Summary
Chromatin featuring the H3 variant CENP-A at the centromere is critical for its mitotic function and epigenetic maintenance. Assembly of centromeric chromatin is restricted to G1 phase through inhibitory action of Cdk1/2 kinases in other phases of the cell cycle. Here, we identify the two key targets sufficient to maintain cell cycle control of CENP-A assembly. We uncovered a single phosphorylation site in the licensing factor M18BP1 and a cyclin A binding site in the CENP-A chaperone, HJURP, mediating specific inhibitory phosphorylation. Simultaneous expression of mutant proteins lacking these residues, results in complete uncoupling from the cell cycle. Consequently, CENP-A assembly is fully recapitulated under high Cdk activities, indistinguishable from G1 assembly. We find that Cdk-mediated inhibition is exerted by sequestering active factors away from the centromere. Finally, we show that displacement of M18BP1 from the centromere is critical for the assembly mechanism of CENP-A.
Introduction
Centromeres are chromosomal loci that drive faithful genome segregation during mitotic division (Allshire and Karpen, 2008). The functional foundation of the centromere is established by a specialized chromatin structure that features the histone H3 variant CENP-A (Black and Cleveland, 2011). This CENP-A-based chromatin domain provides a structural platform for formation of the kinetochore which links chromosomes to spindle microtubules during mitosis (Cheeseman and Desai, 2008; Foltz et al., 2006; Okada et al., 2006). In addition, CENP-A ensures stable maintenance of centromere position through an epigenetic, self-propagating feedback loop (Black and Cleveland, 2011; Gómez-Rodríguez and Jansen, 2013). Support for the epigenetic nature of the centromere comes from naturally occurring neocentromeres (Amor et al., 2004; Marshall et al., 2008), where centromere proteins vacate the original centromeric DNA sequence and assemble heritably on previously naïve chromatin. In addition, ectopic targeting of CENP-A or proteins of the centromere complex to a non-centromeric locus was shown to be sufficient to initiate a functional and heritable centromere (Barnhart et al., 2011; Hori et al., 2013; Mendiburo et al., 2011). Consistent with a key role at the core of a positive epigenetic feedback loop, CENP-A nucleosomes are long lived and are maintained through multiple cell divisions (Bodor et al., 2013; Jansen et al., 2007). The unusually slow turnover of CENP-A at each centromere (Falk et al., 2015) indicates that replenishment is either equally slow or is limited in time and tied to CENP-A redistribution following DNA replication. Indeed, in metazoans, assembly of newly synthesized CENP-A is directly linked to cell cycle progression and is initiated during mitotic exit and restricted to early G1 phase of the cell cycle (Jansen et al., 2007; Schuh et al., 2007).
Previously we showed that brief inhibition of cyclin dependent kinase 1 and 2 (Cdk1/2) activities is sufficient to drive CENP-A deposition prior to mitotic exit (Silva et al., 2012). This has led to a model where the CENP-A assembly machinery is present and poised for activity but is kept inactive throughout S, G2 and M phase, until mitotic exit when activities of Cdk1/2 drop, concomitant with the onset of CENP-A deposition. Key proteins necessary for the process of CENP-A deposition include the Mis18 complex and the CENP-A chaperone HJURP which bears CENP-A-specific nucleosome assembly activity (Dunleavy et al., 2009; Foltz et al., 2009; Fujita et al., 2007). HJURP and M18BP1 (also known as HsKNL2), a member of the Mis18 complex, are phosphoproteins (Bailey et al., 2016; Dephoure et al., 2008; Kato et al., 2007; McKinley and Cheeseman, 2014; Müller et al., 2014; Silva et al., 2012; Wang et al., 2014) and localize to centromeres in a cell cycle controlled manner, in early G1 phase (Dunleavy et al., 2009; Foltz et al., 2009; Fujita et al., 2007; Maddox et al., 2007), indicating they are putative targets for Cdk regulation. In addition, recent work has identified the mitotic kinase Plk1 as a critical component to drive CENP-A assembly (McKinley and Cheeseman, 2014). However, while Plk1 is itself a cell cycle controlled kinase, it does not restrict CENP-A assembly to G1 phase as it is required for both canonical assembly in G1 phase as well as for premature assembly upon Cdk inhibition. In addition, several residues on CENP-A itself are phosphorylated (Bailey et al., 2016; Yu et al., 2015; Zeitlin et al., 2001). One of these, serine 68, is proposed to phosphorylated by mitotic Cdk activity (Yu et al., 2015) but the relevance of this is being disputed (Fachinetti et al., 2017) and mutation of this residue does not lead to a change in the timing of CENP-A deposition. In contrast, mutations of phospho-residues in HJURP or artificial recruitment of M18α to centromeres has been reported to result in premature centromere recruitment of CENP-A (McKinley and Cheeseman, 2014; Müller et al., 2014). While these studies point to a contributing role for these factors, they leave open the critical question of which factors are necessary, which are sufficient, how Cdk-mediated control is exerted, and how key proteins are functionally inhibited.
To resolve the specific molecular steps that ensure cell cycle restricted CENP-A assembly, we report full uncoupling of CENP-A assembly from the cell cycle/Cdk regulation. To achieve this, we identified a functional cyclin-interacting domain in HJURP and a critical phospho-site in M18BP1. Simultaneous uncoupling of these factors from cell cycle progression results in a complete reconstitution of CENP-A assembly process prematurely in G2 phase, prior to mitotic exit. Our results identify a dual inhibitory mechanism that is sufficient to maintain cell cycle restricted centromere propagation and define the molecular underpinnings of how assembly is turned on and subsequently turned off.
Results
HJURP is phosphorylated in a cell cycle dependent manner
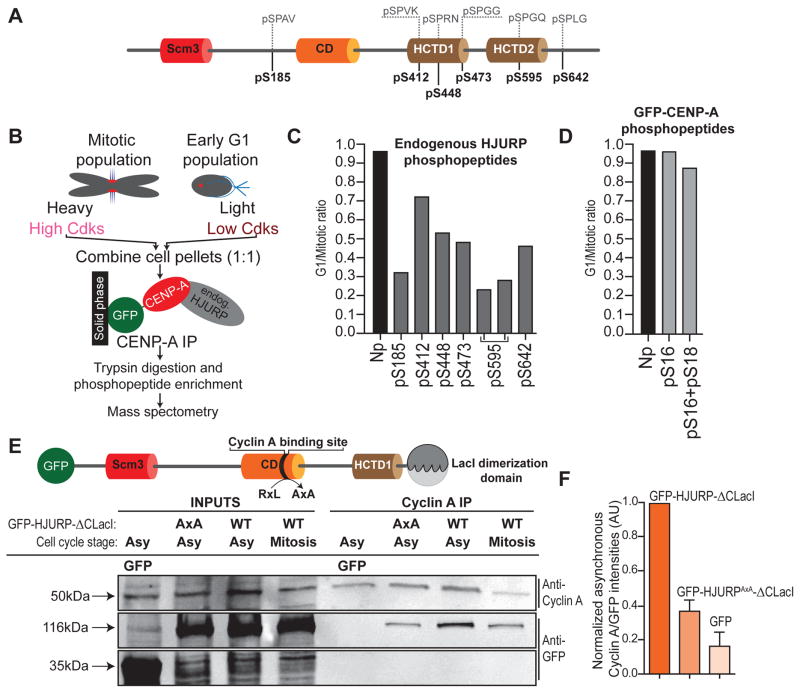
HJURP, the CENP-A specific chaperone, is a phospho-protein and features several putative Cdk sites (Figure 1A and (Bailey et al., 2016; Dephoure et al., 2008; Kato et al., 2007; Müller et al., 2014; Wang et al., 2014), making it a prime candidate for cell cycle control of CENP-A assembly. To quantitatively measure HJURP phosphorylation we used stable isotope labeling by amino acids in cell culture (SILAC) coupled to mass spectrometry. This allowed us, in an unbiased manner, to precisely determine which residues are phosphorylated under high Cdk conditions and how these respond to changes in Cdk activity. Cdk1 levels differ most dramatically between mitosis and G1 phase. We therefore compared levels of phospho-peptides on the prenucleosomal GFP-CENP-A/HJURP complex between populations of mitotically arrested cells and cells that are released from mitotic arrest by Roscovitine-mediated Cdk inhibition (Figure 1B). Normal timing and efficiency of CENP-A assembly is preserved under these conditions (Figure S1). We detected 6 phosphorylated residues corresponding to putative Cdk consensus sites within HJURP, all of which were dephosphorylated upon mitotic exit, ranging from 25–70% decrease relative to mitotic values (Figures 1C, S2). Although three of these sites (S412, S448, S473) correspond to reported phospho-sites (Müller et al., 2014; Wang et al., 2014), our analysis shows that these are neither the sole nor the most responsive sites to inactivation of Cdks, at least in mitosis. In contrast, no change is observed at unphosphorylated peptides of HJURP (Figure 1C) nor at Cdk-consensus phospho-sites on the CENP-A N-terminal tail (Bailey et al., 2013) after forced mitotic exit (Figure 1D), indicating that protein levels of CENP-A and HJURP remain unaffected (see also Figure S1F, G) and that HJURP is selectively dephosphorylated.
Figure 1. HJURP is phosphorylated in a Cdk-dependent manner and interacts with cyclin A (related to Figures S1 and S2).
(A) Schematic representation of HJURP protein (Scm3: CENP-A binding domain; CD: Conserved Domain (CD), HCTD: (HJURP C-Terminal Domain). Position of phospho-sites identified by SILAC in C are indicated. Amino acid sequences flanking phospho-sites are annotated in grey.
(B) Schematic of SILAC experiment (see supplemental experimental procedures for details). Light cells were released into G1 by Roscovitine treatment for 30 min. At this stage HJURP is partially dephosphorylated (See Figure S1F, G).
(C) The L/H ratios of phosphorylated Cdk sites detected on endogenous HJURP are listed. A representative non-phosphorylated peptide (Np) is shown as internal control. Note: pS595 was detected on two independent peptides.
(D) L/H ratios of Cdk consensus sites within the N-terminal tail of CENP-A (See Figure S2 for data from two additional replicate experiments).
(E) HJURP CD mediates interaction with Cyclin A. (Top) Schematic representation of HJURP protein. Mutation of conserved RxL motif to AxA is annotated with black arrow. Experiments are performed with an HJURP construct in which the C-terminal homodimerization domain is replaced with that of LacI to prevent dimerization with wild type HJURP. (Bottom) co-IP of extracts expressing indicated constructs, either from asynchronous or mitotically enriched cells. Bound complexes were separated using SDS-PAGE followed by immunoblotting with indicated antibodies.
(F) Quantification of IP experiments. GFP signal from each IP was normalized to corresponding cyclin A signal and input GFP signal in order to control for IP efficiency and GFP fusion protein expression level, respectively. GFP-HJURP signals were set to 1. Error bars indicate SEM (standard error of mean) from 3 independent experiments.
The HJURP conserved domain interacts with Cyclin A and controls timing of CENP-A assembly
Our findings from SILAC experiments led us to focus on HJURP in particular, and determine how its phospho-regulation is coupled to the control of cell cycle timing of CENP-A chromatin assembly. Although the canonical consensus site for Cdks is (S/T)PX(K/R)(Hagopian et al., 2001; Holmes and Solomon, 1996), 5 of the 6 phospho-sites in HJURP that are affected by Cdk inactivation display a shorter (S/T)P motif (Figure 1A) (Errico et al., 2009). Phosphorylation of such truncated motifs often requires additional cyclin binding sites for enhanced substrate recognition (Adams et al., 1996; Russo et al., 1996). Indeed, we found a typical cyclin A binding RxL motif (Brown et al., 2007) within a vertebrate conserved domain (CD) of HJURP, which has no previously described function (Sanchez-Pulido et al., 2009). We tested whether HJURP interacts with cyclin A and B, the major drivers of Cdk activity in S/G2 phase and mitosis, respectively, all stages at which CENP-A assembly is inhibited (Silva et al., 2012). We performed either Cyclin B or Cyclin A co-immunoprecipitation from HEK293T cells in which we ectopically expressed either GFP-tagged HJURP with a mutated RxL motif (RLL>ALA, henceforth referred to as HJURPAxA), or with a wild type CD. HJURP forms a homodimer (Zasadzińska et al., 2013). To avoid cross-dimerization with endogenous HJURP, we replaced its C-terminal domain with that of LacI, which does not interfere with the CENP-A chaperoning and assembly activity of HJURP, as described (Zasadzińska et al., 2013) (henceforth named HJURP-ΔCLacI). Cyclin A robustly co-immunoprecipitated GFP-tagged HJURP-ΔCLacI (Figure 1E and supplemental Figure S3A). In contrast, GFP-HJURPAxA-ΔCLacI pulldown was reduced by 70% compared to HJURP-ΔCLacI, carrying a wild type CD (Figure 1F). Mitotically enriched cells (low cyclin A) were used as a control to demonstrate that HJURP pulldown is cyclin A dependent. Consistent with the fact that inhibition of CENP-A assembly is maintained in mitosis (Jansen et al., 2007), even though cyclin A is degraded in early mitosis (den Elzen and Pines, 2001; Geley et al., 2001), we find that like cyclin A, cyclin B can interact with HJURP (Figure S3B). However, this interaction is not dependent on an intact CD within HJURP, indicating inhibitory control in mitosis is exerted through a different mechanism.
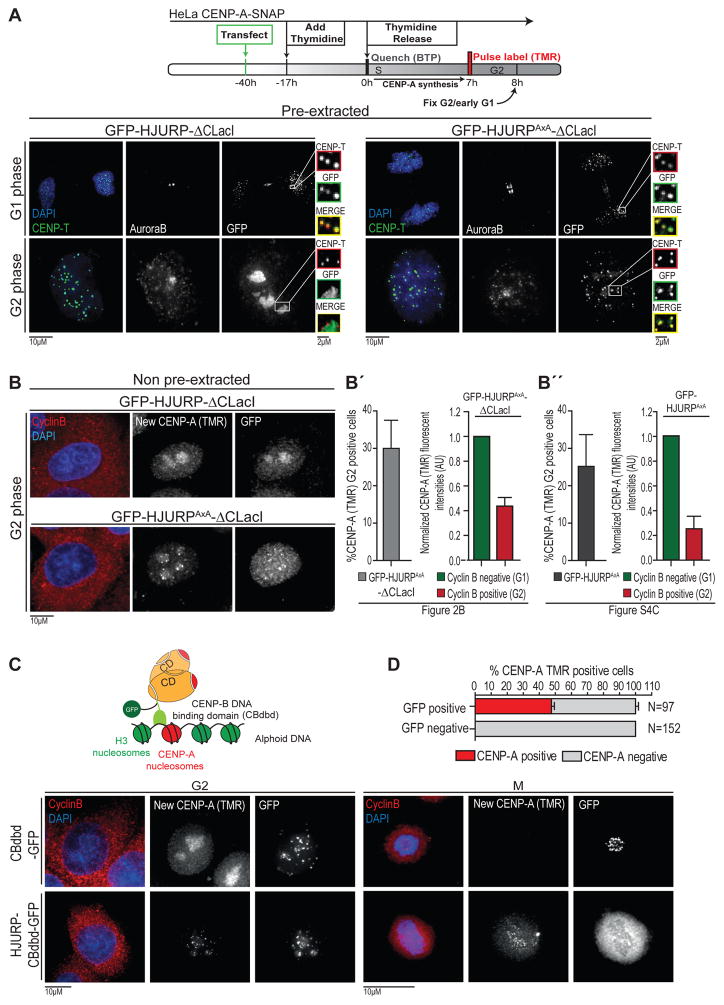
Our mapping of the principal cyclin A interaction site on HJURP allowed us to determine the consequences of the loss of this interaction for the timing of its localization along the cell cycle. Upon removal of soluble HJURP by pre-extraction we revealed that the stably chromatin bound, pre-mitotic HJURP-ΔCLacI is enriched in nucleoli [as observed previously (Dunleavy et al., 2009)]. In contrast, HJURPAxA-ΔCLacI targeted to centromeres prematurely in G2 phase, the time of the cell cycle in which cyclin A is the principal cyclin (Figure 2A). In addition, we analyzed CENP-A deposition using a SNAP tag-based, fluorescent quench-chase-pulse labeling protocol that we described previously (Figure 2B)(Bodor et al., 2012; Silva et al., 2012). Remarkably, expression of the cyclin A binding mutant of HJURP, but not its wild type counterpart resulted in a precocious deposition of nascent CENP-A in G2 phase. We performed these experiments using HJURP-ΔCLacI to force homodimerization of HJURPAxA. In this way, we show that HJURPAxA itself is a functional assembly factor, independent of wildtype HJURP copies. Consistent with this, downregulation of endogenous HJURP showed no effect on either efficiency or frequency of premature CENP-A loading following GFP-HJURPAxA–ΔCLacI expression (Figure S4A, B). Either GFP-HJURPAxA–ΔCLacI (Figure 2B) or GFP-HJURPAxA (carrying the endogenous C-terminal HJURP dimerization domain) (Figure 2B″, S4C) expression result in a similar level of precocious deposition of CENP-A demonstrating that uncoupling is not an artifact of LacI mediated dimerization. Quantitative analysis showed that precocious CENP-A assembly at the centromere reached ~40% of G1 levels (Figure 2B, see also S4D)[i.e. 20% assembly per centromere, considering the replicated state of sister centromeres in G2 phase, unresolvable by microscopy]. We conclude that the CD of HJURP is a cell cycle control element that interacts with cyclin A. Disruption of this site is sufficient to alleviate at least part of the Cdk-mediated inhibition of HJURP.
Figure 2. Timing of HJURP targeting and CENP-A deposition is controlled by HJURP CD. (related to Figure S3 and S4).
(A) HeLa CENP-A-SNAP cells were transiently transfected with indicated constructs and Thymidine synchronized to enrich cells in G2 phase. Cells were permeabilized prior to fixation and counterstained for Aurora B, CENP-T and DAPI to distinguish between G2 and early G1 cell cycle phases, centromere localization and DNA, respectively. GFP booster was used to amplify GFP-HJURP fluorescent signal.
(B) Experiments were performed as in Figure 2A except here CENP-A assembly was assayed using SNAP TMR-labeling of its S phase synthesized pool. Following fixation, cells were counterstained for cyclin B and DAPI to indicate G2 status and DNA, respectively. (See Figure S4C for extended analysis of GFP-HJURP and GFP-HJURPAxA induced assembly).
(B′) Left: Quantification of frequency of premature CENP-A loading in Cyclin B positive cells expressing GFP-HJURPAxA-ΔCLacI. Right: Quantification of CENP-A-SNAP (TMR) fluorescent signal intensities of cells from experiment on the left in G2 phase (Cyclin B positive) and G1 phase (Cyclin B negative), using CENP-T signal as a centromere reference (not depicted). Centromeric CENP-A-SNAP fluorescent signals were normalized to average of G1 cells signals in each experiment (not considering the difference in replicated sister G2 centromeres vs. segregated G1 centromeres). 3 replicates, error bars indicate SEM.
(B″) Left: Quantification of frequency of premature CENP-A loading in Cyclin B positive cells expressing GFP-HJURPAxA from 3 replicate experiments (see Figure S4C for images). Right: Quantification of CENP-A-SNAP (TMR) fluorescent signal intensities from the same experiment.
(C) Top: Schematic of relevant domains in centromere targeted HJURP. Bottom: HeLa CENP-A-SNAP cells were transfected with indicated constructs. 7 hours post Thymidine release cells were either fixed in G2 phase or collected in Nocodazole to enrich for mitotic cells. Cells were counterstained for cyclin B and DAPI to indicate G2 status and DNA, respectively.
(D) Quantification of frequency of premature CENP-A-SNAP deposition in Cyclin B positive cells, driven by expression of Cbdb-HJURP-GFP. Error bars indicate SEM.
Cdk activity controls HJURP localization not its chaperoning activity
Phosphorylation of HJURP could directly interfere with its chaperoning activity, thereby inactivating the key function of the protein. Alternatively, it may sequester an otherwise active HJURP away from the centromere, preventing its untimely recruitment. To distinguish between these possibilities, we fused HJURP to the DNA binding domain of CENP-B (CBdbd)(Figure 2C). This domain binds specifically to centromeric α-satellite DNA and allows us to drive HJURP to centromeres in G2 synchronized cells, while likely bearing inhibitory phosphorylation due to high Cdk activity. We detected nascent CENP-A-SNAP at G2 centromeres after expression of HJURP-CBdbd-GFP (Figure 2C, D) but not CBdbd-GFP alone, indicating centromeric localization of HJURP is sufficient to enable unscheduled CENP-A loading. Although HJURP is removed from mitotic chromatin (a process that apparently overrides the DNA binding activity of the CENP-B DNA binding domain), newly loaded CENP-A-SNAP remained associated with centromeres upon entry into mitosis, suggesting it is assembled into centromeric nucleosomes rather than part of an HJURP-associated prenucleosomal complex (Figure 2C, right). Based on these results, we conclude that Cdk-driven phosphorylation does not interfere with HJURP chaperoning activity, rather it results in sequestering HJURP away from the centromere, preventing its untimely recruitment.
HJURP Serine 210/211 is functionally phosphorylated in G2 phase cells
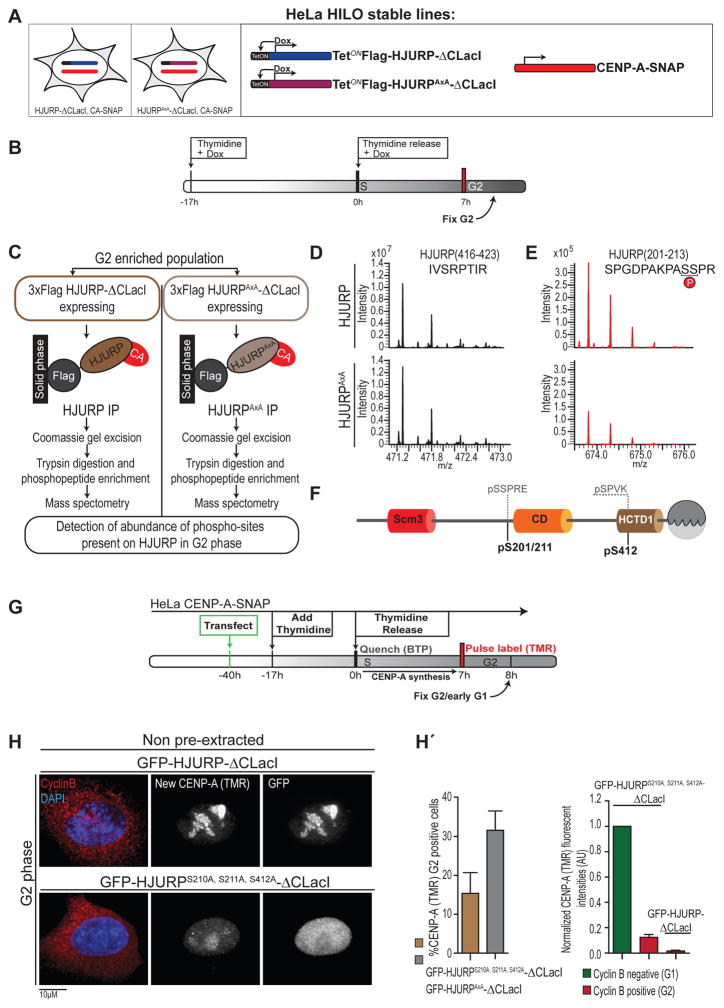
Next, we determined whether the uncoupling of HJURP from its cell cycle control involves specific phosphorylation sites. Expression of HJURP in which the 6 identified putative mitotic Cdk phospho-residues (Figure 1C) were mutated to alanine (either all 6 or combinations thereof) did not result in changes in the timing of CENP-A assembly (Figure S5B), despite previous reports implicating three of these residues [S412, S448, and S472 (Müller et al., 2014)]. Because we observe premature CENP-A assembly in G2 phase during which cyclin A is the major cyclin, we aimed to identify additional, potentially relevant, phospho-residues in this cell cycle window. We expressed Doxycycline (Dox) inducible 3xFlag-HJURP-ΔCLacI or 3xFlag-HJURPAxA-ΔCLacI in G2 phase enriched HeLa HILO cells (Khandelia et al., 2011)[Figure 3A, B (see also section below)]. Following 3xFlag-HJURP-ΔCLacI immunoprecipitation, TiO2 phospho-enrichment and mass spectrometry (Figure 3C), we identified S210/S211 phosphopeptides (the proximity of these residues prevented us from differentiating S210 vs. S211 as the site of phosphorylation). These phosphopeptides were not detected in mitotically synchronized cells (Figure 1C), suggesting differential phosphorylation of HJURP, consistent with our finding that cyclin B also interacts with HJURP but in a CD-independent manner (Figure S3B). Further, we found S412 to be the only common phospho-residue between G2 and mitotically synchronized cells (Figure 3F). Importantly, the relative abundance of S210/S211 phospho-peptides was substantially reduced on the HJURPAxA mutant in which cyclin A binding is reduced compared to wild type (Figure 3E). This suggests that the cyclin A/Cdk complex interaction with HJURP results in phosphorylation of this site.
Figure 3. HJURP Serine 210/211 is functionally phosphorylated in G2 phase cells. (related to Figure S5).
(A) Schematic of cell lines used for a label free mass spec analysis.
(B) HeLa HILO cells carrying indicated Doxycycline-inducible HJURP constructs were enriched in G2 cells by Thymidine arrest and release during Dox induction.
(C) Cell pellets obtained from experiment in (B) were subjected to immunoprecipitation using Flag-coupled agarose beads to isolate 3xFlag-HJURP-ΔCLacI, separated on SDS-PAGE followed by Coomassie-based excision of HJURP proteins. Purified proteins were subjected to trypsin digestion, phosho-peptide enrichment, followed by LC-MS/MS analysis.
(D) Mass spectra of a representative non-phosphorylated HJURP peptide from the flow through of the phospho-enrichment, of samples from cells containing WT HJURP-ΔCLacI (top) and HJURPAxA -ΔCLacI (bottom).
(E) Mass spectra of the phosphopeptide containing pS210/pS211, from the elution of the phospho-enrichment from cells expressing indicated constructs. Because the two serines are adjacent, it was not possible to differentiate between S210 and S211 as the site of phosphorylation.
(F) Schematic representation of Cdk-consensus phospho-sites detected on HJURP in G2 phase.
(G) Experiment analogous to Figure 2A assaying indicated HJURP constructs for localization and CENP-A assembly in G2 phase.
(H) Representative images of cells from experiment in (G). CENP-A assembly was assayed using SNAP TMR-labeling of its S phase synthesized pool. Following fixation, cells were counterstained for cyclin B and DAPI to indicate G2 status and DNA, respectively.
(H′) Left: Quantification of frequency of premature CENP-A loading in Cyclin B positive cells expressing indicated constructs from 3 replicate experiments. Right: Quantification of CENP-A-SNAP (TMR) fluorescent signal intensities.
To test the functional significance of these residues we mutated serines 210 and 211 in combination with serine 412 and expressed HJURPS210A,S211A,S412A-ΔCLacI mutants in G2 phase cells. Quench-chase-pulse labeling of CENP-A-SNAP showed that mutation of these residues to alanine results in low, but detectable levels of nascent CENP-A at centromeres (Figure 3G, H). This indicates that cyclin A binding to HJURP in G2 phase results in phosphorylation, at least on serines S210/211 and S412 and that these modifications contribute to preventing premature CENP-A assembly.
HJURPAxA-induced CENP-A assembly in G2 phase is Mis18-dependent
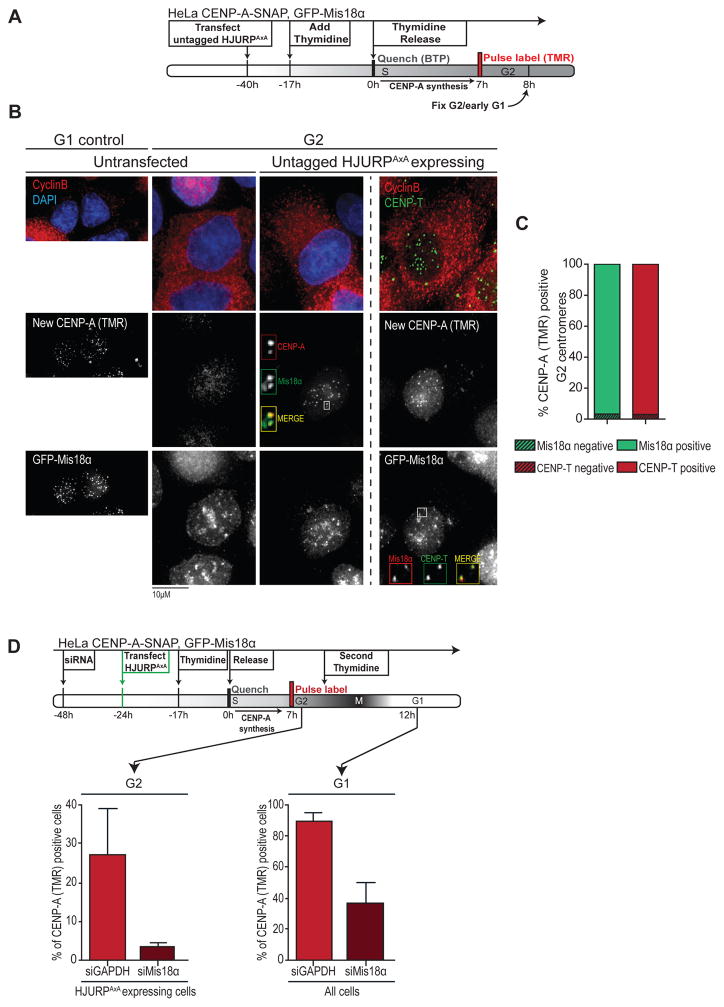
Although HJURPAXA is capable of inducing unscheduled CENP-A assembly, it does so with a relatively low efficiency and centromere specificity as compared to canonical G1 loading (Figure 2B′, B″). This indicates that an additional level of cell cycle control exists. A candidate for this is the Mis18 complex, which includes Mis18α, Mis18β and the associated protein M18BP1 (Fujita et al., 2007). All subunits share a common localization pattern, with highly enriched and centromere specific localization in anaphase, followed by disappearance in mid-G1 (Fujita et al., 2007; Silva and Jansen, 2009). Interestingly, we found that premature, HJURPAXA driven CENP-A assembly in G2 phase correlates with low levels of stably expressed GFP-Mis18α at centromeres (Figure 4A–C). Moreover, siRNA-mediated depletion of Mis18α leads to a loss of both canonical assembly in G1 phase as well as premature assembly of CENP-A in G2 phase (Figure 4D). This demonstrates that HJURPAxA-induced assembly occurs through the canonical assembly pathway and suggests that the partial nature of this assembly is possibly due to low levels of Mis18 complex members at G2 centromeres.
Figure 4. HJURPAxA induced CENP-A assembly is Mis18α dependent.
(A) Stable GFP-Mis18α, CENP-A-SNAP double transgenic HeLa cells were transfected with untagged HJURPAxA, synchronized and assayed for nascent CENP-A assembly by SNAP quench-chase-pulse labeling, followed by immunostaining for cyclin B and DAPI to indicate G2 status and DNA, respectively.
(B) Representative images of experiment described in (A).
(C) Quantification of frequency of CENP-A (TMR) positive G2 centromeres of experiment described in (A). Cells were scored in relation to whether GFP-M18α (green) or CENP-T (red) signals are simultaneously detected together with CENP-A (TMR) or not.
(D) Top: Scheme outlining RNAi against Mis18α or GAPDH, synchronization and Quench-Chase-Pulse labeling of CENP-A-SNAP, GFP-Mis18α cells. (Bottom) Quantification of CENP-A-SNAP (TMR) positive cells from 3 independent experiments. Error bars indicate SEM.
Recruitment of the Mis18 complex to the centromere is controlled by phosphorylation of M18BP1T653
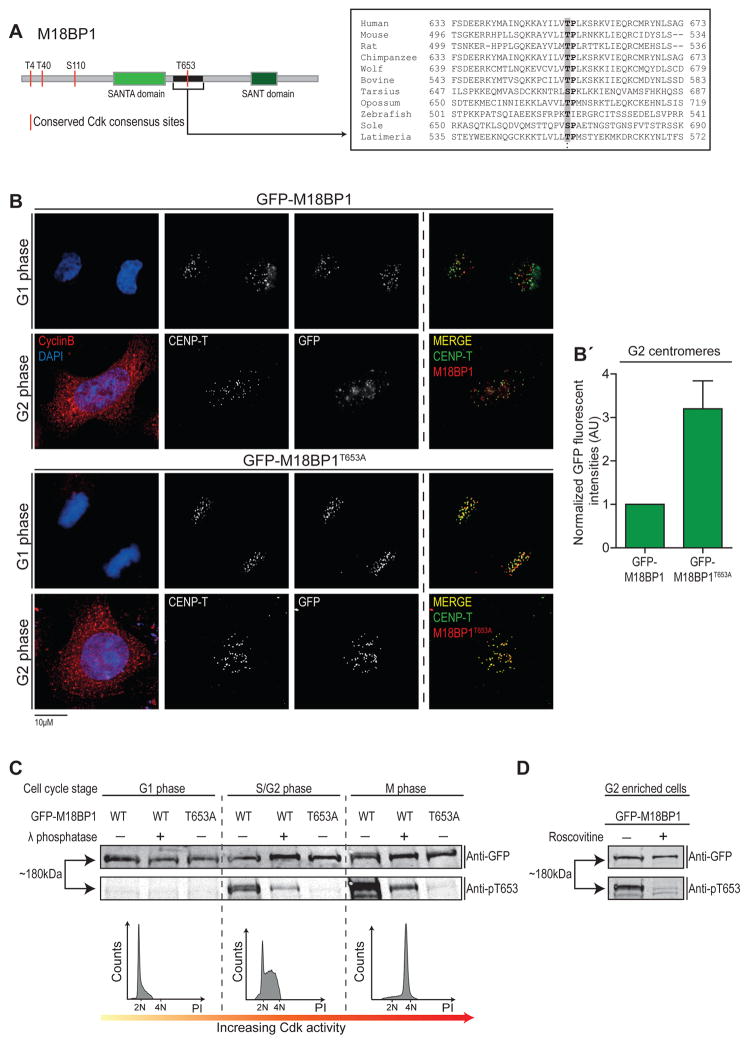
Previously, we reported that a phospho-dead M18BP1 mutant in which 24 known phospho-sites are mutated to alanine, resulted in its premature centromere targeting (Silva et al., 2012), suggesting that at least one of these sites is regulated by Cdks. We now identified four putative Cdk motifs that are highly conserved among vertebrates, three of which are clustered close to the N-terminus of M18BP1 (T4, T40 and S110), while a fourth (T653) is located between the highly conserved SANTA and SANT domains (Maddox et al., 2007) (Figure 5A). Mutation of all 4 sites to alanine leads to a loss of cell cycle controlled localization of M18BP1 (Figure S6A). Interestingly, mutation of T653 alone was sufficient to result in premature centromere targeting of M18BP1 with a ~3-fold increase in centromeric levels relative to wild type protein (Figure 5B). We generated a phospho- and site-specific antibody against the T653 site and show that pT653 levels rise as cells accumulate in S/G2 and mitosis, correlating with increasing levels of Cdk1 and 2 activities (Figure 5C). A brief treatment with Cdk1/2 inhibitor of cells expressing GFP-M18BP1 caused a strong reduction in phosphorylation of T653, suggesting that M18BP1 is a direct target of these kinases (Figure 5D).
Figure 5. Cdk-mediated T653 phosphorylation of M18BP1 controls its centromere recruitment (related to Figure S6).
(A) M18BP1 T653 is conserved amongst vertebrates. Left: Schematic of M18BP1 protein. Relevant domains and conserved Cdk sites are indicated. Right: Conservation of human T653 residue across species. Conserved Threonine or Serine is highlighted in grey.
(B) T653 residue controls cell cycle-dependent M18BP1 centromere recruitment. Indicated constructs were transfected into asynchronous HeLa cells 48hr prior to fixation, followed by counterstaining for cyclin B, CENP-T and DAPI to indicate G2 status, centromeres and DNA, respectively.
(B′) Average centromeric GFP fluorescent signals from Cyclin B positive cells were determined using the Centromere Recognition and Quantification (CRaQ) method (Bodor et al., 2012) and normalized to GFP-M18BP1. Error bars indicate standard error of the mean (SEM) from 3 replicates.
(C) T653 is phosphorylated in a cell cycle dependent manner. Hek293T cells were transiently transfected with GFP-Mis18BP1 (WT) or GFP-Mis18BP1T653A as a non-phosphorylatable control. 24h later, cells were synchronized in indicated cell cycle stages and lysed. Extracts were either left untreated or treated with lambda phosphatase, separated by SDS-PAGE followed by immunoblotting with indicated antibodies (see also supplemental experimental procedures). Apparent molecular weight is indicated. Cells were assayed for cell cycle position by FACS using propidium iodide (PI) to indicated DNA content.
(D) T653 is phosphorylated by Cdk1/2. Hek293T cells were transiently transfected with GFP-Mis18BP1 and enriched in G2 phase by a single thymidine block followed by 7h of release. 30min before fixation, cell were treated with 100μM Roscovitine. Extracts were separated by SDS-PAGE followed by immunoblotting with anti-GFP and anti-pT653 antibodies.
Further, the M18BP1T653A mutant co-recruited Mis18α to G2 centromeres, indicative of ongoing Mis18 complex formation independent of T653 phosphorylation (Figure S6B). An N-terminal 490 amino acid fragment of M18BP1 was reported to be functional in supporting CENP-A assembly in G1 phase (McKinley and Cheeseman, 2014), consistent with our finding that mutation of the T653 residue does not abrogate M18BP1 localization, but we now add that this residue controls cell cycle dependent localization. To test whether M18BP1 phosphorylation of T653 results in disruption of the Mis18α interaction, we expressed a translational fusion of wild type or mutant M18BP1 to the CBdbd in cells synchronized in G2 phase (analogous to artificial HJURP tethering, Figure 2C). Forced recruitment of M18BP1 to centromeres leads to strong co-recruitment of Mis18α to G2 centromeres, suggesting that the Mis18 complex can form under inhibitory Cdk activity, at least at this stage in the cell cycle (Figure S6C), although not in mitosis as observed previously (McKinley and Cheeseman, 2014). Similarly, forced recruitment of a phosphomimetic M18BP1T653D (Figure S6C) or M18BP1T653E (not shown) mutant is capable of co-recruitment of Mis18α. Thus, we find that mutation of the T653 residue does not disrupt the M18BP1/Mis18α interaction. Rather, its phosphorylation prevents centromere targeting of the Mis18 complex in G2 phase until mitotic exit when Cdk1/2 activities are low.
Cdk-mediated control of M18BP1 and HJURP is sufficient to ensure tight cell cycle timing of centromere propagation
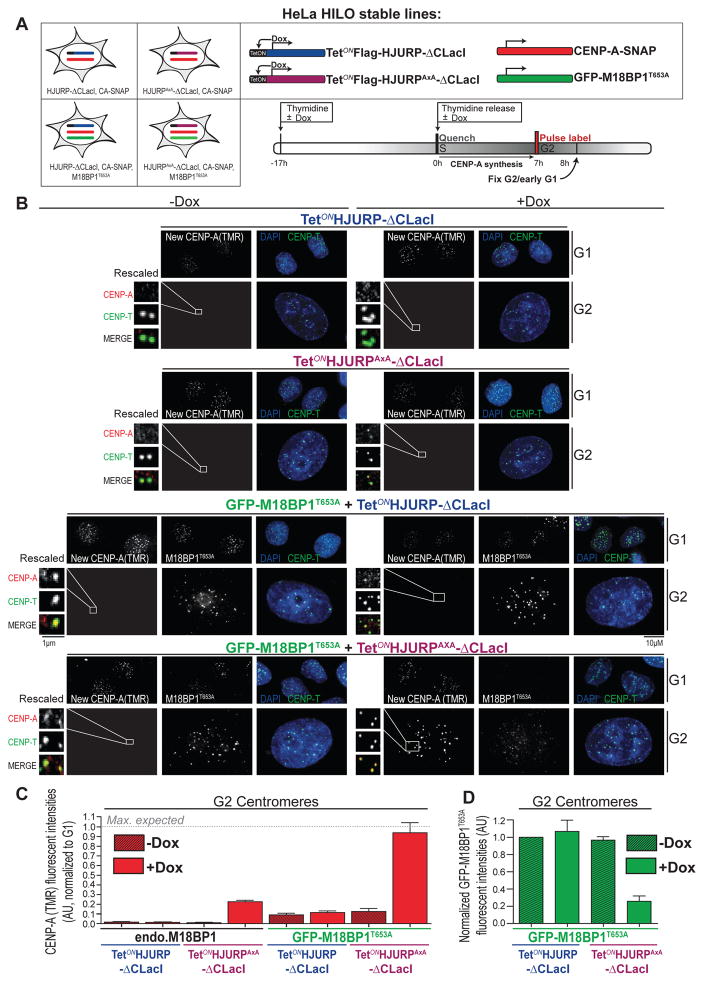
Our results indicate that centromere localization of both HJURP and M18BP1 is blocked by Cdk-mediated phosphorylation, suggesting that combined phospho-control of these protein complexes contributes to cell cycle specific loading of CENP-A. To directly test this, we constructed HeLa HILO cells expressing equal levels of either HJURP-ΔCLacI or HJURPAxA-ΔCLacI (Figure S7A) under the control of a doxycycline-inducible promoter at a defined locus using recombination-mediated cassette exchange (RMCE)(Khandelia et al., 2011). HJURP induction was performed either in cells stably expressing GFP-tagged M18BP1T653A or expressing endogenous M18BP1 along with CENP-A-SNAP to assay for CENP-A assembly (Figure 6A). We compared the efficiency of G2 phase loading to the normal level of assembly in G1 phase. CENP-A assembly in uninduced control G1 cells was equal across all cell lines and essentially completed at the time of fixation (Figure S7B, C, respectively). As observed after transient expression, induction of HJURPAxA alone resulted in low levels (~20% of G1, when corrected for centromere replication in G2 phase) of CENP-A assembly (Figure 6B, C, S7D). Force expression of otherwise wild type but GFP-tagged M18BP1 does not enhance the degree of premature CENP-A assembly (Figure S7E, E′). Conversely, constitutive M18BP1T653A expression led to infrequent and inefficient recruitment of nascent CENP-A to G2 centromeres (Figure 6C). Remarkably, induction of HJURPAxA combined with stably expressed M18BP1T653A resulted in highly efficient and centromere restricted CENP-A assembly in G2 phase, reaching 93% of G1 control levels (Figure 6C). In sum, while disrupting the timing of centromere targeting of either HJURP or M18BP1 results in a limited deregulation of CENP-A assembly, as has been shown previously (McKinley and Cheeseman, 2014; Müller et al., 2014), we now show that simultaneous uncoupling of both of these proteins leads to full-fledged CENP-A assembly, indistinguishable from canonical G1 phase assembly. These findings strongly suggest that M18BP1 and HJURP are the two principal targets of Cdk-mediated inhibition.
Figure 6. A dual inhibitory mechanism restricts CENP-A deposition to G1 phase (related to Figure S7).
(A) Schematic representation of Hela HILO cells carrying low levels of constitutively expressed CENP-A-SNAP (red), with or without stable expression of GFP M18BP1T653A (green) along with Doxycycline-inducible 3xFlag-HJURP-ΔCLacI (blue) or 3xFlag-HJURPAxA-ΔCLacI (purple). Cells were processes as indicated in the scheme.
(B) Representative images of the experiment described above. Following fixation, cells were counterstained for CENP-T and DAPI to indicate centromeres and DNA, respectively. Cell cycle status was determined by measuring total DAPI area (see supplemental experimental procedures).
(C) Quantification of CENP-A-SNAP fluorescent signals from (B). Average CENP-A-SNAP signals from G2 centromeres were normalized to respective G1 centromeres and corrected for centromere number (assuming signal intensity per focus represents 1 and 2 centromeres in G1 and G2, respectively). Error bars indicate SEM of 4 independent experiments.
(D) CENP-A assembly drives M18BP1 displacement from centromeres. Quantification of centromeric GFP-M18BP1T653A fluorescent signals from (B) using CRaQ method. Average GFP-M18BP1T653A signals were normalized to uninduced 3xFlag-HJURP-ΔCLacI expressing cells. Error bars indicate SEM of 4 independent experiments.
Efficient CENP-A assembly requires displacement of M18BP1 from the centromere
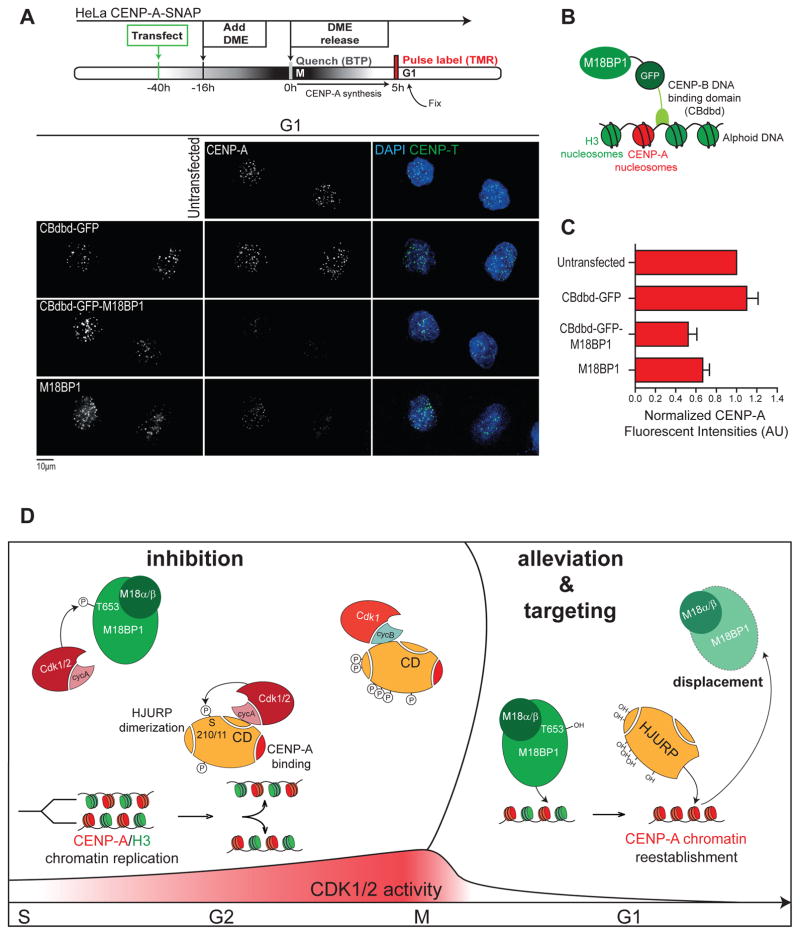
During the course of these experiments, we observed that induction of CENP-A assembly in G2 phase resulted in concomitant loss of centromeric GFP-M18BP1T653A levels to under 30%, on average, relative to the uninduced control (Figure 6C, D). Expression of HJURPAxA-ΔCLacI, but not wild-type HJURP results in GFP-M18BP1T653A loss, showing that displacement is directly dependent on CENP-A assembly. This suggests that M18BP1 removal is an active, CENP-A loading-dependent process and not a passive consequence of cell cycle progression. To test this directly in G1 cells, we either over-expressed wild type M18BP1 or artificially tethered it to G1 centromeres (using the CBdbd tether) while measuring nascent CENP-A chromatin assembly (Figure 7A, B). We observe a ~40% reduction in nascent CENP-A fluorescent intensities in either of these conditions (Figure 7C). We conclude that while M18BP1 is an essential positive regulator of CENP-A assembly, preventing its turnover by overexpression or by rendering it unable to be removed from G1 centromeres results in defects in CENP-A assembly.
Figure 7. M18BP1 removal from G1 centromeres is necessary for efficient canonical CENP-A assembly.
(A) HeLa CENP-A-SNAP cells were transfected with indicated constructs, and synchronized in mitosis by an overnight treatment with Eg5 inhibitor (DMEIII). Newly synthesized CENP-A pool was quenched in mitosis, followed by 5h of release in early G1 when nascent CENP-A-SNAP was labeled with TMR (G1 specific pool).
(B) Schematic of relevant domains in centromere targeted M18BP1.
(C) GFP positive cells were selected and CENP-A TMR fluorescent intensities were determined using CRaQ, with the exception of the untransfected control where all cell were analyzed.
(D) Model summarizing the key molecular steps that are sufficient to restrict CENP-A assembly to G1 phase. CD: HJURP vertebrate conserved domain.
Discussion
We have identified the licensing factor M18BP1 and the CENP-A chaperone HJURP as the two key targets of Cdk-based inhibition sufficient for maintenance of strict cell cycle control of CENP-A assembly (Figure 7D). However, we do not exclude that additional levels of regulation exists, e.g. in chromatin maturation steps or in mitotic inhibition (which we find to be controlled in a distinct manner from G2 phase).
Inhibition of CENP-A assembly prior to mitosis at the level of HJURP or M18BP1 alone is incomplete. This is in agreement with previous studies that showed that mutation of HJURP phospho-sites within the HJURP C-Terminal Domain 1 (HCTD1)(Müller et al., 2014) or forced recruitment of Mis18α resulted in precocious CENP-A assembly (McKinley and Cheeseman, 2014). We note that in our system, mutation of the HCTD1 phosphosites did not result in precocious CENP-A assembly (Figure S5B). This discrepancy is likely the result of expression level differences between the cell types used in each study [Figure S5C and (Bodor et al., 2014)].
We provide evidence that the primary mechanism of Cdk-mediated inhibition is to prevent, otherwise active, factors from reaching the centromere (Figures 2C, S6B, C and 7D). We propose that phosphorylation blocks the ability of M18BP1 and HJURP to bind to a partner(s) already docked at the centromere. M18BP1 interacts with CENP-C which is a constitutive core component of the centromere (Dambacher et al., 2012; Hori et al., 2013; Moree et al., 2011; Nardi et al., 2016; Stellfox et al., 2016). In turn, the prenucleosomal HJURP/CENP-A complex binds to the Mis18 complex (Nardi et al., 2016; Wang et al., 2014). Our proposal is consistent with a recent report describing an interaction between HJURP and the Mis18 complex subunit Mis18β, that is reduced upon Cdk phosphorylation, in vitro (Wang et al., 2014).
Recent studies have reported cell cycle regulated phosphorylation of CENP-A itself (Yu et al., 2015) or Plk1-mediated modification of M18BP1 (McKinley and Cheeseman, 2014). While, the latter is required for Mis18 complex localization upon mitotic exit, none of these modifications directly dictates the G1 restricted CENP-A assembly. Therefore, while key positive regulatory events also involve phospho-regulation (which may include some of the novel phosphorylation sites that we identified on HJURP), we defined the specific targets and mechanisms of the inhibitory control that is responsible for limiting CENP-A assembly to G1 phase. Rather than relying on a single tightly regulated factor, the combinatorial action of two layers of control synergizes to efficiently restrict CENP-A assembly to early G1 phase.
The designation of the Mis18 complex as a priming (licensing) factor was originally inspired by its temporal centromere localization that initiates in anaphase, before the onset of CENP-A assembly (Fujita et al., 2007). This is analogous to licensing of DNA replication by the assembly of the pre-replication complex (pre-RC) in early G1 phase (Nishitani and Lygerou, 2002), the S-phase removal of which ensures a single round of genome duplication per cell cycle (Blow and Dutta, 2005; Blow and Hodgson, 2002). Analogously, we find that removal of M18BP1 from the centromere is directly coupled to the onset of CENP-A deposition, at least under induced conditions in G2 phase, providing a causal link between efficient CENP-A assembly and M18BP1 displacement from the centromere.
These results reveal novel parallels between DNA replication and CENP-A-chromatin, manifested in consumption of the licensing factor which is directly instigated by the start of duplication of the heritable mark. These findings are consistent with a recent study showing that nascent CENP-A/HJURP binding to the Mis18 complex in vitro leads to the disassembly of this complex (Nardi et al., 2016), suggesting that Mis18 complex disassembly could be a mechanism to turn off CENP-A chromatin assembly. We show that not only does CENP-A assembly result in Mis18 complex removal (as shown by (Nardi et al., 2016)) but that this is a requirement for efficient loading of CENP-A. Two possible implications follow from these observations. First, while M18BP1 is required for recruitment of nascent CENP-A to centromeres, its presence may physically block completion of the assembly process. By direct binding to CENP-C (Dambacher et al., 2012; Moree et al., 2011; Shono et al., 2015; Westhorpe et al., 2015) which in turn interacts with CENP-A (Falk et al., 2015; Guse et al., 2011; Kato et al., 2013; Logsdon et al., 2015), it is possibly that M18BP1 physically marks the site of incorporation for nascent CENP-A. Inability to remove M18BP1 would therefore provoke steric inhibition, resulting in low rates of CENP-A incorporation. Secondly, given the key role in initiation of CENP-A loading, removal of M18BP1 from centromeres provides an “OFF” switch for the process of assembly, thereby contributing to a tight cell cycle window ensuring a single round of CENP-A incorporation per cell cycle.
Experimental Procedures
Cell synchronization
Double Thymidine-based synchronization was performed as described (Bodor et al., 2012). For Mitotic synchronization, 2,4 μM of EG5 inhibitor III Dimethylenastron-DMEIII (Calbiochem) was used for 24h. For synchronous mitotic exit, following DMEIII washout, HeLa and Hek293T were released for 5h and 7h, respectively. For Figure 2C, Nocodazole was used at 100ng/ml.
Co-Immunoprecipitation
HEK293T cells were transiently transfected with GFP-tagged constructs. 24h post-transfection cells were either allowed to continue to cycle or were treated overnight in DME III to induce mitotic arrest. Cells were harvested 48h post-transfection and subjected to cyclin A or B immunoprecipitation (See also supplemental experimental procedures). Isolated complexes were separated by SDS-PAGE followed by immunoblotting with anti-Cyclin A, (Figure 1E) or anti-Cyclin B (both Santa Cruz) (Figure S3B) and anti-GFP (Chromotek) antibodies and detected on an Odyssey near-infrared scanner and quantified using the Odyssey software (see also (Bodor et al., 2014)).
SILAC and affinity purification of prenucleosomal HJURP/CENP-A/H4 complex
SILAC labeling medium was supplemented with normal lysine and arginine (Sigma-Aldrich) for “light” medium, and 50 mg/L 13C6,15N2-lysine and 50 mg/L 13C6,15N4-arginine (Silantes) for “heavy” medium (See also supplemental experimental procedures). Two parallel cultures of previously characterized HeLaS3 cells stably expressing (LAP)-tagged CENP-A (Bailey et al., 2013) were grown in either heavy or light medium until reaching ~98% labeling efficiency. To enrich for mitotic cells, both cultures were treated with 50 μM S-trityl-L-cysteine for 17 h. Subsequently, the “light” cells were treated with 100 μM R-Roscovitine (AdipoGen) for 30 min while the “heavy” cells were mock-treated with DMSO. Affinity purification of the prenucleosomal HJURP/CENP-A/H4 complexes from 1:1 mixed “light” and “heavy” cells was performed as previously described (Bailey et al., 2013) except that protein elution was performed with 2% SDS and heating at 95°C.
Mass spectrometry and data analysis
Purified CENP-A and associated proteins were precipitated, washed and dried. Following reconstitution, proteins we cleaved with trypsin and phosphopeptides were enriched by TiO2 prior to mass spec analysis (See supplemental experimental procedures for details). Extracted-ion chromatograms (XICs) of each light and heavy peptide pair were used for quantification. L/H ratio represents the ratio of total area under each elution peak.
Affinity purification of 3XFlag-HJURPwt/AxA-ΔCLacI and Mass spectrometry
HeLa HILO RMCE cell lines carrying either 3XFlag-HJURP-ΔCLacI or 3xFlag-HJURPAxA-ΔCLacI were enriched in G2 phase as described (see cell synchronization) and induced with 10μg/ml of Doxycycline (Sigma) for 24h. HJURP was affinity purified using anti-Flag M2 mouse agarose beads (Sigma-Aldrich) as described (see Co-Immunoprecitation), followed by SDS-PAGE separation of bound complexes, staining (by Instant Blue, Expedeon) and subsequent HJURP band excision, in-gel trypsin digestion and phosphopeptide enrichment by TiO2. Samples were run on a Q Exactive mass spectrometer coupled with Easy nLC 1000 HPLC. MaxQuant was used to search the human protein database and identify peptide sequences and extract their ion chromatograms.
SNAP Quench-Chase-Pulse Labeling
Cell lines expressing CENP-A-SNAP were pulse labeled as previously described (Bodor et al., 2012), with exception of HeLa HILO derived cell lines where BTP (New England Biolabs) concentration was adjusted to 0,5 μM.
Immunofluorescence and pre-extraction procedure
Procedures are essentially as described (Bodor et al., 2012) (See also supplemental experimental procedures). To detect GFP-HJURPAxA-ΔCLacI on G2 centromeres, HeLa CENP-A-SNAP cells transiently expressing the construct were pre-extracted for 5min prior to fixation. Cells were counterstained using anti-CENP-T (Barnhart et al., 2011) and anti-Aurora B (1:100; BD transduction laboratories). GFP-HJURPAxA-ΔCLacI signal was amplified using GFP-Booster Atto488 (Chromotek).
Microscopy
Imaging was performed using a DeltaVision Core system (Applied Precision) inverted microscope (Olympus, IX-71), coupled to a Cascade2 EMCCD camera (Photometrics). Images (1024×1024) were acquired at 1 x binning using a 100x oil objective (NA 1.40, UPlanSApo) with 0, 2 μm z sections.
Supplementary Material
Acknowledgments
We thank N. Martins (IGC) for preliminary work on HJURP mutants, E.V. Makeyev NTU, Singapore and Dan Foltz (Northwestern University) for reagents, W. Siwek (IGC) for help with large scale cell culture, S. Muller and G. Almouzni (Curie) for reagents and help with pre-extraction procedure and T. Panchenko (UPenn) for guidance with culturing LAP-tagged CENP-A cells and helpful discussions. We thank Monica Bettencourt Dias, Alekos Athanasiadis, Raquel Oliveira and Jörg Becker (IGC) for critically reading the manuscript. Salary support to AS and DLB is provided by Fundação para a Ciência e a Tecnologia (FCT) fellowships SFRH/BD/51878/2012 and SFRH/BD/74284/2010, respectively and an “Investigador FCT” position to LETJ. This work is supported by NIH/NIGMS R01-GM082989 (BEB), NIH/NIGMS T32-GM008275 (UPenn Structural Biology and Molecular Biophysics Training Grant; supported LYG), NIH/NCI F30-CA186430 (LYG), NIH Grant GM 037537 (DFH) and GM 110174 (BAG). Work is further supported by the Fundação para a Ciência e a Tecnologia (FCT) grant BIA-BCM/100557/2008 (LETJ), an EMBO installation grant 1818 (LETJ) and an ERC-consolidator grant ERC-2013-CoG-615638 awarded to LETJ.
Footnotes
Supplemental Information includes Supplemental Experimental Procedures and 8 figures that can be found with this article online at XX.
Author contributions
AS performed the experiments. LYG performed CENP-A/HJURP co-purifications for SILAC experiments and helped write the manuscript. JFM generated constructs and performed experiments in Figure 4. DLB generated constructs for experiments in Figure 6 and S7. AOB, JS and DFH performed initial mass spec analysis of HJURP phosphorylation sites. LYG, XC, BEB and BAG performed mass-spec experiments and analysis. BEB helped write the manuscript. AS and LETJ conceived of the study and wrote the manuscript.
References
- Adams PD, Sellers WR, Sharma SK, Wu AD, Nalin CM, Kaelin WG. Identification of a cyclin-cdk2 recognition motif present in substrates and p21-like cyclin-dependent kinase inhibitors. Mol Cell Biol. 1996;16:6623–6633. doi: 10.1128/mcb.16.12.6623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allshire RC, Karpen GH. Epigenetic regulation of centromeric chromatin: old dogs, new tricks? Nat Rev Genet. 2008;9:923–937. doi: 10.1038/nrg2466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amor DJ, Bentley K, Ryan J, Perry J, Wong L, Slater H, Choo KHA. Human centromere repositioning “in progress. Proc Natl Acad Sci U S A. 2004;101:6542–6547. doi: 10.1073/pnas.0308637101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bailey AO, Panchenko T, Sathyan KM, Petkowski JJ, Pai PJ, Bai DL, Russell DH, Macara IG, Shabanowitz J, Hunt DF, et al. Posttranslational modification of CENP-A influences the conformation of centromeric chromatin. Proc Natl Acad Sci. 2013;110:11827–11832. doi: 10.1073/pnas.1300325110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bailey AO, Panchenko T, Shabanowitz J, Lehman SM, Bai DL, Hunt DF, Black BE, Foltz DR. Identification of the Post-translational Modifications Present in Centromeric Chromatin. Mol Cell Proteomics. 2016;15:918–931. doi: 10.1074/mcp.M115.053710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barnhart MC, Kuich PHJL, Stellfox ME, Ward JA, Bassett EA, Black BE, Foltz DR. HJURP is a CENP-A chromatin assembly factor sufficient to form a functional de novo kinetochore. J Cell Biol. 2011;194:229–243. doi: 10.1083/jcb.201012017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Black BE, Cleveland DW. Epigenetic Centromere Propagation and the Nature of CENP-A Nucleosomes. Cell. 2011;144:471–479. doi: 10.1016/j.cell.2011.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blow JJ, Dutta A. Preventing re-replication of chromosomal DNA. Nat Rev Mol Cell Biol. 2005;6:476–486. doi: 10.1038/nrm1663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blow JJ, Hodgson B. Replication licensing--defining the proliferative state? Trends Cell Biol. 2002;12:72–78. doi: 10.1016/s0962-8924(01)02203-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bodor DL, Rodríguez MG, Moreno N, Jansen LET. Analysis of Protein Turnover by Quantitative SNAP-Based Pulse-Chase Imaging. Curr Protoc Cell Biol. 2012;Chapter 8(Unit 8.8) doi: 10.1002/0471143030.cb0808s55. [DOI] [PubMed] [Google Scholar]
- Bodor DL, Valente LP, Mata JF, Black BE, Jansen LET. Assembly in G1 phase and long-term stability are unique intrinsic features of CENP-A nucleosomes. Mol Biol Cell. 2013;24:923–932. doi: 10.1091/mbc.E13-01-0034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bodor DL, Mata JF, Sergeev M, David AF, Salimian KJ, Panchenko T, Cleveland DW, Black BE, Shah JV, Jansen LET. The quantitative architecture of centromeric chromatin. Elife. 2014;3:1–26. doi: 10.7554/eLife.02137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown NR, Lowe ED, Petri E, Skamnaki V, Antrobus R, Johnson L. Cyclin B and Cyclin A Confer Different Substrate Recognition Properties on CDK2. Cell Cycle. 2007;6:1350–1359. doi: 10.4161/cc.6.11.4278. [DOI] [PubMed] [Google Scholar]
- Cheeseman IM, Desai A. Molecular architecture of the kinetochore-microtubule interface. Nat Rev Mol Cell Biol. 2008;9:33–46. doi: 10.1038/nrm2310. [DOI] [PubMed] [Google Scholar]
- Dambacher S, Deng W, Hahn M, Sadic D, Fröhlich J, Nuber A, Hoischen C, Diekmann S, Leonhardt H, Schotta G. CENP-C facilitates the recruitment of M18BP1 to centromeric chromatin. Nucleus. 2012;3:101–110. doi: 10.4161/nucl.18955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dephoure N, Zhou C, Villén J, Beausoleil Sa, Bakalarski CE, Elledge SJ, Gygi SP. A quantitative atlas of mitotic phosphorylation. Proc Natl Acad Sci U S A. 2008;105:10762–10767. doi: 10.1073/pnas.0805139105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dunleavy EM, Roche D, Tagami H, Lacoste N, Ray-Gallet D, Nakamura Y, Daigo Y, Nakatani Y, Almouzni-Pettinotti G. HJURP is a cell-cycle-dependent maintenance and deposition factor of CENP-A at centromeres. Cell. 2009;137:485–497. doi: 10.1016/j.cell.2009.02.040. [DOI] [PubMed] [Google Scholar]
- den Elzen N, Pines J. Cyclin A is destroyed in prometaphase and can delay chromosome alignment and anaphase. J Cell Biol. 2001;153:121–136. doi: 10.1083/jcb.153.1.121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Errico A, Deshmukh K, Tanaka Y, Pozniakovsky A, Hunt T. Identification of substrates for cyclin dependent kinases. Adv Enzyme Regul. 2009;50:375–399. doi: 10.1016/j.advenzreg.2009.12.001. [DOI] [PubMed] [Google Scholar]
- Fachinetti D, Logsdon GA, Amira A, Selzer EB, Cleveland DW, Black BE. CENP-A modifications on Ser68 and Lys124 are dispensable for establishment, maintenance, and long-term function of human centromeres. Dev Cell. 2017 doi: 10.1016/j.devcel.2016.12.014. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Falk SJ, Guo LY, Sekulic N, Smoak EM, Mani T, Logsdon GA, Gupta K, Jansen LET, Van Duyne GD, Vinogradov SA, et al. CENP-C reshapes and stabilizes CENP-A nucleosomes at the centromere. Science. 2015;348:699–703. doi: 10.1126/science.1259308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foltz DR, Jansen LET, Black BE, Bailey AO, Yates JR, Cleveland DW. The human CENP-A centromeric nucleosome-associated complex. Nat Cell Biol. 2006;8:458–469. doi: 10.1038/ncb1397. [DOI] [PubMed] [Google Scholar]
- Foltz DR, Jansen LET, Bailey AO, Yates JR, Bassett Ea, Wood S, Black BE, Cleveland DW. Centromere-specific assembly of CENP-a nucleosomes is mediated by HJURP. Cell. 2009;137:472–484. doi: 10.1016/j.cell.2009.02.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujita Y, Hayashi T, Kiyomitsu T, Toyoda Y, Kokubu A, Obuse C, Yanagida M. Priming of Centromere for CENP-A Recruitment by Human hMis18α, hMis18β, and M18BP1. Dev Cell. 2007;12:17–30. doi: 10.1016/j.devcel.2006.11.002. [DOI] [PubMed] [Google Scholar]
- Geley S, Kramer E, Gieffers C, Gannon J, Peters JM, Hunt T. Anaphase-promoting complex/cyclosome-dependent proteolysis of human cyclin A starts at the beginning of mitosis and is not subject to the spindle assembly checkpoint. J Cell Biol. 2001;153:137–148. doi: 10.1083/jcb.153.1.137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gómez-Rodríguez M, Jansen LET. Basic properties of epigenetic systems: lessons from the centromere. Curr Opin Genet Dev. 2013;23:219–227. doi: 10.1016/j.gde.2012.11.002. [DOI] [PubMed] [Google Scholar]
- Guse A, Carroll CW, Moree B, Fuller CJ, Straight AF. In vitro centromere and kinetochore assembly on defined chromatin templates. Nature. 2011;477:354–358. doi: 10.1038/nature10379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hagopian JC, Kirtley MP, Stevenson LM, Gergis RM, Russo AA, Pavletich NP, Parsons SM, Lew J. Kinetic basis for activation of CDK2/cyclin A by phosphorylation. J Biol Chem. 2001;276:275–280. doi: 10.1074/jbc.M007337200. [DOI] [PubMed] [Google Scholar]
- Holmes JK, Solomon MJ. A Predictive Scale for Evaluating Cyclin-dependent Kinase Substrates: A COMPARISON OF p34cdc2 AND p33cdk2. J Biol Chem. 1996;271:25240–25246. doi: 10.1074/jbc.271.41.25240. [DOI] [PubMed] [Google Scholar]
- Hori T, Shang WHH, Takeuchi K, Fukagawa T. The CCAN recruits CENP-A to the centromere and forms the structural core for kinetochore assembly. J Cell Biol. 2013;200:45–60. doi: 10.1083/jcb.201210106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jansen LET, Black BE, Foltz DR, Cleveland DW. Propagation of centromeric chromatin requires exit from mitosis. J Cell Biol. 2007;176:795–805. doi: 10.1083/jcb.200701066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kato H, Jiang J, Zhou B-R, Rozendaal M, Feng H, Ghirlando R, Xiao TS, Straight AF, Bai Y. A Conserved Mechanism for Centromeric Nucleosome Recognition by Centromere Protein CENP-C. Science. 2013;340:1110–1113. doi: 10.1126/science.1235532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kato T, Sato N, Hayama S, Yamabuki T, Ito T, Miyamoto M, Kondo S, Nakamura Y, Daigo Y. Activation of Holliday junction recognizing protein involved in the chromosomal stability and immortality of cancer cells. Cancer Res. 2007;67:8544–8553. doi: 10.1158/0008-5472.CAN-07-1307. [DOI] [PubMed] [Google Scholar]
- Khandelia P, Yap K, Makeyev EV. Streamlined platform for short hairpin RNA interference and transgenesis in cultured mammalian cells. Proc Natl Acad Sci U S A. 2011;108:12799–12804. doi: 10.1073/pnas.1103532108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Logsdon Ga, Barrey EJ, Bassett Ea, DeNizio JE, Guo LY, Panchenko T, Dawicki-McKenna JM, Heun P, Black BE. Both tails and the centromere targeting domain of CENP-A are required for centromere establishment. J Cell Biol. 2015;208:521–531. doi: 10.1083/jcb.201412011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maddox PS, Hyndman F, Monen J, Oegema K, Desai AB. Functional Genomics Identifies a Myb Domain–Containing Protein Family Required for Assembly of CENP-A Chromatin. J Cell Biol. 2007;176:757–763. doi: 10.1083/jcb.200701065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marshall OJ, Chueh AC, Wong LH, Choo KHA. Neocentromeres: New Insights into Centromere Structure, Disease Development, and Karyotype Evolution. Am J Hum Genet. 2008;82:261–282. doi: 10.1016/j.ajhg.2007.11.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKinley KLLL, Cheeseman IMMM. Polo-like Kinase 1 Licenses CENP-A Deposition at Centromeres. Cell. 2014;158:397–411. doi: 10.1016/j.cell.2014.06.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mendiburo MJ, Padeken J, Fülöp S, Schepers A, Heun P. Drosophila CENH3 Is Sufficient for Centromere Formation. Science. 2011;334:686–690. doi: 10.1126/science.1206880. [DOI] [PubMed] [Google Scholar]
- Moree B, Meyer CB, Fuller CJ, Straight AF. CENP-C recruits M18BP1 to centromeres to promote CENP-A chromatin assembly. J Cell Biol. 2011;194:855–871. doi: 10.1083/jcb.201106079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Müller S, Montes de Oca R, Lacoste N, Dingli F, Loew D, Almouzni G, Montes de Oca R, Lacoste N, Dingli F, Loew D, et al. Phosphorylation and DNA Binding of HJURP Determine Its Centromeric Recruitment and Function in CenH3(CENP-A) Loading. Cell Rep. 2014;8:190–203. doi: 10.1016/j.celrep.2014.06.002. [DOI] [PubMed] [Google Scholar]
- Nardi IKK, Zasadzińska E, Stellfox MEE, Knippler CMM, Foltz DRR. Licensing of Centromeric Chromatin Assembly through the Mis18α-Mis18β Heterotetramer. Mol Cell. 2016;61:774–787. doi: 10.1016/j.molcel.2016.02.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishitani H, Lygerou Z. Control of DNA replication licensing in a cell cycle. Genes Cells. 2002;7:523–534. doi: 10.1046/j.1365-2443.2002.00544.x. [DOI] [PubMed] [Google Scholar]
- Okada M, Cheeseman IM, Hori T, Okawa K, McLeod IX, Yates JR, 3rd, Desai A, Fukagawa T. The CENP-H–I complex is required for the efficient incorporation of newly synthesized CENP-A into centromeres. Nat Cell Biol. 2006;8:446–457. doi: 10.1038/ncb1396. [DOI] [PubMed] [Google Scholar]
- Russo AA, Jeffrey PD, Patten AK, Massagué J, Pavletich NP. Crystal structure of the p27Kip1 cyclin-dependent-kinase inhibitor bound to the cyclin A-Cdk2 complex. Nature. 1996;382:325–331. doi: 10.1038/382325a0. [DOI] [PubMed] [Google Scholar]
- Sanchez-Pulido L, Pidoux AL, Ponting CP, Allshire RC. Common Ancestry of the CENP-A Chaperones Scm3 and HJURP. Cell. 2009;137:1173–1174. doi: 10.1016/j.cell.2009.06.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schuh M, Lehner CF, Heidmann S. Incorporation of Drosophila CID/CENP-A and CENP-C into Centromeres during Early Embryonic Anaphase. Curr Biol. 2007;17:237–243. doi: 10.1016/j.cub.2006.11.051. [DOI] [PubMed] [Google Scholar]
- Shono N, Ohzeki J-I, Otake K, Martins NMC, Nagase T, Kimura H, Larionov V, Earnshaw WC, Masumoto H. CENP-C and CENP-I are key connecting factors for kinetochore and CENP-A assembly. J Cell Sci. 2015 doi: 10.1242/jcs.180786. jcs.180786- [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silva MCC, Jansen LET. At the right place at the right time: novel CENP-A binding proteins shed light on centromere assembly. Chromosoma. 2009;118:567–574. doi: 10.1007/s00412-009-0227-3. [DOI] [PubMed] [Google Scholar]
- Silva MCC, Bodor DL, Stellfox ME, Martins NMC, Hochegger H, Foltz DR, Jansen LET. Cdk Activity Couples Epigenetic Centromere Inheritance to Cell Cycle Progression. Dev Cell. 2012;22:52–63. doi: 10.1016/j.devcel.2011.10.014. [DOI] [PubMed] [Google Scholar]
- Stellfox ME, Nardi IK, Knippler CM, Foltz DR. Differential Binding Partners of the Mis18α/β YIPPEE Domains Regulate Mis18 Complex Recruitment to Centromeres. Cell Rep. 2016:1–9. doi: 10.1016/j.celrep.2016.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J, Liu X, Dou Z, Chen L, Jiang H, Fu C, Fu G, Liu D, Zhang J, Zhu T, et al. Mitotic Regulator Mis18β Interacts with and Specifies the Centromeric Assembly of Molecular Chaperone Holliday Junction Recognition Protein (HJURP) J Biol Chem. 2014;289:8326–8336. doi: 10.1074/jbc.M113.529958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Westhorpe FG, Fuller CJ, Straight AF. A cell-free CENP-A assembly system defines the chromatin requirements for centromere maintenance. J Cell Biol. 2015 doi: 10.1083/jcb.201503132. jcb.201503132- [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu Z, Zhou X, Wang W, Deng W, Fang J, Hu H, Wang Z, Li S, Cui L, Shen J, et al. Dynamic Phosphorylation of CENP-A at Ser68 Orchestrates Its Cell-Cycle-Dependent Deposition at Centromeres. Dev Cell. 2015;32:68–81. doi: 10.1016/j.devcel.2014.11.030. [DOI] [PubMed] [Google Scholar]
- Zasadzińska E, Barnhart-Dailey MEC, Kuich PHJL, Foltz DR. Dimerization of the CENP-A assembly factor HJURP is required for centromeric nucleosome deposition. EMBO J. 2013;32:2113–2124. doi: 10.1038/emboj.2013.142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeitlin SG, Barber CM, Allis CD, Sullivan K. Differential regulation of CENP-A and histone H3 phosphorylation in G2/M. J Cell Sci. 2001;114:653–661. doi: 10.1242/jcs.114.4.653. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.