ACA GGT TCT ACT AAA CAG G-3′; PKR (antisense), 5′-GAA AAC TTG GCC AAA TCC ACC-3′; ISG56 (sense), 5′-TAG CCA ACA TGT CCT CAC AGA C-3′; ISG56 (antisense), 5′-TCT TCT ACC ACT GGT TTC ATG C-3′; ISG15 (sense), 5′-CTC TCG AGC CAT GGG CTG GGA CCT GAC GG-3′; ISG15 (antisense), 5′-GCT CTA GAT TAG CTC CGC CCG CCA GGC TCT G-3′; ADAR1 (sense), 5′-TGA CTT CCG AGA TGC ACG-3′; ADAR1 (antisense), 5′-AAT GCC TCG CGG GCG CAA TGA ATC-3′. 32P-labeled DNA probes were prepared by using a random primer DNA-labeling system (Invitrogen).
Nuclear extracts and electrophoretic mobility shift assays (EMSA).
Cells were scraped into 1.5 ml of cold phosphate-buffered saline (PBS), pelleted, and resuspended in 400 μl of cold buffer A (10 mM HEPES-KOH [pH 7.9 at 4°C], 1.5 mM MgCl2, 10 mM KCl, 0.5 mM dithiothreitol, 0.2 mM phenylmethylsulfonyl fluoride). Cells were allowed to swell on ice for 10 min and were then vortexed for 10 s. Samples were centrifuged for 10 s, the supernatant fraction was discarded, and the pellet was resuspended in 100 μl of cold buffer C (20 mM HEPES-KOH [pH 7.9], 25% glycerol, 420 mM NaCl, 1.5 mM MgCl2, 0.2 mM EDTA, 0.5 mM dithiothreitol, 0.2 mM phenylmethylsulfonyl fluoride) and incubated on ice for 20 min for high-salt extraction. Cellular debris was removed by centrifugation for 2 min at 4°C, and the supernatant fraction was stored at −70°C. Protein concentration was determined by the Bradford assay.
EMSAs were carried out by incubating 5 μg of each extract in a final volume of 20 μl of 20 mM HEPES (pH 7.9) and 50 mM NaCl containing 3 μg of poly(dI)-poly(dC) for 10 min on ice. 32P-labeled double-stranded oligonucleotides (4 × 104 cpm) were added and incubated for 15 min at room temperature. DNA-protein complexes were separated by electrophoresis on a 4.5% polyacrylamide gel in 0.25× TBE buffer (25 mM Tris, 25 mM boric acid, 0.6 mM EDTA) and visualized by autoradiography. Double-stranded oligonucleotides were labeled by fill-in with Klenow polymerase. The sequence of the upper strands of haSIE (corresponding to a sis-inducible element on the c-fos promoter) is 5′-GATCCATTTCCCGTAAATC-3′.
Antibodies.
A monoclonal antibody against HCV NS3 (C-33) was obtained from Biogenesis. Polyclonal antibodies against neomycin phosphotransferase II (NPTII) and phosphorylated STAT1 (phospho-STAT1) (Ser727) were obtained from Upstate Biotechnology Inc. Monoclonal antibodies against Jak1 and Tyk2 were obtained from BD Transduction Laboratories. Polyclonal antibodies against phospho-STAT1(Tyr701), STAT1, phospho-STAT3(Ser727), phospho-STAT3(Tyr705), STAT3, phospho-Jak1(Tyr1022/Tyr1023), phospho-Tyk2(Tyr1054/Tyr1055), phospho-SHP2(Tyr580), and SHP2 were obtained from Cell Signaling Technology. Anti-TcPTP monoclonal antibody was obtained from Oncogene Research Products. Anti-SOCS-1, anti-SOCS-3, and anti-PIAS1/3 were obtained from Santa Cruz Biotechnology. Anti-β-actin polyclonal antibody was obtained from Sigma.
Immunoblot analysis.
Cells were grown on 35-mm plates, washed once with PBS, and lysed directly in hot Laemmli sample buffer (50 mM Tris HCl [pH 6.8], 2% SDS, 0.1% bromophenol blue, 10% glycerol, 100 mM β-mercaptoethanol). Equal amounts of cell lysates were resolved by SDS-polyacrylamide gel electrophoresis and blotted onto nitrocellulose membranes (PROTRAN; Schleicher & Schuell) by using a semidry blotter (Bio-Rad). Membranes were blocked by using 5% nonfat milk in PBS plus 0.05% Tween 20 for 1 h at room temperature, followed by incubation with primary antibodies overnight at 4°C. Specific binding of antibodies was detected with horseradish peroxidase-conjugated secondary antibodies (anti-mouse and anti-rabbit diluted 1:5,000; Jackson Laboratory) and visualized by an enhanced chemiluminescence reaction (Amersham Bioscience).
Immunofluorescence.
Cells were grown on 35-mm plates, fixed with 4% paraformaldehyde in PBS, and permeabilized with 0.2% Triton in PBS. After blocking for 1 h in 5% bovine serum albumin in PBS, the plates were incubated with the primary antibodies for 1 h at 4°C (rabbit anti-STAT1, 1:100; rabbit anti-STAT3, 1:300; mouse anti-TcPTP, 1:300). The plates were then washed with PBS and incubated with a secondary antibody (Cy3-conjugated anti-mouse serum and a Cy3-conjugated anti-rabbit serum at 1:300; Jackson Laboratory). Immunostained samples were counterstained with 4′,6′-diamidino-2-phenylindole dihydrochloride (DAPI; Boehringer). Image collection was done by using a Nikon Eclipse TE 200 microscope with a DXM 1200F digital camera.
RESULTS
Inhibition of cPKC activity impairs IFN-α-mediated subgenomic HCV RNA clearance.
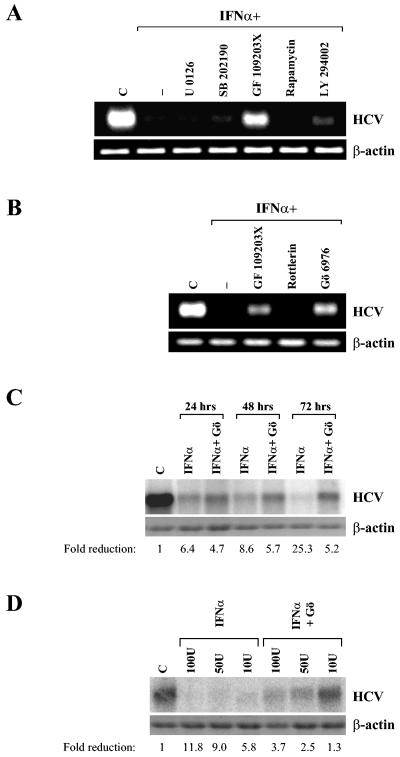
The biological activities of IFNs are known to be mediated by distinct signaling pathways (6). In order to determine which signaling pathways are required for IFN-α-mediated HCV clearance, HCV replicon-carrying cells (Rep60) were incubated for 96 h with IFN-α in the presence of different specific kinase inhibitors: U0126, an inhibitor of MEK1/2 (11); SB202190, an inhibitor of p38 (24); rapamycin, an inhibitor of mTOR (5); GF109203X, an inhibitor of PKC (44); and LY294002, an inhibitor of PI3K (51). RNA was extracted from treated and control cells, and the levels of HCV RNA were determined by RT-PCR analysis. Notably, the general PKC inhibitor GF109203X impaired the IFN-α-mediated HCV clearance as shown by the persistence of a large amount of viral RNA (Fig. 1A). A low but detectable amount of HCV RNA was also present in cells pretreated with a PI3K inhibitor. On the other hand, no effects on IFN-α-mediated HCV RNA clearance were observed in cells pretreated with p38, MEK, or mTOR inhibitors. HCV replication was not directly modulated by the treatment with any kinase inhibitor tested (data not shown), indicating that inhibition of PKC does not influence HCV replication per se but specifically affects the IFN-α-mediated HCV clearance.
FIG. 1.
Effect of different kinase inhibitors on IFN-α-mediated subgenomic HCV RNA clearance. (A) Analysis of IFN-α-mediated HCV replicon clearance in the presence of different kinase inhibitors. Rep60 cells were treated with IFN-α together with the indicated compounds for 96 h. RNA was extracted, and IFN-α-induced HCV clearance was analyzed by measuring viral RNA levels by RT-PCR using specific oligonucleotides (see Materials and Methods). As an RNA loading control, levels of β-actin mRNA were analyzed. (B) Analysis of IFN-α-mediated HCV replicon clearance in the presence of specific PKC inhibitors (see Materials and Methods). Rep60 cells were treated with IFN-α together with the indicated compounds for 96 h, and RNA was analyzed as described in A. (C) Northern blot of time course analysis for IFN-α-mediated HCV replicon clearance in the presence or absence of Gö6976 cPKC inhibitor (Gö). Rep60 cells were treated with 1,000 U of IFN-α/ml for the indicated times and analyzed by Northern blot, and HCV RNA levels were measured by phosphoimage analysis. Numbers indicate the ratio of untreated-to-treated values after β-actin normalization. (D) Northern blot analysis for dose-dependent IFN-α-mediated HCV replicon clearance. Rep60 cells were treated with the indicated doses of IFN-α and analyzed at 72 h as described in panel C. C, control (untreated cells).
PKC enzymes represent a large family of proteins that can be divided into three groups based on their requirements for enzymatic activation: cPKC (cPKCα, -β, and -γ), which requires Ca2+ and diacylglycerol (DAG); novel PKC (nPKCδ, -ɛ, -η, -θ, and -μ), which is dependent on DAG only; and atypical PKC (aPKCζ and -ι/λ), which is insensitive to Ca2+ and DAG (30, 31). We investigated which PKC isoform is required for IFN-α-mediated HCV RNA clearance by using two PKC inhibitors, Gö6976 and rottlerin, specific for cPKC isoforms (27) and PKCδ (16), respectively. As shown in Fig. 1B, the cPKC inhibitor led to an impairment of IFN-α-mediated HCV clearance similarly to that induced by the general PKC inhibitor GF109203X, while the PKCδ inhibitor had no significant effects.
Northern blot analysis was carried out to quantify the relative extent of HCV replicon clearance; while IFN-α leads to a 25-fold reduction of HCV RNA within 72 h, the impairment of cPKC activity limited the reduction to 5-fold (Fig. 1C). Similar results were obtained with IFN-α concentrations spanning from 10 to 100 U/ml (Fig. 1D). Taken together, these results indicate that cPKCs play a role in the IFN-α-induced HCV RNA degradation.
The IFN-α-mediated inhibition of HCV RNA translation is independent of PKC activity.
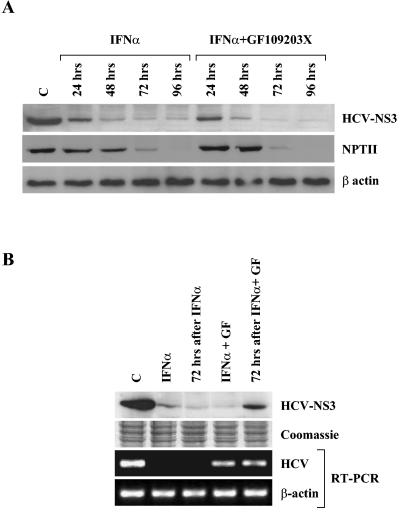
Viral clearance by IFN-α is a complex multistep process, achieved by interfering with the viral life cycle at the level of transcription, replication, or translation (17). It has been previously described, using the HCV replicon system, that IFN-α could have a direct influence on HCV internal ribosome entry site (IRES)-dependent translation (37, 52). We asked, therefore, whether PKC activity is also required for this phenomenon. Rep60 cells were treated for 96 h with IFN-α in the presence or absence of the PKC inhibitor GF109203X, and protein extracts were prepared every 24 h. Unexpectedly, Western blot analysis with an anti-NS3 antibody showed that IFN-α caused HCV protein level reduction even in cells in which PKC activity is inhibited and HCV RNA degradation is impaired (Fig. 2A). We also analyzed NPTII protein levels, since in the HCV replicon, viral protein translation is driven by a heterologous IRES (encephalomyocarditis virus IRES), and HCV IRES controls the synthesis of the selection marker NPTII. Similar to that observed for NS3, NPTII protein amount decreased in IFN-α-stimulated cells irrespective of the presence of GF109203X (Fig. 2A); the observed different kinetics probably reflect the diverse stability of the two proteins. These data indicate that IFN-α inhibits HCV translation independently of PKC activity. Notably, the withdrawal of IFN-α restores viral protein synthesis in PKC inhibitor-treated cells, indicating that in the absence of viral RNA degradation, IFN-α does not cause an irreversible block of viral translation (Fig. 2B).
FIG. 2.
Effect of the PKC inhibitor GF109203X on IFN-α-mediated inhibition of HCV protein synthesis. (A) Western blot for HCV replicon protein levels in IFN-α-stimulated cells in relation to PKC activity. Rep60 cells were treated with IFN-α in the presence or absence of the PKC inhibitor GF109203X. Samples were collected at the indicated times and analyzed for HCV NS3 or NPTII protein levels. Protein loading was controlled by β-actin levels. (B) Analysis of HCV RNA and protein levels after IFN-α withdrawal in relation to PKC activity. Rep60 cells were treated with IFN-α in the presence or absence of the PKC inhibitor GF109203X (GF) for 72 h and further incubated untreated for another 72 h. Samples were collected at the indicated times and analyzed for HCV NS3 protein levels by Western blot and for HCV RNA levels by RT-PCR. Protein and RNA loading were controlled by Coomassie staining and β-actin RT-PCR, respectively. C, control (untreated cells).
Inhibition of cPKC activity impairs IFN-α-inducible gene expression.
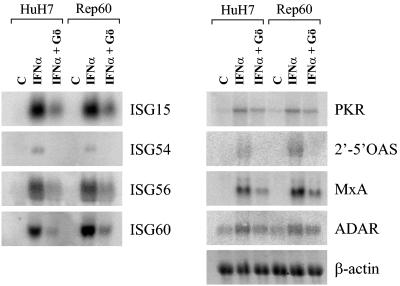
Since IFN-α is known to modulate the expression of a variety of cellular genes (42), some of which are directly involved in its antiviral activity (14), we tested whether IFN-α-inducible transcription requires cPKC activity. Northern blot analysis of the well-known IFN-α target genes ISG15, ISG54, ISG56, ISG60, 2′-5′OAS, MxA, ADAR1, and PKR showed that cPKC inhibition significantly impairs their induction (Fig. 3). Similar results were obtained in both replicon-carrying Rep60 and parental HuH7 cells, indicating that cPKC activity is necessary for full IFN-α-stimulated gene expression in an HCV-independent manner. Furthermore, no significant alterations in IFN-α-inducible gene expression profiles were observed in the presence of PI3K or p38 kinase inhibitors, in agreement with the lack of effects on IFN-α-mediated HCV clearance (data not shown).
FIG. 3.
Effect of the cPKC inhibitor Gö6976 on IFN-α-inducible gene expression. Northern blot analysis for IFN-α target genes in HuH7 and Rep60 cells stimulated with IFN-α in the presence and absence of Gö6976 (Gö). Samples were collected 6 h after IFN-α stimulation, and RNA was extracted and analyzed for the expression of ISG15, ISG54, ISG56, ISG60, PKR, 2′-5′OAS, MxA, and ADAR. RNA loading was controlled by β-actin expression. C, control (untreated cells).
Inhibition of cPKC activity affects IFN-α-induced STAT1 and STAT3 tyrosine phosphorylation.
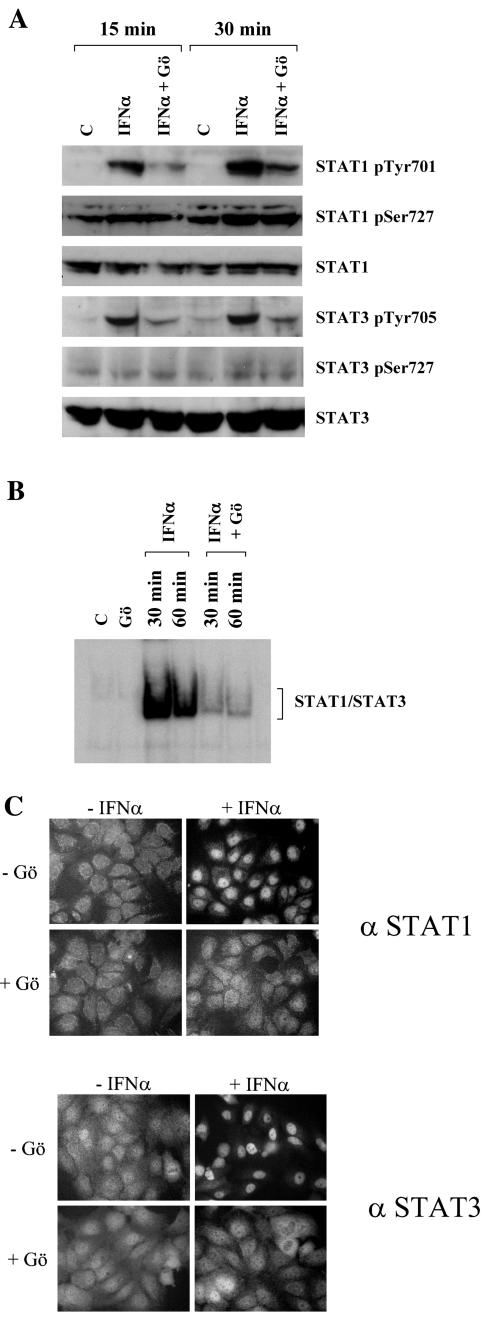
The broad reduction of IFN-α-inducible gene expression by a cPKC inhibitor prompted us to check for changes in early events of IFN-α signaling, such as Jak and STAT phosphorylation. Rep60 cells were stimulated with IFN-α for 15 or 30 min in either the presence or absence of the cPKC inhibitor Gö6976, and protein extracts were prepared for Western blot analysis. Notably, tyrosine phosphorylation of both STAT1 and STAT3 was significantly decreased by the inhibition of cPKC activity, while serine phosphorylation was not influenced (Fig. 4A). As expected, the reduction of STAT1 and STAT3 tyrosine phosphorylation results in the impairment of both DNA binding activity (Fig. 4B) and nuclear translocation (Fig. 4C).
FIG. 4.
Effect of the cPKC inhibitor Gö6976 on IFN-α-inducible STAT activity. (A) Western blot analysis for STAT1 and STAT3 phosphorylation with specific anti-phospho-STAT antibodies [phospho-STAT1(Tyr701) (STAT1 pTyr701), phospho-STAT1(Ser727) (STAT1 pSer727), phospho-STAT3(Tyr705) (STAT3 pTyr705), and phospho-STAT3(Ser727) (STAT3 pSer727)]. Rep60 cells were stimulated for 15 or 30 min with IFN-α in the presence or absence of cPKC inhibitor Gö6976 (Gö). As a protein loading control, blots were stripped and reprobed with the respective anti-total protein antibodies. (B) EMSA analysis of STAT1 and STAT3 DNA binding activity with an oligonucleotide carrying binding sites for STAT1 and STAT3 (corresponding to a sis-inducible element on the c-fos promoter) as a probe. Rep60 cells were stimulated for 30 or 60 min with IFN-α in the presence or absence of cPKC inhibitor Gö6976 (Gö), followed by preparation of nuclear extracts and EMSA analysis as described in Materials and Methods. C, control (untreated cells). (C) Analysis of STAT1 and STAT3 cellular localization by immunofluorescence staining with anti-STAT1 (upper panels) and anti-STAT3 (lower panels) antibodies. Rep60 cells were stimulated for 30 min with IFN-α in the presence or absence of cPKC inhibitor Gö6976 (Gö).
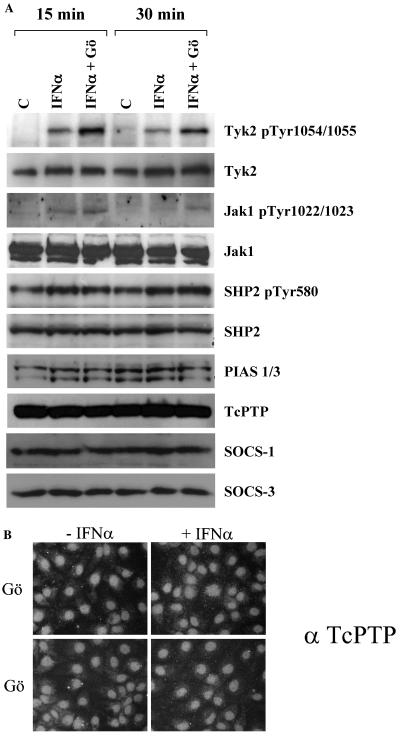
In light of STAT tyrosine phosphorylation reduction, we analyzed the activation state of the upstream kinases. Surprisingly, Tyk2 and Jak1 tyrosine phosphorylation was not impaired by the cPKC inhibitor but rather moderately increased (Fig. 5A). Furthermore, Gö6976 does not induce evident alterations of other Jak/STAT pathway regulatory factors. In fact, the steady-state protein levels of PIAS1/3, TcPTP, SHP2, SOCS-1, and SOCS-3 (Fig. 5A); the phosphorylation of SHP2 (Fig. 5A); and the cellular localization of TcPTP were unaffected (Fig. 5B).
FIG. 5.
Effect of the cPKC inhibitor Gö6976 on the IFN-α-inducible Jak/STAT signaling pathway. (A) Western blot analysis for phospho-Tyk2(Tyr1054/Tyr1055) (Tyk2 pTyr 1054/1055), Tyk2, phospho-Jak1(Tyr1022/Tyr1023) (Jak1 pTyr1022/1023), Jak1, phospho-SHP2(Tyr580) (SHP2 pTyr580), SHP2, PIAS1/3, TcPTP, SOCS-1, and SOCS-3 expression in Rep60 cells stimulated for 15 or 30 min with IFN-α in the presence or absence of cPKC inhibitor Gö6976 (Gö). C, control (untreated cells). (B) Analysis of TcPTP cellular localization by immunofluorescence. Rep60 cells were stimulated for 30 min with IFN-α in the presence or absence of cPKC inhibitor Gö6976.
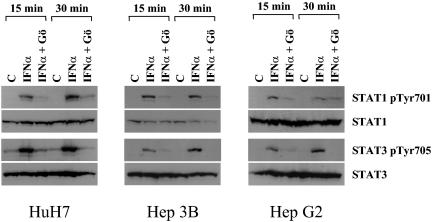
Notably, the impairment of cPKC activity also results in the reduction of STAT1 and STAT3 tyrosine phosphorylation in the parental HuH7 cells, which do not carry the HCV replicon, and in other hepatoma cell lines (HepG2 and Hep3B) (Fig. 6), indicating that cPKC influences IFN-α signaling independently of HCV.
FIG. 6.
Effect of the cPKC inhibitor Gö6976 on IFN-α-inducible STAT activity in HuH7, HepG2, and Hep3B cells. Western blot analysis for STAT1 and STAT3 phosphorylation using specific anti-phospho STAT antibodies [phospho-STAT1(Tyr701) (STAT1 pTyr701) and phospho-STAT3(Tyr705) (STAT3 pTyr705)]. HuH7, HepG2, or Hep3B cells were stimulated for 15 or 30 min with IFN-α in the presence or absence of cPKC inhibitor Gö6976 (Gö). As a control, the blots were stripped and reprobed with the respective anti-total protein antibody. C, control (untreated cells).
DISCUSSION
The cellular responses to IFNs imply the modulation of expression of a vast repertoire of target genes (42). While a pivotal role is played by the activation of the Jak/STAT pathway, many other signaling pathways have been previously described as being involved in the IFN-regulated cellular response (6). The major contribution of this work is the finding that inhibition of cPKC activity impairs IFN-α-mediated STAT activation. By the use of specific inhibitor compounds, we have explored the role of different signaling pathways in IFN-α-mediated HCV replicon clearance. We have found that an inhibitor of PKCs impairs the ability of IFN-α to induce HCV RNA degradation. In contrast, inhibition of p42/p44 MAPK, p38, PI3K, and mTOR kinases did not significantly influence HCV clearance. All compounds were used at doses that have been described in the literature to be fully effective (5, 11, 24, 44, 51). The only compound that was used at a suboptimal concentration was the PI3K inhibitor (10 μM instead of 50 to 100 μM). In fact, higher concentrations of this inhibitor resulted in extensive cell death after 1 day of incubation (data not shown). Therefore, we could not rule out a role of the PI3K-regulated pathway in IFN-α-mediated HCV clearance in our system.
An important role of PKCs in both IFN-α- and IFN-γ-regulated pathways has long been established (35). Recently, specific functions for different PKC isoforms in the regulation of IFN activity have been reported. For example, PKCδ, activated by either IFN-α or IFN-γ, has been proposed to phosphorylate STAT1 in Ser727 modulating its transcriptional activity (8, 47). Moreover, PKCɛ has been shown to be an important regulator of integrin-IFN-γ signaling cross talk (21). Using specific PKC isoform inhibitors, we have shown that conventional PKCs play a role in IFN-α-mediated HCV RNA clearance. On the contrary, PKCδ does not seem to be involved in this process. The lack of specific inhibitors for other PKCs prevents us from exploring their possible role in IFN-α signaling.
We have shown that the block of cPKCs leads to a broad inhibition of IFN-α-stimulated gene expression. MxA, 2′-5′OAS, ISG56, ISG15, ISG54, PKR, ADAR1, and ISG60 induction was significantly reduced when cPKC activity was inhibited. Moreover, we have highlighted a substantial decrease of STAT activity that could explain ISG transcriptional impairment. In fact, inhibition of cPKC activity impairs tyrosine phosphorylation, nuclear import, and DNA binding of both STAT1 and STAT3. On the contrary, STAT serine 727 phosphorylation was not significantly affected by cPKC inhibition. Notably, we found that IFN-α-mediated Jak activation is not impaired by the presence of cPKC inhibitor, pinpointing a crossroad of the two pathways. We conclude, therefore, that cPKC activity is a specific requisite to achieve the efficient Jak-dependent STAT phosphorylation. The extension of this observation to other cell lines demonstrates that the effect of PKC inhibition on IFN-α signaling is independent of HCV replicon and implies that cPKC activity is generally important for IFN-α response in liver cells.
Regulation of STAT phosphorylation by PKCs does not appear to be restricted to IFN-α signaling. In fact, it has been recently reported that mouse embryonic fibroblasts lacking PKCɛ show a significant reduction in STAT1 tyrosine phosphorylation following IFN-γ stimulation (21). The requirement of different PKC isoforms for STAT tyrosine phosphorylation could either depend on the different Jak members activated by specific IFNs or imply that PKC enzymes cooperate in regulating STAT activities; e.g., a PKC isoform activates others.
The molecular mechanism responsible for the regulation of STAT phosphorylation by cPKC remains elusive. A preliminary analysis suggests that the Jak-STAT inhibitor proteins characterized so far, such as PIAS1/3, TcPTP, SHP2, SOCS-1, and SOCS-3, are not involved in the impairment of STAT tyrosine phosphorylation. Moreover, pretreatment of Rep60 cells with a broad tyrosine phosphatase inhibitor (sodium orthovanadate) does not rescue the block of STAT phosphorylation caused by cPKC inhibition (data not shown), suggesting that a dysregulation of tyrosine phosphatase activity is not involved in this phenomenon. In addition, we were not able to show any direct interaction among Jak/STAT and PKCs (our unpublished results). Recently, Usacheva and coworkers showed that the impairment of the interaction between IFNAR2 and RACK1 results in STAT1 phosphorylation inhibition (49). Interestingly, RACK1 is a WD-containing protein originally identified as an activated PKCβ interactor (38). Moreover, RACK1 associates with Jak1, Tyk2, and unphosphorylated STAT1, suggesting that RACK could function as a scaffold protein recruiting STAT1 to the receptor prior to cytokine stimulation (50). Further experiments are required to elucidate whether RACK1 represents the molecular link between IFN-α and cPKC pathways.
Finally, we have shown that inhibition of PKC activity does not prevent IFN-α from shutting off HCV IRES-dependent translation. This observation, which emphasizes the complexity of the IFN-α activity, may suggest that IFN-α hampers viral translation in a STAT-independent manner. Importantly, in the absence of PKC activity, IFN-α withdrawal results in new viral protein synthesis, implying that PKC is important for an effective HCV clearance.
In conclusion, our data suggest that modulation of cPKC activity, which is influenced by many physiological stimuli (9, 30), could contribute to the great variability of IFN-α antiviral therapy outcome. Moreover, the observation that the overexpression of HCV-encoded protease NS3 is able to inhibit the PKC-mediated signal transduction (4) may unveil a further viral mechanism to escape the antiviral action of IFN-α.
Acknowledgments
We are grateful to all the members of the Piacentini and Tripodi laboratories for help, suggestions, and discussion; A. Rinaldi and D. Travaglini for valuable technical assistance; and Penny Lovat for critical reading of the manuscript.
The work was supported by grants from Ricerca Corrente and Ricerca Finalizzata del “Ministero della Salute” and by AIRC (Associazione Italiana Ricerca sul Cancro).
REFERENCES
- 1.Ahmad, S., Y. M. Alsayed, B. J. Druker, and L. C. Platanias. 1997. The type I interferon receptor mediates tyrosine phosphorylation of the CrkL adaptor protein. J. Biol. Chem. 272:29991-29994. [DOI] [PubMed] [Google Scholar]
- 2.Bartenschlager, R., and V. Lohmann. 2000. Replication of hepatitis C virus. J. Gen. Virol. 81:1631-1648. [DOI] [PubMed] [Google Scholar]
- 3.Blight, K. J., A. A. Kolykhalov, and C. M. Rice. 2000. Efficient initiation of HCV RNA replication in cell culture. Science 290:1972-1974. [DOI] [PubMed] [Google Scholar]
- 4.Borowski, P., J. S. zur Wiesch, K. Resch, H. Feucht, R. Laufs, and H. Schmitz. 1999. Protein kinase C recognizes the protein kinase A-binding motif of nonstructural protein 3 of hepatitis C virus. J. Biol. Chem. 274:30722-30728. [DOI] [PubMed] [Google Scholar]
- 5.Brown, E. J., M. W. Albers, T. B. Shin, K. Ichikawa, C. T. Keith, W. S. Lane, and S. L. Schreiber. 1994. A mammalian protein targeted by G1-arresting rapamycin-receptor complex. Nature 369:756-758. [DOI] [PubMed] [Google Scholar]
- 6.David, M. 2002. Signal transduction by type I interferons. BioTechniques 33:S58-S65. [PubMed] [Google Scholar]
- 7.David, M., E. Petricoin III, C. Benjamin, R. Pine, M. J. Weber, and A. C. Larner. 1995. Requirement for MAP kinase (ERK2) activity in interferon alpha- and interferon beta-stimulated gene expression through STAT proteins. Science 269:1721-1723. [DOI] [PubMed] [Google Scholar]
- 8.Deb, D. K., A. Sassano, F. Lekmine, B. Majchrzak, A. Verma, S. Kambhampati, S. Uddin, A. Rahman, E. N. Fish, and L. C. Platanias. 2003. Activation of protein kinase C delta by IFN-gamma. J. Immunol. 171:267-273. [DOI] [PubMed] [Google Scholar]
- 9.Dempsey, E. C., A. C. Newton, D. Mochly-Rosen, A. P. Fields, M. E. Reyland, P. A. Insel, and R. O. Messing. 2000. Protein kinase C isozymes and the regulation of diverse cell responses. Am. J. Physiol. Lung Cell. Mol. Physiol. 279:L429-L438. [DOI] [PubMed] [Google Scholar]
- 10.Enomoto, N., I. Sakuma, Y. Asahina, M. Kurosaki, T. Murakami, C. Yamamoto, Y. Ogura, N. Izumi, F. Marumo, and C. Sato. 1996. Mutations in the nonstructural protein 5A gene and response to interferon in patients with chronic hepatitis C virus 1b infection. N. Engl. J. Med. 334:77-81. [DOI] [PubMed] [Google Scholar]
- 11.Favata, M. F., K. Y. Horiuchi, E. J. Manos, A. J. Daulerio, D. A. Stradley, W. S. Feeser, D. E. Van Dyk, W. J. Pitts, R. A. Earl, F. Hobbs, R. A. Copeland, R. L. Magolda, P. A. Scherle, and J. M. Trzaskos. 1998. Identification of a novel inhibitor of mitogen-activated protein kinase kinase. J. Biol. Chem. 273:18623-18632. [DOI] [PubMed] [Google Scholar]
- 12.Frese, M., T. Pietschmann, D. Moradpour, O. Haller, and R. Bartenschlager. 2001. Interferon-alpha inhibits hepatitis C virus subgenomic RNA replication by an MxA-independent pathway. J. Gen. Virol. 82:723-733. [DOI] [PubMed] [Google Scholar]
- 12a.Giannini, C., and C. Bréchot. 2003. Hepatitis C virus biology. Cell Death Differ. 10(Suppl. 1):S27-S38. [DOI] [PubMed] [Google Scholar]
- 13.Goh, K. C., S. J. Haque, and B. R. Williams. 1999. p38 MAP kinase is required for STAT1 serine phosphorylation and transcriptional activation induced by interferons. EMBO J. 18:5601-5608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Goodbourn, S., L. Didcock, and R. E. Randall. 2000. Interferons: cell signalling, immune modulation, antiviral response and virus countermeasures. J. Gen. Virol. 81:2341-2364. [DOI] [PubMed] [Google Scholar]
- 15.Grandvaux, N., B. R. tenOever, M. J. Servant, and J. Hiscott. 2002. The interferon antiviral response: from viral invasion to evasion. Curr. Opin. Infect. Dis. 15:259-267. [DOI] [PubMed] [Google Scholar]
- 16.Gschwendt, M., H. J. Muller, K. Kielbassa, R. Zang, W. Kittstein, G. Rincke, and F. Marks. 1994. Rottlerin, a novel protein kinase inhibitor. Biochem. Biophys. Res. Commun. 199:93-98. [DOI] [PubMed] [Google Scholar]
- 17.Guidotti, L. G., and F. V. Chisari. 2001. Noncytolytic control of viral infections by the innate and adaptive immune response. Annu. Rev. Immunol. 19:65-91. [DOI] [PubMed] [Google Scholar]
- 18.Guo, J. T., V. V. Bichko, and C. Seeger. 2001. Effect of alpha interferon on the hepatitis C virus replicon. J. Virol. 75:8516-8523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Haller, O., and G. Kochs. 2002. Interferon-induced Mx proteins: dynamin-like GTPases with antiviral activity. Traffic 3:710-717. [DOI] [PubMed] [Google Scholar]
- 20.Hui, D. J., C. R. Bhasker, W. C. Merrick, and G. C. Sen. 2003. Viral stress-inducible protein p56 inhibits translation by blocking the interaction of eIF3 with the ternary complex eIF2.GTP.Met-tRNAi. J. Biol. Chem. 278:39477-39482. [DOI] [PubMed] [Google Scholar]
- 21.Ivaska, J., L. Bosca, and P. J. Parker. 2003. PKCepsilon is a permissive link in integrin-dependent IFN-gamma signalling that facilitates JAK phosphorylation of STAT1. Nat. Cell Biol. 5:363-369. [DOI] [PubMed] [Google Scholar]
- 22.Katze, M. G., Y. He, and M. Gale, Jr. 2002. Viruses and interferon: a fight for supremacy. Nat. Rev. Immunol. 2:675-687. [DOI] [PubMed] [Google Scholar]
- 23.Lauer, G. M., and B. D. Walker. 2001. Hepatitis C virus infection. N. Engl. J. Med. 345:41-52. [DOI] [PubMed] [Google Scholar]
- 24.Lee, J. C., J. T. Laydon, P. C. McDonnell, T. F. Gallagher, S. Kumar, D. Green, D. McNulty, M. J. Blumenthal, J. R. Heys, S. W. Landvatter, et al. 1994. A protein kinase involved in the regulation of inflammatory cytokine biosynthesis. Nature 372:739-746. [DOI] [PubMed] [Google Scholar]
- 25.Lekmine, F., S. Uddin, A. Sassano, S. Parmar, S. M. Brachmann, B. Majchrzak, N. Sonenberg, N. Hay, E. N. Fish, and L. C. Platanias. 2003. Activation of the p70 S6 kinase and phosphorylation of the 4E-BP1 repressor of mRNA translation by type I interferons. J. Biol. Chem. 278:27772-27780. [DOI] [PubMed] [Google Scholar]
- 26.Lohmann, V., F. Korner, J. Koch, U. Herian, L. Theilmann, and R. Bartenschlager. 1999. Replication of subgenomic hepatitis C virus RNAs in a hepatoma cell line. Science 285:110-113. [DOI] [PubMed] [Google Scholar]
- 27.Martiny-Baron, G., M. G. Kazanietz, H. Mischak, P. M. Blumberg, G. Kochs, H. Hug, D. Marme, and C. Schachtele. 1993. Selective inhibition of protein kinase C isozymes by the indolocarbazole G ö 6976. J. Biol. Chem. 268:9194-9197. [PubMed] [Google Scholar]
- 28.Mottola, G., G. Cardinali, A. Ceccacci, C. Trozzi, L. Bartholomew, M. R. Torrisi, E. Pedrazzini, S. Bonatti, and G. Migliaccio. 2002. Hepatitis C virus nonstructural proteins are localized in a modified endoplasmic reticulum of cells expressing viral subgenomic replicons. Virology 293:31-43. [DOI] [PubMed] [Google Scholar]
- 29.Murray, E. M., J. A. Grobler, E. J. Markel, M. F. Pagnoni, G. Paonessa, A. J. Simon, and O. A. Flores. 2003. Persistent replication of hepatitis C virus replicons expressing the β-lactamase reporter in subpopulations of highly permissive Huh7 cells. J. Virol. 77:2928-2935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Parekh, D. B., W. Ziegler, and P. J. Parker. 2000. Multiple pathways control protein kinase C phosphorylation. EMBO J. 19:496-503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Parker, P. J., and J. Murray-Rust. 2004. PKC at a glance. J. Cell Sci. 117:131-132. [DOI] [PubMed] [Google Scholar]
- 32.Pawlotsky, J. M. 2003. Mechanisms of antiviral treatment efficacy and failure in chronic hepatitis C. Antivir. Res. 59:1-11. [DOI] [PubMed] [Google Scholar]
- 33.Pawlotsky, J. M. 2003. The nature of interferon-alpha resistance in hepatitis C virus infection. Curr. Opin. Infect. Dis. 16:587-592. [DOI] [PubMed] [Google Scholar]
- 34.Pfeffer, L. M., J. E. Mullersman, S. R. Pfeffer, A. Murti, W. Shi, and C. H. Yang. 1997. STAT3 as an adapter to couple phosphatidylinositol 3-kinase to the IFNAR1 chain of the type I interferon receptor. Science 276:1418-1420. [DOI] [PubMed] [Google Scholar]
- 35.Pfeffer, L. M., and Y. H. Tan. 1991. Do second messengers play a role in interferon signal transduction? Trends Biochem. Sci. 16:321-323. [DOI] [PubMed] [Google Scholar]
- 36.Platanias, L. C., and E. N. Fish. 1999. Signaling pathways activated by interferons. Exp. Hematol. 27:1583-1592. [DOI] [PubMed] [Google Scholar]
- 37.Rivas-Estilla, A. M., Y. Svitkin, M. L. Lastra, M. Hatzoglou, A. Sherker, and A. E. Koromilas. 2002. PKR-dependent mechanisms of gene expression from a subgenomic hepatitis C virus clone. J. Virol. 76:10637-10653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ron, D., C. H. Chen, J. Caldwell, L. Jamieson, E. Orr, and D. Mochly-Rosen. 1994. Cloning of an intracellular receptor for protein kinase C: a homolog of the beta subunit of G proteins. Proc. Natl. Acad. Sci. USA 91:839-843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Samuel, C. E. 2001. Antiviral actions of interferons. Clin. Microbiol. Rev. 14:778-809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sen, G. C. 2001. Viruses and interferons. Annu. Rev. Microbiol. 55:255-281. [DOI] [PubMed] [Google Scholar]
- 41.Silverman, R. H. 1994. Fascination with 2-5A-dependent RNase: a unique enzyme that functions in interferon action. J. Interferon Res. 14:101-104. [DOI] [PubMed] [Google Scholar]
- 42.Stark, G. R., I. M. Kerr, B. R. Williams, R. H. Silverman, and R. D. Schreiber. 1998. How cells respond to interferons. Annu. Rev. Biochem. 67:227-264. [DOI] [PubMed] [Google Scholar]
- 43.Taniguchi, T., K. Ogasawara, A. Takaoka, and N. Tanaka. 2001. IRF family of transcription factors as regulators of host defense. Annu. Rev. Immunol. 19:623-655. [DOI] [PubMed] [Google Scholar]
- 44.Toullec, D., P. Pianetti, H. Coste, P. Bellevergue, T. Grand-Perret, M. Ajakane, V. Baudet, P. Boissin, E. Boursier, F. Loriolle, et al. 1991. The bisindolylmaleimide GF 109203X is a potent and selective inhibitor of protein kinase C. J. Biol. Chem. 266:15771-15781. [PubMed] [Google Scholar]
- 45.Uddin, S., C. Gardziola, A. Dangat, T. Yi, and L. C. Platanias. 1996. Interaction of the c-cbl proto-oncogene product with the Tyk-2 protein tyrosine kinase. Biochem. Biophys. Res. Commun. 225:833-838. [DOI] [PubMed] [Google Scholar]
- 46.Uddin, S., F. Lekmine, N. Sharma, B. Majchrzak, I. Mayer, P. R. Young, G. M. Bokoch, E. N. Fish, and L. C. Platanias. 2000. The Rac1/p38 mitogen-activated protein kinase pathway is required for interferon alpha-dependent transcriptional activation but not serine phosphorylation of Stat proteins. J. Biol. Chem. 275:27634-27640. [DOI] [PubMed] [Google Scholar]
- 47.Uddin, S., A. Sassano, D. K. Deb, A. Verma, B. Majchrzak, A. Rahman, A. B. Malik, E. N. Fish, and L. C. Platanias. 2002. Protein kinase C-delta (PKC-delta) is activated by type I interferons and mediates phosphorylation of Stat1 on serine 727. J. Biol. Chem. 277:14408-14416. [DOI] [PubMed] [Google Scholar]
- 48.Uddin, S., L. Yenush, X. J. Sun, M. E. Sweet, M. F. White, and L. C. Platanias. 1995. Interferon-alpha engages the insulin receptor substrate-1 to associate with the phosphatidylinositol 3′-kinase. J. Biol. Chem. 270:15938-15941. [DOI] [PubMed] [Google Scholar]
- 49.Usacheva, A., R. Smith, R. Minshall, G. Baida, S. Seng, E. Croze, and O. Colamonici. 2001. The WD motif-containing protein receptor for activated protein kinase C (RACK1) is required for recruitment and activation of signal transducer and activator of transcription 1 through the type I interferon receptor. J. Biol. Chem. 276:22948-22953. [DOI] [PubMed] [Google Scholar]
- 50.Usacheva, A., X. Tian, R. Sandoval, D. Salvi, D. Levy, and O. R. Colamonici. 2003. The WD motif-containing protein RACK-1 functions as a scaffold protein within the type I IFN receptor-signaling complex. J. Immunol. 171:2989-2994. [DOI] [PubMed] [Google Scholar]
- 51.Vlahos, C. J., W. F. Matter, K. Y. Hui, and R. F. Brown. 1994. A specific inhibitor of phosphatidylinositol 3-kinase, 2-(4-morpholinyl)-8-phenyl-4H-1-benzopyran-4-one (LY294002). J. Biol. Chem. 269:5241-5248. [PubMed] [Google Scholar]
- 52.Wang, C., J. Pflugheber, R. Sumpter, Jr., D. L. Sodora, D. Hui, G. C. Sen, and M. Gale, Jr. 2003. Alpha interferon induces distinct translational control programs to suppress hepatitis C virus RNA replication. J. Virol. 77:3898-3912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Williams, B. R. 3. July 2001, posting date. Signal integration via PKR. Sci. STKE 2001:RE2. http://stke.sciencemag.org/cgi/content/full/OC_sigtrans;2001/89/re2. [DOI] [PubMed] [Google Scholar]
- 54.Witherell, G. W., and P. Beineke. 2001. Statistical analysis of combined substitutions in nonstructural 5A region of hepatitis C virus and interferon response. J. Med. Virol. 63:8-16. [PubMed] [Google Scholar]
- 55.Yang, C. H., A. Murti, and L. M. Pfeffer. 1998. STAT3 complements defects in an interferon-resistant cell line: evidence for an essential role for STAT3 in interferon signaling and biological activities. Proc. Natl. Acad. Sci. USA 95:5568-5572. [DOI] [PMC free article] [PubMed] [Google Scholar]