Abstract
Antibodies against hexon, the major coat protein of adenovirus (Ad), are an important component of the neutralizing activity in serum from naturally infected humans and experimentally infected animals. The mechanisms by which antihexon antibodies neutralize the virus have not been defined. As a model system, murine monoclonal antibodies raised against Ad type 5 (Ad5) were screened for antihexon binding and neutralization activity; one monoclonal antibody, designated 9C12, was selected for further characterization. The minimum ratio of 9C12 to Ad5 required for neutralization was 240 antibody molecules per virus particle, or 1 antibody per hexon trimer. Analysis of antibody-virus complexes by dynamic light scattering and negative-stain electron microscopy (EM) showed that the virus particles were coated with electron-dense material but not aggregated at neutralizing ratios. Cryo-EM image reconstruction of the antibody-virus complex showed that the surface of the virus particle was covered by a meshwork of 9C12 antibody density, consistent with bivalent binding at multiple sites. Confocal analysis revealed that viral attachment, cell entry, and intracellular transport to the nuclear periphery still occur in the presence of neutralizing levels of 9C12. A model is presented for neutralization of Ad by an antihexon antibody in which the hexon capsid is cross-linked by antibodies, thus preventing virus uncoating and nuclear entry of viral DNA.
Infection by adenovirus (Ad) elicits a strong antibody (Ab) response against viral proteins, both in humans and in experimental animals (12, 24, 32, 33). Neutralizing Ab (NAb) responses to Ad are directed at components of the virion surface, primarily against fiber, penton base, and hexon (24, 33). Fiber and penton base proteins, present at the vertices of the capsid, are involved in cell attachment and entry (4, 5, 27, 35, 45). Hexon, the major component of the icosahedral virus particle, comprises the facets of the virion and constitutes the bulk of the icosahedral capsid.
NAb responses to Ads of subgroup C, including Ad type 2 (Ad2) and Ad5, have been characterized extensively. Polyclonal and monoclonal antifiber NAbs have been shown to block binding of the Ad5 fiber knob to its cellular attachment receptor, the coxsackievirus-Ad receptor (13, 15, 53). Polyclonal and monoclonal antifiber NAbs have also been shown to aggregate virions by cross-linking fibers on separate virus particles (13-15). NAbs against the penton base have been demonstrated in serum (24, 33). Epitope mapping of antipenton base NAbs by phage display showed that most of the antipenton base NAbs were directed against a variety of epitopes, in addition to the integrin-binding RGD motif (24). In contrast to the antifiber Abs, no antipenton base monoclonal antibodies (MAbs) have been identified that recapitulate the neutralizing activity associated with polyclonal antipenton base NAbs (23).
One antipenton base MAb specific for the integrin-binding RGD peptide loop was found to be neutralizing only in the Fab fragment form and not as an intact immunmoglobulin G (IgG) (43). A cryo-electron microscopy (cryo-EM) study of the Ad:Fab complex suggested that epitope mobility, together with steric hindrance from the Ad fiber and a few bound IgG molecules, likely prevents binding of IgG to all five RGD sites on the penton base, thus precluding neutralization. In contrast to IgG molecules, the neutralizing Fab fragments are narrower and may bind to all five RGD sites simultaneously, thus neutralizing the virus by blocking the interaction with αv integrins and preventing virus internalization.
Antihexon NAbs are a major component of the neutralizing activity in humans and in experimentally infected mice (24, 33, 50). Antihexon NAbs have been described previously (46, 49, 52, 53) that allow virus internalization without concomitant virus-mediated gene expression; however, the mechanisms by which antihexon NAbs neutralize the virus have not been clearly defined. Work by Luftig and Weihing in 1975 (28) suggested that hexon was involved in intracellular transport of the virus particle to the nucleus. Upon entry of Ad into the cytoplasm, hexon has been shown to associate with HSP70 and HSC70 during migration of the capsid to the nucleus (30, 37). Both HSC70 and HSP70 play roles in vesicle recycling and protein transport consistent with their reported interactions with hexon (29, 55). Following entry, the virus capsid, containing hexon and penton base, remains largely intact and protects the viral DNA until the particle docks at the nuclear pore (19). Recently, uncoating of the Ad capsid at the nuclear periphery has been studied and shown to require interactions between hexon, CAN/NUP214, histone H1, and histone H1-associated import factors (18, 47).
To characterize the mechanism of antihexon Ab neutralization, we studied the interaction of a mouse monoclonal antihexon NAb, 9C12, with wild-type Ad5 and an Ad5-green fluorescent protein (GFP) reporter vector. We analyzed the effect of 9C12 Ab binding on virus attachment, entry, and intracellular transport. The data showed that 9C12 remains bound to Ad5 following internalization and that the virus-Ab complex accumulates complexes accumulate at the nuclear periphery in a manner analogous to that of nonneutralized Ad5. Cryo-EM reconstruction of the Ad5-9C12 complex showed that the capsid is coated by a meshwork of bivalently bound Ab molecules. Together, these results suggest that the critical neutralization event likely occurs at the nuclear periphery and involves blocking of virus uncoating, either by physically impeding capsid dissociation or by preventing essential interactions between the Ad capsid and elements of the nuclear import machinery.
MATERIALS AND METHODS
Ab isolation and purification.
MAbs were generated as described previously (3) with Ad5 as the immunogen. Hybridoma cell lines expressing and screened by enzyme-linked immunosorbent assay with purified hexon protein. The MAbs were cultured, and the resultant Abs were then further screened for binding to hexon by Western blot analysis. One Ab from the fusion, 9C12, was selected based on its ability to neutralize Ad5. A second Ab, 27F11, was selected as an isotype-matched nonneutralizing control Ab. Hybridomas 9C12 and 27F11 were cultured, and the Abs were purified by protein G-agarose column purification (QED Biosciences, Inc., San Diego, Calif.).
Western blot analysis.
Recombinant Ad5 (rAd5) (107 particles) was lysed in sodium dodecyl sulfate (SDS) sample buffer, half of the samples were heated to 65°C for 10 min, and then all samples were separated by SDS-polyacrylamide gel electrophoresis (SDS-PAGE) on a precast 1.5-mm NUPage bis-Tris gel with 4 to 12% acrylamide (Invitrogen, Carlsbad, Calif.) and transferred to a polyvinylidene difluoride membrane. The Abs were all diluted in TBST (0.15 M NaCl, 0.01 M Tris-HCl [pH 7.4], 0.05% Tween 20) (Sigma, St. Louis, Mo.) as follows: 9C12 and 27F11 Abs were diluted to a concentration of 0.4 mg/ml, rabbit anti-Ad5 purified IgG was diluted 1:500 (Access Biomedical, San Diego, Calif.), rabbit anti-mouse kappa Ab (Rockland, Gilbertsville, Pa.) was diluted 1:500, and the horseradish peroxidase-conjugated goat anti-rabbit Ab was diluted 1:5,000 (Amersham Biosciences Corp., Piscataway, N.J.). Western blots were analyzed by enhanced chemiluminescence with the SuperSignal kit (Pierce Biotechnology, Inc., Rockford, Ill.).
Immunoprecipitation of Ad5.
rAd5 (2 × 109 particles) was diluted in FAK buffer (50 mM HEPES, 1% Triton X-100, 20 mM Tris-Cl [pH 7.5], 50 mM NaCl, 2.5 mM EDTA) and mixed with 2.5 μg of purified Ab. The concentration of rAd5 particles was determined as described previously (40) and calculated by the American Type Culture Collection Ad5 reference standard (25). The Ad and Ab mixtures were mixed by inversion for 1 h at 4°C. Protein G-agarose beads were added to each Ad5-Ab mixture and mixed as described above. The beads were centrifuged and washed repeatedly in FAK buffer. The bound Ab and virus were eluted by the addition of SDS-PAGE loading buffer supplemented with β-mercaptoethanol. Samples were analyzed by Western blotting with a rabbit anti-Ad5 polyclonal Ab as described below.
Neutralization assay.
Neutralization of Ad-mediated gene expression was assayed by a previously described in vitro neutralization assay (33). In brief, Ab was serially diluted, mixed with a fixed amount of a replication-defective Ad vector expressing GFP (rAdGFP) (4.8 × 107 particles) (51) in each well of a 96-well plate, and incubated at 37°C for 1 h. The mixture was then transferred to wells containing 1 × 104 HeLa cells/well. The cells were incubated with the Ad-containing mixture for a period of no less than 18 h. Fluorescence at 488 nm was assayed with a CytoFluor series 4000 reader (Applied Biosystems, Foster City, Calif.) following excitation at 405 nm. Each sample was analyzed in triplicate. Titration curves were plotted with the mean values and standard deviations from triplicate samples.
Photon correlation spectroscopy.
Ad aggregation was monitored with the N4 Plus (Beckman Coulter, Inc., Hialeah, Fla.). Ad was diluted in Dulbecco's phosphate-buffered saline (PBS) to a final concentration of 2.5 × 1010 particles/ml, and light-scattering measurements were acquired at 90° for 340 s at 20°C with the multimodal industry standard CONTIN algorithm. All samples of virus were assayed prior to the addition of Ab to ensure that a uniform monodispersed peak was detected. The virus was subsequently incubated with Ab at neutralizing levels for 1 h at 37°C. The samples were assayed at 20°C for their ability to shift the monodispersed peak or generate additional peaks. For Ad5 bound by 9C12 anti-mouse IgG1, Ab was added, incubated for 1 h at 37°C, and then assayed as described previously. To control for nonspecific changes in apparent particle size after the addition of Ab, each experiment was repeated with 25 μl of a calibration solution that contained sufficient inert latex beads of approximately 100 nm (catalogue no. 6602336; Beckman Coulter, Inc.) to recapitulate the signal intensity generated by the Ad particles.
EM.
Ad or Ad/Ab complexes were spotted on carbon-Formvar-coated copper grids briefly prerinsed in chloroform (catalogue no. 01821; Ted Pella, Redding, Calif.). The samples were allowed to bind for 10 min, then excess liquid was wicked away, and the samples were briefly air dried. Samples were stained in 4% uranyl acetate and observed with an electron microscope (JOEL 120 EX-II or Philips CM10).
Cryo-EM.
Wild-type Ad5 (1.5 × 1011 particles/ml in 1× PBS with 0.75% sucrose) was incubated at room temperature with 9C12 for 20 min at a ratio of 720 Ab molecules per virus particle, equivalent to 1 Ab molecule per hexon monomer. Cryo-EM sample grids of the Ad5-9C12 complex were prepared immediately after the incubation period.
Frozen hydrated specimen grids were made with carbon grids with holes as described previously (1). Briefly, 3 μl of either the Ad5 or the Ad5-9C12 suspension was applied to a glow-discharged carbon film with holes supported on a 400-mesh copper grid. The grid was allowed to dry briefly, and then an additional 3-μl aliquot of sample was applied. Excess solution was blotted off, and the grid was plunged into ethane slush chilled by liquid nitrogen. The frozen sample grid was then either transferred to a Gatan 626 cryoholder and maintained at liquid nitrogen temperature during image collection or stored under liquid nitrogen for future use. An FEI/Philips CM120 (120 kV, LaB6 filament) transmission EM (TEM) with cryoaccessories was used for the microscopy. Digital cryo-EMs were recorded with a Gatan slow-scan charge-coupled device camera (1,024 by 1,024 by 1,024 pixels; YAG scintillator). Images of Ad5 and the Ad5-9C12 complex were taken with a range of underfocus values (−1.5, −1.0, and −0.7 μm) under low-dose conditions (<20 electrons/Å2) and at a nominal magnification of ×45,000. Calibration with a catalase crystal indicates that this magnification yields a pixel size of 0.41 nm on the molecular scale.
Image processing and three-dimensional reconstruction.
Selection of particle images into individual 320- by 320-pixel fields was done interactively with the QVIEW program (41). Subsequent image processing was performed with the IMAGIC-5 software package (48). The initial orientation of each particle image was determined with defined projections of a preexisting Ad5 reconstruction (10). After the first round of refinement, the reconstruction obtained in the previous round was first masked to select the icosahedral capsid and then used as a search model. The particle images collected at various underfocus values were combined after correction for the contrast transfer function of the microscope (9). Icosahedral symmetry was imposed throughout the refinement process. The final three-dimensional reconstructions of Ad5 and the Ad5-9C12 MAb complex were calculated from 579 and 671 particle images, respectively. The resolution of the icosahedral capsid and bound MAb density were assessed by the Fourier shell correlation method using the 0.5 threshold criterion (31). For the purpose of the resolution assessment, inner and outer soft Gaussian masks were applied so that the resolution of the icosahedral portion of the structures could be assessed independently from the rest of the density map (42). The final resolutions of the Ad5 and Ad5-9C12 MAb reconstructions were calculated to be 2.4 and 2.1 nm, respectively; both density maps are shown filtered to these resolutions.
To separate the bound MAb density from that of Ad5 in the Ad5-9C12 reconstruction, two approaches were tried. A difference map was calculated by subtracting the Ad5 reconstruction from that of Ad5-9C12. Also, the MAb density in the complex reconstruction was selected by applying an inner and outer spherical mask (using radii of 45 and 49 nm). Both approaches yielded essentially the same MAb density with equivalent major bivalent binding combinations. All of the image processing and graphical analysis was performed on HP/Compaq/Digital Equipment Corporation alpha workstations running Tru64 Unix. Isosurface density representations, as well as atomic coordinate Cα polygon representations, were generated with the AVS visualization package (Advanced Visual Systems, Inc., Waltham, Mass.).
Docking of atomic coordinates into cryo-EM density.
The crystallographic coordinates of the Ad5 hexon (Protein Data Bank [PDB] code 1P30) (36) were docked into the reconstructed Ad5 cryo-EM density with the CoLoRes search tool in the Situs 2.0 software package (8). To save computational time, one-eighth of the Ad5 reconstruction was used as the density input. Docked coordinates were found for one copy of each of the four independent hexons in the capsid, and then icosahedral symmetry was applied to generate a full capsid of hexons. The quality of the CoLoRes docking alignment was assessed visually by superimposing the Ad5 reconstruction with the density generated from the calculated capsid of hexons filtered to the same resolution (2.4 nm).
The CoLoRes search tool was also tested for docking the crystallographic coordinates of IgG1κ (PDB code 1IGY) (21) into the reconstructed MAb density. However, presumably because of differences in the IgG hinge angle as well as distortions in one of the two Fab arms in the cryo-EM density, better results were obtained by manual docking with the AVS visualization package. The crystal structures of several other IgGs were also fit within the cryo-EM MAb density: 1AD9 (2), 1GGI (34), 1GIG (6), and 1HZH (38). However, IgG1κ (1IGY) gave the best visual fit and it is also the only one which is of the same IgGκ class (1κ) as 9C12.
Preparation of Cy3B-labeled Ad.
rAdGFP (6.2× 1011 particles) was diluted to 900 μl in PBS, and the pH was adjusted with 50 μl of 1 M sodium carbonate buffer to a pH of 9.3. Cy3B dye (Amersham Biosciences) was solubilized in 200 μl of dimethyl sulfoxide, and 20 μl of the solubilized dye was added to the diluted virus and mixed by inversion. The labeling reaction was carried out for 30 min at room temperature according to the manufacturer's instructions. A total of 100 μl of 1 M glycine in PBS was added to neutralize any free unreacted dye, and the reaction mixture was incubated for an additional 10 min at room temperature.
A Slide-A-Lyzer 10K dialysis cassette (Pierce Biotechnology, Inc.) was used to adjust the pH of the Cy3B-rAdGFP to a pH of 7.4 with several changes of 500 ml of vPBS (3% sucrose, 2 mM MgCl2, 1× PBS) at 4°C over 16 h. The Cy3B-rAdGFP virus was then removed from the dialysis chamber, and aliquots were saved for particle analysis and infectivity comparisons with unlabeled rAd-GFP. All Cy3B-rAdGFP virus was assayed in the neutralization assay to confirm that infectivity was maintained following labeling and that Ab neutralization occurred over a concentration range similar to that of unlabeled virus.
Confocal microscopy.
HeLa cells were grown on 18-mm glass coverslips in 12-well dishes. Cy3B-rAdGFP (1.5 × 1010 particles in 300 μl of medium) was incubated at 37°C in medium with or without 9C12 Ab for 1 h. The cells were cooled on a wet ice bath for 20 min, and the medium was removed immediately prior to the addition of Cy3B-rAdGFP or Cy3B-rAdGFP neutralized with 9C12. Virus was added to the coverslips, incubated on ice for 1 h, and then rinsed with three 1-ml washes of PBS. Cells were either fixed in 300 μl of 4% formalin in dPBS for 20 min on an orbital shaker at room temperature or transferred to 12-well dishes containing 2 ml of fresh complete medium at 37°C and incubated at 37°C for 1 h overnight before the cells were washed and fixed. Fixed cells were neutralized with 500 μl of 0.1 M glycine in PBS for 5 min and then stored at 4°C in PBS for immunofluorescent staining. Cells were blocked for 1 h at room temperature in 2× PBS block (PBS containing 2% bovine serum albumin, 300 μg of normal goat serum/ml, and 0.2% Triton X-100). Triton X-100 was omitted from the blocking solution for studies of unpermeabilized cells. Alexa Fluor 488-rabbit anti-mouse IgG1 (Molecular Probes, Eugene, Oreg.) was diluted 1:500 in 1:1 block to PBS. Each well received 300 μl of diluted Ab that was allowed to bind for 1 h at room temperature with gentle shaking in the dark. The wells were then rinsed with wash buffer (0.1% Triton X-100 in PBS). PBS without detergent was used for studies of unpermeabilized cells. Cells were rinsed with 1:1 PBS block and 300 μl of 4′,6′-diamidino-2-phenylindole (DAPI) (1 μg/ml) was added to each well for 5 min. Coverslips were rinsed in PBS, air dried, and then mounted on glass slides with VectaShield (Vector Laboratories, Burlingame, Calif.). Cells were imaged with the CARV spinning disk confocal system (ATTO Biosciences, Rockville, Md.) and mounted on an Olympus TE-2000 inverted microscope (Olympus Optical, Tokyo, Japan) with a tetramethyl rhodamine isocyanate (TRITC) filter (excitation, 555 nm; emission, 605 nm), a fluorescein isothiocyanate (FITC) filter (excitation, 484 nm; emission, 517 nm), and a DAPI filter (excitation, 395 nm; emission, 457 nm). Confocal fluorescent images were captured starting at the surface of the coverslip and extending to the highest portion of cells within the field of view. Optical planes of either 0.4 or 0.8 μm above the coverslip were chosen for quantitative image analysis to include both the maximum cytoplasmic area and a cross-section of the nucleus. Bright-field images were captured with differential interference contrast (DIC) imaging. Images were captured, analyzed, and overlaid using the MetaMorph 6.02 Imaging System (Universal Imaging, Downingtown, Pa.). MetaMorph software was used to quantify fluorescent signals per cell area, with samples of 20 to 22 cells for each treatment group. Quantification of fluorescent signals in nuclear and cytoplasmic regions was performed on individual optical sections (z sections). The MetaMorph region tool was used to manually outline both the outer edge of the cell based on the DIC image and the periphery of the nucleus with the DAPI image. Fluorescent signals were quantified in two cell areas: one area consisted of the nucleus plus nuclear periphery, and the second area consisted of the cytoplasm plus the border of the plasma membrane included in the 0.4-μm-thick plane of the z section. The graphing and statistical analysis package in GraphPad Prism, version 4.0 (San Diego, Calif.), was used to analyze the quantified fluorescence data. Results were expressed as mean and standard error. One-sided t tests were used for analysis of statistical significance.
RESULTS
MAb 9C12 binds hexon and neutralizes virus particles.
Mouse MAbs raised against Ad5 were screened for binding to the virion protein hexon and for virus neutralization. Two IgG1 Abs from a single hybridoma fusion, designated 9C12 and 27F11, were selected for purification and characterization. As shown in Fig. 1A, both 9C12 and 27F11 specifically bound to hexon protein from Ad5 particles denatured with 1% SDS in a Western blot assay, but this Ab reactivity was abolished when the SDS-disrupted viral particles were heated to 65°C for 10 min prior to electrophoresis. Although SDS treatment results in disruption of the Ad5 particle, the hexon trimers released by SDS are resistant to complete denaturation unless heated (16); unheated hexon trimers migrated at ≥200 kDa, whereas heat-denatured hexon monomers migrated at 110 kDa. The reactivity of affinity-purified anti-Ad polyclonal Abs against heated and unheated SDS-disrupted virions is shown for comparison. The Western blot results suggested that both 9C12 and 27F11 bind to conformation-specific epitopes on Ad5 hexons.
FIG. 1.
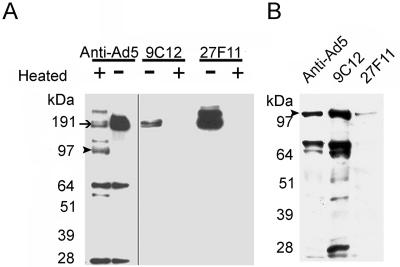
Western analysis and immunoprecipitation of Ad5. (A) SDS-PAGE analysis showed that 9C12 and 27F11 bound specifically to hexon (arrow) while purified polyclonal anti-Ad5 IgGs recognized numerous Ad5 proteins. Heat denaturation (+) of Ad5 abolished binding of both 9C12 and 27F11 to hexon while polyclonal anti-Ad5 IgGs bound to both heat-denatured hexon (∼110 kDa; arrowhead) and residual native hexon (∼190 kDa). (B) Immunoprecipitation of Ad5 proteins demonstrated that mouse anti-Ad5 antisera bound to hexon as well as the fiber-penton base proteins and pulled them out of solution upon the addition of a protein G mixture. 9C12 pulled out the complete set of proteins normally associated with intact Ad5 particles, while 27F11 only pulled down hexon. 9C12 likely bound to the surface of hexon and pulled intact Ad5 particles out of solution, while 27F11 likely bound to free hexon, hence the diminished hexon signal.
To determine whether 9C12 or 27F11 bound to intact virus particles, a pulldown-immunoblotting assay was developed to distinguish binding to intact virus from dissociated proteins. The MAbs were allowed to bind to Ad5, and then the resulting immune complexes were pulled down with protein G-agarose beads. Bound proteins were eluted from washed beads as described in Materials and Methods and then subjected to SDS-PAGE and Western blotting. A mouse polyclonal anti-Ad5 antiserum served as a positive control in the pulldown step. Rabbit polyclonal antiserum against Ad5 was used in the Western blot step as a probe for Ad proteins. As shown in Fig. 1B, a pulldown assay with 9C12 resulted in detection of multiple virion proteins, suggesting that 9C12-bound hexon associated with intact virus particles. The relative abundance of viral protein pulled down by 9C12 was analogous to that expected from intact virus particles (Fig. 1A), while the polyclonal Ab yielded significantly less of the minor 60- and 28-kDa viral proteins. The 27F11 pulldown assay yielded only a faint hexon band, presumably due to a small amount of dissociated hexon in the virus preparation. The results shown in Fig. 1 suggest that 9C12 bound to a conformation-dependent epitope on the surface of the Ad particle and that 27F11, despite also binding to hexon, likely bound an epitope that was not accessible on the exposed surface of intact Ad particles.
The ability of 9C12 to neutralize Ad was analyzed with rAdGFP (33). As shown in Fig. 2, 9C12 neutralization of rAdGFP generated a titration curve that demonstrated a rapid reduction in neutralization over 5 twofold serial dilutions. Interestingly, 9C12 Ab reduced rAdGFP fluorescence by ≥95%, but a residual GFP signal remained detectable, even at 10-fold-higher Ab concentrations (data not shown). The residual signal was reproducible, and the difference from baseline fluorescence was statistically significant. In contrast, treatment with anti-Ad5 polyclonal Ab resulted in 100% inhibition of GFP expression, which could be titrated out over 8 twofold serial dilutions. The control antihexon MAb 27F11 did not neutralize rAdGFP at any concentration tested. Quantitative data from the titration experiment was used to calculate the minimum ratio required for maximum virus neutralization by 9C12. It was calculated from the concentration of purified 9C12 Ab and purified rAdGFP particles, which was found to be 240 Ab molecules per virus particle, equivalent to one Ab per hexon trimer. Interestingly, neutralization of rAdGFP was also tested with the Fab fragment of 9C12 (data not shown) and did not neutralize rAdGFP, even at a 100-fold molar excess of Fab compared to intact Ab at 95% neutralization. The observation that only the intact form of 9C12 can neutralize suggests that bivalent binding might be required for neutralization.
FIG. 2.
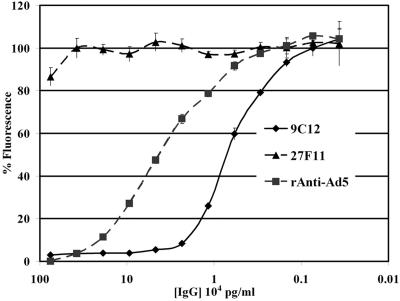
Ad neutralization assay. 9C12 bound to rAdGFP and exhibited a concentration-dependent ability to neutralize rAdGFP-mediated GFP fluorescence. 27F11, a related antihexon MAb, did not neutralize rAdGFP-mediated fluorescence. Purified polyclonal anti-Ad5 IgGs were also able to neutralize rAdGFP.
9C12 does not aggregate virus particles.
Photon correlation spectroscopy was used to determine the particle size of Ad5-9C12 complexes in solution and to determine if virion aggregation was the mechanism of 9C12 neutralization. As shown in Fig. 3A, measurement of untreated Ad5 in solution gives a mean particle diameter of ∼130 nm. The addition of 9C12 to Ad5 does not appreciably alter the size of Ad5 (Fig. 3A). To confirm binding of 9C12 to Ad5, an affinity-purified rabbit Ab against mouse heavy-chain Ig was added (Fig. 3B). The addition of anti-heavy-chain Abs shifted the apparent particle diameter of a portion of the Ad5-9C12 complexes from ∼130 nm to >1,000 nm, suggesting aggregation of Ad5-9C12 complexes and confirming that 9C12 did bind to Ad5. Addition of rabbit anti-mouse Ig alone to Ad5 did not shift the apparent particle diameter of Ad5 (data not shown).
FIG. 3.
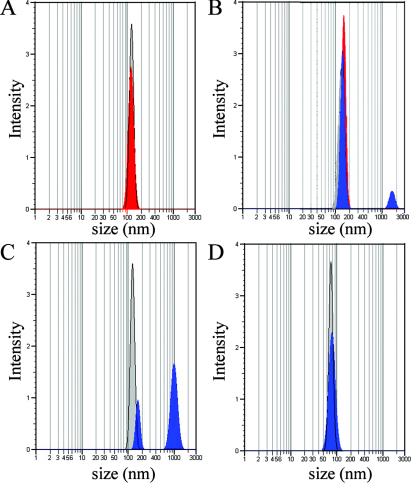
Photon correlation spectroscopy of Ad5 in solution. (A) Ad5 generated a single peak when diluted in PBS (gray). The addition of 9C12 produced only a small shift of the peak (red), indicating no aggregation of Ad5 particles. (B) The addition of an anti-mouse Ab (blue) to the Ad5-9C12 Ab mixture increased the apparent size of a small percentage of Ad5 particles from ∼130 nm to >1,000 nm. (C) The apparent size of Ad5 particles also increased following the addition of purified polyclonal anti-Ad5 IgGs. The addition of anti-Ad5 Abs (blue) (D) to the control 100-nm latex beads had no impact on the apparent size of the beads in solution. All graphs are representative of four independent experiments.
A solution of 9C12 without Ad5 was tested and shown to not form large particles following the addition of the anti-mouse heavy-chain Ab (data not shown). A shift in particle size was also produced by the addition of affinity-purified rabbit polyclonal anti-Ad5 IgGs to Ad5 (Fig. 3C), confirming that the polyclonal anti-Ad5 could aggregate Ad5. To confirm that the increase in particle diameter observed in Fig. 3B and C was dependent on Ab interactions involving Ad5, 100-nm latex beads were substituted for Ad5, and the measurements were repeated. The particle diameter of the beads was not altered by polyclonal anti-Ad5 (Fig. 3D), by 9C12 alone (data not shown), or by 9C12 in combination with anti-heavy-chain Ab (data not shown).
To confirm the photon correlation spectroscopy data, negatively stained Ad5 and Ad5-Ab complexes were imaged by TEM. The negative-stain TEM analysis demonstrated that the samples of Ad5 alone (Fig. 4A) and Ad5 complexed with 9C12 at a neutralizing ratio (Fig. 4B) did not show aggregation. The appearance of some pairs of particles (Fig. 4B) suggests that 9C12 may have caused dimerization of Ad5. Since sample drying for TEM studies can produce particle aggregation, the apparent dimers observed in the results shown in Fig. 4B may be artifacts of sample preparation. This interpretation is supported by the photon correlation spectroscopy data for particles in solution (Fig. 3), which did not show a shift in the Ad5 peak in the presence of 9C12.
FIG. 4.
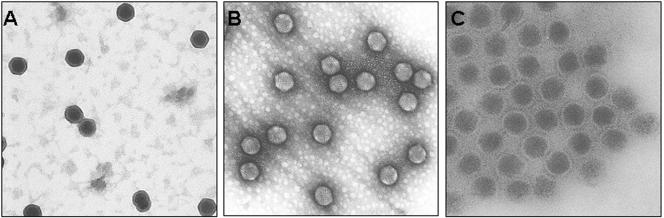
TEMs of negatively stained Ad5. (A) Micrograph of Ad5. (B) Micrograph of Ad5 complexed with 9C12 at a neutralizing ratio. (C) Micrograph of Ad5 complexed with polyclonal anti-Ad5 IgGs. Note the aggregated nature of this sample. All samples are stained with uranyl acetate. These images are representative of at least three fields of view from three independent grids. Magnification, ×55,000.
The sample of Ad5 complexed with polyclonal anti-Ad5 IgGs displayed significant aggregation (Fig. 4C). Note that unlike the Ad5 and Ad5-9C12 grids, the Ad5-polyclonal anti-Ad5 IgG grids were largely devoid of virus except for that contained within the large aggregates. Interestingly, binding of the uranyl acetate to the viral capsid appeared to be greatly diminished in those particles that were incubated with 9C12 (Fig. 4B), suggesting that access to the capsid surface was diminished.
Cryo-EM analysis of the Ad5-9C12 complex shows Ab coating of the virus.
Cryo-EM and three-dimensional image reconstruction were performed to visualize the interaction of 9C12 with Ad5. Cryo-EM images of the Ad5-9C12 complex at ratios of one to five IgG molecules per hexon monomer showed well-separated, nonaggregating virions, supporting the photon correlation spectroscopy and negative-stain TEM results. The density of 9C12 bound to the peripentonal hexons was as strong as the density of the reconstructed icosahedral viral capsid, indicating that Ab occupancy is nearly 100% at these positions. Comparison of cryo-EM images of the Ad5-9C12 complex with those of Ad5 alone revealed that 9C12 coats the virus with a layer of extra density approximately 10 nm thick (data not shown), in agreement with the minor shift observed in the photon correlation spectroscopy experiment. The Ab-coating effect was also observed in the three-dimensional reconstruction of the Ad5-9C12 complex (Fig. 5A). The reconstructed Ad5 virion appeared to be covered with a mesh of Ab density over the entire capsid except within the vicinity of the pentons at the icosahedral fivefold symmetry axes (Fig. 5). Superposition of a reconstruction of Ad5 with that of the Ad5-9C12 complex revealed no detectable conformational change in the virion upon Ab binding. The cryo-EM structure of the Ad5-9C12 complex indicated that the integrin-binding RGD protrusions of the penton base are not blocked by the Ab.
FIG. 5.
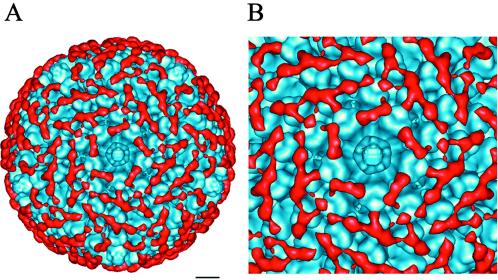
Cryo-EM imaging of the Ad5-9C12 Ab complex. (A) An isosurface representation of the Ad5-9C12 Ab complex viewed along an icosahedral fivefold axis. The scale bar corresponds to 10 nm. (B) An enlarged view of the fivefold region. The Ab density (red) was isolated from the reconstruction of the Ad5-9C12 complex, and the Ad5 density (blue) is that of the uncomplexed Ad5 reconstruction. Note that the Ab density does not obscure the pentons at the fivefold axes.
Clear evidence of bivalent binding was observed for 9C12 between the five peripentonal hexons, which surround the penton (Fig. 5B). In this region, Ab density was observed to bridge adjacent hexons. We interpret this density bridge to be due to the two Fab arms of one IgG molecule. Given the segmental flexibility of IgG molecules (20, 39), it is not surprising that the Fc fragment density was not reconstructed. Some noisy density was observed in the vicinity expected for the Fc fragment, but this noisy, disconnected density has been removed for clarity in the figures. The 10-nm density layer observed in the cryo-EMs (data not shown) can be explained by the bound Fab arms (3 nm) plus the unreconstructed Fc regions (∼7 nm in height) extending further from the viral surface.
Modeling of the interaction between 9C12 and the Ad5 hexon.
Closer examination of the Ab density bridging two peripentonal hexons revealed one well-shaped Fab arm and one somewhat distorted Fab arm (Fig. 6A and B). The well-shaped Fab arm has a prototypical diamond shape that agrees well with the characteristic shape of crystallographic Fab structures.
FIG. 6.
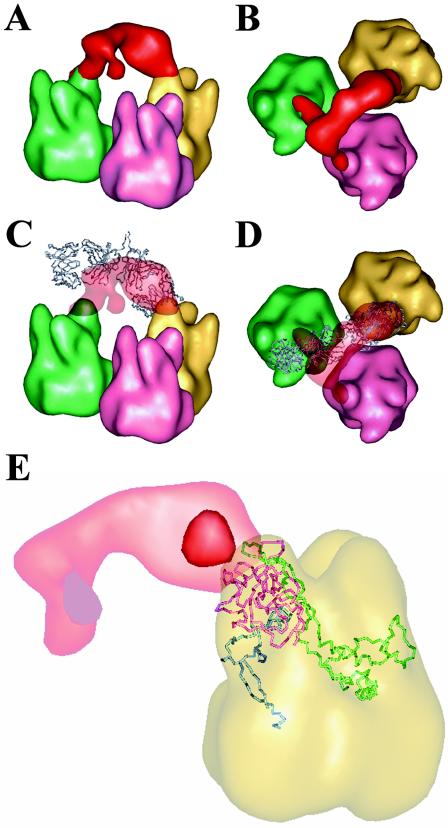
Cryo-EM indicates bivalent binding of 9C12 to Ad5. (A) Side view of a bridge of Ab density (red) observed to link two adjacent peripentonal hexons (green and yellow). Note that one Fab arm (right) has a characteristic diamond shape, while the other Fab arm (left) is somewhat distorted. (B) Top view of the density shown in panel A. Note that a small portion of cryo-EM Ab density extends over a hexon towerof a third, nonperipentonal hexon (pink). (C) The two Fab arms from the crystal structure of a murine IgG1κ MAb (blue) (PDB code 1IGY) (21) manually docked, so that the right Fab arm fits within the diamond-shaped cryo-EM Ab density. (D) Top view of the density and coordinates shown in panel C. (E) A transparent view of a peripentonal hexon shown with a Cα representation of the three loops that comprise one hexon tower. Loop DE1 (residues 115 to 322) is shown in pink; loop FG1 (residues 394 to 553) is shown in green; and loop FG2 (residues 772 to 867) is shown in blue. Note that two isosurface levels for the Ab density are shown: the strong red density indicates the center of the epitope on the hexon; the faint red density indicates the outline of the two Fab arms as shown in panels A to D. The hexons in this figure are 2.4-nm filtered representations of the Ad5 hexon crystal structure (PDB code 1P30) (36) docked within the Ad5 cryo-EM reconstruction.
To model the interaction between 9C12 and the Ad5 hexon capsid, both the Ab density and Ad5 hexon density were fit with atomic coordinates. The crystallographic coordinates for an Ab of the same class as 9C12, the intact murine IgG1κ (MAb 61.1.3; PDB code 1IGY) (21), was used for interactive fitting into the well-defined diamond-shaped Ab density. One Fab arm of the murine IgG1κ molecule was found to fit reasonably well, given the moderate resolution (2.1 nm) of the cryo-EM density (Fig. 6C and D). The other more-distorted arm of Ab density has a small density tail that extends to a third hexon. Since the reconstructed density represents the average of a population of particles, the density tail can be interpreted as a minor alternate binding site for one of the two Fab arms of the IgG molecule. It appears that the well-shaped Fab arm was bound to a unique site within the asymmetric unit of the virus, as this position has no nearby alternate binding sites for 9C12. If there were a hexon at the icosahedral fivefold axis rather than a penton base, then this Fab arm might have an alternate binding site. No attempt was made to adjust the hinge angle of the crystallographic IgG1κ coordinates; however, it was clear that the angle between the long axes of the Fab fragments was significantly more acute than the 115° reported for the uncomplexed IgG1κ (21). Thus, it appears that bivalent binding of 9C12 was facilitated by the inherent segmental flexibility of IgG molecules.
The Ad5 hexon coordinates (PDB code 1P30) (36) were positioned within the Ad5 capsid density with the CoLoRes search tool in the Situs package (8). The distinctive triangular tops of the crystallographic hexon fit well within the cryo-EM density, as had been found in a previous Ad difference-mapping study (44). Each hexon tower is formed by three loops (DE1, FG1, and FG2), with one loop from each of the three hexon monomers in the molecular trimer. Fitting of the atomic hexon coordinates into the cryo-EM density indicates that residues from both loops DE1 and FG1 are likely to form the 9C12 epitope, while loop FG2 is too far away to be involved (Fig. 6E).
Modeling of the Ad5 hexon coordinates within the Ad5 cryo-EM density provided a convenient way to measure the distances between bivalently bound epitopes on the viral surface. Approximate distances were measured between bivalently bound hexon epitopes using the calculated capsid of hexons and considering the midpoint between hexon residues 178 and 179 in the DE1 loop as the center of the epitope. Visual inspection of the docked Ad5 hexon coordinates in the peripentonal hexon positions together with the strongest cryo-EM 9C12 density indicated that these two residues are close to the center of the epitope region. The distance spanned between epitopes on two peripentonal hexons (Fig. 6) is 6.9 nm, which is within the range reported in the literature for bivalent Ab binding (6 to 15 nm) (22, 26). When considering the minor alternate binding site for one Fab arm, the distance spanned by the Ab is 8.0 nm (Fig. 6). However, in this case the angular orientation of the two epitopes appears to be less favorable for bivalent binding.
The rest of the Ab density encapsulating the virion appeared more like a mesh of interconnections than discrete density bridges connecting pairs of epitopes. We interpret the mesh-like density to be the spatial average of many alternate bivalent binding combinations for 9C12 on the surface of the Ad5 capsid. Two specific bivalent binding combinations can be discerned within the mesh-like density. These binding combinations are diagrammed in Fig. 7, along with the major and minor alternate binding combinations of the IgG bound to the peripentonal hexons described above. The distances between the epitopes that appear to be bivalently connected within the mesh-like density are 11.4 and 11.7 nm. These distances are within the range reported for bivalent binding of Abs. They are also approximately the same distance as the span observed in the crystallographic IgG1κ structure between the two complementarity-determining region H3 loops (11.8 nm) (21, 39). Given the extreme flexibility observed in crystal structures of intact human and mouse Abs (39), it seems likely that 9C12 can adapt its conformation to bind epitopes spaced by various distances (in the range of 6.9 to 11.7 nm) on the viral surface. Additional bivalent binding combinations probably occur with lower occupancy values; however, these are not as well represented in the reconstruction.
FIG. 7.
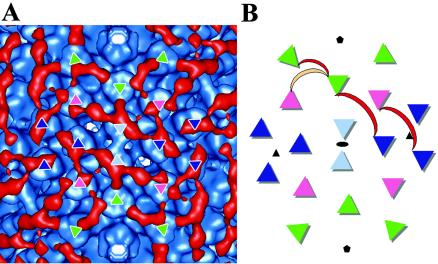
Schematic diagram of the observed pattern of bivalent binding for 9C12 on Ad5. A density region of the Ad5-9C12 complex surrounding an icosahedral twofold axis (A) shown together with a diagrammatic representation (B). The four independent hexon positions are colored as follows: peripentonal hexons are in green; hexons next to the icosahedral twofold axes are in light blue; hexons next to the icosahedral threefold axes are in dark blue; and the fourth hexon position is in pink. The two Fab arms of 9C12 are diagrammed as a crescent with the major bivalent binding combinations in red and one minor alternate binding combination in peach. Only unique bivalent binding combinations are shown without symmetry-related copies. The positions of the icosahedral twofold (black circle), threefold (black triangle), and fivefold (black pentagon) symmetry axes are indicated with crystallographic symbols.
As diagrammed in Fig. 7, the cryo-EM reconstruction indicates that there are at least three intact Abs bound per asymmetric unit of the viral capsid. Given that we cannot completely interpret the mesh-like Ab density, we suspect that the actual number is closer to four intact Abs per asymmetric unit. The Ad asymmetric unit is composed of four hexon trimers. Hence, one Ab molecule per hexon trimer is sufficient to create the observed meshwork of Ab molecules covering the capsid. This ratio deduced from the cryo-EM reconstruction is consistent with our neutralization assay finding of a minimum of one Ab per hexon trimer for neutralization. A ratio of one Ab per hexon trimer is equivalent to two Fab arms per three hexon towers, which leaves some hexon towers unbound as is observed in the cryo-EM density. Nevertheless, this ratio is sufficient to link every hexon trimer with two neighboring hexons via two bound Fab arms from two distinct Abs. Although simple numeric calculation suggests that 120 Ab molecules would be sufficient to bind bivalently to the 240 hexons in the virus particle, consideration of the geometrical constraints for interaction of Ab molecules with the hexon capsid leads to the prediction that 1 Ab per hexon trimer would be required for neutralization via a hexon cross-linking mechanism. A lower ratio of Abs per hexon trimer would mean that some hexons would only be joined to a single neighboring hexon, thus creating pairs of linked hexons but not the full “locked-tight” effect of a complete meshwork of Abs. A higher ratio of Abs per hexon trimer could be accommodated, but it would have no increased cross-linking effect.
It is important to note that all of the observed bivalent binding combinations are between epitopes on different hexon trimers. The geometric arrangement of the three epitopes presented on a single hexon trimer appears to preclude bivalent binding of a single Ab to a single hexon trimer. The cryo-EM reconstruction of the Ad5-9C12 complex indicates that the 9C12 Ab effectively cross-links the hexon capsid by creating multiple bridges between hexons.
9C12 binding does not prevent virus attachment and internalization.
The effect of 9C12 neutralization on virus attachment and entry was analyzed by confocal microscopy with Cy3B-labeled rAdGFP to visualize virus particles (red fluorescence). 9C12-Cy3B-rAdGFP complexes were prepared by incubation at 37°C in medium for 1 h prior to addition to cells. Attachment of virus or virus-Ab complexes to HeLa cells was analyzed following incubation in the cold (0°C; 1 h); virus entry was analyzed after the coverslips were warmed to 37°C and incubated for 1 h. Following incubation, cells were fixed in formalin, followed by permeabilization with Triton X-100 unless otherwise specified. Alexa Fluor 488 anti-mouse IgG (green fluorescence) was used to visualize 9C12; colocalized Cy3B and Alexa Fluor 488, indicating 9C12-Cy3B-rAdGFP complexes, appeared as yellow signals in the merged confocal micrographs. Detergent permeabilization was omitted in some experiments prior to the addition of the Alexa Fluor anti-mouse Ab. Omission of the detergent permeabilization step allowed visualization of surface-accessible 9C12-Cy3B-rAdGFP complexes as yellow signals and internalized 9C12-Cy3B-rAdGFP as red signals. A single z section was used for each analysis to allow visualization and quantification of the two-dimensional distribution of fluorescent signal within different areas of the cell.
After the cold attachment step, the confocal images showed that essentially all Cy3B-rAdGFP was in complexes containing 9C12 and that nearly all of these complexes remained on the cell surface (Fig. 8A and B). The merged fluorescent image from a single z section is shown in Fig. 8A. In Fig. 8B, the z-section image was overlaid on a DIC image to show the distribution of fluorescence relative to each cell in the field. Most of the yellow signal (from colocalized 9C12-Cy3B-rAdGFP) was distributed toward the periphery of the cells where the plane of the z section passed through the cell membrane, indicating that 9C12 binding to Cy3B-rAdGFP did not interfere with virus attachment to the cell. Detergent treatment prior to addition of the Alexa Fluor anti-mouse Ab did not affect the pattern of fluorescence following the attachment step (data not shown). This is in agreement with the accepted notion that little viral cell entry will occur during the cold attachment period. The confocal observation of viral cell attachment even in the presence of 9C12 is consistent with the cryo-EM results indicating that 9C12 did not obstruct the receptor binding domains of penton base and fiber.
FIG. 8.
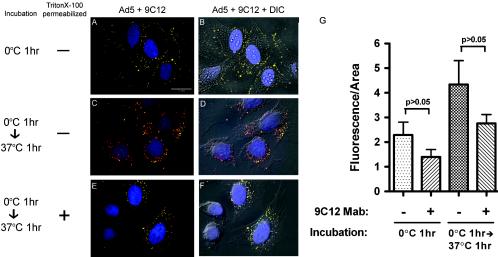
Binding and entry of 9C12-bound Ad5. (A, C, and E) Three-color merged fluorescent images of Ad-infected HeLa cells. All images represent a single 0.4-μm-layer overlay obtained using filters for DAPI (nucleus), Cy3B (Ad5), and Alexa Fluor 488-GFP-filtered (9C12) images. (B, D, and F) Images shown in panels A, C, and E were overlaid with the corresponding nonconfocal brightfield DIC images. (A) Cy3B-rAdGFP was preincubated with neutralizing levels of 9C12, then incubated with HeLa cells for 1 h at 0°C, and then washed to remove unbound virus. Cells were then fixed with formalin and incubated at 37°C for 1 h with Alexa Fluor 488 anti-mouse IgG to detect 9C12 bound to virus. (C) HeLa cells were treated as described in the legend to panel A, then warmed to 37°C and incubated for 1 h. After 18 h at 37°C, the cells were then fixed with formalin and incubated with Alexa Fluor 488 anti-mouse IgG to detect 9C12 bound to virus. (E) HeLa cells were treated as described for panel C, except that the cells were permeabilized with Triton X-100 prior to incubation with Alexa Fluor 488 anti-mouse IgG to permit detection of internalized 9C12. Images are representative samples of three fields of view from at least four independent experiments. (G) Quantification of Cy3B-rAdGFP fluorescence (red channel) from the cytoplasm-plus-plasma membrane border area of z sections. Histograms show the means and standard error for fluorescence per area from 20 to 22 cells incubated with either Cy3B-rAdGFP or 9C12-Cy3B-rAdGFP under the conditions indicated.
After the entry step (37°C; 1-h incubation) and in the absence of detergent permeabilization, the confocal images showed that most of the 9C12-Cy3B-rAdGFP complexes were inaccessible to the Alexa Fluor 488 anti-mouse Ab. Under these experimental conditions, internalized virus-Ab complexes will have only red Cy3B fluorescence (Fig. 8C and D). Most of the red fluorescent signals were observed in the cytoplasmic and perinuclear regions of the z section, whereas the yellow signals (colocalized red plus green signals) indicating surface-accessible 9C12-Cy3B-rAdGFP complexes were distributed toward the outer edges of the cell. In contrast, when detergent permeabilization was used, nearly complete colocalization of 9C12 and Cy3B-rAdGFP was observed (yellow signals) (Fig. 8E and F). This indicates that the intracellular Cy3-rAdGFP (red) remained complexed with 9C12 (green).
The amount of Cy3B fluorescence per area, a measure of Cy3B-rAdGFP in each z section, was quantified for the cytoplasm-plus-plasma membrane border of each z section from fixed, detergent-permeabilized cells, with samples of 20 to 22 cells for each treatment and time point. Statistical analysis showed that neither attachment nor entry of 9C12-Cy3B-rAdGFP was significantly reduced compared to untreated Cy3B-rAdGFP, although lower mean values for 9C12-Cy3B-rAdGFP at both steps suggested a possible trend (Fig. 8G). The mean signal for both Cy3B-rAdGFP and 9C12-Cy3B-rAdGFP increased approximately twofold from the attachment step to the entry step; in each case, the difference was statistically significant (P < 0.004 and P < 0.002, respectively). This supports the idea that appreciable viral cell entry does occur even in the presence of 9C12.
9C12-complexed Ad5 migrates to the nucleus.
We also quantified the Cy3B fluorescence in the region of each z section corresponding to the nucleus and nuclear periphery as a measure of intracellular transport of Cy3B-rAdGFP and 9C12-Cy3B-rAdGFP. As shown in Fig. 9A, relatively little Cy3B fluorescence was detected in the nucleus and nuclear periphery following the 0°C attachment step. The amount of fluorescence in the nucleus and perinucleus increased approximately 40-fold following incubation at 37°C for 1 h for both untreated virus and 9C12-complexed virus, indicating that significant intracellular transport occurred. Comparing fluorescence after the attachment and entry steps showed that mean values increased 43 fold for untreated Cy3B-rAdGFP (P < 0.005) and 37 fold for 9C12-Cy3B-rAdGFP (P < 0.0025). The mean nuclear-plus-perinuclear fluorescence per area for 9C12-Cy3B-rAdGFP was lower than that for Cy3B-rAdGFP at the attachment step, but this difference was not statistically significant (P > 0.05). After the entry step, the reduction in the mean fluorescence value for 9C12-Cy3B-rAdGFP compared to untreated Cy3B-rAdGFP reached statistical significance (P < 0.02). Although the perinuclear signal for 9C12-Cy3B-rAdGFP was significantly reduced (∼60%) compared to that for untreated rAdGFP, 9C12 Ab neutralized ≥95% of GFP expression from Cy3B-rAdGFP at the same Ab:virus ratio used in the confocal experiment (data not shown). This suggests that the difference in nuclear-plus-perinuclear signal cannot fully account for the difference in GFP expression.
FIG. 9.
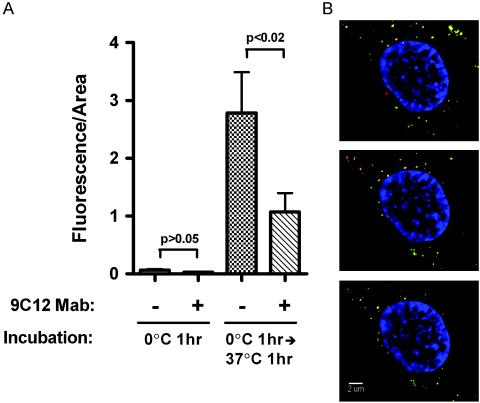
Transport of 9C12-Ad complexes to the nuclear periphery. (A) Quantification of Cy3B fluorescence in the nucleus and nuclear periphery. Histograms show the mean and standard error for fluorescence per area from 20 to 22 cells incubated with either Cy3B-rAdGFP or 9C12-Cy3B-rAdGFP under the conditions indicated. (B) Merged three-color fluorescent images from 0.4-μm z sections were obtained with filters for DAPI (nucleus), Cy3B (Ad5), and Alexa Fluor 488 (9C12). The nucleus and perinuclear region from three serial z sections from a representative cell are shown.
The 37-fold increase in the nuclear-plus-perinuclear Cy3B signal for 9C12-Cy3B-rAdGFP indicated that large numbers of complexes were transported to the nucleus and nuclear periphery. Analysis of the nuclear region from serial z sections of detergent permeabilized cells showed that 9C12-Cy3B-rAdGFP complexes could be observed at the nuclear periphery as yellow signals in the merged red and green fluorescent images shown in Fig. 9B. These data indicated that neutralization of Cy3B-rAdGFP by 9C12 did not prevent intracellular virus transport to the outside of the nucleus. Together with the cryo-EM reconstruction of the Ad5-9C12 complex, these results suggest a model for neutralization of Ad by an antihexon Ab in which the hexon capsid is cross-linked by Abs, thus preventing virus uncoating and nuclear entry of viral DNA.
DISCUSSION
Ab-mediated neutralization of Ad has been studied for several decades. The mechanisms associated with antifiber and antipenton base Ab-mediated neutralization have been previously described (23, 43, 52, 53); however, a clear mechanism for neutralization by antihexon Abs has been lacking. In this study, we present the first structural evidence that neutralization of Ad by an antihexon Ab is mediated by coating of the viral capsid and blocking of a postentry step during infection.
The prevalence of antihexon Ab-mediated neutralization has been reported for polyclonal antihexon Abs as both the major neutralizing component of anti-Ad antisera (54) as well as a very minor contributor to neutralization (17, 24). Considering the large proportion of the capsid surface occupied by hexon, it seems likely that hexon would be a prevalent antigen. We appear to be the first to provide structural data in support of a mechanism for antihexon Ab neutralization of Ad5 by a single Ab species. The 9C12 Ab specifically recognizes hexon and binds an epitope accessible on the surface of the virion. The cryo-EM reconstruction of the Ad5-9C12 complex indicates that the epitope is likely comprised of two hexon loops, the DE1 and FG1 loops, from separate hexon monomers within a trimer. This description of the epitope is consistent with the Western blot data demonstrating epitope stability in the presence of SDS, which is known to disrupt the capsid without disrupting the hexon trimer (16), and the observed sensitivity of the epitope to heat denaturation, which is known to convert hexon trimers to monomers (11, 16). The epitopes for other antihexon Abs have also been mapped to surface locations (46), suggesting that the coating mechanism we describe for 9C12-mediated neutralization of Ad5 may be a general mechanism for other antihexon Abs.
The cryo-EM reconstruction of the Ad5-9C12 complex clearly depicts the viral capsid coated with largely bivalently bound Abs. The cryo-EM findings are consistent with the lack of aggregation observed by TEM of negatively stained samples and by solution dynamic light-scattering experiments. Bivalent binding of 9C12 Abs to the capsid is likely to limit the ability of 9C12 to cross-link adjacent virions. Steric hindrance generated by fiber proteins on adjacent virions would also make the close contact required for aggregation by an antihexon Ab unlikely. Interestingly, aggregation was consistently observed in both TEM and in solution dynamic light-scattering experiments when polyclonal anti-Ad5 sera or purified IgGs were tested. A recent study demonstrated that anti-Ad sera typically contain a high percentage of antifiber Abs (24), which are probably the main cause of the observed aggregation. Clearly aggregation is a common mechanism of virus neutralization, but our results indicate that it is not a factor contributing to neutralization by the antihexon Ab 9C12.
The minimum ratio required for maximal neutralization was 240 Ab molecules per virus particle, equivalent to 1 9C12 Ab per hexon timer. This ratio represents two Fab arms for every three epitopes on hexon, suggesting that not every epitope on hexon monomers would be bound. When 9C12 was incubated with Ad5 at supraneutralizing ratios, the maximum level of neutralization plateaued at 95%. Cryo-EMs of supraneutralizing levels of 9C12 bound to Ad5 showed the viral particles covered by a dense coat of Ab obscuring the virion and suggesting that higher levels of Ab occupancy could be achieved without increased neutralization. The ∼5% persistent gene expression seen in 9C12-neutralized rAdGFP might be explained by transient release of the Abs from the coated viral surface.
Neutralization of other nonenveloped viruses has been described to occur via an Ab-coating mechanism; however, in those cases Abs blocked virus binding to receptors at the cell surface (7). In contrast, Ad5 neutralization by the 9C12 antihexon Ab occurs at the nuclear periphery. Our results indicate that the mechanism of neutralization by 9C12 is either stabilization of the capsid by interhexon cross-linking, which prevents release of the Ad genome at the nuclear envelope, or obstruction of specific interactions with cellular proteins required for import of the viral genome into the nucleus of the host cell.
In conclusion, we propose that neutralization of Ad by antihexon Abs requires bivalent binding to the exterior surface of the virion and that this binding need not achieve 100% occupancy of all epitopes. Ab coating of the viral capsid may provide a physical barrier between the viral surface and cellular proteins that participate in capsid interactions with the nucleus. Alternatively, the Ab cross-linking of the capsid surface may prevent uncoating of the virion at the nuclear periphery. Since the mechanism of neutralization of Ad5 by the antihexon Ab 9C12 does not appear to be based on unique structural features of Ad5, the mechanisms proposed are likely broadly applicable to Ad's in general. These studies provide experimental evidence that Abs are capable of neutralizing Ad's at a postentry step.
Acknowledgments
We thank Glen Nemerow (Scripps Research Institute) for helpful discussions and suggestions and John Abrams (DNAX Research Institute) for generously providing a panel of antihexon monoclonal antibodies from which the 9C12 and 27F11 antibodies were selected.
This work was supported in part by NIH grant AI42929 to P.L.S.
REFERENCES
- 1.Adrian, M., J. Dubochet, J. Lepault, and A. W. McDowall. 1984. Cryo-electron microscopy of viruses. Nature 308:32-36. [DOI] [PubMed] [Google Scholar]
- 2.Banfield, M. J., D. J. King, A. Mountain, and R. L. Brady. 1997. VL:VH domain rotations in engineered antibodies: crystal structures of the Fab fragments from two murine antitumor antibodies and their engineered human constructs. Proteins 29:161-171. [DOI] [PubMed] [Google Scholar]
- 3.Barnstable, C. J., W. F. Bodmer, G. Brown, G. Galfre, C. Milstein, A. F. Williams, and A. Ziegler. 1978. Production of monoclonal antibodies to group A erythrocytes, HLA and other human cell surface antigens—new tools for genetic analysis. Cell 14:9-20. [DOI] [PubMed] [Google Scholar]
- 4.Bergelson, J. M. 1999. Receptors mediating adenovirus attachment and internalization. Biochem. Pharmacol. 57:975-979. [DOI] [PubMed] [Google Scholar]
- 5.Bergelson, J. M., J. A. Cunningham, G. Droguett, E. A. Kurt-Jones, A. Krithivas, J. S. Hong, M. S. Horwitz, R. L. Crowell, and R. W. Finberg. 1997. Isolation of a common receptor for coxsackie B viruses and adenoviruses 2 and 5. Science 275:1320-1323. [DOI] [PubMed] [Google Scholar]
- 6.Bizebard, T., R. Daniels, R. Kahn, B. Golinelli-Pimpaneau, J. J. Skehel, and M. Knossow. 1994. Refined three-dimensional structure of the Fab fragment of a murine IgGl,λ antibody. Acta Crystallogr. D Biol. Crystallogr. 50:768-777. [DOI] [PubMed] [Google Scholar]
- 7.Burton, D. R., E. O. Saphire, and P. W. Parren. 2001. A model for neutralization of viruses based on antibody coating of the virion surface. Curr. Top. Microbiol. Immunol. 260:109-143. [DOI] [PubMed] [Google Scholar]
- 8.Chacon, P., and W. Wriggers. 2002. Multi-resolution contour-based fitting of macromolecular structures. J. Mol. Biol. 317:375-384. [DOI] [PubMed] [Google Scholar]
- 9.Chiu, C. Y., P. Mathias, G. R. Nemerow, and P. L. Stewart. 1999. Structure of adenovirus complexed with its internalization receptor, αvβ5 integrin. J. Virol. 73:6759-6768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chiu, C. Y., E. Wu, S. L. Brown, D. J. Von Seggern, G. R. Nemerow, and P. L. Stewart. 2001. Structural analysis of a fiber-pseudotyped adenovirus with ocular tropism suggests differential modes of cell receptor interactions. J. Virol. 75:5375-5380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Crawford-Miksza, L., and D. P. Schnurr. 1996. Analysis of 15 adenovirus hexon proteins reveals the location and structure of seven hypervariable regions containing serotype-specific residues. J. Virol. 70:1836-1844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.D'Ambrosio, E., N. Del Grosso, A. Chicca, and M. Midulla. 1982. Neutralizing antibodies against 33 human adenoviruses in normal children in Rome. J. Hyg. (London) 89:155-161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Douglas, J. T., B. E. Rogers, M. E. Rosenfeld, S. I. Michael, M. Feng, and D. T. Curiel. 1996. Targeted gene delivery by tropism-modified adenoviral vectors. Nat. Biotechnol. 14:1574-1578. [DOI] [PubMed] [Google Scholar]
- 14.Everitt, E., A. de Luca, and Y. Blixt. 1992. Antibody-mediated uncoating of adenovirus in vitro. FEMS Microbiol. Lett. 77:21-27. [DOI] [PubMed] [Google Scholar]
- 15.Fender, P., A. H. Kidd, R. Brebant, M. Oberg, E. Drouet, and J. Chroboczek. 1995. Antigenic sites on the receptor-binding domain of human adenovirus type 2 fiber. Virology 214:110-117. [DOI] [PubMed] [Google Scholar]
- 16.Fortsas, E., M. Petric, and M. Brown. 1994. Electrophoretic migration of adenovirus hexon under non-denaturing conditions. Virus Res. 31:57-65. [DOI] [PubMed] [Google Scholar]
- 17.Gahery-Segard, H., F. Farace, D. Godfrin, J. Gaston, R. Lengagne, T. Tursz, P. Boulanger, and J. G. Guillet. 1998. Immune response to recombinant capsid proteins of adenovirus in humans: antifiber and anti-penton base antibodies have a synergistic effect on neutralizing activity. J. Virol. 72:2388-2397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Greber, U. F., M. Suomalainen, R. P. Stidwill, K. Boucke, M. W. Ebersold, and A. Helenius. 1997. The role of the nuclear pore complex in adenovirus DNA entry. EMBO J. 16:5998-6007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Greber, U. F., M. Willetts, P. Webster, and A. Helenius. 1993. Stepwise dismantling of adenovirus 2 during entry into cells. Cell 75:477-486. [DOI] [PubMed] [Google Scholar]
- 20.Harris, L. J., S. B. Larson, K. W. Hasel, and A. McPherson. 1997. Refined structure of an intact IgG2a monoclonal antibody. Biochemistry 36:1581-1597. [DOI] [PubMed] [Google Scholar]
- 21.Harris, L. J., E. Skaletsky, and A. McPherson. 1998. Crystallographic structure of an intact IgG1 monoclonal antibody. J. Mol. Biol. 275:861-872. [DOI] [PubMed] [Google Scholar]
- 22.Hewat, E. A., and D. Blaas. 1996. Structure of a neutralizing antibody bound bivalently to human rhinovirus 2. EMBO J. 15:1515-1523. [PMC free article] [PubMed] [Google Scholar]
- 23.Hong, S. S., M. Bardy, M. Monteil, B. Gay, C. Denesvre, J. Tournier, G. Martin, M. Eloit, and P. Boulanger. 2000. Immunoreactive domains and integrin-binding motifs in adenovirus penton base capsomer. Viral Immunol. 13:353-371. [DOI] [PubMed] [Google Scholar]
- 24.Hong, S. S., N. A. Habib, L. Franqueville, S. Jensen, and P. A. Boulanger. 2003. Identification of adenovirus (ad) penton base neutralizing epitopes by use of sera from patients who had received conditionally replicative ad (addl1520) for treatment of liver tumors. J. Virol. 77:10366-10375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hutchins, B. 2002. Development of a reference material for characterizing adenovirus vectors. Bioprocess 1:25-28. [Google Scholar]
- 26.Icenogle, J., H. Shiwen, G. Duke, S. Gilbert, R. Rueckert, and J. Anderegg. 1983. Neutralization of poliovirus by a monoclonal antibody: kinetics and stoichiometry. Virology 127:412-425. [DOI] [PubMed] [Google Scholar]
- 27.Li, E., S. L. Brown, D. G. Stupack, X. S. Puente, D. A. Cheresh, and G. R. Nemerow. 2001. Integrin αvβ1 is an adenovirus coreceptor. J. Virol. 75:5405-5409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Luftig, R. B., and R. R. Weihing. 1975. Adenovirus binds to rat brain microtubules in vitro. J. Virol. 16:696-706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Newmyer, S. L., and S. L. Schmid. 2001. Dominant-interfering Hsc70 mutants disrupt multiple stages of the clathrin-coated vesicle cycle in vivo. J. Cell Biol. 152:607-620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Niewiarowska, J., J. C. D'Halluin, and M. T. Belin. 1992. Adenovirus capsid proteins interact with HSP70 proteins after penetration in human or rodent cells. Exp. Cell Res. 201:408-416. [DOI] [PubMed] [Google Scholar]
- 31.Orlova, E. V., P. Dube, J. R. Harris, E. Beckman, F. Zemlin, J. Markl, and M. van Heel. 1997. Structure of keyhole limpet hemocyanin type 1 (KLH1) at 15 Å resolution by electron cryomicroscopy and angular reconstitution. J. Mol. Biol. 271:417-437. [DOI] [PubMed] [Google Scholar]
- 32.Piedra, P. A., G. A. Poveda, B. Ramsey, K. McCoy, and P. W. Hiatt. 1998. Incidence and prevalence of neutralizing antibodies to the common adenoviruses in children with cystic fibrosis: implication for gene therapy with adenovirus vectors. Pediatrics 101:1013-1019. [DOI] [PubMed] [Google Scholar]
- 33.Rahman, A., V. Tsai, A. Goudreau, J. Y. Shinoda, S. F. Wen, M. Ramachandra, R. Ralston, D. Maneval, D. LaFace, and P. Shabram. 2001. Specific depletion of human anti-adenovirus antibodies facilitates transduction in an in vivo model for systemic gene therapy. Mol. Ther. 3:768-778. [DOI] [PubMed] [Google Scholar]
- 34.Rini, J. M., R. L. Stanfield, E. A. Stura, P. A. Salinas, A. T. Profy, and I. A. Wilson. 1993. Crystal structure of a human immunodeficiency virus type 1 neutralizing antibody, 50.1, in complex with its V3 loop peptide antigen. Proc. Natl. Acad. Sci. USA 90:6325-6329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Roelvink, P. W., A. Lizonova, J. G. Lee, Y. Li, J. M. Bergelson, R. W. Finberg, D. E. Brough, I. Kovesdi, and T. J. Wickham. 1998. The coxsackievirus-adenovirus receptor protein can function as a cellular attachment protein for adenovirus serotypes from subgroups A, C, D, E, and F. J. Virol. 72:7909-7915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Rux, J. J., P. R. Kuser, and R. M. Burnett. 2003. Structural and phylogenetic analysis of adenovirus hexons by use of high-resolution X-ray crystallographic, molecular modeling, and sequence-based methods. J. Virol. 77:9553-9566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Saphire, A. C., T. Guan, E. C. Schirmer, G. R. Nemerow, and L. Gerace. 2000. Nuclear import of adenovirus DNA in vitro involves the nuclear protein import pathway and hsc70. J. Biol. Chem. 275:4298-4304. [DOI] [PubMed] [Google Scholar]
- 38.Saphire, E. O., P. W. Parren, R. Pantophlet, M. B. Zwick, G. M. Morris, P. M. Rudd, R. A. Dwek, R. L. Stanfield, D. R. Burton, and I. A. Wilson. 2001. Crystal structure of a neutralizing human IgG against HIV-1: a template for vaccine design. Science 293:1155-1159. [DOI] [PubMed] [Google Scholar]
- 39.Saphire, E. O., R. L. Stanfield, M. D. Crispin, P. W. Parren, P. M. Rudd, R. A. Dwek, D. R. Burton, and I. A. Wilson. 2002. Contrasting IgG structures reveal extreme asymmetry and flexibility. J. Mol. Biol. 319:9-18. [DOI] [PubMed] [Google Scholar]
- 40.Shabram, P. W., D. D. Giroux, A. M. Goudreau, R. J. Gregory, M. T. Horn, B. G. Huyghe, X. Liu, M. H. Nunnally, B. J. Sugarman, and S. Sutjipto. 1997. Analytical anion-exchange HPLC of recombinant type-5 adenoviral particles. Hum. Gene Ther. 8:453-465. [DOI] [PubMed] [Google Scholar]
- 41.Shah, A. K., and P. L. Stewart. 1998. QVIEW: software for rapid selection of particles from digital electron micrographs. J. Struct. Biol. 123:17-21. [DOI] [PubMed] [Google Scholar]
- 42.Stewart, P. L., R. B. Cary, S. R. Peterson, and C. Y. Chiu. 2000. Digitally collected cryo-electron micrographs for single particle reconstruction. Microsc. Res. Tech. 49:224-232. [DOI] [PubMed] [Google Scholar]
- 43.Stewart, P. L., C. Y. Chiu, S. Huang, T. Muir, Y. Zhao, B. Chait, P. Mathias, and G. R. Nemerow. 1997. Cryo-EM visualization of an exposed RGD epitope on adenovirus that escapes antibody neutralization. EMBO J. 16:1189-1198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Stewart, P. L., S. D. Fuller, and R. M. Burnett. 1993. Difference imaging of adenovirus: bridging the resolution gap between X-ray crystallography and electron microscopy. EMBO J. 12:2589-2599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Tomko, R. P., C. B. Johansson, M. Totrov, R. Abagyan, J. Frisen, and L. Philipson. 2000. Expression of the adenovirus receptor and its interaction with the fiber knob. Exp. Cell Res. 255:47-55. [DOI] [PubMed] [Google Scholar]
- 46.Toogood, C. I., J. Crompton, and R. T. Hay. 1992. Antipeptide antisera define neutralizing epitopes on the adenovirus hexon. J. Gen. Virol. 73:1429-1435. [DOI] [PubMed] [Google Scholar]
- 47.Trotman, L. C., N. Mosberger, M. Fornerod, R. P. Stidwill, and U. F. Greber. 2001. Import of adenovirus DNA involves the nuclear pore complex receptor CAN/Nup214 and histone H1. Nat. Cell Biol. 3:1092-1100. [DOI] [PubMed] [Google Scholar]
- 48.van Heel, M., G. Harauz, E. V. Orlova, R. Schmidt, and M. Schatz. 1996. A new generation of the IMAGIC image processing system. J. Struct. Biol. 116:17-24. [DOI] [PubMed] [Google Scholar]
- 49.Varga, M. J., T. Bergman, and E. Everitt. 1990. Antibodies with specificities against a dispase-produced 15-kilodalton hexon fragment neutralize adenovirus type 2 infectivity. J. Virol. 64:4217-4225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Vincent, T., B. G. Harvey, S. M. Hogan, C. J. Bailey, R. G. Crystal, and P. L. Leopold. 2001. Rapid assessment of adenovirus serum neutralizing antibody titer based on quantitative, morphometric evaluation of capsid binding and intracellular trafficking: population analysis of adenovirus capsid association with cells is predictive of adenovirus infectivity. J. Virol. 75:1516-1521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Wills, K. N., D. C. Maneval, P. Menzel, M. P. Harris, S. Sutjipto, M. T. Vaillancourt, W. M. Huang, D. E. Johnson, S. C. Anderson, S. F. Wen, R. Bookstein, M. H. Shepard, and R. J. Gregory. 1994. Development and characterization of recombinant adenoviruses encoding human p53 for gene therapy of cancer. Hum. Gene Ther. 5:1079-1088. [DOI] [PubMed] [Google Scholar]
- 52.Wohlfart, C. 1988. Neutralization of adenoviruses: kinetics, stoichiometry, and mechanisms. J. Virol. 62:2321-2328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Wohlfart, C. E., U. K. Svensson, and E. Everitt. 1985. Interaction between HeLa cells and adenovirus type 2 virions neutralized by different antisera. J. Virol. 56:896-903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Wu, H., I. Dmitriev, E. Kashentseva, T. Seki, M. Wang, and D. T. Curiel. 2002. Construction and characterization of adenovirus serotype 5 packaged by serotype 3 hexon. J. Virol. 76:12775-12782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Young, J. C., J. M. Barral, and F. Ulrich Hartl. 2003. More than folding: localized functions of cytosolic chaperones. Trends Biochem. Sci. 28:541-547. [DOI] [PubMed] [Google Scholar]


