Abstract
Increasing evidences demonstrated that long noncoding RNAs (lncRNAs) are frequently dysregulated and have critical roles in many tumors. However, the roles and functional mechanisms of lncRNAs in melanoma remain largely unknown. In this study, we identified a novel lncRNA MHENCR which was upregulated in melanoma tissues and further upregulated in metastatic melanoma. Increased expression of MHENCR indicted poor survival of melanoma patients. Functional experiments revealed that MHENCR knockdown significantly inhibited melanoma cells proliferation, induced cell cycle arrest and apoptosis, and also attenuated melanoma cells migration in vitro. Furthermore, we identified MHENCR as a competitively endogenous RNA, which specifically bound to miR-425 and miR-489, upregulated their target genes IGF1 and SPIN1 expression, and further activated PI3K-Akt pathway. Statistically significant correlations were observed between MHENCR expression and IGF1 and SPIN1 in melanoma tissues. In vivo functional experiments further confirmed the pro-growth and pro-metastasis roles of MHENCR. Collectively, our findings revealed that MHENCR functions as an oncogene in melanoma via activating miR-425/489-mediated PI3K-Akt pathway, and may be a therapeutic target for melanoma.
Keywords: Melanoma, long noncoding RNA, competing endogenous RNA, miR-425, miR-489, PI3K-Akt pathway
Introduction
With great progressions in cancer prevention and early diagnosis, the incidence for all cancers combined is reducing, but the incidence for melanoma continues to increase for the past 30 years in the world [1,2]. Although early melanoma can be cured by surgical resection, the late metastatic melanoma has high mortality rate for the lack of effective treatment [3,4]. Therefore, it is urgent to identify molecular drivers of melanoma tumorigenesis and progression [5]. Several reports have revealed some molecules contributing to the progression of melanoma [6,7]. However, the precise mechanisms governing melanoma progression are not fully understood [8]. Further studies are needed to reveal the underlying molecular mechanisms and develop novel methods for melanoma treatment.
Recently, increasing evidences indicate that in addition to proteins, noncoding RNAs also have critical roles in tumor progression [9]. An important portion of these noncoding transcripts is microRNA (miRNA), which is a class of small noncoding RNA with 21-25 nucleotides in lengths [10,11]. miRNA directly binds to 3’-untranslated region of its target genes and induces the translation inhibition and/or gradation of target genes mRNA [12]. Through regulating critical oncogenes or tumor suppressors, miRNA also exerts significant effects on tumor progression [13-15], including melanoma [16], such as the roles of miR-425 in inhibiting melanoma metastasis through targeting IGF1 [17]. Long noncoding RNA (lncRNA) is another important portion of functional noncoding RNAs with greater than 200 nucleotides in length [18-21]. Its dysregulation has recently been reported in various diseases including tumors [22,23]. lncRNAs also regulate many oncogenes and tumor suppressors expression transcriptionally or post-transcriptionally with complex regulation mechanisms [24,25]. The pivotal roles of lncRNAs in tumors including melanoma have been revealed in many reports [26-28]. Although several studies have investigated lncRNAs in melanoma, their completely biological roles and molecular mechanisms are still obscure [29].
In a previous study identifying differently expressed lncRNAs in human melanoma using RNA sequencing [27], we noted that lncRNA XLOC_013615 (ENST00000449500; Refseq NR_132417.1) was significantly upregulated in melanoma, and was further increased in metastatic lesion. In our own clinical melanoma tissues, we confirmed the upregulation of XLOC_013615 in melanoma tissues and metastatic melanomas and hence named MHENCR (melanoma highly expressed noncoding RNA). In this study, we analyzed its association with clinical characteristics and melanoma patients’ prognosis, investigated its biological roles in melanoma using in vitro and In vivo functional experiments. Moreover, we explored the underlying molecular mechanisms contributing to the functions of MHENCR in melanoma.
Materials and methods
Clinical samples
Thirty malignant melanoma tissues, twenty age and gender-matched skin tissues with melanocytic nevus, and sixteen pairs of primary melanoma tissues and lymph node metastatic lesions were obtained from patients who underwent surgery at the 253rd Hospital of PLA (Hohhot, Inner Mongolia, China). The clinical characteristics of melanoma patients and controls are shown in Supplementary Tables 1, 2, 3. All samples were confirmed by histological diagnosis. The Review Board of the 253rd Hospital of PLA approved the use of clinical samples, and all the patients signed written informed consent.
Cell cultures
The human melanoma cell lines A375 and SK-MEL-2 were purchased from Cell Resource Center, Shanghai Institutes for Biological Sciences (Shanghai, China). A375 cells were cultured in DMEM, and SK-MEL-2 cells were cultured in MEM medium, both supplemented with 10% fetal bovine serum (Gibco, CA, USA) in 5% CO2 atmosphere at 37°C.
RNA extraction and quantitative PCR (qPCR)
Total RNA was isolated using Trizol Reagent (Invitrogen, CA, USA) following the manufacturer’s instructions. Reverse transcription was carried out using PrimeScriptTM II 1st Strand cDNA Synthesis Kit (Takara, Dalian, China) according to the manufacturer’s instructions. qPCR was subsequently carried out using SYBR® Premix Ex TaqTM II (Takara, Dalian, China) on ABI StepOne Plus system (Applied Biosystems, CA, USA) following the manufacturer’s manual. The gene expression was calculated using 2-ΔΔCt method. β-actin was used as endogenous control. The primers sequences are as follows: MHENCR, 5’-ATGGTCAGTGGACGGACG-3’ (forward) and 5’-ACAGCAAGCACAAGGGTG-3’ (reverse); IGF1, 5’-GTGTGTGGAGACAGGGGCTT-3’ (forward) and 5’-ACTTGGCAGGCTTGAGGGG-3’ (reverse); SPIN1, 5’-AAAGAGACACTTGGATGG-3’ (forward) and 5’-CGATGTTTTTTGTGGGATG-3’ (reverse); β-actin, 5’-GGGAAATCGTGCGTGACATTAAG-3’ (forward) and 5’-TGTGTTGGCGTACAGGTCTTTG-3’ (reverse).
5’ and 3’ Rapid Amplification of cDNA Ends (RACE)
5’-RACE and 3’-RACE analyses were performed to determine the full-length sequences of MHENCR using a SMARTerTM RACE cDNA Amplification Kit (Clontech, Palo Alto, CA, USA) following the manufacturer’s manual. The primers sequences are as follows: 5’-RACE, 5’-TCCGTCCACTGACCATCGTCCCTCG-3’; 3’-RACE, 5’-CGAGTGCGGTGGCTCATGCCTTTTAC-3’.
Isolation of cytoplasmic and nuclear RNA
Cytoplasmic and nuclear RNA were isolated with the Cytoplasmic & Nuclear RNA Purification Kit (Norgen, Belmont, CA, USA) following the manufacturer’s manual.
miRNAs and plasmids transfection
The miRNAs mimics and inhibitors of miR-425 and miR-489 were purchased from GenePharma (Shanghai, China). miRNAs and plasmids were transfected into melanoma cells using Lipofectamine 3000 (Invitrogen, CA, USA) following the manufacturer’s manual.
Generation of cells stably depleting MHENCR
To inhibit MHENCR expression, three oligonucleotides for shRNAs were synthesized and inserted into the shRNA expression vector pGPH1/Neo (GenePharma, Shanghai, China). The shRNAs sequences are as follows: shRNA #1, 5’-AGGATCCCGGATTCCGTATCA-3’; shRNA #2, 5’-GGTGTCATCGAACACCCTTGT-3’; shRNA #3, 5’-GCTGTAATTAGAGTTGCACAT-3’. A scrambled shRNA was used as negative control. To obtain MHENCR stably depleted melanoma cells, A375 and SK-MEL-2 cells were transfected with the shRNAs expression plasmid, and selected with neomycin for four weeks.
Cell proliferation, cell cycle analysis, and apoptosis assays
Cell Counting Kit-8 (CCK-8) assays were performed to assess cell proliferation using the cell counting kit 8 (Dojindo Laboratories, Kumamoto, Japan) following the manufacturer’s manual. Briefly, a total of approximately 5.0 × 103 cells/well was plated in 96-well plate. After culture for 24, 48, and 72 hours, cell viability was measured by a microplate reader. Cell cycle distribution was measured using the Cell Cycle Analysis Kit (Biyuntian, Jiangsu, China) on a flow cytometer following the manufacturer’s manual. Terminal deoxynucleotidyl transferase (TdT)-mediated dUTP nick end labeling (TUNEL) assays were performed to assess cell apoptosis using the TUNEL Cell Apoptosis Detection Kit (Beyotime, Jiangsu, China) following the manufacturer’s manual. Representative images were acquired by Zeiss axiophot photomicroscope (Carl Zeiss, Oberkochen, Germany) and the results were quantified by Image-Pro plus 6.0 software.
Transwell and scratch assays
To assess the mobility capability of melanoma cells, transwell assays and scratch assays were performed. For transwell assays, a total of 1 × 105 cells in serum-free medium with 1 μg/ml Mitomycin C were seeded into the upper well of a poly-carbonate transwell chamber (BD Biosciences, USA) plated in a 24-well plate. After incubation for 24 hours, cells on the upper surface of the well were scraped off with a cotton swab, and cells on the lower surface were fixed, stained and counted. For scratch assays, cells were plated in 6-well plate and incubated at 37°C until 90% to 95% confluent. Then the cells were scratched with a pipette tip to form a wound in the middle of the plates, followed by rinsed with PBS and replaced with serum-free medium. After incubation for 1, 2, and 3 days, the fractions of cell coverage across the scratched surface were quantified.
Western blot
Protein lysates were prepared in a 1 × sodium dodecyl sulfate loading buffer. Equal amounts of proteins were separated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) and transferred to polyvinylidene fluoride membranes. After incubated with blocking solution, the membranes were blotted with primary antibodies specific for IGF1, SPIN1, PIK3CA, phosphorylated Akt, cyclinD1, BCL2, or β-actin (Abcam, Hong Kong, China). After washed for three times, the membranes were blotted with HRP-conjugated goat anti-rabbit or anti-mouse secondary antibody (Abcam) and detected with enhanced chemiluminescence.
RNA Immunoprecipitation (RIP)
The full-length MHENCR transcript was PCR amplified with the Ex Taq® Hot Start Version DNA Polymerase (Takara, Dalian, China) and subcloned into the Hind III and EcoR I sites of pcDNA3.1 plasmid (Invitrogen), named pcDNA3.1-MHENCR. The primers sequences are as follows: 5’-CCCAAGCTTGTCAGCTCCTAACGCCGCA-3’ (forward) and 5’-GGAATTCCGACTTTATTGACATTTATTTCC-3’ (reverse). pSL-MS2-12X (Addgene) was double digested with EcoR I and Xho I, and the MS2-12X fragment was subcloned into pcDNA3.1 and pcDNA3.1-MHENCR, named pcDNA3.1-MS2 or pcDNA3.1-MS2-MHENCR, respectively. The pcDNA3.1-MS2-MHENCR with point mutations in miRNAs binding sites were synthesized by GenScript (Nanjing, China), named pcDNA3.1-MS2-MHENCR-Mut. pcDNA3.1-MS2, pcDNA3.1-MS2-MHENCR, or pcDNA3.1-MS2-MHENCR-Mut was cotransfected with pGFP-MS2 into A375 cells. Forty-eight hours later, RIP assays were carried out using the Magna RIPTM RNA-Binding Protein Immunoprecipitation Kit (Millipore, Bedford, MA, USA) and anti-GFP antibody (Roche, Mannheim, Germany) according to the manufacturer’s protocols. The retrieved miRNAs were quantified by qPCR using TaqMan miRNA assays following the manufacturer’s manual (Applied Biosystems, CA, USA). For anti-AGO2 RIP, miR-425 or miR-489 mimics and negative control were transfected into A375 or SK-MEL-2 cells. Forty-eight hours later, RIP assays were carried out using the Magna RIPTM RNA-Binding Protein Immunoprecipitation Kit (Millipore, Bedford, MA, USA) and anti-AGO2 antibody (Millipore). The retrieved RNAs were quantified by qPCR.
In vivo xenograft assays
MHENCR stably depleted and control A375 cells (3.0 × 106) were subcutaneously injected into the flanks of athymic BALB/c nude mice. Subcutaneous tumor growth was measured and tumor volume was calculated according to V = 0.5 × LW2 (L, tumor length; W, tumor width). After labeled with luciferase, 1.0 × 106 MHENCR stably depleted or control A375 cells were injected into the tail vein of nude mouse to assess lung metastases. At 6th week after injection, the metastases were monitored using the IVIS Lumina II Imaging System (Caliper Life Sciences, Hopkinton, MA, USA). The Review Board of the 253rd Hospital of PLA approved the animal studies.
Statistical analysis
All statistical analyses were performed using GraphPad Prism Software. For comparisons, Pearson chi-square test, Mann-Whitney U test, Kruskal-Wallis test, Dunn’s multiple comparison test, Wilcoxon signed-rank test, Log-rank test, Student’s t test, and Pearson correlation analysis were performed as indicated. A p value < 0.05 was defined as statistically significant.
Results
MHENCR is upregulated in melanoma and associated with poor prognosis of melanoma patients
MHENCR expression in 30 malignant melanoma tissues and 20 age and gender-matched skin tissues with melanocytic nevus was measured by qPCR. As shown in Figure 1A, MHENCR expression levels were significantly upregulated in melanoma tissues compared with that in control skin tissues. The melanoma patients were grouped according to TNM stages, and MHENCR expression levels were significantly upregulated in later stages melanoma tissues compared with that in early stages melanoma tissues (Figure 1B). However, MHENCR expression levels did not change with age or between males and females (Supplementary Figure 1). We further measured MHENCR expression in 16 pairs of primary melanoma tissues and lymph node metastatic lesions. Significant upregulated expressions of MHENCR were observed in metastatic lesions compared with that in primary melanoma tissues (Figure 1C). Furthermore, we performed Kaplan-Meier survival analysis to investigate the association between MHENCR expression and patients’ prognosis. As shown in Figure 1D, increased MHENCR expression in melanoma tissues indicated poor overall survival. Collectively, these data suggested that MHENCR is upregulated in melanoma tissues and further upregulated in later stage and metastatic lesions. High MHENCR expression is associated with poor prognosis of melanoma patients.
Figure 1.
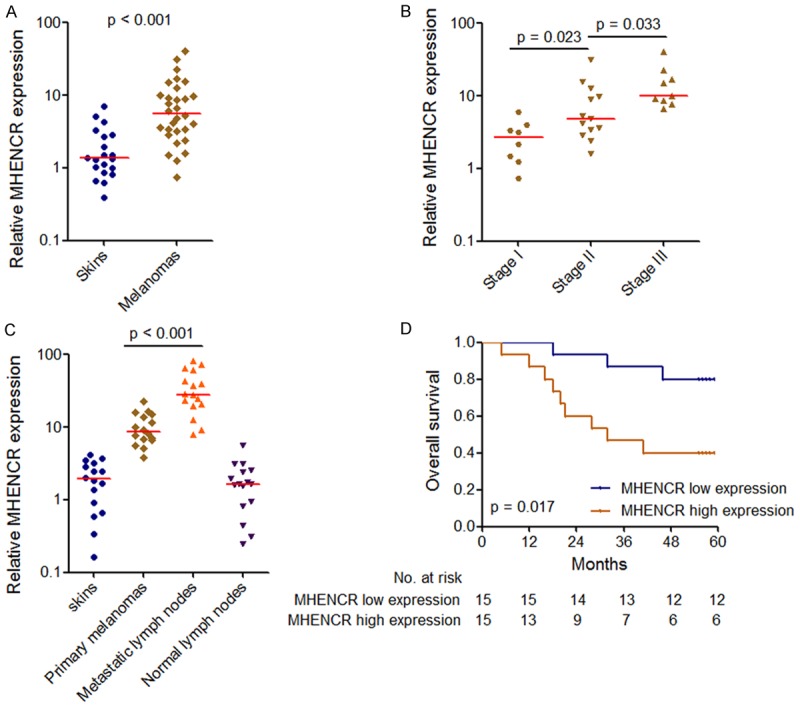
Expression of MHENCR in melanoma and its association with melanoma patients’ prognosis. A: MHENCR expression levels in 30 malignant melanoma tissues and 20 skin tissues were measured by qPCR. ***P < 0.001 by Mann-Whitney U test. B: MHENCR expression levels in 30 melanoma tissues with different clinical stages were measured by qPCR. χ2 = 14.186, P < 0.001 by Kruskal-Wallis test. The p values for the comparisons of different clinical stages by Dunn’s multiple comparison test are shown on the graph. C: MHENCR expression levels in 16 pairs of primary melanoma tissues and lymph node metastatic lesions were measured by qPCR. P < 0.001 by Wilcoxon signed-rank test. D: Kaplan-Meier survival analysis of the association between MHENCR expression and overall survival of melanoma patients. MHENCR median expression level was used as the cutoff. p value was acquired by Log-rank test.
Knockdown of MHENCR attenuates melanoma cells proliferation in vitro
The full-length sequences of MHENCR were confirmed by RACE analyses and are shown in Supplementary Figure 2A. Subcellular fractionation assays revealed that MHENCR was predominantly localized in the cytoplasm of melanoma cells (Supplementary Figure 2B). To explore the biological functions of MHENCR in melanoma, we stably knocked down MHENCR in melanoma cells A375 and SK-MEL-2 using three independent MHENCR specific shRNAs. As shown in Figure 2A and 2B, shRNA #2 and shRNA #3 had significant knockdown efficiencies in both A375 and SK-MEL-2 cells, and were used in subsequent experiments. Cell proliferation was evaluated using CCK-8 assays, and the results showed that knockdown of MHENCR by both shRNAs significantly attenuated A375 and SK-MEL-2 cells proliferation (Figure 2C and 2D). To investigate whether the effects of MHENCR on melanoma cells proliferation are dependent on the regulation of cell cycle or cell apoptosis, we measured cell cycle distribution by flow cytometry. As shown in Figure 2E and 2F, knockdown of MHENCR by both shRNAs increased G1/G0 phase proportion and reduced S phase and G2/M phase proportion in A375 and SK-MEL-2 cells. TUNEL staining showed that knockdown of MHENCR by both shRNAs induced A375 and SK-MEL-2 cells apoptosis (Figure 2G). These data demonstrated that knockdown of MHENCR attenuated melanoma cells proliferation and induced cell cycle arrest and apoptosis.
Figure 2.
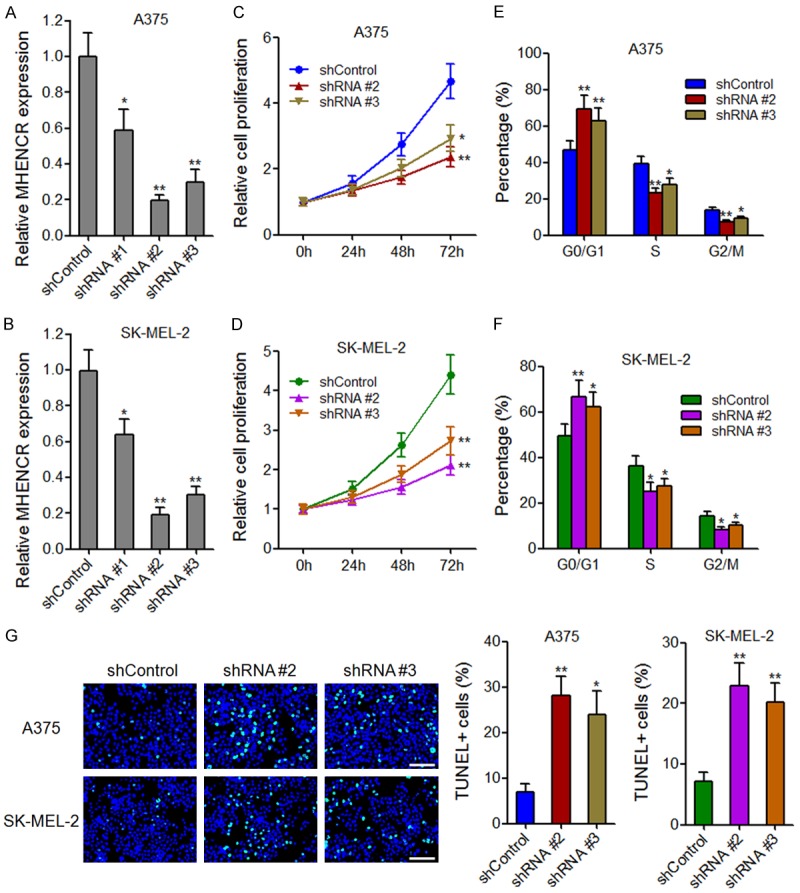
The effects of MHENCR on melanoma cell proliferation, cell cycle and apoptosis. A: MHENCR expression in MHENCR stably depleted and control A375 cells. B: MHENCR expression in MHENCR stably depleted and control SK-MEL-2 cells. C: The effect of MHENCR knockdown on A375 cells proliferation was measured by CCK-8 assays. D: The effect of MHENCR knockdown on SK-MEL-2 cells proliferation was measured by CCK-8 assays. E: The effect of MHENCR knockdown on A375 cell cycle distribution was measured by flow cytometry. F: The effect of MHENCR knockdown on SK-MEL-2 cell cycle distribution was measured by flow cytometry. G: The effect of MHENCR knockdown on A375 and SK-MEL-2 cells apoptosis was measured by TUNEL staining. Scale bars, 100 µm. For all panels, results are shown as mean ± SD. n = 3, *P < 0.05, **P < 0.01 by Student’s t test.
Knockdown of MHENCR attenuates melanoma cells migration in vitro
To investigate the effects of MHENCR on melanoma cells migration, transwell assays and scratch assays were performed. Transwell assays showed that knockdown of MHENCR by both shRNAs significantly attenuated A375 and SK-MEL-2 cells migration (Figure 3A). Scratch assays showed that the wounded area coverage was much slower in MHENCR depleted A375 and SK-MEL-2 cells (Figure 3B and 3C). These data suggested that knockdown of MHENCR attenuated melanoma cells mobility in vitro.
Figure 3.
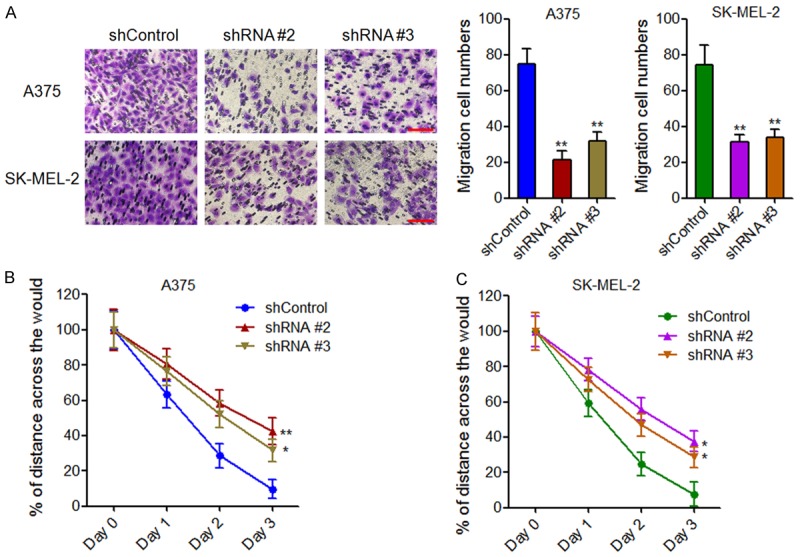
The effects of MHENCR on melanoma cell migration. A: The effect of MHENCR knockdown on A375 and SK-MEL-2 cells migration was measured by transwell assays. Scale bars, 100 µm. B: The effect of MHENCR knockdown on A375 cells mobility was measured by scratch assays. C: The effect of MHENCR knockdown on SK-MEL-2 cells mobility was measured by scratch assays. For all panels, results are shown as mean ± SD. n = 3, *P < 0.05, **P < 0.01 by Student’s t test.
MHENCR directly associates with miR-425 and miR-489
Recently, many lncRNAs are reported to function as competing endogenous RNA via binding miRNAs and to further regulate miRNAs target genes expression [30-32]. Because MHENCR was predominantly localized in the cytoplasm, we hypothesized that MHENCR could also directly bind miRNAs and function as competing endogenous RNA. miRcode program was used to predict potential miRNAs binding sites on MHENCR [33]. The program predicted two miR-425 binding sites and one miR-489 binding sites on MHENCR (Figure 4A). To investigate the association between miRNAs and MHENCR, we performed RIP assays using MS2 vector system (Figure 4B). The results showed that MHENCR specifically bound to miR-425 and miR-489, which was abolished by the mutation of the predicted miR-425 binding sites or the predicted miR-489 binding sites, respectively (Figure 4C). It is well known that miRNAs bind and recruit AGO2 to target genes to induce translation inhibition and/or gradation of target mRNAs [34,35]. So we performed anti-AGO RIP in miR-425 or miR-489 overexpressed A375 and SK-MEL-2 cells. As shown in Figure 4D and 4E, overexpression of miR-425 or miR-489 both significantly increased the association between AGO2 and MHENCR, supporting the specific association between miR-425, miR-489 and MHENCR. Furthermore, we measured MHENCR expression in miR-425 or miR-489 overexpressed A375 and SK-MEL-2 cells. The results showed that miR-425 and miR-489 did not change MHENCR expression (Figure 4F and 4G). Collectively, these data suggested that MHENCR physically associated with miR-425 and miR-489, but was not degraded by miR-425 and miR-489, and implied that MHENCR may function as competing endogenous RNA for miR-425 and miR-489 in melanoma.
Figure 4.
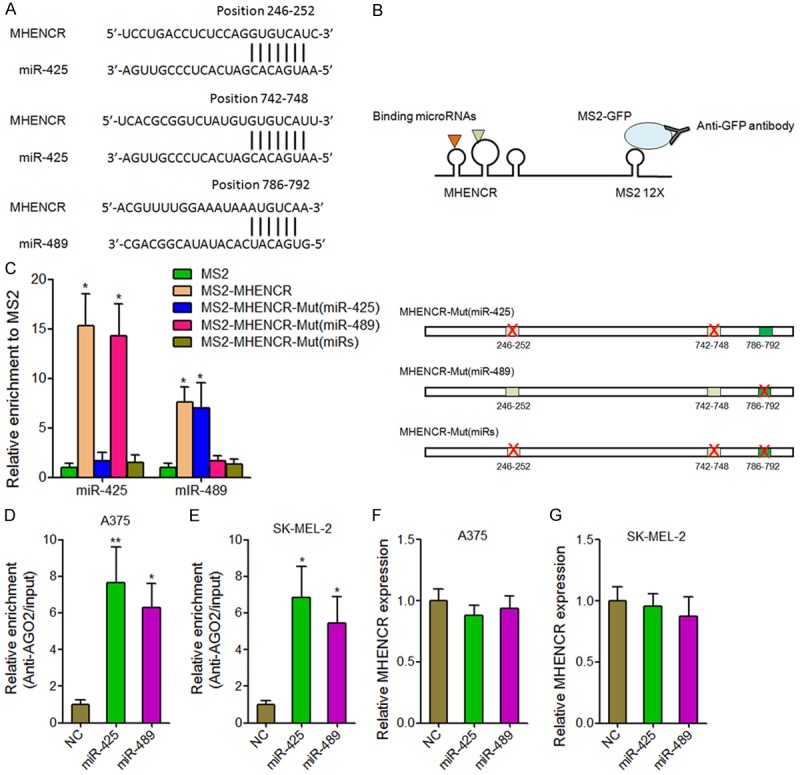
MHENCR physically associated with miR-425 and miR-489. (A) The predicted miR-425 and miR-489 binding sites on MHENCR. (B) Schematic presentation of the RIP assays using MS2 vector system. (C) The specific bindings of miR-425 and miR-489 to MHENCR were measured by MS2-RIP assays, followed by qPCR analysis. (D) Anti-AGO2 RIP assays were performed in miR-425 or miR-489 overexpressed A375 cells, followed by qPCR analysis to measure MHENCR associated with AGO2. (E) Anti-AGO2 RIP assays were performed in miR-425 or miR-489 overexpressed SK-MEL-2 cells, followed by qPCR analysis to measure MHENCR associated with AGO2. (F) The effects of miR-425 and miR-489 overexpression on MHENCR expression in A375 cells were measured by qPCR analysis. (G) The effects of miR-425 and miR-489 overexpression on MHENCR expression in SK-MEL-2 cells were measured by qPCR analysis. For (C-G), results are shown as mean ± SD. n = 3, *P < 0.05, **P < 0.01 by Student’s t test.
Knockdown of MHENCR inhibits IGF1 and SPIN1 expression, and inactivates PI3K-Akt pathway through competitively binding miR-425 and miR-489
We next explore the effects of MHENCR on miR-425 and miR-489 targets. As miR-425 has been reported to inhibit PI3K-Akt pathway via targeting IGF1 [17], and miR-489 has also been reported to suppress PI3K-Akt pathway via targeting SPIN1 [36], we next investigate the influences of MHENCR on IGF1 and SPIN1. As shown in Figure 5A and 5B, knockdown of MHENCR by both shRNAs significantly inhibited IGF1 and SPIN1 mRNA levels. Knockdown of MHENCR by both shRNAs also significantly inhibited IGF1 and SPIN1 protein levels (Figure 5C and 5D). Furthermore, the depletion of miR-425 abolished IGF1 suppression caused by MHENCR knockdown (Figure 5E and 5F). The depletion of miR-489 abolished SPIN1 suppression caused by MHENCR knockdown (Figure 5G and 5H). We then investigated the effects of MHENCR on PI3K-Akt pathway. As shown in Figure 5I and 5J, knockdown of MHENCR significantly inhibited PI3K-AKT pathway, including PIK3CA, phosphorylated Akt, cyclinD1, and BCL2. These data suggested that MHENCR activated IGF1 and SPIN1 mediated PI3K-AKT pathway via associating with miR-425 and miR-489.
Figure 5.
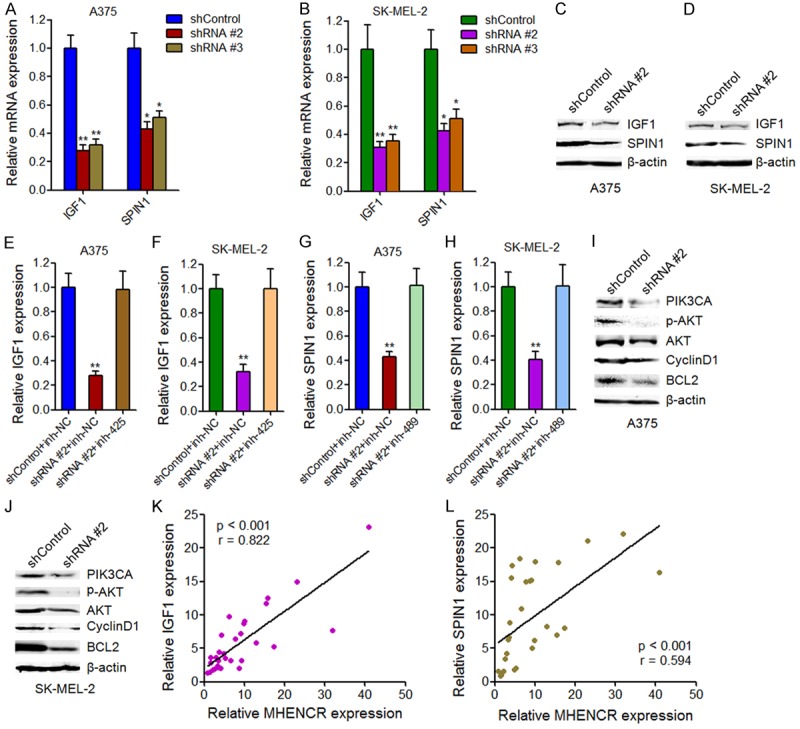
The effects of MHENCR on IGF1 and SPIN1 expression and PI3K-Akt pathway. (A) The effects of MHENCR knockdown on IGF1 and SPIN1 mRNA levels in A375 cells were measured by qPCR. (B) The effects of MHENCR knockdown on IGF1 and SPIN1 mRNA levels in SK-MEL-2 cells were measured by qPCR. (C) The effects of MHENCR knockdown on IGF1 and SPIN1 protein levels in A375 cells were measured by western blot. (D) The effects of MHENCR knockdown on IGF1 and SPIN1 protein levels in SK-MEL-2 cells were measured by western blot. (E) Depletion of miR-425 in A375 cells abolished the effects of MHENCR knockdown on IGF1 mRNA levels. (F) Depletion of miR-425 in SK-MEL-2 cells abolished the effects of MHENCR knockdown on IGF1 mRNA levels. (G) Depletion of miR-489 in A375 cells abolished the effects of MHENCR knockdown on SPIN1 mRNA levels. (H) Depletion of miR-489 in SK-MEL-2 cells abolished the effects of MHENCR knockdown on SPIN1 mRNA levels. (I) The effects of MHENCR knockdown in A375 cells on PI3K-Akt pathway were measured by western blot. (J) The effects of MHENCR knockdown in SK-MEL-2 cells on PI3K-Akt pathway were measured by western blot. For (A-J), results are shown as mean ± SD. n = 3, *P < 0.05, **P < 0.01 by Student’s t test. (K) The association between MHENCR expression and IGF1 expression in melanoma tissues was detected by Pearson correlation analysis. (L) The association between MHENCR expression and SPIN1 expression in melanoma tissues was detected by Pearson correlation analysis.
To investigate whether the regulation of IGF1 and SPIN1 by MHENCR exist In vivo, we analyzed the correlation between the expression of MHENCR and IGF1, SPIN1 in clinical melanoma tissues used in Figure 1A. As shown in Figure 5K and 5L, statistically significant correlations were observed between MHENCR expression and IGF1 and SPIN1, supporting the regulation of IGF1 and SPIN1 by MHENCR In vivo.
Knockdown of MHENCR impairs melanoma growth and metastasis in vivo
To confirm the roles of MHENCR In vivo, we subcutaneously injected MHENCR stably depleted and control A375 cells into athymic BALB/c nude mice. Tumor growth was measured every 7 days, and mice were sacrificed and tumors were sectioned and weighed at the 28th day after injection. As shown in Figure 6A and 6B, knockdown of MHENCR by both shRNAs significantly inhibited subcutaneous xenograft growth. The downregulation of IGF1 and SPIN1 were observed in MHENCR stably depleted xenografts (Figure 6C). To investigate the effects of MHENCR on melanoma metastasis, we labeled MHENCR stably depleted and control A375 cells with luciferase, and then injected the cells into tail veins of nude mouse. As shown in Figure 6D, knockdown of MHENCR by both shRNAs significantly inhibited A375 cells lung metastases. Collectively, these data suggested that knockdown of MHENCR inhibits melanoma cells growth and metastasis In vivo.
Figure 6.
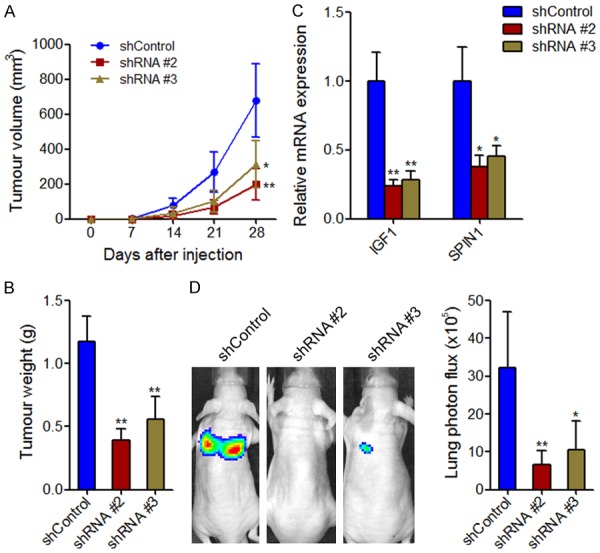
The effects of MHENCR on melanoma xenograft growth and metastasis. A: The subcutaneously xenograft growth curve showed the tumor size derived from MHENCR stably depleted or control A375 cells. B: The tumor weight of xenograft derived from MHENCR stably depleted or control A375 cells at 28th day after injection. C: IGF1 and SPIN1 mRNA levels in xenograft derived from MHENCR stably depleted or control A375 cells. D: The effects of MHENCR on melanoma lung metastasis was detected by the measurement of luciferase signal intensities derived from the lungs of mice at 6 weeks after tail vein injection. For all panels, results are shown as mean ± SD. n = 6 mice in each group, *P < 0.05, **P < 0.01 by Mann-Whitney U test.
Discussion
Every year, there are 160,000 estimated new cases of melanoma and 48,000 estimated deaths from melanoma in the world [4]. Moreover, the incidence of melanoma is increasing quickly and will be a huge challenge for public health [37]. Currently, great progressions have been made in molecularly targeted therapy and immunotherapy [5,38-41], such as the anti-programmed death 1 checkpoint inhibitor and the BRAF inhibitor, which reveal the important clinical values of elucidating the underling molecular mechanisms of melanoma. However, the overall prognosis of melanoma patients, particular that in later stages or with metastases, is still very disappointed. Fully elucidating the underlying molecular mechanisms and developing novel targeted therapy are urgent for the treatment of later stages melanoma.
In this study, we identified a critical lncRNA MHENCR, which functions as an oncogene in melanoma. MHENCR is significantly upregulated in malignant melanoma tissues compared with skin tissues with melanocytic nevus, and is further upregulated in metastatic melanoma lesions. MHENCR expression in melanoma tissues is correlated with clinical stages and poor prognosis of melanoma patients. Functional experiments showed that depletion of MHNCR significantly inhibits melanoma cell proliferation and induces cell cycle arrest and apoptosis in vitro. In vivo xenograft experiments showed that depletion of MHNCR significantly inhibits melanoma growth and lung metastasis. Our results suggest that MHENCR may be a potential prognostic biomarker and therapeutic target for melanoma. In addition to MHENCR, other lncRNAs have also been revealed to have functions in melanoma. lncRNA SLNCR1 increases melanoma invasion through transcriptional activating matrix metalloproteinase 9 [27]. Silencing lncRNA SAMMON drastically decreases melanoma cells viability through disrupting mitochondrial functions [26]. lncRNA BANCR promotes melanoma proliferation via regulating MAPK pathway [42]. All these reports, combining with our results, reveal the important roles of lncRNAs in melanoma, and demonstrate that targeting lncRNAs could be a promising strategy for melanoma therapies.
Recently, many lncRNAs have been reported to function as competing endogenous RNA to protect the genuine targets of miRNAs from degradation and/or translational inhibition [43,44]. In this study, using bioinformatic prediction and experimental verification, we found that MHENCR specifically associated with miRNA-425 and miR-489, both of which regulate PI3K-Akt pathway via targeting IGF1 and SPIN1, respectively. The PI3K-Akt pathway is well known for their important roles in cell proliferation, cell cycle, cell apoptosis, malignant transformation, drug resistance and metastasis of various types of cancer, including melanoma [45-47]. Many molecularly targeted agents against the PI3K-Akt pathway have been developed [48]. In this study, we found that through competitively binding miR-425 and miR-489, MHENCR regulated IGF1 and SPIN1 expression, and further significantly influenced PI3K-Akt pathway, which was abolished by the mutation of miR-425 and miR-489 binding sites. Statistically significant correlations were observed between MHENCR expression and IGF1 and SPIN1 in melanoma tissues, supporting the regulation of IGF1 and SPIN1 by MHENCR In vivo. Besides IGF1 and SPIN1, miR-425 and miR-489 also target PTPN11 and the HER2-SHP2-MAPK signaling [49,50]. Via completely binding miR-425 and miR-489, whether MHENCR also regulates PTPN11 and the HER2-SHP2-MAPK signaling need further investigation. Collectively, our study identifies MHENCR as a critical regulator of PI3K-Akt pathway, supporting the oncogenetic roles of MHENCR in melanoma.
In conclusion, our present study suggests that MHENCR is upregulated in melanoma, predicts poor outcome of melanoma patients. Depletion of MHENCR attenuates melanoma cells proliferation and induces cell cycle arrest and apoptosis in vitro, and reduces melanoma growth and metastasis In vivo. Mechanistically, MHENCR associates with miR-425 and miR-489, upregulates IGF1 and SPIN1 expression, and activates PI3K-Akt pathway. Our study indicates that MHENCR may be a potential prognostic biomarker and therapeutic target for melanoma.
Disclosure of conflict of interest
None.
Supporting Information
References
- 1.Torre LA, Bray F, Siegel RL, Ferlay J, Lortet-Tieulent J, Jemal A. Global cancer statistics, 2012. CA Cancer J Clin. 2015;65:87–108. doi: 10.3322/caac.21262. [DOI] [PubMed] [Google Scholar]
- 2.Siegel RL, Miller KD, Jemal A. Cancer statistics, 2015. CA Cancer J Clin. 2015;65:5–29. doi: 10.3322/caac.21254. [DOI] [PubMed] [Google Scholar]
- 3.Miller AJ, Mihm MC Jr. Melanoma. N Engl J Med. 2006;355:51–65. doi: 10.1056/NEJMra052166. [DOI] [PubMed] [Google Scholar]
- 4.Melanoma research gathers momentum. Lancet. 2015;385:2323. doi: 10.1016/S0140-6736(15)61087-X. [DOI] [PubMed] [Google Scholar]
- 5.Smalley KS, Sondak VK. Inhibition of BRAF and MEK in BRAF-mutant melanoma. Lancet. 2015;386:410–412. doi: 10.1016/S0140-6736(15)60972-2. [DOI] [PubMed] [Google Scholar]
- 6.Luo X, Wei B, Chen A, Zhao H, Huang K, Chen J. Methylation-mediated loss of SFRP2 enhances melanoma cell invasion via Wnt signaling. Am J Transl Res. 2016;8:1502–1509. [PMC free article] [PubMed] [Google Scholar]
- 7.Wu QW. Serpine2, a potential novel target for combating melanoma metastasis. Am J Transl Res. 2016;8:1985–1997. [PMC free article] [PubMed] [Google Scholar]
- 8.Burki TK. Resistance to PD-1 blockade in melanoma. Lancet Oncol. 2016;17:e376. doi: 10.1016/S1470-2045(16)30372-2. [DOI] [PubMed] [Google Scholar]
- 9.Nie L, Wu HJ, Hsu JM, Chang SS, Labaff AM, Li CW, Wang Y, Hsu JL, Hung MC. Long non-coding RNAs: versatile master regulators of gene expression and crucial players in cancer. Am J Transl Res. 2012;4:127–150. [PMC free article] [PubMed] [Google Scholar]
- 10.Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004;116:281–297. doi: 10.1016/s0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- 11.Long JD, Sullivan TB, Humphrey J, Logvinenko T, Summerhayes KA, Kozinn S, Harty N, Summerhayes IC, Libertino JA, Holway AH, Rieger-Christ KM. A non-invasive miRNA based assay to detect bladder cancer in cell-free urine. Am J Transl Res. 2015;7:2500–2509. [PMC free article] [PubMed] [Google Scholar]
- 12.He L, Hannon GJ. MicroRNAs: small RNAs with a big role in gene regulation. Nat Rev Genet. 2004;5:522–531. doi: 10.1038/nrg1379. [DOI] [PubMed] [Google Scholar]
- 13.Yoon AJ, Wang S, Shen J, Robine N, Philipone E, Oster MW, Nam A, Santella RM. Prognostic value of miR-375 and miR-214-3p in early stage oral squamous cell carcinoma. Am J Transl Res. 2014;6:580–592. [PMC free article] [PubMed] [Google Scholar]
- 14.Gee HE, Camps C, Buffa FM, Colella S, Sheldon H, Gleadle JM, Ragoussis J, Harris AL. MicroRNA-10b and breast cancer metastasis. Nature. 2008;455:E8–9. doi: 10.1038/nature07362. author reply E9. [DOI] [PubMed] [Google Scholar]
- 15.Baer C, Squadrito ML, Laoui D, Thompson D, Hansen SK, Kiialainen A, Hoves S, Ries CH, Ooi CH, De Palma M. Suppression of microRNA activity amplifies IFN-gamma-induced macrophage activation and promotes anti-tumour immunity. Nat Cell Biol. 2016;18:790–802. doi: 10.1038/ncb3371. [DOI] [PubMed] [Google Scholar]
- 16.Zhang J, Na S, Liu C, Pan S, Cai J, Qiu J. MicroRNA-125b suppresses the epithelial-mesenchymal transition and cell invasion by targeting ITGA9 in melanoma. Tumour Biol. 2016;37:5941–5949. doi: 10.1007/s13277-015-4409-8. [DOI] [PubMed] [Google Scholar]
- 17.Liu P, Hu Y, Ma L, Du M, Xia L, Hu Z. miR-425 inhibits melanoma metastasis through repression of PI3K-Akt pathway by targeting IGF-1. Biomed Pharmacother. 2015;75:51–57. doi: 10.1016/j.biopha.2015.08.010. [DOI] [PubMed] [Google Scholar]
- 18.Ponting CP, Oliver PL, Reik W. Evolution and functions of long noncoding RNAs. Cell. 2009;136:629–641. doi: 10.1016/j.cell.2009.02.006. [DOI] [PubMed] [Google Scholar]
- 19.Fatica A, Bozzoni I. Long non-coding RNAs: new players in cell differentiation and development. Nat Rev Genet. 2014;15:7–21. doi: 10.1038/nrg3606. [DOI] [PubMed] [Google Scholar]
- 20.Yan X, Hu Z, Feng Y, Hu X, Yuan J, Zhao SD, Zhang Y, Yang L, Shan W, He Q, Fan L, Kandalaft LE, Tanyi JL, Li C, Yuan CX, Zhang D, Yuan H, Hua K, Lu Y, Katsaros D, Huang Q, Montone K, Fan Y, Coukos G, Boyd J, Sood AK, Rebbeck T, Mills GB, Dang CV, Zhang L. Comprehensive genomic characterization of long non-coding RNAs across human cancers. Cancer Cell. 2015;28:529–540. doi: 10.1016/j.ccell.2015.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mercer TR, Dinger ME, Mattick JS. Long non-coding RNAs: insights into functions. Nat Rev Genet. 2009;10:155–159. doi: 10.1038/nrg2521. [DOI] [PubMed] [Google Scholar]
- 22.Yuan JH, Yang F, Wang F, Ma JZ, Guo YJ, Tao QF, Liu F, Pan W, Wang TT, Zhou CC, Wang SB, Wang YZ, Yang Y, Yang N, Zhou WP, Yang GS, Sun SH. A long noncoding RNA activated by TGF-beta promotes the invasion-metastasis cascade in hepatocellular carcinoma. Cancer Cell. 2014;25:666–681. doi: 10.1016/j.ccr.2014.03.010. [DOI] [PubMed] [Google Scholar]
- 23.Lin A, Li C, Xing Z, Hu Q, Liang K, Han L, Wang C, Hawke DH, Wang S, Zhang Y, Wei Y, Ma G, Park PK, Zhou J, Zhou Y, Hu Z, Zhou Y, Marks JR, Liang H, Hung MC, Lin C, Yang L. The LINK-A lncRNA activates normoxic HIF1alpha signalling in triple-negative breast cancer. Nat Cell Biol. 2016;18:213–224. doi: 10.1038/ncb3295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Deng L, Yang SB, Xu FF, Zhang JH. Long noncoding RNA CCAT1 promotes hepatocellular carcinoma progression by functioning as let-7 sponge. J Exp Clin Cancer Res. 2015;34:18. doi: 10.1186/s13046-015-0136-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zhang S, Zhang G, Liu J. Long noncoding RNA PVT1 promotes cervical cancer progression through epigenetically silencing miR-200b. APMIS. 2016;124:649–658. doi: 10.1111/apm.12555. [DOI] [PubMed] [Google Scholar]
- 26.Leucci E, Vendramin R, Spinazzi M, Laurette P, Fiers M, Wouters J, Radaelli E, Eyckerman S, Leonelli C, Vanderheyden K, Rogiers A, Hermans E, Baatsen P, Aerts S, Amant F, Van Aelst S, van den Oord J, de Strooper B, Davidson I, Lafontaine DL, Gevaert K, Vandesompele J, Mestdagh P, Marine JC. Melanoma addiction to the long non-coding RNA SAMMSON. Nature. 2016;531:518–522. doi: 10.1038/nature17161. [DOI] [PubMed] [Google Scholar]
- 27.Schmidt K, Joyce CE, Buquicchio F, Brown A, Ritz J, Distel RJ, Yoon CH, Novina CD. The lncRNA SLNCR1 mediates melanoma invasion through a conserved SRA1-like region. Cell Rep. 2016;15:2025–2037. doi: 10.1016/j.celrep.2016.04.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ding LJ, Li Y, Wang SD, Wang XS, Fang F, Wang WY, Lv P, Zhao DH, Wei F, Qi L. Long noncoding RNA lncCAMTA1 promotes proliferation and cancer stem cell-like properties of liver cancer by inhibiting CAMTA1. Int J Mol Sci. 2016:17. doi: 10.3390/ijms17101617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Guo L, Yao L, Jiang Y. A novel integrative approach to identify lncRNAs associated with the survival of melanoma patients. Gene. 2016;585:216–220. doi: 10.1016/j.gene.2016.03.036. [DOI] [PubMed] [Google Scholar]
- 30.Wei Y, Sun Q, Zhao L, Wu J, Chen X, Wang Y, Zang W, Zhao G. LncRNA UCA1-miR-507-FOXM1 axis is involved in cell proliferation, invasion and G0/G1 cell cycle arrest in melanoma. Med Oncol. 2016;33:88. doi: 10.1007/s12032-016-0804-2. [DOI] [PubMed] [Google Scholar]
- 31.Cesana M, Cacchiarelli D, Legnini I, Santini T, Sthandier O, Chinappi M, Tramontano A, Bozzoni I. A long noncoding RNA controls muscle differentiation by functioning as a competing endogenous RNA. Cell. 2011;147:358–369. doi: 10.1016/j.cell.2011.09.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Salmena L, Poliseno L, Tay Y, Kats L, Pandolfi PP. A ceRNA hypothesis: the Rosetta stone of a hidden RNA language? Cell. 2011;146:353–358. doi: 10.1016/j.cell.2011.07.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Jeggari A, Marks DS, Larsson E. miRcode: a map of putative microRNA target sites in the long non-coding transcriptome. Bioinformatics. 2012;28:2062–2063. doi: 10.1093/bioinformatics/bts344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Schirle NT, MacRae IJ. The crystal structure of human Argonaute2. Science. 2012;336:1037–1040. doi: 10.1126/science.1221551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Challagundla KB, Sun XX, Zhang X, DeVine T, Zhang Q, Sears RC, Dai MS. Ribosomal protein L11 recruits miR-24/miRISC to repress c-Myc expression in response to ribosomal stress. Mol Cell Biol. 2011;31:4007–4021. doi: 10.1128/MCB.05810-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Chen X, Wang YW, Xing AY, Xiang S, Shi DB, Liu L, Li YX, Gao P. Suppression of SPIN1-mediated PI3K-Akt pathway by miR-489 increases chemosensitivity in breast cancer. J Pathol. 2016;239:459–472. doi: 10.1002/path.4743. [DOI] [PubMed] [Google Scholar]
- 37.Jenks S. Melanoma treatment’s changing landscape. J Natl Cancer Inst. 2014;106:dju176. doi: 10.1093/jnci/dju176. [DOI] [PubMed] [Google Scholar]
- 38.Khushalani NI, Sondak VK. Are we there yet? Prolonged MAPK inhibition in BRAFV600-mutant melanoma. Lancet Oncol. 2016;17:1178–9. doi: 10.1016/S1470-2045(16)30368-0. [DOI] [PubMed] [Google Scholar]
- 39.Ascierto PA, McArthur GA, Dreno B, Atkinson V, Liszkay G, Di Giacomo AM, Mandala M, Demidov L, Stroyakovskiy D, Thomas L, de la Cruz-Merino L, Dutriaux C, Garbe C, Yan Y, Wongchenko M, Chang I, Hsu JJ, Koralek DO, Rooney I, Ribas A, Larkin J. Cobimetinib combined with vemurafenib in advanced BRAF(V600)-mutant melanoma (coBRIM): updated efficacy results from a randomised, double-blind, phase 3 trial. Lancet Oncol. 2016;17:1248–60. doi: 10.1016/S1470-2045(16)30122-X. [DOI] [PubMed] [Google Scholar]
- 40.Bowyer S, Lorigan P. The place of PD-1 inhibitors in melanoma management. Lancet Oncol. 2015;16:873–874. doi: 10.1016/S1470-2045(15)00094-7. [DOI] [PubMed] [Google Scholar]
- 41.Ajithkumar T, Parkinson C, Fife K, Corrie P, Jefferies S. Evolving treatment options for melanoma brain metastases. Lancet Oncol. 2015;16:e486–497. doi: 10.1016/S1470-2045(15)00141-2. [DOI] [PubMed] [Google Scholar]
- 42.Li R, Zhang L, Jia L, Duan Y, Li Y, Bao L, Sha N. Long non-coding RNA BANCR promotes proliferation in malignant melanoma by regulating MAPK pathway activation. PLoS One. 2014;9:e100893. doi: 10.1371/journal.pone.0100893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Bak RO, Mikkelsen JG. miRNA sponges: soaking up miRNAs for regulation of gene expression. Wiley Interdiscip Rev RNA. 2014;5:317–333. doi: 10.1002/wrna.1213. [DOI] [PubMed] [Google Scholar]
- 44.Liu L, Yang J, Zhu X, Li D, Lv Z, Zhang X. Long noncoding RNA H19 competitively binds miR-17-5p to regulate YES1 expression in thyroid cancer. FEBS J. 2016;283:2326–2339. doi: 10.1111/febs.13741. [DOI] [PubMed] [Google Scholar]
- 45.Lassen A, Atefi M, Robert L, Wong DJ, Cerniglia M, Comin-Anduix B, Ribas A. Effects of AKT inhibitor therapy in response and resistance to BRAF inhibition in melanoma. Mol Cancer. 2014;13:83. doi: 10.1186/1476-4598-13-83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Govindarajan B, Sligh JE, Vincent BJ, Li M, Canter JA, Nickoloff BJ, Rodenburg RJ, Smeitink JA, Oberley L, Zhang Y, Slingerland J, Arnold RS, Lambeth JD, Cohen C, Hilenski L, Griendling K, Martinez-Diez M, Cuezva JM, Arbiser JL. Overexpression of Akt converts radial growth melanoma to vertical growth melanoma. J Clin Invest. 2007;117:719–729. doi: 10.1172/JCI30102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Villanueva J, Vultur A, Lee JT, Somasundaram R, Fukunaga-Kalabis M, Cipolla AK, Wubbenhorst B, Xu X, Gimotty PA, Kee D, Santiago-Walker AE, Letrero R, D’Andrea K, Pushparajan A, Hayden JE, Brown KD, Laquerre S, McArthur GA, Sosman JA, Nathanson KL, Herlyn M. Acquired resistance to BRAF inhibitors mediated by a RAF kinase switch in melanoma can be overcome by cotargeting MEK and IGF-1R/PI3K. Cancer Cell. 2010;18:683–695. doi: 10.1016/j.ccr.2010.11.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Deuker MM, Marsh Durban V, Phillips WA, McMahon M. PI3’-kinase inhibition forestalls the onset of MEK1/2 inhibitor resistance in BRAF-mutated melanoma. Cancer Discov. 2015;5:143–153. doi: 10.1158/2159-8290.CD-14-0856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Patel Y, Shah N, Lee JS, Markoutsa E, Jie C, Liu S, Botbyl R, Reisman D, Xu P, Chen H. A novel double-negative feedback loop between miR-489 and the HER2-SHP2-MAPK signaling axis regulates breast cancer cell proliferation and tumor growth. Oncotarget. 2016;7:18295–18308. doi: 10.18632/oncotarget.7577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Kikkawa N, Hanazawa T, Fujimura L, Nohata N, Suzuki H, Chazono H, Sakurai D, Horiguchi S, Okamoto Y, Seki N. miR-489 is a tumour-suppressive miRNA target PTPN11 in hypopharyngeal squamous cell carcinoma (HSCC) Br J Cancer. 2010;103:877–884. doi: 10.1038/sj.bjc.6605811. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


