SUMMARY
Antibody somatic hypermutation (SHM) and affinity maturation enhance antigen recognition by modifying antibody paratope structure to improve its complementarity with the target epitope. SHM-induced changes in paratope dynamics may also contribute to antibody maturation, but direct evidence of this is limited. Here, we examine two classes of HIV-1 broadly neutralizing antibodies (bNAbs) for SHM-induced changes in structure and dynamics and delineate the effects of these changes on interactions with the HIV-1 Envelope glycoprotein (Env). In combination with new and existing structures of unmutated and affinity matured antibody Fab fragments, we used hydrogen/deuterium-exchange mass spectrometry to directly measure Fab structural dynamics. Changes in antibody structure and dynamics were positioned to improve complementarity with Env, with changes in dynamics primarily observed at the paratope peripheries. We conclude that SHM optimizes paratope complementarity to conserved HIV-1 epitopes and restricts the mobility of paratope-peripheral residues to minimize clashes with variable features on HIV-1 Env.
eTOC
Hydrogen/deuterium-exchange mass spectrometry (HDX-MS) and X-ray crystallography were combined to study the impact of somatic hypermutation (SHM) on HIV-1 broadly neutralizing antibody structure and dynamics. SHM-induced stabilization and structural changes improve epitope complementarity and may minimize clashes with dynamic glycans on HIV-1 Env.
INTRODUCTION
Antibodies recognize their cognate antigens through specific intermolecular interactions between contact residues in the antibody paratope and the antigen epitope. During antibody affinity maturation, these interactions are refined to enhance binding affinity and specificity for the target antigen. Affinity maturation begins when the unmutated B cell receptor (BCR), encoded by recombined immunoglobulin gene segments and expressed on antigen-naïve B cells, becomes activated as a result of antigen binding and subsequent T cell help. This activation drives sequential rounds of somatic hypermutation (SHM), which optimize contact residues and immunoglobulin variable domain conformation such that the mature BCR recognizes its target antigen with higher affinity than the naïve, unmutated BCR (Victora and Nussenzweig, 2012).
Structural differences between unmutated (sometimes called “germline”) and affinity-matured antibodies have been observed in the context of a number of antibody lineages using x-ray crystallography (Fera et al., 2014; Li et al., 2003; Schmidt et al., 2013; Wedemayer et al., 1997). Paratope flexibility and structural dynamics also impact the affinity and specificity of antibody-antigen interactions (Foote and Milstein, 1994; Jimenez et al., 2004; Manivel et al., 2000), but these features have been difficult to characterize using techniques such as crystallography. Previous work on antibody maturation to model antigens including haptens (Adhikary et al., 2012; Jimenez et al., 2004; Wedemayer et al., 1997; Zimmermann et al., 2006), peptides (Manivel et al., 2000; 2002), and hen egg lysozyme (Mohan et al., 2009) suggests that unmutated antibodies exhibit relatively high levels of paratope flexibility that may improve their ability to recognize diverse antigens (James et al., 2003; Yin et al., 2003), and that the paratope becomes increasingly rigid as the antibody matures to become specific for a given antigen. In this model of antibody maturation to a discrete immunogen, flexibility and poly-reactivity in the immature antibody are exchanged for a well-ordered paratope in the high affinity mature antibody. Here, we test whether this model applies to HIV-1 broadly-neutralizing antibodies (bNAbs) – antibodies capable of recognizing structurally diverse variants of the HIV-1 envelope glycoprotein (Env), which are of substantial interest for HIV-1 vaccine design (Hoxie, 2010; Mascola and Montefiori, 2010). The interplay between antibody paratope flexibility and specificity observed for affinity maturation to model antigens raises a number of considerations for HIV-1 bNAbs. Specifically: how do these antibodies develop high affinity interactions with an exceptionally variable, conformationally dynamic, and continuously evolving glycoprotein antigen? Do they require greater flexibility to accommodate structural variability in diverse Env isolates, or do they exhibit increased stability to favor focused recognition of a single highly conserved epitope, while avoiding variable features?
Many bNAbs undergo extensive somatic hypermutation (SHM) on the path to achieving neutralizing breadth and potency (Klein et al., 2013b; Kwong and Mascola, 2012). The mutations acquired over the course of affinity maturation occur in both the complementarity determining regions (CDR) that make direct contact with the antigen and in the antibody framework (FW) regions, which are generally distal to the antigen-combining site, although some HIV-1 bNAbs use FW regions to contact antigen (Wu et al., 2011). Somatic hypermutation outside of the antibody paratope is critical for the development of neutralization breadth and potency in a number of HIV-1 bNAbs (Georgiev et al., 2014; Klein et al., 2013a). These observations and recent structural studies (Fera et al., 2014; Finton et al., 2014; Garces et al., 2015) suggest that, in addition to optimizing contact residues, affinity maturation may improve bNAb-Env interactions by tailoring CDR loop stability, flexibility, and orientation however no study has directly measured changes in solution-phase local flexibility and structural dynamics in maturing HIV-1 bNAbs.
We used hydrogen/deuterium exchange with mass spectrometry (HDX-MS) in combination with x-ray crystallographic data to study how affinity maturation modifies HIV-1 bNAb paratope structure and dynamics. HDX-MS measures local protein dynamics by monitoring backbone amide deuterium uptake. Dynamic, solvent-exposed regions of a protein take on deuterium rapidly, while stable secondary structures and highly ordered or buried, core regions take on deuterium relatively slowly (Engen, 2009, Skinner et al., 2012). X-ray crystallography was used to study differences in three-dimensional structure between unmutated and mature VRC03, and HDX-MS was used to compare the local dynamics of antigen binding fragments (Fabs) derived from the unmutated and mature sequences for the CD4-binding site-directed bNAbs VRC03 (Wu et al., 2010) and VRC-PG04 (also called PGV04) by HDX-MS (Wu et al., 2011). We also studied the conformational development of the V1/V2-quaternary epitope-specific antibody CAP256-VRC26 (Figure 1) by measuring differences in local dynamics between Fabs derived from the predicted CAP256-VRC26 unmutated common ancestor (UCA) sequence, and three CAP256-VRC26 antibodies from distinct branches of the VRC26 phylogenetic tree, isolated at multiple time points after infection (Doria-Rose et al., 2014). The antibodies chosen for this study were selected in order to explore SHM-induced structural changes in bNAbs with distinct specificities and ontogenies. Our data provide direct evidence that SHM modifies HIV-1 bNAb structural dynamics as well as structure, with changes in dynamics primarily observed at the paratope periphery, at sites adjacent to Env glycans. In this way, SHM may enhance recognition of conserved epitopes on HIV-1 Env by restricting antibody dynamics to minimize non-productive interactions with disordered and variable features of the Env glycoprotein
Figure 1. Overview of bNAb Epitopes and Variable Domain Structure.
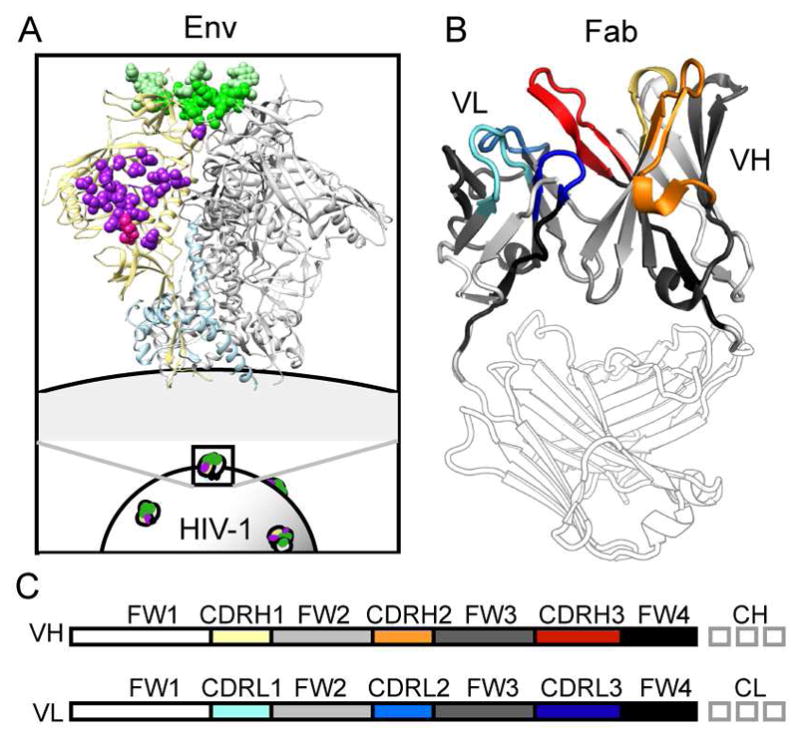
A) The CD4-binding site epitope targeted by VRC-PG04 and VRC03 is shown in purple on the Env trimer structure (PDB 4TVP). The V1V2/quaternary epitope targeted by PG9 - similar to that bound by CAP256-VRC26 - is indicated in green. B) Complementarity determining (CDR) and framework (FW) regions of the Fab heavy and light chain variable domains are indicated in color on the gp120-bound VRC03 crystal structure (PDB 3SE8). C) A linear representation of B, with matching color: CDRH1, 2, and 3 loops are colored yellow, orange, and red, while CDRL1, 2, and 3 loops are colored cyan, blue, and dark blue. Intervening framework regions, FW1, FW2, FW3, and FW4 are shown in white, light gray, dark gray, and black, respectively. Fab heavy and light chain constant regions are shown in gray outline.
RESULTS
Structure of unliganded Fabs from unmutated and mature VRC03
The VRC01 class of antibodies (Zhou et al., 2010; 2013) refers to CD4-binding site-directed antibodies, isolated from multiple donors, that share structural and sequence similarities including: usage of the IGVH1-2*02 heavy chain variable gene segment, light chains containing a five amino acid CDRL3 loop, mimicry of CD4 in binding to gp120, high levels of SHM, and avoidance of clashes with gp120 via SHM sequence changes that decrease the effective size of CDRL1 (Scheid et al., 2011; West et al., 2012; Wu et al., 2011; Zhou et al., 2013). Structural differences between unmutated and mature VRC01-class antibodies VRC01 and NIH45-46 have been previously described and revealed changes primarily within CDRH3 and CDRL1. Notably, unmutated VRC01 and NIH45-46 exhibited CDRH2 loops – which make essential contacts with the gp120 CD4-binding loop - that are essentially identical to their mature counterparts, with a number of critical residues pre-configured to bind gp120 (Jardine et al., 2013; Scharf et al., 2013).
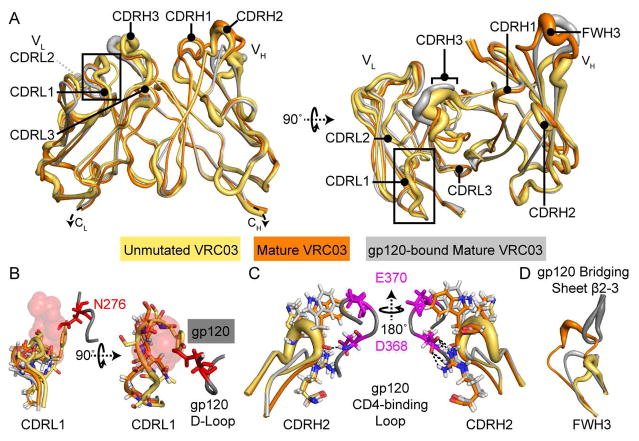
To further define the structural changes that occur during affinity maturation of VRC01-class antibodies, we determined the unliganded structures of mature VRC03 and its unmutated precursor (Figures 2A, 3, S1 and Table S1). The three-dimensional structures were largely similar, with a few notable features. As observed for VRC01 and NIH45-46 (Jardine et al., 2013; Scharf et al., 2013), unmutated VRC03 displays an extended CDRL1, which is positioned to clash with the gp120 N276 glycan (Figure 2B). The CDRL1 loop is truncated in mature VRC03, where it adopts a conformation nearly identical to that of gp120-bound mature VRC03. The CDRH2 loop – which makes critical backbone H-bonds and a salt-bridge with the conserved Asp368 of the gp120 CD4-binding loop - is nearly identical in unmutated and mature VRC03 (with and without gp120) (Figure 2C). Unlike NIH45-46 or VRC01, the FWH3 loop is dramatically extended in mature VRC03 relative to its unmutated precursor. Comparison of the unliganded and gp120-bound mature VRC03 structures reveals that the bridging sheet of gp120 core displaces the elongated FWH3 loop in mature VRC03. While this FWH3/bridging sheet interaction likely does not occur in the context of trimeric Env, it highlights the potential for plasticity within the VRC03 paratope (Figure 2D). Notably, for the unmutated as well as mature VRC03 and the other Fabs discussed in this study, crystal contacts did not appear to influence B factors or loop structures (Figure S1). Overall, we conclude that affinity maturation improves VRC03 paratope complementarity with HIV-1 Env.
Figure 2. Structures of Unmutated and Mature VRC03 (+/− gp120).
A) Unliganded x-ray crystal structures of the variable domain regions of unmutated (yellow) and mature (orange) VRC03 are superimposed with the structure of mature VRC03 bound to gp120 (PDB: 3SE8, light grey). Strand width corresponds to crystal structure B-factors. Complementarity determining regions are indicated. B) Affinity maturation minimizes clashes between VRC03 CDRL1 and the gp120 N276 glycan. C) CDRH2 loop structure and gp120-interactive residue position is conserved throughout affinity maturation. D) FWH3 of mature VRC03 is displaced by the bridging sheet upon binding to gp120 core. This displacement is likely not observed in VRC03 interactions with trimeric Env, but it reveals plasticity within the VRC03 paratope. See also Table S1.
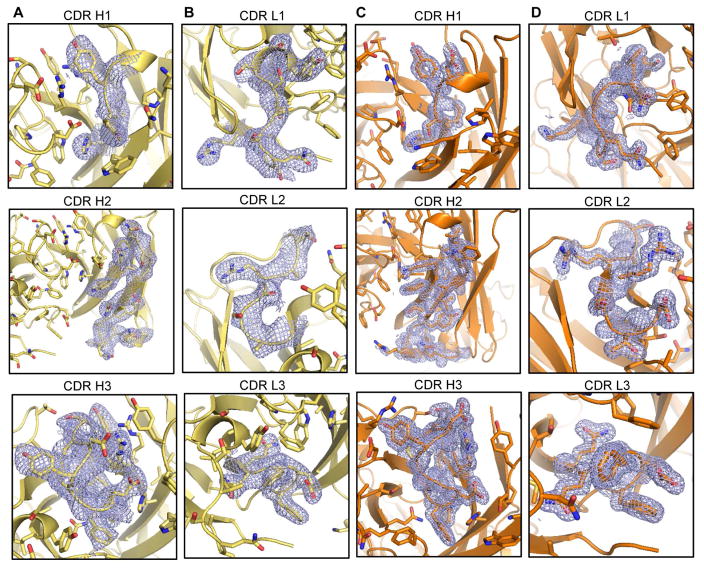
Figure 3. Electron density of complementarity-determining regions (CDRs).
for the unmutated VRC03 (A, B) and mature VRC03 Fab structures (C, D). The electron density for each CDR is shown in mesh format in separate panels for each CDR. Residues that make up each CDR are shown in stick representation with the antibody shown in ribbon representation. See also Figure S1.
Somatic hypermutation stabilizes the light chain of VRC01-class antibodies
To directly measure SHM-induced changes in VRC01-class antibody paratope flexibility, we performed HDX-MS on Fab fragments of the antibodies VRC03 and VRC-PG04 (Wu et al., 2011). These antibodies were isolated from two separate individuals and exhibit substantial sequence differences, but both make critical contacts with gp120 through their CDRH2, CDRH3, CDRL1, and CDRL3 loops (Wu et al., 2011). We identified peptic fragments covering over 94% of the unmutated and mature VRC03 and VRC-PG04 Fab sequences (Figures S2 and S3). While nearly all of the heavy (CH) and light (CL) chain constant domain sequence could be monitored by HDX-MS and compared between Fabs, mutations in the variable domains (VH and VL, for heavy and light variable domains, respectively) led to distinct digestion patterns that hindered a complete comparison of deuterium uptake between unmutated and mature Fabs. In comparing HDX results from proteins with divergent sequences, certain caveats need to be considered, since a peptide sequence can potentially influence its intrinsic rate of exchange (the rate at which unstructured amides exchange) (Wales et al., 2016). For this reason we limited our comparative analysis to peptides with an equal number of exchangeable amides and identical N and C termini in the aligned protein sequence. Furthermore we calculate percent deuteration relative to fully deuterated samples to correct for sequence-dependent differences in rates of D-to-H back exchange. Finally, we calculated intrinsic rates of H-to-D exchange for relevant peptides to ensure that measured differences in deuteration were not an artifact of sequence-dependent differences in rates of intrinsic exchange. By these criteria, we were able to compare the local dynamics of peptides spanning three key regions of the VRC03 VH domain (FW2/CDRH2, CDRH2/FW3, and CDRH3/FW4) and three regions of its VL domain (N-term/FW1, CDRL2/FW3, CDRL3/FW4) (Figure S2). For VRC-PG04, three regions of the VH domain (N-term/FW1, FW3/CDRH3, and FW4) and four regions of the VL domain (N-term/FW1, CDRL2/FW3, FW3, and FW4/CL) could be compared between unmutated and mature Fabs (Figure S3).
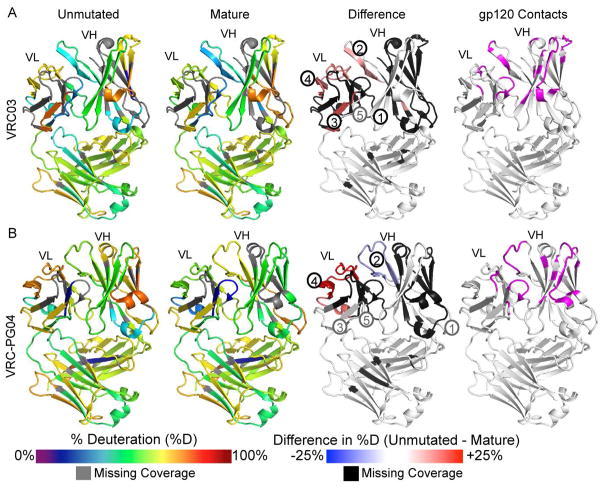
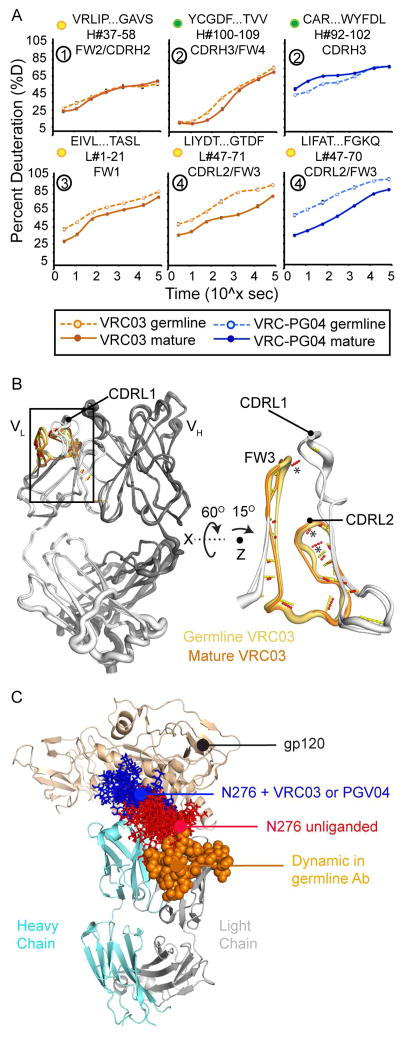
Heatmap summaries of unmutated and mature VRC03 deuterium uptake revealed differences in structural dynamics primarily within the VL domain (Figure 4A). All comparable VH and VL domain peptides were deuterated at a similar or faster rate in unmutated VRC03 as compared to mature VRC03 (Figures 4A and S4). In agreement with the structural conservation of the CDRH2 loop in unmutated and mature VRC03 crystal structures, two peptides spanning FW2/CDRH2 and CDRH2/FW3 showed nearly identical dynamics in unmutated and mature VRC03 (Figure 5A peptide 1). Peptides spanning CDRH3/FW4 were slightly more dynamic in unmutated VRC03 relative to mature VRC03 (Figure 5A peptide 2). The VL peptides showed more dramatic differences: light chain N-terminus, CDRL2/FW3, and CDRL3/FW4 peptides were distinctly more dynamic in unmutated VRC03 (Figures 5A peptides 3 and 4, and S4 peptide 5). Patterns of deuterium uptake correlated well with solvent-accessible surface area (SASA) in unmutated and mature VRC03 crystal structures, and, to a lesser extent, H-bond patterns in unmutated VRC03, but correlated poorly with residue depth and B-factor (Figure S5A and B). Consistent with the stabilization of CDRL2/FW3 in mature VRC03 observed by HDX-MS, mature VRC03 exhibited more predicted H-bonds in this region relative to the unmutated VRC03 crystal structure (Figure 5B).
Figure 4. VRC03 and VRC-PG04 HDX-MS Heatmap Summaries.
A) Deuterium uptake heatmaps for unmutated (far left) and mature (middle left) VRC03. Colors mapped onto the 3SE8 structure indicate % deuteration for VRC03 peptides after 1 minute of incubation in D2O. Grey indicates missing peptide coverage. Difference heatmap (middle right) indicates regions that are more disordered in unmutated relative to mature VRC03 (red), regions that are similarly ordered in unmutated and mature VRC03 (white), and regions that are more ordered in unmutated VRC03 (blue). Black indicates missing coverage on the difference heatmap due to missing or incomparable peptides. Black and grey numbers correspond to peptides in Figure 5 and supplemental information, respectively (see also Figures S2, S3, S4 and S6). Far right: VRC03 residues positioned to contact Env in PDB 3SE8 are highlighted in pink (Wu et al., 2011). B) Corresponding heatmaps for VRC-PG04 mapped onto PDB 3SE9.
Figure 5. Deuterium Uptake and Structure Analysis for Select VRC03 and VRC-PG04 Peptides.
A) Deuterium uptake curves for selected VRC03 (orange) and VRC-PGV04 (blue) variable domain peptides are shown for unmutated (dashed lines) and mature (solid lines) Fabs. The peptide sequence, position, and antibody region are indicated above each graph. Yellow dots indicate that unmutated and mature Fab peptide sequences are not identical; green dots indicate that the compared peptides are identical in sequence. Circled numbers correspond to peptides in Figure 4. Error bars show standard deviation from duplicate samples. B) Focus on the differentially-stabilized CDRL2/FW3 peptide in VRC03 unmutated and mature structures. Strand width indicates B-factor. * indicates predicted H-bonds present in mature – but not unmutated – VRC03 (see also Figure S5). C) The CDRL2/FW3 region (orange spheres) – disordered in unmutated VRC03 and VRC-PG04 - is proximal to the gp120 N276 glycan. Man5 glycans modeled onto gp120 (from PDB 4NCO (Julien et al., 2013)) in the presence (blue) and absence (red) of PGV04 show that displacement of N276 by CDRH3, CDRL1, and CDRL2 is required for PGV04 binding. A similar displacement of the N276 glycan is observed for VRC03 (not shown).
As for VRC03, the CDRL2/FW3 loop of mature VRC-PG04 was dramatically more stable than that of unmutated VRC-PG04 (Figures 4B, and 5A peptide 4). Importantly, these differences in deuterium uptake did not appear to be attributable to differences in rates of intrinsic exchange (Bai et al., 1993) as a result of primary sequence differences (Figure S6C). There were only minor differences in the dynamics of the VRC-PG04 VH domain. A number of peptides including one spanning CDRH3 were slightly more stable in unmutated VRC-PG04 relative to the mature Fab (Figures 5A peptide 2 and S6) - a subtle difference compared to VRC03, where all comparable regions of unmutated VRC03 were either similarly or more disordered than the mature Fab (Figures 4 and S4). Peptides from CH and CL domains were identically deuterated in unmutated and mature VRC-PG04 and were nearly superimposable with CH and CL peptides from VRC03 (Figures S4 and S6), indicating that the dynamics of the Fab constant region are largely uncoupled from the changes in sequence and structural dynamics we observed in the Fab variable domains.
The SHM-induced changes in VRC03 and VRC-PG04 heavy chain stability were relatively subtle, including regions that make direct contact with gp120 (e.g. CDRH2/FW3) – a finding consistent with the structural conservation of these regions in unmutated and mature VRC03, and other VRC01-class antibody crystal structures. By contrast, the magnitude and conservation of the change in structural dynamics within the CDRL2/FW3 region for both VRC03 and VRC-PG04 suggests that these changes may be functionally important in modifying VRC01-class antibody interactions with HIV-1 Env. Indeed, residues in the CDRL2/FW3 region have been shown to be critical for the neutralizing breadth and potency of mature VRC01 (Georgiev et al., 2014). Examination of high-resolution structural data for VRC03 and VRC-PG04 complexed with Env demonstrates that stabilization of the CDRL2/FW3 region is reasonably positioned to improve interactions with Env by minimizing potential clashes with the nearby Env glycan at position N276 (Figure 5C), a glycan that hinders unmutated VRC01-class antibody binding to Env (Jardine et al., 2013). Together, these data support the idea that the CDRL2/FW3 loop, and changes in the structural ordering of this region, may influence VRC01-class antibody interactions with Env.
Changes in structural dynamics within the CAP256-VRC26 antibody family
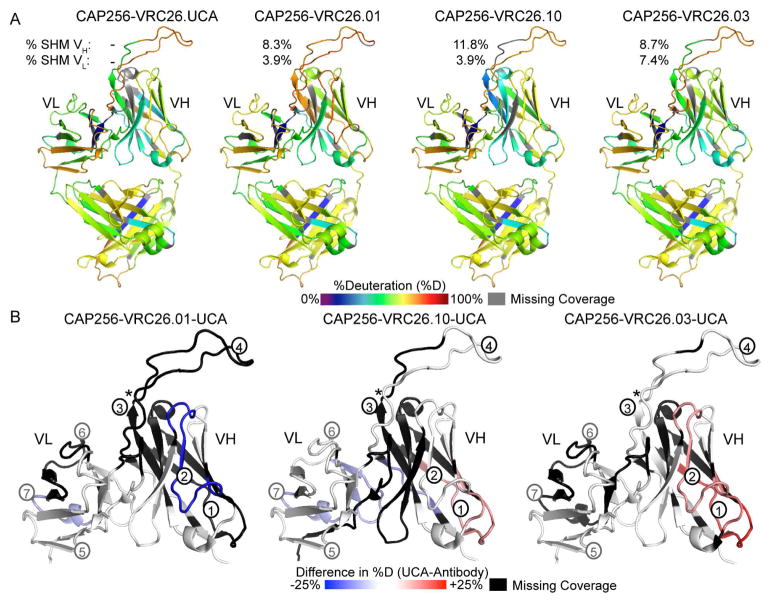
To better understand the generality of changes in bNAb dynamics during maturation, we also explored changes in structural dynamics over the course of affinity maturation in CAP256-VRC26 antibodies isolated from an HIV-1 superinfected individual who developed broadly neutralizing antibodies (Doria-Rose et al., 2014; Moore et al., 2011). CAP256-VRC26 antibodies are descended from the IGHV3-30*18 and IGLV1-51*02 variable gene segments and interact with Env in a manner very similar to the quaternary antibodies PG9/PG16 and CH03, which bind the V2 beta strand of Env through their CDRH3 loops and associate intimately with conserved Env glycans at positions N160 and N156 or N173 (Figure 1) (Gorman et al., 2016; McLellan et al., 2011; Pancera et al., 2013). We studied the dynamics of the predicted unmutated common ancestor (UCA) of CAP256-VRC26 antibodies, as well as three CAP256-VRC26 variants: CAP256-VRC26.01, CAP256-VRC26.03, and CAP256-VRC26.10, which were isolated at 59, 119, and 206 weeks post-infection (wpi), respectively (Doria-Rose et al., 2014). We identified peptic fragments covering over 99% of the antibodies’ primary sequence (Figure S7). While most of the CH and CL domain sequence could be compared by HDX and showed nearly identical deuterium uptake among the CAP256-VRC26 antibodies (Figures 6A and S8), comparisons of deuterium uptake in VH and VL were more limited due to differences in pepsin cleavage. With the exception of the CDRH1 loop, all regions of the variable domains were represented by at least one peptide that could be compared between at least two CAP256-VRC26 Fabs. Comparison of CAP256-VRC26 Fab deuterium uptake patterns with previously determined x-ray crystal structures showed strong correlations with SASA, an inverse correlation with residue depth, and weaker correlations with H-bonding and B-Factor (Figure S5C and D).
Figure 6. CAP256-VRC26 Fab HDX-MS Heatmap Summary.
A) Colors mapped onto the CAP256-VRC26.03 crystal structure (PDB 4OD1) indicate percent deuterium uptake for peptides throughout each CAP256-VRC26 antibody after 1 minute of deuteration as in Figure 4. Values for % mutation (nt) relative to germline VH and VL gene segments are indicated from (Doria-Rose et al., 2014) B) Difference heatmaps focusing on the CAP256-VRC26 variable domains show regions that are more disordered (red), similarly structured (white), or more stable in the UCA (blue) at the 1 minute time point. Regions that could not be compared are indicated in black. Black and grey numbers refer to peptides in Figure 7 and supplemental information, respectively (See also Figures S7 and S8).
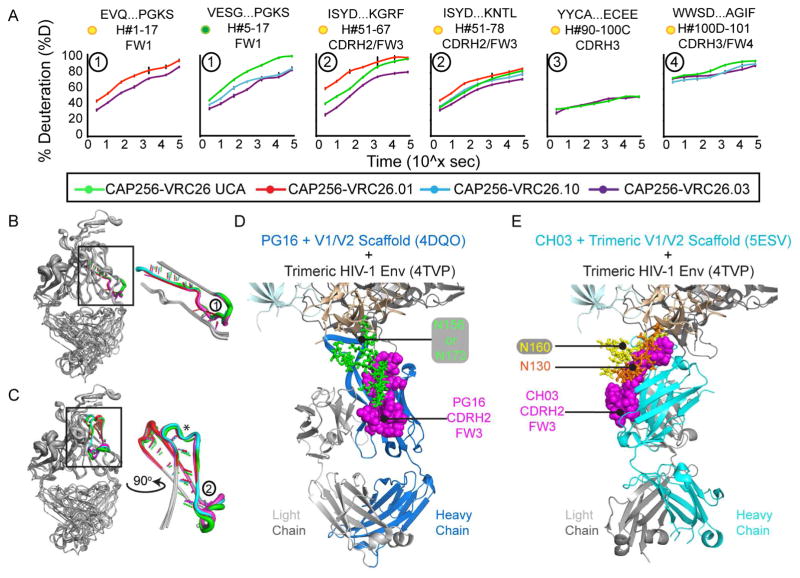
Heatmap summaries of the HDX-MS results for CAP256-VRC26 lineage Fabs revealed that the most dramatic differences in deuterium uptake occurred in the VH domain FW1 and CDRH2/FW3 regions (Figures 6B and 7A, peptides 1 and 2). Notably, crystal structure-predicted backbone H-bonding was nearly identical in these regions (Figure 7B and C), perhaps reflecting a low-energy conformation stabilized in the crystal that is less ordered in solution (as measured by HDX-MS). As for VRC03 and VRC-PG04, the observed differences in deuterium uptake did not appear to be attributable to differences in rates of theoretical intrinsic exchange due to primary sequence differences among the Fabs (Figure S8C and D). Though the rank order of CAP256-VRC26 antibody stability based on FW1 differed slightly from that based on CDRH2/FW3, overall, CAP256-VRC26.UCA and CAP256-VRC26.01 peptides were consistently more disordered, CAP256-VRC26.10 was intermediate, and CAP256-VRC26.03 was the most stable of the antibodies tested here (Figures 7A, peptides 1, 2, 4).
Figure 7. Deuterium Uptake and Crystal Structure Analysis for Select CAP256-VRC26 Peptides.
A) Percent deuterium uptake curves (green = UCA, red = 01, cyan = 10, purple = 03) for select CAP256-VRC26 regions. Data is shown for all antibodies with comparable peptides. Peptide location and length are indicated above each graph. Yellow dots indicate that the compared VRC26 Fab peptide sequences are not identical; green dots indicate that at least two of the compared VRC26 Fab peptides are identical in sequence (see also Figures S7 and S8). Circled numbers correspond to peptides from Figures 6 and S7. Peptide 3 contains one of the emergent disulfide cysteine residues (Doria-Rose et al., 2014), indicated by an asterisk in Figure 6. Error bars show standard deviation from duplicate samples. B) Comparison of structure, B-factors and predicted H-bonds in the heavy chain N-terminus (peptide 1) from the four CAP256-VRC26 crystal structures PDB 4ODH (UCA), 4OCR (01), 4OD1 (03), 4OCS (10). Strand width corresponds to B-factor; each Fab is colored as in A). C) Structure, B-factor and H-bond comparison of the CAP256-VRC26 CDRH2/FW3 region (peptide 2). * indicates an H bond absent in UCA, but present in all other CAP256-VRC26 structures. See also Figure S5. X-ray crystal structures for the CAP256-VRC26-like antibodies D) PG16 (PDB 4DQO) and E) CH03 (PDB 5ESV) bound to glycosylated V1/V2 scaffolds, with V1/V2 strands B and C superimposed. The CDRH2/FW3 region is oriented differently in these structures, but it is positioned to interact with glycans in either orientation.
Given that the CDRH3 is likely to be a critical element of the CAP256-VRC26 paratope (Doria-Rose et al., 2014; Gorman et al., 2016), it was unfortunate that differences in pepsin cleavage limited the direct comparison of deuterium uptake in this immediate region among all four antibodies. Despite this, it was apparent that the extended CDRH3 loop was quite dynamic in all four antibodies (Figure 6A). Additionally, because of similarities in cleavage for CAP256-VRC26.UCA, .10, and .03, it could be seen that the C-terminal half of CDRH3 was similarly - or slightly more - stable in CAP256-VRC26.03 and .10 relative to the UCA (Figures 6B and 7A, peptide 4). It is also notable that the deuteration of a CDRH3 peptide from CAP256-VRC26.03, which contains a disulfide-linked cysteine that emerges during CAP256-VRC26 maturation (Doria-Rose et al., 2014), was similar to that of a matching peptide from CAP256-VRC26.UCA, which lacks this disulfide bond (Figures 6B and 7A, peptide 3). This suggests that although the emergent disulfide bond may constrain the three-dimensional movement of the CDRH3 loop in CAP256-VRC26.03, it does not dramatically alter its stability as probed by HDX-MS.
To understand how differences in stability observed by HDX-MS might relate to CAP256-VRC26 binding and neutralizing activity, we used biolayer interferometry to measure binding of the CAP256-VRC26 Fabs to soluble, trimeric BG505.SOSIP Env (Sanders et al., 2013). The rank order of binding affinity paralleled the stability rankings we observed, with CAP256-VRC26.01 binding weakly, and CAP256-VRC26.10 binding well, though not as well as CAP256-VRC26.03 (Table 1 and Figure S9). CAP256-VRC26.UCA did not bind appreciably to BG505.SOSIP, reflecting the absence of SHM-acquired contact residues. Previously reported CAP256-VRC26 neutralizing activity mirrors the binding affinities measured here: CAP256-VRC26.03 was reported to have the highest breadth and potency of the antibodies; CAP256-VRC26.10 was intermediate, and CAP256-VRC26.01 had the weakest neutralizing activity (Doria-Rose et al., 2014). Though contact residue differences are critical in determining binding and neutralizing activity, these data also support a relationship between CAP256-VRC26 variable domain stability and enhanced recognition of HIV-1 Env.
Table 1.
Binding Constants for CAP256-VRC26 Antibodies to BG505.SOSIP.664 Env Trimer
| Fab | kon (1/M-s) | koff (1/s) | KD (nM) |
|---|---|---|---|
| CAP256-VRC26.UCA | n.b.a | n.b. | n.b. |
| CAP256-VRC26.01 | 1.0(1) x 104 b | 7.1(2) x 10−3 | 680(80) |
| CAP256-VRC26.03 | 6.3(4) x 104 | 4.1(1) x 10−3 | 64(5) |
| CAP256-VRC26.10 | 5.4(3) x 104 | 4.5(1) x 10−3 | 84(6) |
n.b. – no binding was observed at the highest concentration of Env tested. See also Figure S9.
Number in parentheses indicates error on last significant figure (standard error of two experiments)
The magnitude of the differences in stability within FW1 and CDRH2/FW3 and the similar rank orders of stability and binding affinity/neutralizing activity suggest that stabilization of FW1 and CDRH2/FW3 may influence CAP256-VRC26 antibody interactions with Env (Figures 6 and 7 and Table 1). This is supported by the observation that residues in the CDRH2/FW3 region of PG16 make extensive contact with the Env N156/N173 glycan (Pancera et al., 2013), and some of these residues, including Trp64, contribute to the breadth and potency of neutralization for CAP256-VRC26 antibodies (Gorman et al., 2016) as well as PG9 and PG16 (McLellan et al., 2011). Examination of crystal structures for the related quaternary epitope specific antibodies PG16 and CH03 in complex with glycosylated V1/V2 scaffolds shows that the CDRH2/FW3 loops of PG16 and CH03, though oriented differently in complexes with V1/V2 scaffolds, are positioned to make contact with Env glycans in either orientation (Figure 7D,E): N156/173 for PG16 (Pancera et al., 2013), and N160 and N130 for CH03 (Gorman et al., 2016). If CAP256-VRC26 antibodies interact with Env in a manner similar to CH03 or PG9/PG16, as most of the available data suggests, then the differences in dynamics within the CDRH2/FW3 region would be reasonably positioned to impact CAP256-VRC26 binding to Env by modifying the antibody’s interactions with these glycans (Figure 7D,E).
DISCUSSION
In the conventional model of antibody affinity maturation, a flexible, low-affinity, poly-specific unmutated precursor incorporates mutations during SHM that improve its affinity and specificity for the target antigen by optimizing contact residues and rigidifying the paratope through both proximal and distal amino acid changes (Jimenez et al., 2004; Wedemayer et al., 1997; Yin et al., 2003; Zimmermann et al., 2006). Most of the support for the conventional model comes from studies of antibodies that develop in response to a single, invariant immunogen – typically small molecules and peptides. However, antibodies appear to mature to large protein antigens differently than small molecules (Li et al., 2003), and it is therefore critical to re-examine the conformational maturation model in the context of more complex antibody-antigen pairings such as those between HIV-1 bNAbs and the highly variable Env glycoprotein.
A number of studies have explored antibody conformational maturation to viral glycoprotein antigens. Some of these suggest that the conformational equilibrium of the antibody paratope shifts over the course of maturation to favor a single, more stable conformation that is optimized for antigen binding (Schmidt et al., 2013). Other studies indicate that this basic model may not always apply. For example, Finton and colleagues observed that the mature HIV-1 Env-specific bNAb, 4E10, exhibited decreased thermostability relative to a panel of predicted 4E10 precursors (Finton et al., 2014). A similar decrease in thermostability was observed for mature 3BNC60 (an HIV-1 CD4-bs antibody) relative to a partial ‘germline’-revertant 3BNC60 P61A (Klein et al., 2013a). Thermal stability is a global metric of structural ordering, however, and does not provide insight into specific differences in antibody structure under native, physiological conditions. To address this gap, we used HDX-MS to measure changes in structural dynamics with sequence-specific detail. This analysis revealed that SHM alters variable domain flexibility in a site-specific manner, often outside of the paratope, with different patterns of stabilization among distinct antibody specificities. Though patterns of deuterium uptake within a given Fab correlated well with x-ray structure parameters including SASA, residue depth, and H-bonding to a lesser extent, differences in solution-phase Fab structural dynamics between Fabs measured by HDX-MS were not readily apparent from the crystal structures, highlighting the importance of HDX-MS as a complementary structural technique.
The simplest mechanism by which antibody structural dynamics might impact antigen binding would be through stabilization of the paratope in a conformation close to that of the antigen-bound state to increase the rate of association and/or minimize the entropic cost of antigen binding. In light of this, the minimal differences in dynamics within the unmutated and mature antibodies’ primary contact sites (CDRH2 and 3 for VRC03 and VRC-PG04 and CDRH3 for CAP256-VRC26) observed here were somewhat unexpected. The most significant changes in stability we observed for antibodies studied here occurred in sites peripheral to the antibody paratope (CDRL2/FW3 in VRC03 and VRC-PG04, and FW1 and CDRH2/FW3 in CAP256-VRC26). In all three cases, the regions of interest were more disordered in the unmutated/early antibodies and increased in stability at late time points of affinity maturation. These data are consistent with the finding that framework mutations are critical for the neutralizing activity of many HIV-1 bNAbs (Georgiev et al., 2014; Klein et al., 2013a) and the observation that unmutated/early antibodies are conformationally distinct from their mature descendants in sites both central and peripheral to the paratope (Fera et al., 2014; Li et al., 2003; Thomson et al., 2008; Wedemayer et al., 1997; Zimmermann et al., 2006). In combination with the data presented here, these studies suggest that changes at the paratope periphery are likely to impact antibody binding to antigens in general, but changes at the paratope periphery may be especially relevant in the context of antibody affinity maturation to complex antigens, where the antibody must accommodate structures surrounding the epitope, while still making critical binding interactions mediated by the primary paratope.
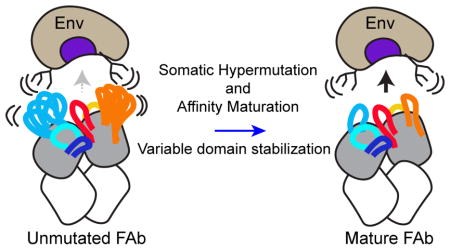
We interpret the observed changes in structural dynamics for VRC03, VRC-PG04, and CAP256-VRC26 under a similar conceptual model: local stabilization of antibody variable domains minimizes clashes with dynamic, structurally heterogenous glycans or variable loops on the Env surface (Figure 8). We note that based on the current data, we cannot exclude the possibility that the peripheral stabilization we observed may reflect epistatic stabilization of the Fab variable domain, as was recently reported for a hapten-specific antibody (Wang et al., 2013). Rather than directly improving Env interactions, stabilization of regions peripheral to the antibody paratope may improve the ‘evolvability’ of the variable domain, facilitating optimization of paratope contact residues. This possibility is intriguing, given the recent evidence that CAP256-VRC26.01 – the most disordered among the VRC26 antibodies tested here – represents a dead-end sub-lineage of the CAP256-VRC26 family (Bhiman et al., 2015). It is conceivable that the relative disorder of the CAP256-VRC26.01 variable domain may have contributed to its diminished capacity for evolution. Stabilization of these regions may also reduce entropic costs of binding if they interact allosterically with the paratope itself.
Figure 8. Model of the Influence of SHM-Induced Changes in Local Stability on Antibody-Env Interactions.
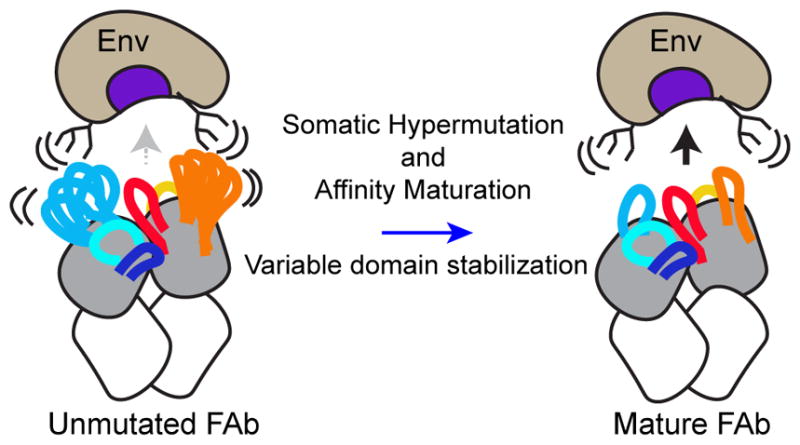
Unmutated bNAbs exhibit increased disorder in regions peripheral to the paratope, which may limit interactions with HIV-1 Env. Hypermutation and affinity maturation stabilize these regions (blue arrow), minimizing clashes with disordered glycans and loops at the epitope perimeter and improving Env binding.
One potential caveat of this study is that germline and UCA sequences are predicted sequences that may differ from the true unmutated precursor antibody sequences. It is important to note, however, that the most dramatic differences in dynamics we observed occurred in regions (CDRH2/FW3 and CDRL2/FW3) where we have high confidence in the true unmutated precursor sequence because these regions are encoded by their respective variable gene segments alleles, not at a gene segment junction. Furthermore, changes in dynamics within the CDRH2/FW3 of CAP256-VRC26 Fabs were observed among real antibody sequences from isolated B cells, which were not predicted. It is thus unlikely that errors in immunoglobulin sequence prediction would alter our observations.
If the date of isolation corresponded with the time that each B cell emerged, it would appear that variable domain flexibility fluctuated over time within the CAP256-VRC26 family. The CAP256-VRC26 family is not a single, successive lineage, however, and there are multiple sub-lineages, which undergo SHM in parallel. It is likely that just as contact residues are optimized by trial-and-error during affinity maturation, so too is the stability of the antibody variable domain. Our data suggest that increases in local stability are favorable for improved Fab-Env interactions and should be positively selected overall because of increased antigen capture and recruitment of T-cell help during affinity maturation.
In future work, it may be informative to examine the effect of stabilizing mutations within the CDRL2/FW3 region of VRC03 and VRC-PG04 and the CDRH2/FW3 of CAP256-VRC26, to test how these changes impact binding and neutralizing activity. It is conceivable that rational modification/optimization of antibody stability may be useful in optimizing therapeutic antibody binding (in addition to pharmacokinetic parameters such as half-life in circulation). Furthermore, these data, combined with Fab-Env co-crystal structures, provide indirect information on the regions of Env that present challenges to the maturing antibody, which should be considered for modification in vaccine immunogens. Overall, this work demonstrates that selective pressures during affinity maturation tailor the structure and structural dynamics of the antibody variable domain to enhance the ability of bNAbs to target the highly variable HIV-1 Env glycoprotein.
EXPERIMENTAL PROCEDURES
Antigen Binding Fragments (Fab)
Antibodies VRC03, VRC-PG04, and CAP256-VRC26 were isolated from HIV-infected individuals with broad neutralizing activity by single-cell sorting, and unmutated “germline” antibody sequences were inferred using next-generation sequencing (NGS) data from donor B cell repertoires (Doria-Rose et al., 2014; Wu et al., 2011). Unmutated VRC03 and VRC-PG04 sequences were approximated by reverting the regions of the variable domain corresponding to the heavy and light V-gene regions. Fab fragments were prepared from full-length IgGs expressed and purified as previously described (Wu et al., 2010).
Crystallization and Structure Determination
Mature and unmutated “germline heavy/germline light” (gHgL) VRC03 Fabs, were robotically screened for crystallization using 0.1 μL of protein and 0.1 μL of Hampton, Wizard and Precipitant-Synergy screen (Majeed et al., 2003). Hits were optimized manually. Data from single cryoprotected crystals were processed by HKL2000 (Otwinowski and Minor, 1997), and the structures determined by molecular replacement using Fab coordinates extracted from previously determined Fab-gp120 complex structures using Phenix (Adams et al., 2010). Methods for correlating crystal structures with HDX-MS data are presented in Supplemental Experimental Procedures.
Hydrogen-Deuterium Exchange with Mass Spectrometry (HDX-MS)
Fabs were buffer-exchanged into an identical phosphate buffered saline solution (150 mM NaCl, 10 mM Phosphate, 1 mM EDTA, 0.02% Sodium Azide, pH 7.4). Fab peptides generated by pepsin digestion were manually identified from tandem mass spectrometry fragmentation data and accurate mass. Deuteration of intact Fabs, including undeuterated, “zero” time-point, and fully-deuterated controls, was carried out as described previously (Guttman et al., 2012) with minor modifications. Exchanges were prepared with 7 μL of Fab at 1 mg/mL, 5 μL of a 20X-concentrated PBS solution (3M NaCl and 200 mM Phosphate) and 88 μL of D2O (Cambridge Isotope Labs). Fabs were allowed to exchange at room temperature for 3 sec, 12 sec, 1 min, 5 min, 30 min, 4 hr, and 20 hr prior to the addition of 100 μL quench solution (3M urea, 500 mM tris(2-carboxyethyl)phosphine (TCEP), 0.1% formic acid, pH 2.5) and pepsin digestion on ice using a 2:1 mass ratio of pepsin:Fab for 5 minutes prior to freezing in liquid nitrogen. Pepsin fragments were resolved and analyzed by LC-MS on a Waters Synapt mass spectrometer (Guttman et al., 2012). Exchanges for mature and unmutated antibodies were prepared and analyzed side-by-side. Data was collected in duplicate for independently prepared deuteration samples and analyzed using HX-Express 2 (Guttman et al., 2013; Weis et al., 2006). Glycan modeling in Figure 5C was performed using allosmod (Weinkam et al., 2012).
Octet Biolayer Interferometry (BLI)
Measurement of CAP256-VRC26 antibody family member binding to BG505.SOSIP.664 soluble HIV-1 Env trimer, purified as described elsewhere (Sanders et al., 2013), was performed on a FortéBio Octet Red96 (Pall Life Sciences). All proteins were diluted into BLI running buffer: 1X HBS-EP (GE Lifesciences) with 0.01% bovine serum albumin, and 0.02% sodium azide. For kinetic analysis of CAP256-VRC26 binding to BG505.SOSIP.664, CAP256-VRC26 Fabs were diluted to 10 μg/mL and were captured on ForteBio Anti-Human Fab-CH1 capture tips (7 min capture, 1000 RPM). After a 3 min baseline step, CAP256-VRC26-captured tips were dipped into seven wells containing BG505.SOSIP.664 (450 nM -7 nM, two-fold dilution series) as well as a buffer reference for baseline subtraction for a 3 min association step (1000 RPM). Dissociation was carried out in BLI running buffer for 3 min at 1000 RPM. Tips were regenerated between experiments using 10 mM glycine pH 1.5 (GE Lifesciences). Binding measurements were carried out in duplicate, with independently prepared dilution series. No binding was observed for CAP256-VRC26.UCA at the highest BG505.SOSIP.664 concentration tested (450 nM). The top five concentrations were fit for CAP256-VRC26.01, whereas the 225 nM – 14 nM curves were fit for CAP256-VRC26.03 and CAP256-VRC26.10. Kinetic data was analyzed globally in ForteBio Data Analysis software using 1:1 langmuir model fits.
HIGHLIGHTS.
X-ray crystallography and HDX-MS reveal SHM-induced changes in HIV-1 bNAbs
HDX-MS revealed changes in bNAb dynamics not apparent in crystal structures
Dynamics of sites peripheral to paratopes are tuned during SHM in two bNAb families
SHM changes bNAb structure and dynamics to improve complementarity with HIV-1 Env
Acknowledgments
We thank members of the Structural Biology Section, VRC, for comments on this manuscript. This work was supported by NIH grants R01-GM099989 and R21-AI112389 (KKL), T32-AI7509 (TMD), and the Bill and Melinda Gates Foundation Collaboration for AIDS Vaccine Discovery (CAVD) grant OPP1033102 (KKL, SLH). Support was also provided by the Intramural Research Program of the Vaccine Research Center, National Institute of Allergy and Infectious Diseases, NIH (JRM, PDK). Use of insertion device 22 (SER-CAT) at the Advanced Photon Source was supported by the U.S. Department of Energy, Basic Energy Sciences, Office of Science, under contract W-31-109-Eng-38.
Footnotes
AUTHOR CONTRIBUTIONS:
Conceptualization, TMD, PDK, SLH, and KKL; Methodology, TMD, MG; Validation, JG and MGJ; Formal Analysis, TMD, MG and CS; Investigation, TMD, MGJ, TZ, MG, and SM; Resources, JG, MGJ, YY, BZ, NADR, and JRM; Writing – Original Draft, TMD and KKL; Writing – Review & Editing, TMD, MG, CS, MGJ, JG, PDK, JRM, SLH and KKL; Visualization, TMD and KKL; Funding Acquisition, PDK, SLH and KKL; Supervision, PDK and KKL.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Adams PD, Afonine PV, Bunkóczi G, Chen VB, Davis IW, Echols N, Headd JJ, Hung LW, Kapral GJ, Grosse-Kunstleve RW, et al. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr D Biol Crystallogr. 2010;66:213–221. doi: 10.1107/S0907444909052925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adhikary R, Yu W, Oda M, Zimmermann J, Romesberg FE. Protein dynamics and the diversity of an antibody response. J Biol Chem. 2012;287:27139–27147. doi: 10.1074/jbc.M112.372698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bai Y, Milne JS, Mayne L, Englander SW. Primary structure effects on peptide group hydrogen exchange. Proteins. 1993;17:75–86. doi: 10.1002/prot.340170110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhiman JN, Anthony C, Doria-Rose NA, Karimanzira O, Schramm CA, Khoza T, Kitchin D, Botha G, Gorman J, Garrett NJ, et al. Viral variants that initiate and drive maturation of V1V2-directed HIV-1 broadly neutralizing antibodies. Nat Med. 2015;21:1332–1336. doi: 10.1038/nm.3963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doria-Rose NA, Schramm CA, Gorman J, Moore PL, Bhiman JN, DeKosky BJ, Ernandes MJ, Georgiev IS, Kim HJ, Pancera M, et al. Developmental pathway for potent V1V2-directed HIV-neutralizing antibodies. Nature. 2014;509:55–62. doi: 10.1038/nature13036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Engen JR. Analysis of Protein Conformation and Dynamics by Hydrogen/Deuterium Exchange MS. Anal Chem. 2009;81:7870–7875. doi: 10.1021/ac901154s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fera D, Schmidt AG, Haynes BF, Gao F, Liao HX, Kepler TB, Harrison SC. Affinity maturation in an HIV broadly neutralizing B-cell lineage through reorientation of variable domains. Proc Natl Acad Sci U S A. 2014;111:10275–10280. doi: 10.1073/pnas.1409954111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finton KAK, Friend D, Jaffe J, Gewe M, Holmes MA, Larman HB, Stuart A, Larimore K, Greenberg PD, Elledge SJ, et al. Ontogeny of recognition specificity and functionality for the broadly neutralizing anti-HIV antibody 4E10. PLoS Pathog. 2014;10:e1004403. doi: 10.1371/journal.ppat.1004403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foote J, Milstein C. Conformational isomerism and the diversity of antibodies. Proc Natl Acad Sci U S A. 1994;91:10370–10374. doi: 10.1073/pnas.91.22.10370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garces F, Lee JH, de Val N, Torrents de la Pena A, Kong L, Puchades C, Hua Y, Stanfield RL, Burton DR, Moore JP, et al. Affinity Maturation of a Potent Family of HIV Antibodies Is Primarily Focused on Accommodating or Avoiding Glycans. Immunity. 2015;43:1053–1063. doi: 10.1016/j.immuni.2015.11.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Georgiev IS, Rudicell RS, Saunders KO, Shi W, Kirys T, McKee K, O’Dell S, Chuang GY, Yang ZY, Ofek G, et al. Antibodies VRC01 and 10E8 neutralize HIV-1 with high breadth and potency even with Ig-framework regions substantially reverted to germline. J Immunol. 2014;192:1100–1106. doi: 10.4049/jimmunol.1302515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gorman J, Soto C, Yang MM, Davenport TM, Guttman M, Bailer RT, Chambers M, Chuang GY, DeKosky BJ, Doria-Rose NA, et al. Structures of HIV-1 Env V1V2 with broadly neutralizing antibodies reveal commonalities that enable vaccine design. Nat Struct Mol Biol. 2016;23:81–90. doi: 10.1038/nsmb.3144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guttman M, Kahn M, Garcia NK, Hu SL, Lee KK. Solution structure, conformational dynamics, and CD4-induced activation in full-length, glycosylated, monomeric HIV gp120. J Virol. 2012;86:8750–8764. doi: 10.1128/JVI.07224-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guttman M, Weis DD, Engen JR, Lee KK. Analysis of Overlapped and Noisy Hydrogen/Deuterium Exchange Mass Spectra. J Am Soc Mass Spectrom. 2013;24:1906–1912. doi: 10.1007/s13361-013-0727-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoxie JA. Toward an Antibody-Based HIV-1 Vaccine. Annu Rev Med. 2010;61:135–152. doi: 10.1146/annurev.med.60.042507.164323. [DOI] [PubMed] [Google Scholar]
- James LC, Roversi P, Tawfik DS. Antibody multispecificity mediated by conformational diversity. Science. 2003;299:1362–1367. doi: 10.1126/science.1079731. [DOI] [PubMed] [Google Scholar]
- Jardine J, Julien JP, Menis S, Ota T, Kalyuzhniy O, McGuire A, Sok D, Huang PS, MacPherson S, Jones M, et al. Rational HIV immunogen design to target specific germline B cell receptors. Science. 2013;340:711–716. doi: 10.1126/science.1234150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jimenez R, Salazar G, Yin J, Joo T, Romesberg FE. Protein dynamics and the immunological evolution of molecular recognition. Proc Natl Acad Sci U S A. 2004;101:3803–3808. doi: 10.1073/pnas.0305745101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Julien JP, Cupo A, Sok D, Stanfield RL, Lyumkis D, Deller MC, Klasse PJ, Burton DR, Sanders RW, Moore JP, et al. Crystal Structure of a Soluble Cleaved HIV-1 Envelope Trimer. Science. 2013;342:1477–1483. doi: 10.1126/science.1245625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klein F, Diskin R, Scheid JF, Gaebler C, Mouquet H, Georgiev IS, Pancera M, Zhou T, Incesu RB, Fu BZ, et al. Somatic mutations of the immunoglobulin framework are generally required for broad and potent HIV-1 neutralization. Cell. 2013a;153:126–138. doi: 10.1016/j.cell.2013.03.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klein F, Mouquet H, Dosenovic P, Scheid JF, Scharf L, Nussenzweig MC. Antibodies in HIV-1 vaccine development and therapy. Science. 2013b;341:1199–1204. doi: 10.1126/science.1241144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwong PD, Mascola JR. Human Antibodies that Neutralize HIV-1: Identification, Structures, and B Cell Ontogenies. Immunity. 2012;37:412–425. doi: 10.1016/j.immuni.2012.08.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y, Li H, Yang F, Smith-Gill SJ, Mariuzza RA. X-ray snapshots of the maturation of an antibody response to a protein antigen. Nat Struct Biol. 2003;10:482–488. doi: 10.1038/nsb930. [DOI] [PubMed] [Google Scholar]
- Majeed S, Ofek G, Belachew A, Huang CC, Zhou T, Kwong PD. Enhancing protein crystallization through precipitant synergy. Structure. 2003;11:1061–1070. doi: 10.1016/s0969-2126(03)00185-0. [DOI] [PubMed] [Google Scholar]
- Manivel V, Sahoo NC, Salunke DM, Rao KV. Maturation of an antibody response is governed by modulations in flexibility of the antigen-combining site. Immunity. 2000;13:611–620. doi: 10.1016/s1074-7613(00)00061-3. [DOI] [PubMed] [Google Scholar]
- Manivel V, Bayiroglu F, Siddiqui Z, Salunke DM, Rao KVS. The primary antibody repertoire represents a linked network of degenerate antigen specificities. J Immunol. 2002;169:888–897. doi: 10.4049/jimmunol.169.2.888. [DOI] [PubMed] [Google Scholar]
- Mascola JR, Montefiori DC. The Role of Antibodies in HIV Vaccines. Annu Rev Immunol. 2010;28:413–444. doi: 10.1146/annurev-immunol-030409-101256. [DOI] [PubMed] [Google Scholar]
- McLellan JS, Pancera M, Carrico C, Gorman J, Julien JP, Khayat R, Louder R, Pejchal R, Sastry M, Dai K, et al. Structure of HIV-1 gp120 V1/V2 domain with broadly neutralizing antibody PG9. Nature. 2011;480:336–343. doi: 10.1038/nature10696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mohan S, Kourentzi K, Schick KA, Uehara C, Lipschultz CA, Acchione M, Desantis ME, Smith-Gill SJ, Willson RC. Association energetics of cross-reactive and specific antibodies. Biochemistry. 2009;48:1390–1398. doi: 10.1021/bi801901d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moore PL, Gray ES, Sheward D, Madiga M, Ranchobe N, Lai Z, Honnen WJ, Nonyane M, Tumba N, Hermanus T, et al. Potent and broad neutralization of HIV-1 subtype C by plasma antibodies targeting a quaternary epitope including residues in the V2 loop. J Virol. 2011;85:3128–3141. doi: 10.1128/JVI.02658-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Otwinowski Z, Minor W. [20] Processing of X-ray diffraction data collected in oscillation mode. Methods in Enzymology. 1997;276:307–326. doi: 10.1016/S0076-6879(97)76066-X. [DOI] [PubMed] [Google Scholar]
- Pancera M, Shahzad-Ul-Hussan S, Doria-Rose NA, McLellan JS, Bailer RT, Dai K, Loesgen S, Louder MK, Staupe RP, Yang Y, et al. Structural basis for diverse N-glycan recognition by HIV-1-neutralizing V1-V2-directed antibody PG16. Nat Struct Mol Biol. 2013;20:804–813. doi: 10.1038/nsmb.2600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanders RW, Derking R, Cupo A, Julien J-P, Yasmeen A, de Val N, Kim HJ, Blattner C, la Peña de AT, Korzun J, et al. A Next-Generation Cleaved, Soluble HIV-1 Env Trimer, BG505 SOSIP.664 gp140, Expresses Multiple Epitopes for Broadly Neutralizing but Not Non-Neutralizing Antibodies. PLoS Pathog. 2013;9:e1003618. doi: 10.1371/journal.ppat.1003618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scharf L, West AP, Gao H, Lee T, Scheid JF, Nussenzweig MC, Bjorkman PJ, Diskin R. Structural basis for HIV-1 gp120 recognition by a germ-line version of a broadly neutralizing antibody. Proc Natl Acad Sci U S A. 2013;110:6049–6054. doi: 10.1073/pnas.1303682110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scheid JF, Mouquet H, Ueberheide B, Diskin R, Klein F, Oliveira TYK, Pietzsch J, Fenyo D, Abadir A, Velinzon K, et al. Sequence and structural convergence of broad and potent HIV antibodies that mimic CD4 binding. Science. 2011;333:1633–1637. doi: 10.1126/science.1207227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt AG, Xu H, Khan AR, O’Donnell T, Khurana S, King LR, Manischewitz J, Golding H, Suphaphiphat P, Carfi A, et al. Preconfiguration of the antigen-binding site during affinity maturation of a broadly neutralizing influenza virus antibody. Proc Natl Acad Sci U S A. 2013;110:264–269. doi: 10.1073/pnas.1218256109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skinner JJ, Lim WK, Bédard S, Black BE, Englader SW. Protein dynamics viewed by hydrogen exchange. Protein Sci. 2012;21:996–1005. doi: 10.1002/pro.2081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomson CA, Bryson S, McLean GR, Creagh AL, Pai EF, Schrader JW. Germline V-genes sculpt the binding site of a family of antibodies neutralizing human cytomegalovirus. EMBO J. 2008;27:2592–2602. doi: 10.1038/emboj.2008.179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Victora GD, Nussenzweig MC. Germinal centers. Annu Rev Immunol. 2012;30:429–457. doi: 10.1146/annurev-immunol-020711-075032. [DOI] [PubMed] [Google Scholar]
- Wales TE, Poe JA, Emert-Sedlak L, Morgan CR, Smithgall TE, Engen JR. Hydrogen Exchange Mass Spectrometry of Related Proteins with Divergent Sequences: A Comparative Study of HIV-1 Nef Allelic Variants. J Am Soc Mass Spectrom. 2016 doi: 10.1007/S23361-016-1365-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang F, Sen S, Zhang Y, Ahmad I, Zhu X, Wilson IA, Smider VV, Magliery TJ, Schultz PG. Somatic hypermutation maintains antibody thermodynamic stability during affinity maturation. Proc Natl Acad Sci U S A. 2013;110:4261–4266. doi: 10.1073/pnas.1301810110. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Wedemayer GJ, Patten PA, Wang LH, Schultz PG, Stevens RC. Structural insights into the evolution of an antibody combining site. Science. 1997;276:1665–1669. doi: 10.1126/science.276.5319.1665. [DOI] [PubMed] [Google Scholar]
- Weinkam P, Pons J, Sali A. Structure-based model of allostery predicts coupling between distant sites. Proc Natl Acad Sci U S A. 2012;109:4875–4880. doi: 10.1073/pnas.1116274109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weis DD, Engen JR, Kass IJ. Semi-automated data processing of hydrogen exchange mass spectra using HX-Express. J Am Soc Mass Spectrom. 2006;17:1700–1703. doi: 10.1016/j.jasms.2006.07.025. [DOI] [PubMed] [Google Scholar]
- West AP, Diskin R, Nussenzweig MC, Bjorkman PJ. Structural basis for germ-line gene usage of a potent class of antibodies targeting the CD4-binding site of HIV-1 gp120. Proc Natl Acad Sci U S A. 2012;109:E2083–E2090. doi: 10.1073/pnas.1208984109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu X, Zhou T, Zhu J, Zhang B, Georgiev I, Wang C, Chen X, Longo NS, Louder M, McKee K, et al. Focused Evolution of HIV-1 Neutralizing Antibodies Revealed by Structures and Deep Sequencing. Science. 2011;333:1593–1602. doi: 10.1126/science.1207532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu X, Yang ZY, Li Y, Hogerkorp CM, Schief WR, Seaman MS, Zhou T, Schmidt SD, Wu L, Xu L, et al. Rational design of envelope identifies broadly neutralizing human monoclonal antibodies to HIV-1. Science. 2010;329:856–861. doi: 10.1126/science.1187659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin J, Beuscher AE, Andryski SE, Stevens RC, Schultz PG. Structural plasticity and the evolution of antibody affinity and specificity. Journal of Molecular Biology. 2003;330:651–656. doi: 10.1016/s0022-2836(03)00631-4. [DOI] [PubMed] [Google Scholar]
- Zhou T, Georgiev I, Wu X, Yang ZY, Dai K, Finzi A, Do Kwon Y, Scheid JF, Shi W, Xu L, et al. Structural Basis for Broad and Potent Neutralization of HIV-1 by Antibody VRC01. Science. 2010;329:811–817. doi: 10.1126/science.1192819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou T, Zhu J, Wu X, Moquin S, Zhang B, Acharya P. Multidonor Analysis Reveals Structural Elements, Genetic Determinants, and Maturation Pathway for HIV-1 Neutralization by VRC01-Class Antibodies. Immunity. 2013;39:245–258. doi: 10.1016/j.immuni.2013.04.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zimmermann J, Oakman EL, Thorpe IF, Shi X, Abbyad P, Brooks CL, Boxer SG, Romesberg FE. Antibody evolution constrains conformational heterogeneity by tailoring protein dynamics. Proc Natl Acad Sci U S A. 2006;103:13722–13727. doi: 10.1073/pnas.0603282103. [DOI] [PMC free article] [PubMed] [Google Scholar]