Abstract
Phylogenetic analysis of foamy virus sequences obtained from Bornean and Sumatran orangutans showed a distinct clustering pattern. One subcluster was represented by both Bornean and Sumatran orangutan simian foamy viruses (SFV). Combined analysis of host mitochondrial DNA and SFV phylogeny provided evidence for the hypothesis of the repopulation of Sumatra by orangutans from Borneo.
Foamy viruses are retroviruses that infect cattle, horses, cats, and primates, including humans (24). Simian foamy viruses (SFV) have been described in Old World and New World monkeys and in apes (3-5, 7, 12, 15, 17, 23, 33, 40). Interestingly, humans are the only hominoids that are not naturally infected, and only zoonotic infections of humans have been described (1, 6, 11, 31, 32). Because of the benign nature of the infections, it is assumed that foamy viruses are ancient viruses that have coevolved with their natural hosts (3, 4, 7, 11, 33). However, these assumptions were primarily drawn based on a limited number of viruses and primate species.
In a recent serosurvey we screened 108 serum samples obtained from wild-caught or locally confiscated Bornean orangutans for anti-SFV antibodies (40). A high percentage, 69.4%, had antibodies that were cross-reactive to antigens from African green monkey foamy virus (SFVagm; SFV-3). We additionally acquired blood samples of Sumatran orangutans, five of which were wild caught and six that were housed in zoos. A foamy virus-specific PCR was performed on DNA isolated from all Sumatran animals and on 15 blood samples from seropositive Bornean individuals. The PCR amplified a fragment of the integrase coding region of the foamy virus pol gene (34). A 425-bp fragment was recovered from all Bornean samples, and from 7 out of 11 Sumatran animals (two wild-caught [Popa_SU76 and Popa_SU78] and five zoo animals [Popa_Du, Popa_Bi, Popa_Pi, Popa_To, and Popa_Wa]). The PCR products were isolated from agarose gel using a QIAquick gel extraction kit (QIAGEN, Hilden, Germany) and directly sequenced. This was done using the ABI PRISM BigDye terminators v. 3.0 cycle sequencing kit on an ABI PRISM 3100 genetic analyzer (Applied Biosystems, Nieuwerkerk aan de IJssel, The Netherlands). Sequences were analyzed using MacVector 6.0 (Oxford Molecular Ltd.) and SeqMan II (DNASTAR, Inc., Madison, Wis.) sequence analysis software packages, and the alignments were manually edited using the sequence alignment editor Se-Al version 2.0a11 (29).
Translation of the integrase sequences revealed that the amino acid sequences fell into three distinct groups (Fig. 1). One group was formed by 12 foamy viruses that were all obtained from Bornean orangutans (Fig. 1, upper 12 sequences). The second and third groups consisted of viruses from Borneo and Sumatra, respectively, and were differentiated from the first group by a glutamine (Q) at position 13 and a threonine (T) residue at position 17 in the alignment. The third group, consisting of two orangutan SFV (SFVora) recovered from Sumatran orangutans (Popa_SU78 and Popa_Du) were further differentiated from the other sequences by specific amino acid residues at positions 37 (V), 42 (N), and 53 (C), an A-A doublet at positions 94 to 95, and an L at position 105. A phylogenetic tree derived from the nucleotide sequences showed a branching pattern for the SFVora that was consistent with our protein sequence analysis (Fig. 2A). The tree was rooted with SFV isolated from a dark-handed gibbon (Hylobates agilis; SFVhpi), a distantly related ape species also inhabiting the islands of Borneo and Sumatra. From the root the tree splits in two branches. A small branch leads to the Sumatran sequences derived from Popa_SU78 and Popa_Du (cluster Sum), while the major branch leads to remaining viruses isolated from both subspecies. This large cluster further separates into two subclusters, one (subcluster Bor) solely consisting of Bornean viruses, while the other (SumBor) contains viruses from a mixed origin.
FIG. 1.

Alignment of the deduced amino acid sequences of 22 SFVora. The published sequence of SFVora_Be was used as a reference (EMBL database accession no. AJ544579) (40). Identical residues are indicated by dashes. Residues that are characteristic of one of the three groups are shaded. Popp, Bornean orangutan (Pongo pygmaeus pygmaeus); Popa, Sumatran orangutan (P. p. abelii.).
FIG. 2.
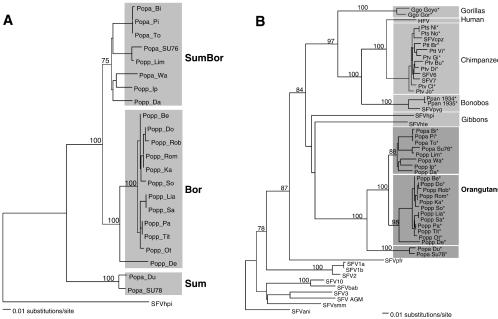
(A) Evolutionary tree derived from SFVora sequences from Bornean (Popp) and Sumatran (Popa) orangutans, and (B) tree constructed with newly obtained and published SFV sequences from different ape species. Phylogenetic analyses of a 425-bp integrase sequence were performed by the neighbor-joining method as implemented in PAUP* version 4.0b10 (37). Bootstrap values (as percentages of 1,000 resamplings) are indicated. The trees were rooted with sequences derived from a dark-handed gibbon (SFVhpi) and an Allen's swamp monkey (SFVani), respectively. All novel SFV sequences described in this article have been deposited in the EMBL and GenBank data libraries, accession numbers AJ544579 and AJ627527 to AJ627560 and are marked by asterisks. Accession numbers from published sequences are as follows: X54482 and X54484, X83290, X83291, X83294 to X83297, AF049079, AF049081, AF049083, AF049086, AF516484 to AF516487, and U04327. Abbreviations: Ggo, Gorilla gorilla gorilla; Pts, Pan troglodytes schweinfurthii; Ptt, P. t. troglodytes; Ptv, P. t. verus; Ppan, Pan paniscus.
The typical clustering of the different variants of SFVora is interesting, as the orangutan subspecies that live on the islands of Borneo (Pongo pygmaeus pygmaeus) and Sumatra (P. p. abelii) have diverged 1.1 to 2.3 million years ago (MYA) (41, 44, 45). If the common ancestor of the two present day orangutan subspecies had been infected with a foamy virus, this long-term spatial separation of the populations would have provided an opportunity for SFVora to coevolve with their hosts. In its simplest form this would have resulted in two virus clusters that are directly related to the host subspecies. Alternatively, new introductions of SFV by transmission across the subspecies may also have contributed to the variation depicted in Fig. 2A. Viral transmission due to mixed housing in zoos or improper reintroduction procedures may also have contributed to the observed clustering. However, our findings are validated by the fact that each (sub)cluster contains a virus from a wild-caught animal (Popa_SU76, Popa_SU78, and Popp_So). In addition, current reintroduction practices include that confiscated animals are reintroduced on their islands of origin (K. S. Warren and E. J. Verschoor, personal observations).
To discern between coevolution and new foamy virus introductions, we performed a comparative phylogenetic analysis of SFV integrase sequences from other primate species and did a thorough analysis of the host genotype. Interspecies transfer of SFV to humans has been described previously (1, 6, 11, 31, 32, 42) and has also been observed from a rhesus macaque to a capuchin monkey (E. J. Verschoor, unpublished observations). In addition, transfer of viruses between gibbons and orangutans has recently been suggested for the hepatitis B virus (35). In order to make a comprehensive analysis of the evolutionary relationships between these viruses, we amplified and sequenced additional SFV sequences from blood samples of various ape species. These sequences, together with published SFV sequences, were used to construct the evolutionary tree that is shown in Fig. 2B. In this tree, all ape SFV form one cluster that segregates into species-specific clusters. The latter finding supports our hypothesis of a strict virus-host coevolution within the apes, in contrast to recent cross-species transmission of foamy viruses in great apes. The latter would have resulted in a different tree topology, with mixtures of SFV from different species within the same cluster, as has been described for primate hepatitis B virus (35). Coevolution due to a single spatial separation event, however, is difficult to reconcile with the finding of a group of SFVora from mixed origin.
To verify the origin of the host, we performed mitochondrial DNA (mtDNA) analysis (41). 12S rRNA gene analysis using conserved primers (18) confirmed the Bornean and Sumatran origin of all samples used (data not shown), but analysis of control region (D-loop) sequences provided additional genetic information (Fig. 3). D-loop fragments were amplified as described previously (41) or by using a set of primers optimized on the Sumatran orangutan mtDNA genome. Sumatran D-loop sequences were amplified in a single-round PCR using the primers SUM-FA (5′-CATACAAAACTCCAACCACACTCG-3′) and ORAREV (5′-CTGAAGTAGGAACCAGATGCCG-3′). Amplification was performed in a 50-μl volume using 5 μl of DNA, 50 pmol of both primers, 0.2 mM each dNTP, 1.5 mM MgCl2, and 2.5 U of AmpliTaqGold (Perkin-Elmer, Nieuwerkerk aan de IJssel, The Netherlands). Samples were preheated for 15 min at 94°C to activate the enzyme and then cycled for 15 s at 94°C, 10 s at 52°C, and 40 s at 72°C for 40 rounds of amplification. While Bornean orangutans form one tight cluster of genotypes, the genetic variability of Sumatran orangutans is much greater. Sumatran orangutans form two well-supported groups A and B (100 and 92% bootstrap), in addition to Popa_Du whose genotype differs from all other Sumatran individuals. The latter finding may be biased due to the limited number of sequences analyzed, but it may also reflect the genetic variability of Sumatran orangutans (25). Interestingly, all four animals forming the Sumatran A genotype cluster were infected with SFVora belonging to subcluster SumBor. Although there is not a 100% correlation (Popa_Wa belongs to genotype B but is infected with a SumBor SFV), this finding suggests a linkage between Sumatran genotype A and infection with this particular SFVora variant.
FIG. 3.
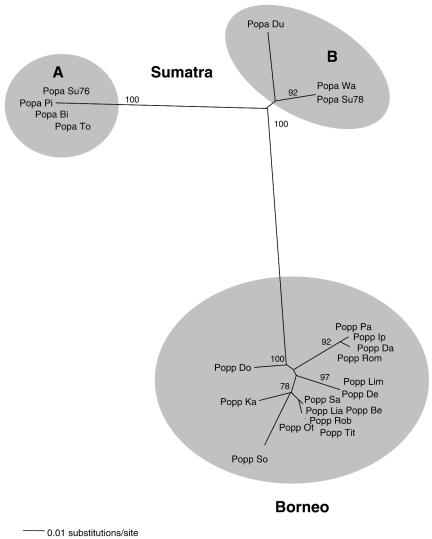
Unrooted neighbor-joining tree constructed using a 130-bp fragment amplified from the mitochondrial control region. All mtDNA sequences have been deposited in the EMBL and GenBank data libraries, accession numbers AJ627498 to AJ627519 for 12S rRNA and AJ627423 to AJ627444 for the D-loop sequences.
The genetic diversity of Sumatran orangutans has also been investigated previously (25). Interestingly, in their study, Muir et al. mention a Sumatran haplotype that was genetically closely related to Bornean haplotypes (25). They hypothesized that Sumatra had been recolonized with Bornean orangutans after the explosion of the volcano Toba, 74,000 years ago, had eradicated most orangutan populations on that island. Repopulation could have occurred during the Wisconsin Glacial Epoch when land bridges connected the islands. The pattern of SFVora genotypes can be explained by this repopulation hypothesis. The SFVora SumBor cluster may be a relic and provides support for this repopulation scenario, when SFV from a Bornean orangutan was introduced on Sumatra. The other fully Bornean and Sumatran SFV clusters likely reflect the viruses infecting founder populations on the islands that evolved 1.1 to 2.3 MYA.
The tight clustering of ape foamy viruses in our analysis (Fig. 2B; 84% bootstrap support) suggests that the lineage that gave rise to the apes was already infected with an ancestral foamy virus before speciation started 12 to 14 MYA (2, 10). However, more data are required, as only two viruses have been characterized from the lesser apes, the family Hylobatidae. In contrast to findings from others (36), we could not conclusively distinguish chimpanzee SFV (SFVcpz) variants linked to chimpanzee subspecies, although SFV from Pan troglodytes schweinfurthii differ from SFVcpz from the other two subspecies analyzed (P. t. troglodytes, n = 2; P. t. verus, n = 5). However, SFVcpz from the latter two subspecies all originated from animals housed in captivity together for over 30 years, and cross-subspecies transmission cannot be excluded.
Combined analysis of virus phylogeny, host phylogeny, and host origin can be an important tool to study the origins and phylogeography of viruses and to trace ancient migration patterns of the hosts (8, 14, 26-28, 30). Studies of retroviruses (8, 9, 13, 19-22, 30, 38, 39, 43, 46) and hepatitis B viruses (16, 35) have provided important information concerning ancient cross-species transmissions and virus-host coevolution in primates. Our phylogeographic analysis of SFVora provides insights in the orangutan's evolution and historical distributions, lending support for the hypothesis concerning ancient migrations of orangutans.
Nucleotide sequence accession numbers.
All novel SFV sequences described in this article have been deposited in the EMBL and GenBank data libraries, accession numbers AJ544579 and AJ627527 to AJ627560. All mtDNA sequences have been deposited under accession numbers AJ627498 to AJ627519 for 12S rRNA and AJ627423 to AJ627444 for the D-loop sequences.
Acknowledgments
We thank Heriyanto and the staff at the Wanariset Orang-utan Reintroduction Centre for assistance with sample collection.
This study was supported by the EU projects “Development of foamy virus vectors for somatic gene therapy” (grant BMH4-CT97-2010) and “INPRIMAT: Research infrastructure to promote primate molecular biology” (grant QLRI-CT-2002-01325).
REFERENCES
- 1.Achong, B. G., P. W. A. Mansell, M. A. Epstein, and P. Clifford. 1971. An unusual virus in cultures from a human nasopharyngeal carcinoma. J. Natl. Cancer Inst. 46:299-302. [PubMed] [Google Scholar]
- 2.Arnason, U., A. Gullberg, and A. Janke. 1998. Molecular timing of primate divergences as estimated by two nonprimate calibration points. J. Mol. Evol. 47:718-727. [DOI] [PubMed] [Google Scholar]
- 3.Bieniasz, P. D., A. Rethwilm, R. Pitman, M. D. Daniel, I. Chrystie, and M. O. McClure. 1995. A comparative study of higher primate foamy viruses, including a new virus from a gorilla. Virology 207:217-228. [DOI] [PubMed] [Google Scholar]
- 4.Blewett, E. L., D. H. Black, N. W. Lerche, G. White, and R. Eberle. 2000. Simian foamy virus infections in a baboon breeding colony. Virology 278:183-193. [DOI] [PubMed] [Google Scholar]
- 5.Blewett, E. L., J. Lewis, E. L. Gadsby, S. R. Neubauer, and R. Eberle. 2003. Isolation of cytomegalovirus and foamy virus from the drill monkey (Mandrillus leucophaeus) and prevalence of antibodies to these viruses amongst wild-born and captive-bred individuals. Arch. Virol. 148:423-433. [DOI] [PubMed] [Google Scholar]
- 6.Brooks, J. I., E. W. Rud, R. G. Pilon, J. M. Smith, W. M. Switzer, and P. A. Sandstrom. 2002. Cross-species retroviral transmission from macaques to human beings. Lancet 360:387-388. [DOI] [PubMed] [Google Scholar]
- 7.Broussard, S. R., A. G. Comuzzie, K. L. Leighton, M. M. Leland, E. M. Whitehead, and J. S. Allan. 1997. Characterization of new simian foamy viruses from African nonhuman primates. Virology 237:349-359. [DOI] [PubMed] [Google Scholar]
- 8.Gessain, A., R. C. Gallo, and G. Franchini. 1992. Low degree of human T-cell leukemia/lymphoma virus type I genetic drift in vivo as a means of monitoring viral transmission and movement of ancient human populations. J. Virol. 66:2288-2295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gessain, A., J. Pecon-Slattery, L. Meertens, and R. Mahieux. 2000. Origins of HTLV-1 in South America. Nat. Med. 6:232-233. (Letter.) [DOI] [PubMed] [Google Scholar]
- 10.Goodman, M., C. A. Porter, J. Czelusniak, S. L. Page, H. Schneider, J. Shoshani, G. Gunnell, and C. P. Groves. 1998. Toward a phylogenetic classification of primates based on DNA evidence complemented by fossil evidence. Mol. Phylogenet. Evol. 9:585-598. [DOI] [PubMed] [Google Scholar]
- 11.Heneine, W., W. M. Switzer, P. Sandstrom, J. Brown, S. Vedapuri, C. A. Schable, A. S. Khan, N. W. Lerche, M. Schweizer, D. Neumann-Haefelin, L. E. Chapman, and T. M. Folks. 1998. Identification of a human population infected with simian foamy viruses. Nat. Med. 4:403-407. [DOI] [PubMed] [Google Scholar]
- 12.Herchenroder, O., R. Renne, D. Loncar, E. K. Cobb, K. K. Murthy, J. Schneider, A. Mergia, and P. A. Luciw. 1994. Isolation, cloning, and sequencing of simian foamy viruses from chimpanzees (SFVcpz): high homology to human foamy virus (HFV). Virology 201:187-199. [DOI] [PubMed] [Google Scholar]
- 13.Herniou, E., J. Martin, K. Miller, J. Cook, M. Wilkinson, and M. Tristem. 1998. Retroviral diversity and distribution in vertebrates. J. Virol. 72:5955-5966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ho, L., S. Chan, R. D. Burk, B. C. Das, K. Fujinaga, J. P. Icenogle, T. Kahn, N. Kiviat, W. Lancaster, P. Mavromara-Nazos, V. Labropoulou, S. Mitriani-Rosenbaum, B. Norrild, M. R. Pillai, J. Stoerker, K. Syrjaenen, S. Syrjaenen, S. Tay, L. L. Villa, C. M. Wheeler, A. Williamson, and H. Bernard. 1993. The genetic drift of human papillomavirus type 16 is a means of reconstructing prehistoric viral spread and the movement of ancient human populations. J. Virol. 67:6413-6423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hooks, J. J., C. J. Gibbs, Jr., S. Chou, R. Howk, M. Lewis, and D. C. Gajdusek. 1973. Isolation of a new simian foamy virus from a spider monkey brain culture. Infect. Immun. 8:804-813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hu, X., A. Javadian, P. Gagneux, and B. H. Robertson. 2001. Paired chimpanzee hepatitis B virus (ChHBV) and mtDNA sequences suggest different ChHBV genetic variants are found in geographically distinct chimpanzee subspecies. Virus Res. 79:103-108. [DOI] [PubMed] [Google Scholar]
- 17.Hussain, A. I., V. Shanmugam, V. B. Bhullar, B. E. Beer, D. Vallet, A. Gautier-Hion, N. D. Wolfe, W. B. Karesh, A. M. Kilbourne, Z. Tooze, W. Heneine, and W. M. Switzer. 2003. Screening for simian foamy virus infection by using a combined antigen Western blot assay: evidence for a wide distribution among Old World primates and identification of four new divergent viruses. Virology 309:248-257. [DOI] [PubMed] [Google Scholar]
- 18.Kocher, T. D., W. K. Thomas, A. Meyer, S. V. Edwards, S. Paabo, F. X. Villablanca, and A. C. Wilson. 1989. Dynamics of mitochondrial DNA evolution in animals: amplification and sequencing with conserved primers. Proc. Natl. Acad. Sci. USA 86:6196-6200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Li, H. C., T. Fujiyoshi, H. Lou, S. Yashiki, S. Sonoda, L. Cartier, L. Nunez, I. Munoz, S. Horai, and K. Tajima. 1999. The presence of ancient human T-cell lymphotropic virus type I provirus DNA in an Andean mummy. Nat. Med. 5:1428-1432. [DOI] [PubMed] [Google Scholar]
- 20.Mang, R., J. Maas, A. C. van Der Kuyl, and J. Goudsmit. 2000. Papio cynocephalus endogenous retrovirus among Old World monkeys: evidence for coevolution and ancient cross-species transmissions. J. Virol. 74:1578-1586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Martin, J., E. Herniou, J. Cook, R. W. O'Neill, and M. Tristem. 1999. Interclass transmission and phyletic host tracking in murine leukemia virus-related retroviruses. J. Virol. 73:2442-2449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Martin, J., E. Herniou, J. Cook, R. Waugh O'Neill, and M. Tristem. 1997. Human endogenous retrovirus type I-related viruses have an apparently widespread distribution within vertebrates. J. Virol. 71:437-443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.McClure, M. O., P. D. Bieniasz, T. F. Schulz, I. L. Chrystie, G. Simpson, A. Aguzzi, J. G. Hoad, A. Cunningham, J. Kirkwood, and R. A. Weiss. 1994. Isolation of a new foamy retrovirus from orangutans. J. Virol. 68:7124-7130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Meiering, C. D., and M. L. Linial. 2001. Historical perspective of foamy virus epidemiology and infection. Clin. Microbiol. Rev. 14:165-176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Muir, C. C., B. M. Galdikas, and A. T. Beckenbach. 2000. mtDNA sequence diversity of orangutans from the islands of Borneo and Sumatra. J. Mol. Evol. 51:471-480. [DOI] [PubMed] [Google Scholar]
- 26.Ong, C., S. Chan, M. S. Campo, K. Fujinaga, P. Mavromara-Nazos, V. Labropoulou, H. Pfister, S. Tay, J. Ter Meulen, L. L. Villa, and H. Bernard. 1993. Evolution of human papillomavirus type 18: an ancient phylogenetic root in Africa and intratype diversity reflect coevolution with human ethnic groups. J. Virol. 67:6424-6431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Pavesi, A. 2003. African origin of polyomavirus JC and implications for prehistoric human migrations. J. Mol. Evol. 56:564-572. [DOI] [PubMed] [Google Scholar]
- 28.Pavesi, A. 2001. Origin and evolution of GBV-C/hepatitis G virus and relationships with ancient human migrations. J. Mol. Evol. 53:104-113. [DOI] [PubMed] [Google Scholar]
- 29.Rambaut, A. 1996. SE-AL Sequence Alignment Editor, v. 2.0a11. Department of Zoology, University of Oxford, Oxford, United Kingdom.
- 30.Salemi, M., M. Lewis, J. F. Egan, W. W. Hall, J. Desmyter, and A. M. Vandamme. 1999. Different population dynamics of human T cell lymphotropic virus type II in intravenous drug users compared with endemically infected tribes. Proc. Natl. Acad. Sci. USA 96:13253-13258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sandstrom, P. A., K. O. Phan, W. M. Switzer, T. Fredeking, L. Chapman, W. Heneine, and T. M. Folks. 2000. Simian foamy virus infection among zoo keepers. Lancet 355:551-552. [DOI] [PubMed] [Google Scholar]
- 32.Schweizer, M., V. Falcone, J. Gange, R. Turek, and D. Neumann-Haefelin. 1997. Simian foamy virus isolated from an accidentally infected human individual. J. Virol. 71:4821-4824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Schweizer, M., and D. Neumann-Haefelin. 1995. Phylogenetic analysis of primate foamy viruses by comparison of pol sequences. Virology 207:577-582. [DOI] [PubMed] [Google Scholar]
- 34.Schweizer, M., R. Turek, H. Hahn, A. Schliephake, K. O. Netzer, G. Eder, M. Reinhardt, A. Rethwilm, and D. Neumann-Haefelin. 1995. Markers of foamy virus infections in monkeys, apes, and accidentally infected humans: appropriate testing fails to confirm suspected foamy virus prevalence in humans. AIDS Res. Hum. Retrovir. 11:161-170. [DOI] [PubMed] [Google Scholar]
- 35.Starkman, S. E., D. M. MacDonald, J. C. Lewis, E. C. Holmes, and P. Simmonds. 2003. Geographic and species association of hepatitis B virus genotypes in non-human primates. Virology 314:381-393. [DOI] [PubMed] [Google Scholar]
- 36.Switzer, W. M., V. Bhullar, V. Shanmugam, M. E. Cong, B. Parekh, N. W. Lerche, J. L. Yee, J. J. Ely, R. Boneva, L. E. Chapman, T. M. Folks, and W. Heneine. 2004. Frequent simian foamy virus infection in persons occupationally exposed to nonhuman primates. J. Virol. 78:2780-2789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Swofford, D. L. 2002. PAUP*. Phylogenetic Analysis Using Parsimony (*and Other Methods), version 4.0b10. Sinauer Associates, Sunderland, Mass.
- 38.van der Kuyl, A. C., J. T. Dekker, and J. Goudsmit. 1995. Distribution of baboon endogenous virus among species of African monkeys suggests multiple ancient cross-species transmissions in shared habitats. J. Virol. 69:7877-7887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Vandamme, A. M., W. W. Hall, M. J. Lewis, P. Goubau, and M. Salemi. 2000. Origins of HTLV-1 in South America. Nat. Med. 6:232-233. (Letter.) [DOI] [PubMed] [Google Scholar]
- 40.Verschoor, E. J., S. Langenhuijzen, S. van den Engel, H. Niphuis, K. S. Warren, and J. L. Heeney. 2003. Structural and evolutionary analysis of an orangutan foamy virus. J. Virol. 77:8584-8587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Warren, K. S., E. J. Verschoor, S. Langenhuijzen, Heriyanto, R. A. Swan, L. Vigilant, and J. L. Heeney. 2001. Speciation and intrasubspecific variation of Bornean orangutans, Pongo pygmaeus pygmaeus. Mol. Biol. Evol. 18:472-480. [DOI] [PubMed] [Google Scholar]
- 42.Wolfe, N. D., W. M. Switzer, J. K. Carr, V. B. Bhullar, V. Shanmugam, U. Tamoufe, A. T. Prosser, J. N. Torimiro, A. Wright, E. Mpoudi-Ngole, F. E. McCutchan, D. L. Birx, T. M. Folks, D. S. Burke, and W. Heneine. 2004. Naturally acquired simian retrovirus infections in central African hunters. Lancet 363:932-937. [DOI] [PubMed] [Google Scholar]
- 43.Yamashita, M., R. Veronesi, M. Menna-Barreto, W. J. Harrington, Jr., C. Sampio, C. Brites, R. Badaro, A. S. Andrade-Filho, S. Okhura, T. Igarashi, J. Takehisa, T. Miura, D. Chamone, O. Bianchini, C. Jardim, S. Sonoda, and M. Hayami. 1999. Molecular epidemiology of human T-cell leukemia virus type I (HTLV-1) Brazil: the predominant HTLV-1s in South America differ from HTLV-ls of Japan and Africa, as well as those of Japanese immigrants and their relatives in Brazil. Virology 261:59-69. [PubMed] [Google Scholar]
- 44.Zhang, Y., and O. A. Ryder. 2001. Genetic divergence of orangutan subspecies (Pongo pygmaeus). J. Mol. Evol. 52:516-526. [DOI] [PubMed] [Google Scholar]
- 45.Zhi, L., W. B. Karesh, D. N. Janczewski, H. Frazier-Taylor, D. Sajuthi, F. Gombek, M. Andau, S. Martenson, and S. J. O'Brien. 1996. Genomic differentiation among natural populations of orang-utan (Pongo pygmaeus). Curr. Biol. 6:326-1336. [DOI] [PubMed] [Google Scholar]
- 46.Zsiros, J., M. F. Jebbink, V. V. Lukashov, P. A. Voute, and B. Berkhout. 1998. Evolutionary relationships within a subgroup of HERV-K-related human endogenous retroviruses. J. Gen. Virol. 79(Pt 1):61-70. [DOI] [PubMed] [Google Scholar]


