Abstract
PAKs, p21-activated kinases, play central roles and act as converging junctions for discrete signals elicited on the cell surface and for a number of intracellular signaling cascades. PAKs phosphorylate a vast number of substrates and act by remodeling cytoskeleton, employing scaffolding, and relocating to distinct subcellular compartments. PAKs affect wide range of processes that are crucial to the cell from regulation of cell motility, survival, redox, metabolism, cell cycle, proliferation, transformation, stress, inflammation, to gene expression. Understandably, their dysregulation disrupts cellular homeostasis and severely impacts key cell functions, and many of those are implicated in a number of human diseases including cancers, neurological disorders, and cardiac disorders. Here we provide an overview of the members of the PAK family and their current status. We give special emphasis to PAK1 and PAK4, the prototypes of groups I and II, for their profound roles in cancer, the nervous system, and the heart. We also highlight other family members. We provide our perspective on the current advancements, their growing importance as strategic therapeutic targets, and our vision on the future of PAKs.
Keywords: p21-activated kinase, PAK, cytoskeleton remodeling, cancer, nervous system, heart
1. Introduction
The p21-activated kinases (PAKs) are serine/threonine kinases and were initially discovered as binding proteins of small GTPases (Manser et al., 1994). PAKs soon emerged as effectors of Cdc42 and Rac1 small GTPases (Martin et al., 1995; Knaus et al., 1995, Bagrodia et al., 1995). The PAK family consists of six members and divided into two groups, group I (PAK1, PAK2, and PAK3) and group II (PAK4, PAK5, and PAK6). PAK5 is sometimes referred to as PAK7. The PAK gene sequences and their structures are conserved from amoeba, yeast, Caenorhabditis elegans, Xenopus, to human (Kumar et al., 2009). The PAK family represents a converging junction for discrete cell surface triggers and affects cytoskeleton remodeling, intracellular signaling, and gene expression (Vadlamudi & Kumar, 2003; Kumar & Li, 2016; Shao et al., 2016). The molecular functions of the PAK family are because of their intrinsic ability to directly phosphorylate effector substrates, physically translocate to distinct subcellular domains, employ scaffolding activity, and modulate target gene expression. These processes are crucial to a large number of cellular functions and many human diseases including cancer.
The initial phase of PAK research primarily focused on studying the role of PAKs as effectors of small GTPases in the context of dynamic remodeling of cytoskeleton motile structures (Sell & Chernoff, 1997; Bokoch, 2003). The next phase of research revealed a definitive contribution to human cancer (Kumar et al., 2006; Molli et al., 2009), with the first report of PAK1 signaling cascade conferring invasiveness to breast cancer cells in response to heregulin-beta1-mediated ErbB2 stimulation (Adam et al., 1998) and its overexpression in breast tumors (Vadlamudi et al., 2000). Around the same time, the PAK researchers also began to move into nuclear functions beginning from the observation that activated PAK1 translocates to the nucleus in human cells (Li et al., 2002), opening up an exciting frontier for PAKs. These developments together set the stage for exploring the significance of PAKs in many cellular processes that confer cancer phenotype, viz., invasion, metastasis, anti-apoptosis, drug resistance, angiogenesis, epithelial-to-mesenchymal transition, DNA-damage repair, modulation of gene expression, and changes in progression of mitosis and cell cycle. PAKs are now known as crucial transducers for signals that originate on the membrane and ultimately confer cancer phenotypes (Fig. 1). The primary focus of the review is PAKs role in cancer, and we delve into PAKs various physiological processes and their significance in the nervous system and the heart. We choose PAK1 and PAK4, the prototypes of groups I and II, to give a detailed outline of the structure, biochemistry, and biology of PAK family. We also discuss other members PAK2, PAK3, PAK5, and PAK6.
Figure 1.
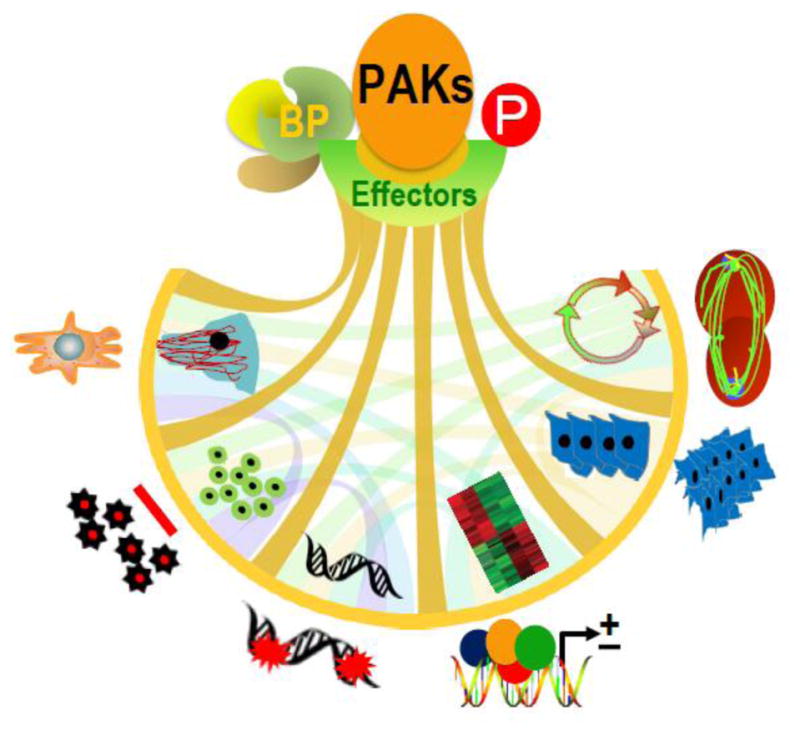
PAK stimulation by upstream signals contributes to phenotypic signaling through binding partners (BP) and phosphorylation of effector substrates. Representative PAK-regulated cancerous phenotypes include – cytoskeleton remodeling, cell motility, inhibition of apoptosis, DNA damage response, gene expression, transformation and invasion, deregulated cell cycle progression and metastasis (from left to the right).
2. PAK’s structural domains and mechanism of action
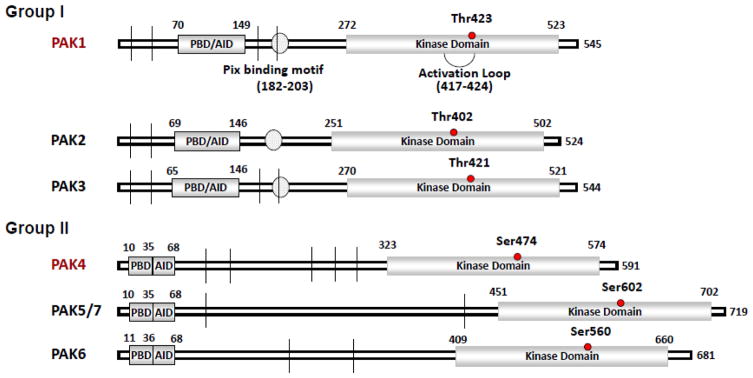
The PAK family contains an auto inhibitory domain (AID), a kinase domain at the carboxyl-terminus, and a p21-binding domain (PBD) at the amino-terminus. The AID domain in group I PAK partly overlap with a modified AID domain in group II PAKs (Fig. 2). The regulatory domains of groups I and II are structurally distinct, and their activation processes are different. The kinase activity of group I PAKs is stimulated when bound to small GTPases or other proteins, but in group II PAKs, the kinase activity is constitutive. The mechanisms of PAK activation have been extensively reviewed before in several reviews (Rane, & Minden, 2014; Jha, & Strauss, 2012; Baker et al., 2014). Here we discuss the principles of PAK activation.
Figure 2.
Structural domains of group I and group II PAKs. For PAK1, detail organization of polypeptide chain was shown. Numerals indicate residue numbers at the boundaries of various subdivisions. The N-terminal auto-regulatory region (70–149aa) and the kinase domain (272–523aa) are shown. The detailed substructure of the auto-regulatory region including overlapped PBD (p21-binding domain, in blue) and AID (auto inhibitory domain, in green) is shown in the expanded upper part of the PAK1 diagram. | indicates proline-rich region and indicates the interaction motif for PIX. For other PAKs, PBD, kinase domain, and proline-rich region are shown for each family member.
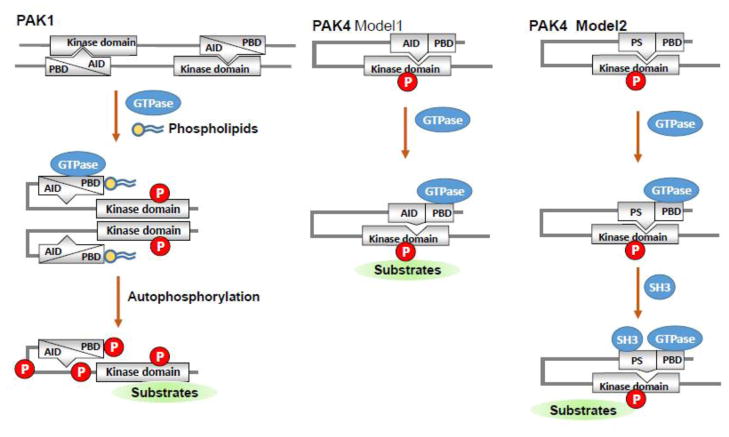
The activity of group I is modified via a reciprocal auto inhibitory mechanism of two PAK molecules acting as a dimer. The PBD domain overlapping with the AID domain binds to the kinase domain of another PAK molecule, and this homodimerization makes them inactive (Fig. 3). Binding of an activated small GTPase to the PBD domain and concomitant interaction with the proximal amino acids and phosphoinositide, trigger distinct changes in the conformation of the catalytic domain. This leads to dissociation of the AID domain from the kinase domain, induce further conformational changes in the dimerized molecules, phosphorylate both PAK molecules, and stimulate their kinase activity (Pirruccello et al., 2006; Buchwald et al., 2001; Strochlic et al., 2010). Upon binding to a substrate, kinase domain also become a monomer, and auto phosphorylation stabilizes the PAK molecule in the activated form. Interestingly, some in vitro studies show that the active PAK1 kinase could continue to remain as a dimer. This aspect of PAK1 activation in physiologically relevant settings awaits further research, and may potentially reshape PAK-directed therapeutic approaches, if proven correct. In addition, AID function could be antagonized by mechanisms independent of small GTPases. For example, group I PAKs are stimulated by interaction of its PXXP motif with the SH3 domain in signaling molecules (Galisteo et al., 1996; Puto et al., 2003), phosphorylation by 3-phospho-inositide dependent kinase-1, AKT and JAK (Bokoch, 2003; Zhou et al., 2003; Tao et al., 2011), and binding of phospholipids, exchange factor β-PIX or SH3 proteins such as NCK1 (Zhou et al., 2003; Fryer et al., 2006) and GRB2 (Strochlic et al., 2010; Banerjee et al., 2002; Thiel et al., 2002; Bokoch et al., 1998; King et al., 2000; Howe and Juliano, 2000; Shin et al., 2013).
Figure 3.
Models of PAK activation. P21-activated kinases (PAKs) have a conserved carboxyl-terminal serine/threonine kinase domain with a single phosphorylation (P) site and an amino-terminal regulatory domain. The regulatory domain of group I PAKs (PAK1–3) contain PBD (p21-binding domain) and overlapped AID (auto-inhibitory domain), which is structurally distinct from that of group II PAKs (PAK4–6). There are two suggested models for PAK4 activation.
As oppose to group I PAKs, group II PAKs remain active constitutively and generally lack an AID domain (Fig. 3). However, recently PAK4 regulatory domain has been shown to contain an AID-like pseudosubstrate sequence that inactivates the kinase activity of Cdc42-bound PBD domain (Baskaran et al., 2012; Ha et al., 2012). It has been suggested that the binding of the PS domain to an SH3 motif-containing protein may help release the catalytic domain and allow stimulation of the PAK kinase activity (Ha et al., 2012). Although our understanding of the mechanisms of PAK activation has improved tremendously over the years, the turn-off mechanism of group II PAKs remains unclear.
3. PAK’s gene structure and splicing
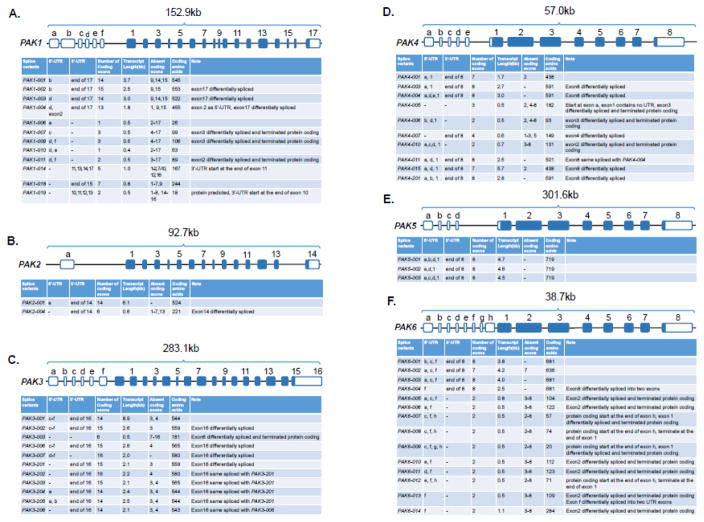
The human PAK1 gene contains 23 exons, of which six exons are for 5′-UTR and seventeen encode protein (Fig. 4A). The 5′-UTR of PAK1 mRNA is differentially expressed. In humans, there are twenty transcripts, ranging from 308-bp to 3.7-kb, generated via alternative splicing. Among these transcripts, only twelve contain open-reading frames and encode ten proteins and two small polypeptides. The sizes other eight are non-coding RNA transcripts range from 308 to 863 nucleotides. In contrast to human PAK1, murine Pak1 generates five transcripts, of which three coding transcripts of 508 to 3.0-kb long, and the other two are non-coding RNAs of about 900 nucleotides with retained-intron sequences.
Figure 4.
Structure and partial transcripts of human PAK genes. The diagram/table and transcripts of PAK1~6 genes are based on the ENSEMBL database (PAK1 ENSG00000149269, PAK2 ENSG00000180370, PAK3 ENSG00000077264, PAK4 ENSG00000130669, PAK5 ENSG00000101349, PAK6 ENSG00000137843). The blues are alternatively spliced coding exons and the white ones are UTRs. The tables are based on the known protein coding transcripts, not including noncoding RNAs.
The PAK2 gene contains 15 exons and one is for 5′-UTR, and the remaining generate three transcripts. The PAK2 gene is much shorter than PAK1, about 92.7-kb (Fig. 4B). Among PAK2 transcripts, two encode proteins of 524 amino acids and 221 amino acids, and the third is a non-coding transcript of 371 nucleotides long. The murine Pak2 gene generates two transcripts. A 5.7-kb protein-encoding transcript for 524 amino acids, and another 1.2-kb non-coding RNA transcript.
The PAK3 gene is the largest among group I members (Fig. 4C). The human PAK3 gene is 283-kb and contains 22 exons of which 6 are for 5′-UTR region, and generates 13 transcripts. Among these transcripts, 11 PAK3 transcripts are predicted to code for proteins of 181 to 580 amino acids, while two transcripts are non-coding RNAs. The murine Pak3 gene generates ten transcripts, which are predicted to encode proteins with 544 amino acids and 559 amino acids, and four small polypeptides.
Group II PAKs have less number of exons compared to group I PAKs, which highlights the structural differences between the two groups. The human PAK4 is 57-kb and has 13 exons (Fig. 4D). There are 12 transcripts in human, of which 10 transcripts encode predicted proteins of 438 to 591 amino acids, while there are two non-coding RNAs. The murine PakK4 gene generates four transcripts. Two coding for proteins of 593 amino acids, and two others are non-coding with one RNA containing retained-intron sequences.
The PAK5 (also known as PAK7) gene is about 301-kb, much longer than other PAKs, and contains 12 exons. Four for 5′-UTR, and eight encode proteins. Alternative splicing of the exons generates three transcripts that encode a 719-amino-acid protein, the largest protein in PAK family (Fig. 4E). The murine Pak5 gene contains two protein-encoding transcripts and a 2.0-kb noncoding transcript with retained intron sequences.
The human PAK6 gene is about 38-kb long, and contains 16 exons, including eight for 5′-UTR and generate 17 transcripts. Fourteen PAK6 transcripts encode four predicted proteins of 681 and 636 amino acids (Fig. 4F), while remaining transcripts are non-coding. The murine Pak6 gene yields five transcripts, of which two are predicted to encode proteins of about 682 amino acids, and three others are non-coding in nature.
4. PAKs and cytoskeleton remodeling
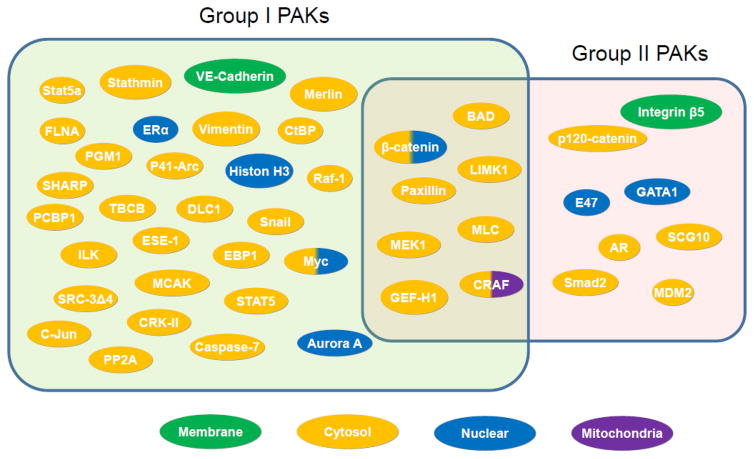
The PAK signaling-initiated cytoskeleton remodeling plays an important role in cellular invasiveness and metastasis due to its phosphorylation of effector substrates and scaffolding (Fig. 5). For example, Cofilin-mediated depolymerization of F-actin is countered by increased ability of PAK1-phosphorylation of LIM-kinase-1 and regulation of its activity Cofilin on Ser3 (Edwards et al., 1999). Interestingly, activated LIMK1 leads to increased secretion of proteases that also have roles in breast cancer invasion (Bagheri-Yarmand et al., 2006), and remodeling of various components of the extracellular matrix (Scott et al., 2010). Similarly, PAK1, PAK4 and PAK5- signaling pathways affect MMP family of proteases in breast, ovarian, and glioma cancer cells (Hammer, Diakonova, 2015; Kesanakurti et al., 2012; Wang et al., 2013). PAK1 also contributes to cytoskeleton remodeling via actin depolymerization and nucleation. For example, the stability and nucleation activity of the Arp2/3 actin nucleation complex and its role in cancer cell motility are modulated by PAK 1 phosphorylation of Arpc1b-Thr21 (Vadlamudi et al., 2004). Further, Arpc1b is also overexpressed in cancer cells (Mahlamäki et al., 2004; Molli et al., 2010). Filamin A (FLAa) is an actin-interacting PAK1 substrate with roles in cytoskeleton remodeling (Vadlamudi et al., 2002) and cancer invasion because of its interaction with CDK4 and MMP9 (Zhong et al., 2010; Sun et al., 2014). The level of FLAa correlates with invasiveness of certain cancer cells such as melanoma (Zhang et al., 2014) and glioblastoma (Chantaravisoot et al., 2015). PAK1 phosphorylates Cortactin, also an actin binding protein, which is upregulated in some cancer (Buday and Downward 2007; Moshfegh et al., 2014). Other PAK1-interacting substrates with roles in tumorigenesis includes dynein light chain 1 (DLC1-Ser88) (Vadlamudi et al., 2004) and integrin-linked kinase-1 (ILK-Thr173/Ser246) (Acconcia et al., 2007). In addition to actin filaments, cancer cell aggressiveness is also affected by microtubule biogenesis, which are regulated by PAK1 by phosphorylating Tubulin Cofactor B (TCoB-Ser65/ Ser128) (Vadlamudi et al., 2005). PAK1 phosphorylation of microtubule destabilizing protein Stathmin inactivates its inhibitory activity against microtubule stabilization (Wittmann et al. 2004).
Figure 5.
Representative substrates of group I and II PAKs and their subcellular localization.
In recent years, similar to PAK1 signaling, there is a growing emphasis on PAK4 substrates with role in cytoskeleton reorganization and cell motility. PAK4 phosphorylates LIMK1 at Thr508 and regulates cell migration (Ahmed et al., 2008). Although PAK4 kinase activity is not stimulated by Cdc42, its interaction with activated Cdc42 is critical for cytoskeletal reorganization. PAK4-Cdc42 interaction has also been shown to support redistribution of PAK4 to the Golgi compartment in porcine endothelial cells (Abo et al., 1998). In addition, PAK4 also signals through LIMK1 and stabilizes actin–myosin filaments (Dan et al., 2001), while PAK4 phosphorylation of integrin β5 on Ser759 and Ser762 promotes cell migration (Li et al., 2010b). Both PAK4 and PAK5 phosphorylate p120-catenin on Ser288 for cytoskeleton reorganization (Wong et al., 2010). Interestingly, PAK4 phosphorylation of superior cervical ganglia 10 (SCG10) facilitates dynamics of microtubules and invasiveness of cancer cells (Guo et al., 2014). PAK4 phosphorylation also inhibits GEF-H1-regulated stress fiber formation in fibroblasts (Callow et al., 2005) (Wells et al., 2010). In addition to its role in cytoskeleton remodeling, PAK4 reduces the level of cyclin-dependent kinase inhibitor 1C with implications in cell-cycle progression in breast cancer cell line (Li et al., 2013). Similarly, PAK4 phosphorylation of β-catenin on Ser675 contributes to its transcriptional function (Li et al., 2012), and Smad2 on Ser465 plays a role in tumorigenesis (Wang et al., 2014).
5. PAKs nuclear functions
In addition to their roles in the cytoplasmic compartment, PAKs also play crucial roles in the nuclear compartment and participate in mitotic events and gene expression (Li et al., 2002; Kumar et al., 2006; Li & Kumar, 2016). Growth factor stimulation of cancer cells triggers redistribution of activated PAK1 to the nucleus in breast cancer cells (Singh et al., 2005). Once in the nucleus, PAK1 interacts with chromatin and modulates target gene expression such as PFK-M (muscle-type isoform), NFAT1 (Singh et al., 2005), and tissue factor (TF) (Sánchez-Solana et al., 2012). Interestingly, PAK1 signaling also modulates chromatin remodeling via phosphorylating MORC2 in cancer cells exposed to ionizing radiation (Li et al., 2012). PAK1 phosphorylation of STAT5 transcription factor on Ser779 modulates its transcriptional activation in mammary epithelial cells during mammary gland differentiation (Wang et al., 2003) and in leukemogenesis in BCR-ABL or FLT3- and KIT pathways (Berger et al., 2014; Chatterjee et al., 2014).
Disease relevance of nuclear PAKs became known after observations showed a close relationship between the level of nuclear PAK1 and resistance to tamoxifen in breast cancer patients (Holm et al., 2006). Consequent to PAK1, other family members are also found to have significant roles in the nucleus and cancer. Nuclear PAK4 has been found in endometrial cancer in postmenopausal women (Siu et al. 2015). Recently, PAK4 has been shown to be methylated by lysine methyltransferase SETD6; and its interaction with SETD6 and β-catenin regulates β-catenin-target genes (Vershinin et al., 2016). Another PAK-signaling-mediated regulation of gene expression is PAK1 phosphorylation-dependent modulation of corepressor functions. For example, PAK1 phosphorylation of C-terminal binding protein 1 (CtBP1) on Ser158 promotes its nucleus-to-cytoplasmic redistribution, relieving CtBP1’s corepressor activity on its target genes (Barnes et al., 2003; Thomas et al., 2015). Activated PAK1 also phosphorylates B-cell lymphoma-6 corepressor in the nuclei of colon cancer cells (Barros et al., 2012). Other examples of PAK1 phosphorylation-mediated regulation of corepressors include SNAIL-Ser246, which affects epithelial-to-mesenchymal transition (Yang et al, 2005), and SHARP-Ser3486/Thr468, which affects regulation of NOTCH target genes (Vadlamudi et al., 2005). In growth factor-stimulated cancer cells, PAK1 phosphorylation of polyC-RNA binding protein 1 (PCBP1) at Thr60/Thr127 promotes its nuclear translocation. PCBP1 activates transcription of eukaryotic translation initiation factor 4E (eIF4E) (Meng et al., 2007).
6. PAKs and mitotic progression
Mitotic events play a fundamental role in normal and diseased cells. PAKs are critical at many levels during mitosis. Overexpression of constitutively activated PAK1–a physiologically relevant setting because PAK1 is hyperactivated in cancer-leads to multipolar spindle phenotypes and defective mitotic segregation of chromosomes (Li et al., 2002). Once PAK1 is on the centrosome, GIT1 stimulates its kinase, and in-turn, PAK1 phosphorylates Aurora-A-Thr288/Ser342 and modulates centrosome maturation (Zhao et al., 2005). PAK1 phosphorylation of histone H3.3A/H3-Ser10 is thought to be involved in the condensation of chromosomes during mitosis. As oppose to PAK1, PAK2 phosphorylation of histone H4 of Ser47 helps its interaction with Histone H3.3 during replication (Kang et al., 2011). Similarly, PAK2 signaling has also been shown to participate in the organization of mitotic spindles (Nekrasova & Minden, 2011; Bompard et al., 2013).
As the process of mitosis is affected by microtubule, PAK1 signaling also regulates astral microtubules during mitosis (Baneerjee et al., 2002). PAK1 signaling affects both polymerization and depolymerization of microtubule dynamics: PAK1 phosphorylates Tubulin Cofactor B on Ser65 and Ser128 and facilitates microtubule polymerization (Vadlamudi et al., 2005), and PAK1 phosphorylation of mitotic centromere-associated kinesin on Ser192 and Ser111 promotes microtubule depolymerization (Pakala et al., 2012). In addition, PAK1 phosphorylation of Arpc1b-Thr21, a component of the Arp2/3 complex, leads to defective centrosome duplication (Vadlamudi et al., 2000). Interestingly, Arpc1b-Thr21 can also be phosphorylated by Aurora-A, which itself is a substrate of PAK1 (Molli et al. 2010). Similarly, Aurora-A phosphorylates mitotic centromere-associated kinesin (Zhang, Ems-McClung, & Walczak, 2008; Braun et al., 2014). Polo-like kinase1 is another important substrate of PAK1 during mitosis (Maroto et al., 2008). The current working model suggests that interactions among the PAK1, Aurora-A, Arpc1b, mitotic centromere-associated kinesin, and Polo-like kinase 1 might be important for the regulation of cell cycle in the mitotic phase.
7. PAKs and steroid receptors
Being a kinase, PAK1 phosphorylates Ser305 of estrogen receptor-α (ER) and modulates its transcriptional activities in a ligand-independent manner (Wang et al., 2002; Rayala et al., 2006; Tharakan et al. 2008). Because ER is transactivated by PAK1 in the absence of estrogen, the breast cancer cells show resistance to anti-estrogenic agents such as tamoxifen (Rayala et al., 2006; Bostner et al., 2007). In this context, ER-positive breast cancer patients with nuclear localization of PAK1 were shown to be tamoxifen insensitive (Holm et al., 2006). In contrast to PAK1, PAK6 interacts androgen receptor (AR) and redistributes to the nucleus, which inhibits the ability of androgen receptor to inhibit its target genes (Yang et al., 2001). Interestingly, AR-Ser578 phosphorylation by PAK6 is also accompanied by AR degradation in a ligase-dependent manner (Liu et al., 2013).
8. PAKs in DNA damage response
The PAK signaling was initially linked to DNA-damage response (DDR) by an observation that ionizing radiation (IR) stimulates PAK2 activity in leukemia cells (Roig & Traugh, 1999). This was followed by the finding that PAK1 signaling regulates the expression of a large number of IR-regulated genes in the DNA repair pathway (Motwani et al., 2013). Further, PAK1 modulates the degree of DNA-damage response via directly phosphorylating microrchidia CW-type zinc finger 2 (MORC2-Ser739) and γH2AX (Li et al., 2012). In addition to radiation, PAK1 is stimulated by genotoxic therapeutic agents such as etoposide, and in-turn, PAK1 phosphorylates CRAF-Ser338 and causes radio-resistance (Advani et al., 2015). In general, PAK activation confers survival advantage to cancer cells against DNA-damaging agents, and hence, targeting PAKs has been thought as potential therapeutic approach to sensitize target cells in radiotherapy.
9. PAKs cross-talk with ErbB2 signaling
ErbB2 overexpression and hyperactivation is a prevalent event in breast tumors and a large number of other solid tumors. In breast cancer cells, stimulation of ErbB2 by heregulin-beta1 activates PAK1 kinase, which in turn causes growth factor-mediated increased motility and invasion of breast cancer cells (Adam et al., 1998). In addition, PAK1 signaling also phosphorylates β-catenin on Ser675 (Zhu et al., 2012) and Ser663 (Park et al., 2012), stimulating β-catenin’s transcriptional activity (Park et al., 2012). As PAK1 phosphorylates β-catenin and both PAK1 and β-catenin are downstream effectors of ErbB2 signaling (Adam et al., 1998; Wang et al., 2006b; Ding et al., 2005), PAK1 connects ErbB2 signaling with β-catenin in breast tumor cells (Arias-Romero et al., 2013). Interestingly, PAK4 also phosphorylates β-catenin-Ser675 and stimulates the transcriptional activity of TCF/LEF (Li et al., 2012). In addition to regulating the invasiveness, ErbB2-PAK1 axis supports ErbB2 regulation of vascular endothelial growth factor and this could be inhibited by Herceptin targeting of ErbB2 (Bagheri-Yarmand et al., 2000). Further PAK1 signaling is stimulated by Angiopoietin-1 via regulatory interactions among Dok, Nck, and Tek receptors in endothelial cells (Master et al., 2001). Another PAK1 target that has role in vascular system is Tie1 endothelial receptor tyrosine kinase Thr794 (Reinardy et al., 2015). Consistent with co-regulatory nature of PAK1 and ErbB2 axis in breast cancer, a recent large-scale phosphoproteomic study of breast cancer specimens found that ErbB2 overexpression closely correlates with activated PAK1 in addition to other kinases (Mertins et al., 2016). In brief, ErbB2-PAK1 axis may regulate many facets of signaling in breast cancer, and perhaps, in other hormone-regulated cancers.
10. PAKs and the nervous system
The PAK family plays an important role in a number of neuronal processes, ranging from cytoskeletal remodeling, neuronal apoptosis, axonal guidance, neuronal polarity, synaptic activity, dendritogenesis, to spinal maintenance. The role of PAKs in the nervous system can be traced back to the role of PAK4 in neural tube development in embryos, as PAK4-knockout mice showed defects in axonal growth and neuronal development (Qu et al., 2003). PAK1 signaling participates in brain development because of its ability to induce neuronal migration in select areas of the cortex (Pan et al., 2015). Netrin-1 chemoattractant modulates the growth of axonal cones in a PAK1 signaling-dependent manner wherein PAK1 phosphorylation of shootin1 facilitates the coupling of F-actin dynamics with the formation of motile structures (Toriyama et al., 2013). Similarly, neural cell adhesion molecule (NCAM) stimulation of PAK1 activity supports actin polymerization during neuronal differentiation (Li et al., 2013). In contrast to Netrin-1 and NCAM, endogenous Nischarin suppresses neurite outgrowth by inhibiting PAK1 and PAK2 phosphorylation (Ding et al., 2013). PAK1 pathway also regulates the process of dendritogenesis and spine formation by influencing actin remodeling (Hayashi et al., 2007). In this context, PAK1 signaling participates in the dendritic spine maintenance via phosphorylation of myosin light chain and cofilin (Rubio et al., 2012). Interestingly, targeting PAK1 pathway in cerebellar granule neurons mimics the effect of FOXO transcriptional factors on neuronal morphology. Since this can be rescued by PAK1, it suggests a potential role of FOXO regulation of PAK1 in neuronal biology (de la et al., 2010). Studies from PAK1-knockout mice suggest that PAK1 signaling contributes to long-term potentiation and synaptic plasticity in hippocampus, presumably, due to the effects of its downstream substrate(s) (Asrar et al., 2009). Studies from PAK1 and PAK3-knockout mice suggest that the ability of PAK to regulate neuronal plasticity and synaptic activities play an important role in the size and growth of brain (Huang et al., 2011).
Because of an inherent role of the PAK family in the neuronal biology, it is not surprising that dysregulation of PAKs and their downstream substrates are associated with a number of brain disorders (Chan & Manser, 2012; section on PAK3 this review). For example, low levels of PAK1 and PAK3 mRNAs were seen in the brains of individuals with depression (Fuchsova et al., 2016). The relevance of PAK signaling in neuronal biology is also evident from the observation that inhibition of PAK activity provides neuroprotective effects and accelerate restoration of spinal cord functions in a murine model of spinal cord injury (Ji et al., 2016). Similarly, PAK6 is upregulated following spinal cord injury in a rat model (Zhao et al., 2011), and it interacts with leucine-rich repeat kinase 2 (LRRK2), which was previously shown to be involved in Parkinson’s disease (Civiero L et al, 2015). In addition, defects in the PAK pathway has been linked to Alzheimer’s disease (AD) (Arsenault et al., 2013; Zhao et al., 2006). PAK expression studies in specific hippocampus regions of AD individuals show an increase in PAK expression in early stages of the disease followed by decreased expression in severe subjects (Nguyen et al., 2008). There are also indications that mutants of the amyloid precursor protein pathway interacts with PAK3 during apoptosis of neurons in familial Alzheimers (McPhie et al., 2003). All these clearly point to the fact that targeting the PAK pathway has potential therapeutic advantages for neurodegenerative diseases. In this context, more recently, chemical inhibition of PAK1 prevented accumulation of F-actin at the disruption of myelin junctions as well as normalized myelin permeability in a murine model of hereditary neuropathy, raising the possibility of targeting PAK pathway in diseases associated with demyelination (Hu, et al., 2016).
11. PAKs and the heart
PAK signaling is central to normal and diseased cardiac physiology because of its effects on ion channels, sarcomeric proteins, and excitation and contraction. Results from knockout mice support the notion that PAK1 may be essential for cardiac growth and optimal functioning, which is associated in part with its ability to regulate calcineurin signaling (Davis et al., 2015). Similarly, studies from PAK4-knockout mice show an essential role in actin remodeling in cardiomyocytes, and in the development of various compartments of heart (Nekrasova & Minden, 2012). In a zebrafish model of heart injury, targeting PAK1 has been shown to impair the growth of cardiomyocytes, while targeting PAK4 was found to affect cardiovascular regeneration, highlighting the significance of PAKs in the heart (Peng et al., 2016; Tian et al., 2009; Qu et al., 2003). Mechanistically, the ability of PAK1 to modulate the activity of protein phosphatase PP2A and affect Ca++ homeostasis plays a major role in cardiac physiology. Antagonizing beta-adrenergic and hypertrophic stress and exerting cardiac protective effects by PAK1 signaling involve activation of protein phosphatase 2A (PP2A) (Sheehan et al., 2007; Taglieri, et al., 2011). The PAK1 kinase-dependent interaction with PP2A potentiates PP2A subunit assembly and activation (Staser et al., 2013). PAK1 signaling also regulates Ca++ homeostasis via sarcoplasmic reticulum Ca++-ATPase type 2a (SERCA2a) expression (Wang et al., 2014). These observations led to the notion that PAK1 is a therapeutic target to treat stress-induced cardiac hypertrophy. Experiments are ongoing and some have been tested in murine models using a synthetic PAK1 activator (Liu et al., 2011).
12. PAK1 and PAK4 in cancer progression
Among PAK family members, PAK1 and PAK4 are mostly upregulated in human cancers. In general, elevated PAK expression and its kinase activity support a variety of cancerous phenotypes and correlates well with the progression to more invasiveness. As we had summarized PAK1 overexpression in cancer (Kumar et al., 2006; Kumar & Li, 2016), here will limit our discussion to PAK4. Like PAK1 in human cancer, overexpression of PAK4 also correlates with poor prognosis, tumor aggressiveness (Li et al., 2015a; Begum et al., 2009), metastasis, and infiltration (Song et al., 2015). PAK4 is upregulated in pancreatic cancer, endometrioid ovarian cancer, basal-like breast cancer, and oral squamous cell carcinoma(Begum et al., 2009; Mahlamaki et al., 2004; Davis et al., 2013; Chen et al., 2008; Jiang et al., 2016). PAK4 upregulation in cancer cell lines lead to an increased cell survival, anchorage-independent growth, and transformation in experimental models (Liu et al., 2008; Menzel et al., 2008). In general, PAK4 overexpressing cancer cells are sensitive to growth inhibition by its depletion (Davis et al., 2013; Ahn et al., 2011; Kimmelman et al., 2008; Callow et al., 2002), suggesting an important role for PAK4-driven pathways. There are also examples of mutations that activate PAK4 kinase in certain cancers (Greenman et al., 2007; Whale et al., 2013; Parsons et al., 2005) but such mutations await functional validation. PAK4 signaling supports the growth of cancer cells through AKT- and ERK-regulated signaling (Tyagi et al., 2014; Tabusa et al., 2013). PAK4 modulation of CRAF and BAD phosphorylation inhibits apoptosis in endothelial cells (Alavi et al., 2003). The biology of PAK4 overexpressing cancer cells has also been shown to be regulated by PAK4 targeting by miR-199a/b-3p (Hou et al., 2011), miR-433 (Xue et al., 2015), and miR-224 (Li et al., 2014) in hepatocellular carcinoma cells. PAK4 also phosphorylates Smad2 on Ser465 in growth factor-stimulated cancer cells (Wang et al., 2014). PAK4 is also a Met-receptor-signaling integrator that confers invasive phenotypes (Paliouras et al., 2009). Furthermore, PAK4 interaction with DGCR6L leads to an increased LIMK1 expression and increased motility of gastric cancer cells (Li et al., 2010a), and modulation of MMP expression in certain cancer cell types (Wang et al., 2013; Kesanakurti et al., 2012; Jiang et al., 2016). PAK4 also control the dynamics of cell-adhesion turnover by protecting RhoU from ubiquitation (Dart et al., 2015).
13. PAK2 biology
As oppose to PAK1 and PAK3, Pak2-knockout is embryonic lethal in mice (Hoffmann, Shepeley & Chernoff, 2004). Upstream activators of the PAK2 activity include transforming growth factor β (Wilkes et al., 2003), alpha2-macroglobulin binding to GRP78 (Mishra et al., 2005) and AMP-activated protein kinase (Bank et al., 2011), while miR-23b and miR-137 regulate expression of PAK2 (Pellegrino et al., 2013; Hao et al., 2015). PAK2 phosphorylation of merlin-Ser518 affects tumor suppressive function (Rong et al, 2004), c-Jun-Thr2/Thr8/Thr89/Thr93/Thr286 alter growth regulatory pathways (Li et al., 2011), Caspase-7-Ser30/Thr173/Ser239 modulates apoptosis (Li et al., 2011), Paxillin-Ser272/Ser274 modulates protease activation (Lee et al., 2013), and STAT5-Ser779 regulates leukemogenesis (Berger et al., 2014). PAK2 signaling also interact with c-Myc pathway; c-Myc-Thr358/Ser373/Thr400 phosphorylation antagonizes the ability of c-Myc to stimulate its target genes while Pak2 knockdown upregulates c-Myc in monocytes (Huang et al., 2004; Zeng et al., 2015). Interestingly, cellular stress such as hyperosmotic shock (Chan et al., 1999), ultraviolet light (Tang et al., 1998), and ionizing radiation (Roig et al., 1999) cleave PAK2 in a caspase-dependent manner, contributing to apoptosis in a variety of cell-types. More recently, insulin signaling has been shown to inhibit PAK2 activity, which leads to an increase in GLUT4-mediated uptake of glucose in neuronal cells (Varshney and Dey, 2016).
14. PAK3 biology
PAK3 is predominantly expressed in the nervous system, and is stimulated by Cool-2, Dbl, Cdc42 (Bagrodia et al., 1998; Bagrodia S et al., 1999), and transcription factor AP1 (Holderness et al., 2013), while GTPase-dependent stimulation of PAK3 is inhibited by p50 (Cool-1) (Bagrodia et al., 1998). PAK3 plays an important role in synapse plasticity (Boda et al., 2004). In this context, PAK3 phosphorylation of AMPA excitatory receptor subunit GluA1, involved in synaptic functions, on Ser863 regulates receptor trafficking in cortical neurons (Hussain et al., 2015). Because AMPAR receptors are important in synaptic plasticity, PAK3-AMPAR signaling has been suggested to be involved in cognition function. A point mutation in PAK3 gene is intimately linked to a form of mental retardation (Allen et al., 1998). Further, a hemizygous PAK3 variant has been identified as a putative disease-linked gene for Cerebral palsy (McMichael et al. 2015). More recently, PAK3 has been shown to functionally interact with Scribble, a signaling protein with a role in the physiology of forgetting in Drosophila, as a component of forgetting signalosome, and participate in the regulation of forgetting (Cervantes-Sandoval et al., 2016). Consistent with a predominantly neuronal functional role, Pak3-deficient mice have a compromised late long term potentiation response and memory retention (Meng et al., 2005). These phenotypes are similar to Pak1-knockout mouse, which is viable but suffers from defects in the immune, neuronal, and metabolic systems (Arias-Romero & Chernoff, 2008). However, deletion of both Pak1 and Pak3 genes in mice leads to stronger defect in memory and learning (Huang et al., 2011).
15. PAK5/7 biology
PAK5, also known as PAK7, is upregulated by Aurora-A (He et al., 2014) and downregulated by miR-129 and miR-186 in certain cancer cells (Zhai J et al., 2015; Zheng J et al., 2015). Studies from Pak5-knockout mice show PAK5 has roles in mobility, learning, and memory related functions (Nekrasova et al., 2008; Furnari et al., 2013). In addition, PAK5 is also believed to be involved in synaptic plasticity because of its coexpression with a psychosis risk gene DISC1 (Morris et al., 2014). Neuronal function of PAK5 may be associated with its ability to phosphorylate two critical regulators of synaptic vesicle trafficking, the Pacsin-1 and Synaptojanin-1 (Strochlic et al., 2012). PAK5-dependent cytoskeletal remodeling also contributes to neurite development and this may involve suppression of Rho activation (Wu and Frost, 2006; Dan et al., 2002). The level of PAK5 is high in many cancers (Li & Kumar, 2016) including liver (Feng et al., 2014), esophageal (He et al., 2016), and gastric cancer (Gu et al., 2013). There are also examples of gain-of-function PAK5 mutations in small number of lung tumors (Fawdar et al., 2013). PAK5 phosphorylation of BAD on Ser112 inhibits apoptosis (Cotteret et al., 2003; Wang et al., 2010). PAK5 phosphorylation of GATA-1 on Ser161 and Ser187 causes epithelial-to-mesenchymal transition in breast cancer cells, phosphorylation of E47 on Ser39 promotes colon cancer metastasis (Li et al., 2015b; Zhu et al., 2016), and phosphorylation of p120-catenin-Ser288 facilitates cytoskeleton reorganization with implications in cancer cell motility and invasion (Wong et al., 2012).
16. PAK6 biology
The PAK6 is unique among PAK family as it is positively regulated by androgen receptor but not Rac or Cdc42 (Lee et al., 2002; Liu et al., 2013). PAK6 is an effector kinase of the Rho family GTPase Chp/RhoV (Shepelev et al., 2012). PAK6 may also be involved in stress-signaling as it is stimulated by p38-MAP kinase (Kaur et al., 2005). Expression of PAK6 is targeted by miR-328 and miR-23a in prostate cancer cells (Liu et al., 2015; Cai et al., 2015), and by miR-429 in colon cancer cells (Tian et al., 2015). PAK6 interacts with the cell junction protein IQGAP1 in a kinase activity-dependent manner and regulates the dissociation of cell-cell junctions in growth factor-stimulated cells (Fram et al., 2014; Morse et al., 2016). PAK6 phosphorylates AR (Ser578 (Liu et al., 2013a), Ser308, Ser346, Ser360, Thr326 and Thr354 (Liu et al., 2013b), MDM2 (Thr158 and Ser186) (Liu et al., 2013a), and LIMK1 (Thr508) (Cai et al., 2015) in prostate cancer cells and is involved in cell growth and migration. PAK6 phosphorylates androgen receptor on Ser-578 and marks its degradation via E3 ligase-Mdm2 pathway (Liu et al., 2013). Interestingly, PAK6 phosphorylation of Mdm2 on Thr-158 and Ser-186 drives AR ubiquitin-mediated degradation.
Genetic studies suggest that Pak6 deletion in mouse leads to a gain in the body weight (Furnari et al., 2013), while combined deletion of PakK5 and Pak6 genes leads to a significant impairment of mobility, memory, and learning as compared to deletion of individual genes (Nekrasova et al, 2008; Furnari et al., 2013). As PAK6 is predominantly expressed in the brain, PAK6 has been recognized as a therapeutic candidate gene for epileptic encephalopathy (Oliver et al., 2016). PAK6 is upregulated in colon cancer (Chen et al., 2015), hepatocellular carcinoma (Chen et al., 2014), and prostate cancer (Kaur et al., 2008; Wen et al., 2009). However, PAK6 downregulation in clear renal cell carcinoma is associated with unfavorable survival of patients (Liu et al., 2014). In human therapeutic treatments, PAK6 upregulation promotes resistance to 5- fluorouracil treatment in colon cancer patients (Chen et al., 2015), and its inhibition enhances the sensitivity of prostate cancer cells to radiation (Zhang et al., 2010) and docetaxel (Wen, et al., 2009).
17. PAKs as modifiers of therapeutic sensitivity
Beginning from the initial demonstration of a critical role for PAK1 in breast cancer, by acting downstream of ErbB2 and promoting aggressiveness, and a widespread observation of PAKs overexpression in human cancer, PAKs have emerged as bona-fide therapeutic targets for cancer. The first generation was PAK inhibitors, which were directed against the ATP-binding domain, and led to the development of ATP-competitive inhibitors, i.e., K252a and CEP-1347 (Kaneko et al., 1997; Nheu et al., 2002; Yi, et al., 2010), or OSU-03012 (Zhu et al., 2004). One such ATP-competitive PAK inhibitor PF-3758309, originally designed as a PAK4 inhibitor, targets both group I and II PAKs as well as other off-target kinases, advanced into Phase I clinical trials (Murray et al., 2010). However, PF-3758309 was eventually withdrawn from clinical use. It may be because of their undesirable pharmacological properties such as drug efflux (Bradshaw-Pierce, et al., 2013). More recently, a derivative of FRAX-597, the FRAX-1306, has been shown to have high selectivity against PAK4, but may also have off-target receptor tyrosine kinases (Ong, et al., 2015). Similarly, another PAK1 inhibitor IPA3 was found to be not suitable for clinical use, but now serves as a good tool to dissect the intricacies of PAK1 signaling in model systems (Singhal et al., 2012). A newer generation of PAK1-specific inhibitors, such as G-5555, are being developed and tested in PK/PD studies (Ndubaku, et al., 2015).
High levels of PAKs cause better cell survival, hence render cancer cells resistance to chemotherapeutic drugs. For example, overexpression of PAK1 confers chemo-resistance to PI-3K inhibitors in colon cancer (Qing et al., 2012) and B-cell lymphomas (Walsh et al., 2013), and resistance to MET inhibitors in pancreatic cancer cells (Zhou et al., 2014). Similar to PAK1, upregulation of PAK4 promotes insensitivity to chemotherapies such as doxorubicin or paclitaxel (Park et al., 2013), cisplatin (Shu et al., 2015; Fu et al., 2014) in different cancer types, and gemcitabine in cancer cells (Moon et al., 2015). Likewise, PAK5 overexpression confers resistance to paclitaxel (Li et al., 2013) and cisplatin in cancer cells (He et al., 2014; Zhang et al., 2015). PAK6 overexpression promotes therapeutic resistance to 5-fluorouracil in some patients (Chen et al., 2015). These representative examples highlight the potential use of PAK-inhibitors in cancer chemotherapies to reverse resistance. Collectively, these observations catalyzed the development of PAK-inhibitor drugs, and advanced structure-functional studies to refine effectiveness and selectivity. Nevertheless, one must take a cautious approach because PAK signaling has profound effect in cells; ErbB3, an upstream activator of PAKs, has vital roles in organs such as the hear. Hence, potential side effects may be inherent of targeting PAKs for therapy. With ongoing intense pharmacokinetics and toxicology studies yielding promising results in animal models, the authors hope that these issues will be resolved.
18. Future of PAKs
Since the initial finding of PAKs as downstream effectors of small GTPases over twenty five years ago, the field of PAK research has made significant advances in both mechanistic and functional studies. More of these studies in the future are likely to shape a better understanding of PAKs. Our appreciation for the role of PAKs in vast number of human diseases such as cancer (Kumar and Li, 2016), inflammation (Dammann, Khare, Gasche, 2014), neurological disorders (Ma et al., 2012), cardiovascular disorders (Ke et al., 2014), and viral diseases (Van den Broeke et al., 2010) is likely to grow. The breadth and scope of developing and improving next generation PAK modifiers will likely gain momentum and may extend to many diseases. The authors see further insights into co-expressing PAKs in tumor specimens, an atlas of co-regulated PAK-intracellular signaling networks within the context of selective or shared effector substrates, aligning specific arms of the PAKs cytoplasmic signaling with the nuclear arms, discovering new PAK nuclear functions, and further refinement of PAK inhibitors. It will be important to learn more about the functions of spliced PAK transcripts, if any. Given that there is an emerging importance for non-coding RNAs in cancer, the authors hypothesize that PAK’s non-coding RNA transcripts will influence the biology of PAK proteins.
Highlights.
PAKs act as modifiers of the cell surface and intracellular signaling cascades.
PAKs function both in the cytoplasm and nucleus and regulation of gene expression.
Relevance of PAKs range from cellular homeostasis to pathobiology of human disease.
PAKs are dysregulated in cancer, and neurological, cardiac and other disorders.
Modifying PAK activity has emerged as a therapeutic approach for many disorders.
Acknowledgments
This review and the corresponding Gene Wiki article are written as part of the Gene Wiki Review series--a series resulting from a collaboration between the journal GENE and the Gene Wiki Initiative. The Gene Wiki Initiative is supported by National Institutes of Health (GM089820). Additional support for Gene Wiki Reviews is provided by Elsevier, the publisher of GENE. The authors are sorry for not citing the work of many of our colleagues due to space limitations. The authors thank their numerous trainees for their work and contributions to the authors’ laboratories over the years, and Dr. Xiaodong Li for his outstanding effort to prepare the figures. Over the years, PAK signaling studies in Rakesh Kumar laboratory have been supported by the National Institutes of Health grant CA090970, and Research in Feng Li laboratory is supported by the National Natural Science Foundation of China, Number 31371424 and 31571457. Rahul Sanawar is supported by a Research Fellowship from the University Grants Commission Award number 23/06/2013(i)EU-V. The corresponding Gene Wiki entry for this review can be found here.
https://en.wikipedia.org/wiki/PAK1
https://en.wikipedia.org/wiki/PAK2
https://en.wikipedia.org/wiki/PAK3
https://en.wikipedia.org/wiki/PAK4
Abbreviations
- PAK
p21-activated kinase
- AID
auto-inhibitory domain
- PBD
p21-binding domain
Footnotes
Conflict of interest statement: The authors have no conflicts of interest to declare.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Abo A, et al. PAK4, a novel effector for Cdc42Hs, is implicated in the reorganization of the actin cytoskeleton and in the formation of filopodia. EMBO J. 1998;17:6527–6540. doi: 10.1093/emboj/17.22.6527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Acconcia F, et al. Phosphorylation-dependent regulation of nuclear localization and functions of integrin-linked kinase. Proc Natl Acad Sci U S A. 2007;104:6782–6787. doi: 10.1073/pnas.0701999104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adam L, et al. Heregulin regulates cytoskeletal reorganization and cell migration through the p21-activated kinase-1 via phosphatidylinositol-3 kinase. J Biol Chem. 1998;273:28238–28246. doi: 10.1074/jbc.273.43.28238. [DOI] [PubMed] [Google Scholar]
- Advani SJ, et al. Kinase-independent role for CRAF-driving tumour radioresistance via CHK2. Nat Commun. 2015;6:8154. doi: 10.1038/ncomms9154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ahmed T, et al. A PAK4-LIMK1 pathway drives prostate cancer cell migration downstream of HGF. Cell Signal. 2008;20:1320–1328. doi: 10.1016/j.cellsig.2008.02.021. [DOI] [PubMed] [Google Scholar]
- Ahn HK, et al. P21-activated kinase 4 overexpression in metastatic gastric cancer patients. Transl Oncol. 2011;4:345–349. doi: 10.1593/tlo.11145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alavi A, et al. Role of Raf in vascular protection from distinct apoptotic stimuli. Science. 2003;301:94–96. doi: 10.1126/science.1082015. [DOI] [PubMed] [Google Scholar]
- Allen, et al. PAK3 mutation in nonsyndromic X-linked mental retardation. Nat Genet. 1998;20:25–30. doi: 10.1038/1675. [DOI] [PubMed] [Google Scholar]
- Arias-Romero LE, Chernoff J. A tale of two PAKs. Biology of the Cell. 2008;100:97–108. doi: 10.1042/BC20070109. [DOI] [PubMed] [Google Scholar]
- Arias-Romero LE, et al. Pak1 kinase links ErbB2 to β-catenin in transformation of breast epithelial cells. Cancer Res. 2013;73:3671–3682. doi: 10.1158/0008-5472.CAN-12-4453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arsenault D, et al. PAK inactivation impairs social recognition in 3xTg-AD Mice without increasing brain deposition of tau and Aβ. J Neurosci. 2013;33:10729–10740. doi: 10.1523/JNEUROSCI.1501-13.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Asrar S, Meng Y, Zhou Z, Todorovski Z, Huang WW, Jia Z. Regulation of hippocampal long-term potentiation by p21-activated protein kinase 1 (PAK1) Neuropharmacology. 2009;56:73–80. doi: 10.1016/j.neuropharm.2008.06.055. [DOI] [PubMed] [Google Scholar]
- Bagheri-Yarmand R, et al. LIM kinase 1 increases tumor metastasis of human breast cancer cells via regulation of the urokinase-type plasminogen activator system. Int J Cancer. 2006;118:2703–2710. doi: 10.1002/ijc.21650. [DOI] [PubMed] [Google Scholar]
- Bagheri-Yarmand R, et al. Vascular endothelial growth factor up-regulation via p21-activated kinase-1 signaling regulates heregulin-beta1-mediated angiogenesis. J Biol Chem. 2000;275:39451–39457. doi: 10.1074/jbc.M006150200. [DOI] [PubMed] [Google Scholar]
- Bagrodia S, et al. Identification of a mouse p21Cdc42/Rac activated kinase. J Biol Chem. 1995;270:22731–22737. doi: 10.1074/jbc.270.39.22731. [DOI] [PubMed] [Google Scholar]
- Bagrodia, et al. A novel regulator of p21-activated kinases. J Biol Chem. 1998;273:23633–23636. doi: 10.1074/jbc.273.37.23633. [DOI] [PubMed] [Google Scholar]
- Bagrodia S, et al. A tyrosine-phosphorylated protein that binds to an important regulatory region on the cool family of p21-activated kinase-binding proteins. J Biol Chem. 1999;274:22393–22400. doi: 10.1074/jbc.274.32.22393. [DOI] [PubMed] [Google Scholar]
- Begum A, et al. Identification of PAK4 as a putative target gene for amplification within 19q13.12-q13.2 in oral squamous-cell carcinoma. Cancer Sci. 2009;100:1908–1916. doi: 10.1111/j.1349-7006.2009.01252.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baker NM, et al. Molecular pathways: targeting RAC-p21-activated serine-threonine kinase signaling in RAS-driven cancers. Clin Cancer Res. 2014;20:4740–4746. doi: 10.1158/1078-0432.CCR-13-1727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Banerjee M, et al. Pak1 phosphorylation on t212 affects microtubules in cells undergoing mitosis. Curr Biol. 2002;12:1233–1239. doi: 10.1016/s0960-9822(02)00956-9. [DOI] [PubMed] [Google Scholar]
- Barnes CJ, et al. Functional inactivation of a transcriptional corepressor by a signaling kinase. Nat Struct Biol. 2003;10:622–628. doi: 10.1038/nsb957. [DOI] [PubMed] [Google Scholar]
- Barros P, et al. Rac1 signalling modulates a STAT5/BCL-6 transcriptional switch on cell-cycle-associated target gene promoters. Nucleic Acids Res. 2012;40:7776–7787. doi: 10.1093/nar/gks571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baskaran Y, et al. Group I and II mammalian PAKs have different modes of activation by Cdc42. EMBO Rep. 2012;13:653–659. doi: 10.1038/embor.2012.75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bradshaw-Pierce, et al. Tumor P-Glycoprotein Correlates with Efficacy of PF-3758309 in in vitro and in vivo Models of Colorectal Cancer. Frontiers Pharmacol. 2013;4:22. doi: 10.3389/fphar.2013.00022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boda, et al. The mental retardation protein PAK3 contributes to synapse formation and plasticity in hippocampus. J Neurosci. 2004;24:10816–10825. doi: 10.1523/JNEUROSCI.2931-04.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bokoch GM, et al. A GTPase-independent mechanism of p21-activated kinase activation. Regulation by sphingosine and other biologically active lipids. J Biol Chem. 1998;273:8137–8144. doi: 10.1074/jbc.273.14.8137. [DOI] [PubMed] [Google Scholar]
- Bokoch GM. Biology of the p21-activated kinases. Annu Rev Biochem. 2003;72:743–781. doi: 10.1146/annurev.biochem.72.121801.161742. [DOI] [PubMed] [Google Scholar]
- Bompard G, et al. P21-activated kinase 4 (PAK4) is required for metaphase spindle positioning and anchoring. Oncogene. 2013;32:910–919. doi: 10.1038/onc.2012.98. [DOI] [PubMed] [Google Scholar]
- Bostner J, et al. Amplification of CCND1 and PAK1 as predictors of recurrence and tamoxifen resistance in postmenopausal breast cancer. Oncogene. 2007;26:6997–7005. doi: 10.1038/sj.onc.1210506. [DOI] [PubMed] [Google Scholar]
- Braun A, et al. Rac1 and Aurora A regulate MCAK to polarize microtubule growth in migrating endothelial cells. J Cell Biol. 2014;206:97–112. doi: 10.1083/jcb.201401063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buchwald G, et al. Conformational switch and role of phosphorylation in PAK activation. Mol Cell Biol. 2001;21:5179–5189. doi: 10.1128/MCB.21.15.5179-5189.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buday L, Downward J. Roles of cortactin in tumor pathogenesis. Biochim Biophys Acta. 2007;1775:263–273. doi: 10.1016/j.bbcan.2006.12.002. [DOI] [PubMed] [Google Scholar]
- Cai S, et al. Downregulation of microRNA-23a suppresses prostate cancer metastasis by targeting the PAK6-LIMK1 signaling pathway. Oncotarget. 2015;6:3904–3917. doi: 10.18632/oncotarget.2880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Callow MG, et al. Requirement for PAK4 in the anchorage-independent growth of human cancer cell lines. J Biol Chem. 2002;277:550–558. doi: 10.1074/jbc.M105732200. [DOI] [PubMed] [Google Scholar]
- Callow MG, et al. PAK4 mediates morphological changes through the regulation of GEF-H1. J Cell Sci. 2005;118:1861–1872. doi: 10.1242/jcs.02313. [DOI] [PubMed] [Google Scholar]
- Chantaravisoot N, et al. Significance of filamin A in mTORC2 function in glioblastoma. Mol Cancer. 2015;14:127. doi: 10.1186/s12943-015-0396-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan PM, Manser E. PAKs in human disease. Prog Mol Biol Transl Sci. 2012;106:171–187. doi: 10.1016/B978-0-12-396456-4.00011-0. [DOI] [PubMed] [Google Scholar]
- Chen H, et al. Expression and prognostic significance of p21-activated kinase 6 in hepatocellular carcinoma. Surg Res. 2014;189:81–88. doi: 10.1016/j.jss.2014.01.049. [DOI] [PubMed] [Google Scholar]
- Chen J, et al. PAK6 increase chemoresistance and is a prognostic marker for stage II and III colon cancer patients undergoing 5-FU based chemotherapy. Oncotarget. 2015;6:355–367. doi: 10.18632/oncotarget.2803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen S, et al. Copy number alterations in pancreatic cancer identify recurrent PAK4 amplification. Cancer Biol Ther. 2008;7:1793–802. doi: 10.4161/cbt.7.11.6840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cervantes-Sandoval I, et al. Scribble Scaffolds a Signalosome for Active Forgetting. Neuron. 2016;90:1230–1242. doi: 10.1016/j.neuron.2016.05.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Civiero L, et al. Leucine-rich repeat kinase 2 interacts with p21-activated kinase 6 to control neurite complexity in mammalian brain. J Neurochem. 2015;135:1242–1256. doi: 10.1111/jnc.13369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cotteret S, et al. p21-Activated kinase 5 (Pak5) localizes to mitochondria and inhibits apoptosis by phosphorylating BAD. Mol Cell Biol. 2003;23:5526–5539. doi: 10.1128/MCB.23.16.5526-5539.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dan C, et al. Cytoskeletal changes regulated by the PAK4 serine/threonine kinase are mediated by LIM kinase 1 and cofilin. J Biol Chem. 2001;276:32115–32121. doi: 10.1074/jbc.M100871200. [DOI] [PubMed] [Google Scholar]
- Dan C, et al. PAK5, a new brain-specific kinase, promotes neurite outgrowth in N1E-115 cells. Mol Cell Biol. 2002;22:567–577. doi: 10.1128/MCB.22.2.567-577.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dart AE, et al. PAK4 promotes kinase-independent stabilization of RhoU to modulate cell adhesion. J Cell Biol. 2015;211:863–879. doi: 10.1083/jcb.201501072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dammann K, Khare V, Gasche C. Tracing PAKs from GI inflammation to cancer. Gut. 2014;63:1173–1184. doi: 10.1136/gutjnl-2014-306768. [DOI] [PubMed] [Google Scholar]
- Davis RT3rd, et al. Knockout of p21-activated kinase-1 attenuates exercise-induced cardiac remodelling through altered calcineurin signalling. Cardiovasc Res. 2015;108:335–347. doi: 10.1093/cvr/cvv234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis SJ, et al. Functional analysis of genes in regions commonly amplified in high-grade serous and endometrioid ovarian cancer. Clin Cancer Res. 2013;19:1411–1421. doi: 10.1158/1078-0432.CCR-12-3433. [DOI] [PubMed] [Google Scholar]
- de la TL, et al. A FOXO-Pak1 transcriptional pathway controls neuronal polarity. Genes Dev. 2010;24:799–813. doi: 10.1101/gad.1880510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding Q, et al. Erk associates with and primes GSK-3beta for its inactivation resulting in upregulation of beta-catenin. Mol Cell. 2005;19:159–170. doi: 10.1016/j.molcel.2005.06.009. [DOI] [PubMed] [Google Scholar]
- Ding Y, et al. Inhibition of Nischarin Expression Promotes Neurite Outgrowth through Regulation of PAK Activity. PLoS One. 2015;10(12):e0144948. doi: 10.1371/journal.pone.0144948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edwards DC, et al. Activation of LIM-kinase by Pak1 couples Rac/Cdc42 GTPase signalling to actin cytoskeletal dynamics. Nat Cell Biol. 1999;1:253–259. doi: 10.1038/12963. [DOI] [PubMed] [Google Scholar]
- Fawdar S, et al. Targeted genetic dependency screen facilitates identification of actionable mutations in FGFR4, MAP3K9, and PAK5 in lung cancer. Proc Natl Acad Sci U S A. 2013;110:12426–12431. doi: 10.1073/pnas.1305207110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fang ZP, et al. P21-activated kinase 5 plays essential roles in the proliferation and tumorigenicity of human hepatocellular carcinoma. Acta Pharmacol Sin. 2014;35:82–88. doi: 10.1038/aps.2013.31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fram, et al. A PAK6-IQGAP1 complex promotes disassembly of cell-cell adhesions. Cell Mol Life Sci. 2014;71:2759–2773. doi: 10.1007/s00018-013-1528-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fryer BH, et al. cGMP-dependent protein kinase phosphorylates p21-activated kinase (Pak) 1, inhibiting Pak/Nck binding and stimulating Pak/vasodilator-stimulated phosphoprotein association. J Biol Chem. 2006;281:11487–11495. doi: 10.1074/jbc.M600279200. [DOI] [PubMed] [Google Scholar]
- Fu X, et al. PAK4 confers cisplatin resistance in gastric cancer cells via PI3K/Akt- and MEK/Erk-dependent pathways. Biosci Rep. 2014;34(2):e00094. doi: 10.1042/BSR20130102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fuchsova B, Alvarez Juliá A, Rizavi HS, Frasch AC, Pandey GN. Expression of p21-activated kinases 1 and 3 is altered in the brain of subjects with depression. Neuroscience. 2016;333:331–344. doi: 10.1016/j.neuroscience.2016.07.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Furnari MA, et al. Functional deficits in PAK5, PAK6 and PAK5/PAK6 knockout mice. PLoS One. 2013;8:e61321. doi: 10.1371/journal.pone.0061321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galisteo ML, et al. The adaptor protein Nck links receptor tyrosine kinases with the serine-threonine kinase Pak1. J Biol Chem. 1996;271:20997–21000. doi: 10.1074/jbc.271.35.20997. [DOI] [PubMed] [Google Scholar]
- Greenman C, et al. Patterns of somatic mutation in human cancer genomes. Nature. 2007;446:153–158. doi: 10.1038/nature05610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu J, et al. A role for p21-activated kinase 7 in the development of gastric cancer. FEBS J. 2013;280:46–55. doi: 10.1111/febs.12048. [DOI] [PubMed] [Google Scholar]
- Guo Q, et al. PAK4 kinase-mediated SCG10 phosphorylation involved in gastric cancer metastasis. Oncogene. 2014;33:3277–3287. doi: 10.1038/onc.2013.296. [DOI] [PubMed] [Google Scholar]
- Ha BH, et al. Type II p21-activated kinases (PAKs) are regulated by an autoinhibitory pseudosubstrate. Proc Natl Acad Sci USA. 2012;109:16107–16112. doi: 10.1073/pnas.1214447109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hammer A, Diakonova M. Tyrosyl phosphorylated serine-threonine kinase PAK1 is a novel regulator of prolactin-dependent breast cancer cell motility and invasion. Adv Exp Med Biol. 2015;846:97–137. doi: 10.1007/978-3-319-12114-7_5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayashi K, Ohshima T, Hashimoto M, Mikoshiba K. Pak1 regulates dendritic branching and spine formation. Dev Neurobiol. 2007;67:655–669. doi: 10.1002/dneu.20363. [DOI] [PubMed] [Google Scholar]
- He S, et al. P21-activated kinase 7 mediates cisplatin-resistance of esophageal squamous carcinoma cells with Aurora-A overexpression. PLoS One. 2014;9:e113989. doi: 10.1371/journal.pone.0113989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He S, et al. Over expression of p21-activated kinase 7 associates with lymph node metastasis in esophageal squamous cell cancers. Cancer Biomark. 2016;16:203–209. doi: 10.3233/CBM-150557. [DOI] [PubMed] [Google Scholar]
- Hofmann C, Shepelev M, Chernoff J. The genetics of Pak. J Cell Sci. 2004;117:4343–4354. doi: 10.1242/jcs.01392. [DOI] [PubMed] [Google Scholar]
- Holm C, et al. Association between Pak1 expression and subcellular localization and tamoxifen resistance in breast cancer patients. J Natl Cancer Inst. 2006;98:671–680. doi: 10.1093/jnci/djj185. [DOI] [PubMed] [Google Scholar]
- Hou J, et al. Identification of miRNomes in human liver and hepatocellular carcinoma reveals miR-199a/b-3p as therapeutic target for hepatocellular carcinoma. Cancer Cell. 2011;19:232–243. doi: 10.1016/j.ccr.2011.01.001. [DOI] [PubMed] [Google Scholar]
- Howe AK, Juliano RL. Regulation of anchorage-dependent signal transduction by protein kinase A and p21-activated kinase. Nat Cell Biol. 2000;2:593–600. doi: 10.1038/35023536. [DOI] [PubMed] [Google Scholar]
- Hu B, et al. Tuning PAK Activity to Rescue Abnormal Myelin Permeability in HNPP. PLoS Genet. 2016 Sep 1;12(9):e1006290. doi: 10.1371/journal.pgen.1006290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang W, et al. p21-Activated kinases 1 and 3 control brain size through coordinating neuronal complexity and synaptic properties. Mol Cell Biol. 2011;31:388–403. doi: 10.1128/MCB.00969-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hussain NK, et al. Regulation of AMPA receptor subunit GluA1 surface expression by PAK3 phosphorylation. Proc Natl Acad Sci USA. 2015;12:E5883–5890. doi: 10.1073/pnas.1518382112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huynh N, et al. Up-regulation of Stem Cell Markers by P21-Activated Kinase 1 Contributes to 5-Fluorouracil Resistance of Colorectal Cancer. Cancer Biol Ther. 2016 Jun;3:0. doi: 10.1080/15384047.2016.1195045. Epub ahead of print. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jha RK, Strauss CE. 3D structure analysis of PAKs: A clue to the rational design for affinity reagents and blockers. Cell Logist. 2012;2:69–77. doi: 10.4161/cl.21883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang YY, et al. Targeting super-enhancer-associated oncogenes in oesophageal squamous cell carcinoma. Gut. 2016 doi: 10.1136/gutjnl-2016-311818. pii: gutjnl-2016–311818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaneko M, et al. Neurotrophic 3,9-bis[(alkylthio)methyl]-and-bis(alkoxymethyl)-K-252a derivatives. J Med Chem. 1997;40:1863–1869. doi: 10.1021/jm970031d. [DOI] [PubMed] [Google Scholar]
- Kang B, et al. Phosphorylation of H4 Ser 47 promotes HIRA-mediated nucleosome assembly. Genes Dev. 2011;25:1359–1364. doi: 10.1101/gad.2055511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaur R, et al. Activation of p21-activated kinase 6 by MAP kinase kinase 6 and p38 MAP kinase. J Biol Chem. 2005;280:3323–3330. doi: 10.1074/jbc.M406701200. [DOI] [PubMed] [Google Scholar]
- Kaur R, et al. Increased PAK6 expression in prostate cancer and identification of PAK6 associated proteins. Prostate. 2008;68:1510–1516. doi: 10.1002/pros.20787. [DOI] [PubMed] [Google Scholar]
- Ke Y, et al. PAK1 is a novel cardiac protective signaling molecule. Front Med. 2014;8:399–403. doi: 10.1007/s11684-014-0380-9. [DOI] [PubMed] [Google Scholar]
- Kelly ML, Chernoff J. Mouse models of PAK function. Cell Logist. 2012;2:84–88. doi: 10.4161/cl.21381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kesanakurti D, et al. Functional cooperativity by direct interaction between PAK4 and MMP-2 in the regulation of anoikis resistance, migration and invasion in glioma. Cell Death Dis. 2012;3:e445. doi: 10.1038/cddis.2012.182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimmelman AC, et al. Genomic alterations link Rho family of GTPases to the highly invasive phenotype of pancreas cancer. Proc Natl Acad Sci U S A. 2008;105:19372–19377. doi: 10.1073/pnas.0809966105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- King CC, et al. p21-activated kinase (PAK1) is phosphorylated and activated by 3-phosphoinositide-dependent kinase-1 (PDK1) J Biol Chem. 2000;275:41201–41209. doi: 10.1074/jbc.M006553200. [DOI] [PubMed] [Google Scholar]
- Kiosses WB, et al. A dominant-negative p65 PAK peptide inhibits angiogenesis. Circ Res. 2002;90:697–702. doi: 10.1161/01.res.0000014227.76102.5d. [DOI] [PubMed] [Google Scholar]
- Koh W, et al. Formation of endothelial lumens requires a coordinated PKCepsilon-, Src-, Pak- and Raf-kinase-dependent signaling cascade downstream of Cdc42 activation. J Cell Sci. 2009;122:1812–1822. doi: 10.1242/jcs.045799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knaus UG, et al. Regulation of human leukocyte p21-activated kinases through G protein--coupled receptors. Science. 1995;269:221–223. doi: 10.1126/science.7618083. [DOI] [PubMed] [Google Scholar]
- Kumar R, et al. p21-activated kinases in cancer. Nat Rev Cancer. 2006;6:459–571. doi: 10.1038/nrc1892. [DOI] [PubMed] [Google Scholar]
- Lee SR, et al. AR and ER interaction with a p21-activated kinase (PAK6) Mol Endocrinol. 2002;16:85–99. doi: 10.1210/mend.16.1.0753. [DOI] [PubMed] [Google Scholar]
- Lei M, et al. Structure of PAK1 in an autoinhibited conformation reveals a multistage activation switch. Cell. 2000;102:387–397. doi: 10.1016/s0092-8674(00)00043-x. [DOI] [PubMed] [Google Scholar]
- Li D, et al. The overexpression of P21-activated kinase 5 (PAK5) promotes paclitaxel-chemoresistance of epithelial ovarian cancer. Mol Cell Biochem. 2013;383:191–199. doi: 10.1007/s11010-013-1767-7. [DOI] [PubMed] [Google Scholar]
- Li D, et al. Activated Pak4 expression correlates with poor prognosis in human gastric cancer patients. Tumour Biol. 2015;36:9431–9436. doi: 10.1007/s13277-015-3368-4. [DOI] [PubMed] [Google Scholar]
- Li DQ, et al. MORC2 signaling integrates phosphorylation-dependent, ATPase-coupled chromatin remodeling during the DNA damage response. Cell Rep. 2012;2:1657–1669. doi: 10.1016/j.celrep.2012.11.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li F, et al. p21-activated kinase 1 interacts with and phosphorylates histone H3 in breast cancer cells. EMBO Rep. 2002;3:767–773. doi: 10.1093/embo-reports/kvf157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li S, et al. The neural cell adhesion molecule (NCAM) associates with and signals through p21-activated kinase 1 (Pak1) J Neurosci. 2013;33:790–803. doi: 10.1523/JNEUROSCI.1238-12.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Q, et al. miR-224 promotion of cell migration and invasion by targeting Homeobox D 10 gene in human hepatocellular carcinoma. J Gastroenterol Hepatol. 2014;29:835–842. doi: 10.1111/jgh.12429. [DOI] [PubMed] [Google Scholar]
- Li X, et al. DGCR6L, a novel PAK4 interaction protein, regulates PAK4-mediated migration of human gastric cancer cell via LIMK1. Int J Biochem Cell Biol. 2010a;42:70–79. doi: 10.1016/j.biocel.2009.09.008. [DOI] [PubMed] [Google Scholar]
- Li Y, et al. GATA1 induces epithelial-mesenchymal transition in breast cancer cells through PAK5 oncogenic signaling. Oncotarget. 2015b;6:4345–4356. doi: 10.18632/oncotarget.2999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y, et al. Nucleo-cytoplasmic shuttling of PAK4 modulates beta-catenin intracellular translocation and signaling. Biochim Biophys Acta. 2012;1823:465–475. doi: 10.1016/j.bbamcr.2011.11.013. [DOI] [PubMed] [Google Scholar]
- Li Y, et al. P21-activated kinase 4 regulates the cyclin-dependent kinase inhibitor p57(kip2) in human breast cancer. Anat Rec (Hoboken) 2013;296:1561–1567. doi: 10.1002/ar.22754. [DOI] [PubMed] [Google Scholar]
- Liu J, et al. N-acetylglucosaminyltransferase V confers hepatoma cells with resistance to anoikis through EGFR/PAK1 activation. Glycobiology. 2013a;23:1097–1109. doi: 10.1093/glycob/cwt049. [DOI] [PubMed] [Google Scholar]
- Li Y, et al. GATA1 induces epithelial-mesenchymal transition in breast cancer cells through PAK5 oncogenic signaling. Oncotarget. 2015;6:4345–4356. doi: 10.18632/oncotarget.2999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Z, et al. p21-activated kinase 4 phosphorylation of integrin beta5 Ser-759 and Ser-762 regulates cell migration. J Biol Chem. 2010b;285:23699–23710. doi: 10.1074/jbc.M110.123497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu C, et al. MicroRNA 328 directly targets p21 activated protein kinase 6 inhibiting prostate cancer proliferation and enhancing docetaxel sensitivity. Mol Med Rep. 2015;12:7389–7395. doi: 10.3892/mmr.2015.4390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu T, et al. p21-Activated kinase 6 (PAK6) inhibits prostate cancer growth via phosphorylation of androgen receptor and tumorigenic E3 ligase murine double minute-2 (Mdm2) J Biol Chem. 2013a;288:3359–3369. doi: 10.1074/jbc.M112.384289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu X, et al. Direct interaction between AR and PAK6 in androgen-stimulated PAK6 activation. PLoS One. 2013b;8:e77367. doi: 10.1371/journal.pone.0077367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y, et al. The pak4 protein kinase plays a key role in cell survival and tumorigenesis in athymic mice. Mol Cancer Res. 2008;6:1215–1224. doi: 10.1158/1541-7786.MCR-08-0087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu W, et al. Pak1 as a novel therapeutic target for antihypertrophic treatment in the heart. Circulation. 2011;124:2702–2715. doi: 10.1161/CIRCULATIONAHA.111.048785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu W, et al. Prognostic significance of p21-activated kinase 6 expression in patients with clear cell renal cell carcinoma. Ann Surg Oncol Suppl. 2014;4:S575–583. doi: 10.1245/s10434-014-3680-z. [DOI] [PubMed] [Google Scholar]
- Liu X, et al. Direct interaction between AR and PAK6 in androgen-stimulated PAK6 activation. PLoS One. 2013;8:e77367. doi: 10.1371/journal.pone.0077367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma QL, Yang F, Frautschy SA, Cole GM. PAK in Alzheimer disease, Huntington disease and X-linked mental retardation. Cell Logist. 2012;2:117–125. doi: 10.4161/cl.21602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mahlamaki EH, et al. High-resolution genomic and expression profiling reveals 105 putative amplification target genes in pancreatic cancer. Neoplasia. 2004;6:432–439. doi: 10.1593/neo.04130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manser E, et al. A brain serine/threonine protein kinase activated by Cdc42 and Rac1. Nature. 1994;367:40–46. doi: 10.1038/367040a0. [DOI] [PubMed] [Google Scholar]
- Maroto B, et al. P21-activated kinase is required for mitotic progression and regulates Plk1. Oncogene. 2008;27:4900–4908. doi: 10.1038/onc.2008.131. [DOI] [PubMed] [Google Scholar]
- Martin GA, et al. A novel serine kinase activated by rac1/CDC42Hs-dependent autophosphorylation is related to PAK65 and STE20. EMBO J. 1995;14:1970–1978. doi: 10.1002/j.1460-2075.1995.tb07189.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Master Z, et al. Dok-R plays a pivotal role in angiopoietin-1-dependent cell migration through recruitment and activation of Pak. EMBO J. 2001;20:5919–5928. doi: 10.1093/emboj/20.21.5919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McPhie DL, et al. DNA synthesis and neuronal apoptosis caused by familial Alzheimer disease mutants of the amyloid precursor protein are mediated by the p21 activated kinase PAK3. J Neurosci. 2003;23:6914–6927. doi: 10.1523/JNEUROSCI.23-17-06914.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Menzel N, et al. The Drosophila p21-activated kinase Mbt modulates DE-cadherin-mediated cell adhesion by phosphorylation of Armadillo. Biochem J. 2008;416:231–341. doi: 10.1042/BJ20080465. [DOI] [PubMed] [Google Scholar]
- Mertins P, et al. Proteogenomics connects somatic mutations to signalling in breast cancer. Nature. 2016;534:55–62. doi: 10.1038/nature18003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McMichael G, et al. Whole-exome sequencing points to considerable genetic heterogeneity of cerebral palsy. Mol Psychiatry. 2015;20:176–182. doi: 10.1038/mp.2014.189. [DOI] [PubMed] [Google Scholar]
- Meng J, et al. Abnormal long-lasting synaptic plasticity and cognition in mice lacking the mental retardation gene Pak3. J Neurosci. 2005;25:6641–6650. doi: 10.1523/JNEUROSCI.0028-05.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meng Q, et al. Signaling-dependent and coordinated regulation of transcription, splicing, and translation resides in a single coregulator, PCBP1. Proc Natl Acad Sci U S A. 2007;104:5866–5871. doi: 10.1073/pnas.0701065104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Molli PR, et al. PAK signaling in oncogenesis. Oncogene. 2009;28:2545–2555. doi: 10.1038/onc.2009.119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Molli PR, et al. Arpc1b, a centrosomal protein, is both an activator and substrate of Aurora A. J Cell Biol. 2010;190:101–114. doi: 10.1083/jcb.200908050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morris DW, et al. An inherited duplication at the gene p21 Protein-Activated Kinase 7 (PAK7) is a risk factor for psychosis. Hum Mol Genet. 2014;23:3316–3326. doi: 10.1093/hmg/ddu025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morse EM, et al. PAK6 targets to cell-cell adhesions through its N-terminus in a Cdc42-dependent manner to drive epithelial colony escape. J Cell Sci. 2016;129:380–393. doi: 10.1242/jcs.177493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moon SU, et al. p21-Activated Kinase 4 (PAK4) as a Predictive Marker of Gemcitabine Sensitivity in Pancreatic Cancer Cell Lines. Cancer Res Treat. 2015;47:501–518. doi: 10.4143/crt.2014.054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moshfegh Y, et al. A Trio-Rac1-Pak1 signalling axis drives invadopodia disassembly. Nat Cell Biol. 2014;16:574–586. doi: 10.1038/ncb2972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murray, et al. Small-molecule p21-activated kinase inhibitor PF-3758309 is a potent inhibitor of oncogenic signaling and tumor growth. Proc Natl Acad Sci USA. 2010;107:9446–9451. doi: 10.1073/pnas.0911863107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ndubaku, et al. Design of Selective PAK1 Inhibitor G-5555: Improving Properties by Employing an Unorthodox Low-pK a Polar Moiety. ACS Med Chem Lett. 2015;6:1241–1246. doi: 10.1021/acsmedchemlett.5b00398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nekrasova T, Minden A. Role for p21-activated kinase PAK4 in development of the mammalian heart. Transgenic Res. 2012;21:797–811. doi: 10.1007/s11248-011-9578-7. [DOI] [PubMed] [Google Scholar]
- Nekrasova T. Targeted disruption of the Pak5 and Pak6 genes in mice leads to deficits in learning and locomotion. Dev Biol. 2008;322:95–108. doi: 10.1016/j.ydbio.2008.07.006. [DOI] [PubMed] [Google Scholar]
- Nekrasova T, Minden A. PAK4 is required for regulation of the cell-cycle regulatory protein p21, and for control of cell-cycle progression. J Cell Biochem. 2011;112:1795–1806. doi: 10.1002/jcb.23092. [DOI] [PubMed] [Google Scholar]
- Nguyen TV, et al. Signal transduction in Alzheimer disease: p21-activated kinase signaling requires C-terminal cleavage of APP at Asp664. J Neurochem. 2008;104:1065–1080. doi: 10.1111/j.1471-4159.2007.05031.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nheu TV, et al. The K252a derivatives, inhibitors for the PAK/MLK kinase family selectively block the growth of RAS transformants. Cancer J. 2002;8:328–836. doi: 10.1097/00130404-200207000-00009. [DOI] [PubMed] [Google Scholar]
- Oliver KL, et al. In silico prioritization based on coexpression can aid epileptic encephalopathy gene discovery. Neurol Genet. 2016;2(1):e51. doi: 10.1212/NXG.0000000000000051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ong, et al. Small molecule inhibition of group I p21-activated kinases in breast cancer induces apoptosis and potentiates the activity of microtubule stabilizing agents. Breast Cancer Res. 2015;17:59. doi: 10.1186/s13058-015-0564-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paliouras GN, et al. Pak4, a novel Gab1 binding partner, modulates cell migration and invasion by the Met receptor. Mol Cell Biol. 2009;29:3018–3032. doi: 10.1128/MCB.01286-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan X, et al. PAK1 regulates cortical development via promoting neuronal migration and progenitor cell proliferation. Mol Brain. 2015;5(8):36. doi: 10.1186/s13041-015-0124-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pakala SB, et al. Signaling-dependent phosphorylation of mitotic centromere-associated kinesin regulates microtubule depolymerization and its centrosomal localization. J Biol Chem. 2012;287:40560–40569. doi: 10.1074/jbc.M112.399576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park MH, et al. Phosphorylation of β-catenin at serine 663 regulates its transcriptional activity. Biochem Biophys Res Commun. 2012;419:543–549. doi: 10.1016/j.bbrc.2012.02.056. [DOI] [PubMed] [Google Scholar]
- Park MH, et al. p21-Activated kinase 4 promotes prostate cancer progression through CREB. Oncogene. 2013;32:2475–2482. doi: 10.1038/onc.2012.255. [DOI] [PubMed] [Google Scholar]
- Parsons DW, et al. Colorectal cancer: mutations in a signalling pathway. Nature. 2005;436:792. doi: 10.1038/436792a. [DOI] [PubMed] [Google Scholar]
- Peng X, He Q, Li G, Ma J, Zhong TP. Rac1-PAK2 pathway is essential for zebrafish heart regeneration. Biochem Biophys Res Commun. 2016;472:637–642. doi: 10.1016/j.bbrc.2016.03.011. [DOI] [PubMed] [Google Scholar]
- Pirruccello M, et al. A dimeric kinase assembly underlying autophosphorylation in the p21 activated kinases. J Mol Biol. 2006;361:312–326. doi: 10.1016/j.jmb.2006.06.017. [DOI] [PubMed] [Google Scholar]
- Puto LA, et al. p21-activated kinase 1 (PAK1) interacts with the Grb2 adapter protein to couple to growth factor signaling. J Biol Chem. 2003;278:9388–9393. doi: 10.1074/jbc.M208414200. [DOI] [PubMed] [Google Scholar]
- Qing H, et al. PAK1-dependent MAPK pathway activation is required for colorectal cancer cell proliferation. Tumour Biol. 2012;33:985–994. doi: 10.1007/s13277-012-0327-1. [DOI] [PubMed] [Google Scholar]
- Qu J, et al. PAK4 kinase is essential for embryonic viability and for proper neuronal development. Mol Cell Biol. 2003;23:7122–7133. doi: 10.1128/MCB.23.20.7122-7133.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rane CK, Minden A. P21 activated kinases: structure, regulation, and functions. Small GTPases. 2014;5 doi: 10.4161/sgtp.28003. pii: e28003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rayala SK, et al. Nuclear p21-activated kinase 1 in breast cancer packs off tamoxifen sensitivity. Cancer Res. 2006a;66:5985–5988. doi: 10.1158/0008-5472.CAN-06-0978. [DOI] [PubMed] [Google Scholar]
- Rayala SK, et al. P21-activated kinase 1 regulation of estrogen receptor-alpha activation involves serine 305 activation linked with serine 118 phosphorylation. Cancer Res. 2006b;66:1694–1701. doi: 10.1158/0008-5472.CAN-05-2922. [DOI] [PubMed] [Google Scholar]
- Reinardy JL, et al. Phosphorylation of threonine 794 on Tie1 by Rac1/PAK1 reveals a novel angiogenesis regulatory pathway. PLoS One. 2015;10:e0139614. doi: 10.1371/journal.pone.0139614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roig J, Traugh JA. p21-activated protein kinase gamma-PAK is activated by ionizing radiation and other DNA-damaging agents. Similarities and differences to alpha-PAK. J Biol Chem. 1999;274:31119–31122. doi: 10.1074/jbc.274.44.31119. [DOI] [PubMed] [Google Scholar]
- Rubio MD, Haroutunian V, Meador-Woodruff JH. Abnormalities of the Duo/Ras-related C3 botulinum toxin substrate 1/p21-activated kinase 1 pathway drive myosin light chain phosphorylation in frontal cortex in schizophrenia. Biol Psychiatry. 2012;71:906–914. doi: 10.1016/j.biopsych.2012.02.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanchez-Solana B, et al. p21-activated kinase-1 signaling regulates transcription of tissue factor and tissue factor pathway inhibitor. J Biol Chem. 2012;287:39291–39302. doi: 10.1074/jbc.M112.404061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scott RW, et al. LIM kinases are required for invasive path generation by tumor and tumor-associated stromal cells. J Cell Biol. 2010;191:169–185. doi: 10.1083/jcb.201002041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sells MA, Chernoff J. Emerging from the Pak: the p21-activated protein kinase family. Trends Cell Biol. 1997;7:162–167. doi: 10.1016/S0962-8924(97)01003-9. [DOI] [PubMed] [Google Scholar]
- Shao YG, Ning K, Li F. Group II p21-activated kinases as therapeutic targets in gastrointestinal cancer. World J Gastroenterol. 2016;22:1224–1235. doi: 10.3748/wjg.v22.i3.1224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheehan KA, Ke Y, Solaro RJ. P21-Activated kinase-1 and its role in integrated regulation of cardiac contractility. Am J Physiol Regul Integr Comp Physiol. 2007;293:R963–973. doi: 10.1152/ajpregu.00253.2007. [DOI] [PubMed] [Google Scholar]
- Shepelev MV, Korobko IV. Pak6 protein kinase is a novel effector of an atypical Rho family GTPase Chp/RhoV. Biochemistry (Mosc) 2012;77:26–32. doi: 10.1134/S0006297912010038. [DOI] [PubMed] [Google Scholar]
- Shin YJ, Kim YB, Kim JH. Protein kinase CK2 phosphorylates and activates p21-activated kinase 1. Mol Biol Cell. 2013;24:2990–2999. doi: 10.1091/mbc.E13-04-0204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shu XR, et al. PAK4 confers the malignance of cervical cancers and contributes to the cisplatin-resistance in cervical cancer cells via PI3K/AKT pathway. Diagn Pathol. 2015;10:177. doi: 10.1186/s13000-015-0404-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh RR, Song C, Yang Z, Kumar R. Nuclear localization and chromatin targets of p21-activated kinase 1. J Biol Chem. 2005;280:18130–18137. doi: 10.1074/jbc.M412607200. [DOI] [PubMed] [Google Scholar]
- Siu MK, et al. p21-activated kinases 1, 2 and 4 in endometrial cancers: effects on clinical outcomes and cell proliferation. PLoS One. 2015;10:e0133467. doi: 10.1371/journal.pone.0133467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singhal R, Kandel ES. The response to PAK1 inhibitor IPA3 distinguishes between cancer cells with mutations in BRAF and Ras oncogenes. Oncotarget. 2012;3:700–708. doi: 10.18632/oncotarget.587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Staser, et al. A Pak1-PP2A-ERM signaling axis mediates F-actin rearrangement and degranulation in mast cells. Exp Hematol. 2013;41:56–66. doi: 10.1016/j.exphem.2012.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strochlic TI, et al. Identification of neuronal substrates implicates Pak5 in synaptic vesicle trafficking. Proc Natl Acad Sci U S A. 2012;109:4116–4121. doi: 10.1073/pnas.1116560109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strochlic TI, et al. Phosphoinositides are essential coactivators for p21-activated kinase 1. Mol Cell. 2010;40:493–500. doi: 10.1016/j.molcel.2010.10.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song B, et al. P21-activated kinase 1 and 4 were associated with colorectal cancer metastasis and infiltration. J Surg Res. 2015;196:130–135. doi: 10.1016/j.jss.2015.02.035. [DOI] [PubMed] [Google Scholar]
- Sun GG, et al. Interactions between filamin A and MMP-9 regulate proliferation and invasion in renal cell carcinoma. Asian Pac J Cancer Prev. 2014;15:3789–3795. doi: 10.7314/apjcp.2014.15.8.3789. [DOI] [PubMed] [Google Scholar]
- Tabusa H, Brooks T, Massey AJ. Knockdown of PAK4 or PAK1 inhibits the proliferation of mutant KRAS colon cancer cells independently of RAF/MEK/ERK and PI3K/AKT signaling. Mol Cancer Res. 2013;11:109–121. doi: 10.1158/1541-7786.MCR-12-0466. [DOI] [PubMed] [Google Scholar]
- Taglieri DM, et al. Ablation of p21-activated kinase-1 in mice promotes isoproterenol-induced cardiac hypertrophy in association with activation of Erk1/2 and inhibition of protein phosphatase 2A. J Mol Cell Cardiol. 2011:51988–996. doi: 10.1016/j.yjmcc.2011.09.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tao J, et al. PAK1-Nck regulates cyclin D1 promoter activity in response to prolactin. Mol Endocrinol. 2011;25:1565–1578. doi: 10.1210/me.2011-0062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tharakan R, et al. Phosphorylation of estrogen receptor alpha, serine residue 305 enhances activity. Mol Cell Endocrinol. 2008;295:70–78. doi: 10.1016/j.mce.2008.07.018. [DOI] [PubMed] [Google Scholar]
- Thiel DA, et al. Cell cycle-regulated phosphorylation of p21-activated kinase 1. Curr Biol. 2002;12:1227–1232. doi: 10.1016/s0960-9822(02)00931-4. [DOI] [PubMed] [Google Scholar]
- Thomas JL, et al. PAK1 and CtBP1 Regulate the Coupling of Neuronal Activity to Muscle Chromatin and Gene Expression. Mol Cell Biol. 2015;35:4110–4120. doi: 10.1128/MCB.00354-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toriyama M, Kozawa S, Sakumura Y, Inagaki N. Conversion of a signal into forces for axon outgrowth through Pak1-mediated shootin 1 phosphorylation. Curr Biol. 2013;23:529–534. doi: 10.1016/j.cub.2013.02.017. [DOI] [PubMed] [Google Scholar]
- Tian X, et al. MicroRNA-429 inhibits the migration and invasion of colon cancer cells by targeting PAK6/cofilin signaling. Oncol Rep. 2015;34:707–714. doi: 10.3892/or.2015.4039. [DOI] [PubMed] [Google Scholar]
- Tyagi N, et al. p-21 activated kinase 4 promotes proliferation and survival of pancreatic cancer cells through AKT- and ERK-dependent activation of NF-kappaB pathway. Oncotarget. 2014;5:8778–8789. doi: 10.18632/oncotarget.2398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vadlamudi RK, et al. Regulatable expression of p21-activated kinase-1 promotes anchorage-independent growth and abnormal organization of mitotic spindles in human epithelial breast cancer cells. J Biol Chem. 2000;275:36238–36244. doi: 10.1074/jbc.M002138200. [DOI] [PubMed] [Google Scholar]
- Vadlamudi RK, et al. Dynein light chain 1, a p21-activated kinase 1-interacting substrate, promotes cancerous phenotypes. Cancer Cell. 2004;5:575–585. doi: 10.1016/j.ccr.2004.05.022. [DOI] [PubMed] [Google Scholar]
- Vadlamudi RK, et al. p21-activated kinase 1 regulates microtubule dynamics by phosphorylating tubulin cofactor B. Mol Cell Biol. 2005a;25:3726–3736. doi: 10.1128/MCB.25.9.3726-3736.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vadlamudi RK, et al. An essential role of Pak1 phosphorylation of SHARP in Notch signaling. Oncogene. 2005b;24:4591–4596. doi: 10.1038/sj.onc.1208672. [DOI] [PubMed] [Google Scholar]
- Vadlamudi RK, Kumar R. P21-activated kinases in human cancer. Cancer Metastasis Rev. 2003;22:385–393. doi: 10.1023/a:1023729130497. [DOI] [PubMed] [Google Scholar]
- Vadlamudi RK, et al. Filamin is essential in actin cytoskeletal assembly mediated by p21-activated kinase 1. Nat Cell Biol. 2002;4:681–690. doi: 10.1038/ncb838. [DOI] [PubMed] [Google Scholar]
- Van den Broeke C, et al. An emerging role for p21-activated kinases (Paks) in viral infections. Trends Cell Biol. 2010;20:160–169. doi: 10.1016/j.tcb.2009.12.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varshney P, Dey CS. P21-activated kinase 2 (PAK2) regulates glucose uptake and insulin sensitivity in neuronal cells. Mol Cell Endocrinol. 2016;429:50–61. doi: 10.1016/j.mce.2016.03.035. [DOI] [PubMed] [Google Scholar]
- Vershinin Z, Feldman M, Chen A, Levy D. PAK4 Methylation by SETD6 Promotes the Activation of the Wnt/beta-Catenin Pathway. J Biol Chem. 2016;291:6786–6795. doi: 10.1074/jbc.M115.697292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang C, et al. Oncogenic PAK4 regulates Smad2/3 axis involving gastric tumorigenesis. Oncogene. 2014;33:3473–3484. doi: 10.1038/onc.2013.300. [DOI] [PubMed] [Google Scholar]
- Wang RA, et al. P21-activated kinase-1 phosphorylates and transactivates estrogen receptor-alpha and promotes hyperplasia in mammary epithelium. EMBO J. 2002;21:5437–5447. doi: 10.1093/emboj/cdf543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang SE, et al. HER2/Neu (ErbB2) signaling to Rac1-Pak1 is temporally and spatially modulated by transforming growth factor beta. Cancer Res. 2006;66:9591–9600. doi: 10.1158/0008-5472.CAN-06-2071. [DOI] [PubMed] [Google Scholar]
- Wang X, et al. p21-activated kinase 5 inhibits camptothecin-induced apoptosis in colorectal carcinoma cells. Tumour Biol. 2010;31:575–582. doi: 10.1007/s13277-010-0071-3. [DOI] [PubMed] [Google Scholar]
- Wang XX, et al. PAK5-Egr1-MMP2 signaling controls the migration and invasion in breast cancer cell. Tumour Biol. 2013;34:2721–2729. doi: 10.1007/s13277-013-0824-x. [DOI] [PubMed] [Google Scholar]
- Wang Y, et al. Pak1 is required to maintain ventricular Ca2+ homeostasis and electrophysiological stability through SERCA2a regulation in mice. Circ Arrhythm Electrophysiol. 2014;7:938–948. doi: 10.1161/CIRCEP.113.001198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wells CM, et al. PAK4: a pluripotent kinase that regulates prostate cancer cell adhesion. J Cell Sci. 2010;123:1663–1673. doi: 10.1242/jcs.055707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wen X, et al. Knockdown of p21-activated kinase 6 inhibits prostate cancer growth and enhances chemosensitivity to docetaxel. Urology. 2009;73:1407–1411. doi: 10.1016/j.urology.2008.09.041. [DOI] [PubMed] [Google Scholar]
- Whale AD, et al. PAK4 kinase activity and somatic mutation promote carcinoma cell motility and influence inhibitor sensitivity. Oncogene. 2013;32:2114–20. doi: 10.1038/onc.2012.233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong LE, et al. p120-catenin is a binding partner and substrate for Group B Pak kinases. J Cell Biochem. 2010;110:1244–1254. doi: 10.1002/jcb.22639. [DOI] [PubMed] [Google Scholar]
- Wu X, Frost JA. Multiple Rho proteins regulate the subcellular targeting of PAK5. Biochem Biophys Res Commun. 2006;351:328–335. doi: 10.1016/j.bbrc.2006.09.172. [DOI] [PubMed] [Google Scholar]
- Wittmann T, et al. Regulation of microtubule destabilizing activity of Op18/stathmin downstream of Rac1. J Biol Chem. 2004;279:6196–6203. doi: 10.1074/jbc.M307261200. [DOI] [PubMed] [Google Scholar]
- Xue J, et al. MicroRNA-433 inhibits cell proliferation in hepatocellular carcinoma by targeting p21 activated kinase (PAK4) Mol Cell Biochem. 2015;399:77–86. doi: 10.1007/s11010-014-2234-9. [DOI] [PubMed] [Google Scholar]
- Yang F, et al. Androgen receptor specifically interacts with a novel p21-activated kinase, PAK6. J Biol Chem. 2001;276:15345–15353. doi: 10.1074/jbc.M010311200. [DOI] [PubMed] [Google Scholar]
- Yang Z, Rayala S, Nguyen D, Vadlamudi RK, Chen S, Kumar R. Pak1 phosphorylation of snail, a master regulator of epithelial-to-mesenchyme transition, modulates snail’s subcellular localization and functions. Cancer Res. 65:3179–184. doi: 10.1158/0008-5472.CAN-04-3480. [DOI] [PubMed] [Google Scholar]
- Zhai J, et al. miR-129 suppresses tumor cell growth and invasion by targeting PAK5 in hepatocellular carcinoma. Biochem Biophys Res Commun. 2015;464:161–167. doi: 10.1016/j.bbrc.2015.06.108. [DOI] [PubMed] [Google Scholar]
- Zhang DG, Zhang J, Mao LL, Wu JX, Cao WJ, Zheng JN, Pei DS. p21-Activated kinase 5 affects cisplatin-induced apoptosis and proliferation in hepatocellular carcinoma cells. Tumour Biol. 2015;36:3685–3691. doi: 10.1007/s13277-014-3007-5. [DOI] [PubMed] [Google Scholar]
- Zhang M, et al. Inhibition of p21-activated kinase 6 (PAK6) increases radiosensitivity of prostate cancer cells. Prostate. 2010;70:807–816. doi: 10.1002/pros.21114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang K, et al. Filamin A expression correlates with proliferation and invasive properties of human metastatic melanoma tumors: implications for survival in patients. J Cancer Res Clin Oncol. 2014;140:1913–1926. doi: 10.1007/s00432-014-1722-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X, et al. Aurora A phosphorylates MCAK to control ran-dependent spindle bipolarity. Mol Biol Cell. 2008;19:2752–2765. doi: 10.1091/mbc.E08-02-0198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao L, et al. Role of p21-activated kinase pathway defects in the cognitive deficits of Alzheimer disease. Nat Neurosci. 2006;9:234–242. doi: 10.1038/nn1630. [DOI] [PubMed] [Google Scholar]
- Zhao W, et al. Upregulation of p21-activated Kinase 6 in rat brain cortex after traumatic brain injury. J Mol Histol. 2011;42:195–203. doi: 10.1007/s10735-011-9324-8. [DOI] [PubMed] [Google Scholar]
- Zheng J, et al. CRNDE affects the malignant biological characteristics of human glioma stem cells by negatively regulating miR-186. Oncotarget. 2015;6:25339–25355. doi: 10.18632/oncotarget.4509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhong Z, et al. Cyclin D1/cyclin-dependent kinase 4 interacts with filamin A and affects the migration and invasion potential of breast cancer cells. Cancer Res. 2010;70:2105–2014. doi: 10.1158/0008-5472.CAN-08-1108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu G, et al. PAK5-mediated E47 phosphorylation promotes epithelial-mesenchymal transition and metastasis of colon cancer. Oncogene. 2016;35:1943–1954. doi: 10.1038/onc.2015.259. [DOI] [PubMed] [Google Scholar]
- Zhou GL, et al. Akt phosphorylation of serine 21 on Pak1 modulates Nck binding and cell migration. Mol Cell Biol. 2003;23:8058–8069. doi: 10.1128/MCB.23.22.8058-8069.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou W, et al. PAK1 mediates pancreatic cancer cell migration and resistance to MET inhibition. J Pathol. 2014;234:502–513. doi: 10.1002/path.4412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu G, et al. A Rac1/PAK1 cascade controls β-catenin activation in colon cancer cells. Oncogene. 2012;31:1001–1012. doi: 10.1038/onc.2011.294. [DOI] [PubMed] [Google Scholar]
- Zhu J, et al. From the cyclooxygenase-2 inhibitor celecoxib to a novel class of 3-phosphoinositide-dependent protein kinase-1 inhibitors. Cancer Res. 2004;64:4309–4318. doi: 10.1158/0008-5472.CAN-03-4063. [DOI] [PubMed] [Google Scholar]