Abstract
Mass cytometry offers the advantage of allowing the simultaneous measurement of a greater number parameters than conventional flow cytometry. However, to date, mass cytometry has lacked a reliable alternative to the light scatter properties that are commonly used as a cell size metric in flow cytometry (forward scatter intensity—FSC). Here, we report the development of two plasma membrane staining assays to evaluate mammalian cell size in mass cytometry experiments. One is based on wheat germ agglutinin (WGA) staining and the other on Osmium tetroxide (OsO4) staining, both of which have preferential affinity for cell membranes. We first perform imaging and flow cytometry experiments to establish a relationship between WGA staining intensity and traditional measures of cell size. We then incorporate WGA staining in mass cytometry analysis of human whole blood and show that WGA staining intensity has reproducible patterns within and across immune cell subsets that have distinct cell sizes. Lastly, we stain PBMCs or dissociated lung tissue with both WGA and OsO4 ; mass cytometry analysis demonstrates that the two staining intensities correlate well with one another. We conclude that both WGA and OsO4 may be used to acquire cell size-related parameters in mass cytometry experiments, and expect these stains to be broadly useful in expanding the range of parameters that can be measured in mass cytometry experiments.
Keywords: cell size, WGA, CyTOF, flow cytometry, mass cytometry, whole blood, U87, HEK293
Introduction
Mass cytometry, although a very recently developed technology (1-3), has already begun to have significant impact on molecular, cell and clinical science (4-24). It shares many similarities with flow cytometry, in that a mass cytometer is able to perform high-throughput multiparametric single cell analysis. However, while flow cytometry uses light and fluorescence-based detection of probes within intact cells (live or fixed), mass cytometry uses metal-conjugated reagents to label cells, which are analyzed by inductively coupled plasma (ICP), time-of-flight mass spectrometry. By overcoming limitations of fluorescence spectra, namely excitation and emission overlap across stains, mass cytometry allows the simultaneous measurement of a greater number of parameters than conventional flow cytometry. Current reagents allow the detection of over 40 lanthanide-conjugated antibodies, together with nucleic acid intercalators and cell viability reagents.
Mass cytometry analysis necessitates the atomization and ionization of cells in the ICP, which eliminates their physical characteristics and reduces each cell to an ion cloud derived from its associated metal-labeled tags. While current mass cytometry measurements include a measure of “Event Length”, this parameter is independent of cell size and is instead largely related to overall metal ion content, a function of total bound tags and ion cloud size, which is largely driven by gas expansion kinetics. Thus, although mass cytometry has distinct advantages over flow cytometry, to date one drawback has been the inability to obtain a metric for cell size in mass cytometry. In flow cytometry, two detectors are generally dedicated to measuring the intensity of the excitation laser light that is refracted by particles passing through the laser; light that is detected in the same axis as the excitation laser is called forward scatter (FSC), while light that is detected orthogonal to the laser is called side scatter (SSC). FSC is a well-established metric that is related to the size and refractive index of a particle and essentially “comes for free” with any flow cytometry experiment. FSC has many established uses in flow cytometry including discrimination of cells vs. debris and identification of cell types of varying sizes (25). Despite the ability of mass cytometry to greatly increase single cell multiplexing relative to flow cytometry, we lack a basic mass cytometer-compatible assay for cell size, which is of universal use in many types of experiments.
Here, we report the development of two such mass cytometer-compatible assays for metrics of cell size, based on staining with wheat germ agglutinin (WGA) or Osmium tetroxide) (OsO4), both of which have been previously used to stain plasma membranes in other assays (26-30,33). We first perform imaging and flow cytometry experiments to demonstrate that WGA staining intensity in multiple cell types correlates well with established measures of cell size. We then adapt both WGA and OsO4 staining to mass cytometry experiments and show that their intensities correlate with one another, and that median WGA and OsO4 staining intensities in various immunological cell types correlate with expected differences in cell size. We conclude that WGA and OsO4 staining provide novel cell size metrics for mass cytometry experiments, and are broadly useful to further expand the already impressive repertoire of possibilities with this technique.
Results and Discussion
Wheat Germ Agglutinin Staining Intensity Correlates with Imaging-Based Cell Size Measurements
Wheat Germ Agglutinin (WGA) is a lectin that binds to sialic acid and N-acetylglucosamine residues and is widely known to bind to cell membranes (26-30). As a first step in determining a quantitative relationship between WGA staining intensity and cell size, we fixed then stained suspension HEK293 cells with Alexa Fluor 488-conjugated WGA, and collected 20X images of 3,602 individual cells (see Methods and Fig. 1A). We used automated image analysis to extract the integrated Alexa Fluor 488-WGA fluorescence intensity for each individual cell, and correlated it with cell size from the same extracted images (Figs. 1B and 1C). This analysis revealed an excellent correlation over a wide range of cell sizes. These results suggest that the intensity of WGA staining is a suitable metric of cell size.
Figure 1.
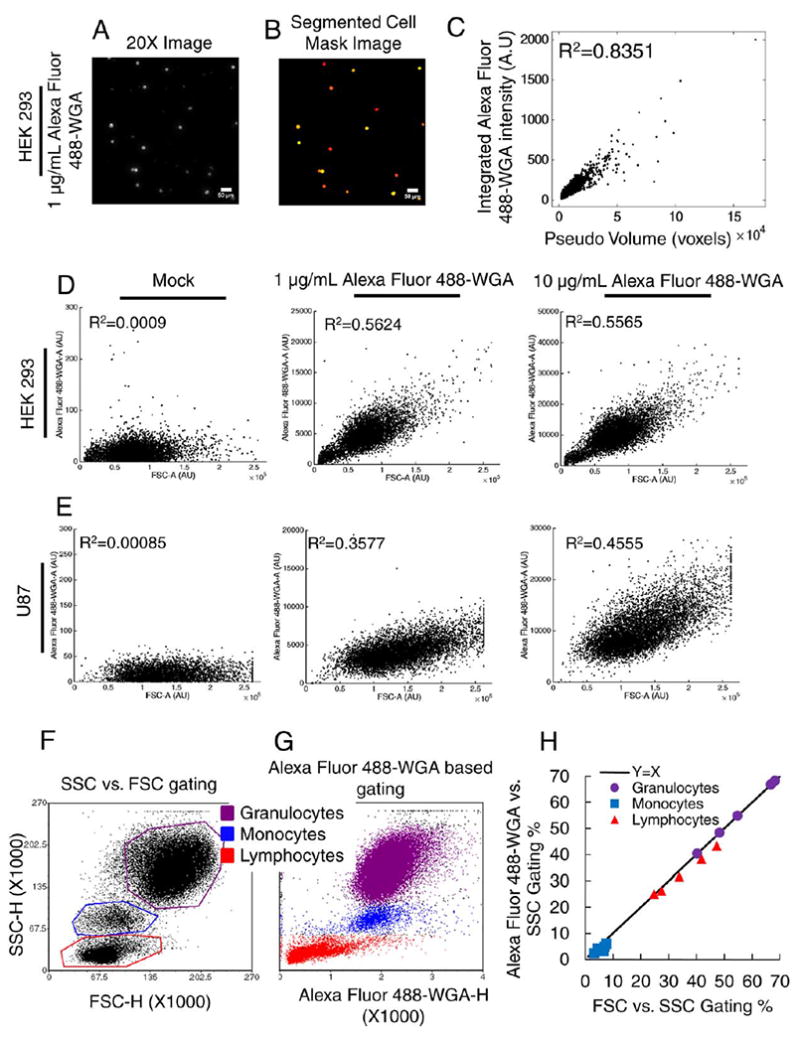
Establishing Wheat Germ Agglutinin Staining Intensity as a Metric for Cell Size. A-C: Imaging of A488-WGA-stained HEK293 cells. Representative fields of view are shown for 20X fluorescence images (intensity adjusted image displayed for illustration purposes only) (A) or processed images (B). Individual cells were identified from the processed images and the integrated fluorescence intensity and area were calculated. Data for each cell are shown in (C). Pseudo volume is cell area raised to the 3/2 power. D, E: WGA-stained HEK293 (D) and U87 cells (E) were subjected to flow cytometry (see Methods), and the correlation between FSC and WGA staining intensity is shown. F–H: Blood samples were subjected to flow cytometry analysis and gating to discriminate between granulocytes, monocytes and lymphocytes was applied based on FSC vs. SSC data (F) or WGA vs. SSC data (G). Consistency of cell type proportions between the two gating strategies are shown in (H). [Color figure can be viewed at wileyonlinelibrary.com]
Wheat Germ Agglutinin Staining Intensity Correlates with Forward Scatter Intensity in Flow Cytometry
In flow cytometry, forward scatter intensity (FSC) is widely regarded as a metric that is related to (but not an exact measure of) cell size. To further examine the suitability of WGA staining intensity as a metric related to cell size, we performed flow cytometry experiments to correlate the two (Figs. 1D and 1E). We stained two different human cell lines, HEK293 and U87, with two different concentrations of Alexa Fluor 488-conjugated WGA. For both lines at both concentrations, we observed a strongly statistically significant correlation between FSC and WGA intensities, whereas there was no correlation in unstained cells.
HEK293 cells are smaller than U87 cells, so we expected that U87 cells may need a higher concentration of WGA during staining. This is what we observed, as quantified by improved R2 values for high WGA staining concentrations in U87 cells, vs. similar R2 values for both WGA staining concentrations in HEK293 cells. This shows that optimal staining concentrations for WGA should be determined independently for different cell lines. In this case, that may be due to differences in cell size. But in general, this may be due to differences in the various cell membrane components that bind WGA (which can vary as a function of cell type), or perhaps even the refractive index of the cells, which FSC also detects. We conclude that WGA staining intensity is a reasonable surrogate for FSC in flow cytometry applications.
Wheat Germ Agglutinin Staining Can Substitute for Forward Scatter in Conventional Flow Cytometry-Based Whole Blood Analysis
A typical flow cytometry application is quantification of the proportions of granulocytes, monocytes and lymphocytes in whole blood samples based on FSC and SSC scatterplots. Since we found that WGA staining intensity correlates well with both cell size and FSC, we wanted to investigate whether it could be used as a surrogate in such whole blood analyses. We stained whole blood samples with Alexa Fluor 488-WGA and subjected them to flow cytometry analysis (see Methods). We gated cells according to conventional standards to determine granulocytes, monocytes and lymphocytes based on FSC vs. SSC (Fig. 1F). The cells within these conventional gates mapped very cleanly onto a scatter plot where FSC was replaced with WGA staining intensity (Fig. 1G). Cell type proportions for each experimental replicate were plotted for the two different gating strategies along a x = y line. Our results show that the population proportions for each gating strategy fall along the x = y line, indicating that these two strategies give similar population proportions (Fig. 1H). These data further support the notion that WGA staining intensity is a suitable metric for cell size, and suggest that it can be used interchangeably with FSC in routine flow cytometry analyses, such as quantification of granulocyte, monocyte and lymphocyte proportions in whole blood samples.
Using WGA Staining as a Metric of Cell Size in Mass Cytometry
The above results suggest that WGA staining intensity is a metric related to cell size. To test the applicability of this metric to mass cytometry, we stained human whole blood with a panel of antibodies to identify major circulating immune cell types (Table 1), followed by Alexa Fluor 488-WGA and a Ho165-conjugated anti-Alexa-488 antibody. SPADE analysis was used to map the multi-dimensional data to 2-dimensional space, and WGA intensity was evaluated across immune subtypes as identified by their canonical marker expression patterns (Fig. 2—color indicates WGA staining intensity) (31). Despite not being used as a clustering parameter, we found that WGA staining intensity nicely self-organizes within this map (Fig. 2A—color indicates WGA staining intensity). We found that relative WGA staining intensity was quite consistent within specific immune cell subsets, and that patterns of relative expression across cell subsets varied reproducibly across individuals (Fig. 2B). We also found that WGA staining intensity across cell subsets generally correlated with published cell diameter (Table 2 and Fig. 2C), with smaller lymphocytes exhibiting significantly lower WGA intensity than larger monocytes and granulocytes. Notably, WGA intensity also differed consistently between larger CD14hi monocytes, which have a diameter of ~18 μm and smaller CD16hi monocytes, which have a diameter of ~14 μm (32).
Table 1.
| a. Whole blood staining panel
| |||
|---|---|---|---|
| ISOTOPE | ANTIBODY | CLONE | SOURCE |
| In113 | CD57 | HCD57 | Biolegend |
| In115 | CD45 | HI130 | Biolegend |
| Nd142 | CD19 | HIB19 | Biolegend |
| Nd143 | CD45RA | HI100 | Biolegend |
| Nd145 | CD4 | RPA-T4 | Biolegend |
| Nd146 | CD8 | RPA-T8 | Biolegend |
| Nd148 | CD16 | 3G8 | Biolegend |
| Sm149 | CD127 | A019D5 | Biolegend |
| Nd150 | CD1c | L161 | Biolegend |
| Eu151 | CD123 | 6H6 | Biolegend |
| Sm152 | CD66b | G10F5 | Biolegend |
| Gd155 | CD27 | O323 | Biolegend |
| Gd160 | CD14 | M5E2 | Biolegend |
| Dy161 | CD56 | B159 | BD Biosciences |
| Er166 | CD25 | M-A251 | Biolegend |
| Er168 | CD3 | UCHT1 | Biolegend |
| Er170 | CD38 | HB-7 | Biolegend |
| Yb171 | CD161 | HP-3G10 | Biolegend |
| Yb174 | HLADR | L243 | Biolegend |
| Ho165 | anti-Alexa Fluor 488 | Life Technologies | |
|
| |||
|
b. Osmium validation panel
| |||
| ISOTOPE | ANTIBODY | CLONE | SOURCE |
|
| |||
| Nd142 | CD19 | HIB19 | Biolegend |
| Nd143 | CD45RA | HI100 | Biolegend |
| Nd145 | CD4 | RPA-T4 | Biolegend |
| Nd146 | CD8 | RPA-T8 | Biolegend |
| Nd148 | CD16 | 3G8 | Biolegend |
| Nd150 | CD1c | L161 | Biolegend |
| Eu151 | CD123 | 6H6 | Biolegend |
| Sm152 | CD66b | G10F5 | Biolegend |
| Gd155 | CD27 | O323 | Biolegend |
| Gd156 | p-p38 | D3F9 | Fluidigm |
| Gd160 | CD14 | M5E2 | Biolegend |
| Dy161 | CD56 | B159 | BD Biosciences |
| Er168 | CD3 | UCHT1 | Biolegend |
| Er170 | CD38 | HB-7 | Biolegend |
| Yb174 | HLADR | L243 | Biolegend |
|
| |||
|
c. Lung staining panel
| |||
| ISOTOPE | ANTIBODY | CLONE | SOURCE |
|
| |||
| In113 | CD57 | HCD57 | Biolegend |
| In115 | HLA-ABC | W6-32 | Biolegend |
| Pr141 | CD326 | 9C4 | Fluidigm |
| Nd142 | CD19 | HIB19 | Biolegend |
| Nd143 | CD45RA | HI100 | Biolegend |
| Nd145 | CD4 | RPA-T4 | Biolegend |
| Nd146 | CD8 | RPA-T8 | Biolegend |
| Nd148 | CD16 | 3G8 | Biolegend |
| Nd150 | CD1c | L161 | Biolegend |
| Eu151 | CD123 | 6H6 | Biolegend |
| Sm152 | CD66b | G10F5 | Biolegend |
| Gd155 | CD27 | O323 | Biolegend |
| Gd158 | CD33 | WM53 | Fluidigm |
| Gd160 | CD14 | M5E2 | Biolegend |
| Dy161 | CD56 | B159 | BD Biosciences |
| Dy162 | CD64 | 10.1 | Biolegend |
| Er167 | CD11c | Bu15 | Biolegend |
| Er168 | CD3 | UCHT1 | Biolegend |
| Er170 | CD38 | HB-7 | Biolegend |
| Yb171 | CD161 | HP-3G10 | Biolegend |
| Yb172 | CD206 | 15-2 | Biolegend |
| Yb174 | HLADR | L243 | Biolegend |
| Pt198 | CD45 | HI130 | Biolegend |
| Bi209 | CD11b | ICRF44 | Fluidigm |
| Ho165 | anti-Alexa Fluor 488 | Life Technologies | |
Figure 2.
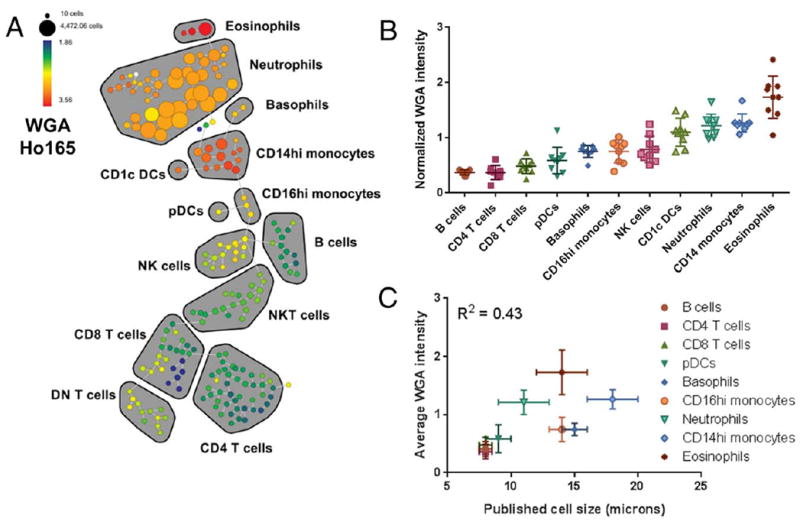
WGA intensity in mass cytometry analysis of human whole blood. Whole blood was stained with a panel of antibodies to identify major immune subsets together with WGA-A488 detected with an Ho165-tagged anti-A488 secondary antibody, and the data were visualized using SPADE. WGA staining in a representative sample is shown across the SPADE tree as fold change relative to MMO control (A). Major immune populations are delineated on the tree based on canonical marker expression. The median normalized WGA staining intensity is shown for the identified immune cell subsets in 8 separate individuals (B), and the average intensity is correlated with the published diameters of the corresponding cell subsets (C and Table 2). [Color figure can be viewed at wileyonlinelibrary.com]
Table 2.
Published cell sizes
These results suggest that WGA staining intensity can be used as a proxy for cell size in mass cytometry. Of course, certain immunological cell types, as well as nonimmunological cell types, may differ in the relative glycoprotein composition of their plasma membranes, which may lead to differences in WGA-binding capacity. This may explain why certain populations, such as eosinophils, exhibited higher WGA staining intensity than other cell of similar expected size. Therefore, while these data suggest WGA staining intensity to be an indicator of cell size, caution should be taken when comparing across cell types quantitatively. Within a population of cells under similar conditions, however, our data do support the notion that WGA staining intensity is a reasonable quantitative measure of cell size that may serve as an analogue for FSC in mass cytometry experiments. In addition, the finding that defined cellular populations express distinct WGA profiles, suggests that the incorporation of this feature may facilitate population identification and doublet discrimination using automated clustering algorithms.
Establishing Osmium Tetroxide as a Novel, Versatile Staining Reagent for Mass Cytometry
The data above are all focused on establishing WGA staining to determine plasma membrane content as a metric for cell size in mass cytometry. However, in recognizing that differences across cell types may result from differences in membrane glycoprotein composition, we sought to confirm these findings with a second independent measurement of cell membrane content that could be applied to mass cytometry. Osmium tetroxide (OsO4) is a lipid-reactive reagent that has been widely used in electron microscopy applications as a cell membrane-selective contrast agent (33). Since OsO4 staining results in the deposition of elemental osmium into the plasma membrane, and osmium falls within the detectable mass range of the CyTOF2, we hypothesized that OsO4 could also serve as mass-cytometry compatible measure of plasma membranes. We found that staining single cell suspensions with a 300 nM solution of OsO4 resulted in a strong cellular osmium signal that was optimally detected in the Os-192 channel, consistent with the expected natural isotopic abundance ratios (Fig. 3A). To further validate the utility and versatility of OsO4 as a staining reagent for mass cytometry, we tested OsO4 labeling prior to and following antibody staining (Fig. 3B), and found that both methods resulted in robust OsO4 labeling across all cells (Fig. 3C). Furthermore, the addition of OsO4 to the samples either before or after antibody staining did not noticeably alter staining for any of the antibodies tested, as visualized by the consistent position of all populations on a viSNE map (Figs. 3C and 3D). We also found that osmium labeling was compatible with methanol permeabilization and intracellular detection of dynamic changes in protein phosphorylation, as shown by consistent detection of changes in p38 phosphorylation in response to PMA stimulation when PBMC were subjected to OsO4 staining prior to or after antibody staining (Fig. 3E).
Figure 3.
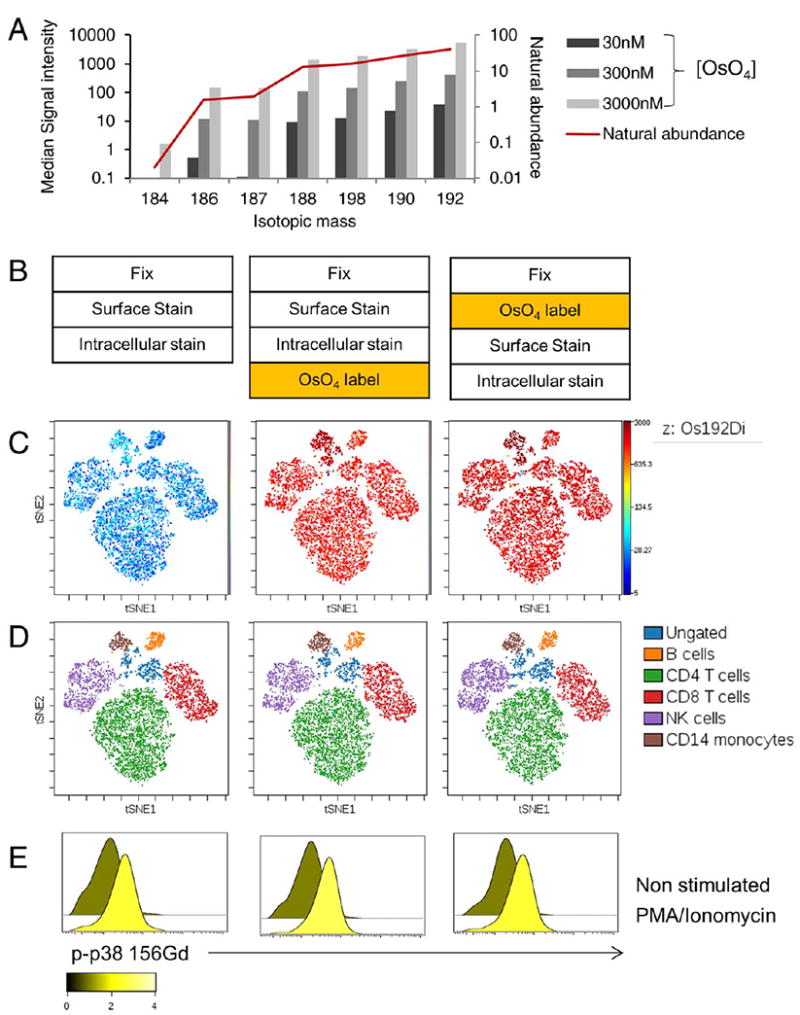
Validation of OsO4 in Mass Cytometry (A) Fixed PBMCs were stained with increasing concentrations of OsO4 and median signal intensity for all gated cell singlets was evaluated for all osmium isotopic mass channels and compared to natural isotopic abundance. B-E: Resting and PMA-stimulated PBMCs were fixed and stained with a panel of antibodies against surface markers and intracellularly with an antibody stained against phospho-p38. OsO4 staining was performed either before or after antibody staining (B). The data were visualized using viSNE to map the multi-dimensional data to two-dimensional space (C-D). Os192 expression is shown across all populations on the map (C) and major immune populations on the map were manually identified using global gates based on canonical marker expression patterns (D). Phospho-p38 expression is shown for the gated T cell population (E), and the overlaid histograms are colored based on the fold change between the stimulated and non-stimulated control samples. [Color figure can be viewed at wileyonlinelibrary.com]
Conveniently, the 192 mass channel is not currently used for any mass tag labels, and when using a 300 nM labeling solution we did not observe any problematic crosstalk or interference with any other channels used for antibody labeling or detectable background persistence on the instrument (data not shown). Together, these data establish that OsO4 staining can be easily incorporated as an additional parameter to supplement current antibody and nucleic acid staining without sacrificing existing antibody detection channels.
Using WGA and OsO4 as Complementary Metrics of Cell Size in Mass Cytometry
We next evaluated OsO4 staining as a measurement of cell size for mass cytometry by cross referencing it with WGA staining as an independent measure, based on our earlier experiments. We performed similar experiments to those in Figure 2, using a panel of antibodies to identify major immune cell subsets in human whole blood and co-staining with both WGA and OsO4. Similar as above, both WGA staining and OsO4 staining show a range of staining intensities across cell types but consistent median intensity within immunological clades as identified by SPADE analyses (4A, B). This could also be visualized using a traditional biaxial plot of WGA against OsO4 (Fig. 4C). The similarity in staining profiles between WGA and Os192 implies that both stains detect plasma membrane content, in turn implying that OsO4 staining intensity also relates to cell size. We also observed a statistically significant correlation between the median staining intensities of WGA and OsO4 for the various cell types across multiple individuals (Fig. 4D). We did note some changes in relative staining intensities for specific cell subsets using these two technique; for example, both stains reliably showed differences between lymphocytes and monocytes, though neutrophils exhibited a relatively lower osmium intensity than WGA intensity. With both stains, eosinophils exhibited high staining intensity at a level greater than would be anticipated solely based on predicted cell size. In the case of osmium intensity, this may relate to the presence of eosinophil lipid bodies, which are known to be characteristically electron dense when visualized using transmission electron microscopy following osmium labeling (34). This attribute may also relate to the characteristically high SSC of eosinophils in conventional flow cytometry, a measurement that is proportional to cell granularity.
Figure 4.
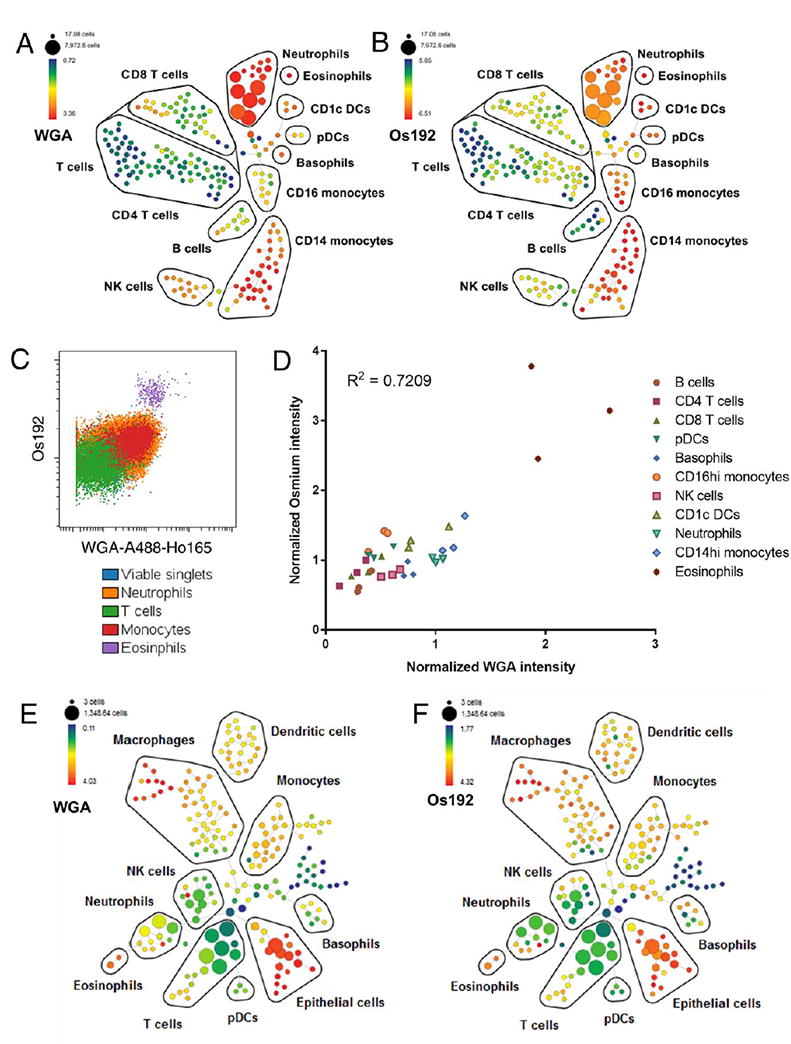
Using WGA and OsO4 in Mass Cytometry. A-D: Whole blood was stained with a panel of antibodies to identify major immune subsets and co-stained with WGA-A488-Ho165 and OsO4, and the data were analyzed using SPADE. Ho165-WGA (A) and Os192 (B) staining intensity are shown across the SPADE tree for a representative sample, with major immune populations delineated based on canonical marker expression. (C) Immune population from a representative sample overlaid on biaxial plot dot plot scaled to show comparative expression of WGA against Os192. (D) Correlation of normalized median WGA-Ho165 and Os192 signal intensity across immune cell subsets in 3 individuals. E,F: A dissociated human lung sample was stained with a panel of antibodies to identify major cell subsets and co-stained with WGA-A488-Ho165 and OsO4, and the data were analyzed using SPADE. Ho165-WGA (E) and Os192 (F) staining intensity are shown across the SPADE tree with major populations delineated based on canonical marker expression. [Color figure can be viewed at wileyonlinelibrary.com]
As a further proof-of-principle of the potentially broad utility of these reagents in characterizing samples by mass cytometry, we applied them to a dissociated human lung sample, comprising a more heterogeneous cellular sample than peripheral blood. As in whole blood, both WGA and Os-192 showed similar distributions of staining intensity across cell subsets (Figs. 4E and 4F) with distinct populations exhibiting characteristic intensity patterns. Relative expression patterns across CD45 hematopoietic cell subsets were similar to those observed among analogous subsets in whole blood, with myeloid cells showing higher expression than T cells. Notably, we found that CD45-CD3261+ lung epithelial cells expressed much higher levels of WGA and Os-192 staining than hematopoietic cells, consistent with their larger expected cell size. Interestingly, some broadly defined populations, such as macrophages, showed heterogeneous WGA and Os-192 expression, suggesting the potential utility of these reagents in revealing additional heterogeneity in complex cellular samples beyond that identified by traditional antibody staining. We conclude that WGA and OsO4 are versatile and complementary assays that can be used to characterize size-related cellular heterogeneity in mass cytometry experiments.
Methods
Subjects and Samples
All samples were collected under protocols approved by Institutional Review Board of the Icahn School of Medicine at Mt. Sinai. Whole blood was collected from healthy donors in sodium heparin vacutainer tubes under a protocol of the Human Immune Monitoring Core and stained within 4 hrs from the time of blood draw. Lung tissue was collected from a patient undergoing resection for stage I non-small cell lung carcinoma. Non-involved lung tissue was collected from a site distal from the tumor and processed for mass cytometry as described below.
Cell Culture
HEK293 (Life Technologies) and U87 (gift from James Gallo) cells were cultured in Dulbecco’s modified eagle medium supplemented with 2 mM L-glutamine and 10% heat inactivated fetal bovine serum at 37°C with 5% CO2. Cells were subcultured every 2–3 days to maintain subconfluency.
Preparation of Cells for WGA Staining
WGA staining stock solution was prepared by dissolving 5 mg of wheat germ agglutinin Alexa Fluor 488 conjugate (A488-WGA) purchased from Life Technologies in 5 mL of PBS. Prior to dilution, A488-WGA stock solution was centrifuged for 5 minutes at 1100xG. Dilutions of 1 μg/mL and 10 μg/mL A488-WGA were freshly prepared by diluting the A488-WGA staining stock solution in PBS. Cells that were to be stained with A488-WGA were lifted with trypsin and fixed using a final concentration of 2% formaldehyde at 37°C for 15 minutes. Cells were spun at 500 × g and washed twice with 5 mL PBS and placed in a FACS tube. Cells were centrifuged at 500 × g for 5 minutes and were re-suspended in 1 μL A488-WGA staining solution per 1000 cells at the desired A488-WGA staining solution concentration. Cells were stained in the dark at room temperature for 15 minutes. Following staining, cells were washed twice with 5 mL PBS, before final suspension in 500 μL of PBS for flow cytometry analysis.
OsO4 Staining
Osmium tetroxide (99.9%+ trace metal basis, ACROS Organics) was first dissolved to a 2% (w/vol) solution and then diluted to a 0.1% stock solution using deionized water and stored in frozen aliquots in glass vials at −20°C. Stocks were diluted 10,000× to produce a 10-5% (300 nM) solution in PBS. To stain cell samples, 1–3 million fixed cells were resuspended in 100 μL of the staining solution, incubated for 10 mins at room temperature, and then washed 2× with PBS +0.2% (g/100 mL) BSA. In the initial validation experiments shown in Figure 3, OsO4 staining was tested either prior to or after surface/intracellular antibody staining; however, in the experiments shown in Figure 4, OsO4 staining was performed after antibody staining. All procedures involving OsO4 were performed in a chemical fume cabinet using appropriate safety precautions to account for the toxicity and volatility of OsO4, and osmium waste solutions were neutralized with corn oil prior to disposal.
Imaging
HEK293 cells were stained with 1μg/mL A488-WGA as outlined in Preparation of Cells for WGA Staining. After staining, 2000 cells were plated on a Corning 3603 96 well plate. The plate was then centrifuged at 100 × g for 5 minutes to settle the stained cells to the bottom of the plate. Cells were then imaged with a GE In-Cell Analyzer 2200 20× objective using the FITC1 channel (Ex 475/25, Em 511.5/23). Images were background subtracted using ImageJ and quantified utilizing a custom CellProfiler pipeline, where cell area and integrated intensity were determined. All scripts are available upon request.
Flow Cytometry
Cultured HEK293 and U87 cells were stained with A488-WGA as outlined in Preparation of Cells for WGA Staining. Following staining, the cells were analyzed with the BD LSRFortessa flow cytometer. The 488 excitation laser and 530/30m (FITC) emission filter were used. Events were gated by FSC and SSC to eliminate debris, and singlets were identified by gating based on FSC-A vs FSC-W. Data were exported to MATLAB, and the correlation between A488-WGA staining intensity and FSC-A was calculated with the function fit.
Whole blood samples from healthy donors (see above) were stained with fluorochrome—conjugated antibodies CD45-BV510 (Clone HI130), CD3-FITC (Clone UCHT1), CD14-APC Cy7 (Clone M5E2) and CD16-BV650 (Clone 3G8) (Biolegend). Antibodies were added directly to the blood, incubated for 20 minutes, and then the blood was treated using BD FACS Lysing Solution (according to manufacturer’s protocol), washed with FACS buffer and then stained with A488-WGA using the method outlined in Preparation of Cells for WGA Staining. Stained blood cells were analyzed with the BD LSR Fortessa flow cytometer with the following channels: Alexafluor 700 (Ex 640 Em 730/45), APC (Ex 640, Em 660/20), FITC (Ex 488, Em 530/30), Qdot655 (Ex 405, Em 670/30), AmCyan (Ex 405, Em 525/20), and APCCy7 (Ex 640, Em 780/60). FITC PMT voltage was adjusted for cells stained with A488-WGA to avoid detector saturation. The data was then processed with FCS Express 5 without compensation, where cells were gated into three populations of cell types based on the conventional method of SSC-H and FSC-H scatter plots: lymphocytes, monocytes and granulocytes. Population percentages for each cell type were calculated for each patient sample using the conventional gating methods. The populations that were determined conventionally were mapped to SSC-H vs. WGA 488-H scatter plots, where the population percentages for each cell type were determined. The population percentages were then compared between the two gating methods: SSC-H, FSC-H and SSC-H, WGA488-H.
Mass Cytometry Sample Processing
All mass cytometry antibodies used in this study were either purchased from Fluidigm or conjugated in house using X8 MaxPar Conjugation kits (Fluidigm Inc.). For the whole blood staining experiments in Figures 2 and 4, a titrated antibody panel (Table 1a) was added directly to 500 μL of whole blood and incubated for 20 minutes at room temperature, after which the blood was lysed and fixed using BD FACS Lysis buffer (BD Biosciences). The antibody–stained blood samples were then divided and either stained with A488-WGA, as described above, or left as a “metal-minus-one” (MMO) staining control. The samples were then washed, treated with 100 U/ml heparin to prevent non-specific antibody binding (35) and stained with a Ho165-conjugated anti-Alexa488 antibody (Life Technologies). The samples were then washed, divided and either stained with OsO4 as described above, or left as a MMO staining control.
For the validation experiments shown in Figures 3B–3E, PBMCs were left unstimulated or stimulated for 15 minutes using a PMA-based Lymphocyte Activation Cocktail (Biolegend) and then fixed with 1.6% formaldehyde, stained with a panel of antibodies against cell surface markers (Table 1b), permeabilized with ice-cold methanol and then intracellularly stained with a 156Gd-conjugated antibody against phosphorylated p38 (T180/Y182, Fluidigm). The cells were treated with OsO4 as described above either prior to or after antibody staining.
For the experiment in Figure 4E, resected human lung tissue was cut into small pieces, enzymatically digested using collagenase type IV (Sigma Aldrich) for 45 minutes at 378C and then passed through a 16-gauge needle. Red blood cells were lysed using RBC lysis buffer (Biolegend) and the sample was filtered through a 70 μm strainer to create a dissociated single cell suspension. The sample was incubated with Rh103 nucleic acid intercalator for 15 minutes at 37°C as a viability dye, and then stained with an antibody panel against cell surface markers (Table 1c). The sample was then fixed and stained with A488-WGA, followed by a Ho165-conjugated anti-Alexa488 antibody, followed by OsO4 as described above.
After staining, all samples were incubated with 0.125 nM Ir nucleic acid intercalator (Fluidigm) to enable cell identification based on DNA-content, and stored in PBS with freshly diluted 2% formaldehyde (Electron Microscopy Sciences) until acquisition. Immediately prior to acquisition, samples were washed once with PBS, once with deionized water and resuspended at a concentration of 600,000 cells/ml in water containing a 1/20 dilution of EQ 4 element beads (Fluidigm). Following routine autotuning according to the manufacturer’s recommendations, the samples were acquired on a CyTOF2 mass cytometer (Fluidigm) equipped with a SuperSampler system (Victorian Airships) at a flow rate of 0.045 ml/min. For quality control, the acquisition event rate was maintained under 400 events/s, and the EQ beads were confirmed to have a median Eu151 intensity of over 1000 to ensure appropriate mass sensitivity.
Mass Cytometry Data Analysis
FCS files were normalized using the bead-based normalization tool in the CyTOF2 software and uploaded to Cyto-bank for analysis. Cell events were identified as Ir191/193 DNA+. Ce140- (viability stain) events and doublets were excluded on the basis of higher DNA content and longer event length. Gated singlets were analyzed using Spanning-tree Progression Analysis of Density-normalized Events (SPADE) (31) or viSNE (23) to map multi-dimensional data onto two-dimensional space. Whole blood SPADE trees in Figures 2A and 4A and viSNE maps in Figures 3C and D were generated using CD45, CD19, CD45RA, CD3, CD4, CD8, CD16, CD66b, CD1c, CD123, CD14, CD56 and HLA-DR. The SPADE trees in Figures 4E and F additionally used expression of CD326, CD33, CD64 and CD206 as clustering parameters. In all cases, WGA and Os192 were excluded as clustering parameters. Major immune populations were identified based on expressed patterns of canonical cell surface markers. Intensity of WGA-A488-Ho165 and Os192 were visualized on the SPADE map relative to MMO staining controls. To account for experimental variability and differences in absolute staining intensity between experiments, when comparing WGA and Os192 staining intensity across experiments and individuals, the median intensity of each immune population was normalized to the median intensity of the respective parameter for all singlets in the sample. Gated populations on the spade tree were exported to generate statistics, and aggregate data were analyzed using Graphpad Prism.
Conclusions
To our knowledge, most mass cytometry experiments to date have been limited to the detection of proteins and nucleic acids. While “Event length” is collected as a default feature during mass cytometry acquisition, this parameter is largely driven by gas expansion kinetics and total ion cloud metal content and does not correlate with cell size, so there is not yet a reliable metric for cell size in mass cytometry experiments. Here, we have adapted two membrane staining assays to mass cytometry, based on either WGA or OsO4 staining, and we provide evidence that they offer useful features related to cell size, and may serve as surrogate parameters for the light scatter properties measured in conventional flow cytometry. Furthermore, while staining resolution of these parameters is likely insufficient to allow robust population identification alone, we believe they may serve as useful additional features to improve accurate population identification and doublet discrimination using automated clustering algorithms, which is a subject of ongoing investigation in our group. Overall, we expect these versatile assays to be broadly useful and to further add to the dimensions that may be analyzed by mass cytometry.
Acknowledgments
The authors thank Soma Kobayashi and Miriam Merad for providing human lung tissue, and Xinzheng Guo, Oksana Mayovska, Seunghee Kim-Schulze and the Mt. Sinai Human Immune Monitoring Core and Flow Cytometry Core for technical assistance.
Grant sponsor: NIH, Grant numbers:
P50GM071558; R21CA196418;
R01GM104184; U54HG008098;
T32GM062754; U24AI118644
Grant sponsor: Icahn School of Medicine at Mount Sinai
Footnotes
Additional Supporting Information may be found in the online version of this article.
Literature Cited
- 1.Bandura DR, Baranov VI, Ornatsky OI, Antonov A, Kinach R, Lou X, Pavlov S, Vorobiev S, Dick JE, Tanner SD. Mass cytometry: Technique for real time single cell multitarget immunoassay based on inductively coupled plasma time-of-flight mass spectrometry. Anal Chem. 2009;81:6813–6822. doi: 10.1021/ac901049w. [DOI] [PubMed] [Google Scholar]
- 2.Bendall SC, Simonds EF, Qiu P, Amir el AD, Krutzik PO, Finck R, Bruggner RV, Melamed R, Trejo A, Ornatsky OI, et al. Single-cell mass cytometry of differential immune and drug responses across a human hematopoietic continuum. Science. 2011;332:687–696. doi: 10.1126/science.1198704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Majonis D, Herrera I, Ornatsky O, Schulze M, Lou X, Soleimani M, Nitz M, Winnik MA. Synthesis of a functional metal-chelating polymer and steps toward quantitative mass cytometry bioassays. Anal Chem. 2010;82:8961–8969. doi: 10.1021/ac101901x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ermann J, Rao DA, Teslovich NC, Brenner MB, Raychaudhuri S. Immune cell profiling to guide therapeutic decisions in rheumatic diseases. Nat Rev Rheumatol. 2015;11:541–551. doi: 10.1038/nrrheum.2015.71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bendall SC, Nolan GP, Roederer M, Chattopadhyay PK. A deep profiler’s guide to cytometry. Trends Immunol. 2012;33:323–332. doi: 10.1016/j.it.2012.02.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ornatsky O, Bandura D, Baranov V, Nitz M, Winnik MA, Tanner S. Highly multi-parametric analysis by mass cytometry. J Immunol Methods. 2010;361:1–20. doi: 10.1016/j.jim.2010.07.002. [DOI] [PubMed] [Google Scholar]
- 7.Nassar AF, Wisnewski AV, Raddassi K. Mass cytometry moving forward in support of clinical research: advantages and considerations. Bioanalysis. 2016;8:255–257. doi: 10.4155/bio.15.257. [DOI] [PubMed] [Google Scholar]
- 8.Frei AP, Bava FA, Zunder ER, Hsieh EW, Chen SY, Nolan GP, Gherardini PF. Highly multiplexed simultaneous detection of RNAs and proteins in single cells. Nat Methods. 2016;13:269–275. doi: 10.1038/nmeth.3742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Spada F, Fuoco C, Pirro S, Paoluzi S, Castagnoli L, Gargioli C, Cesareni G. Characterization by mass cytometry of different methods for the preparation of muscle mononuclear cells. N Biotechnol. 2016 doi: 10.1016/j.nbt.2015.12.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Winter DR, Ledergor G, Amit I. From mass cytometry to cancer prognosis. Nat Biotechnol. 2015;33:931–932. doi: 10.1038/nbt.3346. [DOI] [PubMed] [Google Scholar]
- 11.Mason GM, Lowe K, Melchiotti R, Ellis R, de Rinaldis E, Peakman M, Heck S, Lombardi G, Tree TI. Phenotypic complexity of the human regulatory T cell compartment revealed by mass cytometry. J Immunol. 2015;195:2030–2037. doi: 10.4049/jimmunol.1500703. [DOI] [PubMed] [Google Scholar]
- 12.Levine JH, Simonds EF, Bendall SC, Davis KL, Amir el AD, Tadmor MD, Litvin O, Fienberg HG, Jager A, Zunder ER, et al. Data-driven phenotypic dissection of AML reveals progenitor-like cells that correlate with prognosis. Cell. 2015;162:184–197. doi: 10.1016/j.cell.2015.05.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wong MT, Chen J, Narayanan S, Lin W, Anicete R, Kiaang HT, De Lafaille MA, Poidinger M, Newell EW. Mapping the diversity of follicular helper T cells in human blood and tonsils using high-dimensional mass cytometry analysis. Cell Rep. 2015;11:1822–1833. doi: 10.1016/j.celrep.2015.05.022. [DOI] [PubMed] [Google Scholar]
- 14.Karr JR, Guturu H, Chen EY, Blair SL, Irish JM, Kotecha N, Covert MW. Network-Painter: dynamic intracellular pathway animation in Cytobank. BMC Bioinform. 2015;16:172. doi: 10.1186/s12859-015-0602-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lujan E, Zunder ER, Ng YH, Goronzy IN, Nolan GP, Wernig M. Early reprogramming regulators identified by prospective isolation and mass cytometry. Nature. 2015;521:352–356. doi: 10.1038/nature14274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zunder ER, Lujan E, Goltsev Y, Wernig M, Nolan GP. A continuous molecular road-map to iPSC reprogramming through progression analysis of single-cell mass cytometry. Cell Stem Cell. 2015;16:323–337. doi: 10.1016/j.stem.2015.01.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Becher B, Schlitzer A, Chen J, Mair F, Sumatoh HR, Teng KW, Low D, Ruedl C, Riccardi-Castagnoli P, Poidinger M, et al. High-dimensional analysis of the murine myeloid cell system. Nat Immunol. 2014;15:1181–1189. doi: 10.1038/ni.3006. [DOI] [PubMed] [Google Scholar]
- 18.Mingueneau M, Krishnaswamy S, Spitzer MH, Bendall SC, Stone EL, Hedrick SM, Pe’er D, Mathis D, Nolan GP, Benoist C. Single-cell mass cytometry of TCR signaling: Amplification of small initial differences results in low ERK activation in NOD mice. Proc Natl Acad Sci USA. 2014;111:16466–16471. doi: 10.1073/pnas.1419337111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bendall SC, Davis KL, Amir el AD, Tadmor MD, Simonds EF, Chen TJ, Shenfeld DK, Nolan GP, Pe’er D. Single-cell trajectory detection uncovers progression and regulatory coordination in human B cell development. Cell. 2014;157:714–725. doi: 10.1016/j.cell.2014.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Fienberg HG, Nolan GP. Mass cytometry to decipher the mechanism of nongenetic drug resistance in cancer. Curr Top Microbiol Immunol. 2014;377:85–94. doi: 10.1007/82_2014_365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Newell EW, Sigal N, Nair N, Kidd BA, Greenberg HB, Davis MM. Combinatorial tetramer staining and mass cytometry analysis facilitate T-cell epitope mapping and characterization. Nat Biotechnol. 2013;31:623–629. doi: 10.1038/nbt.2593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Horowitz A, Strauss-Albee DM, Leipold M, Kubo J, Nemat-Gorgani N, Dogan OC, Dekker CL, Mackey S, Maecker H, Swan GE, et al. Genetic and environmental determinants of human NK cell diversity revealed by mass cytometry. Sci Transl Med. 2013;5:208ra145. doi: 10.1126/scitranslmed.3006702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Amir el AD, Davis KL, Tadmor MD, Simonds EF, Levine JH, Bendall SC, Shenfeld DK, Krishnaswamy S, Nolan GP, Pe’er D. viSNE enables visualization of high dimensional single-cell data and reveals phenotypic heterogeneity of leukemia. Nat Biotechnol. 2013;31:545–552. doi: 10.1038/nbt.2594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bodenmiller B, Zunder ER, Finck R, Chen TJ, Savig ES, Bruggner RV, Simonds EF, Bendall SC, Sachs K, Krutzik PO, et al. Multiplexed mass cytometry profiling of cellular states perturbed by small-molecule regulators. Nat Biotechnol. 2012;30:858–867. doi: 10.1038/nbt.2317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.MacDonald HR, Zaech P. Light scatter analysis and sorting of cells activated in mixed leukocyte culture. Cytometry. 1982;3:55–58. doi: 10.1002/cyto.990030112. [DOI] [PubMed] [Google Scholar]
- 26.Aub JC, Sanford BH, Cote MN. Studies on reactivity of tumor and normal cells to a wheat germ agglutinin. Proc Natl Acad Sci U S A. 1965;54:396–399. doi: 10.1073/pnas.54.2.396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Liske R, Franks D. Specificity of the agglutinin in extracts of wheat germ. Nature. 1968;217:860–861. doi: 10.1038/217860a0. [DOI] [PubMed] [Google Scholar]
- 28.Gonatas NK, Avrameas S. Detection of plasma membrane carbohydrates with lectin peroxidase conjugates. J Cell Biol. 1973;59:436–443. doi: 10.1083/jcb.59.2.436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Greenaway PJ, LeVine D. Binding of N-acetyl-neuraminic acid by wheat-germ agglutinin. Nat New Biol. 1973;241:191–192. doi: 10.1038/newbio241191a0. [DOI] [PubMed] [Google Scholar]
- 30.Lotan R, Gussin AE, Lis H, Sharon N. Purification of wheat germ agglutinin by affinity chromatography on a sepharose-bound N-acetylglucosamine derivative. Biochem Biophys Res Commun. 1973;52:656–662. doi: 10.1016/0006-291x(73)90763-8. [DOI] [PubMed] [Google Scholar]
- 31.Qiu P, Simonds EF, Bendall SC, Gibbs KD, Jr, Bruggner RV, Linderman MD, Sachs K, Nolan GP, Plevritis SK. Extracting a cellular hierarchy from high-dimensional cytometry data with SPADE. Nat Biotechnol. 2011;29:886–891. doi: 10.1038/nbt.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Serbina NV, Jia T, Hohl TM, Pamer EG. Monocyte-mediated defense against microbial pathogens. Annu Rev Immunol. 2008;26:421–452. doi: 10.1146/annurev.immunol.26.021607.090326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Thiery G, Bernier J, Bergeron M. A simple technique for staining of cell membranes with imidazole and osmium tetroxide. J Histochem Cytochem. 1995;43:1079–1084. doi: 10.1177/43.10.7560886. [DOI] [PubMed] [Google Scholar]
- 34.Melo RC, D’Avila H, Wan HC, Bozza PT, Dvorak AM, Weller PF. Lipid bodies in inflammatory cells: structure, function, and current imaging techniques. J Histochem Cytochem. 2011;59:540–556. doi: 10.1369/0022155411404073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Rahman AHTL, Berin CM. Heparin reduces non-specific eosinophil staining artifacts in mass cytometry experiments. Cytometry A. 2016;7:601–689. doi: 10.1002/cyto.a.22826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ovalle WK, Nahirney PC, Netter FH. Netter’s Essential Histology. Philadelphia, PA: Elsevier/Saunders; 2013. Blood and Bone Marrow; p. 15772. Print. [Google Scholar]
- 37.Tang F, Du Q, Liu Y-J. Plasmacytoid dendritic cells in antiviral immunity and auto-immunity. Science China Life Sci. 2010;53:172–182. doi: 10.1007/s11427-010-0045-0. [DOI] [PMC free article] [PubMed] [Google Scholar]


