Abstract
miR-34a is an important molecule that can inhibit the tumor growth. This study aimed to investigate the functional role of miR-34a in hepatocellular carcinoma (HCC) and explore the interaction between miR-34a and histone deacetylase 1 (HDAC1). RT-qPCR was employed to detect the mRNA expression of miR-34a and HDAC1 in 60 HCC tissues. Results showed miR-34a expression in HCC tissues was significantly lower than in normal tissues (P<0.05), but HDAC1 expression in HCC tissues was markedly higher than in normal tissues (P<0.05). In addition, miR-34a expression was negatively related to HDAC1 expression. miR-34a mimic was transfected into HCC cell lines (HepB3 and HepG2). CCK8 assay, colony formation assay and flow cytometry showed miR-34a over-expression could inhibit the proliferation of HCC cells and induce their apoptosis. Western blotting indicated miR-34a over-expression down-regulated the expression of Bcl-2, procaspase-3, procaspase-9 and c-Myc, but up-regulate p21 expression. Bioinformatics analysis indicated HDAC1 was a target gene of miR-34a. Dual Luciferase Reporter Gene Assay and retrieval assay showed miR-34a could act at the 3’UTR of HDAC1 gene to regulate its expression. Thus, miR-34a may inhibit the proliferation of HCC cells and induce their apoptosis via regulating HDAC1 expression. Our findings provide evidence for the diagnosis and therapeutic target of HCC.
Keywords: Hepatocellular carcinoma, miR-34a, HDAC1, proliferation
Introduction
Liver cancer has been the second leading cause of cancer related death world wide [1] and mainly includes hepatocellular carcinoma (HCC) and cholangiocarcinoma. HCC accounts for about 90% of primary liver malignancy and has high morbidity and high mortality [2]. The early diagnosis of HCC is usually difficult, it is often highly malignant and frequently diagnosed at advanced stage, therefore the respectable HCC accounts for only 20-30% of HCC, the 5-year survival rate of HCC patients is only 30-50%, and more than 60% of HCC patients will develop metastasis and/or recurrence [3,4]. Other non-surgical treatments for HCC (such as chemotherapy, ablation, transcatheter hepatic arterial chemoembolization [TACE]) usually fail to prevent the recurrence and metastasis of HCC [5,6]. Thus, the early diagnosis of HCC and the identification of therapeutic target are crucial for the prognosis of HCC.
microRNA (miRNAs) are a group of endogenous small non-coding RNA molecules containing 19-22 nucleotides. miRNAs widely exist in eukaryotic cells. miRNA may bind to the specific bases of target mRNA to involve the post-transcriptional regulation, which plays important role in a variety of biological processes [7-9]. Studies have shown that miRNA can regulate the expression of some proteins related to the occurrence and development of cancers, exerting anti-tumor effects. The relationship between miRNA and tumorigenesis has been a hot topic in recent studies [10,11]. There is evidence showing that miR-34a may exert anti-tumor effects on esophageal cancer [12], breast cancer [13], pancreatic cancer [14] and gastric cancer [15]. In HCC, miR-34a expression reduces [16]. To further understand the role of miR-34a in the pathogenesis of HCC, bioinformatics analysis (TargetScan, miRanda target gene prediction database) showed HDAC1 might be a target gene of miR-34a. Whether miR-34a is able to inhibit the HDAC1 expression to affect the proliferation and apoptosis of HCC cells has never been reported. In this study, the expression of miR-34a and HDAC1 was detected in HCC tissues and cells, and their relationship was further evaluated, aiming to explore the role of miR-34a in the occurrence and development of HCC. Our findings may provide a new target for the diagnosis and therapy of HCC.
Materials and methods
Clinical samples
HCC tissues and adjacent normal tissues were collected from 60 patients with HCC who were hospitalized in the Provincial People’s Hospital of He’nan between June 2014 and December 2015. There were 51 males and 9 females with the median age of 51 years (range: 38-65 years). Patients did not receive radiotherapy and immune therapy before study. Informed consent was obtained from each patient before recruitment, and the whole study was approved by the Ethics Committee of our hospital. HCC was diagnosed by ultrasound examination, CT, and pathological examination together with blood AFP level and history of hepatitis. Adjacent normal tissues were collected at the site >2 cm away from the cancer and pathological examination confirmed absence of cancer cells.
Cell lines and materials
HCC cell lines (HepB3 cells [HB-8064] and HepG2 [HB-8065]) were purchased from the Type Culture Collection of the Chinese Academy of Sciences (Shanghai, China). Cells were maintained in high glucose DMEM containing 10% fetal bovine serum, 1000 U/ml penicillin and 100 mg/ml streptomycin (Gibco BRL, Gaithersburg, MD, USA) at 37°C in a humidified environment with 5% CO2.
RNA extraction and RT-qPCR
Real time quantitative PCR (RT-qPCR) was employed to detect the mRNA expression of miR-34a and HDAC1 in HCC tissues and adjacent normal tissues. Total RNA was extracted with a RNA extraction kit (Qiagen, Venlo, Netherlands), and then the concentration and purity of RNA were determined by NanoDrop 1000 spectrophotometer (Thermo Fisher Scientific, Wilmington, DE). RNA was reversely transcribed into cDNA with MMLV RTasecDNA Synthesis Kit (TaKaRa, Dalian, China) according to manufacturer’s instructions. PCR was performed with ABI Power SYBR-Green PCR Master Mix (Applied Biosystems, Foster City, CA). The copies of target genes were normalized to those of U6 gene, and 2-ΔΔCT method was employed to calculate the mRNA expression of miR-34a and HDAC1 with β-actin as a reference control.
miRNA transfection
HCC cells were seeded into 6-well plates at a density of 5×104 cells/well before transfection. When the cell confluence reached 50-80%, transfection was done with LipofectamineTM 2000 kit (Invitrogen, Carlsbad, CA, USA) according to the manufacturer’s instructions. Cells were divided into 3 groups: miR-21 mimic group, scramble miRNA group and blank control group. At 6 h after trasnfection, the medium was refreshed with complete medium, followed by incubation for another 24 h. Then, cells were harvested for detections.
Western blotting
At 24 h after transfection, total protein was extracted from each group. In brief, cells were washed in PBS and then lysed in RIPA, followed by protein quantification with BCA assay kit (Beyotime, Haimen, China). 50 μ of protein was loaded for SDS-PAGE and then transferred onto PVDF membrane. After blocking, the membrane was incubated with primary antibodies (1:500; mouse anti-human HDAC1, Bcl-2, procaspase-3, procaspase-9, c-Myc and P21; Santa Cruz) at 4°C over night. After washing in TBS thrice (15 min for each), the membrane was treated with horseradish peroxidase conjugated goat anti-rabbit secondary antibody (1:1000; Santa Cruz) in TBS at room temperature for 1 h. Detection was done with chemiluminescence detection kit (Amersham Pharmacia Biotech, Piscataway, NJ). GAPDH (Santa Cruz) served as a reference control, and the relative protein expression of target genes was determined.
Detection of cell proliferation by CCK-8 assay
CCK-8 kit (Dojindo Laboratories, Japan) was used to detect cell viability. After transfection, HCC cells were seeded into 96-well plates at a density of 0.5×104 cells/well. Detection was done in 6 wells per group, and blank controls were also detected. At 0, 24 h, 48 h, 72 h and 96 h, 10 μL of CCK-8 solution was added, followed by incubation for 3 h at 37°C. The optical density (OD) was measured at 450 nm to reflect the cell viability.
Colony formation assay
At 24 h after transfection, cells were digested with trypsin and then seeded into 6-well plates, followed by incubation at 37°C in an environment with 5% CO2 for 2 weeks. After crystal violet staining, cell colonies were counted with QCapture Pro software under a microscope (Olympus SZX12). A cell colony includes at least 50 cells. The mean number of cell colonies was used to reflect the capability of colony formation.
Detection of cell apoptosis by flow cytometry
Annexin V-FITC/PI cell apoptosis detection kit (BD Biosciences, SanJose, CA) was used for the detection of apoptotic cells. Cells in logarithmic growth phase were used to prepare single cell suspension at 1×106 cells/ml. After addition of propidium iodide (PI) and FITC Annexin V, the apoptotic cells were detected by flow cytometry (BD Biosciences), and apoptosis rate was calculated. Experiment was done thrice, and data were analyzed with Cell Quest software (BD Biosciences).
Dual luciferase reporter gene assay
The wild type (Wt) and mutant type (Mt) 3’UTR of HDAC1 were independently amplified with PCR. Genomic DNA was extracted from the blood of healthy controls, and the Wt 3’UTR of HDAC1 containing miR-34a binding site was amplified. The overlap PCR was employed for the mutation of seed region of 3’UTR of HDAC1, and then the mutant sequence of 3’UTR of Wt HDAC1 was obtained. The PCR products were inserted into pmirGLO vector to acquire recombinant HDAC1-wt and HDAC1-mut. HCC cells were seeded into 12-well plates, and the vectors (HDAC1-wt and HDAC1-mut) were independently trasnfected into HCC cells with miR-34a mimics or scramble miRNA by using LipofectamineTM 2000 kit (Invitrogen, Carlsbad, CA, USA). 24 h later, dual luciferase reporter gene assay kit (Promega) was used to detect the dual luciferase signals.
Rescue experiment
HDAC1 mRNA without 3’UTR was amplified by PCR and then connected to pcDNA3.1 eukaryotic expression vector. pcDNA3.1-HDAC1 vector expression HDAC1 was obtained. pcDNA3.1-HDAC1 vector was subsequently transfected into HepB3 cells or HepG2 cells together with miR-34a mimic or scramble miRNA. 48 h later, cells were harvested, and apoptotic cells were detected by flow cytometry. Western blotting was done to detect HDAC1 protein expression.
Statistical analysis
Statistical analysis was done with SPSS version 18.0. Quantitative data are expressed as mean ± standard deviation (SD). Comparisons were performed with one way analysis of variance (ANOVA), and correlation was evaluated with Pearson correlation analysis. A value of P<0.05 was considered statistically significant.
Results
Low miR-34a expression and high HDAC1 expression in HCC tissues
RT-qPCR was employed to detect the mRNA expression of miR-34a and HDAC1 in HCC tissues from 60 patients. Results showed miR-34a expression in HCC tissues was significantly lower than in normal tissues (P<0.05; Figure 1A), but HDAC1 expression in HCC tissues increased significantly when compared with normal tissues (P<0.05; Figure 1D). Moreover, the miR-34a expression was negatively related to HDAC1 expression (P<0.05; Figure 1F). Further analysis revealed that miR-34a expression in HCC tissues was related to the clinical stage and lymph node metastasis (P<0.05; Table 1; Figure 1B, 1C), but had no relationship with age, gender, cell differentiation, tumor size and AFP (P>0.05, Table 1). In HCC tissues, HDAC1 mRNA expression was associated with clinical stage (P<0.05; Table 1; Figure 1E), but had no association with age, gender, cell differentiation, lymph node metastasis, tumor size and AFP (P>0.05; Table 1).
Figure 1.
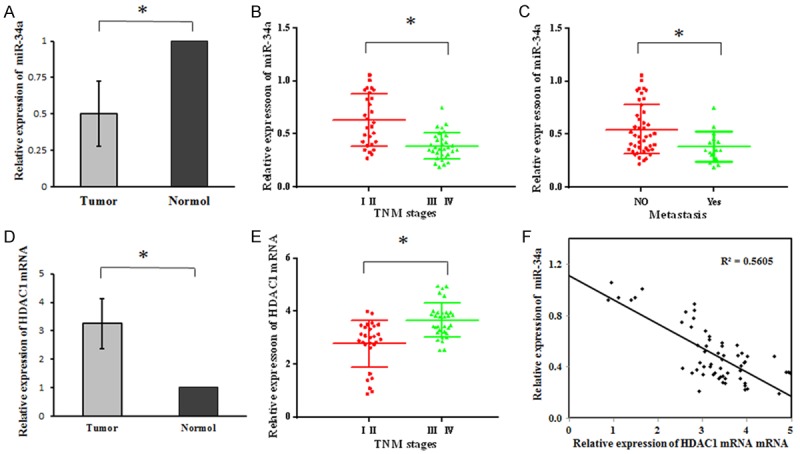
miR-34a and HDAC1 mRNA expression in HCC tissues (RT-qPCR). A: miR-34a expression in HCC tissues reduced significantly as compared to normal tissues (P<0.05); B: In HCC tissues, the higher the TNM stage, the lower the miR-34a expression was (P<0.05); C: miR-34a in HCC tissues with lymph node metastasis was significantly lower than in HCC tissues without lymph node metastasis (P<0.05); D: HDAC1 mRNA expression in HCC tissues was markedly higher than in normal tissues (P<0.05); E: In HCC tissues, the higher the TNM stage, the higher the HDAC1 mRNA expression was (P<0.05); F: miR-34a expression was negatively related to HDAC1 expression in HCC tissues.
Table 1.
Association of miR-34a and HDAC1 mRNA expression in HCC tissues with clinicopathological parameters
| Clinicopathological | n | miR-34a | P value | HDAC1 | P value |
|---|---|---|---|---|---|
|
|
|
|
|||
| Characteristics | Expression level (median ± SD) | Expression level (median ± SD) | |||
| Age (yr) | |||||
| ≤50 | 22 | 0.5545±0.2618 | 0.198 | 3.1805±1.2260 | 0.654 |
| >50 | 38 | 0.4703±0.1966 | 3.2884±0.6320 | ||
| Gender | |||||
| Male | 51 | 0.4732±0.1963 | 0.141 | 3.2402±0.8808 | 0.872 |
| Female | 9 | 0.5783±0.2920 | 3.2978±0.9804 | ||
| Cell differentiation | |||||
| Well | 12 | 0.4733±0.2182 | 0.914 | 3.4944±0.9526 | 0.565 |
| Intermediately | 39 | 0.5087±0.2314 | 3.1641±0.9526 | ||
| Poorly | 9 | 0.4975±0.2226 | 3.3400±0.7959 | ||
| Clinical stage | |||||
| I-II | 28 | 0.6300±0.2440 | 0.000* | 2.7693±0.8893 | 0.000* |
| III-IV | 32 | 0.3884±0.1258 | 3.6684±0.6495 | ||
| Lymph node metastasis | |||||
| No | 44 | 0.5441±0.2340 | 0.013* | 3.1548±0.9384 | 0.123 |
| Yes | 16 | 0.3831±0.1438 | 3.5075±0.6919 | ||
| AFP (μg/L) | |||||
| <400 | 23 | 0.4748±0.2104 | 0.467 | 3.3035±0.8800 | 0.709 |
| ≥400 | 37 | 0.5176±0.2339 | 3.2149±0.9031 | ||
| Tumor size (cm) | |||||
| <5 | 38 | 0.5153±0.2324 | 0.518 | 3.1284±0.8410 | 0.185 |
| ≥5 | 22 | 0.4768±0.2127 | 3.4568±0.9471 |
P<0.05.
miR-34a over-expression inhibits proliferation of HCC cells in vitro
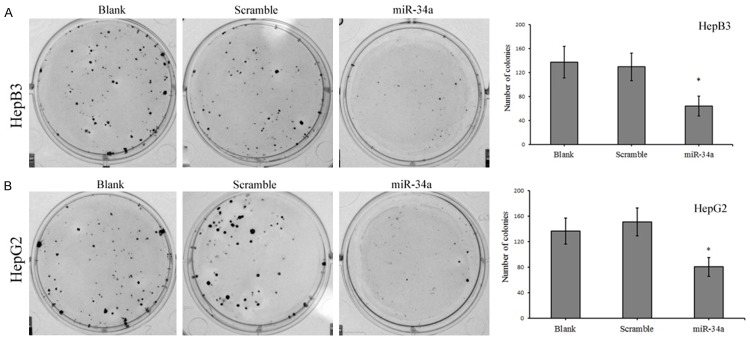
miR-34a mimic or Scramble miRNA was transfected into cells, and CCK-8 assay and colony formation assay were performed to evaluate the effect of miRNA-34a on cell proliferation. At 24 h after transfection with miRNA-34a mimic, CCK-8 assay showed the proliferation rate of HepB3 cells and HepG2 cells reduced markedly as compared to Blank control group and scramble group (P<0.05), and this difference was more obvious at 48 h and 72 h after trasnfection. However, no significant difference was observed between blank control group and scramble group (P>0.05) (Figure 2). Colony formation assay showed the number of cell colonies after transfection with miRNA-34a mimic reduced dramatically when compared with blank control group and scramble group (P<0.05) (Figure 3). These suggest that miRNA-34a over-expression is able to inhibit the proliferation of HepB3 cells and HepG2 cells.
Figure 2.
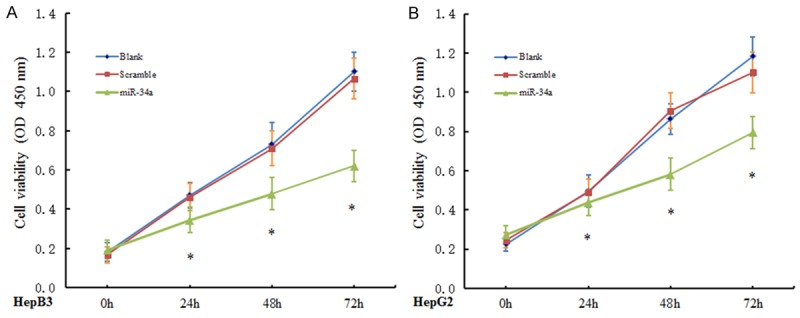
miRNA-34a over-expression inhibits the proliferation of HepB3 cells and HepG2 cells. CCK-8 assay showed the proliferation rate of HepB3 cells (A) and HepG2 cells (B) at 24 h after transfection with miRNA-34a mimic reduced markedly as compared to blank control group and scramble group (P<0.05), and this difference was more obvious at 48 h and 72 h after transfection. *P<0.05.
Figure 3.
The number of cell colonies of HepB3 cells (A) and HepG2 cells (B) after transfection with miRNA-34a mimic reduced markedly when compared with blank control group and scramble group (P<0.05). *P<0.05.
miR-34a over-expression induces apoptosis of HCC cells in vitro
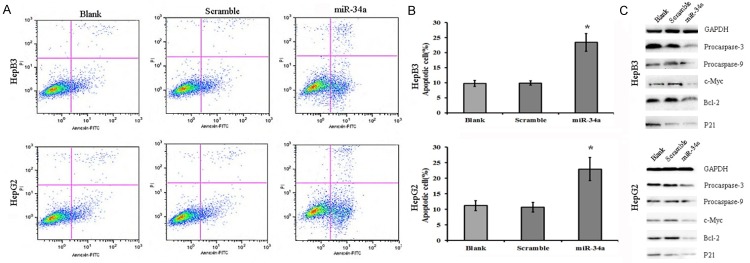
We speculated that the miRNA-34a induced inhibition of HCC cell proliferation was related to the induction of their apoptosis. Thus, flow cytometry was performed to detect apoptotic cells after transfection with miRNA-34a. Results showed, after transfection with miRNA-34a mimic, the apoptosis rate of HepB3 cells and HepG2 cells increased significantly, when compared with blank control group and scramble group (P<0.05; Figure 4A, 4B). This indicates that miRNA-34a is able to promote the apoptosis of HepB3 cells and HepG2 cells.
Figure 4.
miRNA-34a over-expression induces apoptosis of HCC cells. A, B: After transfection with miRNA-34a mimic, the apoptosis of HepB3 cells and HepG2 cells increased markedly as compared to blank control group and scramble group (P<0.05). *P<0.05. C: After transfection with miRNA-34a mimic, caspase-9 and caspase-3 expression reduced, Bcl-2 expression increased, c-Myc expression reduced, but p21 expression increased significantly as compared to blank control group and scramble group.
Western blotting was performed to detect the expression of cell apoptosis related proteins and cell cycle related p21 after transfection with miRNA-34a. Results indicated that miR-34a over expression was able to reduce the expression of caspase-9 and caspase-3, increase Bcl-2 expression, down-regulate c-Myc expression and up-regulate p21 expression in HepB3 cells and HepG2 cells (Figure 4C). This further indicates that miRNA-34a over-expression promotes the apoptosis of HCC cells.
miR-34a acts on 3’UTR of HDAC1 mRNA to inhibit HDAC1 expression
Bioinformatics analysis with TargetScan, miRanda and microRNA.org showed HDAC1 mRNA was a potential target gene of miR-34a, and the binding site of HDAC1 mRNA for miR-34a is hsown in Figure 5A. To confirm the interaction between miR-34a and HDAC1, Western blotting was employed to detect the HDAC1 protein expression in HCC cells after transfection with miR-34a mimic. Results showed the HDAC1 protein expression reduced significantly in HepB3 cells and HepG2 cells after transfection with miR-34a mimic (Figure 5B). To further confirm HDAC1 mRNA as a target of miR-34a, Wt 3’UTR and Mt 3’UTR of HDAC1 were cloned and then connected to dual luciferase reporter vector pmirGLO which was then independently transfected into HCC cells together with miR-34a mimic or Scramble miRNA, followed by detection of luciferase signals. Results showed the luciferase activity after transfection with HDAC1-mut and miR-34a mimic was comparable to that in control group (transfection with scramble miRNA and HDAC1-mut (P>0.05, Figure 5C). After transfection with miR-34a mimic and HDAC1-wt, the luciferase activity was inhibited significantly as compared to control group (transfection with scramble miRNA and HDAC1-wt (P<0.05, Figure 5C). This indicates that miR-34a may binds to HDAC1 mRNA to inhibit HDAC1 expression.
Figure 5.
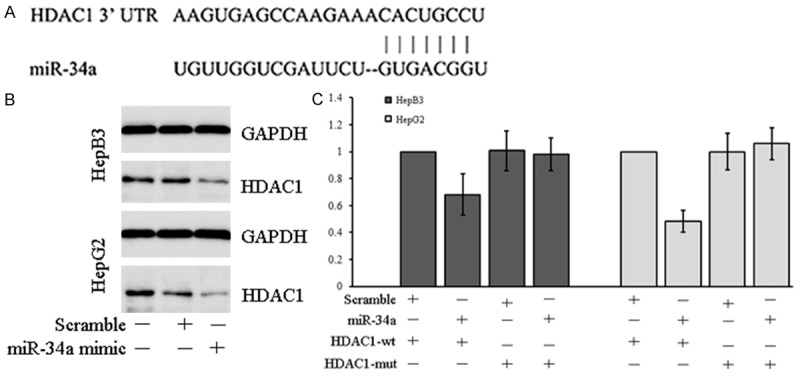
miR-34a acts on 3’UTR of HDAC1 mRNA to regulate HDAC1 expression. A. Seed sequence of HDAC1 3’UTR for miR-34a; B. Western blotting showed miR-34a mimic transfection was able to inhibit HDAC1 protein expression in HepB3 cells and HepG2 cells. C. Dual luciferase reporter gene assay showed, after transfection with miR-34a mimic and HDAC1-mut, the luciferase activity was similar to control group (transfection with scramble miRNA and HDAC1-mut) (P>0.05). After transfection with miR-34a mimic and HDAC1-wt, the luciferase activity reduced significantly as compared to control group (transfection with scramble miRNA and HDAC1-wt) (P<0.05). *P<0.05.
HDAC1 over-expression rescues the miR-34a induced apoptosis of HCC cells
Eukaryotic expression vector pcDNA3.1-HDAC1 without 3’UTR of HDAC1 mRNA was constructed and then transfected into HCC cells together with miR-34a mimic or scramble miRNA. Cells transfected with miR-34a mimic or scramble miRNA served as controls. Western blot assay was performed to detect the HDAC1 protein expression. Results showed HDAC1 expression increased significantly in HepB3 cells and HepG2 cells after transfection with pcDNA3.1-HDAC1 and miR-34a mimic, but transfection with miR-34a mimic alone reduced HDAC1 protein expression. This suggests that pcDNA3.1-HDAC1 rescues the miR-34a induced inhibition of HDAC1 expression (Figure 6A). Cell apoptosis detection showed the apoptosis rate increased significantly after transfection with miR-34a mimic (P<0.05; Figure 6B), but the apoptosis rate reduced markedly after transfection with pcDNA3.1-HDAC1 alone or in combination with miR-34a mimic (P<0.05; Figure 6B). This indicates that pcDNA3.1-Bcl-w transfection in HCC cells with miR-34a over-expression rescues the miR-34a induced apoptosis of HCC cells. This further indicates that miR-34a acts on the 3’UTR seed region of HDAC1 to regulate HDAC1 expression, exerting apoptosis inducing effect.
Figure 6.
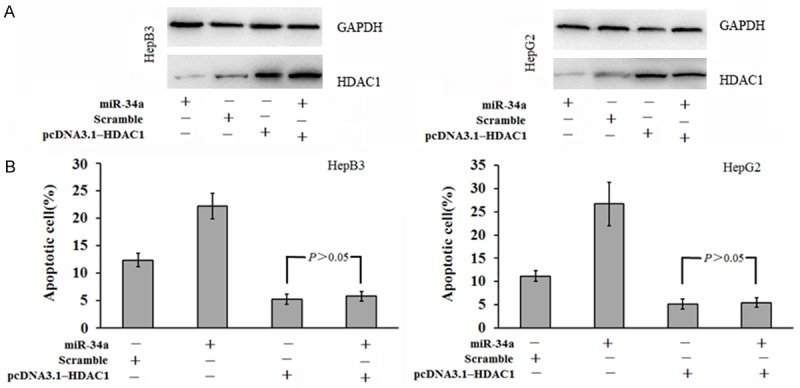
HDAC1 over-expression rescues miR-34a induced apoptosis of HCC cells. A. Western blotting showed transfection of pcDNA3.1-HDAC1 without 3’UTR of HDAC1 together with miR-34a mimic significantly increased HDAC1 expression in HepB3 cells and HepG2 cells; B. Transfection of pcDNA3.1-HDAC1 without 3’UTR of HDAC1 together with miR-34a mimic markedly reduced apoptosis rate of HepB3 ‘cells and HepG2 cells (P<0.05).
Discussion
In recent years, some studies show some non-encoding RNAs play important roles in the normal development of cells and the pathogenesis of diseases, and miRNAs have been intensively studied in recent studies among these non-encoding RNAs [17]. Studies have confirmed that some miRNAs are involved in the occurrence and development of HCC [18], of which miR-34a is the most common miRNA regulated by p53 [19]. In addition, miR-34a is an important molecule that can inhibit the growth of some cancers, and the abnormal miR-34a expression has been revealed in breast cancer [13], colon cancer [20], cervical cancer [21], prostate cancer [22], esophageal squamous cell carcinoma [23], and lung cancer [24]. miR-34a may act on its target genes to regulate the proliferation, apoptosis, invasion, metastasis and epithelial mesenchymal transition of cancer cells, exerting inhibitory effects on the growth and metastasis of cancers [25].
The role of miR-34a in the pathogenesis of liver cancer is still controversial. Some studies indicate that miR-34a acts as an oncogene [26]. Pineau et al [27] found that miR-34a was highly expression in liver cancer, and its expression was positively related to the progression from hepatic cirrhosis to liver cancer. Pogribny et al [28] found miR-34a expression increased significantly in tamoxifen induced liver cancer of mice as compared to controls, indicating that miR-34a acts as an oncogene. However, other studies reveal that miR-34a may act as a tumor suppressor gene in the occurrence and development of liver cancer [29-31]. Dang Y et al [32] found miRNA expression reduced in HCC tissues, and their further investigation indicated that miR-34a mimic could inhibit the growth, migration and invasion of HCC cells, induce cell apoptosis and increase caspase activity. In this study, miR-34a expression was detected in HCC tissues and adjacent normal tissues from 60 patients diagnosed with HCC. Results showed miRNA-34a expression in HCC tissues was lower than in normal tissues and closely related to the clinical stage and lymph node metastasis (P<0.05). Then, miR-34a mimic was independently transfected into HepB3 cells and HepG2 cells, and results showed the cell proliferation reduced, cell apoptosis increased, expression of apoptosis related proteins (caspase-9 and caspase-3) increased, expression of anti-apoptosis protein Bcl-2 reduced, c-Myc expression was down-regulated, and P21 expression was up-regulated. These indicate that miR-34a may act as a tumor suppressor gene to inhibit proliferation of HCC cells.
Bioinformatics analysis showed HDAC1 might be a target gene of miR-34a. HDAC1 is involved in the deacetylation of histones and non-histone, and low acetylation and high methylation are major characteristics of cancer cells [33]. Thus, HDAC1 may serve as a new target in the therapy of cancers. HDAC1 over-expression is related to the development of some cancers such as gastric cancer, breast cancer, lung cancer, colorectal cancer and cervical cancer [34]. Senese et al [35] found human cancer cells with HDAC1 deficiency had no mitosis and the growth of these cancer cells was inhibited. In the present study, RT-qPCR was employed to detect the mRNA expression of HDAC1 in HCC tissues from 60 HCC patients. Results showed HDAC1 mRNA expression in HCC tissues was markedly higher than in normal tissues and closely related to clinical stage (P<0.05). Moreover, miR-34a expression was negatively associated with HDAC1 expression (P<0.05). Western blotting showed HDAC1 protein expression reduced markedly in HCC cells after transfection with miR-34a mimic. Thus, we speculate that miR-34a may exert anti-tumor effect via regulating HDAC1 expression.
To further confirm HDAC1 as a target gene of miR-34a and the interaction between miR-34a and HDAC1, dual luciferase reporter gene assay was performed. HDAC1 3’UTR was cloned into pmirGLO. Results showed, after transfection with HDAC1-wt and miR-34a mimic, the luciferase activity reduced significantly; after transfection with miR-34a mimic and HDAC1-mut, the luciferase activity remained unchanged. This indicates that miR-34a is able to bind to the 3’UTR seed region of HDAC1, which inhibits the HDAC1 expression. Rescue experiment further indictaed that HDAC1 over-expression in HCC cells could rescue the miR-34a induced apoptosis of HCC cells. These findings suggest that miR-34a is able to act on 3’UTR of HDAC1 mRNA to regulate HDAC1 expression, which inhibits cell proliferation and induces cell apoptosis.
Our study indicates low miR-34a expression and high HDAC1 expression in HCC are closely related to the occurrence and development of HCC, and miR-34a may act as a tumor suppressor gene to regulate HDAC1 expression. We further investigate the functions of miR-34 in view of the important role of HDAC1 in cancers, which may provide new evidence for the diagnosis and therapy of HCC.
Disclosure of conflict of interest
None.
References
- 1.Lozano R, Naghavi M, Foreman K, Lim S, Shibuya K, Aboyans V, Abraham J, Adair T, Aggarwal R, Ahn SY, Alvarado M, Anderson HR, Anderson LM, Andrews KG, Atkinson C, Baddour LM, Barker-Collo S, Bartels DH, Bell ML, Benjamin EJ, Bennett D, Bhalla K, Bikbov B, Bin Abdulhak A, Birbeck G, Blyth F, Bolliger I, Boufous S, Bucello C, Burch M, Burney P, Carapetis J, Chen H, Chou D, Chugh SS, Coffeng LE, Colan SD, Colquhoun S, Colson KE, Condon J, Connor MD, Cooper LT, Corriere M, Cortinovis M, de Vaccaro KC, Couser W, Cowie BC, Criqui MH, Cross M, Dabhadkar KC, Dahodwala N, De Leo D, Degenhardt L, Delossantos A, Denenberg J, Des Jarlais DC, Dharmaratne SD, Dorsey ER, Driscoll T, Duber H, Ebel B, Erwin PJ, Espindola P, Ezzati M, Feigin V, Flaxman AD, Forouzanfar MH, Fowkes FG, Franklin R, Fransen M, Freeman MK, Gabriel SE, Gakidou E, Gaspari F, Gillum RF, Gonzalez-Medina D, Halasa YA, Haring D, Harrison JE, Havmoeller R, Hay RJ, Hoen B, Hotez PJ, Hoy D, Jacobsen KH, James SL, Jasrasaria R, Jayaraman S, Johns N, Karthikeyan G, Kassebaum N, Keren A, Khoo JP, Knowlton LM, Kobusingye O, Koranteng A, Krishnamurthi R, Lipnick M, Lipshultz SE, Ohno SL, Mabweijano J, MacIntyre MF, Mallinger L, March L, Marks GB, Marks R, Matsumori A, Matzopoulos R, Mayosi BM, McAnulty JH, McDermott MM, McGrath J, Mensah GA, Merriman TR, Michaud C, Miller M, Miller TR, Mock C, Mocumbi AO, Mokdad AA, Moran A, Mulholland K, Nair MN, Naldi L, Narayan KM, Nasseri K, Norman P, O’Donnell M, Omer SB, Ortblad K, Osborne R, Ozgediz D, Pahari B, Pandian JD, Rivero AP, Padilla RP, Perez-Ruiz F, Perico N, Phillips D, Pierce K, Pope CA 3rd, Porrini E, Pourmalek F, Raju M, Ranganathan D, Rehm JT, Rein DB, Remuzzi G, Rivara FP, Roberts T, De Leon FR, Rosenfeld LC, Rushton L, Sacco RL, Salomon JA, Sampson U, Sanman E, Schwebel DC, Segui-Gomez M, Shepard DS, Singh D, Singleton J, Sliwa K, Smith E, Steer A, Taylor JA, Thomas B, Tleyjeh IM, Towbin JA, Truelsen T, Undurraga EA, Venketasubramanian N, Vijayakumar L, Vos T, Wagner GR, Wang M, Wang W, Watt K, Weinstock MA, Weintraub R, Wilkinson JD, Woolf AD, Wulf S, Yeh PH, Yip P, Zabetian A, Zheng ZJ, Lopez AD, Murray CJ, AlMazroa MA, Memish ZA. Global and regional mortality from 235 causes of death for 20 age groups in 1990 and 2010: a systematic analysis for the Global Burden of Disease Study 2010. Lancet. 2012;380:2095–2128. doi: 10.1016/S0140-6736(12)61728-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Njei B, Rotman Y, Ditah I, Lim JK. Emerging trends in hepatocellular carcinoma incidence and mortality. Hepatology. 2015;61:191–199. doi: 10.1002/hep.27388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Yang T, Zhang J, Lu JH, Yang LQ, Yang GS, Wu MC, Yu WF. A new staging system for resectable hepatocellular carcinoma: comparison with six existing staging systems in a large Chinese cohort. J Cancer Res Clin Oncol. 2011;137:739–750. doi: 10.1007/s00432-010-0935-3. [DOI] [PubMed] [Google Scholar]
- 4.Fan ST, Mau Lo C, Poon RT, Yeung C, Leung Liu C, Yuen WK, Ming Lam C, Ng KK, Ching Chan S. Continuous improvement of survival outcomes of resection of hepatocellular carcinoma: a 20-year experience. Ann Surg. 2011;253:745–758. doi: 10.1097/SLA.0b013e3182111195. [DOI] [PubMed] [Google Scholar]
- 5.Germano D, Daniele B. Systemic therapy of hepatocellular carcinoma: current status and future perspectives. World J Gastroenterol. 2014;20:3087–3099. doi: 10.3748/wjg.v20.i12.3087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Shi M, Chen JA, Lin XJ, Guo RP, Yuan YF, Chen MS, Zhang YQ, Li JQ. Transarterial chemoembolization as initial treatment for unresectable hepatocellular carcinoma in southern China. World J Gastroenterol. 2010;16:264–269. doi: 10.3748/wjg.v16.i2.264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ebert MS, Sharp PA. Roles for microRNAs in conferring robustness to biological processes. Cell. 2012;149:515–524. doi: 10.1016/j.cell.2012.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bartel DP. MicroRNAs: target recognition and regulatory functions. Cell. 2009;136:215–233. doi: 10.1016/j.cell.2009.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Friedman RC, Farh KK, Burge CB, Bartel DP. Most mammalian mRNAs are conserved targets of microRNAs. Genome Res. 2009;19:92–105. doi: 10.1101/gr.082701.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hata A, Kashima R. Dysregulation of microRNA biogenesis machinery in cancer. Crit Rev Biochem Mol Biol. 2016;51:121–134. doi: 10.3109/10409238.2015.1117054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zhang S, Lu Z, Unruh AK, Ivan C, Baggerly KA, Calin GA, Li Z, Bast RC Jr, Le XF. Clinically relevant microRNAs in ovarian cancer. Mol Cancer Res. 2015;13:393–401. doi: 10.1158/1541-7786.MCR-14-0424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Shi H, Zhou S, Liu J, Zhu J, Xue J, Gu L, Chen Y. miR-34a inhibits the in vitro cell proliferation and migration in human esophageal cancer. Pathol Res Pract. 2016;212:444–449. doi: 10.1016/j.prp.2016.02.019. [DOI] [PubMed] [Google Scholar]
- 13.Li L, Yuan L, Luo J, Gao J, Guo J, Xie X. MiR-34a inhibits proliferation and migration of breast cancer through down-regulation of Bcl-2 and SIRT1. Clin Exp Med. 2013;13:109–117. doi: 10.1007/s10238-012-0186-5. [DOI] [PubMed] [Google Scholar]
- 14.Xia J, Duan Q, Ahmad A, Bao B, Banerjee S, Shi Y, Ma J, Geng J, Chen Z, Rahman KM, Miele L, Sarkar FH, Wang Z. Genistein inhibits cell growth and induces apoptosis through up-regulation of miR-34a in pancreatic cancer cells. Curr Drug Targets. 2012;13:1750–1756. doi: 10.2174/138945012804545597. [DOI] [PubMed] [Google Scholar]
- 15.Wu H, Huang M, Liu Y, Shu Y, Liu P. Luteolin induces apoptosis by up-regulating miR-34a in human gastric cancer cells. Technol Cancer Res Treat. 2015;14:747–755. doi: 10.7785/tcrt.2012.500434. [DOI] [PubMed] [Google Scholar]
- 16.Xu X, Chen W, Miao R, Zhou Y, Wang Z, Zhang L, Wan Y, Dong Y, Qu K, Liu C. miR-34a induces cellular senescence via modulation of telomerase activity in human hepatocellular carcinoma by targeting FoxM1/c-Myc pathway. Oncotarget. 2015;6:3988–4004. doi: 10.18632/oncotarget.2905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.He L, Hannon GJ. MicroRNAs: small RNAs with a big role in gene regulation. Nat Rev Genet. 2004;5:522–531. doi: 10.1038/nrg1379. [DOI] [PubMed] [Google Scholar]
- 18.Callegari E, Gramantieri L, Domenicali M, D’Abundo L, Sabbioni S, Negrini M. MicroRNAs in liver cancer: a model for investigating pathogenesis and novel therapeutic approaches. Cell Death Differ. 2015;22:46–57. doi: 10.1038/cdd.2014.136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hermeking H. p53 enters the microRNA world. Cancer Cell. 2007;12:414–418. doi: 10.1016/j.ccr.2007.10.028. [DOI] [PubMed] [Google Scholar]
- 20.Mohan M, Kumar V, Lackner AA, Alvarez X. Dysregulated miR-34a-SIRT1-acetyl p65 axis is a potential mediator of immune activation in the colon during chronic simian immunodeficiency virus infection of rhesus macaques. J Immunol. 2015;194:291–306. doi: 10.4049/jimmunol.1401447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Geng D, Song X, Ning F, Song Q, Yin H. MiR-34a inhibits viability and invasion of human papillomavirus-positive cervical cancer cells by targeting E2F3 and regulating survivin. Int J Gynecol Cancer. 2015;25:707–713. doi: 10.1097/IGC.0000000000000399. [DOI] [PubMed] [Google Scholar]
- 22.Kashat M, Azzouz L, Sarkar SH, Kong D, Li Y, Sarkar FH. Inactivation of AR and Notch-1 signaling by miR-34a attenuates prostate cancer aggressiveness. Am J Transl Res. 2012;4:432–442. [PMC free article] [PubMed] [Google Scholar]
- 23.Nie J, Ge X, Geng Y, Cao H, Zhu W, Jiao Y, Wu J, Zhou J, Cao J. miR-34a inhibits the migration and invasion of esophageal squamous cell carcinoma by targeting Yin Yang-1. Oncol Rep. 2015;34:311–317. doi: 10.3892/or.2015.3962. [DOI] [PubMed] [Google Scholar]
- 24.Garofalo M, Jeon YJ, Nuovo GJ, Middleton J, Secchiero P, Joshi P, Alder H, Nazaryan N, Di Leva G, Romano G, Crawford M, Nana-Sinkam P, Croce CM. Correction: MiR-34a/c-dependent PDGFR-alpha/beta downregulation Inhibits tumorigenesis and enhances TRAIL-induced apoptosis in lung cancer. PLoS One. 2015;10:e0131729. doi: 10.1371/journal.pone.0131729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Li L. Regulatory mechanisms and clinical perspectives of miR-34a in cancer. J Cancer Res Ther. 2014;10:805–810. doi: 10.4103/0973-1482.146084. [DOI] [PubMed] [Google Scholar]
- 26.Sukata T, Sumida K, Kushida M, Ogata K, Miyata K, Yabushita S, Uwagawa S. Circulating microRNAs, possible indicators of progress of rat hepatocarcinogenesis from early stages. Toxicol Lett. 2011;200:46–52. doi: 10.1016/j.toxlet.2010.10.013. [DOI] [PubMed] [Google Scholar]
- 27.Pineau P, Volinia S, McJunkin K, Marchio A, Battiston C, Terris B, Mazzaferro V, Lowe SW, Croce CM, Dejean A. miR-221 overexpression contributes to liver tumorigenesis. Proc Natl Acad Sci U S A. 2010;107:264–269. doi: 10.1073/pnas.0907904107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Pogribny IP, Tryndyak VP, Boyko A, Rodriguez-Juarez R, Beland FA, Kovalchuk O. Induction of microRNAome deregulation in rat liver by long-term tamoxifen exposure. Mutat Res. 2007;619:30–37. doi: 10.1016/j.mrfmmm.2006.12.006. [DOI] [PubMed] [Google Scholar]
- 29.Li N, Fu H, Tie Y, Hu Z, Kong W, Wu Y, Zheng X. miR-34a inhibits migration and invasion by down-regulation of c-Met expression in human hepatocellular carcinoma cells. Cancer Lett. 2009;275:44–53. doi: 10.1016/j.canlet.2008.09.035. [DOI] [PubMed] [Google Scholar]
- 30.Tryndyak VP, Ross SA, Beland FA, Pogribny IP. Down-regulation of the microRNAs miR-34a, miR-127, and miR-200b in rat liver during hepatocarcinogenesis induced by a methyl-deficient diet. Mol Carcinog. 2009;48:479–487. doi: 10.1002/mc.20484. [DOI] [PubMed] [Google Scholar]
- 31.Yang F, Li QJ, Gong ZB, Zhou L, You N, Wang S, Li XL, Li JJ, An JZ, Wang DS, He Y, Dou KF. MicroRNA-34a targets Bcl-2 and sensitizes human hepatocellular carcinoma cells to sorafenib treatment. Technol Cancer Res Treat. 2014;13:77–86. doi: 10.7785/tcrt.2012.500364. [DOI] [PubMed] [Google Scholar]
- 32.Dang Y, Luo D, Rong M, Chen G. Underexpression of miR-34a in hepatocellular carcinoma and its contribution towards enhancement of proliferating inhibitory effects of agents targeting c-MET. PLoS One. 2013;8:e61054. doi: 10.1371/journal.pone.0061054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ropero S, Esteller M. The role of histone deacetylases (HDACs) in human cancer. Mol Oncol. 2007;1:19–25. doi: 10.1016/j.molonc.2007.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Nakagawa M, Oda Y, Eguchi T, Aishima S, Yao T, Hosoi F, Basaki Y, Ono M, Kuwano M, Tanaka M, Tsuneyoshi M. Expression profile of class I histone deacetylases in human cancer tissues. Oncol Rep. 2007;18:769–774. [PubMed] [Google Scholar]
- 35.Senese S, Zaragoza K, Minardi S, Muradore I, Ronzoni S, Passafaro A, Bernard L, Draetta GF, Alcalay M, Seiser C, Chiocca S. Role for histone deacetylase 1 in human tumor cell proliferation. Mol Cell Biol. 2007;27:4784–4795. doi: 10.1128/MCB.00494-07. [DOI] [PMC free article] [PubMed] [Google Scholar]