Abstract
Background
Overweight and obesity are becoming more widespread with alarming projections for the coming years. Obesity may increase susceptibility to the adverse effects of PM exposure, exacerbating the effects on cardiovascular diseases and altering the biomarkers of vascular inflammation. The associated biological mechanisms have not been fully understood yet; the common denominator in the pathogenesis of the co-morbidities of obesity is the presence of an active, low-grade inflammatory process. DNA methylation has been shown to regulate inflammatory pathways that are responsible for the development of cardiovascular diseases.
Objectives
The aim of the study was to investigate, in a population of overweight/obese subjects, the effects of PM on blood DNA methylation in genes associated to inflammatory response.
Methods
Using bisulfite pyrosequencing, we measured DNA methylation in peripheral blood mononuclear cells from 186 overweighted/obese subjects. In particular, we quantified DNA methylation in a set of 3 candidate genes, including CD14, TLR4 and TNF-α, because of the important roles that these genes play in the inflammatory pathway. Personal exposure to PM10 was estimated for each subject based on the local PM10 concentrations, measured by monitoring stations at residential address. Repeated measure models were used to evaluate the association of PM10 with each genes, accounting for possible correlations among the genes that regulate the same inflammatory pathway.
Results
We found an inverse association between the daily PM10 exposure and the DNA methylation of inflammatory genes, measured in peripheral blood of healthy overweight/obese subjects. Considering different exposure time-windows, the effect on CD14 and TLR4 methylation was observed, respectively, in days 4–5-6, and days 6–7-8. TNF-α methylation was not associated to PM10.
Conclusions
Our findings support a picture in which PM10 exposure and transcriptional regulation of inflammatory gene pathway in obese subjects are associated.
Abbreviations: BMI, Body mass index; CD14, cluster of differentiation 14; LPS, lipopolysaccharide; NF-кB, nuclear factor κB; PM, particulate matter; PM10, particulate matter with aerodynamic diameters <10 µm; TLR4, toll-like receptor 4; TNF, tumor necrosis factor
Keywords: PM exposure, DNA methylation, Obesity, Inflammation, Pyrosequencing
Highlights
-
•
Overweight/obese subjects has been proposed as susceptible population for PM related effects.
-
•
DNA methylation is a key molecular mechanisms linking PM exposure to systemic pro-inflammatory effects
-
•
PM10 exposure resulted associated to DNA methylation of inflammatory genes in a population of obese patients.
-
•
The relationship between PM10 and DNA methylation of inflammation pathway-genes was confirmed in obese subjects.
1. Introduction
Previous studies have shown that the exposure to particulate matter (PM) is associated with increased morbidity and mortality, primarily from cardiovascular disease (CVD) (Dai et al., 2014; Nasser et al., 2015; Pope et al., 2015; Talbott et al., 2014; Weichenthal et al., 2014). To date, the underlying molecular mechanism responsible for this consistently observed association between air pollution and CVD risk is still poorly understood. It has been proposed that ambient particles might trigger pulmonary oxidative stress and inflammatory responses, leading to the release of molecular signals into the circulatory system (Bollati et al., 2015).
Several investigators have hypothesized that oxidative stress directly induced in the lungs after PM inhalation (Sofer et al., 2013), might cause a systemic inflammatory cascade, increasing cardiovascular risk among susceptible individuals (Bertazzi et al., 2014; Dubowsky et al., 2006; Weichenthal et al., 2013).
Obesity may increase susceptibility to PM10 exposure, exacerbating its effects on cardiovascular disease (Qin et al., 2015) and on biomarkers of vascular inflammation (Matsuda and Shimomura, 2014); in fact, it has been suggested that obese subjects inhale more air-per-day than normal-weight individuals (Brochu et al., 2014, WHO, 2013), thus potentially increasing their overall dose (Weichenthal et al., 2014). Recent studies have further demonstrated that populations characterized by a high body mass index (BMI), when compared to normal weight populations, exhibit an increased risk of CVDs, due to a higher exposure to air pollutants (Chen et al., 2007, Huang et al., 2012; Jung CC and Liang, 2016)
The biological mechanisms behind these associations have not been fully elucidated yet, but many of the obesity-related diseases are thought to be linked to a state of chronic oxidative stress and inflammation. Obese subjects have indeed increased systemic oxidative stress and impaired oxidant defense (De Pergola and Silvestris, 2013, Keaney et al., 2003, Matsuda and Shimomura, 2014, Savini et al., 2013).
To try to improve our understanding on the role of obesity as a susceptibility factor, we designed an epidemiological study where we investigated the effects of PM10 on blood DNA methylation in a population of 186 overweight/obese subjects (Wang et al., 2010).
DNA methylation has been shown to regulate biological processes underlying CVDs, such as inflammation and immune response. (Baccarelli et al., 2010, Bayarsaihan, 2011) By regulating inflammatory pathways, that are key elements in the onset of CVDs, pro-inflammatory gene methylation might be related to cardiovascular health (Alexeeff et al., 2013).
In detail, we measured DNA methylation in a set of candidate genes, including of CD14, TLR4 and TNF-α (Bollati et al., 2014a), selected for the important roles that they play in the inflammatory pathway. CD14 is a pattern recognition receptor in the innate immune response against microorganisms and other exogenous and endogenous stress factors. The most important CD14 signaling co-receptor is TLR4, required to trigger the downstream signaling pathways that lead to the activation of the nuclear factor κB (NF-кB) inflammatory pathway that in turn promotes the increase in pro-inflammatory cytokine production including tumor necrosis factor (TNF) (Bes-Houtmann et al., 2007; De Loera-Rodriguez et al., 2014; Hoareau et al., 2010). In addition CD14/TLR4 are over-expressed in obese subjects, reflecting a state of low-grade chronic inflammation (Devevre et al., 2015, Hoareau et al., 2010).
2. Materials and methods
2.1. Study subjects
Overweight/obese subjects were recruited at the Center for Obesity and Work (Department of Preventive Medicine, IRCCS Fondazione Ca’Granda – Ospedale Maggiore Policlinico) in the period between September 2010 and April 2011. These subjects were the first 186 subjects consecutively enrolled in the context of the larger study SPHERE (ERC-2011-StG 282413), (Bollati et al., 2014b) which was aimed at examining the possible molecular mechanisms underlying the effects of PM exposure in relation to health. Each participant signed a written informed consent, approved by the Ethic Committee of the Fondazione Ca’Granda – Ospedale Maggiore Policlinico (Approval no. 1425), in accordance with the Helsinki Declaration principles.
All subjects willing to participate in the study had to unchanged body weight and constant dietary habits in the last year. In addition, they had constant physical activity for at least one month before entry into the study. Exclusion criteria was the presence of a major disease such as cancer, autoimmune disease, cardiovascular disease.
Subjects who agreed to participate were asked to complete lifestyle and dietary questionnaires and to donate a blood sample. Information about lifestyle factors, including current and past smoking habits, alcohol consumption and physical activity, were collected with a self-administered questionnaire.
Body mass index (BMI) was calculated as the ratio between the subject’s weight (kg) and height squared (m2). According to the current definition ((WHO), 2000) , subjects who had a BMI ranging from 25 to 29.9 were classified as overweight, and subjects who had a BMI of 30 or higher were classified as obese.
Waist circumference (cm) was also assessed as marker of central fat accumulation. Fasting level of total cholesterol was measured from serum samples by routine methods (Modular, Roche- Basel, Switzerland). Blood pressure was measured with the participant supine, after 5 min of rest.
2.2. Assessment of PM10 exposure
PM10 concentrations were assigned to each participant's residential address from September 1, 2010 to April 11, 2011. The PM10 concentrations were recorded by the Regional Environmental Protection Agency (ARPA Lombardia) through monitoring stations located throughout Lombardy and available online as daily means. To assign a PM10 exposure level to each subject, the addresses of the monitoring stations and the study subjects were geocoded and the PM10 measured from the nearest monitor to residential address was assigned. For each subject, we used the daily mean of PM10 concentrations measured 1 day before the date of recruitment and back to 14 days before enrollment,. In case of incomplete series, each missing value was imputed by using an algorithm that integrates the annual average of the incomplete series and the PM10 concentrations of the nearest and more correlated monitors (Cattani et al., 2010).
2.3. Sample collection, DNA extraction and bisulfite treatment
Seven milliliters of whole blood were collected into EDTA tubes from each participant by venous phlebotomy. Blood was centrifuged at 2500 rpm for 15 min. The buffy coat fraction was transferred to a cryovial and immediately frozen at −80 °C until use. DNA was extracted by the Wizard Genomic DNA purification kit (Promega, Madison, WI, USA), according to the manufacturer’s instructions.
500 ng of genomic DNA was treated with the EZ DNA Methylation-Gold™ Kit (Zymo Research, Orange, CA, USA), in accordance with the manufacturer’s protocol. Bisulfite-treated DNA was eluted in 30 μL of M-Elution Buffer and kept at −80 °C until use.
2.4. DNA Methylation
Analysis of DNA methylation was performed by previously published methods (Bollati et al., 2007, Tarantini et al., 2013), with minor modifications. Briefly, a 50-μL PCR reaction was carried out with 25 μL of Hot Start GoTaq Green Master mix (Promega), 1 pmol of forward primer, 1 pmol of biotinylated reverse primer and 25 ng of bisulfite-treated genomic DNA. Biotin-labeled primers were used to purify the final PCR product with Sepharose beads. The PCR product was bound to a Streptavidin Sepharose HP (Amersham Biosciences, Uppsala, Sweden). Sepharose beads containing the immobilized PCR product were purified, washed, denatured with 0.2 M NaOH and washed again with the Pyrosequencing Vacuum Prep Tool (Pyrosequencing, Inc., Westborough, MA, USA), according to the manufacturer’s instructions. Pyrosequencing primer (0.3 µm) was annealed to the purified single-stranded PCR product, and pyrosequencing was performed with the PyroMark MD System (Pyrosequencing, Inc. Westborough, MA, USA). PCR cycling conditions and primer sequences are reported in Table 1. The methylation level at CpG positions within each gene’s promoter region, was expressed as the percentage of cytosines that were methylated, determined as the number of methylated cytosines divided by the sum of methylated and unmethylated cytosines, multiplied by 100% (% 5-methyl-Cytosine). Every sample was tested two times for each marker to confirm reproducibility and to increase the precision of the findings.
Table 1.
Pyrosequencing assays information.
| Gene | Chromosome | CpG positiona | Primers: Forward (F), Reverse (R) and Sequencing (S) |
|---|---|---|---|
| CD14 | 5 | 140632047 | F:GGGTTTATAGAGGAGGGAATTGA |
| 140632043 | R:Biotin-AAACCCCATCCAACCCCTAT | ||
| 140632041 | S:TGTAGGGTTTTGGGG | ||
| 140632031 | |||
| TLR4 | 9 | 117704398 | F: Biotin-AGGAAGAGAAGATATTAGTGTTTT |
| 117704406 | R: CCTAACATCATCCTCACTACTTCT | ||
| 117704415 | S: ACAACAATAACCCTATAAAT | ||
| 117704422 | |||
| TNF-α | 6 | 2874672 | F:Biotin-TGAGGGGTATTTTTGATGTTTGT |
| 2874678 | R: TTAATAATTATTTTTATATATTTT | ||
| 2874680 | S: ATAAATTTTATATTTTTTAT | ||
| 2874695 | |||
According to UCSC Genome Browser (https://genome.ucsc.edu/), Dec 2013 release (GRch38/hg38).
Intra-observer coefficient of variability between the two replicates of DNA methylation measurements was 7.9% (SD=9.1%) for CD14, 3% (SD=2.7%) for TLR4 and 5% (SD=5.1%) for TNF-α.
2.5. Statistical analysis
DNA methylation values exhibited non-normal distribution, and were thus log-transformed. We expressed them as variations in term of Z-score for each gene, in order to standardize values according to the population’s mean and standard deviation. DNA methylation measurements for each subject were performed for four CpG dinucleotide positions replicated in two measurements. To consider intra-individual correlation due to repeated-measure data structure we designed linear mixed models. The CpG dinucleotide position was considered introducing a random effect model. The intra-observer coefficient of variability between the two replicate of DNA methylation was measured using the mean of the four positions for each gene.
We then evaluated the association of daily mean concentration of PM10 with specific methylation.
DNA methylation Z-scores were modelled both as singles genes and as a pathway which identifies a common inflammatory system pathway. Methylation gene Z-scores for multiple genes within the pathway were modelled as a vector of 24 responses within each sample (i.e., three genes measured on each sample; each sample measured at four positions in two runs) as a function of PM10 concentrations and other selected covariates. Mixed effect models allowed us to analyze each gene separately while taking into account differences between the three genes within the pathway, and simultaneously adjusting for the correlation between the repeated measured on a single subject. Scaled regression coefficients were used to represent the increase in DNA methylation estimated for a 1-SD increment in the independent variable.
Daily mean PM10 concentrations were considered both as continuous measures, assuming linearity of the relationship pollutant-gene, and as categorized into quartiles. We reported beta regression coefficient (β) expressing: a) the logarithmic change in target methylation Z-scores associated with a standard deviation increase in PM10 concentration level; b) the logarithmic change in methylation Z-scores associated with change in PM10 switching from the first quartile to the others, in the context of quartile model approach. We used the fitted mixed effect model to calculate the predicted mean Z-score and standard errors for each quartile of PM10 exposure, in order to show dose-response relationships. All models were adjusted for covariates that were selected a priori: body mass index, age (years), sex, percentage of neutrophils, smoking habits (never smokers, ex-smokers actual smokers), position, run, batch, interaction terms among position, run and batch. Pathway model was also adjusted for singles methylation genes. The role of further possible covariates (diabetes lymphocytes, physical activity, week, season, apparent temperature) was checked by univariate analyses, but none of the them resulted significantly associated with DNA methylation. We finally checked the possible modification of PM10 effect on DNA methylation testing the interaction between PM and BMI and in case of significance we stratified subjects according to their BMI level. A p-value of 0.05 was considered as statistically significant. All analyses were performed by SAS 9.3 (SAS Institute Inc., Cary, NC, USA).
3. Results
Subjects included in our study were a subset of the SPHERE participants, whose characteristics have been detailed in previous works . (Bollati, 2014a, Bollati et al., 2014b) The study subjects were predominantly female (80% of the population), 122 were obese (66%) and 64 were overweight (34%), with a mean age of 51 years.
51% of the subjects had never smoked, 30% were former smokers and approximately 16% were current smokers; mean value of BMI was 33.4±6 kg/cm2 and of total cholesterol was 217.8±41.8 mg/dl (Table 2).
Table 2.
Baseline subjects characteristics.
| Characteristics of subjects | N | |
|---|---|---|
| Age, Years | 186 | 50.7 ± 11.7 |
| Sex, n(%) | ||
| Male | 37 (20.0%) | |
| Female | 149 (80.0%) | |
| BMI, kg/cm2 | 186 | 33.4 ± 6.0 |
| Overweight (25≤BMI<30) ) | 64 (34.4) | |
| Class I obesity (30≤BMI<35) | 60 (32.3) | |
| Class II-III obesity (BMI≥35) | 62 (33.3.) | |
| Waist circumference, cm | 183 | 99.9 ± 13.3 |
| Smoking, n(%) | ||
| Never smoker | 95 (51.1%) | |
| Ex-smoker | 56 (30.1%) | |
| Actual smoker | 29 (15.6%) | |
| Missing | 6 (3.2%) | |
| Percentage of neutrophils | 186 | 58.8 ± 7.9 |
| Total Cholesterol, mg/dl | 183 | 217.8 ± 41.8 |
| Diabetes, n(%) | ||
| Yes | 18 (9.7%) | |
| No | 148 (79.6%) | |
| Missing | 20 (10.8%) | |
| Hypertension, n(%) | ||
| Yes | 53 (28.5%) | |
| No | 116 (62.4%) | |
| Missing | 17 (9.1%) | |
| Physical Activity Frequency, n(%) | ||
| Never | 98 (52.7%) | |
| < 2 times a week | 28 (15.1%) | |
| 2–4 times a week | 20 (10.8%) | |
| > 4 times a week | 22 (10.8.%) | |
| Missing | 18 (9.7%) |
Continuous variables are expressed as mean±SD, categorical variables are expressed as absolute numbers and frequencies.
Table 3 shows the mean daily levels of the individual PM10 values attributed to each study subject and evaluated within 1 day to 14 days before the subject recruitment: PM10 averaged from 48.0 (2 days before recruitment) to 54.8 ng/m3 (4 and 13 days before recruitment).
Table 3.
Distribution of the daily mean levels of ambient particulate matter ≤10 µm in aerodynamic diameter (PM10), within 1 day to 14 days before recruitment'.
| Days before recruitment | Mean (µg/m3) | SD |
Quartiles (µg/m3) |
|||
|---|---|---|---|---|---|---|
| 1th | 2nd | 3th | 4th | |||
| 1 day | 49.1 | 32.6 | ≤24 | 25–39 | 40–69 | >154 |
| 2 days | 48.0 | 31.6 | ≤26 | 27–42 | 43–64 | >163 |
| 3 days | 51.3 | 33.2 | ≤28 | 29–45 | 46–67 | >174 |
| 4 days | 54.8 | 34.3 | ≤28 | 29–48 | 49–70 | >174 |
| 5 days | 53.9 | 30.2 | ≤31 | 32–47 | 48–68 | >145 |
| 6 days | 53.3 | 29.1 | ≤33 | 34–46 | 47–70 | >157 |
| 7 days | 54.1 | 33.2 | ≤30 | 31–45 | 46–67 | >174 |
| 8 days | 51.5 | 33.6 | ≤27 | 28–42 | 43–68 | >159 |
| 9 days | 48.6 | 31.1 | ≤26 | 27–40 | 41–66 | >157 |
| 10 days | 49.9 | 32.6 | ≤27 | 28–42 | 43–64 | >174 |
| 11 days | 53.4 | 29.7 | ≤31 | 32–51 | 52–70 | >157 |
| 12 days | 54.6 | 30.5 | ≤32 | 33–48 | 49–70 | >164 |
| 13 days | 54.8 | 30.7 | ≤31 | 32–45 | 46–71 | >157 |
| 14 days | 52.4 | 34.6 | ≤26 | 27–40 | 41–72 | >157 |
Values in table are mean, standard deviation (SD) and 1th to 4th quartiles of daily means levels of the individual values attributed to each study subject.
The mean methylation levels (%5mC) of CD14, TLR4 and TNF-α genes are reported in Table 4, together with the %5mC values for the log-transformed genes and their corresponding Z-scores. Results of the models estimating the association of the PM10 exposure as a continuous measure with single methylation genes, grouped by inflammatory pathway, are shown in Table 5. From the evaluation of the linear association of the PM10 exposure with the single gene methylation, we found that CD14 was associated with PM10 exposure between 4 and 6 days before recruitment, (P4days=0.029; P5days=0.028; P6days=0.006). TLR4 was associated with the PM10 exposure between 6 and 8 days before recruitment (P6days=0.048; P7days=0.006; P7days=0.059), while no significant association was observed with TNF-α (Table 5).
Table 4.
Mean values of genes, log-transformed genes and their corresponding Z-scores grouped by common inflammatory pathway.
| Measures |
Methylation genes (%5mC) |
||
|---|---|---|---|
| CD14 | TLR4 | TNF-α | |
| Inflammatory pathway | |||
| Mean±SD | 3.82±1.98 | 4.58±1.43 | 11.41±5.36 |
| Mean±SD, log transformed | 1.26±0.39 | 1.47±0.32 | 2.32±0.51 |
| Mean Z-score±SD | −0.01±1.01 | 0.01±0.99 | −0.01±1.01 |
Table 5.
Association between PM10 exposures, singles methylation genes and genes grouped by common inflammatory pathway.
| Exposure to PM10 before recruitment | β-coefficienta (SE), p-values |
|||||||
|---|---|---|---|---|---|---|---|---|
| CD14 | TLR4 | TNF-α | Inflammatory pathway: CD14, TLR4, TNF-αb | |||||
| 1 day | −0.030 (0.056) | 0.591 | −0.156 (0.088) | 0.080 | 0.015 (0.035) | 0.673 | −0.002 (0.032) | 0.942 |
| 2 days | −0.061 (0.055) | 0.269 | 0.048 (0.083) | 0.570 | 0.002 (0.035) | 0.957 | 0.007 (0.032) | 0.833 |
| 3 days | −0.091 (0.054) | 0.093 | 0.041 (0.078) | 0.599 | 0.015 (0.034) | 0.670 | −0.006 (0.032) | 0.848 |
| 4 days | −0.120 (0.054) | 0.029 | −0.017 (0.085) | 0.839 | −0.034 (0.035) | 0.327 | −0.052 (0.032) | 0.109 |
| 5 days | −0.120 (0.054) | 0.028 | 0.011 (0.096) | 0.905 | −0.047 (0.035) | 0.180 | −0.047 (0.032) | 0.149 |
| 6 days | −0.155 (0.056) | 0.006 | −0.169 (0.084) | 0.048 | −0.011 (0.036) | 0.768 | −0.075 (0.032) | 0.019 |
| 7 days | −0.041 (0.055) | 0.453 | −0.260 (0.093) | 0.006 | −0.004 (0.036) | 0.918 | −0.035 (0.032) | 0.279 |
| 8 days | 0.047 (0.055) | 0.400 | −0.187 (0.098) | 0.059 | −0.005 (0.036) | 0.902 | 0.014 (0.033) | 0.677 |
| 9 days | 0.018 (0.054) | 0.741 | −0.060 (0.091) | 0.511 | 0.017 (0.036) | 0.640 | 0.018 (0.033) | 0.580 |
| 10 days | 0.051 (0.056) | 0.365 | 0.012 (0.088) | 0.890 | −0.011 (0.037) | 0.768 | 0.043 (0.034) | 0.204 |
| 11 days | 0.002 (0.056) | 0.972 | −0.020 (0.075) | 0.787 | 0.015 (0.035) | 0.671 | 0.011 (0.032) | 0.726 |
| 12 days | −0.032 (0.057) | 0.574 | −0.077 (0.079) | 0.332 | 0.019 (0.036) | 0.590 | −0.011 (0.033) | 0.726 |
| 13 days | −0.053 (0.059) | 0.367 | −0.003 (0.102) | 0.975 | 0.016 (0.037) | 0.666 | −0.005 (0.034) | 0.893 |
| 14 days | −0.083 (0.057) | 0.146 | −0.007 (0.108) | 0.952 | −0.014 (0.036) | 0.703 | −0.015 (0.034) | 0.657 |
Beta coefficients expressing the logarithmic change in DNA methylation associated with a standard deviation change in PM10 level. Models adjusted for: body mass index, age, sex, percentage of neutrophils, smoking habits, position, run, batch, interaction terms among position, run and batch.
Also adjusted for singles methylation genes.
When we jointly analyzed the three genes as a whole pathway, we found a significant association between the daily mean PM10 concentration measured six days before recruitment (P=0.019) and the inflammatory pathway.
In order to assess whether the BMI could influence the relationship between the PM10 and singles DNA methylation genes, we tested with a sensitivity analysis the interaction between the BMI and the PM10 exposure. We noticed that the BMI levels seem not to affect differentially the relationship between PM10 and each single methylation gene level but they have differential effects on the inflammatory gene pathway. In the analysis stratified according to three levels of BMI (25≤BMI<30; 30≤BMI<35; BMI≥35) the effect of PM10 on reducing the methylation of the inflammatory pathway was confirmed in the subgroup of overweight patients (β25≤BMI<30=−0.191,P<0.001), but not in the subgroup of obese (β30≤BMI<35=0.012; P=0.856) and severely obese patients (βBMI≥35=−0.030; P=0.910).
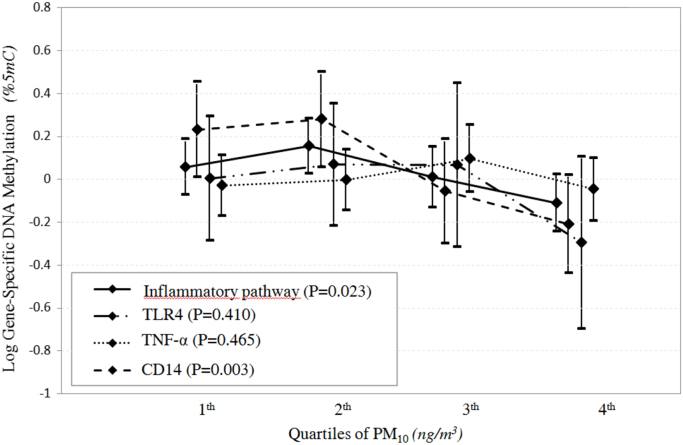
In the quartile model approach, focusing on the 6 days before recruitment , we observed differences in the levels of methylation genes across quartiles of exposure for the CD14 gene (P=0.003) and for the inflammatory pathway (P=0.023). (Table 6; Fig. 1).
Table 6.
Association of PM10 exposure, the sixth days before recruitment, with singles methylation genes/inflammatory system pathway, using a quartile model approach.
| Methylation geneb | β-coefficient (SE)a |
|||
|---|---|---|---|---|
| Quartile 2 | Quartile 3 | Quartile 4 | p-valuec | |
| Exposure to PM10 6 days before | ||||
| CD14 | 0.047 (0.145) | −0.288 (0.153) | −0.441 (0.152) | 0.003 |
| TLR4 | 0.065 (0.181) | 0.063 (0.222) | −0.300 (0.231) | 0.410 |
| TNF-α | 0.027 (0.094) | 0.128 (0.1) | −0.017 (0.099) | 0.465 |
| Inflammatory pathway: | 0.098 (0.084) | −0.048 (0.09) | −0.168 (0.089) | 0.023 |
| CD14, TLR4, TNF-αd | ||||
Beta coefficients expressing the logarithmic change in DNA methylation Z-scores associated with change in PM10 switching from the first quartile to the others.
Adjusted for: body mass index, age, sex, percentage of neutrophils, smoking habits, position, run, batch, interaction terms among positions, run and batch.
Also adjusted for single methylation genes.
The type 3 test of fixed effects, testing whether there is any difference among the 4 group. Quartile 1 is the referent group.
Fig. 1.
Quantile plot of estimated Z-score (±SD) of gene-specific DNA methylation and inflammatory pathway in relation to exposure to PM10 six days before withdrawal, depicts a significant association between PM10 quartiles and CD14 methylation gene (P=0.003) and inflammatory pathway (P=0.023) and a significant different slopes of the among genes in relation to exposure to PM10 quartiles the sixth day before withdrawal(P<0.001).
4. Discussion
Our results describe an inverse association between measured level of daily PM10 exposure and DNA methylation of inflammatory genes, assessed in peripheral blood of overweight/obese subjects.
The effect was maximum for the exposure of 6 days before recruitment, and spanned from day 4 to day 8. In particular, the association between PM10 and CD14 methylation was observed in days 4–5-6, between PM10 and TLR4 methylation in days 6–7-8. TNF-α methylation was not associated to PM10.
Numerous recent studies about the pathogenesis of different types of diseases have reported associations between PM exposure and alterations in DNA methylation. Investigated outcomes include cardiovascular and respiratory disease, lung cancer, type 2 diabetes, and diseases of the central nervous system (Alzheimer’s and Parkinson’s diseases) (Ho et al., 2012). The pathogenic process of most diseases begins with the inflammatory cascade. In this study we analyzed candidate genes involved in the inflammatory pathway, regulated by DNA methylation and whose individual alterations were associated to PM exposure (Bind et al., 2012, Jiang et al., 2014, Lepeule et al., 2014).
The inflammation cytokine CD14 is mainly produced by monocytes (Li et al., 2015) and it performs as a component of the innate immune system (Ulevitch and Tobias, 1995). CD14 plays a key role in the process of inflammation as it is the receptor of lipopolysaccharide (LPS), a cytoderm ingredient of gram-negative bacteria, with which it could combine and lead to the cell activation and the release of the pro-inflammatory cytokines (Nabih et al., 2015). The expression levels of CD14 can be modulated by epigenetic factors, especially DNA methylation (Cardoso et al., 2014, Munthe-Kaas et al., 2010, Slaats et al., 2012).
TLR4, along with the CD14 receptor, is associated with the recognition of LPS and the activation of the immune response (Tukhvatulin et al., 2010). In a recent study Kim T.W. et al. suggest that the expression of TLR4 is suppressed due to epigenetic events such as methylation of DNA (Kim et al., 2015, Takahashi et al., 2009).
TNF-α is a highly pleiotropic cytokine (Bradley, 2008, Olmos and Llado, 2014) produced by macrophages/monocytes during acute inflammation (Wajant et al., 2003) playing a critical role in the pathogenesis of chronic inflammatory diseases along with cell proliferation, differentiation, necrosis and apoptosis (Liu, 2005). TNF-α promoter methylation has also been related to the adipogenesis process (Zhu et al., 2012a) and to age-related inflammatory diseases (Gowers et al., 2011).
Our findings, taken together, may reflect the biological mechanism underlying the beginning of the inflammatory cascade. CD14 could represent a key actor of the inflammatory pathway because it is essential in the activation of TLR4 which, in turn, triggers NF-кB, the central element in the PM-induced inflammatory pathway promoting the cytokines cascade (Dagher et al., 2007, Yun et al., 2015). Taking into account this biological consideration, we could explain the time-line of the observed associations: CD14 was associated with the level of PM10 recorded 4–6 days before the blood drawing, TLR4 was associated with PM10 level recorded 6–8 days before, whereas the genes grouped in the inflammatory pathway showed an association 6 days before.
Although in literature it is widely recognized that PM exposure is associated to an increase of gene expression of cytokines, such as TNF-α (Mostafavi et al., 2015, Wu et al., 2014, Zhang et al., 2015), in our study we didn’t find any association between TNF-α, methylation and PM10 exposure. It is plausible that, in the regulation of TNF-α expression, it may be involved either a pre-transcriptional regulation, such as other epigenetic markers (e.g. histone modifications), or post-transcriptional mechanisms (e.g. microRNAs). Future investigations regarding other possible regulation markers are needed in order to better understand biological events linking PM exposure to TNF-α gene expression.
Epidemiological studies have shown that obesity may confer greater susceptibility to the adverse effects of PM exposure, since adipocytes have increased particle accumulation (Bollati et al., 2014b; Dubowsky et al., 2006) A positive correlation between exhaled nitric oxide, a marker of pulmonary inflammation, and BMI has been shown in healthy adults (De Winter-de Groot et al., 2005) Moreover, a differential autonomic cardiac response (measured as heart rate variability) to metal particles has been observed between obese and non-obese individuals (Chen et al., 2007). Obesity represents a pro-inflammatory state (Jacobsen et al., 2016, Karkeni et al., 2016) which itself may increase susceptibility to air pollution by increasing the response to inflammatory stimuli.
Our study was based on a population of overweight/obese subjects recruited at the Center for Obesity and Work and it did not include any normal weight subjects. Notably, this population is characterized by a higher BMI than the Italian average one. Hence, the study group may be quite different from the general population, and these differences could inhibit the generalization of our findings. Although the prevalence of overweight/obese males is higher than the prevalence of overweight/obese females in the general population, our study group included a higher proportion of overweight/obese women. One possible explanation is that women are, in general, more concerned about their appearance than men, so they may be more likely to seek out personalized diet protocols and diet centers that provide periodic medical checks (Bollati et al., 2014a).
Because of the limited number of study subjects, it is possible that the associations observed were due to chance. However, we controlled for several potential confounders by fitting multivariable models that included several individual characteristics as independent variables, in order to provide a good setting for evaluating mechanistic questions and reduce bias and chance findings.
The methylation analyses were performed on white blood cell DNA. Blood leukocytes include several cell type subsets and data from previous DNA methylation studies have suggested that small differences in DNA methylation in blood may be localized in a cell subtype with a specific immune or inflammatory function (Koestler et al., 2013, Tarantini et al., 2013, Zhu et al., 2012b). In this work, further analysis showed that the addition in multivariable regression models of percent monocytes and lymphocytes did not modify the associations of DNA methylation of CD14, TLR4 and TNF-α. However, since we used unfractionated leukocytes, we cannot determine which leukocyte subtypes were sensitive to the effects of the exposure on DNA methylation. Since white blood cells regulate the systemic response to inflammation, which is the biological key mechanism in the relation we examined, the measurements of DNA methylation of CD14, TLR4 and TNF-α in white blood cells may be appropriate. We also noticed that the effects observed in blood leukocytes cannot be extended to other relevant cell types, such as endothelia and other vascular tissues. Further researches are needed to identify the effects of PM on specific isolated leukocyte subtypes.
Our statistical approach modelled the overall DNA methylation over multiple CpG sites measured in each of the three genes we evaluated. In addition, our data showed a high correlation among individual CpG sites of each gene. This approach is consistent with a recent study regarding the correlation of methylation states between adjacent CpG sites, which showed that the methylation status at adjacent CpG sites tends to be correlated and may act in concert to control gene expression (Bibikova et al., 2011, Huh et al., 2014).
It is difficult to draw conclusions with a limited set of gene targets per pathway. These targets and their relationships with PM should be pursued by including additional gene targets and posttranscriptional markers.
In conclusion, our findings confirm the relationship among PM exposure, obesity and transcriptional regulation of inflammatory gene pathway, even though it will be required to include a larger number of genes targets for inflammatory pathway, in future studies.
Human subjects
This research project was approved by the ‘Maggiore Policlinico Hospital, Mangiagalli and Regina Elena Foundation’ Institutional Review Board and the participants gave their informed consent.
Acknowledgments
This study was supported by the EU Programme “Ideas” (ERC-2011-StG 282413, to Valentina Bollati).
We wish to thank the Occupational Medicine Medical Residents for their help in examining and recruiting the study subjects. We are grateful to the nurses of the “Medicina del Lavoro” Unit, Fondazione IRCCS Ca’ Granda Ospedale Maggiore Policlinico, the nutritionist Daniela Sommaruga, Elisabetta Angiolino from Regional Environmental Protection Agency (ARPA), as well as the volunteers who participated in the study.
References
- Alexeeff S.E. Association between blood pressure and DNA methylation of retrotransposons and pro-inflammatory genes. Int. J. Epidemiol. 2013;42:270–280. doi: 10.1093/ije/dys220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baccarelli A. Cardiovascular epigenetics: basic concepts and results from animal and human studies. Circ. Cardiovasc. Genet. 2010;3:567–573. doi: 10.1161/CIRCGENETICS.110.958744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bayarsaihan D. Epigenetic mechanisms in inflammation. J. Dent. Res. 2011;90:9–17. doi: 10.1177/0022034510378683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bertazzi P.A. Does enhancement of oxidative stress markers mediate health effects of ambient air particles? Antioxid. Redox Signal. 2014;21:46–51. doi: 10.1089/ars.2013.5694. [DOI] [PubMed] [Google Scholar]
- Bes-Houtmann S. Presence of functional TLR2 and TLR4 on human adipocytes. Histochem. Cell Biol. 2007;127:131–137. doi: 10.1007/s00418-006-0230-1. [DOI] [PubMed] [Google Scholar]
- Bibikova M. High density DNA methylation array with single CpG site resolution. Genomics. 2011;98:288–295. doi: 10.1016/j.ygeno.2011.07.007. [DOI] [PubMed] [Google Scholar]
- Bind M.A. Air pollution and markers of coagulation, inflammation, and endothelial function: associations and epigene-environment interactions in an elderly cohort. Epidemiology. 2012;23:332–340. doi: 10.1097/EDE.0b013e31824523f0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bollati V. Changes in DNA methylation patterns in subjects exposed to low-dose benzene. Cancer Res. 2007;67:876–880. doi: 10.1158/0008-5472.CAN-06-2995. [DOI] [PubMed] [Google Scholar]
- Bollati V. Nutrients intake is associated with DNA methylation of candidate inflammatory genes in a population of obese subjects. Nutrients. 2014;6:4625–4639. doi: 10.3390/nu6104625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bollati V. Susceptibility to particle health effects, miRNA and exosomes: rationale and study protocol of the SPHERE study. BMC Public Health. 2014;14:1137. doi: 10.1186/1471-2458-14-1137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bollati V. Microvesicle-associated microRNA expression is altered upon particulate matter exposure in healthy workers and in A549 cells. J. Appl. Toxicol. 2015;35:59–67. doi: 10.1002/jat.2987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bradley J.R. TNF-mediated inflammatory disease. J. Pathol. 2008;214:149–160. doi: 10.1002/path.2287. [DOI] [PubMed] [Google Scholar]
- Brochu P. Physiological daily inhalation rates for health risk assessment in overweight/obese children, adults, and elderly. Risk Anal. 2014;34:567–582. doi: 10.1111/risa.12125. [DOI] [PubMed] [Google Scholar]
- Cardoso F.P. Methylation pattern of the CD14 and TLR2 genes in human dental pulp. J. Endod. 2014;40:384–386. doi: 10.1016/j.joen.2013.11.024. [DOI] [PubMed] [Google Scholar]
- Cattani G. Evaluation of the temporal variation of air quality in Rome, Italy, from 1999 to 2008. Ann. Ist. Super. Sanita. 2010;46:242–253. doi: 10.4415/ANN_10_03_04. [DOI] [PubMed] [Google Scholar]
- Chen J.C. Obesity is a modifier of autonomic cardiac responses to fine metal particulates. Environ. Health Perspect. 2007;115:1002–1006. doi: 10.1289/ehp.9609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dagher Z. Role of nuclear factor-kappa B activation in the adverse effects induced by air pollution particulate matter (PM2.5) in human epithelial lung cells (L132) in culture. J. Appl. Toxicol. 2007;27:284–290. doi: 10.1002/jat.1211. [DOI] [PubMed] [Google Scholar]
- Dai L. Associations of fine particulate matter species with mortality in the United States: a multicity time-series analysis. Environ. Health Perspect. 2014;122:837–842. doi: 10.1289/ehp.1307568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Loera-Rodriguez C.O. Over-expression of TLR4-CD14, pro-inflammatory cytokines, metabolic markers and NEFAs in obese non-diabetic Mexicans. J. Inflamm. 2014;11:39. doi: 10.1186/s12950-014-0039-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Pergola G., Silvestris F. Obesity as a major risk factor for cancer. J. Obes. 2013;2013:291546. doi: 10.1155/2013/291546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Devevre E.F. Profiling of the three circulating monocyte subpopulations in human obesity. J. Immunol. 2015;194:3917–3923. doi: 10.4049/jimmunol.1402655. [DOI] [PubMed] [Google Scholar]
- Dubowsky S.D. Diabetes, obesity, and hypertension may enhance associations between air pollution and markers of systemic inflammation. Environ. Health Perspect. 2006;114:992–998. doi: 10.1289/ehp.8469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gowers I.R. Age-related loss of CpG methylation in the tumour necrosis factor promoter. Cytokine. 2011;56:792–797. doi: 10.1016/j.cyto.2011.09.009. [DOI] [PubMed] [Google Scholar]
- Ho S.M. Environmental epigenetics and its implication on disease risk and health outcomes. ILAR J. 2012;53:289–305. doi: 10.1093/ilar.53.3-4.289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoareau L. Signaling pathways involved in LPS induced TNFalpha production in human adipocytes. J. Inflamm. 2010;7:1. doi: 10.1186/1476-9255-7-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang W. Air pollution and autonomic and vascular dysfunction in patients with cardiovascular disease: interactions of systemic inflammation, overweight, and gender. Am. J. Epidemiol. 2012;176:117–126. doi: 10.1093/aje/kwr511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huh I. Bis-class: a new classification tool of methylation status using bayes classifier and local methylation information. BMC Genom. 2014;15:608. doi: 10.1186/1471-2164-15-608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacobsen M.J. Altered methylation profile of lymphocytes is concordant with perturbation of lipids metabolism and inflammatory response in obesity. J. Diabetes Res. 2016;2016:8539057. doi: 10.1155/2016/8539057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang R. Short-term diesel exhaust inhalation in a controlled human crossover study is associated with changes in DNA methylation of circulating mononuclear cells in asthmatics. Part Fibre Toxicol. 2014;11:71. doi: 10.1186/s12989-014-0071-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jung CC S.H., Liang H.H. Association between indoor air pollutant exposure and blood pressure and heart rate in subjects according to body mass index. Sci. Total Environ. 2016;539:271–276. doi: 10.1016/j.scitotenv.2015.08.158. [DOI] [PubMed] [Google Scholar]
- Karkeni E. Obesity-associated inflammation induces microRNA-155 expression in adipocytes and adipose tissue: outcome on adipocyte function. J. Clin. Endocrinol. Metab. 2016:jc20153410. doi: 10.1210/jc.2015-3410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keaney J.F., Jr. Obesity and systemic oxidative stress: clinical correlates of oxidative stress in the Framingham Study. Arterioscler. Thromb. Vasc. Biol. 2003;23:434–439. doi: 10.1161/01.ATV.0000058402.34138.11. [DOI] [PubMed] [Google Scholar]
- Kim T.W. Epigenetic modification of TLR4 promotes activation of NF-kappaB by regulating methyl-CpG-binding domain protein 2 and Sp1 in gastric cancer. Oncotarget. 2015 doi: 10.18632/oncotarget.6549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koestler D.C. Blood-based profiles of DNA methylation predict the underlying distribution of cell types: a validation analysis. Epigenetics. 2013;8:816–826. doi: 10.4161/epi.25430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lepeule J. Epigenetic influences on associations between air pollutants and lung function in elderly men: the normative aging study. Environ. Health Perspect. 2014;122:566–572. doi: 10.1289/ehp.1206458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y.Y. CD14 gene-159C/T polymorphism and coronary artery disease: a meta-analysis involving 4467 subjects. Int. J. Clin. Exp. Med. 2015;8:12149–12160. [PMC free article] [PubMed] [Google Scholar]
- Liu Z.G. Molecular mechanism of TNF signaling and beyond. Cell Res. 2005;15:24–27. doi: 10.1038/sj.cr.7290259. [DOI] [PubMed] [Google Scholar]
- Matsuda M., Shimomura I. Roles of adiponectin and oxidative stress in obesity-associated metabolic and cardiovascular diseases. Rev. Endocr. Metab. Disord. 2014;15:1–10. doi: 10.1007/s11154-013-9271-7. [DOI] [PubMed] [Google Scholar]
- Mostafavi N. Inflammatory markers in relation to long-term air pollution. Environ. Int. 2015;81:1–7. doi: 10.1016/j.envint.2015.04.003. [DOI] [PubMed] [Google Scholar]
- Munthe-Kaas M.C. CD14 polymorphisms and serum CD14 levels through childhood: a role for gene methylation? J. Allergy Clin. Immunol. 2010;125:1361–1368. doi: 10.1016/j.jaci.2010.02.010. [DOI] [PubMed] [Google Scholar]
- Nabih E.S. Association between CD14 polymorphism (-1145G/A) and childhood bronchial asthma. Biochem. Genet. 2015 doi: 10.1007/s10528-015-9699-4. [DOI] [PubMed] [Google Scholar]
- Nasser Z. Outdoor particulate matter (PM) and associated cardiovascular diseases in the Middle East. Int. J. Occup. Med Environ. Health. 2015;28:641–661. doi: 10.13075/ijomeh.1896.00186. [DOI] [PubMed] [Google Scholar]
- Olmos G., Llado J. Tumor necrosis factor alpha: a link between neuroinflammation and excitotoxicity. Mediat. Inflamm. 2014;2014:861231. doi: 10.1155/2014/861231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pope C.A., 3rd Relationships between fine particulate air pollution, cardiometabolic disorders, and cardiovascular mortality. Circ. Res. 2015;116:108–115. doi: 10.1161/CIRCRESAHA.116.305060. [DOI] [PubMed] [Google Scholar]
- Qin X.D. Gender-specific differences of interaction between obesity and air pollution on stroke and cardiovascular diseases in Chinese adults from a high pollution range area: a large population based cross sectional study. Sci. Total Environ. 2015;529:243–248. doi: 10.1016/j.scitotenv.2015.05.041. [DOI] [PubMed] [Google Scholar]
- Savini I. Obesity-associated oxidative stress: strategies finalized to improve redox state. Int J. Mol. Sci. 2013;14:10497–10538. doi: 10.3390/ijms140510497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slaats G.G. DNA methylation levels within the CD14 promoter region are lower in placentas of mothers living on a farm. Allergy. 2012;67:895–903. doi: 10.1111/j.1398-9995.2012.02831.x. [DOI] [PubMed] [Google Scholar]
- Sofer T. Exposure to airborne particulate matter is associated with methylation pattern in the asthma pathway. Epigenomics. 2013;5:147–154. doi: 10.2217/epi.13.16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi K. Epigenetic regulation of TLR4 gene expression in intestinal epithelial cells for the maintenance of intestinal homeostasis. J. Immunol. 2009;183:6522–6529. doi: 10.4049/jimmunol.0901271. [DOI] [PubMed] [Google Scholar]
- Talbott E.O. A case-crossover analysis of the impact of PM(2.5) on cardiovascular disease hospitalizations for selected CDC tracking states. Environ. Res. 2014;134:455–465. doi: 10.1016/j.envres.2014.06.018. [DOI] [PubMed] [Google Scholar]
- Tarantini L. Blood hypomethylation of inflammatory genes mediates the effects of metal-rich airborne pollutants on blood coagulation. Occup. Environ. Med. 2013;70:418–425. doi: 10.1136/oemed-2012-101079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tukhvatulin A.I. Toll-like receptors and their adapter molecules. Biochemistry. 2010;75:1098–1114. doi: 10.1134/s0006297910090038. [DOI] [PubMed] [Google Scholar]
- Ulevitch R.J., Tobias P.S. Receptor-dependent mechanisms of cell stimulation by bacterial endotoxin. Annu. Rev. Immunol. 1995;13:437–457. doi: 10.1146/annurev.iy.13.040195.002253. [DOI] [PubMed] [Google Scholar]
- Wajant H. Tumor necrosis factor signaling. Cell Death Differ. 2003;10:45–65. doi: 10.1038/sj.cdd.4401189. [DOI] [PubMed] [Google Scholar]
- Wang X. Obesity related methylation changes in DNA of peripheral blood leukocytes. BMC Med. 2010;8:87. doi: 10.1186/1741-7015-8-87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weichenthal S. Obesity and the cardiovascular health effects of fine particulate air pollution. Obesity. 2014;22:1580–1589. doi: 10.1002/oby.20748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weichenthal S.A. PM2.5, oxidant defence and cardiorespiratory health: a review. Environ. Health. 2013;12:40. doi: 10.1186/1476-069X-12-40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- WHO, 2013. World Health Organization Fact Sheet (Number 311) for Worldwide Prevalence of Obesity
- WHO W.H.O. Obesity: preventing and managing the global epidemic. Rep. a WHO Consult. World Health Organ Tech. Rep. Ser. 2000;894(i-xii):1–253. [PubMed] [Google Scholar]
- De Winter-de Groot K.M. Exhaled nitric oxide: the missing link between asthma and obesity? J. Allergy Clin. Immunol. 2005;115:419–420. doi: 10.1016/j.jaci.2004.11.025. [DOI] [PubMed] [Google Scholar]
- Wu W. Inflammatory response of monocytes to ambient particles varies by highway proximity. Am. J. Respir. Cell Mol. Biol. 2014;51:802–809. doi: 10.1165/rcmb.2013-0265OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yun Y. Synergistic effects of particulate matter (PM10) and SO2 on human non-small cell lung cancer A549 via ROS-mediated NF-kappaB activation. J. Environ. Sci. 2015;31:146–153. doi: 10.1016/j.jes.2014.09.041. [DOI] [PubMed] [Google Scholar]
- Zhang Y. Inflammatory response and endothelial dysfunction in the hearts of mice co-exposed to SO , NO , and PM. Environ. Toxicol. 2015 doi: 10.1002/tox.22200. [DOI] [PubMed] [Google Scholar]
- Zhu J.G. Differential DNA methylation status between human preadipocytes and mature adipocytes. Cell Biochem. Biophys. 2012;63:1–15. doi: 10.1007/s12013-012-9336-3. [DOI] [PubMed] [Google Scholar]
- Zhu Z.Z. Predictors of global methylation levels in blood DNA of healthy subjects: a combined analysis. Int. J. Epidemiol. 2012;41:126–139. doi: 10.1093/ije/dyq154. [DOI] [PMC free article] [PubMed] [Google Scholar]