Abstract
Altered promoter DNA methylation is one of the most important epigenetic abnormalities in human cancer. DNMT3B, de novo methyltransferase, is clearly related to abnormal methylation of tumour suppressor genes, DNA repair genes and its overexpression contributes to oncogenic processes and tumorigenesis in vivo. The purpose of this study was to assess the effect of the overexpression of DNMT3B in HaCaT cells on global gene expression and on the methylation of selected genes to the identification of genes that can be target of DNMT3B. We found that the overexpression of DNMT3B in HaCaT cells, modulate the expression of genes related to cancer, downregulated the expression of 151 genes with CpG islands and downregulated the expression of the VAV3 gene via methylation of its promoter. These results highlight the importance of DNMT3B in gene expression and human cancer.
Keywords: Methylation, de novo methyltransferase, overexpression of DNMT3B, cancer, cancer-related genes, VAV3, CpG island
Introduction
Epigenetic and genetic alterations are common in the genesis and progression of various types human cancer. The abnormal expression of genes related to cell cycle, DNA repair, cellular metabolism and tumor suppressor are frequent defects that contribute to development of cancer [1]. Abnormal DNA methylation is one of the most important epigenetic factors directly involved in tumourigenesis, because methylation can induce repression of tumor suppressor genes or activation of oncogenes [2].
In human cancer the patterns of DNA methylation are altered: the overall level of DNA methylation is lower in normal cells than in cancer cells and the methylation of CpG islands of tumor suppressor and DNA repair is higher in cancer than normal cells [3]. DNA methylation at the 5’ cytosine of CpG sites is catalyzed by DNA methyltranferases (DNMTs). The DNMT family includes three enzymes, DNMT1 responsible for maintaining pre-existing methylation patterns after DNA replication and DNMT3A and DNMT3B, de novo methyltransferases that are required to establish methylation during development and imprinting [4,5]. Genetic abnormalities and aberrant overexpression of DNMTs contribute to DNA hypermethylation in cancer [6,7]. Inhibition of these enzymes in cancer can decrease DNA methylation, reactivate silence genes and diminish tumorigenicity [8]. Furthermore, it has been showed that DNMT3B is overexpressed in cell lines of cancer and in several types of primary tumors [9-14]. In several works of cancer, it has has been reported that there is a positive correlation between DNMT3B expression and promoter DNA methylation [11,13,15,16]. Interestingly, DNMT3B contributes to oncogenic processes and tumorigenesis in vivo by gene-specific de novo methylation and transcriptional silencing [17]. Overexpression of DNMT3B protein significantly contributes to elevated methyltransferase activity and hypermethylation in breast cancer cells [13]. Although, the important role of DNMT3B in cancer development is clear, at present only a few genes have been identified as targets for transcriptional regulation by this enzyme [18-21].
Therefore, the purpose of this study was to assess the effect of the overexpression of DNMT3B in HaCaT cells on global gene expression and on the methylation of selected genes to the identification of genes that can be target of DNMT3B. We found that the overexpression of DNMT3B in HaCaT cells downregulated the expression of VAV3, SORBS2, and GPR137 genes by microarray and RT-qPCR and a clear increase in DNA methylation was detected in VAV3 promoter.
Materials and methods
Cell culture and cervical samples
The HaCaT (human skin keratinocyte), C-33A (cervical cancer), HeLa (cervical cancer), SiHa (cervical cancer), A549 (lung adenocarcinoma) and MCF-7 (breast adenocarcinoma) cells lines were obtained from American Type Culture Collection (ATCC, USA), cultured in DMEM and F-12 1:1, medium supplemented with 10% fetal bovine serum, 100 U/ml penicillin and 100 µg/ml streptomycin. The cells were grown at 37°C in 5% CO2. The samples were collected at the Cancer Institute of the State of Guerrero located in southern Mexico. The population consisted of 25 healthy women and 25 women with cervical cancer. The diagnosis of normal cervix was done by cytomorphological examination through conventional Papanicolaou test and cervical cancer by histological diagnosis, according to the classification system of the International Federation of Gynecology and Obstetrics (FIGO). All samples were obtained after the patients gave their informed consent and the Bioethics and Research Committee of the Cancer Institute of the State of Guerrero, Mexico, approved the study, which followed the ethical guidelines of the 2008 Helsinki Declaration.
Transient transfection
Complementary DNA encoding DNMT3B was cloned into pcDNA3.1(+) plasmid (Invitrogent, Carlsbad, CA USA) to generate the pcDNA-DNMT3B expression plasmid that was confirmed by sequencing. The HaCaT cells (25 × 103 cells, 6-well plates) were transfected with Lipofectamine 2000 Reagent (Invitrogent) according to the manufacturer’s protocol. The cells were transfected with 3.5 µg of pcDNA-DNMT3B plasmid or empty vector pcDNA3.1(+) and after 48 h the cells were harvested for RNA and DNA extraction.
RNA and DNA extraction
Total RNA was isolated and purified from the cell lines and cervical tissue with Direct-zol RNA MiniPrep (ZYMO Research, Irvine, USA) according to the manufacturer’s instructions including DNase I treatment. RNA integrity was determined by electrophoresis in a 1% agarose gel. Genomic DNA was extracted from the cells using a standard phenol chloroform method [22]. The concentration of RNA and DNA was evaluated by spectrophotometry using NanoDrop 2000c (Thermo Scientific, Wilmington, DE USA).
Microarray analysis
H35K array was performed in Microarray Unit of Cellular Physiology Institute, UNAM, Mexico City. H35K contains 70-mer oligonucleotide probes representing 35764 human transcripts. Total RNA was extracted of HaCaT cells transfected with pcDNA-DNMT3B and of HaCaT cells transfected with pcDNA3.1(+) (empty vector). Equimolar concentrations of total RNA from of 3 independent experiments were mixed. Ten µg of RNA were used for cDNA synthesis and equal quantities of Cy3-labeled cDNA from control cells and Cy5-labeled cDNA from experimental cells were hibridized to the H35K array. Each hybridization was carried out in duplicate. Array signal intensities were analyzed with ScanArray 4000 from Packard BioChips. Microarray data analysis, background correction, normalization and selection of differentially expressed genes were performed with GenArise software (http://www.ifc.unam.mx/genarise/). Differentially expressed genes were selected according to the Z-score value [23]. Differential expressed genes were considered upregulated when Z-score > 1.5 standard deviation or downregulated when Z-score < 1.5 standard deviation.
Bioinformatics analysis
Gene ontology (GO) analysis of the differentially expressed genes was performed with PANTHER (http://www.pantherdb.org/) and according to the program an enrichment score of P < 0.05 was considered as significant. For promoter prediction we considered 3000 pb (-2000 pb to +1000 pb) relative to ATG using the ExPASy Bioinformatics Resource Portal (http://www.expasy.org/genomics). For CpG island prediction the criteria was regions > 200 bp with a GC content ≥ 50% with an observed CpG/expected CpG > 0.6 [24]. CpG islands prediction was done using the Methprimer Program (http://www.urogene.org/methprimer/). The prediction of transcription factors that can bind to VAV3 promoter was done with CONSITE database (http://consite.genereg.net/).
RT-qPCR
One hundred ng of total RNA were used in each RT-qPCR assay. Reverse transcription and quantitative PCR were performed with KAPA SYBR FAST One-Step qRT-PCR kit (Kapa Biosystems, Boston, Massachusetts, USA), according to the manufacturer’s protocol. In all cases, the conditions of reverse transcription and amplifications were: 30 s at 37°C, 42°C for 5 min and 95°C for 5 min; 40 cycles of amplification: 5 s at 95°C, 30 s at 60°C and 30 s at 72°C; melt curve: 15 s at 95°C, 1 min at 60°C and 15 s 95°C. The reactions were done in Real Time ABI-PRISM 7500 SDS (Applied Biosystems, Foster City, CA). Data were normalized using GAPDH as an internal control and relative expression differences were calculated using the 2-ΔΔCt method. Primers sequences are shown in Table 1.
Table 1.
Primer sequences used in this study
| Gene | Sequence | Tm °C |
|---|---|---|
| RT-qPCR | ||
| MSH2 | F5’-TTCATGGCTGAAATGTTGGA | 59 |
| R5’-ATGCTAACCCAAATCCATCG | ||
| NSD1 | F5’-TGAAGGCAGACATCAATTCG | 55 |
| R5’-CCAACTTGATTGAACCAGGAA | ||
| SORBS2 | F5’-AAGCACAGCCTGCAAGACCA | 60 |
| R5’-TGGGGTATTGGAGGGTCAGG | ||
| ARHGAP29 | F5’-TTAGAGGATGTTGTACGCC | 58 |
| R5’-TTCGATGAAAGTCTCCTGG | ||
| VAV3 | F5’-ACAAGGAGCCAGAACATTCAG | 58 |
| R5’-TTGCACAGAAGTCATACCGAG | ||
| GPR137 | F5’-TCAGCTATCAGACGGTGTTC | 52 |
| R5’-AGCAGTAGAGAAGCCAGAAG | ||
| C1ORF201 | F5’-CTTGTGAAGCAGTCGCCAAATACAT | 58 |
| F5’-CACGATCTCATACTGACCAGGACCT | ||
| THSD1 | F5’-GGAGGCCAACACCAATCAGA | 59 |
| R5’-CAGTAGTCACCAGCCTCCTT | ||
| ST6GALNAC2 | F5’-GGGTCGTTCTTCTGGCTGCT | 59 |
| R5’-TGATGTGGTGTCCCTGGCTC | ||
| MSX1 | F5’-CCAGAAGATGCGCTCGTCAA | 59 |
| R5’-TCGTCTTGTGTTTGCGGAGG | ||
| GAPDH | F5’-CCGGGAAACTGTGGCGTGATGG | 60 |
| R5’-AGGTGGAGGAGTGGGTGTCGCTGTT | ||
| MSP | ||
| VAV3-R1 M | F5’-GTTTTGGGGGATTTTATCGTATTAT | 58 |
| R5’-GACCCGCCACTAAACATACCCAAC | ||
| VAV3-R1 U | F5’-TGGGGGATTTTATTGTATTATAGTA | 55 |
| R5’-AACCCACCACTAAACATACCCAACA | ||
| VAV3-R2 M | F5’GGCGTTGGAGTCGGAAGTTTGTG | 60 |
| R5’-CACTACTTCCACGACTCCATACC | ||
| VAV3-R2 U | F5’-GGTGTTGGAGTTGGAAGTTTGTGT | 59 |
| R5’-CACACACTACTTCCACAACTCCATACC | ||
| SORBS2-R1 M | F5’-ATAATAAAAGAATAAATTTAGGTCGGG | 58 |
| R5’-CTATCGCCCAAACTAAAATACAAT | ||
| SORBS2-R1 U | F5’-TATAATAAAAGAATAAATTTAGGTTGGG | 54 |
| R5’-AAAATAAAATCTCACTCTATCAC | ||
| SORBS2-R2 M | F5’-GGGAATTATGTGTTAATTTAATTCG | 52 |
| R5’-AAATCATAAATACTAAACGCTCC | ||
| SORBS2-R2 U | F5’-GGAATTATGTGTTAATTTAATTTGATG | 56 |
| R5’-ATAAAATCATAAATACTAAACACTCC | ||
| BSP | ||
| VAV3-R1 | F5’-AGGGGGTTTTGGGGGATTTTAT | 56 |
| R5’-CCACTAAACATACCCAACA | ||
| VAV3-R2 | F5’-GGCGTTGGAGTCGGAAGTTTGTG | 60 |
| R5’-CACTACTTCCACGACTCCATACC | ||
| SORBS2-R1 | F5’-AGTTATAAAATTTTGATTGGTTGA | 58 |
| F5’-AACCTACAAACTTACTCTAAATCCTAT | ||
| SORBS2-R2 | F5’-GGAATGATGTTTATAGGGAATTATGTG | 59 |
| F5’-CCCTAAAAATAAAATCATAAATACTAAA | ||
| GPR137-R1 | F5’-GGGGGTATTGGAGATAAGGAAAGG | 59 |
| F5’-CTCCTCTCTCCTATACCCAAATC | ||
| GPR137-R2 | F5’-TTTTTTTTTTTTGAGGTTGGAG | 59 |
| F5’-CAAACCCCTCACTCAAAAACA | ||
Methylation-specific PCR (MSP) and bisulfite sequencing (BSP)
For MSP, 1 µg of DNA was treated with sodium bisulfite using the EpiTect Bisulfite kit (QUIAGEN, Hilden, Germany) according to the manufacturer’s instructions. MSP primer sequences are shown in Table 1. MSP was performed in a total of 10 µL, containing 1 µL of bisulfite-treated DNA, 250 nM of each primers and AmpliTaq Gold360 Master Mix (Applied Biosystems) and under the following amplification conditions: denaturation 95°C for 10 min, 40 cycles of amplification: 30 s at 95°C, 30 s at 60°C and 30 s at 72°C, and a final extension of 72°C for 10 min. Bisulfite sequencing was done for VAV3, SORBS2, and GPR137 genes. The promoters of these genes were divided into two regions to facilitate the methylation analysis. One hundred ng of bisulfite-treated DNA was used as a template, and PCR was performed using specific primers (Table 1). The reactions were done in Eppendorf Mastercycler EP Gradient 96 Thermal cycler (Applied Biosystems). The PCR products were gel purified and cloned into the pJET1.2/blunt vector (Thermo Scientific). Five independent clones were subjected to automated sequencing (ABI Prism 310 Genetic Analyzer (Applied Biosystems).
Statistical analysis
The data are shown as mean ± standard deviation. The P value was determined using Student’s t-test. P values below 0.05 were considered statistically significant.
Results
DNMT3B has an important role in aberrant DNA methylation to repress transcription. To identify downregulated genes by DNMT3B, we overexpressed DNMT3B in the HaCaT cell line, and H35K microarray that interrogated 35764 genes was used to identify changes in gene expression. We found 1085 downregulated genes, 1741 upregulated genes and 32938 unchanged genes (Figure 1A). To gain insights into the biological processes where 1085 downregulated genes are implicated, we carried out a gene ontology (GO) analysis using Protein Analysis Through Evolutionary Relationships (PANTHER). This analysis revealed that an important part of the 1085 downregulated genes are involved in the immune system, development processes, cell communication, cellular processes and metabolic processes (Figure 1B). The GO analysis for the 1741 upregulated genes is shown in Supplementary Figure 1.
Figure 1.
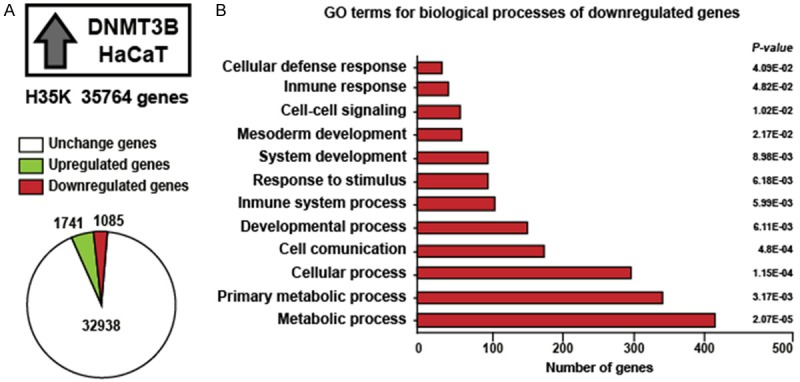
Gene ontology analysis of downregulated genes by overexpression of DNMT3B in HaCaT cells. A: We used H35K array of 35764 genes, the graph shows the number of genes that change their expression by overexpression of DNMT3B. B: Gene ontology (GO) analysis for downregulated genes by overexpression of DNMT3B.
The 1085 downregulated genes were classified according to Z-score value (Figure 2A). We narrowed down this group of genes by the selection of gene subsets with Z-scores of -2 to -6.8 (252 genes). Hypermethylation of CpG islands found within promoters is clearly related to transcriptional repression. Therefore, to relate the 252 downregulated genes with the methylation of its promoter by overexpression DNMT3B, we used MethPrimer to prediction of CpG islands for 252 genes. We found 151 genes with CpG islands, 73 genes without CpG islands and 28 genes with absent data (Figure 2B). To know the biological processes where 151 genes with CpG islands are involved, we carried out GO analysis. We found that some of these genes are implicated in molecular and cellular processes altered in cancer such as adhesion, apoptosis, response to stimulus, development, biological regulation and metabolic processes (Figure 2C). Among the 151 genes with CpG islands, we find genes with previous reports of abnormal methylation in several types human tumors, many genes putative or tumor suppressor and genes related with cancer. The complete list of 151 genes with CpG islands is shown in Supplementary Table 1.
Figure 2.
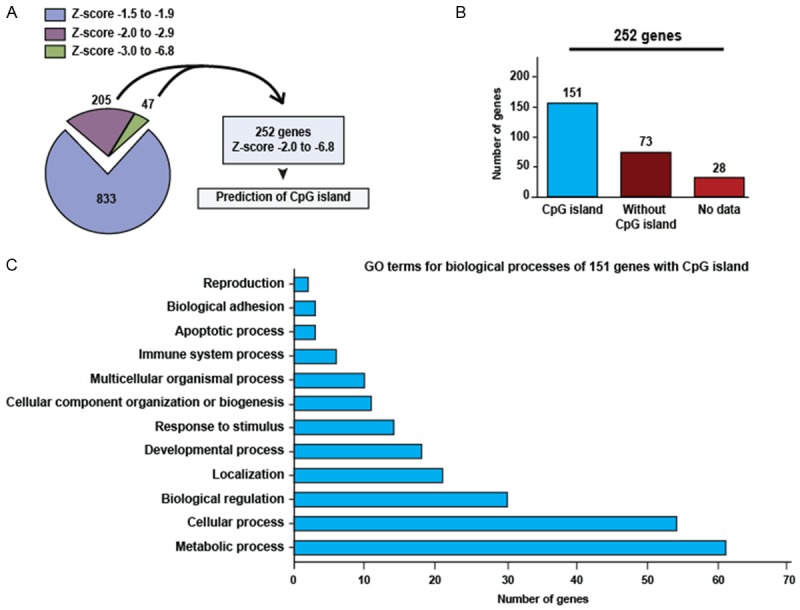
Prediction of CpG island in downregulated genes by overexpression of DNMT3B in HaCaT cells. A: Classification of downregulated genes according to Z-score value, the graph shows the number of genes for each Z-score range. B: Number of genes with and without CpG island. C: Gene ontology (GO) analysis for 151 genes with CpG island.
To validate the results of the microarray, we analyzed the level of expression of 10 genes by RT-qPCR. These 10 genes were selected for further validation because 1) they were downregulated by overexpression of DNMT3B, 2) they have CpG islands and 3) they are involved in regulating important molecular and cellular functions which are disrupted in cancer. The function of 10 genes is shown in Supplementary Table 2. The level of expression of 7 genes was consistent with data from microarray analysis and inconsistent in three genes (Figure 3). The analysis by RT-qPCR showed that expression levels of SORBS2, VAV3 and GPR137 mRNAs were significantly downregulated by the overexpression of DNMT3B.
Figure 3.
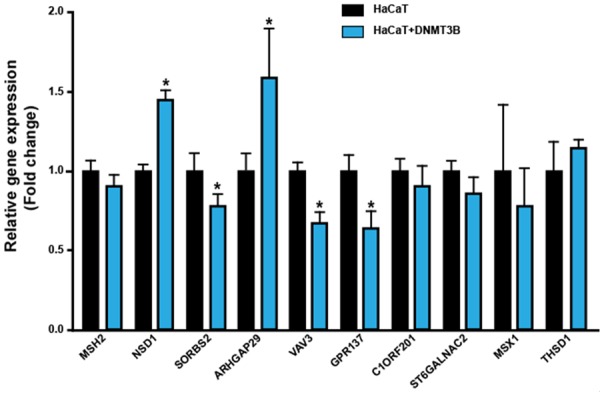
Validation of microarray data by RT-qPCR. mRNA quantification of 10 genes in HaCaT cells with overexpression of DNMT3B and control HaCaT cells. The bars represent the mean ± standard deviation from at least three independent experiments. *P < 0.05
To clarify whether downregulation of VAV3, SORBS2, and GPR137 is mediated by DNA hypermethylation in overexpression of DNMT3B HaCaT cells, we analyzed the methylation status of its promoters by using methylation-specific PCR (MSP) and bisulfite conversion and sequencing. For the VAV3 gene, its CpG island spanning from -599 pb to +20 pb of the transcription start site, within of this region we found 95 CpGs sites (Figure 4A). No obvious methylation changes were observed between HaCaT cells and HaCaT cells with overexpression of DNMT3B by MSP analysis (Figure 4B). To make a more detailed analysis of methylation status, we analyzed the methylation in the 95 CpGs sites of the VAV3 promoter. We found two small, more densely methylated regions (15, 16, 17, 18, 19 and 21 CpG sites of region 1 and 52, 53, 54, 55, 56, 57, 58 and 59 CpG sites of region 2) of the VAV3 promoter in HaCaT cells with overexpression of DNMT3B in comparison with HaCaT cells (Figure 4C). These results suggest that the overexpression of DNMT3B in HaCaT cells probably has a role in the methylation of the VAV3 promoter. The MSP and bisulfite conversion and sequencing analysis was done for SORBS2, and GPR137 genes but no methylation changes were observed between HaCaT cells with overexpression of DNMT3B and HaCaT cells (Supplementary Figures 2 and 3).
Figure 4.

Methylation analysis of VAV3 promoter in HaCaT cells. A: Schematic representation of the CpG island and CpG sites in the VAV3 promoter. For methylation analysis the VAV3 promoter was divided into 2 regions: R1 -599 to -307 with 34 CpGs and R2 -299 to +20 with 61 CpGs, the positions are relative to the transcription start site. The primers for MSP and BSP are indicated by black and red arrows, respectively. Each CpG site is represented by a vertical bar. B: The methylation status of the VAV3 promoter (R1 and R2) was determined by MSP in HaCaT cells with overexpression of DNMT3B and control HaCaT cells. U showed unmethylation-specific primer amplification, M showed methylation-specific primer amplification. C: BSP analysis of the VAV3 promoter (R1 and R2) in HaCaT cells with overexpression of DNMT3B and control HaCaT cells. Black circles represent methylated CpG site and white circles represent unmethylated CpG site. The red box shows the two regions more densely methylated by overexpression of DNMT3B.
Finally, to correlate our results with what occurs in human cancer, we analyzed the expression of DNMT3B in cervical cancer samples and normal cervical tissue. As well as DNMT3B, VAV3, SORBS2 and GPR137 expression in cervical, lung and breast cancer cell lines. RT-qPCR analysis showed that mRNA level of DNMT3B in cervical cancer samples was significantly higher that in normal tissue (Figure 5A). In general, in the analyzed cell lines, we found overexpression of DNMT3B and low levels VAV3, SORBS2 and GPR137 (Figure 5B). These results suggest that overexpression of DNMT3B can be a common event in human cancer and expression of VAV3, SORBS2 and GPR137 could be regulated by DNMT3B.
Figure 5.
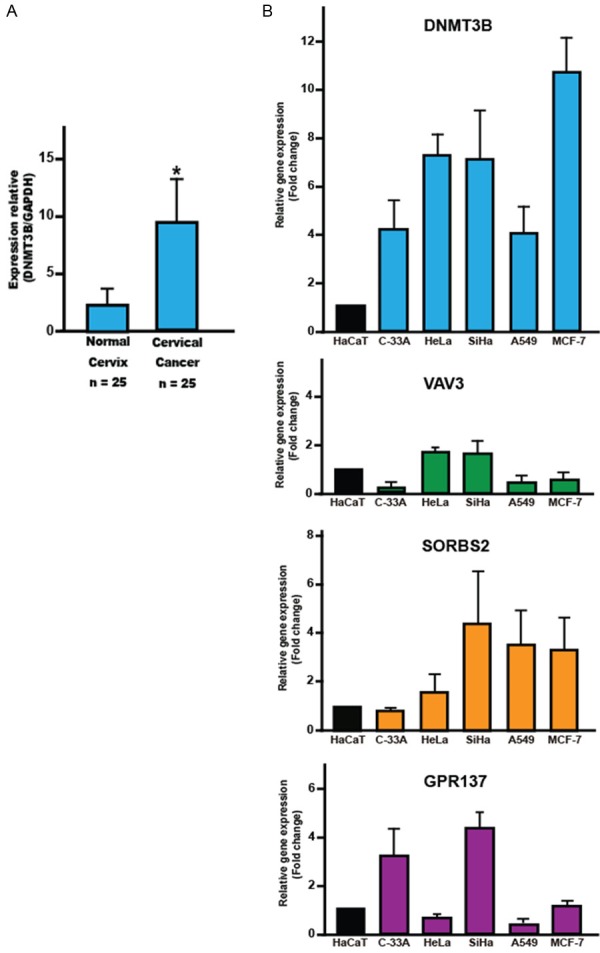
Expression of DNMT3B, VAV3, SORBS2 and GPR137 in human cancer by RT-qPCR. A: mRNA expression levels of DNMT3B in cervical cancer and normal cervix. The mRNA expression levels of GAPDH were used as internal control. B: mRNA expression levels of DNMT3B, VAV3, SORBS2 and GPR137 in cervical, lung and breast cancer cell lines. The data are presented as the fold change in cancer cell line relative to HaCaT cell line. The bars represent the mean ± standard deviation from at least three independent experiments. *P < 0.05.
Discussion
DNMT3B overexpression and abnormal methylation of tumour suppressor and DNA repair genes are common alterations in several types of human cancer [6,25]. There is evidence indicating the involvement of DNMT3B in the initiation and progression of cancer [20,26]. In addition DNMT3B is clearly related to the abnormal methylation in cancer [21,27]. Although only 5 genes have been identified as targets for transcriptional repression by DNMT3B [18-21].
In this work the overexpression of DNMT3B in HaCaT cells downregulated 151 genes with CpG islands. This result suggests that the downregulated genes could be result from the methylation of its promoter by DNMT3B overexpression. In this sense, it has been reported that DNMT3B preferably to methylate CpG-dense promoter regions and is excluded from active promoters [28]. Also, downregulation or repression by methylation requires promoters with high methylated-cytocines [29-31]. In initiation and progression of cancer, DNMT3B has directly or indirectly been associated with abnormal expression and methylation [8,26,27]. An similar scenario it could be also seen in our study in which of the downregulated 151 genes by DNMT3B were found 22 genes with previous reported of abnormal methylation in several types of human cancer, 9 reported as putative or tumor suppressors genes and 61 genes related to many aspects of human cancer.
The overexpression of DNMT3B in HaCaT cells, downregulated the expression of VAV3, SORBS2, and GPR137 genes by RT-qPCR, but a clear increase in DNA methylation was only detected in the VAV3 promoter. Therefore it is possible that the VAV3 gene is regulated by DNMT3B via methylation of its promoter. VAV3 is a guanine nucleotide exchange factor involved in the regulation of Rho GTPases and in several cellular processes, including regulation of cytoskeleton organization, cell transformation and oncogenesis [32-34]. In addition, abnormal methylation of the VAV3 promoter has been reported in breast cancer cell lines and in gastric cancer the methylation of its promoter is considered as a marker to estimate the fraction of cancer cells in primary gastric cancer [35,36]. On the other hand, we detected methylation of the VAV3 promoter in HaCaT cells without overexpression of DNMT3B. Although this result is unexpected, previously methylation of the VAV3 promoter in normal cells of the gastric mucosa has been reported [36]. By in silico analysis with CONSITE we detected that the transcription factors: Sp1, AP2 alpha, MZF, E2F, Hen-1 and Thing1-E47 can bind to localized sites in the more densely methylated regions of the VAV3 promoter. It is well known that the methylation of CpG in the Sp1 binding site generally interferes with its binding and can affect the transcription [37,38]. The E2F transcription factor, does not bind DNA when their site recognition is methylated [39]. To some promoters AP2 alpha can act as a suppressor for Sp1 binding, also the AP2 alpha binding to DNA may initiate transcriptional silencing by recruiting of DNMTs [40,41]. Therefore it is possible that the methylation of binding sites Sp1, AP2 alpha and E2F located in the two more densely methylated regions of VAV3 promoter can inhibit its binding and its subsequent transcriptional activation. This event could explain the expression decrease of the VAV3 gene in HaCaT cells with overexpression of DNMT3B.
The overexpression of DNMT3B in HaCaT cells, downregulates the expression of SORBS2 and GPR137 genes, but the methylation of its promoters do not increase. SORBS2 is a scaffold protein involved in the assembly of signaling complexes in stress fibers and actin cytoskeleton [42,43]. This gene is considered as putative tumour suppressor and although there is evidence of the loss or decrease of its expression in cervical and pancreatic cancer [44,45], there is no evidence that this is due to promoter methylation. GPR137 is an integral membrane protein that belongs to the GPR137 family of cell mediators of signal transduction [46,47]. Although the role of GPR137 in cancer is little known, several reports indicate that this gene is important a regulator of cell growth, apoptosis, invasion and migration in different types of human cancer [48-52]. Similar to SORBS2 there are no reports of abnormal methylation of the GPR137 promoter in human cancer. It is therefore likely that additional events are causing the downregulation the expression of SORBS2 and GPR137 genes. For example, methylation-independent repressor activities of DNMT3B [53].
In the current study, we found overexpression of DNMT3B in cervical cancer and various cancer cell lines. This event has been previously reported in various types of human cancer [8,9,13]. We also reported overexpression of DNMT3B and low levels of VAV3, SORBS2 and GPR137 in cervical, lung and breast cancer cell lines. This could indicate that the findings in the DNMT3B overexpression in HaCaT cells model also occur in primary human tumors and human cancer cell lines.
In conclusion, our results suggest that the overexpression of DNMT3B in HaCaT cells, modulate the expression of genes related to cancer, downregulate the expression of 151 genes with CpG islands and downregulate the expression of the VAV3 gene via methylation of its promoter. These findings highlight the importance of DNMT3B in the gene expression and human cancer.
Acknowledgements
We thank Lorena Chávez, José Luis Santillán, Simón Guzmán and Jorge Ramírez Salcedo for technical assistance in the microarray data analysis. This study was supported by grant from CONACYT (242812), México.
Disclosure of conflict of interest
None.
Supporting Information
References
- 1.Hanahan D, Weinberg RA. Hallmarks of cancer: the next generation. Cell. 2011;144:646–674. doi: 10.1016/j.cell.2011.02.013. [DOI] [PubMed] [Google Scholar]
- 2.Feinberg AP, Tycko B. The history of cancer epigenetics. Nat Rev Cancer. 2004;4:143–153. doi: 10.1038/nrc1279. [DOI] [PubMed] [Google Scholar]
- 3.Sharma S, Kelly TK, Jones PA. Epigenetics in cancer. Carcinogenesis. 2010;31:27–36. doi: 10.1093/carcin/bgp220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Okano M, Bell DW, Haber DA, Li E. DNA methyltransferases Dnmt3a and Dnmt3b are essential for de novo methylation and mammalian development. Cell. 1999;99:247–257. doi: 10.1016/s0092-8674(00)81656-6. [DOI] [PubMed] [Google Scholar]
- 5.Robert MF, Morin S, Beaulieu N, Gauthier F, Chute IC, Barsalou A, MacLeod AR. DNMT1 is required to maintain CpG methylation and aberrant gene silencing in human cancer cells. Nat Genet. 2003;33:61–65. doi: 10.1038/ng1068. [DOI] [PubMed] [Google Scholar]
- 6.Esteller M. Epigenetics in cancer. N Engl J Med. 2008;358:1148–1159. doi: 10.1056/NEJMra072067. [DOI] [PubMed] [Google Scholar]
- 7.Ley TJ, Ding L, Walter MJ, McLellan MD, Lamprecht T, Larson DE, Kandoth C, Payton JE, Baty J, Welch J, Harris CC, Lichti CF, Townsend RR, Fulton RS, Dooling DJ, Koboldt DC, Schmidt H, Zhang Q, Osborne JR, Lin L, O’Laughlin M, McMichael JF, Delehaunty KD, McGrath SD, Fulton LA, Magrini VJ, Vickery TL, Hundal J, Cook LL, Conyers JJ, Swift GW, Reed JP, Alldredge PA, Wylie T, Walker J, Kalicki J, Watson MA, Heath S, Shannon WD, Varghese N, Nagarajan R, Westervelt P, Tomasson MH, Link DC, Graubert TA, DiPersio JF, Mardis ER, Wilson RK. DNMT3A mutations in acute myeloid leukemia. N Engl J Med. 2010;363:2424–2433. doi: 10.1056/NEJMoa1005143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Rhee I, Bachman KE, Park BH, Jair KW, Yen RW, Schuebel KE, Cui H, Feinberg AP, Lengauer C, Kinzler KW, Baylin SB, Vogelstein B. DNMT1 and DNMT3b cooperate to silence genes in human cancer cells. Nature. 2002;416:552–556. doi: 10.1038/416552a. [DOI] [PubMed] [Google Scholar]
- 9.Choi MS, Shim YH, Hwa JY, Lee SK, Ro JY, Kim JS, Yu E. Expression of DNA methyltransferases in multistep hepatocarcinogenesis. Hum Pathol. 2003;34:11–17. doi: 10.1053/hupa.2003.5. [DOI] [PubMed] [Google Scholar]
- 10.Girault I, Tozlu S, Lidereau R, Bieche I. Expression analysis of DNA methyltransferases 1, 3A, and 3B in sporadic breast carcinomas. Clin Cancer Res. 2003;9:4415–4422. [PubMed] [Google Scholar]
- 11.Mizuno S, Chijiwa T, Okamura T, Akashi K, Fukumaki Y, Niho Y, Sasaki H. Expression of DNA methyltransferases DNMT1, 3A, and 3B in normal hematopoiesis and in acute and chronic myelogenous leukemia. Blood. 2001;97:1172–1179. doi: 10.1182/blood.v97.5.1172. [DOI] [PubMed] [Google Scholar]
- 12.Robertson KD, Jones PA. DNA methylation: past, present and future directions. Carcinogenesis. 2000;21:461–467. doi: 10.1093/carcin/21.3.461. [DOI] [PubMed] [Google Scholar]
- 13.Roll JD, Rivenbark AG, Jones WD, Coleman WB. DNMT3b overexpression contributes to a hypermethylator phenotype in human breast cancer cell lines. Mol Cancer. 2008;7:15. doi: 10.1186/1476-4598-7-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Saito Y, Kanai Y, Sakamoto M, Saito H, Ishii H, Hirohashi S. Overexpression of a splice variant of DNA methyltransferase 3b, DNMT3b4, associated with DNA hypomethylation on pericentromeric satellite regions during human hepatocarcinogenesis. Proc Natl Acad Sci U S A. 2002;99:10060–10065. doi: 10.1073/pnas.152121799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ahluwalia A, Hurteau JA, Bigsby RM, Nephew KP. DNA methylation in ovarian cancer. II. Expression of DNA methyltransferases in ovarian cancer cell lines and normal ovarian epithelial cells. Gynecol Oncol. 2001;82:299–304. doi: 10.1006/gyno.2001.6284. [DOI] [PubMed] [Google Scholar]
- 16.Rahman MM, Qian ZR, Wang EL, Yoshimoto K, Nakasono M, Sultana R, Yoshida T, Hayashi T, Haba R, Ishida M, Okabe H, Sano T. DNA methyltransferases 1, 3a, and 3b overexpression and clinical significance in gastroenteropancreatic neuroendocrine tumors. Hum Pathol. 2010;41:1069–1078. doi: 10.1016/j.humpath.2010.01.011. [DOI] [PubMed] [Google Scholar]
- 17.Wang J, Bhutani M, Pathak AK, Lang W, Ren H, Jelinek J, He R, Shen L, Issa JP, Mao L. Delta DNMT3B variants regulate DNA methylation in a promoter-specific manner. Cancer Res. 2007;67:10647–10652. doi: 10.1158/0008-5472.CAN-07-1337. [DOI] [PubMed] [Google Scholar]
- 18.Fan H, Chen L, Zhang F, Quan Y, Su X, Qiu X, Zhao Z, Kong KL, Dong S, Song Y, Chan TH, Guan XY. MTSS1, a novel target of DNA methyltransferase 3B, functions as a tumor suppressor in hepatocellular carcinoma. Oncogene. 2012;31:2298–2308. doi: 10.1038/onc.2011.411. [DOI] [PubMed] [Google Scholar]
- 19.Ghoshal K, Motiwala T, Claus R, Yan P, Kutay H, Datta J, Majumder S, Bai S, Majumder A, Huang T, Plass C, Jacob ST. HOXB13, a target of DNMT3B, is methylated at an upstream CpG island, and functions as a tumor suppressor in primary colorectal tumors. PLoS One. 2010;5:e10338. doi: 10.1371/journal.pone.0010338. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 20.Linhart HG, Lin H, Yamada Y, Moran E, Steine EJ, Gokhale S, Lo G, Cantu E, Ehrich M, He T, Meissner A, Jaenisch R. Dnmt3b promotes tumorigenesis in vivo by gene-specific de novo methylation and transcriptional silencing. Genes Dev. 2007;21:3110–3122. doi: 10.1101/gad.1594007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Teneng I, Tellez CS, Picchi MA, Klinge DM, Yingling CM, Snider AM, Liu Y, Belinsky SA. Global identification of genes targeted by DNMT3b for epigenetic silencing in lung cancer. Oncogene. 2015;34:621–630. doi: 10.1038/onc.2013.580. [DOI] [PubMed] [Google Scholar]
- 22.Davis LG, Kuehl WM, Battey JF. Basic methods in molecular biology. Norwalk, Conn.: Appleton & Lange; 1994. [Google Scholar]
- 23.Cheadle C, Vawter MP, Freed WJ, Becker KG. Analysis of microarray data using Z score transformation. J Mol Diagn. 2003;5:73–81. doi: 10.1016/S1525-1578(10)60455-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Takai D, Jones PA. Comprehensive analysis of CpG islands in human chromosomes 21 and 22. Proc Natl Acad Sci U S A. 2002;99:3740–3745. doi: 10.1073/pnas.052410099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Jones PA, Baylin SB. The epigenomics of cancer. Cell. 2007;128:683–692. doi: 10.1016/j.cell.2007.01.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Saito Y, Kanai Y, Sakamoto M, Saito H, Ishii H, Hirohashi S. Expression of mRNA for DNA methyltransferases and methyl-CpG-binding proteins and DNA methylation status on CpG islands and pericentromeric satellite regions during human hepatocarcinogenesis. Hepatology. 2001;33:561–568. doi: 10.1053/jhep.2001.22507. [DOI] [PubMed] [Google Scholar]
- 27.Kim GD, Ni J, Kelesoglu N, Roberts RJ, Pradhan S. Co-operation and communication between the human maintenance and de novo DNA (cytosine-5) methyltransferases. EMBO J. 2002;21:4183–4195. doi: 10.1093/emboj/cdf401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Baubec T, Colombo DF, Wirbelauer C, Schmidt J, Burger L, Krebs AR, Akalin A, Schubeler D. Genomic profiling of DNA methyltransferases reveals a role for DNMT3B in genic methylation. Nature. 2015;520:243–247. doi: 10.1038/nature14176. [DOI] [PubMed] [Google Scholar]
- 29.Boyes J, Bird A. Repression of genes by DNA methylation depends on CpG density and promoter strength: evidence for involvement of a methyl-CpG binding protein. EMBO J. 1992;11:327–333. doi: 10.1002/j.1460-2075.1992.tb05055.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hsieh CL. Dependence of transcriptional repression on CpG methylation density. Mol Cell Biol. 1994;14:5487–5494. doi: 10.1128/mcb.14.8.5487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Weber M, Hellmann I, Stadler MB, Ramos L, Paabo S, Rebhan M, Schubeler D. Distribution, silencing potential and evolutionary impact of promoter DNA methylation in the human genome. Nat Genet. 2007;39:457–466. doi: 10.1038/ng1990. [DOI] [PubMed] [Google Scholar]
- 32.Bustelo XR. Regulatory and signaling properties of the Vav family. Mol Cell Biol. 2000;20:1461–1477. doi: 10.1128/mcb.20.5.1461-1477.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Bustelo XR. Vav proteins, adaptors and cell signaling. Oncogene. 2001;20:6372–6381. doi: 10.1038/sj.onc.1204780. [DOI] [PubMed] [Google Scholar]
- 34.Uen YH, Fang CL, Hseu YC, Shen PC, Yang HL, Wen KS, Hung ST, Wang LH, Lin KY. VAV3 oncogene expression in colorectal cancer: clinical aspects and functional characterization. Sci Rep. 2015;5:9360. doi: 10.1038/srep09360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Loss LA, Sadanandam A, Durinck S, Nautiyal S, Flaucher D, Carlton VE, Moorhead M, Lu Y, Gray JW, Faham M, Spellman P, Parvin B. Prediction of epigenetically regulated genes in breast cancer cell lines. BMC Bioinformatics. 2010;11:305. doi: 10.1186/1471-2105-11-305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zong L, Hattori N, Yoda Y, Yamashita S, Takeshima H, Takahashi T, Maeda M, Katai H, Nanjo S, Ando T, Seto Y, Ushijima T. Establishment of a DNA methylation marker to evaluate cancer cell fraction in gastric cancer. Gastric Cancer. 2016;19:361–9. doi: 10.1007/s10120-015-0475-2. [DOI] [PubMed] [Google Scholar]
- 37.Cao YX, Jean JC, Williams MC. Cytosine methylation of an Sp1 site contributes to organ-specific and cell-specific regulation of expression of the lung epithelial gene t1alpha. Biochem J. 2000;350:883–890. [PMC free article] [PubMed] [Google Scholar]
- 38.Kim TW, Lee SJ, Oh BM, Lee H, Uhm TG, Min JK, Park YJ, Yoon SR, Kim BY, Kim JW, Choe YK, Lee HG. Epigenetic modification of TLR4 promotes activation of NF-kappaB by regulating methyl-CpG-binding domain protein 2 and Sp1 in gastric cancer. Oncotarget. 2016;7:4195–4209. doi: 10.18632/oncotarget.6549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Campanero MR, Armstrong MI, Flemington EK. CpG methylation as a mechanism for the regulation of E2F activity. Proc Natl Acad Sci U S A. 2000;97:6481–6486. doi: 10.1073/pnas.100340697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Bennett KL, Romigh T, Arab K, Teresi RE, Tada Y, Eng C, Plass C. Activator protein 2 alpha (AP2alpha) suppresses 42 kDa C/CAAT enhancer binding protein alpha (p42(C/EBPalpha)) in head and neck squamous cell carcinoma. Int J Cancer. 2009;124:1285–1292. doi: 10.1002/ijc.24087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Liu H, Tan BC, Tseng KH, Chuang CP, Yeh CW, Chen KD, Lee SC, Yung BY. Nucleophosmin acts as a novel AP2alpha-binding transcriptional corepressor during cell differentiation. EMBO Rep. 2007;8:394–400. doi: 10.1038/sj.embor.7400909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Flynn DC. Adaptor proteins. Oncogene. 2001;20:6270–6272. doi: 10.1038/sj.onc.1204769. [DOI] [PubMed] [Google Scholar]
- 43.Kioka N, Ueda K, Amachi T. Vinexin, CAP/ponsin, ArgBP2: a novel adaptor protein family regulating cytoskeletal organization and signal transduction. Cell Struct Funct. 2002;27:1–7. doi: 10.1247/csf.27.1. [DOI] [PubMed] [Google Scholar]
- 44.Backsch C, Rudolph B, Steinbach D, Scheungraber C, Liesenfeld M, Hafner N, Hildner M, Habenicht A, Runnebaum IB, Durst M. An integrative functional genomic and gene expression approach revealed SORBS2 as a putative tumour suppressor gene involved in cervical carcinogenesis. Carcinogenesis. 2011;32:1100–1106. doi: 10.1093/carcin/bgr093. [DOI] [PubMed] [Google Scholar]
- 45.Taieb D, Roignot J, Andre F, Garcia S, Masson B, Pierres A, Iovanna JL, Soubeyran P. ArgBP2-dependent signaling regulates pancreatic cell migration, adhesion, and tumorigenicity. Cancer Res. 2008;68:4588–4596. doi: 10.1158/0008-5472.CAN-08-0958. [DOI] [PubMed] [Google Scholar]
- 46.Daub H, Weiss FU, Wallasch C, Ullrich A. Role of transactivation of the EGF receptor in signalling by G-protein-coupled receptors. Nature. 1996;379:557–560. doi: 10.1038/379557a0. [DOI] [PubMed] [Google Scholar]
- 47.Vanti WB, Nguyen T, Cheng R, Lynch KR, George SR, O’Dowd BF. Novel human G-protein-coupled receptors. Biochem Biophys Res Commun. 2003;305:67–71. doi: 10.1016/s0006-291x(03)00709-5. [DOI] [PubMed] [Google Scholar]
- 48.Cui X, Liu Y, Wang B, Xian G, Liu X, Tian X, Qin C. Knockdown of GPR137 by RNAi inhibits pancreatic cancer cell growth and induces apoptosis. Biotechnol Appl Biochem. 2015;62:861–867. doi: 10.1002/bab.1326. [DOI] [PubMed] [Google Scholar]
- 49.Ren J, Pan X, Li L, Huang Y, Huang H, Gao Y, Xu H, Qu F, Chen L, Wang L, Hong Y, Cui X, Xu D. Knockdown of GPR137,G protein-coupled receptor 137, inhibits the proliferation and migration of human prostate cancer cells. Chem Biol Drug Des. 2016;87:704–13. doi: 10.1111/cbdd.12704. [DOI] [PubMed] [Google Scholar]
- 50.Shao X, Liu Y, Huang H, Zhuang L, Luo T, Ge X. Down-regulation of G protein-coupled receptor 137 by RNA interference inhibits cell growth of two hepatoma cell lines. Cell Biol Int. 2015;39:418–426. doi: 10.1002/cbin.10412. [DOI] [PubMed] [Google Scholar]
- 51.Wang C, Liang Q, Chen G, Jing J, Wang S. Inhibition of GPR137 suppresses proliferation of medulloblastoma cells in vitro. Biotechnol Appl Biochem. 2015;62:868–873. doi: 10.1002/bab.1331. [DOI] [PubMed] [Google Scholar]
- 52.Wang Z, Zhang H, Wang J, Yang Y, Wu Q. RNA interference-mediated silencing of G protein-coupled receptor 137 inhibits human gastric cancer cell growth. Mol Med Rep. 2015;11:2578–2584. doi: 10.3892/mmr.2014.3091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Haney SL, Hlady RA, Opavska J, Klinkebiel D, Pirruccello SJ, Dutta S, Datta K, Simpson MA, Wu L, Opavsky R. Methylation-independent repression of Dnmt3b contributes to oncogenic activity of Dnmt3a in mouse MYC-induced T-cell lymphomagenesis. Oncogene. 2015;34:5436–5446. doi: 10.1038/onc.2014.472. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


