Abstract
We have previously described the RNA replication properties of poliovirus transcripts harboring chimeric RNA polymerase sequences representing suballelic exchanges between poliovirus type 1 (PV1) and coxsackievirus B3 (CVB3) utilizing an in vitro translation and RNA replication assay (C. Cornell, R. Perera, J. E. Brunner, and B. L. Semler, J. Virol. 78:4397-4407, 2004). We showed that three of the seven chimeras were capable of RNA replication in vitro, although replication levels were greatly reduced compared to that of wild-type transcripts. Interestingly, one of the replication-competent transcripts displayed a strand-specific RNA synthesis defect suggesting (i) a differential replication complex assembly mechanism involving 3D and/or precursor molecules (i.e., 3CD) required for negative- versus positive-strand RNA synthesis or (ii) effect(s) on the ability of the 3D polymerase to form higher-ordered structures required for positive-strand RNA synthesis. In this study, we have attempted to rescue defective RNA replication in vitro by cotranslating nonstructural proteins from a transcript encoding a large precursor polyprotein (P3) to complement 3D polymerase and/or precursor polypeptide functions altered in each of the chimeric constructs. Utilization of a wild-type P3 construct revealed that all transcripts containing chimeric PV1/CVB3 polymerase sequences can be complemented in trans for both negative- and positive-strand RNA synthesis. Furthermore, data from experiments utilizing genetically modified forms of the P3 polyprotein, containing mutations within 3C or 3D sequences, strongly suggest the existence of different protein-protein and protein-RNA interactions required for positive- versus negative-strand RNA synthesis. These results, combined with data from in vitro RNA elongation assays, indicate that the delivery of active 3D RNA polymerase to replication complexes requires a series of macromolecular interactions that rely on the presence of specific 3D amino acid sequences.
The intracellular replication cycles of picornaviruses involve multiple gene products of viral and host origins to alter the intracellular environment to make it suitable for rapid genome amplification and virion production. This process results in the shutdown of protein synthesis (21, 39) and host transcription (57, 59-61), a rearrangement and compartmentalization of host cell membranes (8, 13, 50), an alteration of nuclear import-export and membrane trafficking (18, 19, 23), and the inhibition of major histocompatibility complex class I expression on the cell surface (16). To achieve such complex functions, viral proteins and precursor polypeptides (Fig. 1A) interact with themselves or host proteins to mediate different, highly specific functions in the viral life cycle (54).
FIG. 1.
(A) Processing map of poliovirus. Shown is a schematic of the VPg-linked genome of PV1. Regions encoding both capsid (structural) and noncapsid proteins are indicated, with nucleotide numbers given. The 247-kDa viral polyprotein is shown, with sites of proteolytic cleavage and precursor or mature cleavage products indicated (with estimated molecular masses). The diamond indicates the primary cleavage site liberating the P2-P3 polypeptide from the P1 polyprotein. Triangles indicate Q-G dipeptides cleaved by the 3C and 3CD proteinases, and the star indicates a cleavage necessary for virion maturation that occurs by an undefined mechanism. (B) Schematic of 5′ ribozyme PV1 cDNA (RzPV1) and PV1/CVB3 chimeras generated in this study. The schematic indicates the genetic organization of RzPV1, which allows in vitro transcription by bacteriophage T7 RNA polymerase and the production of a full-length PV1 transcript with a precise 5′ end. Below the cDNA schematic are the eight transcripts (wild type and chimeric) generated for in vitro translation and RNA replication studies presented in this work. (C) Schematic of pT7-5′-NCR-P3 transcripts utilized for in vitro RNA replication rescue experiments. These constructs produce T7-derived, polyadenylated transcripts possessing the P3 (wild-type or mutant) coding region translated by the PV1 internal ribosome entry site and were previously described by Towner et al. (53). When present, the hatched region in each schematic indicates the location of the described point mutation.
Poliovirus type 1 (PV1), the prototypic member of the family Picornaviridae, possesses a positive-sense single-stranded RNA (ssRNA) genome of ∼7.4 kb containing one open reading frame from which a 247-kDa viral polyprotein is translated by a cap-independent mechanism (internal ribosome entry). This viral polyprotein is processed by the two viral proteinases 2A and 3C (and the 3CD precursor polypeptide) (for a review, see reference 37). The 2A proteinase is responsible for cleavage between the structural precursor and the nonstructural precursors and cleaves host factor eukaryotic initiation factor 4G, resulting in the shutdown of cap-dependent host protein synthesis (10, 34). The majority of the processing events within the viral polyprotein are carried out by the 3C and/or 3CD proteinases (24, 49). It has been shown that the presence of 3D polymerase sequences in the context of the 3CD polypeptide enhances the ability of 3C to cleave the capsid (P1) precursor protein (42, 62), centering on recognition and cleavage of the VP0-VP3 junction within P1 (32, 42).
Due to the limited number of genes contained within a typical picornavirus genome, some precursor polypeptides have biochemical activities that differ greatly from those of their mature cleavage products. In addition, several precursor and mature cleavage products have been shown to be multifunctional. For example, the poliovirus 3CD polypeptide, a polyprotein containing the amino acid sequences of the 3C proteinase and the 3D RNA-dependent RNA polymerase, is an active proteinase that does not possess measurable RNA synthesis activity in vitro (25, 55). In addition to proteolytic activity, the 3C and 3CD proteins contain determinants that mediate binding to viral RNA (1, 2, 9, 43). 3CD has been shown to interact with the 5′ and 3′ noncoding regions of the poliovirus genome (2, 26) and could serve as a source of 3D polymerase within replication complexes utilized for RNA synthesis. Furthermore, the 3CD polypeptide has been shown to dramatically stimulate the process of VPg uridylylation, which may involve protein-protein and protein-RNA contacts between 3CD and the 3D polymerase (which catalyzes the reaction) (46), VPg (or the 3AB precursor polypeptide), and the cis-acting replication element within 2C (2C-cre) (45, 48). 3CD could have additional roles critical for RNA synthesis initiation that involve contacts with host proteins poly(rC) binding protein 2 (PCBP2) and poly(A) binding protein (28). This has been hypothesized to mediate long-range contacts within the genomic RNA, which is then utilized as a template for negative-strand RNA synthesis (5, 28).
In infected cells, poliovirus RNA synthesis may require the formation of replication complexes containing viral proteins that have been translated from the RNA around which they assemble (22, 52). Novak and Kirkegaard demonstrated a coupling between translation and RNA replication by showing that viral genomes not actively translating are not utilized as templates for RNA synthesis, even in the presence of fully replication-competent viral proteins translated from other genomes in the same cell (41). Membranous vesicle formation, providing a platform on which RNA synthesis occurs, has been shown to be coupled to viral RNA replication, translation, and encapsidation (20). The linkage of protein synthesis to RNA replication could provide a mechanism by which poliovirus, and likely other picornaviruses, amplify only those RNA molecules that yield fully active viral proteins to ensure the production of progeny virions that contain viable genomes.
The requirement for replication proteins in cis is not absolute, as several groups have successfully trans complemented the replication of RNAs containing one or more mutations within various nonstructural gene products. To date, it has been shown that mutations in 2A, 2C, 3A, 3B (VPg), and 3C coding regions can be complemented in trans, whereas lesions within 2B cannot (7, 11, 17, 31, 38, 51, 53). Interestingly, the 3D RNA polymerase gene is differentially responsive to trans rescue, depending on the lesion present (7, 12, 51).
Using an in vitro translation and RNA replication assay (3, 4, 40), our laboratory found that efficient complementation of a 3AB lesion required a cotranslating RNA that expressed the entire P3 precursor protein, whereas processed wild-type (wt) 3AB alone was not sufficient to rescue RNA replication in trans (53). This finding demonstrated that early steps in the assembly of RNA replication complexes utilize gene products in their precursor forms, with complete proteolysis of these precursors occurring prior to RNA synthesis initiation. A recent report suggests that proteins within the P2 region of the genome also assemble in precursor forms (33).
Studies aimed at analyzing the role of the 3D polymerase domain of the poliovirus 3CD polypeptide in modulating 3CD biochemical activities have been previously described (6, 15, 42). Chimeric recombinant proteins with suballelic exchanges of polymerase subdomain sequences between PV1 and coxsackievirus B3 (CVB3) were utilized to understand the RNA binding and protein-processing activities of 3CD (15). The present study analyzed the effects of these 3D mutations on viral RNA replication in vitro, providing not only an indication of polymerase activity but more importantly the ability of the 3D polymerase and polymerase precursor polypeptides to assemble into functional complexes required for RNA synthesis. Three of the seven chimeric constructs showed detectable levels of RNA synthesis in vitro; however, not all constructs were capable of both negative- and positive-strand RNA synthesis. Although in vitro 3D elongation assays confirmed that several of the polymerases themselves are somewhat defective at the level of RNA chain elongation, complementation experiments utilizing wild-type P3 precursor protein provided in trans demonstrated that all of our chimeric transcripts are responsive to a rescue of both negative- and positive-strand RNA synthesis. Additional experiments utilizing mutated forms of P3 revealed differences in macromolecular interactions (protein-protein and protein-RNA) between proteins in cis and those in trans.
MATERIALS AND METHODS
Plasmids and cloning.
Chimeric PV1/CVB3 polymerase sequences were originally cloned as luciferase replicon constructs (14) based on a modified version of a previously published plasmid, pRib(+)RLuc (27). pRib(+)RLuc was first digested with BglII and MluI in the presence of alkaline phosphatase (Promega). The ∼8.0-kb vector fragment was gel purified and incubated with the ∼1.8-kb fragment from an equivalent digest of pT7PV1(MluI) (56) in the presence of T4 DNA ligase to generate pRib(+)RLucM. This construct was then redigested with BglII and MluI in the presence of alkaline phosphatase (Promega), and the ∼8.0-kb vector fragment was gel purified. The vector fragment was incubated in a four-fragment ligation with the BglII-BsaI fragment from pT7PV1(MluI); the BsaI-MlyI fragment from pET15b-3CD(PCP), pET15b-3CD(PCC), pET15b-3CD(CCP), or pET15b-3CD(CCC); and the MlyI-MluI fragment from pRib(+)RLucM to generate the PCP, PCC, CCP, and CCC versions of pRib(+)RLucM. To generate the CPP, PPC, and CPC versions of this construct, four-fragment ligations were assembled with the same vector fragments from pRib(+)RLucM and pT7PV1(MluI); the BsaI-XcmI fragment from pET15b-3CD(CPP), pET15b-3CD(PPC), or pET15b-3CD(CPC); and the XcmI-MluI fragment from pRib(+)RLucM. Once luciferase replicon constructs harboring the PV1/CVB3 chimeric sequences were generated, full-length constructs were made by cloning the capsid coding sequences in place of luciferase. To accomplish this, the chimeric pRib(+)RLucM plasmids were digested with AgeI and BglII, and the ∼5.5-kb vector fragment was then ligated into the corresponding AgeI-BglII fragment from pT7PV1(MluI). All plasmids generated contained a 5′ hammerhead ribozyme sequence immediately upstream of the PV1 sequence and either wild-type [RzPV1(wt)] or chimeric 3D ribozyme PV1 cDNA (RzPV1) polymerase sequences [e.g., RzPV1(CPP)]. All constructs were verified by restriction enzyme digests and nucleotide sequencing (Biotech Diagnostic, Laguna Niguel, Calif.).
pT7-5′-NCR-P3(wt), pT7-5′-NCR-P3(C147A), pT7-5′-NCR-P3(μ61), pT7-5′-NCR-P3(R13N), and pT7-5′-NCR-P3(Y6N) have been previously described (9, 53).
In vitro synthesis of chimeric and mutated transcripts.
To generate templates for in vitro transcriptions, full-length PV1/CVB3 chimeric constructs (RzPV1) were linearized with MluI, whereas pT7-5′-NCR-P3 plasmids were linearized with AatII followed by end-fill repair with the Klenow fragment of DNA polymerase I. Each linearized template was phenol-chloroform extracted, ethanol precipitated, washed with 70% ethanol, and resuspended in diethylpyrocarbonate-treated water. In vitro transcription reactions were carried out by using bacteriophage T7 RNA polymerase at 37°C in a total volume of 20 μl. Following incubation for 1 h, reactions were treated with 2 U of DNase and incubated for an additional 15 min at 37°C. Reactions were quenched with the addition of 375 μl of sodium dodecyl sulfate stop buffer (0.5% sodium dodecyl sulfate, 100 mM NaCl, 10 mM Tris-HCl [pH 7.5], 1 mM EDTA) and 100 μg of predigested proteinase K. This mixture was incubated at 37°C for an additional 30 min, phenol-chloroform extracted, and ethanol precipitated. RNA pellets were washed with 70% ethanol, dried, and resuspended in diethylpyrocarbonate-treated water. All transcripts were quantitated on an ethidium bromide-stained agarose gel by using transcript RNA of the same length and known quantity as a standard.
In vitro translation and RNA replication reactions.
For translation-RNA replication experiments, 50-μl reaction mixtures contained 65% (vol/vol) HeLa S10 cytoplasmic extract, 1.0 μg of wild-type RzPV1 or chimeric RzPV1 RNA, 10% (vol/vol) of 10× replication mix (10 mM ATP, 2.5 mM GTP, 2.5 mM UTP, 600 mM potassium acetate, 300 mM creatine phosphate [Boehringer Mannheim], 4 mg of creatine kinase [Boehringer Mannheim] per ml, 155 mM HEPES-KOH [pH 7.4]), and 2 mM guanidine hydrochloride. Cotranslation reaction mixtures contained 1.0 μg of full-length RzPV1 RNA plus an equimolar amount (400 ng) of 5′ noncoding region (5′NCR)-P3 RNA. For translation analysis, 10 μl of this reaction mixture was added to 10 μCi of [35S]methionine (>1,000 Ci/mmol; Amersham Pharmacia Biotech). The remaining 40 μl was used for RNA replication analysis. Both reaction mixtures were incubated at 30°C for 5 h, at which time 10 μl of 2× Laemmli sample buffer (35) was added to each translation reaction mixture. The samples were boiled and resolved on a 12.5% polyacrylamide gel containing sodium dodecyl sulfate, fluorographed, and subjected to autoradiography on X-MR film (Kodak). The 40-μl RNA replication reaction mixtures were subjected to centrifugation for 20 min at 15,000 × g at 4°C, and the supernatants were removed. Pellets containing replication complexes were resuspended in 9 μl of fresh HeLa S10 cytoplasmic extract, 1.3 μl of 10× replication mix, and 2.5 μl (25 μCi) of [α-32P]CTP (3,000 Ci/mmol; Amersham Pharmacia Biotech) and incubated for 2 h at 34°C. Following incubation, total RNA in each reaction was isolated by RNeasy spin column purification (QIAGEN), subjected to a final ammonium acetate precipitation, washed with 70% ethanol, resuspended in 10 μl of diethylpyrocarbonate-treated water-RNA loading buffer (Ambion), and subjected to gel electrophoresis on a native 1% agarose Tris-borate EDTA gel containing ethidium bromide. The levels of 18S and 28S rRNA present in each lane were used to confirm equal loading of samples before the gel was dried and subjected to PhosphorImager analysis on a Personal Molecular Imager FX (Bio-Rad).
3Dpol in vitro elongation assays.
For in vitro 3Dpol elongation assays, we first carried out in vitro translations with slight modifications of previously published procedures (53). Translation reaction mixtures (25 μl) containing 60% (vol/vol) HeLa S10 cytoplasmic extract, 5% (vol/vol) HeLa ribosomal salt wash, 12 U of RNasin, 10% (vol/vol) 10× replication mix, and 0.7 μg of wild-type RzPV1 or chimeric RzPV1 RNA were assembled. Following incubation at 32°C for 6 h, 3 μl of each translation mixture was used in a subsequent elongation reaction mixture consisting of 10% (vol/vol) 10× elongation buffer (50 mM HEPES-KOH [pH 8.0]; 10 mM dithiothreitol; 3 mM MgCl2; 300 mM each ATP, GTP, and UTP), 2 μg of PV1 virion RNA, 30 ng of oligo(U) (kindly provided by Joan Morasco and James B. Flanegan, University of Florida), and 10 μCi of [α-32P]CTP adjusted to 5 μM with unlabeled CTP in a final volume of 30 μl. The reaction mixtures were incubated at 30°C for 0, 15, 30, and 60 min, and 4 μl was spotted onto a DE 81 paper circle (Whatman). The circles were washed three times in 5% dibasic sodium phosphate buffer, washed once in distilled water, rinsed in methanol, and air dried. Incorporation of labeled nucleotides was measured by liquid scintillation counting, and the counts above background were calculated for each sample.
RESULTS AND DISCUSSION
Infectivity analysis of chimeric PV1/CVB3 transcripts.
Chimeric PV1/CVB3 RNA polymerase sequences were cloned into a full-length PV1 cDNA containing a 5′ hammerhead ribozyme sequence (Fig. 1B). Inclusion of the 5′ hammerhead ribozyme sequence allows the generation of transcripts from these constructs with precise 5′ ends, previously shown by Herold and Andino to significantly increase the ability of PV1 transcripts to synthesize positive-strand RNA (27). Linearization of these plasmids with MluI followed by in vitro transcription with bacteriophage T7 RNA polymerase generated eight different RNA transcripts (Fig. 1B). We first examined the ability of each transcript RNA to yield infectious virus in cell culture at 33 or 37°C by transfection of HeLa cell monolayers. Whereas the wild-type transcript resulted in complete cytopathic effects at approximately 18 h posttransfection, none of the chimeric transcripts yielded cytopathic effects or detectable virus even after harvesting and freeze thawing the monolayers as late as ∼4 days posttransfection (data not shown).
Two biochemical properties (RNA binding and protein processing) of 3CD polypeptides harboring these same PV1/CVB3 chimeric polymerase subdomains have been previously reported (15). It was shown that for several of the chimeras, RNA binding and protein processing occurred with an efficiency equal to or better than that of a wild-type 3CD polypeptide. Thus, the lack of infectivity of the full-length chimeric transcripts suggests that we have disrupted other 3D polymerase or 3CD polypeptide interactions critical for RNA synthesis and/or virion production.
Translation and RNA replication of PV1/CVB3 transcripts with and without P3(wt) RNA.
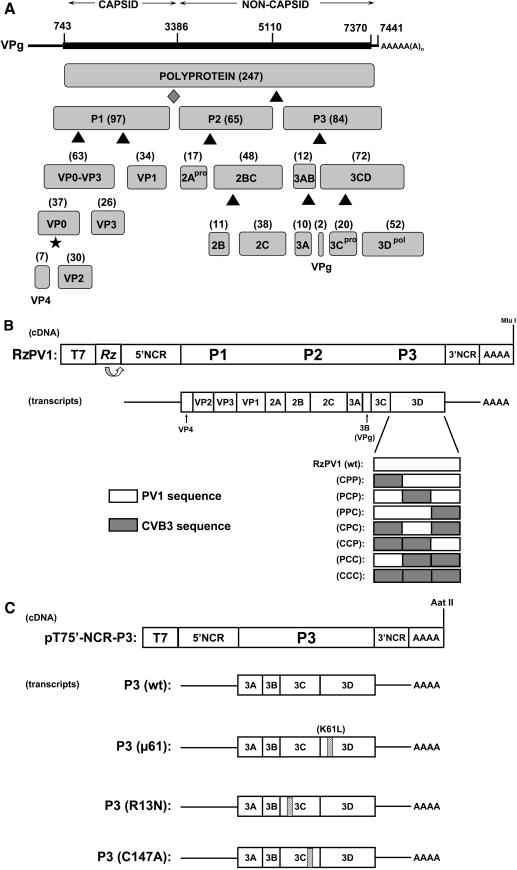
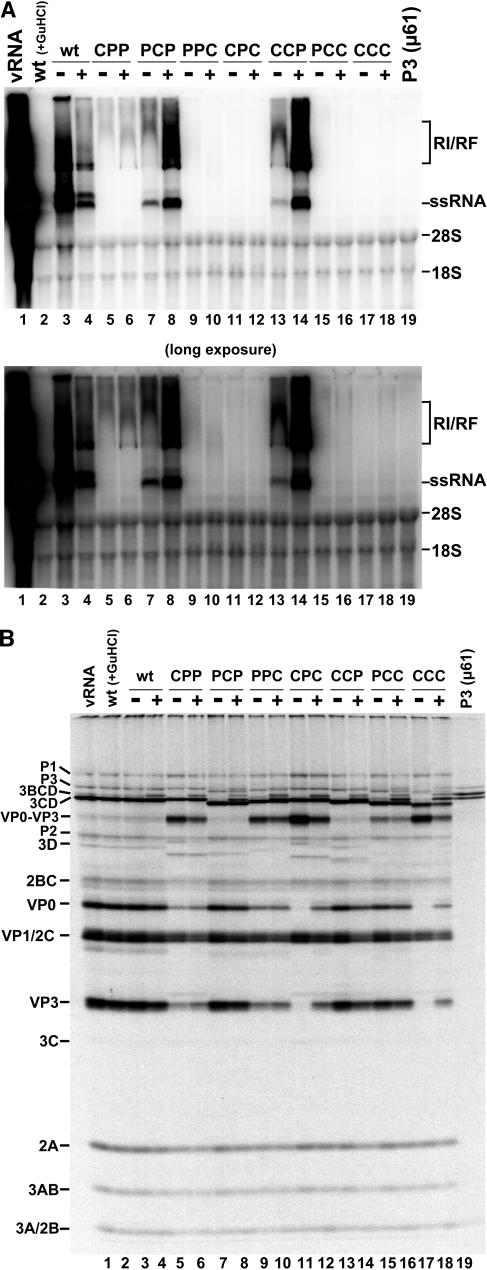
In a previous study, Cornell et al. analyzed the ability of chimeric transcripts to replicate autonomously in an in vitro translation and RNA replication assay (14). In agreement with those results, Fig. 2 shows that the CPP chimera synthesizes only very low levels of negative-strand RNA in the absence of positive-strand RNA synthesis (Fig. 2A, lane 5). Two other chimeras (PCP and CCP) showed both negative- and positive-strand RNA synthesis, albeit at much lower levels compared to replication of wild-type PV1 RNA (Fig. 2A, compare lanes 7 and 13 to lane 3). A positive-strand RNA signal for CCP becomes evident only with an extremely long exposure of the gel shown in Fig. 2B (data not shown) and is apparent in results presented later in this report (e.g., see Fig. 4A, lane 13). The other four chimeras (PPC, CPC, PCC, and CCC) showed no detectable levels of RNA synthesis (Fig. 2A, lanes 9, 11, 15, and 17). These results demonstrate differences in the ability of each chimeric 3D and/or 3CD polypeptide to effectively carry out one or more functions necessary for RNA replication in vitro. Furthermore, since transfected PCP and CCP transcripts do not yield virus in HeLa cell monolayers, there is likely an inability of these chimeras to generate sufficient threshold amounts of positive-strand RNA in cells or perhaps a packaging or assembly defect preventing the production of new virions.
FIG. 2.
Rescue of RNA replication by all chimeras in the presence of 5′-NCR-P3(wt)-cotranslating RNA. (A) [α-32P]-labeled RNA replication reactions were programmed with 1.0 μg of full-length wild-type or chimeric RzPV1 transcript RNA as described in Materials and Methods. The plus signs indicate reactions that were programmed withan equimolar (∼0.4 μg) amount of 5′-NCR-P3(wt) RNA. The figure displays an autoradiograph of a nondenaturing agarose gel. Lane 1 shows a positive control utilizing RNA purified from poliovirus virions (viral RNA). Lane 2 is a negative control in which the wild-type RzPV1 replication reaction was supplemented with 2 mM guanidine hydrochloride (GuHCl). The identity of the full-length RzPV1 transcript RNA utilized in each reaction is indicated above each lane. Lane 19 is a control reaction utilizing ∼0.4 μg of 5′-NCR-P3(wt) transcript alone. The mobilities (visualized by ethidium bromide staining) of ssRNA and the 28S and 18S rRNAs are indicated to the right of the panel. The RNA species migrating slightly slower than the ssRNA species in some lanes (e.g., lanes 1 and 3) in this and other subsequent figures most likely represents a variant isoform of full-length viral RNA. Also shown is the predicted mobility of the RI/RF, corresponding in part to negative-strand RNA synthesis. The autoradiogram shown below is a longer exposure of the same gel. (B) [35S]methionine-labeled translations from the corresponding reactions shown in panel A. Full-length wild-type or chimeric RzPV1 transcripts were incubated in the presence and absence of 5′-NCR-P3(wt) rescue RNA. The identities of the precursor and mature cleavage products generated during translation and processing in vitro are indicated on the left side of the panel. Note the different mobilities of the chimeric 3CD and 3D polypeptides in each lane due to the substitution of PV1 amino acids with those from CVB3. Lane 19 shows the products from the reaction programmed with 5′-NCR-P3(wt) RNA alone (full-length P3, 3BCD, and 3CD precursor polypeptides, respectively). A longer exposure of this gel reveals the presence of mature 3A protein (data not shown).
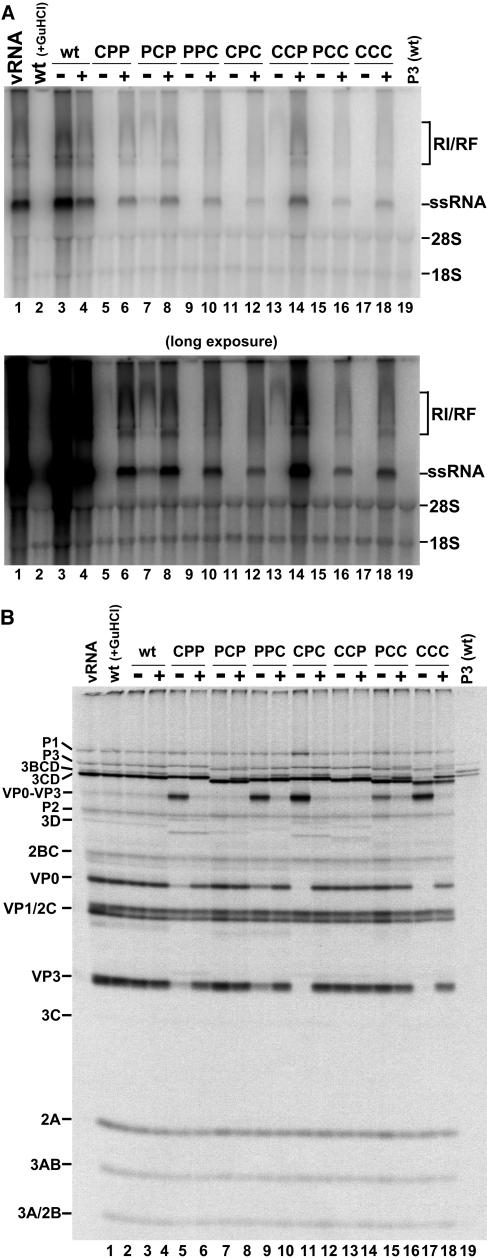
FIG. 4.
RNA replication of wild-type and chimeric transcripts with and without 5′-NCR-P3(C147A)-cotranslating RNA. (A) RNA replication carried out as described in the legend for Fig. 2A, utilizing a 5′-NCR-P3(C147A) transcript in each cotranslation reaction (indicated with a plus sign). The autoradiogram shown below is a longer exposure of the same gel. (B) [35S]methionine-labeled translations from the corresponding reactions shown in panel A carried out as described in the legend for Fig. 2B with and without 5′-NCR-P3(C147A).
We also carried out experiments with wild-type and chimeric transcripts lacking a precise 5′ end generated from cDNAs that do not contain a 5′ ribozyme sequence. The 5′ ends of these transcripts contain two guanosine nucleotides required by the T7 RNA polymerase and previously shown to reduce the levels of positive-strand RNA synthesis (27). By using this additional set of constructs in similar experiments, we were able to confirm that the signal labeled RI/RF (replicative intermediate-replicative form) in Fig. 2A corresponds in part to the synthesis of negative strands (data not shown), verifying the RNA synthesis phenotypes described above.
The range of RNA replication phenotypes of these chimeras raised several questions about the macromolecular interactions involving the 3D polymerase and/or 3CD polypeptide required for positive- versus negative-strand RNA synthesis. We carried out additional translation-RNA replication reactions in the presence of an internal ribosome entry site-driven RNA expressing the entire wild-type P3 precursor polypeptide 5′NCR-P3(wt) (Fig. 1C). Based on previously published results (33, 53) involving in vitro complementation of nonstructural gene functions, we predicted that the P3(wt) precursor polypeptide would be utilized in trans as a source of wild-type 3D polymerase or precursor (i.e., 3CD) proteins and could complement deficient chimeric 3D polymerases that are inactive in RNA recognition and elongation as well as those chimeric 3CD polypeptides defective in RNA binding or protein processing. Importantly, this approach should yield insights into the ability of wild-type proteins provided in trans to interact with and form functional replication complexes with the viral RNA, P2 region polypeptides, and chimeric proteins provided in cis.
Results from complementation studies using the 5′NCR-P3(wt) RNA are shown in Fig. 2. Fig. 2A shows that the defective RNA replication phenotypes of each chimeric transcript can be complemented in trans by the coexpression of a wild-type P3 precursor protein (Fig. 2A, lanes 3 to 18, compare odd- and even-numbered lanes), albeit with different efficiencies (for example, compare PCC to CCC in Fig. 2A, lanes 16 and 18). The presence of wild-type P3 RNA slightly reduced the ability of wild-type RzPV1 transcript (Fig. 1B) to synthesize RNA (Fig. 2A, compare lanes 3 and 4), perhaps reflecting a competition of proteins binding to P3 versus full-length transcripts. An increase in the detectable levels of both RI/RF and ssRNA was observed in each chimera in the presence of the rescue RNA encoding P3(wt), suggesting that wild-type proteins provided in trans are capable of either recognizing the chimeric RNA on their own (in the absence of chimeric proteins) or forming functional heterocomplexes with chimeric proteins capable of initiating RNA synthesis. By itself, the CPP chimera was unable to synthesize positive-strand RNA, but this construct displayed positive-strand RNA synthesis in the presence of the wt P3 precursor protein (Fig. 2A, compare lanes 5 and 6). Rescue occurred to the same level as that with PCP, a chimera capable of low levels of positive-strand RNA synthesis in the absence of wt P3 (Fig. 2A, compare lane 6 to lane 8), indicating that these two chimeras are able to participate in equally productive interactions with wild-type proteins provided in trans. Results with chimeras not as responsive to P3-mediated rescue (i.e., CPC and PCC [Fig. 2A, lanes 12 and 16]) suggest a reduced ability of those chimeric proteins to form functional interactions with their wild-type binding partners or to act as dominant-negative inhibitors by forming some nonfunctional protein-protein or protein-RNA complexes. Fig. 2A (lane 19) shows the inability of the P3(wt) RNA to replicate on its own, demonstrating that the RNA synthesis observed in each reaction represents the amplification of only full-length RzPV1 transcripts.
The translation results for the P3(wt) rescue experiment are shown in Fig. 2B. These results verify that the differences observed in RNA replication among the chimeric transcripts in Fig. 2A are not due to inherent differences in translation levels (Fig. 2B, lanes 5 to 18). Furthermore, the presence of the P3-cotranslating RNA does not inhibit the synthesis of gene products from the full-length RNAs (Fig. 2B, lanes 3 to 18, compare odd- and even-numbered lanes). Fig. 2B (lane 19) shows the proteins generated from a translation mixture incubated with 5′NCR-P3(wt) transcript alone and demonstrates the production of full-length P3 precursor protein and two major processing intermediates, 3BCD and 3CD (the overall translation level and the methionine content of the 3A protein prevent it from being seen here). We therefore conclude that the cotranslated P3 precursor protein provides full-length P3 protein as well as several processing intermediates to each reaction, consistent with previously published results (53).
The biochemical properties of recombinant 3CD polypeptides harboring each of the chimeric polymerase sequences have been previously described (15). Consistent with previously published data, chimeras CPP, PPC, CPC, PCC, and CCC displayed a P1 (capsid)-processing deficiency in this assay, indicated by the level of uncleaved VP0-VP3 precursor protein compared to mature cleavage products VP0 and VP3 observed in the translation reactions (Fig. 2B, lanes 5, 9, 11, 15, and 17). Furthermore, there are no other detectable processing defects that could prohibit RNA replication. In the samples that contained the 5′NCR-P3(wt) RNA, the wild-type 3CD polypeptide provided in trans is capable of processing the uncleaved VP0-VP3 polypeptide to completion (compare levels of VP0-VP3 with and without P3 RNA for CPP, PPC, CPC, PCC, and CCC). This finding provides direct evidence for the ability of proteins encoded by the full-length transcripts to interact with those produced from the subgenomic P3 RNA (i.e., 3CD).
Translation and RNA replication of PV1/CVB3 transcripts in the presence of P3(R13N) RNA.
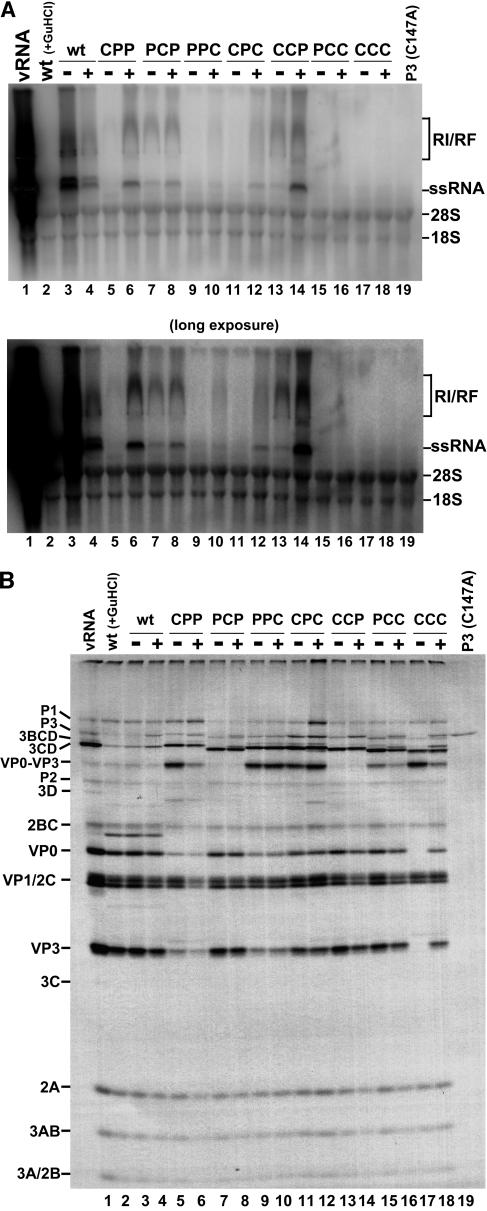
We next determined if the efficiency of RNA replication rescue achieved by cotranslation of a wild-type P3 RNA could be altered by the use of a mutant form of P3. 5′NCR-P3(R13N) (Fig. 1C) encodes a P3 protein harboring a previously described point mutation in 3C that inhibits the ability of 3C to bind the PV1 RNA cloverleaf (stem-loop I) at the very 5′ end of the positive strand without affecting the ability of 3CD to process polyprotein (9). Therefore, this version of P3 can serve as a source of wild-type 3D RNA polymerase and produce a 3CD precursor polypeptide able to process polyprotein but unable to bind RNA. The results of the RNA replication reactions in the presence and absence of P3(R13N) are shown in Fig. 3A. Consistent with our results with the wild-type P3 construct, both CPP and PCP rescued to similar levels of RNA synthesis with the P3(R13N) RNA (Fig. 3A, compare lane 5 to 6 and 7 to 8). The block to positive-strand RNA synthesis by the CPP chimera is relieved in the presence of P3(R13N) (Fig. 3A, compare lanes 5 and 6). RNA replication of the CCP chimera was rescued to a similar level to that observed with the wt P3 protein (compare lanes 13 and 14 in Fig. 2A and 3A). Therefore, rescue of these chimeras is not related to the ability of 3CD provided in trans to bind the cloverleaf structure near the 5′ end of the viral RNA. The accessibility of 3CD (wt or R13N) to sites of negative- and positive-strand RNA synthesis initiation may be mediated completely by protein-protein contacts with chimeric 3CD molecules bound to the RNA in cis. In contrast, PPC and CPC show a dramatically reduced ability to be rescued by P3(R13N) (Fig. 3A, compare lanes 9 and 10 and lanes 11 and 12). This result could reflect a deficiency in protein-protein contacts between 3CD(R13N) and 3CD(PPC) as well as 3CD(R13N) and 3CD(CPC), resulting in an inability of 3CD(R13N) to be recruited to initiation complexes. Furthermore, PPC and CPC could encode polymerases defective in RNA synthesis initiation or elongation which could oligomerize with wild-type 3D provided in trans, resulting in higher-order RNA polymerase structures reduced in overall activity. Finally, protein-protein interactions between PCC and CCC chimeric proteins may not be sufficient for even low levels of trans protein recruitment, resulting in a complete lack of RNA synthesis in the presence of P3(R13N) (Fig. 3A, lanes 15 to 18). As expected, P3(R13N) is incapable of autonomous RNA replication (Fig. 3A, lane 19).
FIG. 3.
RNA replication of wild-type and chimeric transcripts with and without 5′-NCR-P3(R13N)-cotranslating RNA. (A) RNA replication carried out as described in the legend for Fig. 2A, utilizing a 5′-NCR-P3(R13N) transcript in each cotranslation reaction (indicated with a plus sign). The autoradiogram shown below is a longer exposure of the same gel. (B) [35S]methionine-labeled translations from the corresponding reactions shown in panel A carried out as described in the legend for Fig. 2B with and without 5′-NCR-P3(R13N).
The translation and processing results for this rescue experiment appear to mirror those observed in our P3(wt) rescue assay (compare Fig. 2B and 3B). As expected, 3CD(R13N) is able to trans process the VP0-VP3 precursor polypeptide to completion (Fig. 3B, lanes 6, 10, 12, 16, and 18).
Another 5′NCR-P3 mutant construct containing a different lesion in 3C [5′NCR-P3(Y6N)] also known to inhibit RNA binding (9) was utilized in similar translation and RNA replication experiments. Complementation with the P3(Y6N) construct yielded results (data not shown) very similar to the data shown in Fig. 2, thereby strengthening the validity of our conclusions.
Translation and RNA replication of PV1/CVB3 transcripts in the presence of P3(C147A) RNA.
To directly test whether a 3CD polypeptide provided in trans requires proteolytic activity to rescue chimeric RNA replication, we used a construct harboring a mutation in 3C [5′NCR-P3(C147A)] (Fig. 1C) that destroys the proteolytic cleavage activity of the 3CD molecule (36). The processing of this version of P3 would be entirely dependent on the proteolytic activity of each chimeric 3CD polypeptide, since cleavage of P3(C147A) cannot occur in cis (53). Following trans processing by the chimeric 3CD polypeptide, P3(C147A) yields a wild-type 3D polymerase and a proteolytically inactive 3CD molecule (36) that retains its ability to bind cloverleaf RNA (T. B. Parsley and B. L. Semler, unpublished data). Results from translation and replication reactions in the presence and absence of P3(C147A) are shown in Fig. 4. The presence of a cotranslating P3(C147A) transcript reduced the efficiency of wild-type RNA synthesis (Fig. 4A, compare lanes 3 and 4). Approximately equal levels of rescue were observed with the CPP and CCP chimeras in the presence of the P3(C147A) RNA (Fig. 4A, compare lanes 5 and 6 and lanes 13 and 14). This finding suggests that the ability of P3(C147A) proteins to interact with these two sets of chimeric polypeptides is similar, resulting in the recruitment of chimeric 3CD molecules to active sites of negative- and positive-strand RNA synthesis. The processing of 3CD(C147A) by the chimeric 3CD polypeptides must occur, providing a source of active, wild-type polymerase to sites of RNA synthesis initiation. The PCP chimera did not rescue to the same level as CPP and CCP (Fig. 4A, compare lane 8 to lanes 6 and 14), suggesting that protein-protein interactions required to recruit P3(C147A) polypeptides to sites of replication are not efficient with the PCP chimera. By comparing this result to the that of the rescue of PCP using P3(R13N) shown in Fig. 3A, we hypothesize that the ability of the 3CD molecule provided in trans to process itself could be a factor in determining the susceptibility of this chimera to complementation. This conclusion can be extended to our results with the PPC and CPC chimeras, which also do not rescue to the same levels with P3(C147A) compared to P3(R13N) (compare lanes 9 to 12 in Fig. 3A and 4A). Alternatively, our results with the PCP, PPC, and CPC chimeras suggest that a 3CD molecule capable of binding RNA in the context of a replication complex but unable to process itself to yield 3D polymerase inhibits the overall function of that complex. Finally, P3(C147A) does not rescue RNA replication of the PCC and CCC chimeras to levels detectable by this assay (Fig. 4A, lanes 15 to 18). Since this result is independent of the ability of P3 proteins provided in trans to bind RNA (Fig. 3A, lanes 15 to 18), we again conclude that the PCC and CCC chimeric proteins do not participate in productive protein-protein interactions with trans binding partners. As a control, Fig. 4A (lane 19) shows that P3(C147A) is incapable of autonomous RNA replication.
The translation results for the P3(C147A) complementation experiment are displayed in Fig. 4B. The data shown (lane 19) confirm that P3(C147A) is incapable of cis cleavage, indicated by the presence of unprocessed P3 precursor protein and the absence of cleavage products. Consistent with our predictions, the proteolytically inactive 3CD(C147A) protein does not process the VP0-VP3 precursor protein that accumulates from the inefficiency of the CPP, PPC, CPC, and PCC chimeras to cleave at the VP0-VP3 junction. By comparing the levels of VP0-VP3 (Fig. 4B, lanes 5 and 6), one might conclude that additional processing of this precursor polypeptide by 3CD(C147A) occurred. However, this is most likely an effect of a sample loading error (compare levels of 3CD and VP1 polypeptides in Fig. 4, lanes 5 and 6). Surprisingly, one of the chimeras (CCC) showed a bona fide rescue of VP0-VP3 processing (Fig. 4B, compare lanes 17 and 18), as demonstrated by a reduction in the amount of VP0-VP3 and an appearance of VP0 and VP3 in the presence of P3(C147A). This unexpected result suggests that an inactive 3CD(C147A) polypeptide can enhance the ability of a deficient 3CD molecule [e.g., 3CD(CCC)] to process the VP0-VP3 junction. This could occur through multimerization of two defective 3CD polypeptides to form an active complex or by 3CD(C147A) enhancing the recognition of the Q-G dipeptide within the VP0-VP3 precursor polyprotein by binding and recruiting 3CD(CCC) to the site of proteolytic cleavage. The mechanistic implications of this phenomenon are being explored.
Translation and RNA replication of PV1/CVB3 transcripts in the presence of P3(μ61) RNA.
As previously discussed, P3(wt), P3(C147A), and P3(R13N) are likely providing a source of wild-type 3D polymerase, generated via a cis or trans cleavage event within P3, to RNA replication complexes that assemble around full-length transcript RNAs. We therefore wanted to carry out RNA replication complementation experiments utilizing a P3 precursor polypeptide that yields an inactive polymerase. This experiment utilizes a cotranslating 5′NCR-P3(μ61) RNA (Fig. 1C) containing a K61L mutation in the 3D polymerase that renders it incapable of RNA chain elongation (47, 53). Figure 5A shows the RNA replication results in the presence and absence of this cotranslating RNA. Unlike the other forms of P3 tested, P3(μ61) was unable to rescue positive-strand RNA synthesis when incubated with full-length CPP chimeric RNA, although a detectable increase in negative-strand RNA synthesis was observed (Fig. 5A, compare lanes 5 and 6). This result demonstrates that in previous experiments with this chimera, wild-type 3D provided in trans was the primary source of polymerase that allowed positive-strand RNA synthesis to occur. The increase in negative-strand RNA synthesis in the absence of new positive strands suggests that P3(μ61) proteins are capable of recruiting active CPP polymerase to sites of negative-strand RNA synthesis initiation. These data provide additional evidence that the viral proteins that form complexes to initiate negative-strand RNA synthesis are different, in part, from those that assemble for the synthesis of positive strands.
FIG. 5.
RNA replication of wild-type and chimeric transcripts with and without 5′-NCR-P3(μ61)-cotranslating RNA. (A) RNA replication carried out as described in the legend for Fig. 2A, utilizing a 5′-NCR-P3(μ61) transcript in each cotranslation reaction (indicatedwith a plus sign). The autoradiogram shown below is a longer exposure of the same gel. (B) [35S]methionine-labeled translations from the corresponding reactions shown in panel A carried out as described in the legend for Fig. 2B with and without 5′-NCR-P3(μ61).
The PCP and CCP chimeras were equally responsive to the presence of P3(μ61) and showed an increase in both negative- and positive-strand RNA synthesis (Fig. 5A, compare lanes 7 and 8 and lanes 13 and 14). These data indicate that the PCP and CCP chimeric polymerases are equally capable of interacting with P3(μ61) proteins in the formation of RNA synthesis initiation complexes. However, given our results with CPP (Fig. 5A, lanes 5 and 6), we predict that the increase in positive-strand RNA generated by these two chimeras is primarily the result of an increase in negative-strand RNA synthesis in the presence of P3(μ61) proteins, thus providing significant quantities of newly synthesized templates for positive-strand RNA synthesis. In contrast, chimeras PPC and CPC were completely unresponsive to rescue with P3(μ61) (Fig. 5A, lanes 9 to 12), whereas they were rescued with P3(wt) (Fig. 2A), P3(R13N) (Fig. 3A), and to a limited extent P3(C147A) (Fig. 4A). These observations, combined with the fact that P3(μ61) cannot provide active polymerase molecules, suggest that the PPC and CPC 3D polymerases could be enzymatically defective. Finally, chimeras PCC and CCC were also unresponsive to rescue by P3(μ61) (Fig. 5A, compare lanes 15 and 16 and lanes 17 and 18). Since previously presented results with these chimeras utilizing P3(R13N) indicate a deficiency in protein-protein interactions with complementing polypeptides, we cannot draw conclusions regarding the activity of the PCC and CCC chimeric polymerases themselves. The rescue phenotypes of these two chimeras may simply be a result of an inability of chimeric 3D polymerase molecules to be recruited by P3(μ61) polypeptides to RNA replication complexes.
The translation results for the P3(μ61) rescue experiment are shown in Fig. 5B. Interestingly, the presence of 3CD(μ61) provided by the P3(μ61)-cotranslating RNA (Fig. 5B, even-numbered lanes 4 to 18) did not result in the complete processing of VP0-VP3 precursor polypeptide in trans (Fig. 5B, lanes 2 to 18, compare even- and odd-numbered lanes). Previously published results have shown that point mutations within the polymerase subdomain of 3CD can reduce the ability of 3CD to recognize the VP0-VP3 junction (42). Although not directly addressed in this study, these results suggest that the K61L mutation present in context of the 3CD(μ61) polypeptide provided in trans has this same effect. Furthermore, in the absence of complete VP0-VP3 processing, an increase in RNA synthesis is observed in the presence of P3(μ61) for chimeras CPP, PCP, and CCP (Fig. 5A, compare lanes 5 and 6, lanes 7 and 8, and lanes 13 and 14). Combined with results presented earlier in this work, these findings confirm that complete VP0-VP3 processing is not required for efficient RNA synthesis.
3Dpol in vitro elongation assays.
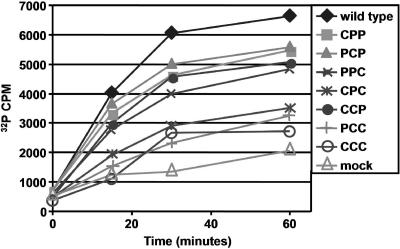
Data presented thus far strongly suggest alterations to 3D RNA polymerase enzymatic activities in each of the PV1/CVB3 chimeras. This hypothesis is supported by both the dramatic differences in autonomous replication (compared to the wild type) as well as a differential responsiveness of each chimera to P3 complementation. To test this possibility, we assayed directly for RNA chain elongation activity encoded by each of the PV1/CVB3 3D polymerases by using a method previously described by Barton and Flanegan (4). First, in vitro translation reactions were carried out with wild-type PV1 RNA or PV1/CVB3 transcripts. Next, a portion of each reaction was used as a source of 3D RNA polymerase to catalyze the elongation of RNA chains from an oligo(U)-primed PV1 virion RNA template in the presence of radiolabeled CTP (see Materials and Methods). Aliquots from each reaction were removed at specific times during the 60-min reaction step, and the incorporated nucleotide was measured and used as an indication of RNA polymerase elongation activity. The results of this experiment are shown in Fig. 6. Wild-type polymerase was capable of incorporating significant amounts of radiolabeled CTP during the time course experiment. As expected, chimeras CPP, PCP, and CCP, each previously shown to be capable of autonomous RNA replication, have similar elongation activities, albeit slightly reduced compared to those of wild-type polymerase. Interestingly, the PPC chimeric polymerase is also able to elongate from the oligo(U)-primed, viral RNA-primed template. This strengthens our initial conclusion that 3D/3CD(PPC) is deficient in protein-protein contacts required for the formation of RNA synthesis complexes and could also indicate a deficiency in RNA synthesis initiation rather than elongation. We also hypothesize that the CPC chimera is deficient in similar protein-protein contacts, exhibited by its reduced ability to be complemented in trans by all forms of P3 tested. The results in Fig. 6 indicate that an additional defect exists in RNA elongation activity as well. Finally, chimeras PCC and CCC displayed the lowest levels of elongation activity compared to the other constructs. This finding was not surprising, since these two chimeras were almost completely unresponsive to RNA synthesis complementation by the P3 constructs used in this study.
FIG. 6.
In vitro RNA polymerase assays. Wild-type and PV1/CVB3 transcripts were translated in vitro, providing a source of 3D RNA polymerase used to elongate nascent RNA chains from an oligo(U)-primed viral RNA template as described in Materials and Methods. Aliquots at 0, 15, 30, and 60 min were counted, and after subtracting background levels of radioactivity, the data were plotted. The graph shown in the figure displays representative data from two independent experiments.
Implications of RNA replication rescue data.
A summary of our P3 complementation data is displayed in Fig. 7. Based upon the differential rescue of the 3D polymerase chimeras, we have divided these chimeras into three functional groups. Members of one group include CPP, PCP, and CCP. These constructs encode an active 3D RNA polymerase, as evidenced by autonomous RNA replication (albeit at low levels). A second group includes PPC and CPC, which encode 3D RNA polymerase molecules with reduced elongation activity, unable to efficiently assemble RNA replication complexes. Support for this conclusion comes from our polymerase elongation assays, the lack of autonomous RNA replication, the inability to be complemented by a P3 precursor protein harboring an enzymatically inactive 3D polymerase, and molecular modeling studies that predict the disruption of one or more protein-protein interfaces shown to be important for 3D RNA polymerase activity (14). Members of the final group, consisting of chimeras PCC and CCC, appear to be incapable of participating in protein-protein interactions critical for RNA replication and encode RNA polymerases with the lowest levels of RNA synthesis activity detected in our elongation assays (Fig. 6).
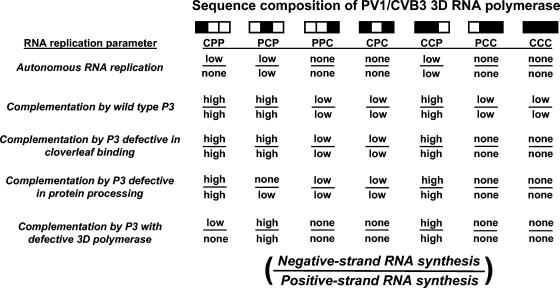
FIG. 7.
Summary of RNA replication and complementation phenotypes of PV1/CVB3 transcripts. The 3D polymerase composition of each full-length chimeric transcript tested is indicated at the top of the figure. Shown below the chimeras is a qualitative assessment (indicated by “high,” “low, ” or “none”) of the ability of each chimera to replicate autonomously or to be complemented in trans by wild-type or genetically altered P3 protein (left side of panel). Each phenotype is shown as negative-strand RNA synthesis over positive-strand RNA synthesis (see bottom of figure). Based on a comparison to RNA synthesis levels observed for wild-type PV1 transcript, all chimeras capable of synthesizing RNA in the absence of complementing P3 protein were given the designation low. For the complementation results with and without P3 protein, the designation high was assigned to RNA replication levels estimated to be at least fivefold greater than those observed in the absence of P3 protein. Low indicates an increase in RNA levels approximately two- to fourfold over those observed without P3, whereas none indicates no detectable increase in RNA synthesis in the presence of P3.
Our attempts to rescue defects in RNA synthesis with a wild-type P3 precursor protein resulted in an increase in RNA replication for each of the seven chimeras tested (Fig. 2). The rescue of chimeras that do not replicate autonomously (PPC, CPC, PCC, and CCC) suggests that either wild-type proteins provided in trans can form complexes with viral RNA independent of chimeric binding partners or there are protein-protein contacts that allow recruitment of polypeptides provided in trans to sites of RNA synthesis initiation. The importance of protein-protein contacts in the recruitment of viral polypeptides to active RNA replication complexes is underscored by our results with P3(R13N) (Fig. 3), since this form of P3 yields a 3CD molecule incapable of binding cloverleaf RNA but capable of rescuing replication in some of the chimeras. These results indicate that sequences within the amino-terminal and carboxy-terminal one-third of the 3D polymerase (or 3D domain of the 3CD polypeptide) are necessary for productive interactions with proteins provided in trans, which could involve 3D-3D, 3D-3CD, 3CD-3CD, 3D-3AB, or 3CD-3AB contacts (29, 30, 44, 58). Previously published biochemical data regarding defective 3CD(CPC) and 3CD(PPC) protein-protein interactions support this hypothesis (15).
A P3 precursor yielding an inactive polymerase (μ61) was able to increase negative-strand RNA synthesis but only in those chimeras capable of autonomous RNA replication (CPP, PCP, and CCP). This finding suggests that a precursor to the polymerase (i.e., 3CD) can recruit active chimeric polymerase molecules (provided in cis) to sites of negative-strand RNA synthesis initiation, possibly via 3CD-3CD or 3CD-3D interactions mediated by 3D polymerase domain contacts. There may be a mechanism to favor assembly of protein-protein and protein-RNA complexes which involve polypeptides provided in cis. However, when polymerase or polymerase precursor polypeptides provided in cis are incapable of forming one or more complexes, proteins provided in trans are no longer kinetically excluded and may participate in replication complex assembly. This is evidenced by our ability to complement the RNA synthesis of the PPC, CPC, PCC, and CCC chimeras (incapable of replicating on their own and therefore of assembling one or more cis-dependent complexes) with a wild-type P3 precursor. For CPP, the chimeric proteins provided in cis are unable to form complexes necessary for positive-strand RNA synthesis, making the replication proteins provided in trans the only source of active polymerase used for this process. When active polymerase is not provided in trans, as is the case for the P3(μ61) construct, no rescue of positive-strand RNA synthesis was observed (Fig. 5A, compare lanes 5 and 6).
For two of the chimeras incapable of autonomous RNA replication (PPC and CPC), we can draw significant conclusions regarding the source of the 3D RNA polymerase provided to RNA synthesis complexes. Previously published biochemical data indicated that these two chimeric 3CD polypeptides are capable of forming ribonucleoprotein complexes with the poliovirus cloverleaf RNA and host protein PCBP2 with an efficiency greater than that of wt 3CD (15). Since neither PPC nor CPC replicates in the absence of a P3 subgenomic RNA, these two chimeras may bind viral RNA in cis but are deficient in other protein-protein interactions necessary for RNA synthesis initiation separate from those involving PCBP2, providing an explanation for their differential responses to complementation using the P3(R13N) and P3(C147A) constructs, both of which yield sufficient levels of wild-type 3D RNA polymerase. Taken together, these results suggest a kinetically favored assembly of highest-affinity ribonucleoprotein complexes in cis.
Results from our complementation analysis of negative- and positive-strand RNA synthesis have yielded insights into possible mechanisms utilized for the assembly of strand-specific RNA replication complexes. These insights include the following: (i) higher-order complexes containing 3CD molecules that could serve as donors of 3D RNA polymerase favor assembly with viral proteins provided in cis, (ii) protein-protein interactions between viral proteins in cis and those in trans can facilitate the recruitment of helper functions derived from proteins that are by themselves deficient in binding viral RNA, and (iii) protein complexes that assemble for negative- versus positive-strand RNA synthesis are not equally capable of recruiting and/or utilizing functional P3 proteins provided in trans, thereby inhibiting the progression of RNA synthesis when such proteins are limiting or defective. These differential effects are mediated, in part, by specific domains within proteins harboring 3D amino acid sequences. However, the roles that these domains have during specific steps of RNA replication (VPg uridylylation, interactions with host proteins, RNA chain elongation, and putative subunit exchange between strand-specific RNA synthesis complexes) remain to be determined.
Acknowledgments
We are grateful to Kristin Bedard for critical reading of the manuscript. We are indebted to Jonathan Towner for the gift of all pT7-5′-NCR-P3 plasmids utilized in this work, Raul Andino for the gift of plasmid pRib(+)RLuc, and Bert Flanegan for providing advice and the oligo(U) used in the in vitro RNA elongation assays.
This work was supported by Public Health Service grant AI 22693 from the National Institutes of Health. C.T.C. and J.E.B. were predoctoral trainees supported by Public Health Service training grants AI 07319 and GM 07311, respectively.
REFERENCES
- 1.Andino, R., G. E. Rieckhof, P. L. Achacoso, and D. Baltimore. 1993. Poliovirus RNA synthesis utilizes an RNP complex formed around the 5′-end of viral RNA. EMBO J. 12:3587-3598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Andino, R., G. E. Rieckhof, and D. Baltimore. 1990. A functional ribonucleoprotein complex forms around the 5′ end of poliovirus RNA. Cell 63:369-380. [DOI] [PubMed] [Google Scholar]
- 3.Barton, D. J., E. P. Black, and J. B. Flanegan. 1995. Complete replication of poliovirus in vitro: preinitiation RNA replication complexes require soluble cellular factors for the synthesis of VPg-linked RNA. J. Virol. 69:5516-5527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Barton, D. J., and J. B. Flanegan. 1993. Coupled translation and replication of poliovirus RNA in vitro: synthesis of functional 3D polymerase and infectious virus. J. Virol. 67:822-831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Barton, D. J., B. J. O'Donnell, and J. B. Flanegan. 2001. 5′ cloverleaf in poliovirus RNA is a cis-acting replication element required for negative-strand synthesis. EMBO J. 20:1439-1448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bell, Y. C., B. L. Semler, and E. Ehrenfeld. 1999. Requirements for RNA replication of a poliovirus replicon by coxsackievirus B3 RNA polymerase. J. Virol. 73:9413-9421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bernstein, H. D., P. Sarnow, and D. Baltimore. 1986. Genetic complementation among poliovirus mutants derived from an infectious cDNA clone. J. Virol. 60:1040-1049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bienz, K., D. Egger, and L. Pasamontes. 1987. Association of polioviral proteins of the P2 genomic region with the viral replication complex and virus-induced membrane synthesis as visualized by electron microscopic immunocytochemistry and autoradiography. Virology 160:220-226. [DOI] [PubMed] [Google Scholar]
- 9.Blair, W. S., T. B. Parsley, H. P. Bogerd, J. S. Towner, B. L. Semler, and B. R. Cullen. 1998. Utilization of a mammalian cell-based RNA binding assay to characterize the RNA binding properties of picornavirus 3C proteinases. RNA 4:215-225. [PMC free article] [PubMed] [Google Scholar]
- 10.Borman, A. M., R. Kirchweger, E. Ziegler, R. E. Rhoads, T. Skern, and K. M. Kean. 1997. eIF4G and its proteolytic cleavage products: effect on initiation of protein synthesis from capped, uncapped, and IRES-containing mRNAs. RNA 3:186-196. [PMC free article] [PubMed] [Google Scholar]
- 11.Cao, X., and E. Wimmer. 1995. Intragenomic complementation of a 3AB mutant in dicistronic polioviruses. Virology 209:315-326. [DOI] [PubMed] [Google Scholar]
- 12.Charini, W. A., C. C. Burns, E. Ehrenfeld, and B. L. Semler. 1991. trans rescue of a mutant poliovirus RNA polymerase function. J. Virol. 65:2655-2665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cho, M. W., N. Teterina, D. Egger, K. Bienz, and E. Ehrenfeld. 1994. Membrane rearrangement and vesicle induction by recombinant poliovirus 2C and 2BC in human cells. Virology 202:129-145. [DOI] [PubMed] [Google Scholar]
- 14.Cornell, C. T., R. Perera, J. E. Brunner, and B. L. Semler. 2004. Strand-specific RNA synthesis determinants in the RNA-dependent RNA polymerase of poliovirus. J. Virol. 78:4397-4407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cornell, C. T., and B. L. Semler. 2002. Subdomain specific functions of the RNA polymerase region of poliovirus 3CD polypeptide. Virology 298:200-213. [DOI] [PubMed] [Google Scholar]
- 16.Deitz, S. B., D. A. Dodd, S. Cooper, P. Parham, and K. Kirkegaard. 2000. MHC I-dependent antigen presentation is inhibited by poliovirus protein 3A. Proc. Natl. Acad. Sci. USA 97:13790-13795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Dewalt, P. G., and B. L. Semler. 1989. Molecular biology and genetics of poliovirus protein processing, p. 73-93. In B. L. Semler and E. Ehrenfeld (ed.), Molecular genetic aspects of picornavirus infection and detection. American Society for Microbiology, Washington, D.C.
- 18.Doedens, J. R., T. H. J. Giddings, and K. Kirkegaard. 1997. Inhibition of endoplasmic reticulum-to-Golgi traffic by poliovirus protein 3A: genetic and ultrastructural analysis. J. Virol. 71:9054-9064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Doedens, J. R., and K. Kirkegaard. 1995. Inhibition of cellular protein secretion by poliovirus proteins 2B and 3A. EMBO J. 14:894-907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Egger, D., N. Teterina, E. Ehrenfeld, and K. Bienz. 2000. Formation of the poliovirus replication complex requires coupled viral translation, vesicle production, and viral RNA synthesis. J. Virol. 74:6570-6580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Etchison, D., S. C. Milburn, I. Edery, N. Sonenberg, and J. W. Hershey. 1982. Inhibition of HeLa cell protein synthesis following poliovirus infection correlates with the proteolysis of a 220,000-dalton polypeptide associated with eucaryotic initiation factor 3 and a cap binding protein complex. J. Biol. Chem. 257:14806-14810. [PubMed] [Google Scholar]
- 22.Giachetti, C., S. S. Hwang, and B. L. Semler. 1992. cis-acting lesions targeted to the hydrophobic domain of a poliovirus membrane protein involved in RNA replication. J. Virol. 66:6045-6057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gustin, K. E., and P. Sarnow. 2001. Effects of poliovirus infection on nucleo-cytoplasmic trafficking and nuclear pore complex composition. EMBO J. 20:240-249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hanecak, R., B. L. Semler, C. W. Anderson, and E. Wimmer. 1982. Proteolytic processing of poliovirus polypeptides: antibodies to polypeptide P3-7c inhibit cleavage at glutamine-glycine pairs. Proc. Natl. Acad. Sci. USA 79:3973-3977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Harris, K. S., S. R. Reddigari, M. J. Nicklin, T. Hammerle, and E. Wimmer. 1992. Purification and characterization of poliovirus polypeptide 3CD, a proteinase and a precursor for RNA polymerase. J. Virol. 66:7481-7489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Harris, K. S., W. Xiang, L. Alexander, W. S. Lane, A. V. Paul, and E. Wimmer. 1994. Interaction of poliovirus polypeptide 3CDpro with the 5′ and 3′ termini of the poliovirus genome. Identification of viral and cellular cofactors needed for efficient binding. J. Biol. Chem. 269:27004-27014. [PubMed] [Google Scholar]
- 27.Herold, J., and R. Andino. 2000. Poliovirus requires a precise 5′ end for efficient positive-strand RNA synthesis. J. Virol. 74:6394-6400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Herold, J., and R. Andino. 2001. Poliovirus RNA replication requires genome circularization through a protein-protein bridge. Mol. Cell 7:581-591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hobson, S. D., E. S. Rosenblum, O. C. Richards, K. Richmond, K. Kirkegaard, and S. C. Schultz. 2001. Oligomeric structures of poliovirus polymerase are important for function. EMBO J. 20:1153-1163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hope, D. A., S. E. Diamond, and K. Kirkegaard. 1997. Genetic dissection of interaction between poliovirus 3D polymerase and viral protein 3AB. J. Virol. 71:9490-9498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Johnson, K. L., and P. Sarnow. 1991. Three poliovirus 2B mutants exhibit noncomplementable defects in viral RNA amplification and display dosage-dependent dominance over wild-type poliovirus. J. Virol. 65:4341-4349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Jore, J., B. De Geus, R. J. Jackson, P. H. Pouwels, and B. E. Enger-Valk. 1988. Poliovirus protein 3CD is the active protease for processing of the precursor protein P1 in vitro. J. Gen. Virol. 69:1627-1636. [DOI] [PubMed] [Google Scholar]
- 33.Jurgens, C., and J. B. Flanegan. 2003. Initiation of poliovirus negative-strand RNA synthesis requires precursor forms of p2 proteins. J. Virol. 77:1075-1083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Krausslich, H. G., M. J. Nicklin, H. Toyoda, D. Etchison, and E. Wimmer. 1987. Poliovirus proteinase 2A induces cleavage of eucaryotic initiation factor 4F polypeptide p220. J. Virol. 61:2711-2718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Laemmli, U. K. 1970. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227:680-685. [DOI] [PubMed] [Google Scholar]
- 36.Lawson, M. A., and B. L. Semler. 1991. Poliovirus thiol proteinase 3C can utilize a serine nucleophile within the putative catalytic triad. Proc. Natl. Acad. Sci. USA 88:9919-9923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Leong, L. E., C. T. Cornell, and B. L. Semler. 2002. Processing determinants and functions of cleavage products of picornavirus polyproteins, p. 187-197. In B. L. Semler and E. Wimmer (ed.), Molecular biology of picornaviruses. ASM Press, Washington, D.C.
- 38.Li, J. P., and D. Baltimore. 1988. Isolation of poliovirus 2C mutants defective in viral RNA synthesis. J. Virol. 62:4016-4021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lloyd, R. E., H. G. Jense, and E. Ehrenfeld. 1987. Restriction of translation of capped mRNA in vitro as a model for poliovirus-induced inhibition of host cell protein synthesis: relationship to p220 cleavage. J. Virol. 61:2480-2488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Molla, A., A. V. Paul, and E. Wimmer. 1991. Cell-free, de novo synthesis of poliovirus. Science 254:1647-1651. [DOI] [PubMed] [Google Scholar]
- 41.Novak, J. E., and K. Kirkegaard. 1994. Coupling between genome translation and replication in an RNA virus. Genes Dev. 8:1726-1737. [DOI] [PubMed] [Google Scholar]
- 42.Parsley, T. B., C. T. Cornell, and B. L. Semler. 1999. Modulation of the RNA binding and protein processing activities of poliovirus polypeptide 3CD by the viral RNA polymerase domain. J. Biol. Chem. 274:12867-12876. [DOI] [PubMed] [Google Scholar]
- 43.Parsley, T. B., J. S. Towner, L. B. Blyn, E. Ehrenfeld, and B. L. Semler. 1997. Poly (rC) binding protein 2 forms a ternary complex with the 5′-terminal sequences of poliovirus RNA and the viral 3CD proteinase. RNA 3:1124-1134. [PMC free article] [PubMed] [Google Scholar]
- 44.Pata, J. D., S. C. Schultz, and K. Kirkegaard. 1995. Functional oligomerization of poliovirus RNA-dependent RNA polymerase. RNA 1:466-477. [PMC free article] [PubMed] [Google Scholar]
- 45.Paul, A. V., E. Rieder, D. W. Kim, J. H. van Boom, and E. Wimmer. 2000. Identification of an RNA hairpin in poliovirus RNA that serves as the primary template in the in vitro uridylylation of VPg. J. Virol. 74:10359-10370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Paul, A. V., J. H. van Boom, D. Filippov, and E. Wimmer. 1998. Protein-primed RNA synthesis by purified poliovirus RNA polymerase. Nature 393:280-284. [DOI] [PubMed] [Google Scholar]
- 47.Richards, O. C., S. Baker, and E. Ehrenfeld. 1996. Mutation of lysine residues in the nucleotide binding segments of the poliovirus RNA-dependent RNA polymerase. J. Virol. 70:8564-8570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Rieder, E., A. V. Paul, D. W. Kim, J. H. van Boom, and E. Wimmer. 2000. Genetic and biochemical studies of poliovirus cis-acting replication element cre in relation to VPg uridylylation. J. Virol. 74:10371-10380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Semler, B. L., C. W. Anderson, N. Kitamura, P. G. Rothberg, W. L. Wishart, and E. Wimmer. 1981. Poliovirus replication proteins: RNA sequence encoding P3-1b and the sites of proteolytic processing. Proc. Natl. Acad. Sci. USA 78:3464-3468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Teterina, N. L., A. E. Gorbalenya, D. Egger, K. Bienz, and E. Ehrenfeld. 1997. Poliovirus 2C protein determinants of membrane binding and rearrangements in mammalian cells. J. Virol. 71:8962-8972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Teterina, N. L., W. D. Zhou, M. W. Cho, and E. Ehrenfeld. 1995. Inefficient complementation activity of poliovirus 2C and 3D proteins for rescue of lethal mutations. J. Virol. 69:4245-4254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Todd, S., J. S. Towner, D. M. Brown, and B. L. Semler. 1997. Replication-competent picornaviruses with complete genomic RNA 3′ noncoding region deletions. J. Virol. 71:8868-8874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Towner, J. S., M. M. Mazanet, and B. L. Semler. 1998. Rescue of defective poliovirus RNA replication by 3AB-containing precursor polyproteins. J. Virol. 72:7191-7200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Towner, J. S., H. H. Roehl, S. Todd, T. P. Parsley, T. V. Ho, and B. L. Semler. 1996. Complex formation and template recognition in poliovirus RNA replication, p. 3-22. In S. Jameel and E. K. Wagner (ed.), Current developments in animal virology. Oxford and IBH Publishing Co. Pvt. Ltd., New Delhi, India.
- 55.Van Dyke, T. A., and J. B. Flanegan. 1980. Identification of poliovirus polypeptide P63 as a soluble RNA-dependent RNA polymerase. J. Virol. 35:732-740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Walter, B. L., T. B. Parsley, E. Ehrenfeld, and B. L. Semler. 2002. Distinct poly(rC) binding protein KH domain determinants for poliovirus translation initiation and viral RNA replication. J. Virol. 76:12008-12022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Weidman, M. K., P. Yalamanchili, B. Ng, W. Tsai, and A. Dasgupta. 2001. Poliovirus 3C protease-mediated degradation of transcriptional activator p53 requires a cellular activity. Virology 291:260-271. [DOI] [PubMed] [Google Scholar]
- 58.Xiang, W., A. Cuconati, D. Hope, K. Kirkegaard, and E. Wimmer. 1998. Complete protein linkage map of poliovirus P3 proteins: interaction of polymerase 3Dpol with VPg and with genetic variants of 3AB. J. Virol. 72:6732-6741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Yalamanchili, P., R. Banerjee, and A. Dasgupta. 1997. Poliovirus-encoded protease 2APro cleaves the TATA-binding protein but does not inhibit host cell RNA polymerase II transcription in vitro. J. Virol. 71:6881-6886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Yalamanchili, P., U. Datta, and A. Dasgupta. 1997. Inhibition of host cell transcription by poliovirus: cleavage of transcription factor CREB by poliovirus-encoded protease 3Cpro. J. Virol. 71:1220-1226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Yalamanchili, P., K. Weidman, and A. Dasgupta. 1997. Cleavage of transcriptional activator Oct-1 by poliovirus encoded protease 3Cpro. Virology 239:176-185. [DOI] [PubMed] [Google Scholar]
- 62.Ypma-Wong, M. F., P. G. Dewalt, V. H. Johnson, J. G. Lamb, and B. L. Semler. 1988. Protein 3CD is the major poliovirus proteinase responsible for cleavage of the P1 capsid precursor. Virology 166:265-270. [DOI] [PubMed] [Google Scholar]