Abstract
The long non-coding RNA (lncRNA) NKILA has been reported to participate in the development of human cancers. The purpose of this study was to explore the potential role of lncRNA-NKILA, which acts through NF-ĸB, in the process of melanoma development. Real-time PCR (qRT-PCR) showed that NKILA was expressed at low levels in human melanoma tissues. The area under the ROC curve of NKILA was 0.875, which indicated that NKILA may be a potential diagnostic biomarker of melanoma. Our results also indicated that NKILA inhibited the progression of cell proliferation, migration, and invasion, and promoted apoptosis of melanoma cells. Furthermore, qRT-PCR showed that NF-κB, which was negatively correlated with NKILA, was highly expressed in human melanoma tissues. Moreover, our results indicated that NKILA inhibited the carcinogenesis of melanoma cells through the inhibition of NF-ĸB in vitro. More importantly, we found that NKILA suppressed the growth of melanoma tumors via NF-ĸB in vivo. In conclusion, NKILA suppressed the development of malignant melanoma via the regulation of NF-ĸB and may be a potential therapeutic target in patients with melanoma.
Keywords: LncRNA-NKILA, NF-ĸB, melanoma, metastasis
Introduction
Melanoma is one of the most common cancers worldwide. It is second in incidence only to breast cancer in women, and in male cancer patients under 40 years of age, it is second only to leukemia [1]. It has been reported that approximately 200,000 new cases of malignant melanoma occur globally each year and that an estimated 46,000 deaths occur from the disease worldwide each year [2]. Recently, the incidence of melanoma has continued to increase in many countries such as Australia and New Zealand [3], where the incidence rates are the highest worldwide, but its incidence has also increased in Europe [4-6] and North America [7,8] over the last 50 years [3,9,10]. Melanoma is regarded as the most malignant skin neoplasm, and distant metastasis is common. Therefore, early diagnosis and prompt treatment are urgent. Presently, effective biomarkers are scarce, and the diagnostic technology for melanoma is limited. Therefore, it is important that the molecular and functional mechanisms of melanoma be investigated.
Long non-coding RNAs (lncRNAs), such as Xist [11], NEAT1 [12], MALAT-1 [13] and H19 [14,15], have been found to be of crucial functional importance in the pathogenesis of some diseases. With the development of sequencing technologies and the characterization of functional genomic elements, approximately 9640 lncRNA loci have been found in the human genome [16,17], and this number continues to grow. Accumulating evidence has indicated that lncRNAs play important roles in tumorigenesis through the regulation of gene expression, such as the regulation of transcription, post-transcription events, and chromatin modification [18-20]. Therefore, lncRNAs are regarded as important therapeutic targets for cancer. However, functional and mechanistic studies of the lncRNA NKILA in melanoma have not been widely conducted.
One study found that NF-κB serves as a nuclear factor that binds to the enhancer element of activated B cells [21,22]. Other studies indicated that NF-κB possesses specific DNA binding activity and that it can regulate a massive number of target genes with a variety of functions [23]. At present, growing evidence suggests that NF-κB plays a crucial role in multiple steps of cancer initiation and progression and that it is regarded as a cancer treatment target [24]. Recent studies have also indicated that NF-ĸB can interact with NKILA during cancer development [25]. It has also been found that NKILA combines with the NF-κB/IκB complex to suppress NF-κB signaling via the inhibition of IκB, which stabilizes the complex [26]. However, the effects of the interaction of NKILA and NF-ĸB on the development of melanoma are not entirely clear.
To better understand the mechanism of melanoma occurrence, we detected the mRNA expression levels of NKILA and NF-ĸB in human melanoma tissues and examined the correlation between NKILA or NF-ĸB and the clinical features of the disease. We also examined the prognosis of melanoma and the effects of NKILA on the cell cycle progression, proliferation, apoptosis, migration, and invasiveness of melanoma cells. In addition, the regulatory relationship between NKILA and NF-ĸB in melanoma tissues and cells, and the roles of NKILA on the growth of melanoma via NF-ĸB in vivo were also studied. Consequently, it is well established that melanoma is driven by the inhibition of NKILA, which occurs most frequently through the activation of NF-ĸB.
Materials and methods
Clinical specimens
In this study, we collected tissue samples from 92 patients with melanoma at the General Hospital of Jinan Military Command between 2007 and 2016. Each patient provided informed consent. This study was also approved by the Ethics Committee of The General Hospital of Jinan Military Command. The histological diagnosis of melanoma was evaluated according to the World Health Organization (WHO) criteria. All tissue samples were stored at -80°C.
Cell lines
Human Epidermal Melanocytes, neonatal (HEMn) cells were purchased from American Type Culture Collection (ATCC, Manassas, VA, USA), and melanoma cell lines (M21, B16, MEL-RM, MM200, A375 and A2058) were purchased from the Type Culture Collection of the Chinese Academy of Sciences, Shanghai, China. HEMn cells were cultured in Melanocyte Growth Media (PromoCell, Shanghai, China), A2058, B16, and A375 cells were cultured in Dulbecco’s Modified Eagle’s medium (DMEM) and M21, MEL-RM and MM200 cells were maintained in RPMI 1640 medium (Invitrogen, Carlsbad, CA, USA). All media were supplemented with 10% fetal bovine serum (FBS) (Sigma Aldrich) and 100 U/mL penicillin/streptomycin (Invitrogen, Carlsbad, CA). All cells were cultured at 37°C in an appropriate incubator in an atmosphere of 5% CO2.
Lentiviral vector construction, production and transfection
Human NKILA and NF-κB full-length cDNAs were amplified by PCR from the mRNA of A2058 cells. Then, the PCR products were each inserted into a lentiviral vector. A lentiviral vector expressing Enhanced Green Fluorescent Protein (EGFP) was used as a control. The objective products were cloned into a pcDNA3.1 vector (Invitrogen). In addition, the shNKILA sequences were designed, and shLUC was used as the negative control (NC). We synthesized DNA fragments of shRNA and cloned the shRNA fragments into a human U6 promoter-containing pBluescript SK (+) plasmid (pU6) after annealing. Then, the U6-shRNA was cloned into a lentiviral vector [27,28]. The constructed vectors and the lentiviral packaging vectors (pMD2.G, pMDL-G/P-RRE, pRSV-REV) were cotransfected into HEK293T cells for 48 hrs. Lentiviruses were produced, harvested, and purified by ultracentrifugation. A2058 and M21 cells (1 × 104 cells/well) were seeded in 24-well plates. A2058 cells were transfected with NKILA or the control using 8 μg/mL polybrene (Sigma), and similarly, M21 cells were transfected with shNKILA or the control; 800 μg/ml G418 (Sigma) was then used to screen the stably transfected cells.
SiRNA transfection
An siRNA that targets the human NF-κB gene was designed based on the public GenBank and was purchased from GenePharma (GenePharma Co., Ltd., Shanghai, China). The sequences of NF-κB siRNAs were as follows: (sense) 5’-GGA CAU AUG AGA CCU UCA AdT dT-3’, and (antisense) 5’-UUG AAG GUC UCA UAU GUC CdT dT-3’. A2058 and M21 cells (2 × 104 cells/well) were seeded in 6-well plates and were transfected with 50 nM scrambled siRNA (Negative control, NC) or NF-κB-siRNA using Lipofectamine 3000 (Invitrogen, Carlsbad, CA, USA) according to the manufacturer’s protocol.
Quantitative real-time reverse transcription PCR (qRT-PCR)
According to the manufacturer’s instructions, total RNA was isolated from melanoma tissues, HEMn cells, melanoma cells and the treated A2058 and M21 cells using TRIzol (Invitrogen, CA, USA). The RevertAid First Strand cDNA Synthesis kit (Thermo Fisher) along with random primers and corresponding total RNA was used to synthesize cDNA. As described previously [28], the cDNA template was amplified by qRT-PCR using a SYBR-Green PCR Master Mix kit (Takara). The primer sequences for NKILA were: 5’-TGG ATT GTT GGG TAT ATT TTG GA-3’ (the forward primer) and 5’-TGT ATG AAG AGG ATG CTG AAG GC-3’ (the reverse primer). The primer sequences for NF-κB were: 5’-ACA AGT GGC CAT TGT GTT CC-3’ (the forward primer) and 5’-ACG TTT CTC CTC AAT CCG GT-3’ (the reverse primer). The primer sequences for GAPDH were: 5’-CCT CGT CTC ATA GAC AAG ATG GT-3’ (the forward primer) and 5’-GGG TAG AGT CAT ACT GGA ACA TG-3’ (the reverse primer) (internal control).
Western blot assay
Tissue samples and the treated cells were lysed in Radio Immunoprecipitation Assay (RIPA) buffer (Thermo Scientific, Rockford, IL, USA) and Protease Inhibitor (Thermo Fisher Scientific, Waltham, MA, USA). The total protein (30 μg) in equal concentration was separated by 8% SDS/PAGE gels and transferred to a polyvinylidene fluoride membrane (PVDF, Millipore, Billerica, MA, USA). The PVDF membranes were blocked for 2 hrs in 5% skim milk at room temperature and were then incubated with the appropriate primary antibodies at 4°C overnight. The following day, the membranes were incubated with HRP-conjugated secondary antibody for 1 h at room temperature. All blots were visualized using an enhanced chemiluminescence (ECL) substrate kit (Amersham Biosciences) and an enhanced chemiluminescence detection system (Amersham Biosciences). The antibodies used were rabbit anti-GAPDH (1:1000; Santa Cruz Biotechnology, Santa Cruz, CA, USA) and rabbit anti-NF-κB (1:1000; Cell Signaling Technology, Beverly, MA, USA).
Proliferation assay
The treated A2058 and M21 cells (4000 cells/well) were seeded in 96-well plates and incubated at 37°C for 1, 2, 3, 4, or 5 days, after which 10 μl of MTT solution (5 mg/ml) was added to each well at the indicated time points. After 4 hrs, 200 μl dimethyl sulfoxide (DMSO) was added to each well to dissolve the precipitates. Then, the absorbance value was measured at 490 nm using a micro-plate reader (Bio Tek Instruments, Inc., Winooski, VT, USA).
Flow cytometric analysis
For the cell cycle assay, the treated A2058 and M21 cells were collected, fixed in 70% ethanol, and stained with propidium iodide solution. For the cell apoptosis assay, cells (1 × 106 cells/mL) were digested, dispersed, centrifuged, collected, washed with cold PBS and resuspended in 1 × binding buffer. The cells were then double-stained with a phosphatidylethanolamine (PE) and annexin-V-FITC staining kit (BD Biosciences). The flow cytometry images of the cell cycle were obtained and analyzed using a FACSCalibur system (BD Biosciences) and FlowJo software (Tree Star Corp, Ashland, OR, USA).
Migration and invasion assays
Transwell chambers (Corning Costar Corp., Cambridge, MA, USA) were used in the migration and invasion assays. In all, 200 μl of the treated A2058 and M21 cells (5 × 105) incubated in serum-free medium were seeded in the upper chamber, while 600 μl complete medium supplemented with 10% FBS was added to the lower chamber. For the invasion assays, the upper chambers were pre-coated with Matrigel (BD Biosciences, San Diego, CA, USA). After 24 hrs, the migratory and invasive cells were fixed in 4% paraformaldehyde and stained in 0.1% crystal violet solution, and then cells on the upper membrane were removed. Finally, the migratory and invasive cells were photographed and counted.
Tumor formation in nude mice
All animal studies were approved by the Laboratory Animal Ethics Committee and were performed according the Institutional Animal Care and Use Committee. BALB/c nude mice (male, 16-18 g, 4-6 weeks old) were purchased from the Shanghai LAC Laboratory Animal Co. Ltd. and were housed in a specific pathogen-free (SPF) grade animal center. The treated A2058 or M21 cells (1 × 107 cells in 100 μl) were subcutaneously injected into nude mice, and then the tumor volumes were recorded every 10 days.
Statistical analysis
All experiments were performed in triplicate, and all data are expressed as the mean ± S.D. All data were analyzed with GraphPad Prism 5.0 (GraphPad Software, La Jolla, CA, USA) and IBM SPSS Statistics 21 (IBM SPSS Inc., Chicago, Illinois). Statistical analyses were performed using Student’s t test or one-way analysis of variance (ANOVA). The statistical significance was set at P < 0.05.
Results
NKILA is downregulated in human melanoma tissues
In order to study the expression status of lncRNA-NKILA in human melanoma tissues, 92 pairs of melanoma tissues and adjacent normal tissues were subjected to qRT-PCR. The results indicated that NKILA was downregulated in melanoma tissues compared with adjacent noncancerous tissues (P < 0.001) (Figure 1A). The correlations between NKILA and the clinical characteristics of patients with melanoma were also analyzed. We found that the expression level of NKILA was significantly associated with the invasion and metastasis of melanoma (Table 1). Furthermore, receiver operating characteristic (ROC) curve analysis was performed. The results indicated a good diagnostic potential of NKILA in melanoma based on the finding that the area under the ROC curve of NKILA was 0.875 (P < 0.0001, Figure 1B).
Figure 1.
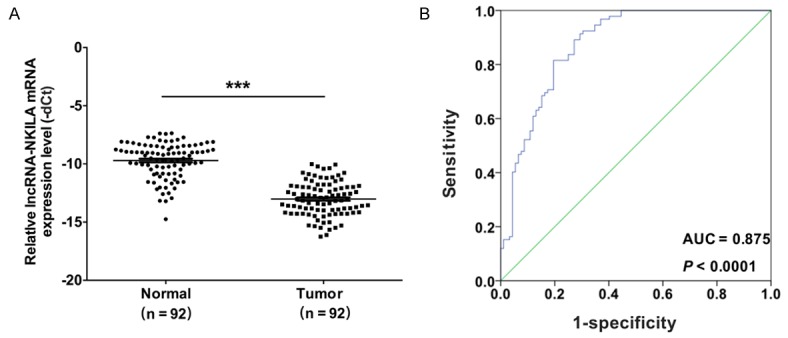
LncRNA-NKILA was downregulated in human melanoma tissues. A. Quantitative RT-PCR (qRT-PCR) was performed to analyze the mRNA expression level of NKILA in 92 pairs of melanoma tissues and adjacent normal tissues (***P < 0.001). B. A receiver operating characteristic (ROC) curve was used to analyze the performance of NKILA in the prediction of prognosis; the area under the ROC curves of NKILA was 0.875 (P < 0.0001).
Table 1.
Correlation analysis between lncRNA-NKILA expression and clinicopathological characteristics of patients with melanoma
| Characteristics | No. of patients | Mean ± SD | P value |
|---|---|---|---|
| Total no. of patients | 92 | ||
| Age (year) | |||
| > 60 | 41 (44.6%) | 12.72 ± 1.02 | 0.265 |
| ≤ 60 | 51 (55.4%) | 13.01 ± 1.38 | |
| Gender | |||
| Male | 43 (46.7%) | 13.46 ± 1.68 | 0.602 |
| Female | 49 (53.3%) | 13.62 ± 1.24 | |
| Invasion | |||
| T0-T2 | 63 (68.5%) | 12.23 ± 1.42 | 0.001** |
| T3-T4 | 29 (31.5%) | 13.24 ± 1.21 | |
| Lymphatic metastasis | |||
| N0 | 69 (75%) | 12.74 ± 1.89 | 0.0003*** |
| N1-N3 | 23 (25%) | 14.79 ± 1.07 | |
| Distal metastasis | |||
| M0 | 73 (79.3%) | 12.87 ± 2.12 | 0.020* |
| M1 | 19 (20.7%) | 14.07 ± 1.19 | |
| TNM stage | |||
| 0 & I | 65 (70.7%) | 12.64 ± 1.65 | 0.001** |
| II & III & IV | 27 (29.3%) | 13.99 ± 2.01 |
Indicated statistical significance (P < 0.05);
Indicated statistical significance (P < 0.01);
Indicated statistical significance (P < 0.001).
NKILA inhibits the progression of the cell cycle and the proliferation of melanoma cells
First, the expression level of NKILA was detected by qRT-PCR in human epidermal melanocytes, neonatal (HEMn) and in melanoma cell lines (M21, B16, MEL-RM, MM200, A375 and A2058). The results showed that NKILA had a lower expression level in melanoma cell lines compared with HEMn (Figure 2A). To further study the mechanism of NKILA in melanoma, lentiviral vectors of NKILA and shNKILA were produced and then separately transfected into A2058 and M21 cells. The qRT-PCR results indicated that the expression level of NKILA was obviously increased in A2058 cells transfected with NKILA (Figure 2B), while the expression level was dramatically decreased in M21 cells transfected with shNKILA (Figure 2C). We also found that the ectopic expression of NKILA inhibited the proliferation ability of A2058 cells (Figure 2D), while the knockdown of NKILA accelerated the proliferation ability of M21 cells (Figure 2E). In addition, the flow cytometric analysis showed a significant increase in the proportion of G1-phase A2058 cells that were transfected with NKILA (94.89% of total cells) compared with the control (77.08% of total cells). The analysis also showed a significant decrease in S-phase A2058 cells that were transfected with NKILA (6.01% of total cells) compared with the control (20.31% of total cells), which suggests that NKILA induced cell cycle arrest in A2058 cells (Figure 2F). Similarly, a significant decrease in the proportion of G1-phase M21 cells that were transfected with shNKILA (58.84% of total cells) was observed relative to the control (76.3% of total cells), which indicates that the silencing of NKILA inhibited cell cycle arrest in M21 cells (Figure 2G).
Figure 2.
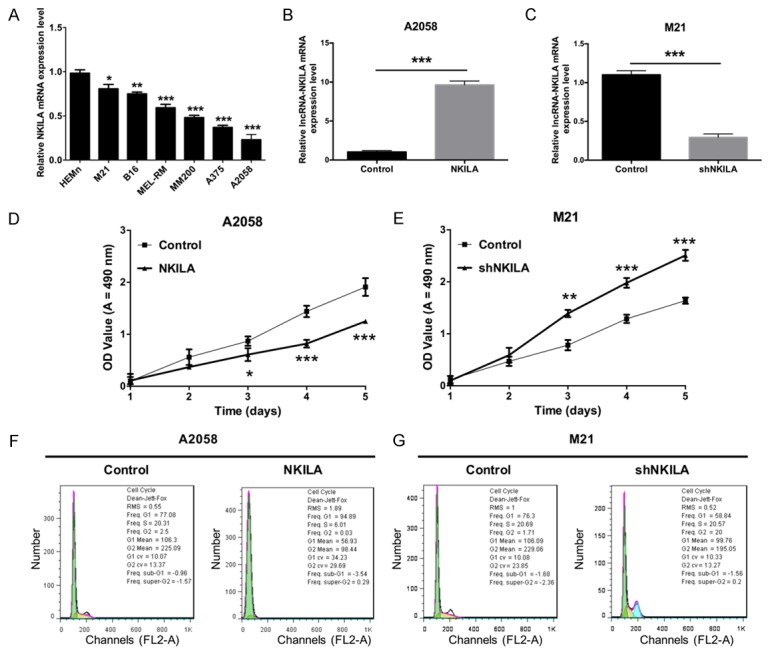
NKILA inhibits the progression of melanoma cell cycle and proliferation. A. qRT-PCR was used to detect the mRNA expression level of NKILA in Human Epidermal Melanocytes, neonatal (HEMn), and melanoma cell lines (M21, B16, MEL-RM, MM200, A375 and A2058) (*P < 0.05, **P < 0.01, ***P < 0.001). B. The mRNA expression level of NKILA was measured by qRT-PCR in A2058 cells that were transfected with NKILA or the control using Lipofectamine 3000 (***P < 0.001). C. The mRNA expression level of NKILA was measured by qRT-PCR in M21 cells that were transfected with shNKILA or the control using Lipofectamine 3000 (***P < 0.001). D. The proliferation ability was determined by MTT assay in A2058 cells treated as in B (***P < 0.001). E. The proliferation ability was determined by MTT assay in M21 cells treated as in C (***P < 0.001). F. The cell cycle distribution was determined by flow cytometry in A2058 cells treated as in B. G. The cell cycle distribution was determined by flow cytometry in M21 cells treated as in C.
NKILA promotes apoptosis, and inhibits migration and invasiveness of melanoma cells
Furthermore, in order to investigate the effect of NKILA on the apoptosis of melanoma cells, we used flow cytometry to detect the level of apoptosis of A2058 cells transfected with NKILA or the control. The results showed that the average percentages of apoptotic A2058 cells that were transfected with NKILA or the control were 23.764% and 2.018%, respectively, which suggests that NKILA significantly promoted apoptosis of A2058 cells (Figure 3A). Moreover, we measured the levels of apoptosis of M21 cells transfected with shNKILA or the control. The results showed that the average percentages of apoptotic M21 cells that were transfected with shNKILA or the control were 2.509% and 20.355%, respectively, which suggests that the silencing of NKILA significantly inhibited apoptosis of M21 cells (Figure 3B). In addition, we found that NKILA significantly inhibited the migration and invasion abilities of A2058 cells (P < 0.01) (Figure 3C) and that the silencing of NKILA significantly promoted the migration and invasion abilities of M21 cells (P < 0.001) (Figure 3D).
Figure 3.
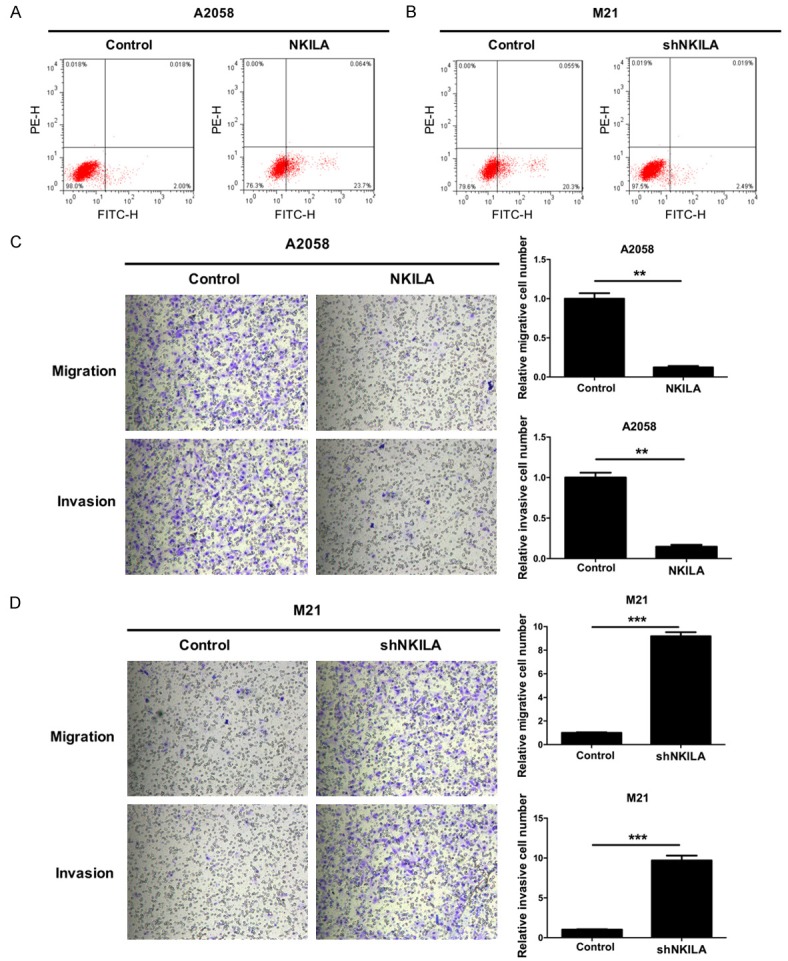
NKILA promotes apoptosis and inhibits the migration and invasiveness of melanoma cells. A. A2058 cells were transfected with either NKILA or the control, and then apoptotic cell death was measured by flow cytometric analysis with Annexin V-FITC and PE staining. B. M21 cells were transfected with shNKILA or the control, and then apoptotic cell death was measured by flow cytometric analysis with Annexin V-FITC and PE staining. C. NKILA significantly inhibited the migration and invasion abilities of A2058 cells. The number of cells that migrated and invaded was quantified, magnification 100 ×, **P < 0.001. D. The silencing of NKILA significantly promoted the migration and invasion abilities of M21 cells, magnification 100 ×, ***P < 0.001.
NKILA inhibits the proliferation, migration, and invasion abilities of melanoma cells through the inhibition of NF-ĸB expression
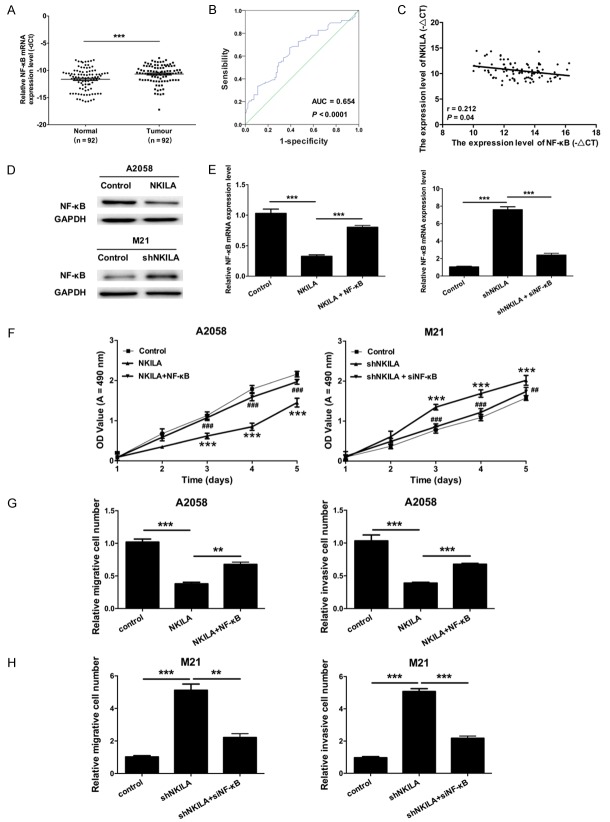
To further study the role of NF-ĸB in NKILA-induced melanoma tumorigenesis, we first assessed the expression level of NF-ĸB in 92 pairs of melanoma tissues and adjacent normal tissues by qRT-PCR. The results demonstrated that the expression level of NF-ĸB was higher in melanoma tissues compared with adjacent noncancerous tissues (P < 0.001) (Figure 4A). We also found that the expression level of NF-ĸB was significantly associated with invasion and metastasis (Table 2). Furthermore, the results showed that the area under the ROC curves of NF-ĸB was 0.654 (P < 0.0001, Figure 4B). The Pearson’s correlation algorithm was used to analyze the correlation coefficient and the significance between the expression of NKILA and NF-ĸB, which revealed a negative correlation between NKILA and NF-ĸB expression (r = 0.212, P = 0.04, Figure 4C). The western blot results revealed that the protein level of NF-κB was downregulated in A2058 cells transfected with NKILA compared with the control, while the protein level of NF-κB was up-regulated in M21 cells transfected with shNKILA compared with the control (Figure 4D). qRT-PCR results also revealed that NKILA inhibited the expression level of NF-ĸB mRNA, while the overexpression of NF-ĸB rescued the phenotype. The silencing of NKILA increased the expression level of NF-ĸB, while the knockdown of NF-ĸB rescued the phenotype (P < 0.001) (Figure 4E). Based on the results discussed above, an MTT assay indicated that NKILA inhibited the proliferation abilities of A2058 and M21 cells though NF-ĸB (P < 0.001) (Figure 4F). Likewise, Transwell assays also showed that NKILA inhibited the migration and invasion abilities of A2058 and M21 cells though NF-ĸB (P < 0.001) (Figure 4G, 4H).
Figure 4.
NKILA inhibits proliferation, migration, and invasion abilities of melanoma cells through the inhibition of NF-ĸB expression. A. qRT-PCR was performed to measure the mRNA expression level of NF-ĸB in 92 pairs of melanoma tissues and paired adjacent normal tissues (***P < 0.001). B. A receiver operating characteristic (ROC) curve was used to analyze the performance of NF-ĸB in the prediction of prognosis; the area under the ROC curve of NKILA was 0.654 (P < 0.0001). C. Pearson’s correlation algorithm was used to analyze the correlation coefficient and the significance between the mRNA expression levels of NKILA and NF-ĸB (r = 0.212, P = 0.04). D. Western blot was used to measure the protein expression level of NF-κB in the treated A2058 and M21 cells; GAPDH was used as a reference protein. E. A2058 cells were transfected with control, NKILA, or NKILA and NF-κB, and M21 cells were transfected with control, shNKILA or shNKILA and NF-κB siRNAs. qRT-PCR was performed to analyze the mRNA expression level of NF-ĸB (***P < 0.001). F. An MTT assay was performed to determine the proliferation abilities of A2058 and M21 cells treated as in E. G. A Transwell assay was used to detect the migration and invasion abilities of A2058 cells treated as in E. H. A Transwell assay was used to detect the migration and invasion abilities of M21 cells treated as in E.
Table 2.
Correlation analysis between NF-κB expression and clinicopathological characteristics of patients with melanoma
| Characteristics | No. of patients | Mean ± SD | P value |
|---|---|---|---|
| Total no. of patients | 92 | ||
| Age (year) | |||
| > 60 | 41 (44.6%) | 12.94 ± 2.45 | 0.996 |
| ≤ 60 | 51 (55.4%) | 13.11 ± 1.97 | |
| Gender | |||
| Male | 43 (46.7%) | 12.75 ± 1.97 | 0.485 |
| Female | 49 (53.3%) | 13.03 ± 1.86 | |
| Invasion | |||
| T0-T2 | 63 (68.5%) | 13.59 ± 1.97 | 0.039* |
| T3-T4 | 29 (31.5%) | 12.69 ± 1.79 | |
| Lymphatic metastasis | |||
| N0 | 69 (75%) | 13.16 ± 1.83 | 0.100 |
| N1-N3 | 23 (25%) | 12.47 ± 1.35 | |
| Distal metastasis | |||
| M0 | 73 (79.3%) | 12.97 ± 1.49 | 0.011* |
| M1 | 19 (20.7%) | 12.01 ± 1.19 | |
| TNM stage | |||
| 0 & I | 65 (70.7%) | 13.73 ± 1.89 | 0.002* |
| II & III & IV | 27 (29.3%) | 12.17 ± 2.52 |
Indicated statistical significance (P < 0.05).
NKILA suppresses the growth of melanoma tumors via NF-ĸB in vivo
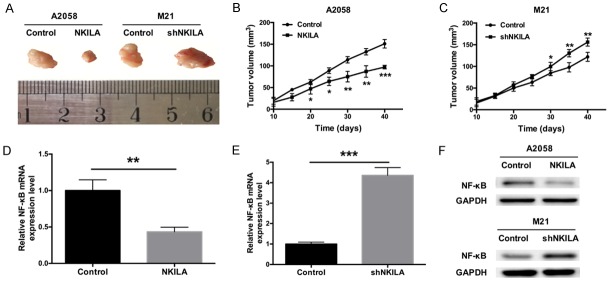
To explore the effect of NKILA on tumorigenesis in vivo, stable NKILA-overexpressing A2058 cells or stable NKILA knockdown M21 cells were implanted subcutaneously into nude mice; mice in the control groups received no cells. The tumors were then excised after 40 days (Figure 5A), and the tumor volumes were calculated at 10, 20, 30 and 40 days. The tumor volume was smaller in mice that received NKILA-overexpressing A2058 cells, while the tumor volume was larger in mice that received M21 cells in which NKILA was silenced; the tumor volume was smallest in mice from the control group (Figure 5B, 5C). The total RNA and protein of the tumor were extracted, and the expression levels of NF-ĸB mRNA and protein were detected by qRT-PCR and western blot, respectively. We found that the expression of NF-ĸB was dramatically decreased in mice that were injected with NKILA-overexpressing A2058 cells compared with the control group. In contrast, the expression of NF-ĸB was dramatically increased in mice that were injected with M21 cells, in which NKILA was silenced, compared with the control group (Figure 5D-F). These results further support that NKILA suppresses the growth of melanoma tumors via NF-ĸB in vivo.
Figure 5.
NKILA suppresses the growth of melanoma tumors via NF-ĸB in vivo. A. Nude mice were injected with A2058 and M21 cells stably transfected with NKILA, and the tumors were removed at a particular point in time. B. NKILA reduced the volume of the tumors induced in nude mice (*P < 0.05, **P < 0.01, ***P < 0.001). C. Silencing of NKILA expression increased the size of the tumors induced in nude mice (**P < 0.01, ***P < 0.001). D. Nude mice were injected with A2058 cells stably transfected with the NKILA overexpression vector or control cells. The tumors were removed, and the mRNA expression level of NF-ĸB was determined by qRT-PCR (**P < 0.01). E. Nude mice were injected with stable NKILA knockdown M21 cells or control cells. The tumors were removed, and the mRNA expression level of NF-ĸB was determined by qRT-PCR (***P < 0.001). F. Western blot was used to measure the protein expression level of NF-ĸB in the tumors of nude mice. GAPDH was used as a reference protein.
Discussion
An increasing number of lncRNAs are regarded as oncogenes or tumor suppressors, which will add new aspects to the molecular mechanisms of human cancers. Recent studies have shown the following: lncRNA-UCA1 promotes cell proliferation through Sox4 in esophageal cancer [29]; lncRNA-MA-linc1 regulates cell cycle progression, and increases the sensitivity of cancer cells to Paclitaxel [30]; lncRNA-NEAT1 is related to estrogen receptor alpha and is considered an important modulator of prostate cancer [12]; lncRNA-ATB participates in the progression of colorectal cancer and is related to its prognosis [31]; lncRNA PCAT29 inhibits the oncogenic phenotypes of prostate cancer [32]; lncRNA DSCAM-AS1 plays a crucial role in breast cancer progression [33]; androgen-induced lncRNA SOCS2-AS1 promotes cell proliferation and inhibits apoptosis of prostate cancer cells [34]. Among these studied lncRNAs, the lncRNA NKILA has been found to suppress the migration and invasion abilities of tongue squamous cell carcinoma cells through the inhibition of epithelial-mesenchymal transition (EMT) [35]. However, its role in melanoma is unclear. In our study, we found that NKILA was downregulated in human melanoma tissues and cell lines. We also found that NKILA was significantly associated with the invasion and metastasis of melanoma. The area under the ROC curve of NKILA was 0.875, which suggests that NKILA has a good prognostic value for melanoma. The results also indicated that NKILA inhibited cell proliferation, migration, and invasion, and promoted apoptosis of melanoma cells. Therefore, we speculated that NKILA may be a potential tumor suppressor in melanoma.
Research has suggested that NF-κB can be constitutively activated in various cancers and that it serves a number of pro-tumorigenic functions [36,37]. For example, the silencing of MARCH1 inhibits proliferation, migration and invasiveness of ovarian cancer through NF-κB and Wnt/β-catenin pathways [38]. The PRKD1 promoter acts on the KRas-NF-κB pathway in pancreatic cancer [39], and inhibition of NF-κB increases the sensitivity of breast cancer that is resistant to tamoxifen [40]. The inhibition of the NF-κB pathway induces the deregulation of EMT and neural invasion in pancreatic cancer [41]. Furthermore, studies have detected the characteristics of the NF-κB transcription factor in uveal melanoma [42], and other studies have shown that NF-κB is involved in the development of melanoma. For example, NF-κB mediates the anti-melanoma activity of Cynanchi atrati Radix [43] and regulates TNF-related apoptosis in melanoma cells [44]. Interestingly, it has been reported that the interaction of NF-κB and NKILA can reverse PMEPA1 expression in cancer [25], which reveals that NKILA may regulate the expression level of NF-κB. In our study, our results showed that NF-κB was highly expressed in human melanoma tissues. A negative correlation was also observed between NKILA and NF-κB expression. We also demonstrated that NKILA inhibited the progression of melanoma through the inhibition of NF-ĸB. Therefore, we speculated that NKILA has an important effect in the development and progression of melanoma via NF-ĸB.
In summary, we found that NKILA was downregulated and that NF-κB was upregulated in human melanoma tissues. NKILA and NF-κB were significantly associated with invasion and metastasis, and NKILA and NF-κB are both good prognostic biomarkers for melanoma. A negative correlation was observed between NKILA and NF-κB expression. Our results also indicated that NKILA inhibited the proliferation, migration, and invasion abilities of melanoma cells through the inhibition of NF-ĸB expression in vitro. More importantly, we found that NKILA suppressed the growth of melanoma tumors via NF-ĸB in vivo. Taken together, our study shows that NKILA suppressed the development of malignant melanoma via the regulation of NF-ĸB, and thus NKILA may be a potential therapeutic target in patients with melanoma (Figure 6).
Figure 6.
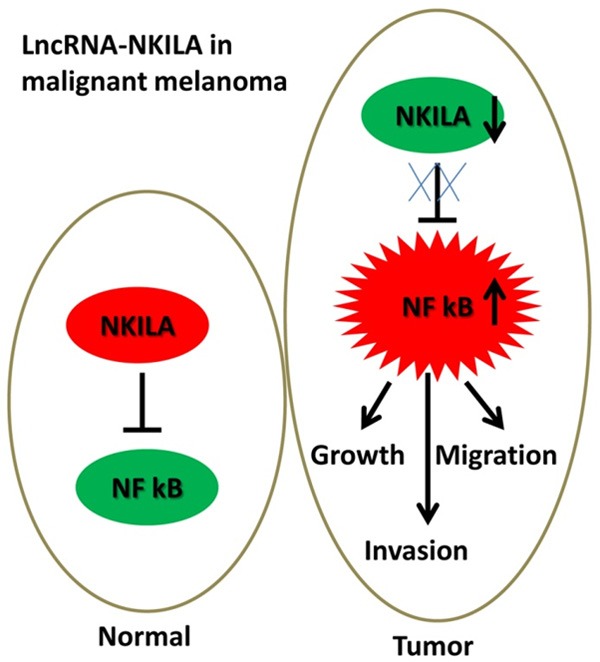
A Schematic model of NKILA in malignant melanoma. In normal cells, NKILA can regulate NF κB. In tumor cells, downregulated NKILA caused the upregulation of NF κB, which induced an increase in melanoma cell growth, invasion and migration.
Disclosure of conflict of interest
None.
References
- 1.Siegel R, Naishadham D, Jemal A. Cancer statistics, 2012. CA Cancer J Clin. 2012;62:10–29. doi: 10.3322/caac.20138. [DOI] [PubMed] [Google Scholar]
- 2.Ferlay J, Soerjomataram I, Dikshit R, Eser S, Mathers C, Rebelo M, Parkin DM, Forman D, Bray F. Cancer incidence and mortality worldwide: sources, methods and major patterns in GLOBOCAN 2012. Int J Cancer. 2015;136:E359–386. doi: 10.1002/ijc.29210. [DOI] [PubMed] [Google Scholar]
- 3.Coory M, Baade P, Aitken J, Smithers M, McLeod GR, Ring I. Trends for in situ and invasive melanoma in Queensland, Australia, 1982-2002. Cancer Causes Control. 2006;17:21–27. doi: 10.1007/s10552-005-3637-4. [DOI] [PubMed] [Google Scholar]
- 4.Holterhues C, Vries E, Louwman MW, Koljenovic S, Nijsten T. Incidence and trends of cutaneous malignancies in the Netherlands, 1989-2005. J Invest Dermatol. 2010;130:1807–1812. doi: 10.1038/jid.2010.58. [DOI] [PubMed] [Google Scholar]
- 5.Downing A, Newton-Bishop JA, Forman D. Recent trends in cutaneous malignant melanoma in the Yorkshire region of England; incidence, mortality and survival in relation to stage of disease, 1993-2003. Br J Cancer. 2006;95:91–95. doi: 10.1038/sj.bjc.6603216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Mansson-Brahme E, Johansson H, Larsson O, Rutqvist LE, Ringborg U. Trends in incidence of cutaneous malignant melanoma in a Swedish population 1976-1994. Acta Oncol. 2002;41:138–146. doi: 10.1080/028418602753669508. [DOI] [PubMed] [Google Scholar]
- 7.Hall HI, Miller DR, Rogers JD, Bewerse B. Update on the incidence and mortality from melanoma in the United States. J Am Acad Dermatol. 1999;40:35–42. doi: 10.1016/s0190-9622(99)70562-1. [DOI] [PubMed] [Google Scholar]
- 8.Bulliard JL, Cox B, Semenciw R. Trends by anatomic site in the incidence of cutaneous malignant melanoma in Canada, 1969-93. Cancer Causes Control. 1999;10:407–416. doi: 10.1023/a:1008964621225. [DOI] [PubMed] [Google Scholar]
- 9.Tryggvadottir L, Gislum M, Hakulinen T, Klint A, Engholm G, Storm HH, Bray F. Trends in the survival of patients diagnosed with malignant melanoma of the skin in the Nordic countries 1964-2003 followed up to the end of 2006. Acta Oncol. 2010;49:665–672. doi: 10.3109/02841861003702528. [DOI] [PubMed] [Google Scholar]
- 10.Garbe C, Leiter U. Melanoma epidemiology and trends. Clin Dermatol. 2009;27:3–9. doi: 10.1016/j.clindermatol.2008.09.001. [DOI] [PubMed] [Google Scholar]
- 11.Rincic M, Iourov IY, Liehr T. Thoughts about SLC16A2, TSIX and XIST gene like sites in the human genome and a potential role in cellular chromosome counting. Mol Cytogenet. 2016;9:56. doi: 10.1186/s13039-016-0271-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chakravarty D, Sboner A, Nair SS, Giannopoulou E, Li R, Hennig S, Mosquera JM, Pauwels J, Park K, Kossai M, MacDonald TY, Fontugne J, Erho N, Vergara IA, Ghadessi M, Davicioni E, Jenkins RB, Palanisamy N, Chen Z, Nakagawa S, Hirose T, Bander NH, Beltran H, Fox AH, Elemento O, Rubin MA. The oestrogen receptor alpha-regulated lncRNA NEAT1 is a critical modulator of prostate cancer. Nat Commun. 2014;5:5383. doi: 10.1038/ncomms6383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ying L, Chen Q, Wang Y, Zhou Z, Huang Y, Qiu F. Upregulated MALAT-1 contributes to bladder cancer cell migration by inducing epithelial-to-mesenchymal transition. Mol Biosyst. 2012;8:2289–2294. doi: 10.1039/c2mb25070e. [DOI] [PubMed] [Google Scholar]
- 14.Feil R, Walter J, Allen ND, Reik W. Developmental control of allelic methylation in the imprinted mouse Igf2 and H19 genes. Development. 1994;120:2933–2943. doi: 10.1242/dev.120.10.2933. [DOI] [PubMed] [Google Scholar]
- 15.Li H, Yu B, Li J, Su L, Yan M, Zhu Z, Liu B. Overexpression of lncRNA H19 enhances carcinogenesis and metastasis of gastric cancer. Oncotarget. 2014;5:2318–2329. doi: 10.18632/oncotarget.1913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Derrien T, Johnson R, Bussotti G, Tanzer A, Djebali S, Tilgner H, Guernec G, Martin D, Merkel A, Knowles DG, Lagarde J, Veeravalli L, Ruan X, Ruan Y, Lassmann T, Carninci P, Brown JB, Lipovich L, Gonzalez JM, Thomas M, Davis CA, Shiekhattar R, Gingeras TR, Hubbard TJ, Notredame C, Harrow J, Guigo R. The GENCODE v7 catalog of human long noncoding RNAs: analysis of their gene structure, evolution, and expression. Genome Res. 2012;22:1775–1789. doi: 10.1101/gr.132159.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Consortium EP. An integrated encyclopedia of DNA elements in the human genome. Nature. 2012;489:57–74. doi: 10.1038/nature11247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Guttman M, Rinn JL. Modular regulatory principles of large non-coding RNAs. Nature. 2012;482:339–346. doi: 10.1038/nature10887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Khalil AM, Guttman M, Huarte M, Garber M, Raj A, Morales DR, Thomas K, Presser A, Bernstein BE, van Oudenaarden A. Many human large intergenic noncoding RNAs associate with chromatin-modifying complexes and affect gene expression. Proc Natl Acad Sci U S A. 2009;106:11667–11672. doi: 10.1073/pnas.0904715106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Tsai MC, Spitale RC, Chang HY. Long intergenic noncoding RNAs: new links in cancer progression. Cancer Res. 2011;71:3–7. doi: 10.1158/0008-5472.CAN-10-2483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sen R, Baltimore D. Multiple nuclear factors interact with the immunoglobulin enhancer sequences. Cell 1986. 46: 705-716. J Immunol. 2006;177:7485–7496. [PubMed] [Google Scholar]
- 22.Sen R, Baltimore D. Multiple nuclear factors interact with the immunoglobulin enhancer sequences. Cell. 1986;46:705–716. [PubMed] [Google Scholar]
- 23.May MJ, Ghosh S. Signal transduction through NF-kappa B. Immunol Today. 1998;19:80–88. doi: 10.1016/s0167-5699(97)01197-3. [DOI] [PubMed] [Google Scholar]
- 24.Hoesel B, Schmid JA. The complexity of NF-kappaB signaling in inflammation and cancer. Mol Cancer. 2013;12:86. doi: 10.1186/1476-4598-12-86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Dijkstra JM, Alexander DB. The “NF-κB interacting long noncoding RNA” (NKILA) transcript is antisense to cancer-associated gene PMEPA1. F1000Res. 2015;4:96. doi: 10.12688/f1000research.6400.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Liu B, Sun L, Liu Q, Gong C, Yao Y, Lv X, Lin L, Yao H, Su F, Li D, Zeng M, Song E. A cytoplasmic NF-kappaB interacting long noncoding RNA blocks IkappaB phosphorylation and suppresses breast cancer metastasis. Cancer Cell. 2015;27:370–381. doi: 10.1016/j.ccell.2015.02.004. [DOI] [PubMed] [Google Scholar]
- 27.Chen Y, Lin MC, Yao H, Wang H, Zhang AQ, Yu J, Hui CK, Lau GK, He ML, Sung J, Kung HF. Lentivirus-mediated RNA interference targeting enhancer of zeste homolog 2 inhibits hepatocellular carcinoma growth through down-regulation of stathmin. Hepatology. 2007;46:200–208. doi: 10.1002/hep.21668. [DOI] [PubMed] [Google Scholar]
- 28.Jiang L, Lai YK, Zhang J, Wang H, Lin MC, He ML, Kung HF. Targeting S100P inhibits colon cancer growth and metastasis by Lentivirus-mediated RNA interference and proteomic analysis. Mol Med. 2011;17:709–716. doi: 10.2119/molmed.2011.00008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Jiao C, Song Z, Chen J, Zhong J, Cai W, Tian S, Chen S, Yi Y, Xiao Y. lncRNA-UCA1 enhances cell proliferation through functioning as a ceRNA of Sox4 in esophageal cancer. Oncol Rep. 2016;36:2960–2966. doi: 10.3892/or.2016.5121. [DOI] [PubMed] [Google Scholar]
- 30.Bida O, Gidoni M, Ideses D, Efroni S, Ginsberg D. A novel mitosis-associated lncRNA, MA-linc1, is required for cell cycle progression and sensitizes cancer cells to paclitaxel. Oncotarget. 2015;6:27880–27890. doi: 10.18632/oncotarget.4944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Iguchi T, Uchi R, Nambara S, Saito T, Komatsu H, Hirata H, Ueda M, Sakimura S, Takano Y, Kurashige J, Shinden Y, Eguchi H, Sugimachi K, Maehara Y, Mimori K. A long noncoding RNA, lncRNA-ATB, is involved in the progression and prognosis of colorectal cancer. Anticancer Res. 2015;35:1385–1388. [PubMed] [Google Scholar]
- 32.Malik R, Patel L, Prensner JR, Shi Y, Iyer MK, Subramaniyan S, Carley A, Niknafs YS, Sahu A, Han S, Ma T, Liu M, Asangani IA, Jing X, Cao X, Dhanasekaran SM, Robinson DR, Feng FY, Chinnaiyan AM. The lncRNA PCAT29 inhibits oncogenic phenotypes in prostate cancer. Mol Cancer Res. 2014;12:1081–1087. doi: 10.1158/1541-7786.MCR-14-0257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Niknafs YS, Han S, Ma T, Speers C, Zhang C, Wilder-Romans K, Iyer MK, Pitchiaya S, Malik R, Hosono Y, Prensner JR, Poliakov A, Singhal U, Xiao L, Kregel S, Siebenaler RF, Zhao SG, Uhl M, Gawronski A, Hayes DF, Pierce LJ, Cao X, Collins C, Backofen R, Sahinalp CS, Rae JM, Chinnaiyan AM, Feng FY. The lncRNA landscape of breast cancer reveals a role for DSCAM-AS1 in breast cancer progression. Nat Commun. 2016;7:12791. doi: 10.1038/ncomms12791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Misawa A, Takayama K, Urano T, Inoue S. Androgen-induced long noncoding RNA (lncRNA) SOCS2-AS1 promotes cell growth and inhibits apoptosis in prostate cancer cells. J Biol Chem. 2016;291:17861–17880. doi: 10.1074/jbc.M116.718536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Huang W, Cui X, Chen J, Feng Y, Song E, Li J, Liu Y. Long non-coding RNA NKILA inhibits migration and invasion of tongue squamous cell carcinoma cells via suppressing epithelial-mesenchymal transition. Oncotarget. 2016;7:62520–62532. doi: 10.18632/oncotarget.11528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Xia Y, Shen S, Verma IM. NF-kappaB, an active player in human cancers. Cancer Immunol Res. 2014;2:823–830. doi: 10.1158/2326-6066.CIR-14-0112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Viennois E, Chen F, Merlin D. NF-kappaB pathway in colitis-associated cancers. Transl Gastrointest Cancer. 2013;2:21–29. doi: 10.3978/j.issn.2224-4778.2012.11.01. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Meng Y, Hu J, Chen Y, Yu T, Hu L. Silencing MARCH1 suppresses proliferation, migration and invasion of ovarian cancer SKOV3 cells via downregulation of NF-kappaB and Wnt/beta-catenin pathways. Oncol Rep. 2016;36:2463–2470. doi: 10.3892/or.2016.5076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Doppler H, Panayiotou R, Reid EM, Maimo W, Bastea L, Storz P. The PRKD1 promoter is a target of the KRas-NF-kappaB pathway in pancreatic cancer. Sci Rep. 2016;6:33758. doi: 10.1038/srep33758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.deGraffenried LA, Chandrasekar B, Friedrichs WE, Donzis E, Silva J, Hidalgo M, Freeman JW, Weiss GR. NF-kappa B inhibition markedly enhances sensitivity of resistant breast cancer tumor cells to tamoxifen. Ann Oncol. 2004;15:885–890. doi: 10.1093/annonc/mdh232. [DOI] [PubMed] [Google Scholar]
- 41.Nomura A, Majumder K, Giri B, Dauer P, Dudeja V, Roy S, Banerjee S, Saluja AK. Inhibition of NF-kappa B pathway leads to deregulation of epithelial-mesenchymal transition and neural invasion in pancreatic cancer. Lab Invest. 2016;96:1268–1278. doi: 10.1038/labinvest.2016.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Dror R, Lederman M, Umezawa K, Barak V, Pe’er J, Chowers I. Characterizing the involvement of the nuclear factor-kappa B (NF kappa B) transcription factor in uveal melanoma. Invest Ophthalmol Vis Sci. 2010;51:1811–1816. doi: 10.1167/iovs.09-3392. [DOI] [PubMed] [Google Scholar]
- 43.Son SW, Kim HG, Han JM, Lee JS, Choi MK, Lee JS, Son CG. Anti-melanoma activity of Cynanchi atrati Radix is mediated by regulation of NF-kappa B activity and pro-apoptotic proteins. J Ethnopharmacol. 2014;153:250–257. doi: 10.1016/j.jep.2014.02.037. [DOI] [PubMed] [Google Scholar]
- 44.Franco AV, Zhang XD, Van Berkel E, Sanders JE, Zhang XY, Thomas WD, Nguyen T, Hersey P. The role of NF-kappa B in TNF-related apoptosis-inducing ligand (TRAIL)-induced apoptosis of melanoma cells. J Immunol. 2001;166:5337–5345. doi: 10.4049/jimmunol.166.9.5337. [DOI] [PubMed] [Google Scholar]