Abstract
The herpes simplex virus virion host shutoff protein (vhs) is an mRNA-specific RNase that contributes to shutoff of host protein synthesis. We show here that vhs-induced mRNA decay proceeds 5′ to 3′ in an in vitro assay system derived from rabbit reticulocyte lysate.
The herpes simplex virus (HSV) virion host shutoff protein (vhs) encoded by gene UL41 triggers accelerated turnover of cellular and viral mRNAs, thereby contributing to shutoff of host protein synthesis and timely transitions between the successive phases of viral gene expression (reviewed in reference 15). Although vhs is dispensable for virus replication in established cell lines in tissue culture, vhs mutants are severely impaired in mouse models of HSV infection (17, 18). Mounting evidence indicates that the attenuated phenotype of vhs mutants stems (at least in part) from an impaired ability to disarm elements of the host innate immune response including the type I interferon system (3, 11, 13, 19). Thus, vhs is a key determinant of HSV virulence. vhs displays extensive amino acid sequence similarity to a family of cellular nucleases that are involved in DNA replication and repair (2, 7), and Everly and coworkers have provided compelling genetic and biochemical evidence that vhs has inherent RNase activity (7). It therefore seems plausible that most or all of the effects of vhs on cellular and viral gene expression stem from its RNase activity on target mRNAs. Thus, the mode of RNA decay induced by vhs is of considerable interest.
Previous studies have shown that vhs-induced RNA degradation involves endoribonucleolytic cleavage of the target RNA, at least in in vitro assay systems (4, 5, 21). Remarkably, vhs preferentially targets mRNAs over other cytoplasmic RNA species in vivo (10, 14). Some evidence suggests that this selectivity stems from interactions with the host translation initiation machinery: vhs initially targets the 5′ region and sequences immediately downstream of picornavirus internal ribosome entry sites (IRES elements) on several mRNAs in an in vitro translation system (4, 5), and it renders the 5′ end of HSV tk mRNA less abundant than the 3′ end in vivo (9). In addition, vhs binds host translation initiation factors 4B (eIF4B) and eIF4H (1, 8). Taken in combination, these observations have led to the hypothesis that vhs is initially delivered to mRNAs during the process of translation initiation (8) and then degrades the transcript in an overall 5′ to 3′ direction (9). However, it is important to note that the foregoing studies have not examined the mode of vhs-induced decay in detail beyond mapping the initial cleavage events. In addition, Taddeo and colleagues and Esclatine and colleagues have recently presented evidence that vhs-induced decay of some mRNAs bearing AU-rich instability elements initiates at or close to the 3′ end of the transcript (6, 20). Whether the subsequent decay of these AU-rich mRNAs displays directionality remains to be determined.
To gain further insight into the overall pattern of mRNA decay induced by vhs, we conducted a detailed examination of the fate of a 2.3-kb transcript of pCITE-1 in a vhs-dependent in vitro RNA degradation system derived from rabbit reticulocyte lysate (RRL) (4, 16). The test transcript bears the encephalomyocarditis virus IRES at its 5′ end, followed by encephalomyocarditis virus coding sequences (Fig. 1A). Previous work has demonstrated that the vhs-dependent RNase initiates degradation of pCITE-1 RNA through endoribonucleolytic cleavage immediately 3′ to the IRES (5). The resulting 3′ fragment is then subjected to extensive further degradation. Here we asked if these secondary decay events display detectable overall polarity.
FIG. 1.
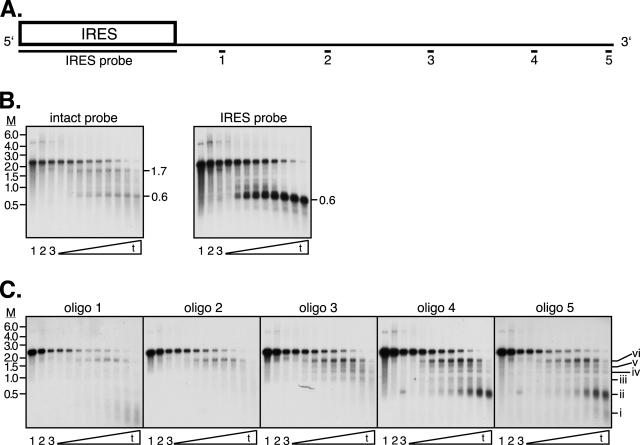
vhs-induced decay of pCITE-1 mRNA. Unlabeled pCITE-1 RNA was added to control RRL or RRL containing pretranslated vhs, and RNA samples recovered at various times were subjected to electrophoresis through a 1% formaldehyde-agarose gel and analyzed by Northern blot hybridization. (A) Diagram of pCITE-1 indicating the location of the IRES element and the locations of the various probes used. (B and C) Northern blot hybridization. Membranes were hybridized to the indicated probes. Lane 1, pCITE-1 RNA; lanes 2 and 3, pCITE-1 incubated with blank RRL for 0 and 20 min, respectively; remaining lanes, pCITE-1 RNA incubated with RRL containing pretranslated vhs for 0, 0.5, 1, 1.5, 2, 3, 5, 10, and 20 min. The mobilities of selected products of vhs action are indicated on the right of several panels. M, marker RNA sizes in kilobases.
Unlabeled pCITE-1 RNA prepared by in vitro transcription of EcoNI-linearized pCITE-1 (1, 5) was added to RRL (Promega) containing pretranslated vhs, and the reaction mixture was incubated at 30°C as previously described (1, 4, 5). Aliquots were removed after 0, 0.5, 1, 1.5, 2, 3, 5, 10, and 20 min, and RNA was extracted using TRIzol reagent (Invitrogen) following the manufacturer's protocol. Samples were then subjected to electrophoresis through duplicate 1% formaldehyde-agarose gels and transferred to nylon membranes. Following UV cross-linking, membranes were sequentially hybridized to 32P-labeled probes complementary to various segments of pCITE-1 RNA to visualize the course of the reaction and characterize the degradation intermediates (Fig. 1).
The data obtained with the full-length and IRES probes confirmed that the vhs-induced degradation reaction proceeded as previously described. Thus, the full-length probe hybridized to the intact 2.3-kb transcript and to two major cleavage products of ca. 0.6 and 1.7 kb which correspond to the 5′ and 3′ products of IRES-directed cleavage, respectively (Fig. 1B). As predicted, the 0.6-kb fragment hybridized to the IRES probe but failed to react with oligonucleotide probes 1 to 5 (Fig. 1B and C); conversely, the 1.7-kb fragment hybridized to oligonucleotide probes 1 to 5 (Fig. 1C) but not to the IRES probe (Fig. 1B). As observed previously, the 0.6-kb IRES-bearing fragment was stable throughout the course of the reaction, while the 1.7-kb 3′ fragment produced at early times decayed later during the reaction.
We used five oligonucleotide probes to monitor the mode of decay of the 1.7-kb 3′ fragment. These probes were designed to detect sequences spanning the length of the 1.7-kb fragment (Fig. 1A; Table 1). Probe 5 is complementary to residues located at the extreme 3′ end of the pCITE-1 transcript, and as such it indirectly end labels all of the degradation intermediates that include the 3′ end of the RNA. As shown in Fig. 1C, probe 5 detected intact pCITE-1 RNA and six discrete degradation intermediates (i to vi) that ranged in size from ca. 200 nucleotides (nt) (i) to the 1.7-kb fragment (vi) produced by IRES-directed cleavage. We have previously demonstrated that fragment vi is produced by endo- rather than exo-RNase action, as shown by the simultaneous production of matching 5′ and 3′ products (5). If this is also the case for the smaller fragments i to v, then the data obtained with probe 5 map the positions of additional sites of endonucleolytic cleavage in the 1.7-kb fragment; alternatively, if decay of the 1.7-kb fragment is mediated via exonucleolytic action, then the sizes of the products map preferred pause sites of a 5′ → 3′ exonuclease. In either case, the fragments detected by probe 5 represent a nested set of 3′ coterminal degradation intermediates. The larger decay products detected by probe 5 declined in abundance by the latest time points analyzed, while the smallest (i and ii) accumulated late during the reaction. A large fraction of the hybridization signal obtained with probe 5 was retained even at the latest time point, a finding that excludes a major role for 3′ → 5′ exonucleases in the overall decay process. Taken in combination, these observations provided a strong indication that the RNA degradation intermediates detected by probe 5 are generated via a process that exhibits overall 5′ → 3′ polarity.
TABLE 1.
Probes used in this study
| Oligonucleotide probe | Sequence | pCITE-1 RNA residues |
|---|---|---|
| 1 | GCCCTGTAACTCGAAAACGACTTCC | 771-795 |
| 2 | CCTTGAAGGTGTAGTACCTTTCCAC | 1177-1201 |
| 3 | GTTTACATATGGCACCTCAAGATCC | 1575-1599 |
| 4 | GTGCGGGACGGCGTTACCAGAAACT | 1973-1997 |
| 5 | GCAGCCCAGTAGTAGGTTGAGGCCG | 2260-2284 |
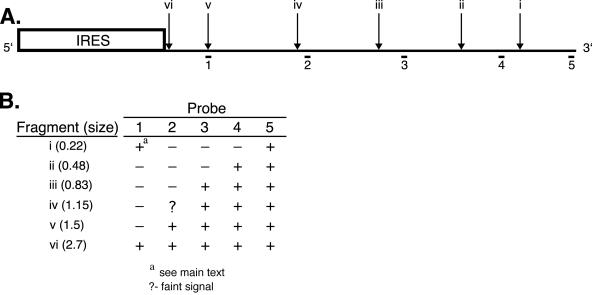
Figure 2 depicts the approximate locations of the 5′ ends of fragments i to vi detected by probe 5 and relates these sites to the sequences represented by oligonucleotide probes 1 to 5. If, as argued above, the 1.7-kb fragment is degraded 5′ → 3′, then one predicts that probes for progressively more 5′ sequences would illuminate ever more restricted subsets of the fragments detected by probe 5. Moreover, these 3′ coterminal fragments should comprise a major fraction of the degradation intermediates detected with each probe. As shown in Fig. 1C and summarized in Fig. 2, the data obtained with oligonucleotide probes 1 to 4 provided strong confirmation of these predictions. Thus, probe 4 illuminated bands that comigrate with fragments ii to vi and probe 3 detected bands that comigrate with fragments iii to iv (data summarized in Fig. 2). Along the same lines, probe 2 clearly detected bands corresponding in size to fragments v and vi and possibly also reacted with fragment iv. Finally, probe 1 detected a band of the mobility of fragment vi as well as a broad smear corresponding to fragments of 200 to 300 nt. We note that although this latter smear overlaps the mobility of fragment i detected by probe 5, it cannot represent the same degradation intermediate as fragment i.
FIG. 2.
Summary of the vhs degradation products detected. The structure of pCITE-1 RNA is diagrammed as in Fig. 1, indicating the locations of the 5′ ends of the RNA fragments i to vi detected by oligonucleotide probe 5. The lower portion of the figure summarizes which of these RNA fragments are detected by each of the oligonucleotide probes in Fig. 1C.
A striking feature of these data is that, with the exception of the 200- to 300-nt fragments detected by probe 1, all of the degradation intermediates detected by probes 1 to 4 correspond to 3′ coterminal fragments detected by probe 5 (summarized in Fig. 2). That is, all of the oligonucleotide probes detected members of the same nested set of 3′ coterminal degradation intermediates, and with one exception, no additional prominent degradation products were observed. Taken in combination, these data provide a compelling demonstration that the 1.7-kb product of IRES-directed cleavage is degraded 5′ → 3′.
The 1.7-kb fragment is itself generated by endonucleolytic cleavage (5), and the vhs-dependent RNase also acts endonucleolytically on an unrelated RNA that lacks an IRES in the RRL assay system (4). It is therefore plausible that the 1.7-kb fragment is degraded via repeated vhs-dependent endonucleolytic cleavage events that proceed 5′ → 3′ across the RNA. For example, the fragment may be subjected to multiple additional cleavage cycles in which the nuclease loads onto the substrate in a 5′ end-directed fashion, tracks to preferred cleavage sites, cleaves the RNA, and is released. Another possibility is that, once loaded onto the substrate via IRES-directed targeting, the vhs endonuclease tracks processively along the transcript, remaining attached to the 3′ fragment produced by each cleavage event. This latter model is attractive, as the initial substrate targeting event would suffice for complete degradation of the mRNA. Alternatively, it is possible that the 1.7-kb fragment is processed by cellular 5′ → 3′ exonucleases rather than by vhs itself. Although this latter scenario may at first glance appear inconsistent with our previous finding that the vhs1 point mutation (Thr 214 → Ile) severely reduces the rate of decay of the 1.7-kb fragment (12), it is possible that the mutant vhs protein interferes with the activity of a required cellular exonuclease, for example by remaining tightly bound to the 1.7-kb fragment after the first endonucleolytic cleavage event. Further experiments are required to distinguish among these possibilities.
As shown in Fig. 2, the 5′ ends of the degradation intermediates detected in this study are spaced at ca. 200- to 400-nt intervals across the 1.7-kb fragment. However, we were unable to detect appreciable quantities of 200- to 400-nt cleavage products with probe 2, 3, or 4 (Fig. 1C). If the 1.7-kb fragment is indeed processed via endo- rather than exonucleolytic cleavage, we can think of two possible explanations for this apparent discrepancy. Perhaps the predicted 200- to 400-nt fragments are rapidly degraded by other nucleases present in the RRL assay system. Alternatively, the sites displayed in Fig. 2 may mark the positions of sequences that serve as kinetic barriers to 5′ → 3′ tracking of the nuclease (i.e., pause sites), rather than preferred cleavage sites per se. According to this model, the endonuclease cleaves the 1.7-kb fragment at many sites in addition to those diagrammed in Fig. 2, but the resulting products are rapidly recleaved and therefore do not accumulate.
Our finding that vhs-induced degradation of pCITE-1 RNA displays overall 5′ → 3′ polarity is consistent with the in vivo data obtained by Karr and Read in studies of HSV tk mRNA (9). However, our data are not as obviously consistent with the suggestion by Taddeo and colleagues and Esclatine and colleagues that vhs-induced decay of cellular IEX-1 mRNA and certain other transcripts bearing AU-rich instability elements starts at or close to the 3′ end of the RNA (6, 20). Perhaps vhs employs multiple mechanisms to induce mRNA turnover, as suggested by these authors. Alternatively, it is possible that the vhs-dependent nuclease is selectively targeted to the 3′ untranslated region of IEX-1 mRNA via interactions with host components, cleaves at this position, and degrades the resulting 3′ fragment in the 5′ → 3′ fashion that we describe here. Further studies are required to resolve these and other questions regarding the mechanism of vhs action.
Acknowledgments
We thank Rob Maranchuk for technical assistance.
This work was supported by an operating grant from the Canadian Institutes for Health Research.
REFERENCES
- 1.Doepker, R. C., W. L. Hsu, H. A. Saffran, and J. R. Smiley. 2004. Herpes simplex virus virion host shutoff protein is stimulated by translation initiation factors eIF4B and eIF4H. J. Virol. 78:4684-4699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Doherty, A. J., L. C. Serpell, and C. P. Ponting. 1996. The helix-hairpin-helix DNA-binding motif: a structural basis for non-sequence-specific recognition of DNA. Nucleic Acids Res. 24:2488-2497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Duerst, R. J., and L. A. Morrison. 2004. Herpes simplex virus 2 virion host shutoff protein interferes with type I interferon production and responsiveness. Virology 322:158-167. [DOI] [PubMed] [Google Scholar]
- 4.Elgadi, M. M., C. E. Hayes, and J. R. Smiley. 1999. The herpes simplex virus vhs protein induces endoribonucleolytic cleavage of target RNAs in cell extracts. J. Virol. 73:7153-7164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Elgadi, M. M., and J. R. Smiley. 1999. Picornavirus internal ribosome entry site elements target RNA cleavage events induced by the herpes simplex virus virion host shutoff protein. J. Virol. 73:9222-9231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Esclatine, A., B. Taddeo, L. Evans, and B. Roizman. 2004. The herpes simplex virus 1 UL41 gene-dependent destabilization of cellular RNAs is selective and may be sequence-specific. Proc. Natl. Acad. Sci. USA 101:3603-3608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Everly, D. N. J., P. Feng, I. S. Mian, and G. S. Read. 2002. mRNA degradation by the virion host shutoff (Vhs) protein of herpes simplex virus: genetic and biochemical evidence that Vhs is a nuclease. J. Virol. 76:8560-8571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Feng, P., D. N. J. Everly, and G. S. Read. 2001. mRNA decay during herpesvirus infections: interaction between a putative viral nuclease and a cellular translation factor. J. Virol. 75:10272-10280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Karr, B. M., and G. S. Read. 1999. The virion host shutoff function of herpes simplex virus degrades the 5′ end of a target mRNA before the 3′ end. Virology 264:195-204. [DOI] [PubMed] [Google Scholar]
- 10.Kwong, A. D., and N. Frenkel. 1987. Herpes simplex virus-infected cells contain a function(s) that destabilizes both host and viral mRNAs. Proc. Natl. Acad. Sci. USA 84:1926-1930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Leib, D. A., T. E. Harrison, K. M. Laslo, M. A. Machalek, N. J. Moorman, and H. W. Virgin. 1999. Interferons regulate the phenotypes of wild-type and mutant herpes simplex viruses in vivo. J. Exp. Med. 189:663-672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lu, P., H. A. Saffran, and J. R. Smiley. 2001. The vhs1 mutant form of herpes simplex virus virion host shutoff protein retains significant internal ribosome entry site-directed RNA cleavage activity. J. Virol. 75:1072-1076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Murphy, J. A., R. J. Duerst, T. J. Smith, and L. A. Morrison. 2003. Herpes simplex virus type 2 virion host shutoff protein regulates alpha/beta interferon but not adaptive immune responses during primary infection in vivo. J. Virol. 77:9337-9345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Oroskar, A. A., and G. S. Read. 1989. Control of mRNA stability by the virion host shutoff function of herpes simplex virus. J. Virol. 63:1897-1906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Smiley, J. R. 2004. Herpes simplex virus virion host shutoff protein: immune evasion mediated by a viral RNase? J. Virol. 78:1063-1068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Smiley, J. R., M. M. Elgadi, and H. A. Saffran. 2001. Herpes simplex virus vhs protein. Methods Enzymol. 342:440-451. [DOI] [PubMed] [Google Scholar]
- 17.Smith, T. J., L. A. Morrison, and D. A. Leib. 2002. Pathogenesis of herpes simplex virus type 2 virion host shutoff (vhs) mutants. J. Virol. 76:2054-2061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Strelow, L. I., and D. A. Leib. 1995. Role of the virion host shutoff (vhs) of herpes simplex virus type 1 in latency and pathogenesis. J. Virol. 69:6779-6786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Suzutani, T., M. Nagamine, T. Shibaki, M. Ogasawara, I. Yoshida, T. Daikoku, Y. Nishiyama, and M. Azuma. 2000. The role of the UL41 gene of herpes simplex virus type 1 in evasion of non-specific host defence mechanisms during primary infection. J. Gen. Virol. 81:1763-1771. [DOI] [PubMed] [Google Scholar]
- 20.Taddeo, B., A. Esclatine, W. Zhang, and B. Roizman. 2003. The stress-inducible immediate-early responsive gene IEX-1 is activated in cells infected with herpes simplex virus 1, but several viral mechanisms, including 3′ degradation of its RNA, preclude expression of the gene. J. Virol. 77:6178-6187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zelus, B. D., R. S. Stewart, and J. Ross. 1996. The virion host shutoff protein of herpes simplex virus type 1: messenger ribonucleolytic activity in vitro. J. Virol. 70:2411-2419. [DOI] [PMC free article] [PubMed] [Google Scholar]