Abstract
The Epstein-Barr virus (EBV) BamHI A transcripts are a family of transcripts that are differentially spliced and can be detected in multiple EBV-associated malignancies. Several of the transcripts may encode proteins. One transcript of interest, RK-BARF0, is proposed to encode a 279-amino-acid protein with a possible endoplasmic reticulum-targeting sequence. In this study, the properties of RK-BARF0 were examined through identification of cellular-interacting proteins through yeast two-hybrid analysis and characterization of its expression in EBV-infected cells and tumors. In addition to the interaction previously identified with cellular Notch, it was determined that RK-BARF0 also bound cellular human I-mfa domain-containing protein (HIC), epithelin, and scramblase. An interaction between RK-BARF0 and Notch or epithelin induced proteasome-dependent degradation of Notch and epithelin but not of HIC or scramblase. Low levels of endogenous Notch expression in EBV-positive cell lines may correlate with RK-BARF0 expression. However, a screen of EBV-positive cell lines and tumors with an affinity-purified α-RK-BARF0 antiserum did not consistently detect RK-BARF0. These data suggest that while RK-BARF0 may have important cellular functions during EBV infection, and while the phenotype of EBV-positive cells suggest its expression, RK-BARF0 levels may be too low to detect.
Epstein-Barr virus (EBV) is associated with the development of several epithelial and lymphoid malignancies, such as nasopharyngeal carcinoma (NPC), gastric carcinoma, Hodgkin lymphoma, endemic Burkitt's lymphoma, and posttransplantation lymphoma (14, 20). In each malignancy, viral infection is latent and each disease state is accompanied by a specific pattern of latent viral gene expression. EBV encodes at least nine latent viral proteins, including six nuclear proteins, EBV nuclear antigens (EBNAs) EBNA1, EBNA2, EBNA3A, EBNA3B, EBNA3C, and EBNALP, and three latent membrane proteins (LMPs), LMP1, LMP2A, and LMP2B (14). In Burkitt's lymphoma, a type I lymphoid malignancy, only EBNA1 is expressed. The malignancies NPC and Hodgkin lymphoma have a type II pattern of gene expression, and the viral proteins EBNA1, LMP1, and LMP2A are expressed (4, 7, 31). Posttransplantation lymphoma and lymphoblastoid cell lines (LCLs) immortalized with EBV express all latent proteins and, thus, have a type III pattern of gene expression.
In vitro studies have demonstrated that EBNA1, EBNA2, EBNA3A, EBNA3C, and LMP1 are essential for B-lymphocyte immortalization (6, 13, 18, 19), while EBNA3B, LMP2A, and LMP2B are dispensable (18, 27). In addition to the latent proteins, a family of rightward transcripts from the BamHI A region are abundantly expressed in several EBV-associated malignancies and EBV-infected cell lines (2, 4, 5, 7, 9, 11, 21, 31). The BamHI A transcripts (also called BARTs or CSTs) are differentially spliced and give rise to several different family members that are 3′ coterminal. Several of these transcripts have open reading frames (ORFs) that possibly encode proteins (21). RK103 (also called RPMS1) is proposed to encode a nuclear protein that interacts with the cellular CSL family [CBF1, Su(H), Lag-1] DNA-binding protein CBF1 (also called RBPJκ) and inhibits the activity of EBNA2 and Notch intracellular fragment (NotchIC) (25, 33). RB2 (also called A73) is proposed to encode a cytoplasmic protein that interacts with the cellular receptor for activated C kinase (RACK) (25).
All of the BamHI A transcripts contain the BARF0 ORF at their 3′ ends, which potentially encodes a 174-amino-acid protein. One cDNA cloned from the xenografted NPC, C15, contains the BARF0 ORF extended at its 5′ end, termed RK-BARF0 (21). RK-BARF0 is proposed to encode a 279-amino-acid protein with a highly hydrophobic N-terminal region that resembles an endoplasmic reticulum (ER)-targeting sequence (8, 21). A yeast two-hybrid analysis of the RK-BARF0 ORF revealed that it binds unprocessed Notch1 and Notch4 and induces nuclear translocation of a portion of full-length Notch, suggesting that the putative protein may be responsible for modulating Notch signaling (17).
Other indirect evidence for RK-BARF0 includes the presence of a functional CTL epitope in RK-BARF0 and antibodies against RK-BARF0 detected in the antisera of NPC patients (9, 16). Anti-BARF0 antisera can immunoprecipitate unprocessed Notch from Raji cells, an EBV-positive cell line, but not DG75 cells, an EBV-negative cell line, providing evidence that RK-BARF0 is expressed in an EBV-positive Burkitt's LCL (17). However, antisera that was used to identify what was thought to be RK-BARF0 in EBV-positive cell lines and tumors (8) was subsequently shown to cross-react with HLA DR β chain, which runs at the same molecular weight as RK-BARF0 as determined by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (15, 23). In this study, additional cellular binding partners of the RK-BARF0 protein have been identified to further characterize its function. The ability of RK-BARF0 to degrade Notch and induce nuclear translocation was also employed for evidence of in vivo expression.
MATERIALS AND METHODS
Plasmid construction.
Flag- and His-tagged RK-BARF0 as well as deletion mutants RK-158 and RK-179 were cloned into pMEP4 (Invitrogen) as previously described (17). Myc-tagged Notch4 was cloned into the pMEP4 vector as previously described (17). Epithelin precursor, scramblase, and HIC cDNA fragments were amplified by Pfx PCR from a human lung cDNA library. Epithelin precursor was cloned into pCDNA3 with three myc epitopes at the C terminus (Invitrogen). HIC and scramblase were cloned into pCDNA3 with three myc epitopes at the N terminus (Invitrogen). Myc-tagged epithelin was also subcloned into the pMEP4 vector (Invitrogen).
Yeast two-hybrid analysis.
The yeast two-hybrid analysis was performed as previously described (17).
Cell culture, tumors, and transfections.
COS-1 and H1299 cells were maintained in Dulbecco's modified Eagle medium (DMEM) (Gibco) containing 10% fetal bovine serum (Sigma) and antibiotics. BL30, BL30 B95-8, BL30 P3HR1, BL41, BL41 B95-8, BL41 P3HR1, B95-8, BJAB, DG75, P3HR1, Raji, Jijoye, Akata, Akata EG, and Akata 3F2 cells were maintained in RPMI 1640 (Gibco) containing 10% fetal bovine serum and antibiotics. C15, C17, and C18 xenografted nasopharyngeal tumors were passaged in nude mice as previously described (3). DG75 and Raji cells stably transfected with myc-tagged Notch4 in the pMEP4 vector were transfected by electroporation and selected with 200 μg of hygromycin B (Roche)/ml. COS-1 cells were transiently transfected with RK-BARF0 using the Fugene6 (Roche) transfection reagent as instructed by the manufacturer.
Cell extracts and Western blots.
DG75 and Raji cells containing inducible myc-tagged Notch4 or the pMEP4 vector were treated with 5 μM CdCl2 for 16 h. MG132-treated cells were treated at a concentration of 20 μM or were treated with dimethyl sulfoxide (DMSO) vehicle control for 8 h. Cell extracts were prepared by lysing cells on ice for 30 min in lysis buffer {20 mM Tris-HCl (pH 7.8), 150 mM NaCl, 1 mM EDTA, 10% glycerol, 0.5% 3-[(3-cholamidopropyl)-dimethylammonia]-1-propanesulfonate (CHAPS)} containing 1 mM phenylmethylsulfonyl fluoride (PMSF), 0.5 mM sodium orthovanadate, and 5 μg of aprotinin per ml. Cell lysates were clarified by centrifugation. Equal amounts of protein were loaded onto SDS-PAGE gels and were transferred to Immobilon-P (Millipore). Polyclonal α-cMyc, α-His, and α-HA were used to detect respectively tagged proteins (Santa Cruz). Polyclonal α-Notch1 that recognizes both the intracellular and extracellular portions of Notch1 was used to detect endogenous Notch1 (Santa Cruz). α-β-Actin was used as a loading control (Santa Cruz). Corresponding horseradish peroxidase-conjugated secondary antibody was used (α-rabbit [Amersham Pharmacia] and α-goat [DAKO]), and proteins were detected with the Supersignal West Pico Chemiluminescence system (Pierce) followed by exposure to X-ray film (Kodak).
Coimmunoprecipitation of RK-BARF0 and cellular proteins.
myc-tagged HIC, scramblase, and epithelin precursor were immunoprecipitated with FLAG-tagged RK-BARF0 by using α-FLAG M2 affinity gel (Sigma) overnight at 4°C. The beads were then washed and proteins were recovered by boiling in SDS sample buffer. Immunoprecipitations were then subjected to SDS-PAGE, transferred to Immobilon-P, and used for Western blot analysis.
Cell fractionation.
Cells were lysed by incubation in a hypotonic buffer (20 mM HEPES, 10 mM KCl, 0.1 mM EDTA, 0.1 mM EGTA, 1 mM dithiothreitol, protease, and phosphatase inhibitor cocktails [Sigma]) for 15 min on ice followed by addition of Nonidet P-40 to a final concentration of 1%. Nuclei were pelleted by low-speed centrifugation at 1,000 × g for 10 min at 4°C. The supernatant was collected as the cytosolic fraction. The crude nuclear pellet was further purified by using Optiprep (Sigma) reagent as directed by the manufacturer. Briefly, crude nuclei were resuspended in a 25% solution of Optiprep and underlaid with 30 and 35% layers of Optiprep. The gradient was spun for 20 min at 10,000 × g. The band of nuclei was collected from the 30%-35% interface, washed, pelleted, and lysed with nuclear extraction buffer (20 mM Tris [pH 8.0], 420 mM NaCl, 1.5 mM MgCl2, 0.2 mM EDTA, 25% glycerol, protease inhibitor cocktail [Sigma], and phosphatase inhibitor cocktail [Sigma]) with the salt concentration adjusted to 400 mM with 5 M NaCl. Insoluble nuclear material was pelleted at high speed for 10 min.
Affinity purification of RK-BARF0 antisera.
Anti-RK-BARF0 antisera were produced by injecting peptides generated from the RK-BARF0 amino acid sequence (8) (Immuno-Dynamics, Inc., La Jolla, Calif.). The antisera were affinity purified against glutathione S-transferase (GST)-fusion proteins immobilized on glutathione beads. Briefly, Escherichia coli DH5α cells were heat shocked with pGEX-2TK encoding GST-BARF0 or GST and the first 158 amino acids of RK-BARF0 (GSTRK-158) (8). A 100-ml starter culture was grown from one colony overnight at 37°C with vigorous shaking. The next morning, the culture was diluted up to 1.1 liters and grown for an additional 2 h. Protein expression was induced with 1 mM IPTG for 3.5 h. The bacteria was pelleted, washed with 1× phosphate-buffered saline (PBS), resuspended in 5 ml of 1× PBS, and frozen overnight at −80°C. The bacteria were thawed and spiked with a protease inhibitor cocktail (Sigma), and a 5-μl aliquot was run on an SDS-PAGE gel and Coomassie stained to confirm induction. The bacteria were lysed by sonication, spiked with Triton X-100 to a final concentration of 1%, shaken for 20 min at 4°C, and clarified by centrifugation. One-and-a-half milliliters of glutathione Sepharose beads (Amersham Biosciences) were prepared by washing in PBS and resuspending in PBS-Triton X-100. Clarified lysate was added to the resin and was shaken for 2 h at 4°C. The beads were spun, washed three times with 200 mM HEPES (pH 8.5), resuspended in 5 ml of cross-linking buffer (20 mM dimethyl pimelimidate, 200 mM HEPES [pH 8.5]), and incubated with continuous mixing for 1 h at 4°C. The beads were spun, and the cross-linking reaction was terminated by incubation in 200 mM ethanolamine for 30 min at 4°C. The ethanolamine was removed by centrifugation, and the resin was resuspended in 200 mM HEPES and loaded into a column. The noncovalently attached molecules were removed by washing the column two times with glycine elution buffer (150 mM NaCl, 200 mM glycine-HCl [pH 2.0]) and then washing them two times with Tris-buffered saline (TBS). Before addition to the column, 2 ml of each serum was diluted with 2 ml of TBS and then was filtered through a 2-μm-pore-size filter. The filtered antisera were applied to their appropriate columns. Serum 2161 was applied to the column with bound GSTRK-158, and serum 2157 was applied to the column with bound GSTBARF0. Antiserum was loaded onto the appropriate column a total of five times. The column was washed once with TBS, once with wash buffer (500 mM NaCl, 20 mM Tris-HCl [pH 7.5], 0.1% Triton X-100), and once more with TBS. The purified antisera were eluted in 500-μl fractions into 100-μl aliquots of 1 M Tris-HCl (pH 8.5) with glycine elution buffer. The fraction containing the purified antibody was dialyzed against 1× TBS with 10% glycerol.
RESULTS
RK-BARF0 binds cellular proteins.
The yeast two-hybrid method was used to identify cellular proteins that interact with RK-BARF0 protein. The Gal4 DNA binding domain was fused to an RK-BARF0 mutant with the highly hydrophobic 12 N-terminal amino acids deleted. The GAL4-activating domain was fused to a human lung cDNA library. Approximately 8.5 × 105 transformants were tested on growth selection medium. As published in an earlier report, RK-BARF0 bound Notch1, -3, and -4 in two-hybrid and coimmunoprecipitation experiments. Colonies that exhibited moderate to fast growth on selective media were tested for β-galactosidase (β-gal) activity, and those with moderate to intense β-gal activity were sequenced. Through the two-hybrid screen it was determined that, in addition to Notch, RK-BARF0 bound human I-mfa domain-containing protein (HIC), scramblase, and epithelin precursor (Table 1).
TABLE 1.
RK-BARF0 Yeast-two-hybrid results
| Interacting protein | β-gal activity (time point [h]) |
|---|---|
| HIC | +++ (<0.5) |
| Notch | ++ (1-2) |
| Scramblase | ++ (1-2) |
| Epithelin precursor | + (2-3) |
HIC was initially identified for its ability to modulate expression from viral and cellular promoters through an interaction with positive transcription elongation factor b or human immunodeficiency virus (HIV) Tat (32). Epithelin precursor (also called acrogranin or granulin) is a growth factor implicated in epithelial cell function that also binds HIV Tat (28). Scramblase is a nonspecific lipid translocase that is responsible for disruption of plasma membrane asymmetry during apoptosis, resulting in the presence of phosphatidyl serine in the outer leaflet. The effects of these interactions have not yet been characterized.
RK-BARF0 induces proteasome-dependent degradation of Notch.
Notch is an unusual family of transmembrane receptors. Notch is synthesized as an unprocessed precursor in the ER, processed in the trans-Golgi network (TGN) into two fragments joined through disulfide bonds and eventually embedded in the plasma membrane (1, 12). When the N-terminal extracellular domain binds ligand such as Delta or Jagged, the cytoplasmic domain is proteolytically processed, released from the plasma membrane, and translocated to the nucleus, where it affects transcription through and interaction with CBF1 (RBPJκ) (24, 26).
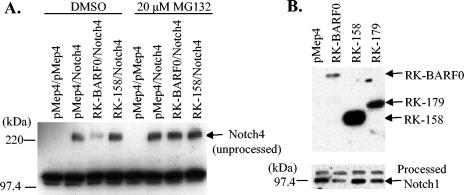
It has been previously published that RK-BARF0 binds the extracellular domain of Notch1, -3, and -4, and coexpression of RK-BARF0 and Notch can induce nuclear translocation of unprocessed Notch4 (17). RK-BARF0 also has an ER-targeting sequence and most likely binds unprocessed Notch in the ER before it is processed into two fragments in the TGN. It was also observed that Notch4 expression levels were reduced when coexpressed with RK-BARF0 compared to levels when expressed alone. Therefore, it was hypothesized that RK-BARF0 may induce proteasome-dependent degradation of Notch4. H1299 cells were transiently transfected with myc-tagged Notch4 and His6-tagged RK-BARF0 or the first 158 amino acids of RK-BARF0 (RK-158), which cannot bind Notch4, both under the control of an inducible promoter. Protein expression was induced with CdCl2, and cells were treated with the proteasome inhibitor MG132 or DMSO vehicle control. Unprocessed Notch4 levels were examined because Notch4 was expressed exogenously in stable cell lines. Notch4 levels were reduced when coexpressed with RK-BARF0 as determined by α-myc Western blot, and this reduction was blocked by MG132 treatment (Fig. 1A). Notch4 was not reduced when coexpressed with RK-158, which indicates that the reduction was dependent upon the interaction between Notch and RK-BARF0. To confirm that this phenomenon was not a by-product of an exogenous expression system, RK-BARF0, RK-158, and the first 179 amino acids of RK-BARF0 (RK-179), which retains the ability to bind Notch, were stably transfected into BJAB cells. Processed Notch levels were examined, because an antibody directed against intracellular Notch was used for Western blot analysis. Endogenous Notch1 levels were reduced in cells expressing RK-BARF0 and were reduced less dramatically in RK-179 cells (Fig. 1B). These data confirmed that the reduction is dependent on an interaction with RK-BARF0 which induces proteasome-mediated degradation of both endogenously and exogenously expressed Notch.
FIG. 1.
RK-BARF0 induces proteasome-dependent degradation of Notch. (A) H1299 cells were transiently transfected with combinations of myc-tagged Notch4 and full-length His6-tagged RK-BARF0 or a mutant encoding the first 158 amino acids of RK-BARF0 (RK-158), which cannot bind Notch. Expression was induced with 5 μM CdCl2, and the samples were treated with DMSO or 20 μM MG132. Notch4 was detected by immunoblot with α-myc. (B) BJAB stable cells expressing full-length His6-tagged RK-BARF0, RK-158, or the first 179 amino acids of RK-BARF0 (RK-179) were induced with 5 μM CdCl2 and immunoblotted with α-His6 to detect RK-BARF0 (top panel) or α-Notch1 to detect endogenous Notch1 (bottom panel).
RK-BARF0 induces proteasome-dependent degradation of epithelin precursor.
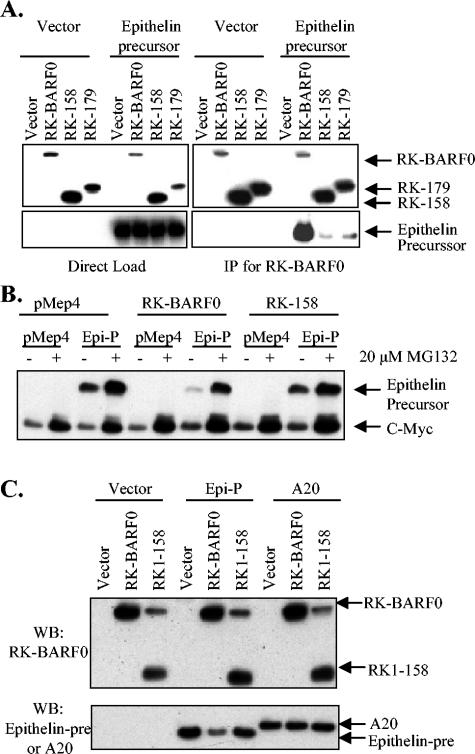
To confirm that RK-BARF0 and epithelin precursor interact in mammalian cells, Flag-His6-tagged RK-BARF0, RK-158, or RK-179 was cotransfected with myc-tagged epithelin precursor and used for coimmunoprecipitations. Lysates were either directly loaded onto an SDS-PAGE gel or immunoprecipitated with α-FLAG beads for RK-BARF0 and were used for Western blot analysis. RK-BARF0, RK-158, and RK-179 were detected with α-His antibody, and epithelin precursor was detected with α-myc. Abundant epithelin precursor immunoprecipitated with RK-BARF0, but little epithelin precursor immunoprecipitated with RK-179 and RK-158 (Fig. 2A). This result was in contrast to those of coimmunoprecipitation experiments with RK-BARF0 and Notch, with the interaction mapping to amino acids 158 to 179 (17). Amino acids 158 to 179 were partially required, but the interaction was not completely eliminated with RK-158 (Fig. 2A). However, like Notch4, epithelin precursor protein expression was reduced when coexpressed with RK-BARF0 but not with RK-158, as determined by α-myc Western blot (Fig. 2B). Furthermore, this reduction in expression was inhibited by MG132 treatment (Fig. 2B). As RK-158 does not efficiently bind epithelin and did not induce degradation of epithelin, these data suggest that an interaction between RK-BARF0 and epithelin is required for proteasome-induced degradation.
FIG. 2.
RK-BARF0 binds and induces proteasome-dependent degradation of epithelin precursor. (A) H1299 cells transiently transfected with Flag and His6-tagged RK-BARF0, RK-158, or RK-179 or with epithelin precursor were induced with CdCl2. Lysates were directly loaded onto an SDS-PAGE gel (left panel) or were immunoprecipitated with α-Flag (right panel). RK-BARF0 was detected with α-His6, and epithelin precursor was detected with α-cMyc. (B) H1299 cells transiently transfected with a combination of RK-BARF0 or RK-158 and epithelin precursor (Epi-P) were induced with 5 μM CdCl2 and treated with 20 μM MG132 or DMSO control. Epithelin precursor was detected by immunoblot with α-cMyc. (C) H1299 cells transiently transfected with RK-BARF0 or RK-158 and epithelin precursor (Epi-P) or A20 were used for immunoblot. RK-BARF0 was detected with α-His6, and epithelin and A20 were detected with α-cMyc. IP, immunoprecipitation; WB, Western blot.
To ensure that RK-BARF0-induced degradation of cellular proteins is specific for proteins that directly bind RK-BARF0, His6-tagged RK-BARF0 or RK-158 was coexpressed with myc-tagged epithelin or myc-tagged A20, which does not bind RK-BARF0. RK-BARF0 was detected by Western blot using α-His, and epithelin precursor and A20 were detected with α-myc. Confirming the earlier data, epithelin precursor levels were reduced when coexpressed with RK-BARF0 but not with RK-158 (Fig. 2C). In contrast, A20 levels were not reduced by coexpression of RK-BARF0 or RK-158, suggesting that RK-BARF0-induced degradation was specific for bound cellular protein and not for the myc epitope tag or noninteracting proteins.
RK-BARF0 binds HIC and scramblase but does not induce proteasome-dependent degradation.
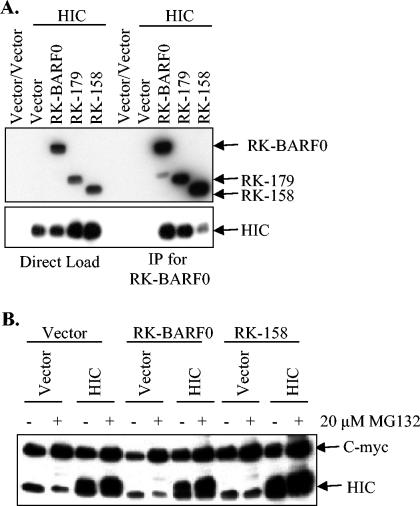
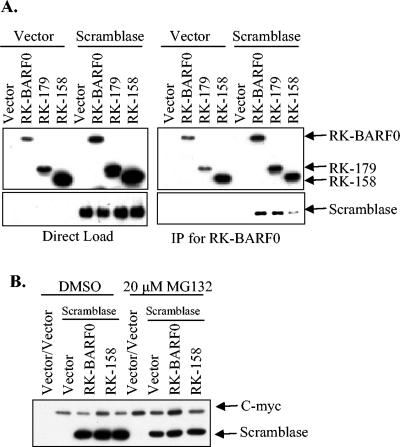
To confirm the yeast two-hybrid results, FLAG- and His6-tagged RK-BARF0, RK-158, or RK-179 was coexpressed with either myc-tagged HIC or scramblase and lysates were used for coimmunoprecipitation. Lysates were either directly loaded onto an SDS-PAGE gel or were immunoprecipitated for RK-BARF0 by using α-FLAG beads. When RK-BARF0 and HIC were coexpressed, myc-tagged HIC coimmunoprecipitated with RK-BARF0 and RK-179 but not with RK-158 (Fig. 3A), confirming the yeast two-hybrid results. To examine the effect of RK-BARF0 expression on degradation of HIC, myc-tagged HIC was coexpressed with His6-tagged RK-BARF0 or RK-158 and cells were treated with the proteasome inhibitor MG132. HIC levels were not reduced when coexpressed with RK-BARF0 or RK-158 (Fig. 3B). Furthermore, levels of protein expression were unchanged by treatment with MG132. When RK-BARF0 and scramblase were coexpressed, myc-tagged scramblase also coimmunoprecipitated with RK-BARF0 and RK-179 but not with RK-158 (Fig. 4A). To examine the effect of RK-BARF0 expression on degradation of scramblase, myc-tagged scramblase was coexpressed with His6-tagged RK-BARF0 or RK-158 and cells were treated with the proteasome inhibitor MG132. Scramblase levels were not reduced when coexpressed with RK-BARF0 or RK-158 (Fig. 4B). In contrast to Notch and epithelin precursor, expression of RK-BARF0 did not induce proteasome-dependent degradation of HIC or scramblase. These data indicate that RK-BARF0 has specific effects on cellular interacting proteins as it targets Notch and epithelin for proteasome degradation, while the RK-BARF0 and HIC-scramblase interaction may affect the function of the proteins in other ways.
FIG. 3.
RK-BARF0 binds HIC but does not induce proteasome-dependent degradation. (A) Flag- and His6-tagged RK-BARF0, RK-179, and RK-158 were transiently transfected into H1299 cells with myc-tagged HIC. Lysates were directly loaded onto an SDS-PAGE gel (left panel) or immunoprecipitated (IP) with α-Flag (right panel). RK-BARF0 was detected with α-His6, and HIC was detected with α-cMyc. (B) RK-BARF0 and RK-158 were transiently transfected into H1299 cells with HIC and were treated with 20 μM MG132 to examine degradation of HIC. HIC was detected with α-cMyc.
FIG. 4.
RK-BARF0 binds scramblase but does not induce proteasome-dependent degradation. (A) Flag- and His6-tagged RK-BARF0, RK-179, and RK-158 were transiently transfected into H1299 cells with myc-tagged scramblase. Lysates were directly loaded onto an SDS-PAGE gel (left panel) or were immunoprecipitated (IP) with α-Flag (right panel). RK-BARF0 was detected with α-His6, and scramblase was detected with α-cMyc. (B) RK-BARF0 and RK-158 were coexpressed with scramblase and were treated with 20 μM MG132 to examine degradation of scramblase. Scramblase was detected with α-cMyc.
Phenotype of EBV-positive cells indicates presence of RK-BARF0 protein.
Definitive evidence for a protein encoded by the RK-BARF0 transcript has been elusive. While a previous publication detected a protein of approximately the same size predicted for RK-BARF0 in only EBV-positive cells by using a rabbit antisera raised against an antigenic peptide (8), subsequent publications indicated that the antiserum also detects the HLA DR β chain, which is upregulated by EBV infection and is approximately the same size as the predicted RK-BARF0 (15, 23). Indirect evidence suggesting the existence of the protein includes the point that antibodies directed against RK-BARF0 are readily detectable in the antisera of NPC patients (9). Furthermore, α-BARF0 serum can be used to immunoprecipitate transfected Notch4 from the EBV-positive cell line Raji but not the EBV-negative cell line DG75 (17).
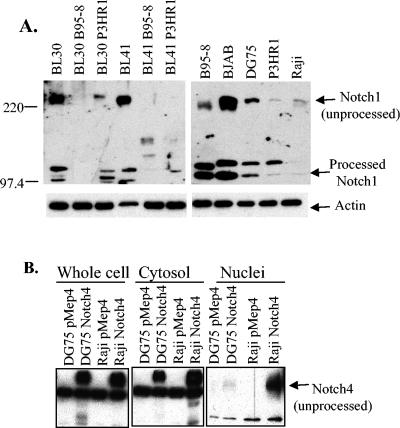
Based on the properties of RK-BARF0, the phenotypes of EBV-positive and -negative cell lines were characterized to evaluate the potential expression of RK-BARF0 protein in vivo. As was demonstrated in Fig. 1, RK-BARF0 induces the degradation of endogenous Notch1. Levels of Notch1 were examined by Western blot analysis in EBV-positive and -negative cell lines to determine if a correlation exists between Notch1 levels and possible RK-BARF0 expression. The cell lines included the EBV-negative Burkitt's lymphoma cell lines BL30 and BL41 and their infected counterparts, BL30 B95-8, BL30 P3HR1, BL41 B95-8, and BL41 P3HR1, which were infected with the B95-8 or P3HR1 strain of EBV. The EBV-negative B-cell lymphoma BJAB and Burkitt's lymphoma, DG75; EBV-positive, immortalized marmoset LCL B95-8; and Burkitt's lymphoma P3HR1 and Raji cell lines were also examined by Western Blot analysis. There was a strong correlation between low levels of both unprocessed and processed Notch1 and EBV infection (Fig. 5A). The two parental Burkitt's lymphoma cell lines BL30 and BL41 had relatively high levels of Notch1, whereas their infected counterparts had relatively low levels of Notch1 (Fig. 5A, left panel). The EBV-negative cell line BJAB had high levels of both unprocessed Notch1 and processed Notch1, whereas the EBV-negative cell line DG75 had intermediate levels of unprocessed Notch1. The EBV-positive cell line B95-8 also had an intermediate level of unprocessed Notch1, and the EBV-positive cell lines P3HR1 and Raji had very low levels of unprocessed and processed Notch1 (Fig. 5A, right panel). Equal protein loading was confirmed by β-actin Western blot analysis (Fig. 5A, bottom panel).
FIG. 5.
Notch levels in EBV-positive and -negative cell lines. (A) A series of EBV-positive and -negative cell lines were used for immunoblot analysis. α-Notch1 was used to detect endogenous Notch1 (top panel), and α-β-actin was used as a loading control (bottom panel). (B) DG75 and Raji cells stably expressing myc-tagged Notch4 were fractionated into cytosolic and nuclear lysates and used for immunoblot analysis. α-cMyc was used to detect Notch4.
It has also been previously published that RK-BARF0 has the unusual ability to induce nuclear accumulation of unprocessed Notch (17). Normally Notch is synthesized in the ER, processed in the TGN, and embedded in the plasma membrane. Upon ligand binding, only the intracellular portion translocates to the nucleus, where it can affect transcription. Full-length unprocessed Notch had never before been detected in the nucleus. Using this information, the subcellular location of exogenously expressed myc-tagged Notch4 was examined in the EBV-negative cell line DG75 and compared to that of the EBV-positive cell line Raji. Both cell lines were fractionated in parallel and used for Western blot analysis. While both cell lines show approximately equal levels of expression of Notch4, Raji has dramatically higher levels of nuclear unprocessed Notch4 (Fig. 5B). Because the ability to translocate unprocessed Notch4 to the nucleus is unique to RK-BARF0, this data supports the hypothesis that RK-BARF0 is expressed in Raji cells. While the presence of Notch4 in the nuclear fraction due to cytosolic contamination could not be completely eliminated, both sets of cell lines were fractionated in parallel and should have approximately equal contamination. It is also possible that additional genetic changes in the Raji lymphoma cell line may induce nuclear accumulation of unprocessed Notch. However, unprocessed Notch has only been detected in the nucleus through an interaction with RK-BARF0, suggesting that the levels of unprocessed nuclear Notch4 are most likely significant.
Antisera raised against RK-BARF0-specific peptides were purified with GST fusion proteins immobilized on glutathione beads
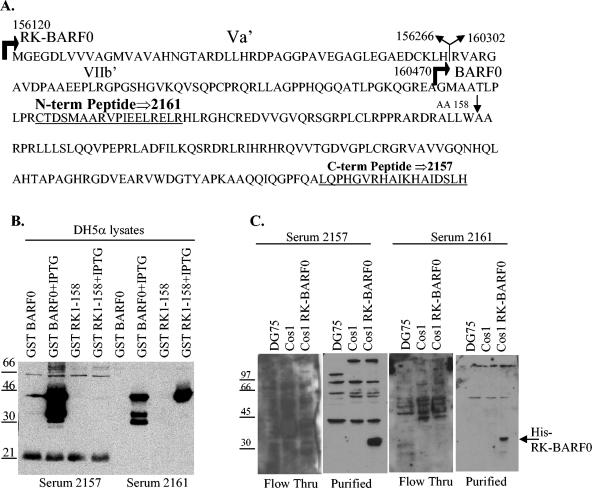
Two peptides from the putative RK-BARF0 protein were synthesized and used to immunize rabbits to produce two RK-BARF0 antisera. The RK-BARF0 amino acid sequence is listed with the two peptide sequences underlined that were used to immunize rabbits (Fig. 6A). Antiserum 2161 was used previously to survey EBV-positive and -negative cells and tumors and identified a protein of the predicted molecular weight in the EBV-positive cell lines (8). However, subsequent publications revealed that the antiserum also detected HLA DR β chain, which is upregulated by EBV infection and is approximately the same molecular weight as the predicted molecular weight of RK-BARF0 (15, 23). In order to reduce the likelihood of detecting cellular proteins with the RK-BARF0 antisera, each antisera was affinity purified with GST-BARF0 or GST-RK-158 fusion protein. The fusion proteins were expressed in and purified from E. coli DH5α. DH5α lysates were used for Western blot analysis with the antisera to confirm fusion protein expression and reactivity to the antisera (Fig. 6B). As expected, the antiserum 2157, made with the more C-terminal peptide, reacted efficiently against GST-BARF0 but not against GST-RK-158. Also, the antiserum 2161, made with the N-terminal peptide, reacted against both GST-BARF0 and GST-RK-158. Serum 2161 reacted more specifically with GST-RK-158, so GST-RK-158 was used to affinity purify 2161 and GST-BARF0 was used to purify 2157. Both antisera were purified against their fusion protein immobilized on glutathione beads. Column flowthrough and purified antisera were used to probe Western blots with DG75, COS-1, and COS-1 transiently transfected with His-tagged RK-BARF0 cell lysates (Fig. 6C). DG75 cells should express HLA DR β chain and, therefore, any HLA DR β chain should be detected in these lysates. The flowthrough from both antisera had multiple cross-reacting bands. Purified antisera had dramatically fewer cross-reacting bands and readily detected His6-tagged RK-BARF0. However, antiserum 2157 reacted more strongly to RK-BARF0 and has not been shown to cross-react with an HLA DR β chain. Therefore, 2157 was determined to be the more useful antiserum to survey EBV-positive and -negative cell lines.
FIG. 6.
Purification of RK-BARF0 specific antisera. (A) The sequence of the peptides used to inoculate rabbits are underlined within the RK-BARF0 amino acid sequence. The EBV coordinates corresponding to the B95-8 genome are indicated along with RK-BARF0 and BARF0 start sites, the RK-BARF0 splice site, its corresponding exons (Va′ and VIIb), and amino acid 158. (B) Lysates from DH5α expressing GST-BARF0 and GST RK-158 were probed with 2157 and 2161 sera to confirm reactivity. (C) Serum 2157 was purified against immobilized GST-BARF0, and serum 2161 was purified against immobilized GST-RK-158. Purified sera and column flowthrough (Flow Thru) were tested against DG75, COS-1, and COS-1 RK-BARF0 cell extracts.
RK-BARF0 is not consistently detected in EBV-positive cells.
The newly purified antiserum, 2157, was used to detect the putative RK-BARF0 protein in EBV-positive cell lines and tumors by Western blot analysis. COS-1 cells and COS-1 cells transiently transfected with His6-tagged RK-BARF0 were used as negative and positive controls. Anti-His antibody was used to detect His6-tagged RK-BARF0 (Fig. 7A, first two lanes). Purified serum 2157 was used to detect His-tagged RK-BARF0 and endogenous RK-BARF0. While serum 2157 detected His-tagged RK-BARF0 as efficiently as the α-His antibody, it did not detect any proteins of the correct molecular weight in either EBV-positive cell line Akata or Akata EG or in the passaged EBV-positive NPCs C15, C17, or C18 (Fig. 7A). Western blot analysis of the EBV-positive cell lines B95-8, Jijoye, and Raji and the EBV-positive C15 NPC tumor also did not consistently detect the putative RK-BARF0 protein (Fig. 7B). However, a protein that is the same size as is predicted for BARF0 was detected approximately 50% of the time in the B95-8 cell line (Fig. 7B).
FIG. 7.
RK-BARF0 is not detected in EBV-positive cell lines and in tumors. The purified serum 2157 was used to screen multiple EBV-positive and -negative cell lines and tumors. COS-1 cells expressing His6-tagged RK-BARF0 were used as a positive control. The serum was used to screen whole-cell lysates (A and B) and fractionated cell lysates (C and D) as well as MG132-treated cells (D). C-term, C terminal; WB, Western blot.
Because RK-BARF0 can be nuclear through utilization of the nuclear localization signal (NLS) from bound Notch, it was possible that RK-BARF0 may be concentrated in the nucleus of infected cells. EBV-positive NPC tumors were fractionated into cytosolic and nuclear extracts and were used for Western blot analysis with the purified 2157 serum. However, RK-BARF0 protein was not detected in either cytosolic or nuclear extracts of C15 or C17 in this immunoblot (Fig. 7C).
As RK-BARF0 has the ability to induce proteasome-dependent degradation of Notch and epithelin, it was possible that in the process of inducing proteasome-dependent degradation of bound proteins RK-BARF0 itself may be degraded, making it difficult to detect. To test this hypothesis, EBV-negative cells of DG75 and EBV-positive cells of Raji were treated with MG132, fractionated, and subjected to Western blot analysis with purified 2157 serum. In the nuclear fraction of MG132-treated Raji cells, a band of approximately the correct molecular weight was detected that was not present in DG75 cells (Fig. 7D). However, the band was not detected every time the experiment was repeated. Collectively, the sporadic appearance of proteins corresponding to the size of BARF0 and RK-BARF0 suggest that both proteins may be expressed in vivo; however, they may be expressed at very low levels or may be controlled by a yet-to-be-determined factor.
DISCUSSION
The data presented in this paper identify new cellular binding partners for the putative viral protein, RK-BARF0, and establish the ability of RK-BARF0 to induce proteasome-dependent degradation of Notch and epithelin precursor. The low levels of Notch may be indicative of RK-BARF0 expression in EBV-positive cells. Western blot analysis using rabbit antiserum raised against an RK-BARF0 C-terminal peptide and affinity purified using a GST-BARF0 fusion protein strongly detected exogenously expressed RK-BARF0 but could only occasionally detect a band that possibly represents RK-BARF0 in EBV-positive cells. The antiserum also detected a band of the same apparent molecular weight as BARF0 in B95-8 cells and RK-BARF0 in nuclear extracts from Raji cells treated with MG132, suggesting that it is possibly expressed at low levels and may be processed by the proteasome.
The existence of multiple unique BamHI A transcripts in many EBV-associated malignancies has been confirmed by a variety of different techniques (2, 5, 21, 25, 29). Distinct splice variants have been detected by RNase protection assay, which indicates that RK-BARF0 is less abundant than RK103 (RPMS1) or RB2 (A73) (25). Conversely, reverse transcription-PCR from size-specific bands of C15 mRNAs that hybridize to BamHI A-specific probes indicate that RK-BARF0 and RB2 transcripts can be amplified from the most abundant mRNA and therefore may be expressed at high levels in vivo (21). However, the majority of RK-BARF0 transcripts lack a translation stop site while only a small minority of transcripts use a stop site that is embedded within the polyadenylation site (21). This would suggest that only a portion of RK-BARF0 transcripts contain the entire ORF to make a protein product. The low abundance of the RK-BARF0 splice variant and the unusual 3′ processing also suggest that its protein product would be expressed at low levels.
The data presented in this study indicate that RK-BARF0 can induce proteasome-dependent degradation of Notch and epithelin precursor. Difficulty in detecting RK-BARF0 protein consistently could be due to this function, as RK-BARF0 may be degraded along with its binding partners. This hypothesis is substantiated by detection of a protein of the correct molecular weight in Raji cells treated with the proteasome inhibitor MG132 (Fig. 7D). Furthermore, using the ability of RK-BARF0 to induce Notch degradation and nuclear translocation of unprocessed Notch, RK-BARF0 expression in EBV-infected cells correlated with reduced levels of endogenous Notch and unprocessed Notch subcellular localization. The difficulties in detecting endogenous RK-BARF0 may reflect its low abundance. RK-BARF0, RK103, and RB2 can all be epitope tagged, efficiently expressed, and bind intriguing cellular partners through exogenous expression. These abilities suggest that they all have the capacity to function within a cell.
From a yeast two-hybrid screen it was determined that the RK-BARF0 protein binds cellular Notch (17). During Notch processing and signaling, Notch is processed in the TGN, is embedded in the plasma membrane, and is cleaved at the plasma membrane upon ligand binding. In addition, the intracellular portion is released and translocated to the nucleus, where it affects transcription. RK-BARF0 binds Notch prior to processing in the ER and induces nuclear translocation of full-length, unprocessed Notch. As a result of this interaction, a portion of unprocessed Notch is translocated to the nucleus (17) while most of the remaining Notch is degraded by the proteasome. Nuclear unprocessed Notch has not been characterized and may not be functional. Furthermore, reduction of overall Notch protein should inhibit Notch signaling. Previous studies have determined that RK103 (RPMS) binds CBF1, another component of the Notch signaling pathway, and blocks Notch signaling (25, 33). Coexpression of the two BamHI A proteins could effectively launch a multipronged attack on Notch signaling to diminish it. The Notch pathway regulates development and cell fate decisions through control of diverse mechanisms, such as proliferation, cell cycle arrest, differentiation, cell survival, and apoptosis. Due to its diverse functions, disruption of controlled Notch activation, either through constitutive activation or through constitutive repression, could promote transformation and oncogenesis (30). Notch appears to act as an oncoprotein in the context of murine mammary epithelium, Reed-Sternberg cells in Hodgkin lymphoma, and anaplastic large-cell lymphoma (30). Notch also appears to act as a tumor suppressor in keratinocytes and squamous cell carcinomas (30). Therefore, disruption of Notch signaling by the BamHI A proteins could contribute to virus-induced oncogenesis.
The yeast two-hybrid screen also identified RK-BARF0-interacting proteins HIC, epithelin, and scramblase. HIC was initially identified through its ability to bind HIV Tat and regulates expression from viral and cellular promoters. Epithelin also binds HIV Tat and is involved in cellular growth. Scramblase is a nonspecific lipid translocase and is responsible for disruption of membrane asymmetry during apoptosis. Interactions between these cellular proteins and RK-BARF0 could affect a wide variety of cellular functions, including transcription, cell growth, and apoptosis.
The BamHI A transcripts are not essential for B-lymphocyte transformation. However, they were originally cloned from NPC and therefore may contribute to the development of epithelial malignancy. Primate epithelial cells have been immortalized by using a cosmid clone from the BamHI A region of EBV that contains none of the latent proteins considered essential for B-lymphocyte transformation (10). Precedence also exists for a nonessential EBV protein promoting transformation, as LMP2A, which is also not essential for B-lymphocyte transformation, was found to transform epithelial cells through its activation of the phosphatidylinositol 3′-kinase/Akt pathway (22). Furthermore, in type II latency, such as in NPC, RK-BARF0 and RK103 may be able to compensate for the absence of EBNA2 and its ability to modulate Notch signaling.
In summary, these data indicate that the RK-BARF0 protein has the capacity to function within a cell and that it may affect important cellular proteins through direct interactions. While the protein has proven to be difficult to detect, these data suggest its expression.
Acknowledgments
We gratefully acknowledge Jennifer Morrison and Bernardo Mainou for critical reviews of the manuscript.
This study was supported by NIH grant CA32979 to N.R.-T.
REFERENCES
- 1.Blaumueller, C. M., H. Qi, P. Zagouras, and S. Artavanis-Tsakonas. 1997. Intracellular cleavage of Notch leads to a heterodimeric receptor on the plasma membrane. Cell 90:281-291. [DOI] [PubMed] [Google Scholar]
- 2.Brooks, L. A., A. L. Lear, L. S. Young, and A. B. Rickinson. 1993. Transcripts from the Epstein-Barr virus BamHI A fragment are detectable in all three forms of virus latency. J. Virol. 67:3182-3190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Busson, P., G. Ganem, P. Flores, F. Mugneret, B. Clausse, B. Caillou, K. Braham, H. Wakasugi, M. Lipinski, and T. Tursz. 1988. Establishment and characterization of three transplantable EBV-containing nasopharyngeal carcinomas. Int. J. Cancer 42:599-606. [DOI] [PubMed] [Google Scholar]
- 4.Busson, P., R. McCoy, R. Sadler, K. Gilligan, T. Tursz, and N. Raab-Traub. 1992. Consistent transcription of the Epstein-Barr virus LMP2 gene in nasopharyngeal carcinoma. J. Virol. 66:3257-3262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chen, H. L., M. M. Lung, J. S. Sham, D. T. Choy, B. E. Griffin, and M. H. Ng. 1992. Transcription of BamHI-A region of the EBV genome in NPC tissues and B cells. Virology 191:193-201. [DOI] [PubMed] [Google Scholar]
- 6.Cohen, J. I., F. Wang, J. Mannick, and E. Kieff. 1989. Epstein-Barr virus nuclear protein 2 is a key determinant of lymphocyte transformation. Proc. Natl. Acad. Sci. USA 86:9558-9562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Fahraeus, R., H. L. Fu, I. Ernberg, J. Finke, M. Rowe, G. Klein, K. Falk, E. Nilsson, M. Yadav, P. Busson, et al. 1988. Expression of Epstein-Barr virus-encoded proteins in nasopharyngeal carcinoma. Int. J. Cancer 42:329-338. [DOI] [PubMed] [Google Scholar]
- 8.Fries, K. L., T. B. Sculley, J. Webster-Cyriaque, P. Rajadurai, R. H. Sadler, and N. Raab-Traub. 1997. Identification of a novel protein encoded by the BamHI A region of the Epstein-Barr virus. J. Virol. 71:2765-2771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gilligan, K. J., P. Rajadurai, J. C. Lin, P. Busson, M. Abdel-Hamid, U. Prasad, T. Tursz, and N. Raab-Traub. 1991. Expression of the Epstein-Barr virus BamHI A fragment in nasopharyngeal carcinoma: evidence for a viral protein expressed in vivo. J. Virol. 65:6252-6259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Griffin, B. E., and L. Karran. 1984. Immortalization of monkey epithelial cells by specific fragments of Epstein-Barr virus DNA. Nature 309:78-82. [DOI] [PubMed] [Google Scholar]
- 11.Hitt, M. M., M. J. Allday, T. Hara, L. Karran, M. D. Jones, P. Busson, T. Tursz, I. Ernberg, and B. E. Griffin. 1989. EBV gene expression in an NPC-related tumor. EMBO J. 8:2639-2651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Honjo, T. 1996. The shortest path from the surface to the nucleus: RBP-J kappa/Su(H) transcription factor. Genes Cells 1:1-9. [DOI] [PubMed] [Google Scholar]
- 13.Kaye, K. M., K. M. Izumi, and E. Kieff. 1993. Epstein-Barr virus latent membrane protein 1 is essential for B-lymphocyte growth transformation. Proc. Natl. Acad. Sci. USA 90:9150-9154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kieff, E., and A. B. Rickinson. 2001. Epstein-Barr virus and its replication, p. 2511-2573. In D. M. Knipe and P. M. Howley (ed.), Field's virology, 4th ed., vol. 2. Lippincott Williams & Wilkins Publishers, Philadelphia, Pa. [Google Scholar]
- 15.Kienzle, N., M. Buck, S. Greco, K. Krauer, and T. B. Sculley. 1999. Epstein-Barr virus-encoded RK-BARF0 protein expression. J. Virol. 73:8902-8906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kienzle, N., T. B. Sculley, L. Poulsen, M. Buck, S. Cross, N. Raab-Traub, and R. Khanna. 1998. Identification of a cytotoxic T-lymphocyte response to the novel BARF0 protein of Epstein-Barr virus: a critical role for antigen expression. J. Virol. 72:6614-6620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kusano, S., and N. Raab-Traub. 2001. An Epstein-Barr virus protein interacts with Notch. J. Virol. 75:384-395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Longnecker, R., C. L. Miller, B. Tomkinson, X. Q. Miao, and E. Kieff. 1993. Deletion of DNA encoding the first five transmembrane domains of Epstein-Barr virus latent membrane proteins 2A and 2B. J. Virol. 67:5068-5074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mannick, J. B., J. I. Cohen, M. Birkenbach, A. Marchini, and E. Kieff. 1991. The Epstein-Barr virus nuclear protein encoded by the leader of the EBNA RNAs is important in B-lymphocyte transformation. J. Virol. 65:6826-6837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Raab-Traub, N. 2002. Epstein-Barr virus in the pathogenesis of NPC. Semin. Cancer Biol. 12:431-441. [DOI] [PubMed] [Google Scholar]
- 21.Sadler, R. H., and N. Raab-Traub. 1995. Structural analyses of the Epstein-Barr virus BamHI A transcripts. J. Virol. 69:1132-1141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Scholle, F., K. M. Bendt, and N. Raab-Traub. 2000. Epstein-Barr virus LMP2A transforms epithelial cells, inhibits cell differentiation, and activates Akt. J. Virol. 74:10681-10689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Schroder, W., N. Kienzle, G. Bushell, and T. Sculley. 2002. Antiserum raised against the Epstein-Barr virus BARF0 protein reacts to HLA-DR beta chain. Arch. Virol. 147:723-729. [DOI] [PubMed] [Google Scholar]
- 24.Schroeter, E. H., J. A. Kisslinger, and R. Kopan. 1998. Notch-1 signalling requires ligand-induced proteolytic release of intracellular domain. Nature 393:382-386. [DOI] [PubMed] [Google Scholar]
- 25.Smith, P. R., O. de Jesus, D. Turner, M. Hollyoake, C. E. Karstegl, B. E. Griffin, L. Karran, Y. Wang, S. D. Hayward, and P. J. Farrell. 2000. Structure and coding content of CST (BART) family RNAs of Epstein-Barr virus. J. Virol. 74:3082-3092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Struhl, G., and A. Adachi. 1998. Nuclear access and action of notch in vivo. Cell 93:649-660. [DOI] [PubMed] [Google Scholar]
- 27.Tomkinson, B., and E. Kieff. 1992. Use of second-site homologous recombination to demonstrate that Epstein-Barr virus nuclear protein 3B is not important for lymphocyte infection or growth transformation in vitro. J. Virol. 66:2893-2903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Trinh, D. P., K. M. Brown, and K. T. Jeang. 1999. Epithelin/granulin growth factors: extracellular cofactors for HIV-1 and HIV-2 Tat proteins. Biochem. Biophys. Res. Commun. 256:299-306. [DOI] [PubMed] [Google Scholar]
- 29.van Beek, J., A. A. Brink, M. B. Vervoort, M. J. van Zijp, C. J. Meijer, A. J. van den Brule, and J. M. Middeldorp. 2003. In vivo transcription of the Epstein-Barr virus (EBV) BamHI-A region without associated in vivo BARF0 protein expression in multiple EBV-associated disorders. J. Gen. Virol. 84:2647-2659. [DOI] [PubMed] [Google Scholar]
- 30.Weng, A. P., and J. C. Aster. 2004. Multiple niches for Notch in cancer: context is everything. Curr. Opin. Genet. Dev. 14:48-54. [DOI] [PubMed] [Google Scholar]
- 31.Young, L., C. Alfieri, K. Hennessy, H. Evans, C. O'Hara, K. C. Anderson, J. Ritz, R. S. Shapiro, A. Rickinson, E. Kieff, et al. 1989. Expression of Epstein-Barr virus transformation-associated genes in tissues of patients with EBV lymphoproliferative disease. N. Engl. J. Med. 321:1080-1085. [DOI] [PubMed] [Google Scholar]
- 32.Young, T. M., Q. Wang, T. Pe'ery, and M. B. Mathews. 2003. The human I-mfa domain-containing protein, HIC, interacts with cyclin T1 and modulates P-TEFb-dependent transcription. Mol. Cell. Biol. 23:6373-6384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zhang, J., H. Chen, G. Weinmaster, and S. D. Hayward. 2001. Epstein-Barr virus BamHi-a rightward transcript-encoded RPMS protein interacts with the CBF1-associated corepressor CIR to negatively regulate the activity of EBNA2 and NotchIC. J. Virol. 75:2946-2956. [DOI] [PMC free article] [PubMed] [Google Scholar]