Abstract
The hepatitis C virus (HCV) RNA-dependent RNA polymerase (RdRp) has several distinct biochemical activities, including initiation of RNA synthesis by a de novo mechanism, extension from a primed template, nontemplated nucleotide addition, and synthesis of a recombinant RNA product from two or more noncovalently linked templates (template switch). All of these activities require specific interaction with nucleoside triphosphates (NTPs). Based on the structure of the HCV RdRp bound to NTP (S. Bressanelli, L. Tomei, F. A. Rey, and R. DeFrancesco, J. Virol. 76:3482-3492, 2002), we mutated the amino acid residues that contact the putative initiation GTP and examined the effects on the various activities. Although all mutations retained the ability for primer extension, alanine substitution at R48, R158, R386, R394, or D225 decreased de novo initiation, and two or more mutations abolished de novo initiation. While the prototype enzyme had a Km for GTP of 3.5 μM, all of the mutations except one had Kms that were three- to sevenfold higher. These results demonstrate that the affected residues are functionally required to interact with the initiation nucleotide. Unexpectedly, many of the mutations also affected the addition of nontemplated nucleotide, indicating that residues in the initiating NTP (NTPi)-binding pocket are required for nontemplated nucleotide additions. Interestingly, mutations in D225 are dramatically affected in template switch, indicating that this residue of the NTPi pocket also interacts with components in the elongation complex. We also examined the interaction of ribavirin triphosphate with the NTPi-binding site.
Hepatitis C Virus (HCV), a member of the flaviviridae family of positive-strand RNA viruses, infects up to 3% of the world's population (1). A prime target for antiviral treatment in HCV is the NS5B protein, the RNA-dependent RNA polymerase (RdRp) responsible for viral RNA synthesis. The RdRp is one of the subunits expected to participate in HCV replication and is responsible for the initiation and elongation of viral RNA synthesis. Initiation of RNA synthesis in infected cells likely starts de novo; that is, by use of a 1-nucleotide (nt) primer (3, 14). This mode of RNA synthesis requires a specialized site in the RdRp, called the I site, that specifically recognizes the initiating nucleoside triphosphate (designated NTPi, with GTP being preferred by the recombinant HCV RdRp) (14, 23, 25). A second NTP-binding pocket, the I + 1 site, binds a nucleotide for phosphoryl transfer as dictated by the template base (14). Cocrystal structures of the HCV RdRp and nucleotides have been reported previously (3, 21), identifying a number of residues at D225, R48, R158, R386, R394, and S367 that interact with specific moieties of the initiation GTP (Fig. 1A). In this structure, it is not known whether the GTP binds to the I site or the I + 1 site. NTPi binding to the I site is base specific, while binding of the second NTP to the I + 1 site should be directed by the template (25). Because the RdRp-GTP structure was determined without a template, it is likely that the residues identified recognized the NTPi.
FIG. 1.
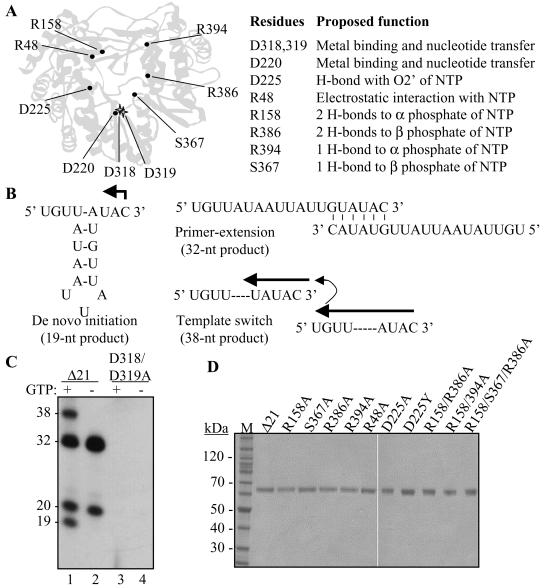
Mutations within the catalytic pocket of the HCV RdRp and assays for activities of the HCV RdRp. (A) Locations of the mutations in the HCV RdRp and their proposed function in interacting with the initiation GTP. The two stars representing D318, D319, and D220 denote the location of the site for nucleotide polymerization. (B) The modes of RNA synthesis that are directed by RNA LE19. A monomer of LE19 could direct de novo initiation from the 3′-most cytidylate, producing a 19-nt product. In the event that two LE19 molecules base pair with sequences at their 3′ ends, the resultant heterodimer could direct the synthesis of a 32-nt primer extension product which, based on the sequence, could be generated in the absence of GTP. Should the ternary complex translocate from one template to another without release of the nascent RNA, a process called template switch, a product of 38 nt is generated. (C) Demonstration of the products from LE19 template by the prototype HCV RdRp, Δ21, and one with mutations at the active site. GTP is needed for de novo initiation, resulting in 19-nt RNA and 38-nt template switch products generated after de novo initiation. GTP is not needed to produce a primer extension product of 32- and 20-nt nontemplated nucleotide addition to LE19. The sizes of the products have been previously characterized (22, 23, 25). (D) Preparation of recombinant proteins used in this study. The proteins were separated by sodium dodecyl sulfate-polyacrylamide electrophoresis (4 to 12% polyacrylamide) and were stained with Coomassie blue. The names of the mutant proteins are derived from the respective amino acid, the position along the NS5B polypeptide, and the final amino acid at that position.
Despite evidence that de novo initiation is the most relevant mechanism for HCV RNA synthesis in cells, a version of the HCV NS5B named Δ21, which lacks the C-terminal 21 hydrophobic residues, has at least three additional activities related to nucleotide polymerization: primer extension, nontemplated nucleotide addition to the 3′ terminus of the RNA, and the ability to synthesize one RNA product from two or more noncovalently linked templates (Fig. 1B). For clarity, we shall call the latter process template switch. Nontemplated nucleotide addition, de novo initiation, and template switch by the HCV RdRp occur at lower levels in reactions lacking Mn2+, but these activities are greatly increased when up to 2 mM Mn2+ is added to the reaction (16, 25). To date, the functional coordination between these activities has not been carefully examined.
We have characterized various activities of the HCV RdRp by using a 19-nt template, named LE19, derived from the 3′ end of the minus-strand RNA of bovine viral diarrhea virus (BVDV). LE19 is predicted to form a simple stem-loop with an unpaired 3-nt initiation sequence at the 3′ end that directs initiation (Fig. 1B). A similar, but more stable, structure is also found at the 3′ end of the minus-strand HCV RNA (15, 26). Several of the products produced by Δ21 with LE19 were previously characterized (23, 25). De novo initiation generates a 19-nt product that is initiated from a 3′-terminal cytidylate. LE19 could also partially anneal as a dimer and be extended to form a 32-nt product (Fig. 1C). Other products included ones with additional nucleotide(s) at the 3′ end of LE19 and template switch products of 38 nt and longer that are thought to be multimeric forms of the 19-nt product. In this work we made mutations in recombinant HCV NS5B that are predicted to affect the I site and characterized the different modes of RNA synthesis in vitro by primarily using LE19. We found that some mutations that affected interaction with the NTPi could differentially alter the various activities of the HCV RdRp.
MATERIALS AND METHODS
Purification of NS5B from Escherichia coli.
NS5B proteins were expressed from pET plasmid derivatives in E. coli BL21(DE3). The prototype HCV NS5B protein lacked the hydrophobic C-terminal 21 amino acids. Six histidines were added to the C termini of each of the proteins to allow affinity purification. Bacteria were grown at 30°C in standard Luria-Bertani medium supplemented with 50 μg of ampicillin/ml and 34 μg of chloramphenicol/ml until the culture reached an optical density at 600 nm of 1.0. The culture temperature was then lowered to 25°C, and expression was induced for 4 h with 1 mM isopropyl-thiogalactoside. Cells were harvested after centrifugation, and recombinant RdRp from HCV was purified in buffer lacking divalent metals by passage through a Talon nickel affinity column (Invitrogen Inc.) followed by a poly(U)-agarose column (Pharmacia Inc). The protein was adjusted to between 1 and 2 mg/ml after quantification by the Lowry assay, with bovine serum albumin as a concentration control.
Site-directed mutagenesis of the HCV NS5B cDNA was made by using the QuickChange kit (Stratagene, San Diego, Calif.) according to the manufacturer's protocol. The entire cDNA was then sequenced to confirm that the specified mutation was made and that no additional changes had inadvertently taken place. The mutant plasmids were transformed into E. coli BL21(DE3), induced for expression, and purified as described above for Δ21.
RdRp activity assays.
RNAs were chemically synthesized by Dharmacon, Inc. (Boulder, Colo.), and were purified by denaturing gel electrophoresis, checked for quality by toluidine blue staining, and quantified by spectrophotometry. Standard RdRp assays consisted of 0.125 μM template with 0.08 μM NS5B in a 20-μl reaction mixture containing 20 mM sodium glutamate (pH 8.2), 4 mM MgCl2, 12.5 mM dithiothreitol, 0.5% (vol/vol) Triton X-100, 200 μM each ATP and UTP, and 250 nM [α-P32]CTP (Amersham, Inc.). GTP was used at a final concentration of 200 μM unless stated otherwise. For titrations with GTP analogs, no GTP was added to the reaction mix, because LE19 used GTP only for initiation. Where present, MnCl2 was added to a final concentration of 1 mM. Modifications to the reaction mixtures will be noted as appropriate. The final reaction mixture contained 10 mM NaCl that derived from the RdRp storage buffer. RNA synthesis reaction mixtures were incubated at 25°C for 60 min (a time at which RNA synthesis was in the linear phase for all of the mutant proteins) and was stopped by phenol-chloroform extraction followed by ethanol precipitation in the presence of 5 μg of glycogen and 0.5 M ammonium acetate. Time course analyses used a master reaction mixture. Aliquots of the reaction mixtures were sampled at 0-, 5-, 10-, 15-, 20-, 30-, 45-, 60-, and 90-min time intervals and were terminated for RNA synthesis by the addition of an equal volume of electrophoresis dye mixture containing 95% formamide. Products were usually separated by electrophoresis on denaturing (7.5 M urea) polyacrylamide gels. Gels were wrapped in plastic and exposed to film at −80°C. Each result shown was reproduced in at least one and usually two to four independent experiments. Quantification of radiolabeled bands was performed with a PhosphorImager (Molecular Dynamics).
The kinetic constant, Km, for the initiation GTP was determined as previously described by Ranjith-Kumar et al. (25). To prevent reinitiation and thereby determine the initial reaction rates, heparin (to 0.5 mg/ml) and 50 μM nonradiolabeled CTP was added to the reaction mixture after 10 min of incubation. Mn2+ was present at 1 mM. The reaction products were quantified and plotted using Microsoft Excel, and the Km values were determined from the X intercept.
RESULTS
In an attempt to better understand the mechanism of de novo initiation by viral RdRps, we used the HCV RdRp, Δ21, as the prototype to construct a series of mutant proteins with one or more alanine substitutions in the putative NTPi site (Fig. 1A). All proteins were purified by use of a metal affinity column followed by a poly(U) column, resulting in greater than 98% purity (Fig. 1D). No obvious differences in either the expression of the mutant proteins or their purification were apparent (data not shown). For nomenclature, the mutant proteins were named by the mutated amino acid followed by the identity of the final mutation. Where more than one amino acid was changed, the protein was named by all of the mutations.
To determine whether the residues identified by Bressanelli et al. (3) affected the I or the I + 1 pocket, we initially examined the effects of these mutations using LE19 as the template (Fig. 2). RNA synthesis conditions, such as the use of Mn2+, the concentration of template, NTPs, etc., were previously optimized and used as described in Ranjith-Kumar et al. (23-25). In the absence of Mn2+, all of the mutant proteins except S367A lost the ability to initiate RNA synthesis by the de novo mechanism (Fig. 2). In contrast, only protein R158A showed significantly less ability to extend from a primer (Fig. 2). These results indicate that most of the mutated residues do affect interaction with the NTPi. Additional evidence consistent with this claim is presented below (for example, see Fig. 3A). For the remainder of this work, we will refer to these residues as affecting interaction with the NTPi.
FIG. 2.
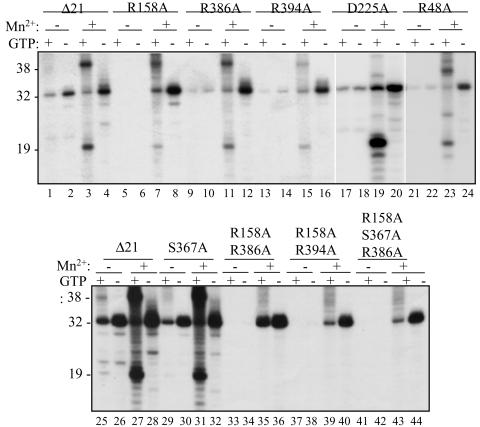
Prototype and mutant proteins used to characterize the HCV RdRp activities and an initial survey of the effects of the mutations. The activities of Δ21 and mutant proteins affected in the residues identified by Bressanelli et al. (3) to bind NTP. Names of the proteins used in the reactions are underlined at the top of the autoradiograms. The presence or absence of Mn2+ at 1 mM and GTP at 200 μM are indicated by plus and minus signs, respectively. The size of each band (in nucleotides) is noted to the left of the autoradiograms.
FIG. 3.
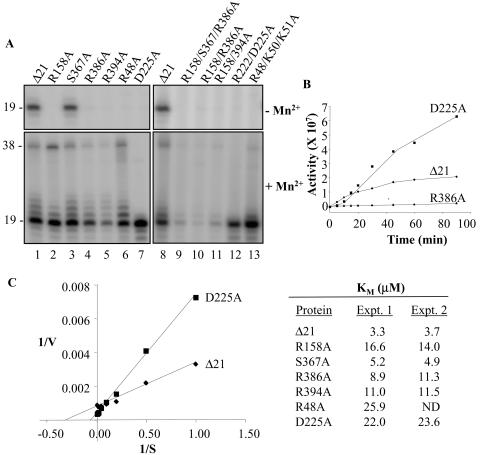
Analysis of de novo initiation by Δ21 and its mutant derivatives. (A) Effect of Δ21 and mutants on de novo RNA synthesis. The template used was LE19P, which contains a 3′-terminal puromycin that prevents nontemplated nucleotide addition to the template. The proteins used in each reaction and the most relevant conditions are listed above the appropriate lanes in the autoradiograms. RNA synthesis resulted in a 19-nt band, but higher molecular size bands likely resulted from nontemplated nucleotide addition to the nascent RNA. Lower molecular size bands may have resulted from premature termination. (B) Time course of de novo RNA synthesis by Δ21, D225A, and R386A. (C) Determination of Km for initiation GTP for Δ21 and single mutants. Representative Lineweaver-Burke double-reciprocal plot for Δ21 and D225A is shown on the left. All estimations are done in duplicates except for R48A. ND, not determined.
All proteins were examined for the activities attributed to the HCV RdRp. The reactions were performed in the presence and absence of GTP, which was required for de novo initiation (25), and with and without Mn2+ at 1 mM, which significantly increases de novo initiation, primer extension, and nontemplated nucleotide addition (22, 25). Mutant S367A, which putatively affects H bonding to the β phosphate, had activities similar to those of Δ21 (Fig. 2, lanes 29 to 32). Activities of the other mutant proteins were dependent on the divalent metal(s) present in the reaction (Fig. 2). In the presence of only Mg2+, R158A was severely affected for all four activities of Δ21. All other single-amino-acid mutants were active for primer extension to various degrees, ranging from levels comparable to those of Δ21 (as in D225A) to reduced levels (e.g., R386A and R48A). To examine the effect on primer extension in the absence of Mn2+ further, we tested double mutants R158A/R386A and R158A/R394A. These two mutant proteins did not produce the 32-nt product (compare lanes 25 to 26 with 33 to 34 and 37 to 38). Together, these results suggest that residues that bind the NTPi can participate in other reactions that follow the initial RNA synthesis.
Activities of the mutant proteins were quite different in the presence of Mn2+. D225A, which putatively forms an H bond with the ribose 2′ OH of the NTP, was unaffected for de novo initiation when assayed at 60 min after the initiation of RNA synthesis. Other single-amino-acid changes had reduced, albeit detectable, amounts of de novo initiation products (Fig. 2, lanes 1 to 32). Proteins R158A/R386A, R158A/R394A, and R158A/R367A/R386A eliminated de novo initiation product even in the presence of Mn2+ but were capable of primer extension (Fig. 2, lanes 33 to 44). These results suggest that Mn2+ likely helps to stabilize the interaction between the active site and the NTPi. This activity will be characterized further with oligonucleotide mimics of the NTPi.
An activity associated with NTPi binding is the inhibition of primer extension (24). GTP is postulated to inhibit primer extension in a Mn2+-independent manner that requires the C-terminal tail and the active-site β loop. This effect is seen by the ca. three- to fivefold difference in the amount of the 32-nt RNA in the presence of GTP (Fig. 2, lanes 1 and 2). In the absence of Mn2+ only S367A had different levels of the 32 mer if GTP was present. These results are consistent with the model proposed in Ranjith-Kumar et al. (24), in that shutoff of primer extension requires the residues that bind the NTPi.
Template switch was also affected by the amino acid substitutions in the presence of Mn2+. As expected, several of the mutations that reduced de novo initiation also reduced the amount of the 38 mer. However, D225A, which is capable of robust de novo initiation in the presence of Mn2+, unexpectedly produced reduced amounts of product of 38 nt and longer that arose from template switch (Fig. 2, lanes 17 to 20, and data not shown). Hence, D225 may also have a role in interacting with the template or the nascent RNA. Lastly, mutations in the NTPi-binding site also affected TNTase activity of the HCV RdRp. A 21-nt product generated by nontemplated nucleotide addition was clearly observed with Δ21 (Fig. 2, lanes 1, 2, 25, and 26) but was significantly reduced with mutations in the NTPi-binding residues.
This survey of the products made from LE19 by Δ21 and the NTPi mutants clearly indicates that, while the mutated residues affect interaction with the NTPi, different activities of the HCV RdRp can be intricately linked. To understand the function of these residues better, we will expand the examination of their roles in de novo initiation, TNTase activity, and template switch in the following sections.
De novo initiation.
To confirm that the mutations caused a defect in de novo initiation, we used LE19P, a chemically synthesized RNA identical to LE19, except with an additional 3′ puromycin. This modification renders the template incapable of nontemplated nucleotide addition but allows de novo initiation (22). In the absence of Mn2+, Δ21 and S367A produced detectable and comparable amounts of the de novo-initiated 19 mer (Fig. 3A, top panel, lanes 1 and 3). However, none of the other mutations produced detectable amounts of the 19 mer. In the presence of Mn2+, all single-amino-acid substitution proteins produced the 19 mer at levels comparable to those of Δ21, except for D225A, which increased de novo initiation by 30%, and R394A, which decreased it by 77%. We then tested proteins with two or three mutations and found that, similar to results with LE19, de novo initiation was further decreased by combining two or more amino acid substitutions that affect NTPi binding (Fig. 3A, lower panel, lanes 9 to 11). An effect was also observed on the production of the 38-nt template switch product, except for D225A, which produces significantly less of this product (Fig. 3A, lane 7). These defects in de novo initiation with LE19P were less obvious than with LE19 (compare Fig. 3, bottom panel, to Fig. 2), likely due to puromycin providing additional contacts for the HCV RdRp or puromycin blocking primer extension activity and increasing the frequency of de novo initiation.
To examine whether there is an effect on the kinetics of RNA synthesis, we compared the synthesis of the 19-nt RNA over time by Δ21 and two mutants, R386A and D225A. In the presence of Mn2+, all the proteins showed an increase in RNA synthesis with time up to 90 min, suggesting that analysis of RNA synthesis for 60 min is still within the linear range. As observed earlier (Fig. 2), RNA accumulation by R386A was significantly lower than that of Δ21 throughout the time course. D225A, on the other hand, showed less RNA synthesis within the first 15 min of the reaction than Δ21 but accumulated higher levels of 19-nt product upon longer incubation. This suggests that D225A cannot form a stable productive initiation complex to the same extent as Δ21, but it is capable of efficient elongation and reinitiation.
To further confirm that the mutations affected de novo initiation, we determined whether there is an effect on the Km for the GTPi. LE19 is designed where GTP is used only as the initiation nucleotide and, hence, the Km value will not affect a requirement during elongative RNA synthesis. Kinetic analysis was done with the addition of 0.5 mg of heparin/ml at 10 min after the start of the reaction to prevent reinitiation of RNA synthesis and, hence, would represent the actual Km for GTP. Δ21 had a Km for GTP of 3.3 μM (Fig. 3C), a value consistent with the one reported by Ranjith-Kumar et al. (25). All the mutant proteins tested, except S367A, showed higher Km values, ranging from 9 to 26 μM (Fig. 3C). The majority of the mutated residues are, therefore, involved in productive ternary complex formation with GTP and RNA.
Specificity for the T1 nucleotide.
De novo initiation requires specific interaction with the template initiation nucleotide, T1, and the NTPi. We examined whether mutations in the NTPi-binding site affected specific recognition of the T1 by comparing the amount of product from LE19 to that of a version of LE19 in which the 3′-most C was changed to a U (Table 1). The HCV RdRp has been reported to initiate RNA synthesis from a T1 uridylate at a low rate (23), and the HCV-N replicon may even switch to using an initiation uridylate preferentially (27). In the absence of Mn2+, the only mutant that retained RNA synthesis was S367A. Like Δ21, S367A clearly preferred a T1 cytidylate for de novo initiation, because both proteins had a reduced rate of synthesis from LE19 + 1U. In the presence of Mn2+, while RNA synthesis was recovered from LE19, there was no increased preference to initiate from a T1 uridylate. Mutant D225A, which had increased de novo initiation in the presence of Mn2+, was also unable to initiate efficiently from a T1 uridylate. These results indicate that the mutations affecting the NTPi pocket do not apparently decrease the specificity for the T1 site.
TABLE 1.
Effects of mutations on the recognition of the template initiation nucleotide
| Protein | With Mn2+
|
Without Mn2+
|
||
|---|---|---|---|---|
| +1C | +1U | +1C | +1U | |
| Δ21 | 100 | 11 | 100 | 12 |
| R158A | 12 | 1 | 0 | 0 |
| S367A | 88 | 8 | 50 | 15 |
| R386A | 75 | 8 | 0 | 0 |
| R394A | 53 | 5 | 0 | 0 |
| R48A | 59 | 1 | 0 | 0 |
| D225A | 205 | 1 | 0 | 0 |
| R158A/R386A | 0 | 0 | 0 | 0 |
| R158A/R394A | 0 | 0 | 0 | 0 |
| R158A/S367A/R386A | 0 | 0 | 0 | 0 |
Use of NTPi analogs.
Several GTP analogs can substitute for the NTPi in vitro (23). We examined the use of GTP analogs, including dGTP, GDP, GMP, and guanosine, to determine whether changes in the NTPi-binding site will affect the recognition of the NTPi (Table 2). Because LE19 contains only a single cytidylate at the T1 position, the analogs should be incorporated only as the 5′-most residue of the newly synthesized RNA and therefore would affect de novo initiation. In the absence of Mn2+, Δ21 and mutant S367A were capable of initiating RNA synthesis with GDP and GMP but not with guanosine or dGTP. The other mutant proteins were unable to direct de novo initiation in the absence of Mn2+ (Table 2), and the substitution of GTP with analogs did not rescue de novo initiation.
TABLE 2.
Effect of mutation in the catalytic pocket of Δ21 on the incorporation of NTPi analogs
| Protein | Without Mn2+
|
With Mn2+
|
||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| GTP | dGTP | GDP | GMP | G | GU | GUAUA | GTP | dGTP | GDP | GMP | G | GU | GUAU | |
| Δ21 | 100 | 5 | 62 | 139 | 6 | 2,480 | 6,630 | 100 | 90 | 288 | 306 | 48 | 537 | 102 |
| R158A | 0 | 0 | 0 | 0 | 0 | 1 | 13 | 18 | 3 | 19 | 5 | 0 | 144 | 154 |
| S367A | 51 | 1 | 30 | 68 | 1 | 978 | 5,751 | 85 | 98 | 292 | 285 | 25 | 381 | 71 |
| R386A | 0 | 0 | 0 | 0 | 0 | 5,207 | 444 | 68 | 2 | 175 | 160 | 139 | 1,044 | 39 |
| R394A | 0 | 0 | 0 | 0 | 0 | 2,253 | 226 | 51 | 6 | 283 | 399 | 125 | 767 | 32 |
| R48A | 0 | 0 | 0 | 0 | 0 | 0 | 1,140 | 60 | 12 | 150 | 66 | 3 | 349 | 250 |
| D225A | 0 | 0 | 0 | 0 | 0 | 0 | 2,810 | 272 | 26 | 226 | 198 | 8 | 755 | 480 |
The presence of Mn2+ altered the use of the GTP analogs by both Δ21 and the mutant proteins. With Δ21, for example, dGTP was used to initiate RNA synthesis in the presence of Mn2+. In addition, reactions with Mn2+ produced more de novo initiation products with GDP and GMP than with GTP (Table 2) (23). We observed a range of activities with the NTPi mutants. R386A, R394A, S367A, and R48A had activity profiles generally similar to that of Δ21, with some minor differences in the use of guanosine (R386A and R394A used it more efficiently than Δ21) and GMP (R48A initiated with GMP less efficiently than Δ21) (Table 2). R158A was unable to efficiently initiate synthesis with any of the analogs and did not exhibit preferential initiation with GDP and GMP. Interestingly, D225A initiated with dGTP less efficiently than GTP even in the presence of Mn2+, and it did not preferentially initiate with GDP and GMP. In this regard, D225A appeared to more specifically require GTP as the NTPi than did Δ21. Small differences in the amount of products initiated with GTP analogs in this work and results reported earlier (23) may be due to the source of NS5B protein (HCV strain J4 was used in the previous study while BK was used in this one).
The HCV RdRp can also use short oligonucleotides as substitutes for the NTPi, as long as the oligonucleotides are able to maintain correct base pairing with the T1 nucleotide and the adjacent template sequence (23). We used these oligonucleotides to examine whether the increased base pairing stability between NTPi and the template can overcome the mutation-associated effects. In the absence of Mn2+, the amount of the de novo-initiated products synthesized increased with the length of the oligonucleotide (Table 2). Relative to GTP, GU and pentamer GUAUA synthesized 25- and 66-fold more product. Mutant protein R158A was unable to use either GU or GUAUA to initiate synthesis in the absence of Mn2+. Mutant S367A had activities quite similar to those of Δ21, consistent with results with single-nucleotide GTP analogs. Other mutant proteins also had notable differences from Δ21. R386A and R394A produced more of the 19 mer with GU in place of GTP, but they were unable to use GUAUA as efficiently as Δ21. Mutants D225A and R48A, which were unable to initiate with GTP in the absence of Mn2+, could use GUAUA, but not GU, for product synthesis.
In the presence of Mn2+, Δ21 still preferred GU to GTP for initiation (by approximately fivefold), but it produced a comparable amount of the product using GTP and GUAUA. This phenomenon may be related to the conformational change that results as a consequence of divalent metal binding to the active site of the HCV RdRp (2, 25), which could make the NTPi pocket more spatially constrained. The differential use of GU and GUAUA was seen with mutants R386A, R394A, and S367A but was less obvious with R158A, D225A, and R48A. All of the analyses with the single and oligonucleotide NTPi analogs confirm our hypothesis that these mutations affected the recognition of the NTPi. Furthermore, the differential use of NTPi mimics suggests that the mutations affect different steps in de novo-initiated RNA synthesis. More specifically, R386 and R394 are required for the formation of the first phosphodiester bond, because their defects could be rescued with the dinucleotide GU while the defects in R48A and D225A are rescued only after the nascent RNA is 5 nt long.
TNTase activity.
HCV RdRp can add nontemplated nucleotides to LE19 (22). This activity is difficult to distinguish relative to template-dependent RNA synthesis in solution-based assays (13). For Δ21, TNTase activity was detectable in reactions with or without Mn2+ but was more abundant in its presence (Fig. 4) (22). Furthermore, TNTase activity was at approximately 150% the amount of nascent RNA synthesis with LE19. It is unclear whether the TNTase activity uses the NTPi pocket, the I + 1 pocket, or both. To examine this, we tested the effects of mutations on the NTPi site on TNTase activity. In the absence of Mn2+, TNTase activity by the HCV RdRp was readily detectable. Of all of the mutants, only S367A (having 85% de novo initiation activity versus that of Δ21) retained TNTase activity at 30% of Δ21. Furthermore, for all other single mutants, the requirements for de novo initiation, TNTase activity, was restored in the presence of Mn2+ (Fig. 4). These results suggest that the residues in the HCV RdRp that are involved in de novo initiation are also involved in TNTase activity. However, mutants R48A and D225A retained robust TNTase activity but were significantly decreased for de novo initiation in the absence of Mn2+. These results indicate that the NTPi-binding residues are required for TNTase activity, but the requirements for the two activities do not completely overlap.
FIG. 4.
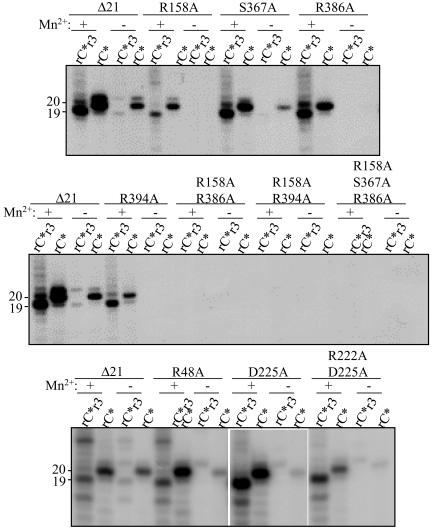
Examination of nontemplated nucleotide addition to the template RNA. The names of the proteins used and the most relevant conditions are indicated above the appropriate lanes in the autoradiogram. The term rC*r3 means that the reaction contains all four nucleotides, with CTP being the radiolabel. rC* denotes a reaction performed with only [α-32P]CTP. The lengths of the products, in nucleotides, are indicated to the left of the autoradiogram. All of the lanes within a panel are from one experiment, but some lanes are juxtaposed to facilitate presentation of the results.
Template switch.
Several of the mutations in the NTPi pocket affected template switch (Fig. 2). A defect in the template switch (generating a 38 mer) is to be expected with many of the NTPi mutants, because fewer ternary complexes are available. However, mutant D225A, which was quite robust for de novo initiation of the 19-nt product, produced only 14% of the 38-nt recombinant product compared to that of Δ21 (Fig. 2). A double mutant R222A/D225A also showed a similar property (Fig. 5A), while mutant R222A behaved like the wild type for de novo initiation and template switch (data not shown). To examine this further, we made mutant D225Y and found that it also had reduced template switch activity (Fig. 5A, lanes 9 to 12). We hypothesize that D225, which recognizes the ribose 3′ hydroxyl of the NTPi, also plays an additional role either in recognition of the second template or in the release of the nascent RNA.
FIG. 5.
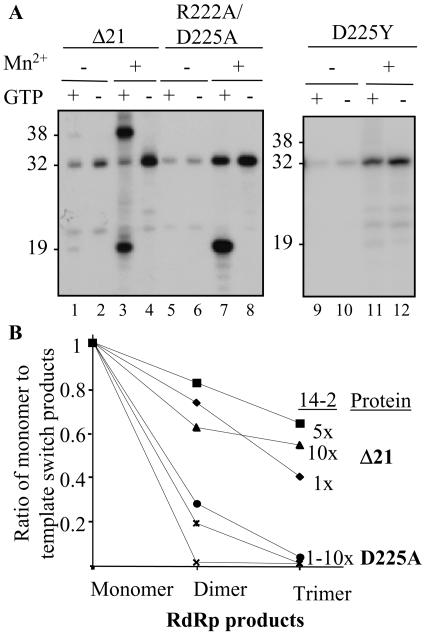
Production of template switch products by mutant D225. (A) Autoradiogram of the products made by Δ21 and mutant R222A/D225A and D225Y. The relevant conditions for the reactions are indicated above the autoradiogram. (B) Quantification of the oligomeric products made by Δ21 and D225A using three concentrations of template 14-2 (12.5, 62.5, and 125 nM represented as 1-, 5-, and 10-, respectively), which was previously characterized for template switch (16). The amounts of the products made were normalized for the amount of radiolabeled CMP incorporated and plotted as ratio of monomer to template switch products.
To characterize the role of D225A further, we used template 14-2, which has been demonstrated to give a high level of template switch with the HCV RdRp (16). The ratios of multimeric products formed relative to monomer by Δ21 and D225A at three different template concentrations were quantified. The reactions were performed in the presence of Mn2+ to increase the frequency of template switch, and the results were normalized for the number of radiolabeled CMP incorporated. The frequency of template switch events by D225A was more than fivefold lower than that of Δ21 at all template concentrations tested (Fig. 5B). Taken together, these data support a model where multiple RdRp conformational intermediates are formed during RNA synthesis, with residue D225 being involved in these functions.
Ribavirin and HCV RNA synthesis.
A medically relevant aspect of NTP use by the HCV RdRp is the incorporation of ribavirin, which is used in combination with interferon to treat HCV infection (10-12, 20). Ribavirin is incorporated in place of purine triphosphates and may result in an increased error rate in the newly synthesized RNAs, leading to a loss of virus infectivity (9, 19). Ribavirin incorporation was reported to involve residue Y415 of the HCV RdRp, which is a residue that lines the template-binding pocket of the HCV RdRp (28). We sought to determine whether incorporation of ribavirin triphosphate (RTP) requires the residues of the NTPi pocket.
We first determined whether the HCV RdRp could incorporate RTP into product RNAs by using template LE19. In reactions with only ATP, CTP, and UTP (each at 100 μM final concentration), no de novo initiation was observed due to the absence of GTP (Fig. 6A, lanes 2 and 12). Increasing RTP in the reactions up to 500 μM did not result in further production of the 19 mer either in the presence or absence of Mn2+ (Fig. 6A, lanes 3 to 6 and 13 to 16), indicating that RTP cannot functionally substitute for the NTPi. Similar results were obtained with LE19P, confirming that RTP was unable to initiate RNA synthesis (Fig. 6B). However, we note that the 32-nt primer extension product and the 20- and 21-nt TNTase products were quite prominent in these reactions and that increasing ribavirin did not significantly inhibit their production unless Mn2+ was present in the reactions (Fig. 6A). These results indicate that ribavirin is incorporated poorly in the presence of ATP, CTP, and UTP during primer extension and terminal nucleotide addition.
FIG. 6.
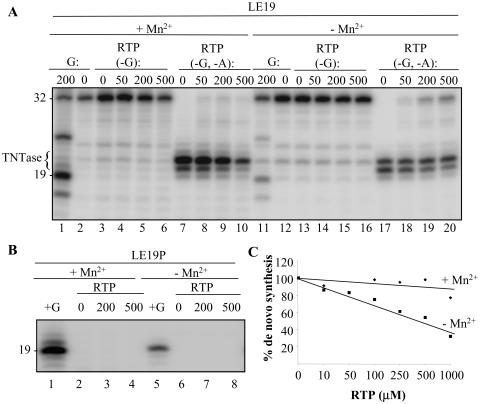
RNA synthesis by the HCV RdRp in the presence of ribavirin triphosphate. (A) Autoradiogram of the products made by Δ21 under different reaction conditions. RTP was added to reactions lacking either GTP or both GTP and ATP, as indicated by the notations −G and −G,−A. The 32-nt product in lanes 17 to 20 demonstrates that RTP was incorporated during elongative synthesis in place of ATP. (B) An assay to confirm that RTP cannot serve as the initiation NTP. The assay used the puromycin-modified LE19P. The control reactions are indicated by +G. (C) Examination of whether RTP inhibits RNA synthesis by the HCV RdRp in reactions containing all of the normal NTPs. The level of synthesis was quantified and normalized to one lacking RTP.
We attempted to incorporate ribavirin in reactions lacking both GTP and ATP. Increasing RTP resulted in production of the 32-nt primer extension product, possibly indicating that RTP was inefficiently incorporated into the newly synthesized RNA in place of ATP (Fig. 6A, lanes 17 to 20). Because GTP is not necessary for the synthesis of the 32-nt elongation product, we were unable to test whether RTP could substitute for GTP during elongation with LE19. Interestingly, incorporation of RMP was slightly higher in the absence of Mn2+ (Fig. 6A, lanes 7 to 10).
Because RTP could not substitute for the initiating GTP, we determined whether it would inhibit de novo initiation with LE19P when the full complement of NTPs was present. In the absence of Mn2+ there was an inhibitory effect of ribavirin, with the 50% inhibitory concentration being approximately 400 μM. In the presence of Mn2+, where there was robust de novo initiation, the effect of up to 1 mM RTP was minimal, with a decrease to only 15% relative to that of a reaction lacking RTP. Any role of ribavirin on de novo-initiated RNA synthesis thus appeared to be indirect.
We also tested whether any of the catalytic mutants are affected in RTP incorporation during RNA synthesis. Incorporation of RTP was not significantly different for any of the mutants compared to that of Δ21 (data not shown). These results show that perturbations of the catalytic site in the HCV RdRp do not necessarily increase the incorporation of ribavirin triphosphate.
DISCUSSION
Cocrystallization of the HCV RdRp with NTP identified a number of contacting residues in the RdRp (Fig. 1A) (3). To functionally analyze the role of these amino acids, we generated several alanine substitution mutants and analyzed their biochemical activities. A change of S367, which putatively hydrogen bonds with the β phosphate of the GTP, had no obvious effect on the activities we tested. The remainder of the Discussion will deal with observations for the other residues (R48, R158, R386, R394, and D225). First, we present evidence that the residues interact with the NTPi, because alanine substitutions in them (i) preferentially affected de novo initiation rather than primer extension; (ii) increased the Km for the NTPi during de novo initiation; (iii) had defects that could be overcome with Mn2+, which likely interacts with the phosphates and helps to neutralize their charges, thereby ameliorating the impact of some mutations in the RdRp; and (iv) could be partially overcome by replacing the GTP with NTPi analogs. We note, however, that despite the effects on interaction with the NTPi, the mutations did not make the HCV RdRp less specific in the use of the NTPi, because the mutants still preferred + 1C for initiation and all of the analogs used possessed a guanine (Tables 1 and 2). Mutational analysis of the NTPi residues also revealed that the different activities of the HCV RdRp could be functionally separated into de novo initiation, primer extension, TNTase activity, and template switch (Table 3). Again, Mn2+ plays an active role in these activities, and its presence can ameliorate the effects of some mutations. The effects of some of the mutant proteins revealed interesting relationships, as will be discussed below.
TABLE 3.
Summary of the activities of the mutations tested with RNA LE19 in this studya
| Protein | Without Mn2+
|
With Mn2+
|
||||||
|---|---|---|---|---|---|---|---|---|
| Primer extension | Template switch | De novo initiation | TNTase | Primer extension | Template switch | De novo initiation | TNTase | |
| Δ21 | ++ | ++ | ++ | ++ | ++ | ++ | ++ | ++ |
| S367A | ++ | + | + | + | ++ | ++ | ++ | ++ |
| R158A | − | − | − | − | ++ | ++ | + | + |
| R386A | + | − | − | − | ++ | ++ | + | ++ |
| R394A | + | − | − | − | ++ | + | + | + |
| R48A | + | − | + | + | ++ | + | + | ++ |
| D225A | + | − | − | + | ++ | − | ++ | ++ |
++, high level of activity; +, detectable; −, not detected.
Relationships between biochemical activities.
Some mutations affect more than one biochemical activity. A summary of how the mutations affected the activities examined is provided in Table 3. D225 is required for de novo initiation (both in terms of the synthesis of the 19 mer and in mediating the GTP-induced repression of primer extension) and in the ability to switch templates. Previously it was determined that template switch was also more efficient when the second template had an initiation cytidylate, suggesting that template switch shared requirements for de novo initiation (16). One model to explain the dual role of the de novo residues is that multiple conformations of the RdRp are associated with RNA synthesis. During initiation, the polymerase must bind two NTPs, but after the formation of the first phosphodiester bond the residues that are required to bind the NTPi may be used for other activities.
In general, mutations that affected de novo initiation also affected TNTase activity. Except for R48A and D225A, mutations that caused the loss of de novo synthesis in the absence of Mn2+ were also unable to add nontemplated nucleotide (Fig. 4). Consistent with this claim, the presence of Mn2+ increased both de novo initiation and nontemplated nucleotide addition (Fig. 2). These results suggest that the NTPi-binding pocket may also be required for TNTase activity, either by providing the NTP for the TNTase activity or allowing a more stable interaction with the template RNA to facilitate nontemplated nucleotide addition. We favor the latter model for the following reasons. First, mutational analysis of the NTPi site shows that the requirements for the two activities are not completely overlapping. Second, the NTPi pocket is highly specific for GTP and GTP analogs, while TNTase activity prefers to add ATP and CTP (22). Third, we could functionally separate the two activities in the presence of ∼100 mM NaCl (22). In addition, several mutations affecting the NTPi pocket also exhibited defects in template switch, suggesting that the defect in template interaction may be the reason behind effects on several activities.
Recognition of the NTPi.
It was observed in a previous work that the use of the NTPi was highly specific (23). In this work, we demonstrated that changes in the RdRp did not increase the range of the NTPi molecules that can be incorporated. Ribavirin triphosphate, a purine nucleotide analog, was not recognized as the NTPi. We have observed that ribavirin can be incorporated during elongative synthesis when ATP concentration is low (data not shown), demonstrating that it is incorporated only after binding to the 1 + 1 site. The use of the GTP analogs and the discrimination against RTP suggest that recognition of the NTPi is not dependent on Watson-Crick interactions with the template. The T7 RNA-dependent RNA polymerase also recognizes the NTPi in a base-specific manner (17). Because recognition of the NTPi is not needed in polymerases that extend from a primer, it is likely a unique property of de novo-initiating polymerases. The identities of the residues that interact with the guanine base in the HCV RdRp need to be determined.
Another aspect in the recognition of the NTPi is the requirement for the ribose sugar. The HCV RdRp appears to discriminate against use of dGTP by forming an H bond with the ribose 2′ hydroxyl. Unlike the base requirements, recognition of the ribose is not absolutely required, because dGTP can still direct initiation when Mn2+ is present in the reaction. Furthermore, mutant D225A could initiate RNA synthesis de novo in the presence of Mn2+. However, when changed to a tyrosine, de novo initiation was severely reduced (Fig. 5A). These results indicate that a bulky tyrosine group at position 225 is incompatible for RNA synthesis. We have not been able to detect incorporation of dNTPs efficiently by the HCV RdRp variants, including D225A and D225Y, except as the NTPi (C. T. Ranjith-Kumar, unpublished observations). In other polymerases, mutations that expand the use of the nucleotides affected the recognition of the N + 1 nucleotide, not the NTPi. We would expect that the recognition of the riboses of the two nucleotides should have different requirements (4, 5).
Several lines of evidence suggest that the mutations affect the stability of the initiation complex. One potential contributor to the stability may be Mn2+, whose presence rescued a number of activities (Table 3). Mn2+-dependent activity was also observed with poliovirus RdRp in cells (8), indicating that even though it is present in only micromolar concentrations, it should not be ruled out as a possible RdRp cofactor, because the HCV RdRp may have higher affinity for Mn2+ than for Mg2+. In addition, mutants R386A and R394A, which were incapable of RNA synthesis with GTP in the absence of Mn2+, were capable of efficient RNA synthesis with GU (Table 2). Also, D225A and R48A, which could not use GTP and GU in the absence of Mn2+, were capable of robust RNA synthesis with GUAUA, a GTPi analog that could form more stable base pairing with the template. This suggests that the catalytic mutants have a differential effect on RNA synthesis, with R386 and R394 being required for the formation of the first phosphodiester bond formation. Mutants R48A and D225A, however, may require additional forces to stabilize the initiation in order to successfully produce RNA products.
Recently, the crystal structure of bovine viral diarrhea virus RdRp in complex with GTP was determined, with assigned i and i + 1 sites (7). This complex did not contain divalent metals that, in the HCV RdRp, caused a detectable conformational change (25). Perhaps due to the lack of divalents, the GTP molecule in complex with the BVDV was 6 Å from the expected site. Nonetheless, several residues that interacted with this GTP and with the initiating NTPs corresponded to the ones that are seen with the HCV RdRp. More specifically, residues S498, R517, D350, and R285 correspond, respectively, to residues S367, R386, D225, and R158 in HCV RdRp. S498 and D350 were shown previously to be necessary for RNA synthesis (28).
In summary, our functional characterization of the effects of mutations in the NTPi pocket revealed a complex relationship between several of the RdRp-associated activities in vitro. Given that de novo initiation is likely used for HCV RNA replication in cells, we suspect that similar mutations will prevent HCV replication in vivo by disrupting one or more activities. Results consistent with this were presented by Cheney et al. (6), who showed that residues that negatively affected RNA synthesis in vitro debilitated HCV replication in a subgenomic replicon. Lastly, the availability of variants of the HCV RdRp that are differentially defective for specific activities will be useful for further dissection of the mechanism of RNA-dependent RNA synthesis.
Acknowledgments
We thank our colleagues at GlaxoSmithKline and Texas A&M University for helpful discussions.
The C. Kao lab is supported by National Science Foundation MCB grant 0332259.
REFERENCES
- 1.Alter, M. J., and E. E. Mast. 1994. The epidemiology of viral hepatitis in the United States. Gastroenterol. Clin. N. Am. 23:437-455. [PubMed] [Google Scholar]
- 2.Bougie, I., S. Charpentier, and M. Bisaillon. 2003. Characterization of the metal ion binding properties of the hepatitis C virus RNA polymerase. J. Biol. Chem. 278:3868-3875. [DOI] [PubMed] [Google Scholar]
- 3.Bressanelli, S., L. Tomei, F. A. Rey, and R. DeFrancesco. 2002. Structural analysis of the hepatitis C virus RNA polymerase in complex with ribonucleotides. J. Virol. 76:3482-3492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Brieba, L. G., and R. Sousa. 2000. Role of histidine 784 and tyrosine 629 in ribose discrimination by T7 RNA polymerase. Biochemistry 39:919-923. [DOI] [PubMed] [Google Scholar]
- 5.Cases-Gonzalez, C. E., M. Gutierrex-Rivas, and L. Medendez-Arias. 2000. Coupling ribose selection to fidelity of DNA synthesis. J. Biol. Chem. 275:19759-19767. [DOI] [PubMed] [Google Scholar]
- 6.Cheney, I. W., S. Naim, V. C. Lai, S. Dempsey, D. Bellows, M. P. Walker, J. H. Shim, N. Horscroft, Z. Hong, and W. Zhong. 2002. Mutations in NS5B polymerase of hepatitis C virus: impacts on in vitro enzymatic activity and viral RNA replication in the subgenomic replicon cell culture. Virology 297:298-306. [DOI] [PubMed] [Google Scholar]
- 7.Choi, K. H., J. M. Groarke, D. C. Young, R. J. Kuhn, J. L. Smith, D. C. Pevear, and M. G. Rossmann. 2004. The structure of the RNA-dependent RNA polymerase from bovine viral diarrhea virus establishes the role of GTP in de novo initiation. Proc. Natl. Acad. Sci. USA 30:4425-4430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Crotty, S., D. Gohara, D. K. Gilligan, S. Karelsky, C. E. Cameron, and R. Andino. 2003. Manganese-dependent polioviruses caused by mutations within the viral polymerase. J. Virol. 77:5378-5388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Crotty, S., D. Maag, J. J. Arnold, W. Zhong, J. Y. Lau, Z. Hong, R. Andino, and C. E. Cameron. 2000. The broad-spectrum antiviral ribonucleoside ribavirin is an RNA virus mutagen. Nat. Med. 6:1375-1379. [DOI] [PubMed] [Google Scholar]
- 10.Cummings, K. J., S. M. Lee, E. S. West, J. Cid-Ruzafa, S. G. Fein, Y. Aoki, M. S. Sulkowski, and S. N. Goodman. 2001. Interferon and ribavirin versus interferon alone in the re-treatment of chronic hepatitis C previously nonresponsive to interferon: a meta-analysis of randomized trials. JAMA 285:193-199. [DOI] [PubMed] [Google Scholar]
- 11.Davis, G. L., R. Esteban-Mur, V. Rustgi, J. Hoefs, S. C. Gordon, C. Trepo, M. L. Shiffman, S. Zeuzem, A. Craxi, M. H. Ling, and J. Albrecht. 1998. Interferon alfa-2b alone or in combination with ribavirin for the treatment of relapse of chronic hepatitis C. International Hepatitis Interventional Therapy Group. N. Engl. J. Med. 339:1493-1499. [DOI] [PubMed] [Google Scholar]
- 12.Di Bisceglie, A. M., J. Thompson, N. Smith-Wilkaitis, E. M. Brunt, and B. R. Bacon. 2001. Combination of interferon and ribavirin in chronic hepatitis C: re-treatment of nonresponders to interferon. Hepatology 33:704-707. [DOI] [PubMed] [Google Scholar]
- 13.Ferrari, E., J. Wright-Minogue, J. W. S. Fang, B. M. Baroudy, J. Y. N. Lau, and Z. Hong. 1999. Characterization of soluble hepatitis C virus RNA-dependent RNA polymerase expressed in Escherichia coli. J. Virol. 73:1649-1654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kao, C. C., P. Singh, and D. Ecker. 2001. De novo initiation of viral RNA-dependent RNA synthesis. Virology 287:252-260. [DOI] [PubMed] [Google Scholar]
- 15.Kashiwagi, T., K. Hara, M. Kohara, J. Iwahashi, N. Hamada, H. Honda-Yoshino, and T. Toyoda. 2002. Promoter/origin structure of the complementary strand of hepatitis C virus genome. J. Biol. Chem. 277:28700-28705. [DOI] [PubMed] [Google Scholar]
- 16.Kim, M.-J., and C. C. Kao. 2001. Factors regulating template switch in vitro by viral RNA-dependent RNA polymerases: implications for RNA-RNA recombination. Proc. Natl. Acad. Sci. USA 98:4972-4977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kuzmine, I., P. A. Gottlied, and C. T. Martin. 2003. Binding of the priming nucleotide in the initiation of transcription by T7 RNA polymerase. J. Biol. Chem. 278:2819-2823. [DOI] [PubMed] [Google Scholar]
- 18.Lai, V. C., C. C. Kao, E. Ferrari, J. Park, A. S. Uss, J. Wright-Minogue, Z. Hong, and J. Y. Lau. 1999. Mutational analysis of bovine viral diarrhea virus RNA-dependent RNA polymerase. J. Virol. 73:10129-10136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Maag, D., C. Castro, Z. Hong, and C. E. Cameron. 2001. Hepatitis C virus RNA-dependent RNA polymerase (NS5B) as a mediator of the antiviral activity of ribavirin. J. Biol. Chem. 276:46094-46098. [DOI] [PubMed] [Google Scholar]
- 20.McHutchison, J. G., S. C. Gordon, E. R. Schiff, M. L. Shiffman, W. M. Lee, V. K. Rustgi, Z. D. Goodman, M. H. Ling, S. Cort, and J. K. Albrecht. 1998. Interferon alfa-2b alone or in combination with ribavirin as initial treatment for chronic hepatitis C. Hepatitis Interventional Therapy Group. N. Engl. J. Med. 339:1485-1492. [DOI] [PubMed] [Google Scholar]
- 21.O'Farrell, D., R. Trowbridge, D. Rowlands, and J. Jager. 2003. Substrate complexes of hepatitis C virus RNA polymerase (HC-J4): structural evidence for nucleotide import and de-novo initiation. J. Mol. Biol. 326:1025-1035. [DOI] [PubMed] [Google Scholar]
- 22.Ranjith-Kumar, C. T., J. Gajewski, L. Gutshall, D. Maley, R. Sarisky, and C. Kao. 2001. Viral RNA-dependent RNA polymerase has terminal transferase activity: implications for viral RNA synthesis J. Virol. 75:8615-8623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ranjith-Kumar, C. T., L. Gutshall, M.-J. Kim, R. T. Sarisky, and C. C. Kao. 2002. Requirements for de novo initiation of rna synthesis by recombinant flaviviral RNA-dependent RNA polymerases. J. Virol. 76:12526-12536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ranjith-Kumar, C. T., L. Gutshall, R. T. Sarisky, and C. C. Kao. 2003. Multiple interactions within the hepatitis C virus RNA polymerase repress primer-dependent RNA synthesis. J. Mol. Biol. 330:675-685. [DOI] [PubMed] [Google Scholar]
- 25.Ranjith-Kumar, C. T., Y. C. Kim, L. Gutshall, C. Silverman, S. Khandekar, R. T. Sarisky, and C. C. Kao. 2002. Mechanism of de novo initiation by the hepatitis C virus RNA-dependent RNA polymerase: role of divalent metals. J. Virol. 76:12513-12525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Schuster, C., C. Isel, I. Imbert, C. Ehresmann, R. Marquet, and M. P. Kieny. 2002. Secondary structure of the 3′ terminus of hepatitis C virus minus-strand RNA. J. Virol. 76:8058-8068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Yi, M., and S. M. Lemon. 2003. Structure-function analysis of the 3′ stem-loop of hepatitis C virus genomic RNA and its role in viral RNA replication. RNA 9:331-345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Young, K. C., K. L. Lindsay, K. J. Lee, W. C. Liu, J. W. He, S. L. Milstein, and M. M. Lai. 2003. Identification of a ribavirin-resistant NS5B mutation of hepatitis C virus during ribavirin monotherapy. Hepatology 38:869-878. [DOI] [PubMed] [Google Scholar]


