Abstract
Recent studies show that many microRNAs (miRNAs) are found to play important roles in breast cancer, however, most of miRNAs are not investigated completely. In the present study, significant down-regulation of miR-520a-3p was found in the breast cancer tissues and cell lines. The restoration of miR-520a-3p expression in breast cancer cells could inhibit cell proliferation, migration and invasion. In addition, miR-520a-3p was able to induce breast cancer cell apoptosis. Luciferase assay was used to confirm that CCND1 and CD44 were the direct target genes of miR-520a-3p. The ectopic expression of miR-520a-3p repressed CCND1 and CD44 expression on post-transcriptional levels in breast cancer cells. This study suggests that miR-520a-3p may act as an optional method for breast cancer therapy.
Keywords: miR-520a-3p, breast cancer, proliferation, metastasis, CD44, CCND1
Introduction
Breast cancer is the most common cancer in women world widely. Although there are great achievements in breast cancer therapy with the development of modern technology, there are still unknown molecular mechanisms in breast cancer. There needs to investigate the molecular mechanism of breast cancer to develop more effective and efficient therapeutic results. MicroRNAs (miRNAs) are a family of non-coding small RNAs which function as the regulators of gene expression in a variety of cells and involve in many biological processes such as cell growth and survival, apoptosis, autophagy, migration, stem cell self-renewal and drug sensitivity by targeting different genes [1-3].
Previous studies have identified a number of miRNAs that were deregulated in breast cancer. For example, mir-17-92 cluster, miR-21, miR-142-3p are up-regulated and miR-98, miR-22, miR-145, miR-378, miR-497, miR-320 and miR-451 are downregulated in breast cancer depending on cell background [4-6]. However, there are no studies to reveal the role of miR-520a-3p in breast cancer. In this study, we will evaluate the potential role of miR-520a-3p in breast cancer, which includes cell growth, apoptosis, cell cycle distribution and metastasis. The predicted results indicated that CCND1 and CD44 were the direct target genes of miR-520a-3p. Functional and molecular tests, bioinformatics and survival analyses suggest that miR-520-3p plays a role in breast cancer development by targeting CCND1 and CD44. In addition, the correlation between the expressions of miR-520a-3p, CD44 and CCND1 in breast cancer samples were analyzed.
Materials and methods
Breast cancer samples
Primary human breast cancer and their matched normal adjacent tissues were obtained from 96 patients with median age 51.5 years at the first affiliated hospital with Nanjing Medical University (Nanjing, China). The diagnosis was based on pathological evidence. The patients were all underwent chemotherapy and the specimens were immediately snap-frozen and stored at -80°C. All samples were collected after the patients provided written informed consent which was approved by the Ethics Boards of the first affiliated hospital with Nanjing Medical University.
Cell culture
All the breast cancer cell lines used in the study were primarily obtained from American Type Culture Collection (ATCC, Manassas, VA, USA). They were grown according to the standard protocols. MCF10A cells were cultured in DMEM-F12 with 10% fetal bovine serum and antibiotics (100 U/ml penicillin and 100 µg/ml streptomycin sulfate) and EGF and insulin. The cells were grown in a humidified incubator at 37°C with 5% CO2. Other media supplies were obtained from Sigma.
MiRNAs, plasmids and transfection
Nuclear acid transfection was performed using Lipofectamine 2000 according to the protocols. The cells were seeded in 12-well plates and were grown up to 40% confluence before the transfection. The RNA were extracted at 48 h after the transfection. The final concentration of miR-520a-3p mimic and anti-miR-520a-3p was 50 nM.
Real time RT-PCR
Total RNA isolated from tissues or cells were performed for poly-A polymerase based First-Strand Synthesis kit (TaKaRa Bio, Japan) following the manufacturer’s protocol. To quantify the CD44 and CCND1 mRNA levels, 1 ug of total RNA was subjected to first-strand cDNA synthesis for 15 min at 37°C and 5 s at 85°C using a PrimeScript RT Reagent kit (TaKaRa, Japan). The qPCR was performed using SYBR Green PCR master mix (TaKaRa, Japan) on the ABI 7500HT System. The U6 or GAPDH were used as an endogenous control. All the primers were ordered from Invitrogen. The relative fold expression was calculated with the 2-ΔΔCT method. All the qRT-PCR reactions were run in triplicate.
Cell proliferation assay
Cell proliferation was determined by MTT assay. Briefly, breast cancer cells (5×103 per well) were plated in 96-well plates in RPMI 1640 with 10% FBS. After 24 h, the cells were transfected with 50 nM miR-520a-3p mimics, and the control using Lipofectamine 2000 (Invitrogen). The cells were then cultured in the medium for another 48 h and were assessed by a colorimetric assay using MTT solution (5 mg/mL) at 570 nm.
Evaluation of apoptosis and cell cycle distribution
For apoptosis analysis, cells were transfected with miR-520-3p mimics for 48 h. The transfected cells were collected, washed twice with cold phosphate buffered saline (PBS), suspended in binding buffer at a cell density of 1×106/mL. Cells were stained with Annexin V-FITC and propodium iodide according to the manufacturer’s protocol. The signal was acquired by a FACS Calibur flow cytometer (BD Biosciences) and the data was analyzed with a Cellquest software. For the cell cycle analysis, cells were harvested, washed twice using cold PBS and fixed in 70% ethanol overnight at -20°C. Then the cells were treated with DNA staining solution containing 3.4 mM Tris-Cl (pH 7.4), propodium iodide, 0.1% triton X-100 buffer and 100 µg/ml RNase A. Cell cycle was analyzed by flow cytometry.
Matrigel invasion assay
The Matrigel invasion chamber was used to assess cell invasion ability (24-well plates, 8 mm pore size, Corning). In brief, cells (5×104) were seeded in the upper chamber at 37°C with the media containing 0.1% bovine serum albumin, while the media containing 20% fetal bovine serum was placed in the lower well. After 48 hours, the noninvading cells were removed with cotton swabs. Invasive cells at the bottom of the membrane were stained with 0.1% crystal violet and were counted under a microscope.
Western blotting
Breast cancer cells were transfected with 50 nM miR-520a-3p mimic or its control and then total protein was extracted after 48 h. SDS-PAGE gel electrophoresis and blotting were performed. Several different primary antibodies were used including CCND1, CD44 and GAPDH (Santa Cruz Biotechnology, Santa Cruz, USA). The secondary antibody incubations were performed for 2 h at room temperature and protein bands were visualized on the X-ray film using an enhanced chemiluminescence substrate.
Dual luciferase assay
Co-transfection experiments were performed in 96-well plates. A total of 1×104 cells were seeded per well in 200 µl medium. A total of 100 ng wild type or mutant reporter constructs were co-transfected with Lipofectamine 2000 transfection reagent into the breast cancer cells with 50 nM miR-520-3p or the controls according to the manufacturer’s instruction. After 48 h, luciferase activity was measured with the Dual-Luciferase reporter assay system (Promega). Firefly luciferase activity was then normalized to the corresponding Renilla luciferase activity.
Xenograft assays in nude mice
Four-week-old BALB/c female nude mice were purchased from the Model Animal Research Center of Nanjing University (Nanjing, China). Hs578T cells were transfected with miR-520a-3p mimics or the control and infected in the fat pads of nude mice. The mice were sacrificed and photographed at 28 days post-implantation. Xenograft tumors were excised, photographed and weighed. All animal care and handling procedures were performed in accordance with the National Institutes of Health’s Guide for the Care and Use of Laboratory Animals.
Statistical analysis
Statistical analyses were performed by using SPSS 15.0 software. The unpaired t-test analysis was used to calculate P values for comparisons of CCND1 and CD44 mRNA and protein levels, and miR-520-3p expression levels between treated animals and respective age-matched controls, as well in cell lines. The unpaired t-test analysis was also used to calculate P values for comparisons of MTT assay, cell invasion and cell apoptosis. A P value <0.05 was considered significant.
Results
Relationship of miR-520a-3p expression and poor breast cancer progression
In order to explore the role of miR-520a-3p in breast cancer, its expression in breast cancer tissues and cell lines was examined using real time RT-PCR. The total RNA was extracted from 96 breast cancer samples real time RT-PCR analysis. The average of miR-520a-3p change folds in breast cancer tissues were lower compared to that of the normal tissues (Figure 1A). We observed significant down-regulation of miR-520a-3p in breast cancer tissues from 65 cases (Figure 1B). Meanwhile, lack of miR-520a-3p expression in the breast cancer cell lines were confirmed, which showed that the miR-520a-3p expression was significantly decreased in most of breast cancer cell lines examined as compared to the miR-520a-3p expression in MCF10A cells (normal epithelial breast cell line) (Figure 1C). So, the results indicated that miR-520-3p may play as a suppressing role in breast cancer.
Figure 1.
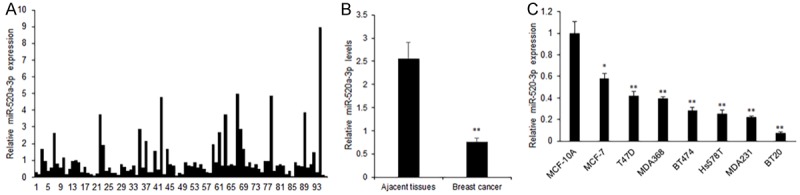
MiR-520a-3p is related to poor breast cancer progression. (A) miR-520a-3p expression was analyzed by real time PCR in breast cancer samples. (B) Data analysis of (A). (C) miR-520a-3p expression was analyzed by real time PCR in breast cancer cell lines. **P<0.01; *P<0.05.
Suppression role of miR-520a-3p on breast cell proliferation in vitro and in vivo
Based on the above data, we speculated miR-520a-3p plays a role in breast cancer as a tumor suppressor. To observe miR-520a-3p on breast cell proliferation, breast cancer cells Hs578T and BT-20 were transfected with synthesized miR-520a-3p mimics. As shown in Figure 2A, the miR-520a-3p mimics transfection was effective in breast cancer cells. MiR-520a-3p mimics transfection led to the inhibition of Hs578T cell proliferation (Figure 2B). MiR-520a-3p could also inhibit BT-20 cell proliferation (Figure 2C). Next, breast cancer nude mice were set up to investigate the role of miR-520a-3p on cell growth in vivo, the data demonstrated that miR-520a-3p inhibited the breast cancer cell growth (Figure 2D and 2E).
Figure 2.
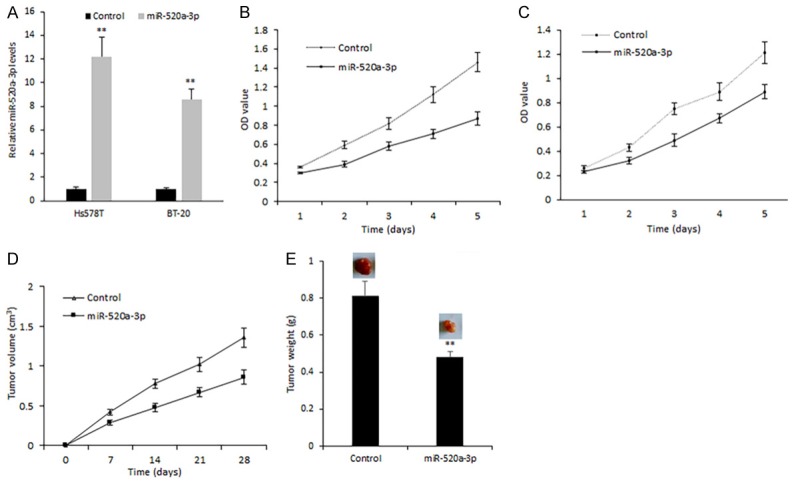
MiR-520a-3p suppresses breast cell proliferation both in vitro and in vivo. (A) miR-520a-3p expression in Hs578T and BT-20 cells with miR-520a-3p mimics transfection. (B, C) Cell proliferation of Hs578T and BT-20 cells with miR-520a-3p mimics transfection was assayed by MTT method. (D) Cell growth curve of nude mice with tumors using Hs578T cells transfected miR-520a-3p mimics or the control. (E) Tumor weight from (D) **P<0.01; *P<0.05.
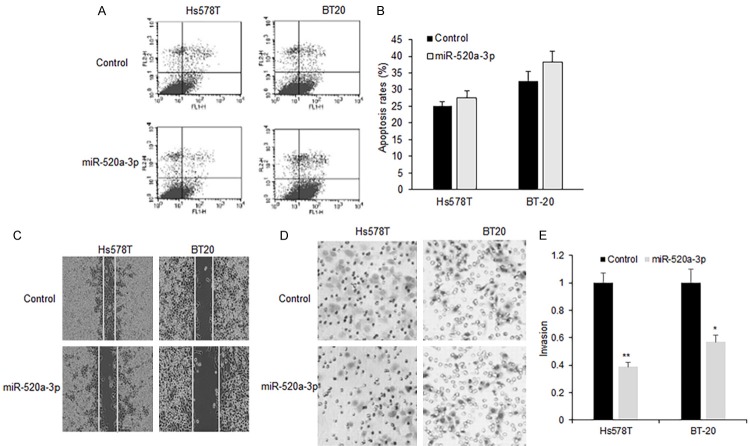
Role of miR-520a-3p in breast cancer cell apoptosis and metastasis
To elucidate the mechanisms of miR-520a-3p inhibition on breast cancer cell proliferation, Hs578T and BT-20 cells were transfected with miR-520a-3p mimics and then treated with paclitaxel, and apoptosis was analyzed using flow cytometry. It was found that apoptosis rates were not changed much in Hs578T and BT-20 cells with miR-520a-3p transfection compared to that of the miRNA control (Figure 3A and 3B). To learn the effect of miR-520a-3p on metastasis of breast cancer cells, Hs578T and BT-20 cells were transfected with miR-520a-3p and migration was analyzed using wound healing method. The founding was shown that miR-520a-3p significantly increased the migration in Hs578T and BT-20 cells with miR-520a-3p transfection comparing with their controls (Figure 3C and 3D). Moreover, miR-520a-3p greatly reduced the invading ability in Hs578T and BT-20 (Figure 3E).
Figure 3.
MiR-520a-3p induced apoptosis and inhibited metastasis of breast cancer cells. (A) Apoptosis was assayed by flow cytometry in the Hs578T and BT-20 cells transfected miR-520a-3p mimics or the control. (B) Qualification of (A). (C) Migration was assayed using wound healing method in the Hs578T and BT-20 cells transfected miR-520a-3p mimics or the control. (D) Invasion was assayed by transwell system in the Hs578T and BT-20 cells transfected miR-520a-3p mimics or the control. (E) Qualification of (D) **P<0.01.
CCND1 and CD44 are the target genes of miR-520a-3p in breast cancer cells
To elucidate the molecular mechanism of suppressing roles of miR-520a-3p on breast cancer cellular functions, we used online software to predict the putative miR-520a-3p target genes. The predicted results showed that CCND1 and CD44 are the predicted target genes with high possibility (Figure 4A). We constructed the reporter plasmids with the wide type of 3’UTRs and the mutated type of 3’UTRs of the predicted target genes including CCND1 and CD44. It was found that miR-520a-3p significantly decreased the firefly luciferase activity in the reporter with wild type 3’UTR, but the activity of mutant 3’UTR vector remained unaffected (Figure 4B and 4C). The breast cancer cells were further transfected with miR-520a-3p and were examined for CCND1 and CD44 mRNA expression by qRT-PCR. It was found that miR-520-3p transfection led to an obvious decrease in CCND1 and CD44 mRNA (Figure 4D), so did protein expression (Figure 4E). When the breast cancer cell were transfected with the inhibitors of miR-520a-3p, CCND1 and CD44 protein levels increased (Figure 4F and 4G). The data demonstrated that CCND1 and CD44 were the target genes of miR-520a-3p.
Figure 4.
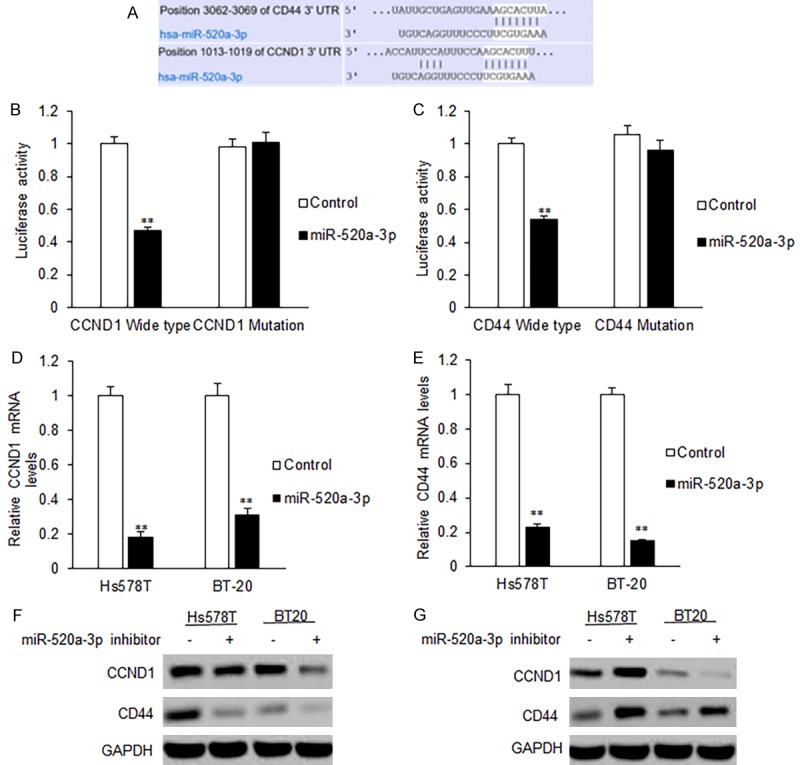
CCND1 and CD44 are the target genes of miR-520a-3p in breast cancer cells. A. Binding sequences of CCND1 and CD44 with miR-520-3p. B. Luciferase activity of CCND1 3’UTR in Hs578T cells with miR-520a-3p mimics transfection. C. Luciferase activity of CD44 3’UTR in Hs578T cells with miR-520a-3p mimics transfection. D, E. miR-520-3p led to an obvious decrease in CCND1 and CD44 mRNA in Hs578T and BT20 cells using qRT-PCR. F. miR-520-3p led to an obvious decrease in CCND1 and CD44 protein levels in Hs578T and BT20 cells using Western blotting. G. miR-520-3p inhibitor led to an obvious increase in CCND1 and CD44 protein levels in Hs578T and BT20 cells using Western blotting. **P<0.01.
Relationship between miR-520-3p expression and CCND1 or CD44 expression in breast cancer tissues
To know the relationship of miR-520a-3p and CCND1 or CD44, CCND1 and CD44 mRNA were measured by real time RT-PCR, and they were both higher in breast cancer tissues (T) than in adjacent normal tissues (N) (Figure 5A and 5B). The analyzed relationship between CCND1 or CD44 and miR-520a-3p expression levels indicated that CCND1 or CD44 were reversely related to miR-520a-3p in breast cancer tissues (Figure 5C and 5D).
Figure 5.
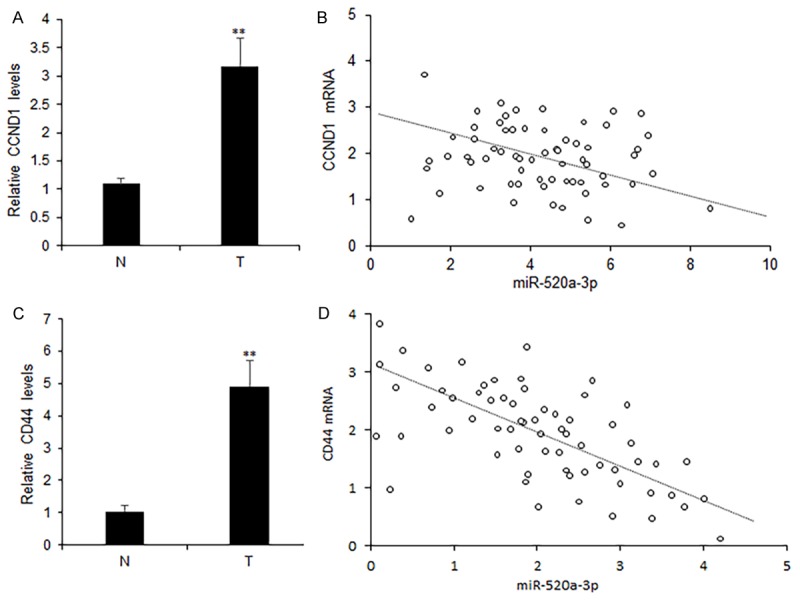
Inverse correlation between miR-520-3p expression and CCND1 and CD44 in breast cancer tissues. A, B. Average CCND1 and CD44 mRNA levels in breast cancer tissues. C. Relationship of CCND1 and miR-520-3p expression in breast cancer tissues (n=96). D. Relationship of CD44 and miR-520-3p expression in breast cancer tissues (n=96). **P<0.01.
Discussion
In the past years, thousands of miRNAs in cancer has been widely found and they participate in many physiological and pathological processes, which widens the knowledge of cancer cell biology [27]. MiR-520a-3p is one of a potential tumor suppressing miRNA [7], however, its expression and roles in breast cancer are unknown. In this study, we attempted to determine the roles of miR-520a-3p in breast cancer. The expression of miR-520a-3p in breast cancer tissue samples and cell lines was down-regulated and further study indicated that miR-520a-3p suppressed breast cancer cell survival ability and metastasis by inhibiting the expression of CCND1 and CD44.
There was a report showed that miR-520a-3p is down-regulated in NSCLC, which indicates it functions as a tumor suppressor by directly targeting MAP3K2 in NSCLC cells [7]. Our study indicated that miR-520a-3p was lack of expression in breast cancer tissues and cells. Forced expression of miR-520a-3p in breast cancer cells suppressed cell proliferation and growth in vivo. MiR-520a-3p could inhibit cancer cell migration and invasion. The results supported that miR-520a-3p plays a suppressing role in breast cancer.
Previous studies indicate CD44, a common marker of cancer stem cells, has multi-function in cancer development and progression. Our study showed that CD44 is a direct target gene of miR-520a-3p in breast cancer cells. CD44 could be regulated by other miRNAs such as miR-199a, miR-199a-3p, miR-9, miR-106b, miR-328, miR-708, miR-21, miR-34a, miR-373 and miR-520c in cancer cells and the cellular functions of them involve in regulating cell proliferation, migration, multidrug resistance, cancer stem cell maintaining and others [8-19]. Our study demonstrated that CD44 was verified as a direct target gene of miR-520a-3p by luciferase assay, real time RT-PCR and western blotting. CCND1 gene encodes the cell cycle G1 regulatory protein cyclinD1, which overexpressed in 90% breast cancers and regulated by miR-326, miR-211, miR-195 and others [20-22]. CCND1 is another identified target gene of miR-520a-3p in breast cancer cells in this study. MiR-520a-3p could inhibit CCND1 mRNA and cyclinD1 protein levels in breast cancer. Data from the breast cancer tissues showed that miR-520a-3p was negatively correlated with CCND1 mRNA.
In a conclusion, our study give an evidence that miR-520a-3p plays an inhibiting role in breast cancer cell proliferation and metastasis. Importantly, we show, to our knowledge, for the first time that miR-520a-3p is a key negative regulator of CCND1 and CD44 of breast cancer cells and that both of them are the direct functional target genes of miR-520a-3p.
Acknowledgements
This work was supported by Jiangsu Province Clinical science and technology projects (Clinical Research Center, BL2012008).
Disclosure of conflict of interest
None.
References
- 1.Ohtsuka M, Ling H, Doki Y, Mori M, Calin GA. MicroRNA processing and human cancer. J Clin Med. 2015;4:1651–1667. doi: 10.3390/jcm4081651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Pichler M, Calin GA. MicroRNAs in cancer: from developmental genes in worms to their clinical application in patients. Br J Cancer. 2015;113:569–573. doi: 10.1038/bjc.2015.253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lan H, Lu H, Wang X, Jin H. MicroRNAs as potential biomarkers in cancer: opportunities and challenges. Biomed Res Int. 2015;2015:125094. doi: 10.1155/2015/125094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Verigos J, Magklara A. Revealing the complexity of breast cancer by next generation sequencing. Cancers (Basel) 2015;7:2183–2200. doi: 10.3390/cancers7040885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Takahashi RU, Miyazaki H, Ochiya T. The roles of MicroRNAs in breast cancer. Cancers (Basel) 2015;7:598–616. doi: 10.3390/cancers7020598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bertoli G, Cava C, Castiglioni I. MicroRNAs: new biomarkers for diagnosis, prognosis, therapy prediction and therapeutic tools for breast cancer. Theranostics. 2015;5:1122–1143. doi: 10.7150/thno.11543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Yu J, Tan Q, Deng B, Fang C, Qi D, Wang R. The microRNA-520a-3p inhibits proliferation, apoptosis and metastasis by targeting MAP3K2 in non-small cell lung cancer. Am J Cancer Res. 2015;5:802–811. [PMC free article] [PubMed] [Google Scholar]
- 8.Wang SH, Zhou JD, He QY, Yin ZQ, Cao K, Luo CQ. MiR-199a inhibits the ability of proliferation and migration by regulating CD44-ezrin signaling in cutaneous squamous cell carcinoma cells. Int J Clin Exp Pathol. 2014;7:7131–7141. [PMC free article] [PubMed] [Google Scholar]
- 9.Cheng W, Liu T, Wan X, Gao Y, Wang H. MicroRNA-199a targets CD44 to suppress the tumorigenicity and multidrug resistance of ovarian cancer-initiating cells. FEBS J. 2012;279:2047–2059. doi: 10.1111/j.1742-4658.2012.08589.x. [DOI] [PubMed] [Google Scholar]
- 10.Song Y, Li J, Zhu Y, Dai Y, Zeng T, Liu L, Li J, Wang H, Qin Y, Zeng M, Guan XY, Li Y. MicroRNA-9 promotes tumor metastasis via repressing E-cadherin in esophageal squamous cell carcinoma. Oncotarget. 2014;5:11669–11680. doi: 10.18632/oncotarget.2581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Henry JC, Park JK, Jiang J, Kim JH, Nagorney DM, Roberts LR, Banerjee S, Schmittgen TD. miR-199a-3p targets CD44 and reduces proliferation of CD44 positive hepatocellular carcinoma cell lines. Biochem Biophys Res Commun. 2010;403:120–125. doi: 10.1016/j.bbrc.2010.10.130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Yu D, Shin HS, Lee YS, Lee YC. miR-106b modulates cancer stem cell characteristics through TGF-β/smad signaling in CD44-positive gastric cancer cells. Lab Invest. 2014;94:1370–1381. doi: 10.1038/labinvest.2014.125. [DOI] [PubMed] [Google Scholar]
- 13.Ishimoto T, Sugihara H, Watanabe M, Sawayama H, Iwatsuki M, Baba Y, Okabe H, Hidaka K, Yokoyama N, Miyake K, Yoshikawa M, Nagano O, Komohara Y, Takeya M, Saya H, Baba H. Macrophage-derived reactive oxygen species suppress miR-328 targeting CD44 in cancer cells and promote redox adaptation. Carcinogenesis. 2014;35:1003–1011. doi: 10.1093/carcin/bgt402. [DOI] [PubMed] [Google Scholar]
- 14.Wang CH, Lee DY, Deng Z, Jeyapalan Z, Lee SC, Kahai S, Lu WY, Zhang Y, Yang BB. MicroRNA miR-328 regulates zonation morphogenesis by targeting CD44 expression. PLoS One. 2008;3:e2420. doi: 10.1371/journal.pone.0002420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Saini S, Majid S, Shahryari V, Arora S, Yamamura S, Chang I, Zaman MS, Deng G, Tanaka Y, Dahiya R. miRNA-708 control of CD44(+) prostate cancer-initiating cells. Cancer Res. 2012;72:3618–3630. doi: 10.1158/0008-5472.CAN-12-0540. [DOI] [PubMed] [Google Scholar]
- 16.Bourguignon LY, Earle C, Wong G, Spevak CC, Krueger K. Stem cell marker (Nanog) and Stat-3 signaling promote MicroRNA-21 expression and chemoresistance in hyaluronan/CD44-activated head and neck squamous cell carcinoma cells. Oncogene. 2012;31:149–160. doi: 10.1038/onc.2011.222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Liu C, Kelnar K, Liu B, Chen X, Calhoun-Davis T, Li H, Patrawala L, Yan H, Jeter C, Honorio S, Wiggins JF, Bader AG, Fagin R, Brown D, Tang DG. The microRNA miR-34a inhibits prostate cancer stem cells and metastasis by directly repressing CD44. Nat Med. 2011;17:211–215. doi: 10.1038/nm.2284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Huang Q, Gumireddy K, Schrier M, le Sage C, Nagel R, Nair S, Egan DA, Li A, Huang G, Klein-Szanto AJ, Gimotty PA, Katsaros D, Coukos G, Zhang L, Puré E, Agami R. The microRNAs miR-373 and miR-520c promote tumour invasion and metastasis. Nat Cell Biol. 2008;10:202–210. doi: 10.1038/ncb1681. [DOI] [PubMed] [Google Scholar]
- 19.Yang K, Handorean AM, Iczkowski KA. MicroRNAs 373 and 520c are downregulated in prostate cancer, suppress CD44 translation and enhance invasion of prostate cancer cells in vitro. Int J Clin Exp Pathol. 2009;2:361–369. [PMC free article] [PubMed] [Google Scholar]
- 20.Sun C, Huang C, Li S, Yang C, Xi Y, Wang L, Zhang F, Fu Y, Li D. Hsa-miR-326 targets CCND1 and inhibits non-small cell lung cancer development. Oncotarget. 2016;7:8341–8359. doi: 10.18632/oncotarget.7071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Xia B, Yang S, Liu T, Lou G. miR-211 suppresses epithelial ovarian cancer proliferation and cell-cycle progression by targeting cyclin D1 and CDK6. Mol Cancer. 2015;14:57. doi: 10.1186/s12943-015-0322-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Han K, Chen X, Bian N, Ma B, Yang T, Cai C, Fan Q, Zhou Y, Zhao TB. MicroRNA profiling identifies MiR-195 suppresses osteosarcoma cell metastasis by targeting CCND1. Oncotarget. 2015;6:8875–8889. doi: 10.18632/oncotarget.3560. [DOI] [PMC free article] [PubMed] [Google Scholar]