Abstract
Genome sequencing has revealed that a far greater number of natural product biosynthetic pathways exist than there are known natural products. To access these molecules directly and deterministically, a new generation of heterologous expression methods is needed. Cell-free protein synthesis has not previously been used to study nonribosomal peptide biosynthesis, and provides a tunable platform with advantages over conventional methods for protein expression. Here, we demonstrate the use of cell-free protein synthesis to biosynthesize a cyclic dipeptide with correct absolute stereochemistry. From a single-pot reaction, we measured the expression of two nonribosomal peptide synthetases larger than 100 kDa, and detected high-level production of a diketopiperazine. Using quantitative LC–MS and synthetically prepared standard, we observed production of this metabolite at levels higher than previously reported from cell-based recombinant expression, approximately 12 mg/L. Overall, this work represents a first step to apply cell-free protein synthesis to discover and characterize new natural products.
Keywords: cell-free protein synthesis, natural products, diketopiperazine, cyclic dipeptide, biosynthesis, synthetic biology
Graphical Abstract
Sequencing of over 10000 microbial genomes has revealed that the diversity of secondary metabolism covers a largely unexplored region of chemical space.1,2 Currently, the rate at which new biosynthetic pathways are identified within biological sequence data greatly outpaces the capacity to characterize the small molecules for which production is encoded. This is at least in part due to the difficulty of cultivating natural product producing organisms in the laboratory, and therefore presents a major opportunity for synthetic biology to establish new methods for robust heterologous expression and characterization of secondary metabolism to unlock access to often bioactive secondary metabolites. Additionally, robust heterologous expression of biosynthetic proteins opens the possibility of rapid design-build-test cycles to re-engineer pathways to produce useful scaffolds beyond those found in nature.3
Proteins involved in the biosynthesis of complex natural products (NPs) provide a challenge for established heterologous expression platforms because of their sheer size, complex multidomain structures, and occurrence within even larger biosynthetic gene clusters (BGCs). Nonribosomal peptide synthetases (NRPSs) and polyketide synthases (PKSs) are often 100 to >300 kDa proteins that operate like assembly lines, loading and passing covalently attached intermediates between catalytic domains (Figure 1, center).4 Production of NPs by these classes of proteins frequently involves exotic precursors that may not be found in the cytosol of a typical heterologous host such as Escherichia coli or Saccharomyces cerevisiae.5,6
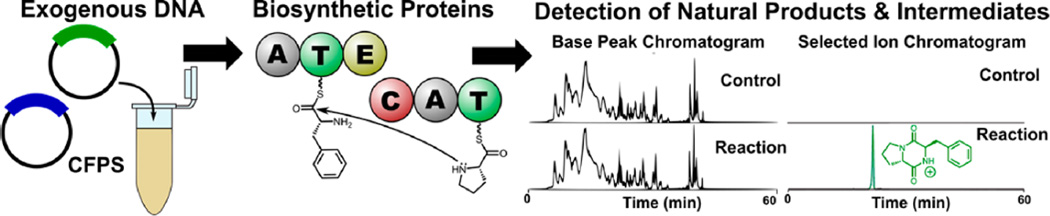
Figure 1.
Overview of a system for cell-free production of natural products via nonribosomal peptide biosynthesis. From left to right, exogenous DNA is used as the input information for the production of biosynthetic enzymes. In the center, nonribosomal peptide synthetase proteins function in concert to select substrates and catalyze the formation of peptide bonds, ultimately resulting in the production of a 2,5-diketopiperazine. Right panel, detection of a d-Phe-l-Pro diketopiperazine natural product by LC–MS as the result of in situ production of biosynthesis proteins.
Cell-free protein synthesis (CFPS) offers an alternative protein expression platform with potential advantages for expressing natural product biosynthetic enzymes (Figure 1).7,8 CFPS is emerging as a robust tool for fundamental and applied research in the areas of synthetic biology and biotechnology.9–11 Recently, cell-free systems have been shown to enable high yielding synthesis of, for example, active metalloenzymes,12–14 and allow rapid prototyping of biological circuits and pathways.15–18 For natural products research, CFPS offers flexibility over cell-based expression because the reaction conditions can be more easily controlled: for example, atypical precursors can simply be fed in during the course of a reaction19,20 or made in situ using all enzymes from a biosynthetic gene cluster. Biosynthetic pathways could potentially be engineered for higher expression without the need to modify a native host strain.21 Reactions can also be performed in the span of a few hours in a “single pot” and have a greater tolerance for producing potentially toxic proteins and metabolites than living cells.
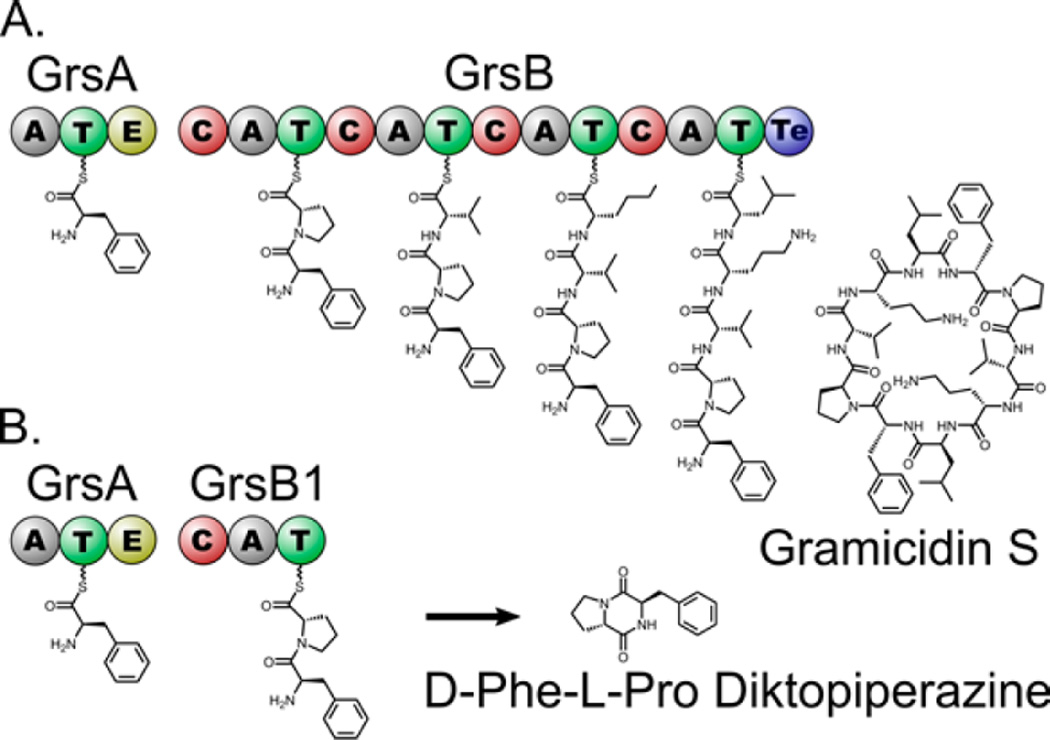
To investigate the potential of CFPS for studying natural product pathways, we utilized the proteins involved in the first steps of gramicidin S biogenesis as a model. Gramicidin S is biosynthesized by two NRPS proteins, GrsA and GrsB, that catalyze thiotemplated peptide bond formation and release by cyclodimerization to form the cyclic decapeptide (Figure 2A). A natural shunt product of the gramicidin S pathway is the d-Phe-l-Pro diketopiperazine (DKP) formed by cyclization of the substrates of the first two NRPS modules in the pathway, GrsA and GrsB1.22 Cyclic dipeptide DKPs have been studied for their diverse biological activity and are produced by many species of bacteria, fungi, and plants.23 Bacterial DKPs have two known general biosynthetic origins: they are either synthesized by NRPS enzymes, as in the case of the d-Phe-l-Pro-DKP presented here,24 or produced by aminoacyl-tRNA mediated cyclodipeptide synthase pathways as in the case of the antibiotic albonoursin.25
Figure 2.
Panel A summarizes the biosynthesis of gramicidin S as it occurs in Brevibacillus brevis. By turning through the assembly line pathway two times, the complete cyclodecapeptide is produced. Panel B shows how the first two modules interact to form d-Phe-l-Pro DKP.
Here, we report in vitro expression of GrsA and GrsB1, encompassing the first two of five modules of gramicidin S biosynthesis in Brevibacillus brevis. Each NRPS module typically consists of the three domains needed to incorporate an amino acid into the nonribosomal peptide: condensation (C), adenylation (A), and thiolation (T). GrsA, as a starter module, does not contain a C-domain; it contains an A and a T-domain, as well as an epimerization (E) domain that converts l-phenylalanine to d-phenylalanine. Previous reports have shown that isolated GrsA and GrsB1 can be harnessed to produce d-Phe-l-Pro DKP (Figure 2B).26 While this diketopiperazine is not the primary product of this pathway in Brevibacillus brevis, it has been found to be produced by other microbes.27 In this report, we demonstrate high-level expression of GrsA and GrsB1, show that they are produced in their functional state when expressed using CFPS, and use these two proteins in a concerted reaction to produce d-Phe-l-Pro DKP with a final yield of about 12 mg/L.
RESULTS AND DISCUSSION
To establish an in vitro platform for d-Phe-l-Pro DKP biosynthesis, NRPS proteins GrsA and GrsB1 were expressed using an E. coli-based cell-free protein expression system reported previously.28–30 GrsA and GrsB1 along with the functionally equivalent tyrocidine synthetases TycA and TycB1 (often referred to as pheATE and proCAT) have served as the model thiotemplated biosynthetic systems for studying substrate specificity31 and peptide bond formation32 in NRPS pathways.
Individual plasmids containing the genes grsA and grsB1 were used to express these enzymes in the combined transcription–translation system. Transcription was driven using a T7 RNA polymerase. We tested protein synthesis yields in 20 h batch reactions across three different E. coli crude extracts (S30 crude extracts).33 On the basis of the expression yields of GrsA and GrsB1, we found that E. coli BL21 Star (DE3) extract synthesized the highest amount of NRPS, yielding full-length, soluble GrsA at ~106 µg/mL and GrsB1 at ~77 µg/mL (Figure S1). Fully assembled GrsA (126 kDa) and GrsB1 (121 kDa) with correct molecular weight bands were observed by SDS-PAGE and further confirmed by autoradiogram analysis (Figure S2).
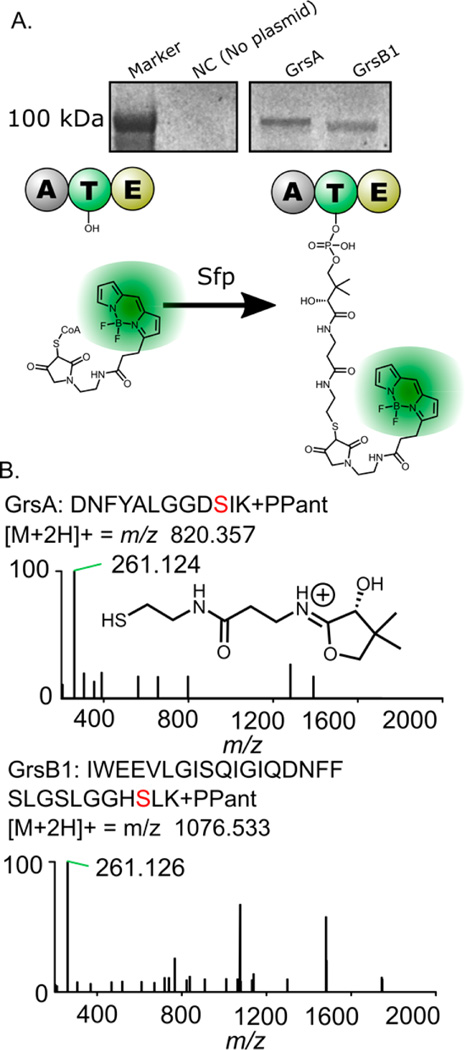
Once we had successfully expressed the two large NRPSs, GrsA and GrsB1 in vitro, we showed that the enzymes could be converted to their functional (holo) form using the 4′-phosphopantetheinyl transferase enzyme Sfp from Bacillus subtilis. To be functional, NRPS proteins require modification by transfer of a phosphopantetheine group from coenzyme A (CoA) to a conserved serine residue in their thiolation (T) domain.34 To verify that this essential modification is possible in our cell-free system, both GrsA and GrsB1 were labeled with a fluorescent Bodipy-CoA analogue by the promiscuous action of Sfp (Figure 3A). Expression of GrsA and GrsB1 continued for 17 h in vitro, allowing nascent proteins to fold properly before labeling. After this incubation, Bodipy-CoA and Sfp were added directly to the cell-free expression system, followed by another 3 h incubation at 30 or 37 °C.
Figure 3.
Experiments showing that GrsA and GrsB1 are present in their active (holo) forms. Panel A shows the fluorescent labeling of GrsA and GrsB1 on the thiolation domain active sites with a conjugated Bodipy-CoA fluorophore (see Figure S3 for complete gel image). Panel B top shows the MS2 spectrum resulting from the fragmentation of a precursor peptide containing the GrsA phosphopantetheine modification. Panel B bottom shows the MS2 spectrum for the corresponding GrsB1 T-domain peptide, indicating the mass of the observed pantetheine-derived ion.
Phosphopantetheinylation was also detectable by LC–MS/MS using the method described in Miller et al. 2005 (Figure 3B).35 GrsA and GrsB1 were prepared in vitro in separate reactions. Following phosphopantetheinylation by Sfp, GrsA with a 6×His tag was captured using cobalt-affinity resin, digested with trypsin, and analyzed by proteomic-style LC– MS/MS. In the GrsA sample, a peptide with the sequence DNFYALGGDSIK and m/z 820.357 was observed, representing the predicted mass of the tryptic peptide with the addition of phosphopantetheine. GrsB1 peptides were detected in a peptide sample prepared by digesting the total lysate with 1:5 trypsin. The T-domain active-site peptide (IWEEVLGISQIGIQDNFFSLGGHSLK, m/z 1076.533) was also observed for GrsB1 with attached phosphopantetheine.
Successful fluorescence labeling of GrsA and GrsB1 with Bodipy-CoA, and direct LC–MS/MS detection of the Ppant modification (Figure S3, Figure 3) demonstrated that, in the current in vitro system, (i) both NRPSs were properly folded with accessible conserved serine residues located on T domains; (ii) the purified recombinant Sfp is active for the phosphopantetheinylation; and (iii) most importantly, the holo-NRPS could be functionally reconstituted for target molecule biosynthesis. In addition, GrsA and GrsB1 were expressed in soluble form and an extended incubation at 37 °C was not necessary for priming the NRPSs by Sfp (Figure S3). Therefore, we posited that the target compound could be biosynthesized via active holo-NRPSs without further optimization of the reaction conditions or incubation temperature.
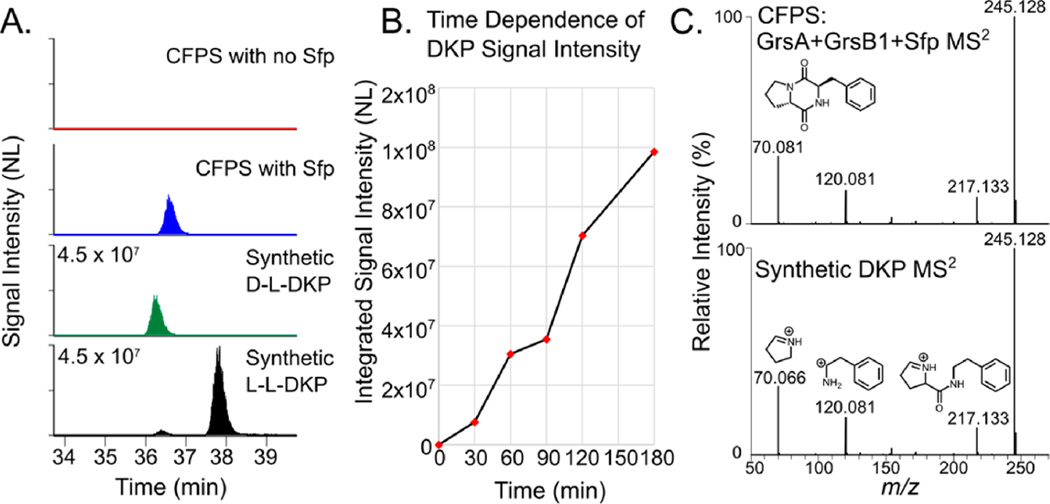
We next sought to biosynthesize d-Phe-l-Pro DKP, and instead of expressing GrsA and GrsB1 separately, the two enzymes were coexpressed in a single-pot mixture allowing reconstitution of the partial NRPS assembly line for product formation in situ. After carrying out the CFPS reaction, we added Sfp to the mixture to initiate biosynthesis, using the same reaction mixture without Sfp as a negative control. The reaction was stopped by extraction with n-butanol and chloroform (4:1, v/v) and the extract was analyzed by metabolomics-style LC–MS/MS. We detected a time-dependent increase in signal intensity of the ion corresponding to predicted DKP ion within 3 ppm mass error tolerance (m/z 245.129) (Figure 4A,B).
Figure 4.
Detection of d-Phe-l-Pro DKP by LC–MS/MS and comparison to synthetically prepared DKP. (A) Retention time comparison of d–l, l–l, and CFPS-produced DKP to determine the stereochemistry of the DKP produced by CFPS. This panel also shows that SFP is required for DKP production. (B) Time-dependent increase in m/z 245.128 signal after Sfp is added to the CFPS reaction. Data points are the average of two technical replicates. (C) Comparison of the fragmentation pattern of the CFPS-produced (top) and synthetically prepared DKP (bottom). The spectrum at the bottom of panel C is annotated with predicted fragment ion structures.
To verify that the ion observed was the expected d-Phe-l-Pro DKP—and with correct d–l stereochemistry—we prepared both the d–l and l–l DKP as synthetic references (Scheme S1). On the basis of earlier reporting, we expected the d–l stereoisomer to elute earlier than l–l on a reversed phase HPLC column,36 and its elution profile matched with the metabolite produced with the CFPS reaction. Synthetic DKP was indistinguishable from the CFPS produced metabolite by MS2 analysis (Figure 4C). We used LC–MS to prepare a standard curve from the integrated intensity of synthetically prepared DKP measured at six different concentrations and used this curve to estimate the quantity of DKP produced by CFPS (Figure S4). Even without optimization of the system for high titers, we estimated our production of d-Phe-l-Pro DKP at ~12 mg/L, a concentration higher than the 9 mg/L previously reported from recombinant protein expression in E. coli.26
We believe this work to be the first example of natural product biosynthesis from NRPS enzymes produced using an E. coli-based in vitro transcription and translation system. CFPS provides a feasible option for exploring natural product biosynthesis, at least for proteins ~130 kDa such as the two presented here. CFPS offers freedom from many of the problematic processes present in cell-based expression systems (e.g., inclusion body formation, protein degradation), and may circumvent the issues of low-expressing or cryptic (nonexpressing) BGCs encountered in native-producing organisms. While more investigation will be needed, the production of these two single-module NRPS proteins in vitro and the demonstration of their concerted function provides a groundwork for the study of increasingly complex natural product biosynthesis pathways using CFPS. Indeed, our discovery-centered cell-free approach sets the stage for high-throughput experimentation in a cell-free environment, where design–build–test iterations can be performed without the need to reengineer organisms, DNA for pathway enzymes is directly input with plasmid refactoring, and substrates and cofactors needed for secondary metabolism can be controlled and maintained at defined concentrations.37
As a resurgence of interest in natural products continues, and the number of sequenced biosynthetic gene clusters continues to grow, we expect that protein expression systems will play an increasingly important role in obtaining and studying new natural products. Especially as the price of DNA synthesis declines, direct expression of entirely synthetic gene clusters (typically 30–120 kilobases in length) will remove barriers to accessing biosynthetic pathways from clusters assembled using metagenomics for uncultivable organisms. By merging bottom-up design principles with innovative cell-free pathway engineering methodologies, our cell-free approach will create a greatly simplified framework for studying and engineering natural product pathways.
METHODS
Preparation of S30 Cell Extracts
E. coli cells were grown in 1 L of 2×YTPG (yeast extract 10 g/L, tryptone 16 g/L, NaCl 5 g/L, K2HPO4 7 g/L, KH2PO4 3 g/L, and glucose 18 g/L, pH 7.2) in a 2.5-L Tunair flask (IBI Scientific, Peosta, IA) at 34 °C and 220 rpm with inoculation of 20 mL overnight cultures (initial OD600 of ~0.05). When the OD600 reached 3.0, cells were collected by centrifugation at 5,000g and 4 °C for 15 min. The pellets were washed thrice with cold S30 buffer (10 mM Tris-acetate pH 8.2, 14 mM magnesium acetate, 60 mM potassium acetate, 2 mM dithiothreitol (DTT)). Cells were suspended in 0.8 mL of S30 buffer per gram wet weight and lysed on ice using a Q125 Sonicator (Qsonica, Newtown, CT) for three pulses (50% amplitude, 45 s on and 59 s off). After sonication, 3 µL of DTT (1 M) was added per milliliter of lysate, followed by centrifugation at 12 000g and 4 °C for 10 min. The supernatant (S30 extract) was flash frozen in liquid nitrogen and stored at −80 °C until use.
Cell-Free Protein Synthesis (CFPS) Reactions
The standard CFPS reactions were performed in 1.5 mL microcentrifuge tubes with 15 µL of mixture composed of the following reagent concentrations: 12 mM magnesium glutamate, 10 mM ammonium glutamate, 130 mM potassium glutamate, 1.2 mM ATP, 0.85 mM each of GTP, UTP, and CTP, 34 µg/mL folinic acid, 170 µg/mL of E. coli tRNA mixture, 2 mM each of 20 standard amino acids, 10 µM of l-[14C(U)]-leucine (used only in protein quantitation experiments, 11.1 GBq mmol−1, PerkinElmer, Waltham, MA), 0.33 mM nicotinamide adenine dinucleotide (NAD), 0.27 mM coenzyme A (CoA), 1.5 mM spermidine, 1 mM putrescine, 4 mM sodium oxalate, 33 mM phosphoenolpyruvate (PEP), 26.7 µg/mL plasmid, 100 µg/mL T7 RNA polymerase, and 27% (v/v) of S30 cell extract. All CFPS reactions were incubated for 20 h at 30 °C unless otherwise described.
Fluorescence Labeling of GrsA and GrsB1
The coenzyme A analogue Bodipy-CoA was prepared as previously described.38 To label thiolation domains of GrsA and GrsB1, standard CFPS reactions with 26.7 µg/mL of each plasmid were incubated at 30 °C for 17 h. Afterward, labeling reactions were performed following three strategies. Strategy #1: the CFPS system was directly supplemented with 1 µL of Bodipy-CoA (1 mg/mL) and 1 µL of Sfp (2 mg/mL) and incubated at 30 °C for another 3 h. Strategy #2: the CFPS reaction was first centrifuged at 12 000g and 4 °C for 10 min. Then, 13 µL of supernatant was transferred to a new 1.5 mL microcentrifuge tube with the addition of 1 µL of Bodipy-CoA and 1 µL of Sfp. The mixture was incubated at 30 °C for 3 h. Strategy #3: the treatment of the CFPS reaction was the same as in Strategy #2, but was incubated at 37 °C for 3 h. After the labeling reaction, 3 µL of each sample was loaded on a 4–12% NuPAGE SDSPAGE gel (Invitrogen). The Bodipy-labeled proteins were visualized by a fluorescence imaging system with 473 nm laser and 520 nm emission filter (Typhoon FLA7000, GE Healthcare Biosciences, Uppsala, Sweden). See Supporting Information for additional details and fluorescent gel images (Figure S3).
DKP Production in Vitro
d-Phe-l-Pro diketopiperazine (DKP) was biosynthesized by GrsA and GrsB1 expressed in situ. Cell-free coexpression of GrsA and GrsB1 was performed in the 15 µL reaction mixture as described above with the addition of both plasmids (each of 26.7 µg/mL). The BL21 Star (DE3) S30 extract was used for the coupled transcription–translation. After incubation of the reaction mixture at 30 °C for 17 h, 1 µL of Sfp (2 mg/mL); 1 µL of Sfp (2 mg/mL) and 1 µL of CoA (5 mM); or 1 µL of Sfp (2 mg/mL), 1 µL of CoA (5 mM), 1 µL of Phe (1 mM) and 1 µL of Pro (1 mM) were added directly to the reactions, followed by another 3 h incubation at 30 °C. Reactions without plasmids and without addition of Sfp were carried out as negative controls. At the end of the production, all DKP samples were immediately extracted from the reaction mixtures for analysis by LC–MS/MS (see Supporting Information for further details).
Supplementary Material
Acknowledgments
This work was supported by the Departments of Chemistry and Molecular Biosciences at Northwestern University and Grants AT009143-13 (to N.L.K.) the Chemistry of Life Processes Predoctoral Training Program at Northwestern University (T32 GM105538) to R.A.M. and the David and Lucile Packard Foundation (2011-37152), ARPA-E (DE-AR0000435), the Camille Dreyfus Teacher Scholar Award, and the DARPA 1KM Program (HR0011-15-C-0084) to M.C.J.
Footnotes
ASSOCIATED CONTENT
Supporting Information
- Quantitation of synthesized GrsA and GrsB1; expression and purification of recombinant Sfp from E. coli autoradiography analysis; synthetic preparation of DKP; bacterial strains and plasmid construction (PDF)
The authors declare no competing financial interest.
References
- 1.Doroghazi JR, Albright JC, Goering AW, Ju KS, Haines RR, Tchalukov KA, Labeda DP, Kelleher NL, Metcalf WW. A roadmap for natural product discovery based on largescale genomics and metabolomics. Nat. Chem. Biol. 2014;10:963–968. doi: 10.1038/nchembio.1659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Charlop-Powers Z, Owen JG, Reddy BV, Ternei MA, Brady SF. Chemical-biogeographic survey of secondary metabolism in soil. Proc. Natl. Acad. Sci. U. S. A. 2014;111:3757–3762. doi: 10.1073/pnas.1318021111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lowry B, Walsh CT, Khosla C. Reconstitution of Metabolic Pathways: Insights into Nature’s Chemical Logic. Synlett. 2015;26:1008–1025. doi: 10.1055/s-0034-1380264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Fischbach MA, Walsh CT. Assembly-line enzymology for polyketide and nonribosomal Peptide antibiotics: logic, machinery, and mechanisms. Chem. Rev. 2006;106:3468–3496. doi: 10.1021/cr0503097. [DOI] [PubMed] [Google Scholar]
- 5.Zemella A, Thoring L, Hoffmeister C, Kubick S. Cell-Free Protein Synthesis: Pros and Cons of Prokaryotic and Eukaryotic Systems. ChemBioChem. 2015;16:2420–2431. doi: 10.1002/cbic.201500340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Li J, Neubauer P. Escherichia coli as a cell factory for heterologous production of nonribosomal peptides and polyketides. New Biotechnol. 2014;31:579–585. doi: 10.1016/j.nbt.2014.03.006. [DOI] [PubMed] [Google Scholar]
- 7.Dudley QM, Karim AS, Jewett MC. Cell-free metabolic engineering: biomanufacturing beyond the cell. Biotechnol. J. 2015;10:69–82. doi: 10.1002/biot.201400330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zawada JF, Yin G, Steiner AR, Yang J, Naresh A, Roy SM, Gold DS, Heinsohn HG, Murray CJ. Microscale to manufacturing scale-up of cell-free cytokine production–a new approach for shortening protein production development timelines. Biotechnol. Bioeng. 2011;108:1570–1578. doi: 10.1002/bit.23103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sun ZZ, Yeung E, Hayes CA, Noireaux V, Murray RM. Linear DNA for rapid prototyping of synthetic biological circuits in an Escherichia coli based TX-TL cell-free system. ACS Synth. Biol. 2014;3:387–397. doi: 10.1021/sb400131a. [DOI] [PubMed] [Google Scholar]
- 10.Carlson ED, Gan R, Hodgman CE, Jewett MC. Cell-free protein synthesis: applications come of age. Biotechnol. Adv. 2012;30:1185–1194. doi: 10.1016/j.biotechadv.2011.09.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hodgman CE, Jewett MC. Cell-free synthetic biology: thinking outside the cell. Metab. Eng. 2012;14:261–269. doi: 10.1016/j.ymben.2011.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kwon YC, Oh IS, Lee N, Lee KH, Yoon YJ, Lee EY, Kim BG, Kim DM. Integrating cell-free biosyntheses of heme prosthetic group and apoenzyme for the synthesis of functional P450 monooxygenase. Biotechnol. Bioeng. 2013;110:1193–1200. doi: 10.1002/bit.24785. [DOI] [PubMed] [Google Scholar]
- 13.Boyer ME, Stapleton JA, Kuchenreuther JM, Wang CW, Swartz JR. Cell-free synthesis and maturation of [FeFe] hydrogenases. Biotechnol. Bioeng. 2008;99:59–67. doi: 10.1002/bit.21511. [DOI] [PubMed] [Google Scholar]
- 14.Li J, Lawton TJ, Kostecki JS, Nisthal A, Fang J, Mayo SL, Rosenzweig AC, Jewett MC. Cell-free protein synthesis enables high yielding synthesis of an active multicopper oxidase. Biotechnol. J. 2016;11:212–218. doi: 10.1002/biot.201500030. [DOI] [PubMed] [Google Scholar]
- 15.Takahashi MK, Chappell J, Hayes CA, Sun ZZ, Kim J, Singhal V, Spring KJ, Al-Khabouri S, Fall CP, Noireaux V, Murray RM, Lucks JB. Rapidly characterizing the fast dynamics of RNA genetic circuitry with cell-free transcription-translation (TX-TL) systems. ACS Synth. Biol. 2015;4:503–515. doi: 10.1021/sb400206c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Garamella J, Marshall R, Rustad M, Noireaux V. The All E. coli TX-TL Toolbox 2.0: A Platform for Cell-Free Synthetic Biology. ACS Synth. Biol. 2016;5:344–355. doi: 10.1021/acssynbio.5b00296. [DOI] [PubMed] [Google Scholar]
- 17.Takahashi MK, Hayes CA, Chappell J, Sun ZZ, Murray RM, Noireaux V, Lucks JB. Characterizing and prototyping genetic networks with cell-free transcription-translation reactions. Methods. 2015;86:60–72. doi: 10.1016/j.ymeth.2015.05.020. [DOI] [PubMed] [Google Scholar]
- 18.de Los Santos EL, Meyerowitz JT, Mayo SL, Murray RM. Engineering Transcriptional Regulator Effector Specificity Using Computational Design and In Vitro Rapid Prototyping: Developing a Vanillin Sensor. ACS Synth. Biol. 2016;5:287–295. doi: 10.1021/acssynbio.5b00090. [DOI] [PubMed] [Google Scholar]
- 19.Salehi AS, Smith MT, Bennett AM, Williams JB, Pitt WG, Bundy BC. Cell-free protein synthesis of a cytotoxic cancer therapeutic: Onconase production and a just-add-water cell-free system. Biotechnol. J. 2016;11:274–281. doi: 10.1002/biot.201500237. [DOI] [PubMed] [Google Scholar]
- 20.Stech M, Brodel AK, Quast RB, Sachse R, Kubick S. Cell-free systems: functional modules for synthetic and chemical biology. Adv. Biochem. Eng./Biotechnol. 2013;137:67–102. doi: 10.1007/10_2013_185. [DOI] [PubMed] [Google Scholar]
- 21.Chappell J, Jensen K, Freemont PS. Validation of an entirely in vitro approach for rapid prototyping of DNA regulatory elements for synthetic biology. Nucleic Acids Res. 2013;41:3471–3481. doi: 10.1093/nar/gkt052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kurotsu T, Hori K, Kanda M, Saito Y. Characterization and location of the L-proline activating fragment from the multifunctional gramicidin S synthetase 2. J. Biochem. 1991;109:763–769. doi: 10.1093/oxfordjournals.jbchem.a123454. [DOI] [PubMed] [Google Scholar]
- 23.Prasad C. Bioactive cyclic dipeptides. Peptides. 1995;16:151–164. doi: 10.1016/0196-9781(94)00017-z. [DOI] [PubMed] [Google Scholar]
- 24.Loria R, Bignell DR, Moll S, Huguet-Tapia JC, Joshi MV, Johnson EG, Seipke RF, Gibson DM. Thaxtomin biosynthesis: the path to plant pathogenicity in the genus Streptomyces. Antonie van Leeuwenhoek. 2008;94:3–10. doi: 10.1007/s10482-008-9240-4. [DOI] [PubMed] [Google Scholar]
- 25.Belin P, Moutiez M, Lautru S, Seguin J, Pernodet JL, Gondry M. The nonribosomal synthesis of diketopiperazines in tRNA-dependent cyclodipeptide synthase pathways. Nat. Prod. Rep. 2012;29:961–979. doi: 10.1039/c2np20010d. [DOI] [PubMed] [Google Scholar]
- 26.Gruenewald S, Mootz HD, Stehmeier P, Stachelhaus T. In vivo production of artificial nonribosomal peptide products in the heterologous host Escherichia coli. Appl. Environ. Microbiol. 2004;70:3282–3291. doi: 10.1128/AEM.70.6.3282-3291.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Abed RM, Dobretsov S, Al-Fori M, Gunasekera SP, Sudesh K, Paul VJ. Quorum-sensing inhibitory compounds from extremophilic microorganisms isolated from a hypersaline cyanobacterial mat. J. Ind. Microbiol. Biotechnol. 2013;40:759–772. doi: 10.1007/s10295-013-1276-4. [DOI] [PubMed] [Google Scholar]
- 28.Hong SH, Kwon YC, Martin RW, Des Soye BJ, de Paz AM, Swonger KN, Ntai I, Kelleher NL, Jewett MC. Improving cell-free protein synthesis through genome engineering of Escherichia coli lacking release factor 1. ChemBioChem. 2015;16:844–853. doi: 10.1002/cbic.201402708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Jewett MC, Calhoun KA, Voloshin A, Wuu JJ, Swartz JR. An integrated cell-free metabolic platform for protein production and synthetic biology. Mol. Syst. Biol. 2008;4:220. doi: 10.1038/msb.2008.57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Jewett MC, Swartz JR. Mimicking the Escherichia coli cytoplasmic environment activates long-lived and efficient cell-free protein synthesis. Biotechnol. Bioeng. 2004;86:19–26. doi: 10.1002/bit.20026. [DOI] [PubMed] [Google Scholar]
- 31.Villiers BR, Hollfelder F. Mapping the limits of substrate specificity of the adenylation domain of TycA. ChemBioChem. 2009;10:671–682. doi: 10.1002/cbic.200800553. [DOI] [PubMed] [Google Scholar]
- 32.Bergendahl V, Linne U, Marahiel MA. Mutational analysis of the C-domain in nonribosomal peptide synthesis. Eur. J. Biochem. 2002;269:620–629. doi: 10.1046/j.0014-2956.2001.02691.x. [DOI] [PubMed] [Google Scholar]
- 33.Kwon YC, Jewett MC. High-throughput preparation methods of crude extract for robust cell-free protein synthesis. Sci. Rep. 2015;5:8663. doi: 10.1038/srep08663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lambalot RH, Gehring AM, Flugel RS, Zuber P, LaCelle M, Marahiel MA, Reid R, Khosla C, Walsh CT. A new enzyme superfamily - the phosphopantetheinyl transferases. Chem. Biol. 1996;3:923–936. doi: 10.1016/s1074-5521(96)90181-7. [DOI] [PubMed] [Google Scholar]
- 35.Miller LM, Mazur MT, McLoughlin SM, Kelleher NL. Parallel interrogation of covalent intermediates in the biosynthesis of gramicidin S using high-resolution mass spectrometry. Protein Sci. 2005;14:2702–2712. doi: 10.1110/ps.051553705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Stachelhaus T, Walsh CT. Mutational analysis of the epimerization domain in the initiation module PheATE of gramicidin S synthetase. Biochemistry. 2000;39:5775–5787. doi: 10.1021/bi9929002. [DOI] [PubMed] [Google Scholar]
- 37.Niederholtmeyer H, Sun ZZ, Hori Y, Yeung E, Verpoorte A, Murray RM, Maerkl SJ. Rapid cell-free forward engineering of novel genetic ring oscillators. eLife. 2015;4:e09771. doi: 10.7554/eLife.09771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.La Clair JJ, Foley TL, Schegg TR, Regan CM, Burkart MD. Manipulation of carrier proteins in antibiotic biosynthesis. Chem. Biol. 2004;11:195–201. doi: 10.1016/j.chembiol.2004.02.010. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.