Abstract
A procedure was developed to assess the biodegradation of micropollutants in cell-free lysates produced from activated sludge of a municipal wastewater treatment plant (WWTP). This proof-of-principle provides the basis for further investigations of micropollutant biodegradation via native enzymes in a solution of reduced complexity, facilitating downstream protein analysis. Differently produced lysates, containing a variety of native enzymes, showed significant enzymatic activities of acid phosphatase, β-galactosidase and β-glucuronidase in conventional colorimetric enzyme assays, whereas heat-deactivated controls did not. To determine the enzymatic activity towards micropollutants, 20 compounds were spiked to the cell-free lysates under aerobic conditions and were monitored via LC-ESI-MS/MS. The micropollutants were selected to span a wide range of different biodegradabilities in conventional activated sludge treatment via distinct primary degradation reactions. Of the 20 spiked micropollutants, 18 could be degraded by intact sludge under assay conditions, while six showed reproducible degradation in the lysates compared to the heat-deactivated negative controls: acetaminophen, N-acetyl-sulfamethoxazole (acetyl-SMX), atenolol, bezafibrate, erythromycin and 10,11-dihydro-10-hydroxycarbamazepine (10-OH-CBZ). The primary biotransformation of the first four compounds can be attributed to amide hydrolysis. However, the observed biotransformations in the lysates were differently influenced by experimental parameters such as sludge pre-treatment and the addition of ammonium sulfate or peptidase inhibitors, suggesting that different hydrolase enzymes were involved in the primary degradation, among them possibly peptidases. Furthermore, the transformation of 10-OH-CBZ to 9-CA-ADIN was caused by a biologically-mediated oxidation, which indicates that in addition to hydrolases further enzyme classes (probably oxidoreductases) are present in the native lysates. Although the full variety of indigenous enzymatic activity of the activated sludge source material could not be restored, experimental modifications, e.g. different lysate filtration, significantly enhanced specific enzyme activities (e.g. >96% removal of the antibiotic erythromycin). Therefore, the approach presented in this study provides the experimental basis for a further elucidation of the enzymatic processes underlying wastewater treatment on the level of native proteins.
Keywords: Activated sludge, Protein extraction, Native enzymes, Enzyme assay, Micropollutant biodegradation
Graphical abstract
Highlights
-
•
Production of cell-free lysates able to directly transform micropollutants.
-
•
96-well plate assay for the biodegradation of multiple micropollutants.
-
•
Insights into the variety of soluble enzymatic activities in environmental samples.
-
•
Foundation for the consecutive assessment of enzyme activity and identity.
1. Introduction
Biological wastewater treatment is targeted towards the removal of phosphorous, nitrogen and organic substances by the metabolic activity of a diverse activated sludge microbial community. More precisely, it is based on the versatile catalytic activity of the microbial enzymes. It is generally recognized that the metabolic potential of microorganisms and especially of bacteria is immense. Despite the manifold experimental evidence that microbial isolates are able to transform a large variety of xenobiotics (Abu Laban et al., 2009, Kolvenbach et al., 2014), analytical surveys have shown that many polar organic micropollutants such as pharmaceuticals, biocides and personal care products are not or incompletely removed in conventional wastewater treatment (Daughton and Ternes, 1999). As a consequence, micropollutants and their transformation products (TPs) are released into rivers and streams, from where some even pass into ground water and drinking water, potentially causing adverse effects on aquatic ecosystems and human health (Garcia-Rodriguez et al., 2014).
This raises the question whether it might be possible to achieve a more complete degradation of micropollutants in biological wastewater treatment and other engineered systems by exploiting the metabolic potential of complex microbial communities. Although it is known, that the expression and kinetics of the metabolic potential are strongly dependent on environmental conditions, there is still little information on the enzymatic processes underlying microbial micropollutant degradation and their dependency on technical and physicochemical parameters (e.g. sludge age, redox conditions, temperature).
Efforts to characterize the microbial community and its function on a molecular level have been hindered by the huge complexity of activated sludge and the dynamics of the wastewater treatment process (wastewater composition, seasonal changes, geographical differences, etc.). In the past decade, at least some of these methodological limitations were overcome through the advent of high-throughput sequencing techniques. As in many other research areas, the rapid increase in sequence information pushed forward techniques which facilitated the elucidation and systemic understanding of biodegradation pathways. These techniques were also applied to wastewater treatment to varying extents, ranging from qPCR (Helbling et al., 2012b) to FISH (Ettwig et al., 2009, Lolas et al., 2012), metagenomics (Martin et al., 2006) or metatranscriptomics (Helbling et al., 2012a), 16S-amplicon based taxonomic community profiling (Vanwonterghem et al., 2014) and metaproteomics (Collado et al., 2013, Hansen et al., 2014, Wilmes et al., 2008).
The abovementioned molecular techniques are state-of-the-art for the identification of candidate organisms, genes or gene products (transcripts, enzymes) likely to be involved in micropollutant biotransformation. The advantage of these molecular techniques is that they avoid culturing biases as they are directly applicable to complex environmental samples. Meta-omics methodologies furthermore aspire to characterize the entire pool of genes or gene products present in a community at a given time, to establish statistical associations with biotransformations. These meta-omics techniques, however, still depend on the results of biochemical experiments with extracted, native enzymes to establish causality to micropollutant biotransformation (Johnson et al., 2015).
Studies with native enzymes originating from specialized or pure cultures help to reliably assign function to unknown proteins, thereby improving database annotations (Mills et al., 2015) and mechanistic understanding of degradation processes (Prior et al., 2009, Xu et al., 2015). However, pure or enriched laboratory cultures cannot provide reliable information about the diversity and activity of the enzymes actually involved in micropollutant degradation under environmental conditions. Therefore, the exact identities, organismal origins and subcellular localizations of the involved candidate enzymes in situ are still largely unknown. Furthermore, the variety of enzymes (and organisms) and the metabolic or co-metabolic nature of the biotransformation of a certain micropollutant remain unclear (Fischer and Majewsky, 2014).
To unravel the complex biodegradation processes in environmental systems, it is therefore essential to complement meta-omic studies with experiments using native, i.e. active enzymes from environmental samples (Arnosti et al., 2014). Studies of native activated sludge enzymes so far mainly focused on enzymes which are thought to convert conventional wastewater substrates like organic and inorganic nitrogen and phosphorous compounds. These are predominantly hydrolytic enzymes such as peptidases (formerly known as proteases (Barrett et al., 2013)), phosphatases, glucosidases, and lipases, which are being monitored via robust colorimetric or fluorometric assays based on the turnover of commercially available substrate derivatives (Burgess and Pletschke, 2008a, Cortés-Lorenzo et al., 2012, Gómez-Silván et al., 2013). Also some non-hydrolytic enzymes, e.g. catechol dioxygenases have been assayed, relying on the use of proxy substrates (Grekova-Vasileva and Topalova, 2009, Khunjar et al., 2011).
Conventional assays rely on a comparable transformation of the target analytes and the structurally similar (but not identical) proxy substrates and are commonly used to measure individual enzyme activities in (pre-treated) intact sludge flocs (Burgess and Pletschke, 2008a, Gessesse et al., 2003, Yu et al., 2007). These heterogeneous samples, consisting of viable and dead cells, EPS and other components (Arnosti et al., 2014, Flemming and Wingender, 2010) are however dynamic micro-environments, which challenges the analysis of enzymatic transformation processes. Cell-free systems produced via cell disruption, centrifugation and filtration steps therefore might be more appropriate to study enzymatic variety and activity as they simplify the complex environment and eliminate dynamic cellular processes.
Studies with extracted native enzymes from activated sludge are rare and so far mainly focused on the extraction of specific enzymes from the sludge EPS, which were then monitored with selected reporter assays (Gessesse et al., 2003, Nabarlatz et al.,). This approach neglects the involvement of intracellular enzymes, which inarguably exhibit a high metabolic versatility for the degradation of micropollutants (Fischer and Majewsky, 2014).
The critical step for the establishment of cell-free systems is the transfer of (native) enzymes from the activated sludge matrix into a buffered, particle-free solution where they are tangible for downstream protein analytical techniques like (native) fractionation and purification, activity testing, enzymatic digestion/modification and identification techniques (e.g. proteomics). The extraction of soluble proteins from disrupted microbial cells naturally leads to a loss of functionality, since extraction efficiencies vary for different proteins, such as e.g. integral membrane enzymes, which cannot be co-extracted without greater experimental efforts (Speers and Wu, 2007).
Therefore, the aims of this study were to:
-
(i)
produce native cell-free lysates from activated sludge with measurable enzymatic activity towards micropollutants and without intentional bias towards intra- or extracellular enzymes, but focusing on soluble enzymes,
-
(ii)
develop an assay method for the degradation of micropollutants allowing for the simultaneous monitoring of a multitude of enzyme activities using the target compounds themselves (not substrate proxies),
-
(iii)
assess the variety of the observable enzymatic reactions for a set of diverse micropollutants in dependence of experimental parameters.
2. Material and methods
2.1. Chemicals
Detailed information on the chemicals, their CAS numbers, standards and solvents used for this study is provided in the supporting information (SI).
2.2. Experimental procedure and modifications
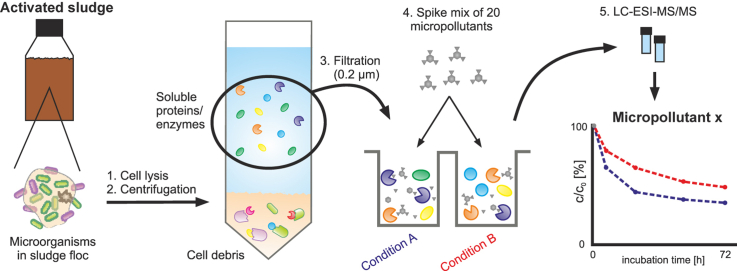
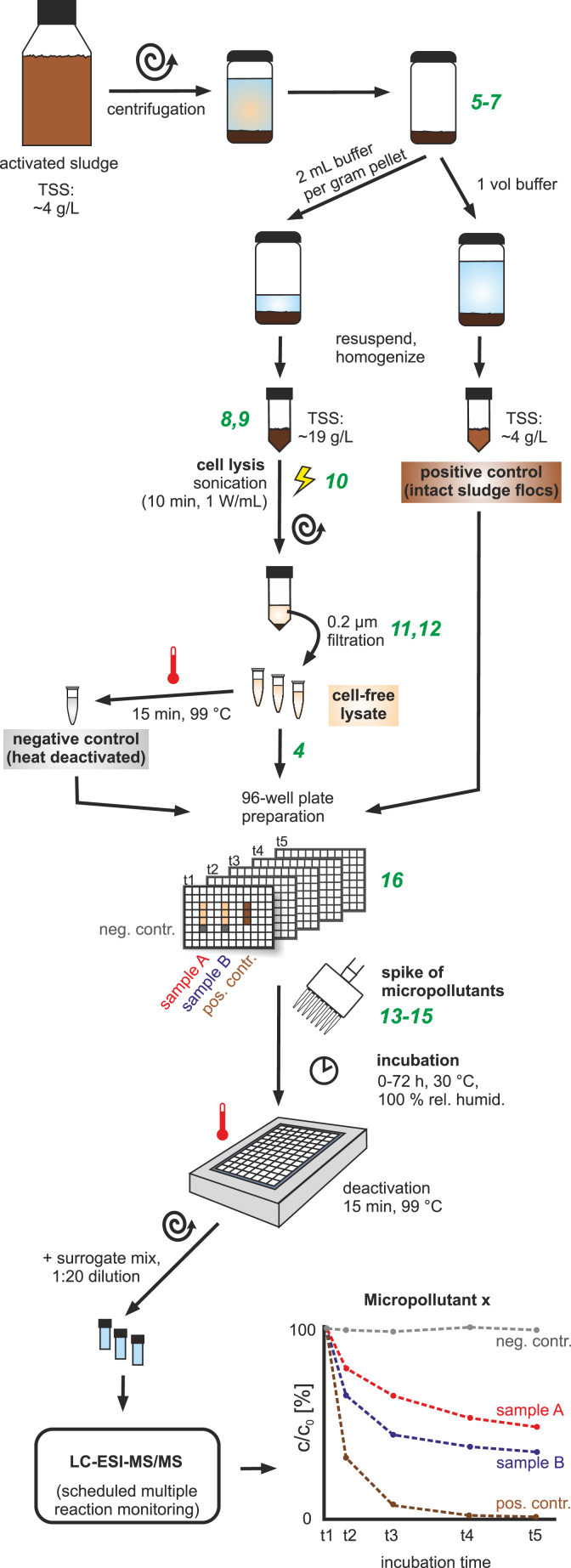
The tested experimental conditions are referred to by the experimental ID (1–16) throughout the document. The basic procedure is described below, modifications of the basic procedure are summarized in Table A.1. Fig. 1 illustrates the basic workflow and the location of the modifications therein. Detailed information on the modifications can be found in the SI.
Fig. 1.
Illustration of the basic workflow and location of the modifications therein. The locations of the modifications (4–16) in the basic workflow (described in sections 2.3, 2.4, 2.5) are marked in green and summarized in Table A.1. More detailed information is provided in the SI. (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
2.3. Sampling & processing of activated sludge
Activated sludge samples were collected from a German municipal wastewater treatment plant serving 320,000 population equivalents (Koblenz-Wallersheim, daily flow rate: 61,000 m3, hydraulic retention time: 12 h, sludge retention time: 12 d). The sludge (total suspended solids (TSS): 4 gTSS L−1, total organic carbon (TOC) per gram dry weight of sludge: 0.3 gTOC gdw sludge−1, pH 6.8) was transported to the laboratory within 30 min and stored at 4 °C or handled on ice from that point on. Cell lysis was carried out within 1 h of sampling and was preceded by a concentration step, in which sludge was centrifuged for 6 min at 3500 g in a swing-bucket rotor (Heraeus Multifuge 4 KR, rotor LH-4000). The supernatant was then discarded and the sludge pellets were washed and resuspended with HN-buffer (50 mmol L−1 HEPES, 50 mmol L−1 NaCl, pH 7.4). For usage in cell lysis the pellets were resuspended in 2 mL HN-buffer per gram pellet resulting in a TSS of ∼19 gTSS L−1. For usage as positive control in a target analyte assay the pellets were diluted to the original sludge TSS with HN-buffer. Afterwards, all sludge samples were mechanically homogenized for 2 min at 20,000 rpm (IKA Works Ultra-Turrax T8).
2.4. Production of a basic cell lysate
Using the reporter assays as indicators, a basic method for lysate production comprising only the most necessary processing steps was established (experiments 1–3, Fig. 1). This method was modified (Table A.1) to test the influence of single experimental parameters on enzymatic activities, which were assessed using the target analyte assay.
Ultrasonication (physical disruption) was applied as a standard method for the disruption of microbial cells from activated sludge. Cell lysis was performed with a sonication microtip (Bandelin MS73 connected to ultrasonic transducer HD3100) at 10 mm immersion depth in 50 mL tubes filled with 25 mL of concentrated sludge (∼19 gTSS L−1 in HN-buffer). Sonication was applied to the ice cooled samples with a power density of 1 W/mL for 10 min (net) in intervals of 15 s with 15 s breaks to avoid sample heating.
Cell debris was removed by centrifugation for 20 min at 14,000 rpm, 4 °C (Sigma 3K30, rotor 19776-H) and the supernatant, i.e. the crude cell lysate, was then filtered through 0.2 μm syringe filters (Whatman 25 mm GD/X polyethersulfone (PES) membrane with glass microfiber prefilter) to remove residual cells or cell debris. The resulting cell-free lysate (in HN-buffer) was kept on ice at all times and analyzed the same day.
2.5. Assays
All assay types (protein, reporter enzyme and target analyte degradation assay) were scaled down and optimized to fit 96-well microplates for a higher sample throughput, parallelization and a more economical use of samples and reagents. Microplates were measured spectrophotometrically in a multimode microplate reader (Tecan infinite 200 pro). For all enzyme assays heat-deactivated samples (15 min incubation at 100 °C) were run as negative controls. If not indicated otherwise, samples were measured in triplicate and controls in duplicate.
2.5.1. Protein concentration
Protein concentrations were measured using the bicinchoninic acid (BCA) method according to Smith et al. (1985). To limit the interference of co-extracted biomolecules like humic substances with protein quantification, cell lysates were diluted five-fold with water (5 μl cell lysate with 20 μl water) prior to measurements (Ras et al., 2008). After the addition of BCA solution (200 μl reagent, A:B ratio: 25:1 (BCA protein quantitation kit, Interchim Uptima)) assays were incubated at 37 °C for 30 min. Bovine serum albumin (BSA) was used as a protein standard and absorbance was measured at 562 nm.
2.5.2. Reporter enzyme activity
All of the applied enzyme activity assays utilize analogues of their natural substrates derivatized with p-nitrophenol (pNP) for spectrophotometric quantification. Solutions of pNP in the respective buffers served as standard. The assays (100 μl substrate solution with 20 μl of cell lysate) were incubated at 37 °C for 1 h and then stopped by addition of 0.5 mol L−1 NaOH (100 μl). Absorbance was measured at 405 nm for all reporter enzyme assays and all samples were analyzed in quadruplicate. The absorbance readings of the negative controls were deducted from the respective samples to yield the corrected activity. One unit (U) of enzyme activity was defined as the amount of enzyme in 1 mL solution forming 1 μg pNP per hour at 37 °C.
Phosphatase activity. For measurements of (acid) phosphatase activity the substrate solution contained 23 mmol L−1 p-nitrophenyl phosphate in 20 mmol L−1 bis-tris buffer (pH 6.0).
Galactosidase activity. For measurements of β-galactosidase activity the substrate solution contained 3 mmol L−1 p-nitrophenyl β-d-galactopyranoside in 50 mmol L−1 phosphate buffer (pH 7.4).
Glucuronidase activity. For measurements of β-glucuronidase activity the substrate solution contained 3 mmol L−1 p-nitrophenyl β-d-glucuronide in 50 mmol L−1 phosphate buffer (pH 7.4).
2.5.3. Target analyte degradation
Target Analytes. A set of 20 selected low sorbing (solid-water partitioning coefficients between 0.01 and 0.5 L gTSS−1 (Falås et al., 2016)) organic micropollutants which are present in conventional WWTPs were used as target analytes in the assays (see Table A.2). These micropollutants were selected to span a wide range of different biodegradabilities in conventional activated sludge systems, including e.g. N-acetyl-sulfamethoxazole (acetyl-SMX) and acetaminophen (>90%), atenolol, codeine, 10,11-dihydro-10-hydroxy-carbamazepine (10-OH-CBZ), iopromide, bezafibrate and diclofenac (30–90%) as well as fluconazole and carbamazepine (<30%) (Joss et al., 2006, Onesios et al., 2009). Since the antiepileptic drug carbamazepine is usually not significantly removed in municipal WWTPs, it served as an “internal negative control”.
Assay procedure. The assays were carried out in low protein binding 96-well plates (Greiner non-binding F-bottom plates) with each sample consisting of 100 μl cell lysate spiked with 20 μl analyte mix (0.1 ng μl−1 per analyte dissolved in water). The resulting spike concentration in the sample was 16.7 μg L−1 per analyte. To prevent changes in analyte concentration due to evaporation of the small sample volumes, the assays were conducted in a climate chamber operated at a temperature of 30 °C and a relative humidity of 100%. The plates were incubated for a duration of 3 days (72 h) with destructive sampling at 5 time points (<30 min, 4–8 h, 24 h, 48 h, 72 h) and gravimetrical control of sample volumes. Exemplary measurements showed that the oxygen saturation of the samples was above 80% during the incubation period. For the samples of each time point a separate 96-well plate was used to enable independent sample handling. Every tested experimental condition was represented through triplicate samples and one negative control (heat-deactivated lysate, incubation for 15 min at 99 °C, 300 rpm) at each time point (i.e. each plate) resulting in 15 samples plus 5 negative controls per condition and assay. Intact activated sludge flocs were used as a positive control (see section 2.3) and analyzed in triplicate for every time point to verify that degradation was still observable under assay conditions and to give a representation of the optimally expectable activities. During sample preparation the plates were kept on ice and after addition of cell lysate, negative and positive controls the analyte mix was added simultaneously to all samples (t0). The plates were then sealed with a breathable sealing film (breathe easy sealing-membrane, Sigma-Aldrich) which allows gas exchange (oxygen, carbon dioxide and water) and was moved to the climate chamber for incubation. At the specified time points deactivation of all samples in the respective plate was carried out simultaneously through incubation of the plate for 15 min at 99 °C at 300 rpm. Therefore, the breathable sealing film was replaced by a non-breathable, heat-stable sealing film (Rotilabo microtest plate sealing film, 50 μm, C. Roth) which was tightly adhered to the plate using a rubber roller. The first sampling (t1) was carried out 5–30 min after t0 and was later used to determine the initial analyte concentration in the respective series of measurements. After sample deactivation each plate was centrifuged for 7 min at 2500 g (Hettich Universal 320 R, Rotor 1460) to collect condensate and precipitate sludge flocs in the positive controls. Exact 100 μl of each sample were then transferred into 1.5 mL tubes and stored at 4 °C until further usage.
2.6. Sample preparation and analysis of target compounds
The deactivated samples were diluted with ultrapure water and a labelled surrogate mix was added (15 μl sample, 270 μl water, 15 μl surrogate mix) to a final concentration of 0.83 μg L−1 analyte mix and 0.2 μg L−1 surrogate mix. A sample aliquot of 80 μl was then analyzed via reversed-phase liquid chromatography (Agilent 1260 Series, Agilent Technologies) coupled to electrospray mass spectrometry (LC-ESI-MS/MS; AB SCIEX TripleQuad™ 5500 mass spectrometer, Applied Biosystems). For chromatographic separation a Zorbax Eclipse XDB-C18 column (2.1 × 150 mm, 3.5 μm) was used following a method described in detail in Rühmland et al. (2015). Human metabolites of carbamazepine and their TPs were analyzed using a method described by Kaiser et al. (2014).
2.7. Statistical analysis
Statistical analysis of TSS, protein yield and assay data was carried out using R 3.2.0. Significant differences between samples and negative controls as well as between basic lysates and different assay conditions were calculated by pairwise comparisons using the “pairwise.t.test” function with Bonferroni correction. For TSS, protein yield and the conventional reporter assays, comparisons are based on replicate values. To increase statistical sensitivity, pairwise comparisons for target analyte degradation assays are based on the degradation rate constants of first order kinetics (indicated as prc) rather than removal values. Degradation rate constants for target analyte assays were determined by regression analysis of log transformed data against time using the “lm” function. The range stated behind mean values (e.g. 59 ± 5%) indicates the standard deviation.
3. Results & discussion
3.1. Characterization of native cell lysates with conventional reporter assays
Four conventional colorimetric assays (see section 2.5) were used as proxies for protein concentration (BCA assay) and general enzymatic activity (acid phosphatase, β-galactosidase, β-glucuronidase), to optimize the lysate production procedure. With increasing duration and intensity of sonication the gained protein concentration increased, while the relative phosphatase activity (per protein) decreased (Figure SI 1). Therefore, as a compromise between maintaining enzymatic activity and maximizing protein yield, an intermediate sonication duration (10 min (net) in intervals of 15 s with 15 s breaks) and an intermediate sonication intensity (1 W/mL) were used to produce a basic cell lysate (see section 2.4). To assess the enzymatic activity of this basic cell lysate, three lysates produced from sludge sampled at different days were tested in separate assays (1–3, Table 1). Whereas technical replicates (intra-day) of all four conventional assays showed little variation (relative standard deviation (RSD) < 5%, also see Figure SI 2), biological replicates (inter-day) showed a higher variation of protein concentration (RSD = 13%), even though TSS values of the concentrated sludge used for cell lysis did not (RSD = 5%). This reflects the dynamic composition of activated sludge, even if originating from the same municipal WWTP. To consider the variations of the protein content during the optimization of the extraction procedure, the results of the reporter enzyme assays were normalized to the respective protein concentration. Details on the variations of TSS values and protein concentrations are being discussed in the SI.
Table 1.
Characterization of differently produced lysates with conventional reporter assays. Significant differences compared to the mean of the basic lysates 1–3 are indicated by stars (p = ns ≥ 0.05 ≥ * > 0.01 ≥ ** > 0.001 ≥ ***). TSS: total suspended solids of the concentrated sludge used for cell lysis (n = 3); Protein: protein concentrations of the lysates determined by the BCA assay (n = 3). All enzyme activities (n = 4) were corrected for their respective negative control and normalized with the respective protein concentration; PI: peptidase inhibitors; AS: ammonium sulfate; US: ultrasonication; PF: pressure filtration.
| ID | Category | Condition | Addition of PI | TSS |
Protein concentration |
Phosphatase activity |
Galactosidase activity |
Glucuronidase activity |
|---|---|---|---|---|---|---|---|---|
| [g/L] | [mg/mL] | [U/mg protein] | ||||||
| 1 | Basic lysate | Sludge A | 19.1 ± 0.6 | 1.06 ± 0.02 | 38.0 ± 1.0 | 12.9 ± 0.4 | 9.6 ± 0.2 | |
| 2 | Basic lysate | Sludge B | 18.0 ± 0.2 | 0.86 ± 0.03 | 40.0 ± 1.8 | 17.4 ± 0.8 | 9.2 ± 0.5 | |
| 3 | Basic lysate | Sludge C | 18.9 ± 0.3 | 1.05 ± 0.04 | 35.3 ± 1.5 | 15.7 ± 0.6 | 11.1 ± 0.6 | |
| Mean (basic lysates 1–3) | 18.7 ± 0.9 | 0.99 ± 0.13 | 37.8 ± 4.7 | 15.3 ± 2.8 | 10.0 ± 1.5 | |||
| 5 | Pre-treatment | +EDTA | x | 24.6 ± 0.5∗∗∗ | 1.29 ± 0.05∗∗∗ | 21.7 ± 0.9∗∗∗ | 6.0 ± 0.4∗∗∗ | 5.5 ± 0.3∗∗∗ |
| 6 | Pre-treatment | −EDTA | x | 28.7 ± 0.9∗∗∗ | 1.20 ± 0.03∗∗ | 25.4 ± 0.7∗∗∗ | 5.7 ± 0.2∗∗∗ | 4.7 ± 0.1∗∗∗ |
| 7 | Pre-treatment | +EDTA/US/PF | x | 26.3 ± 0.6∗∗∗ | 0.98 ± 0.05ns | 23.6 ± 1.2∗∗∗ | 16.2 ± 0.8ns | 15.4 ± 0.9∗∗∗ |
| 8 | Additives | PI | x | 19.1 ± 0.6ns | 0.83 ± 0.01ns | 35.2 ± 0.4∗ | 11.6 ± 0.3∗∗∗ | 7.5 ± 0.1∗∗∗ |
| 9 | Additives | AS | x | 19.1 ± 0.6ns | 1.12 ± 0.01ns | 44.1 ± 0.5∗∗∗ | 7.2 ± 0.2∗∗∗ | 5.9 ± 0.2∗∗∗ |
| 10 | Lysis method | Bead beating | 18.0 ± 0.2ns | 0.44 ± 0.04∗∗∗ | 43.5 ± 3.9∗∗∗ | 25.6 ± 2.3∗∗∗ | 11.3 ± 1.1∗∗ | |
| 11 | Filtration | No prefilter | 18.9 ± 0.3ns | 1.12 ± 0.04ns | 39.4 ± 1.4ns | 16.5 ± 0.6ns | 11.3 ± 0.6∗∗ | |
| 12 | Filtration | Unfiltered | 18.9 ± 0.3ns | 1.25 ± 0.11∗∗∗ | 39.5 ± 3.6ns | 16.3 ± 1.4ns | 11.3 ± 1.0∗∗ | |
All lysates showed significant enzymatic activity in the three reporter enzyme assays, whereas heat-deactivated negative controls did not. Glucuronidase and galactosidase activity largely showed similar trends whereas phosphatase activity showed opposing trends in some lysates. Furthermore, galactosidase and glucuronidase activity generally seemed to be more sensitive to experimental changes (four-fold change in activity) than phosphatase activity (two-fold change) (Table 1). One possible explanation for this varying effect might be that the involved enzymes differ in their (extra-)cellular localizations. In fact, the enzyme assays were initially chosen to distinguish between mostly intracellular (β-galactosidase (Agerkvist and Enfors, 1990) and β-glucuronidase (Bitton, 2005)) and extracellular activity (acid phosphatase (Jansson et al., 1988)) to observe the enrichment of cellular protein in preliminary studies. The hypothesis is further supported by the strong impact of pre-treatment procedures (5–7), which utilize different combinations of EDTA addition, ultrasonication and pressure filtration to deplete extracellular material (EPS), including exoenzymes. After pre-treatments 5 and 6 all three enzyme activities were reduced by 30–60% compared to the basic lysate (p < 0.001), while the third pre-treatment (7) led to a marked increase of glucuronidase activity from 10.0 ± 1.5 to 15.4 ± 0.9 U/mg protein (+54%, p < 0.001). This increase could mean, that in pre-treatment 7 a successful enrichment of cellular biomass was achieved through depletion of EPS in contrast to pre-treatments 5 and 6. While the definite prove of this hypothesis was beyond the scope of this study, the results show, that different sludge pre-treatments change the composition of the sludge and concomitantly the enzyme activity of the resulting lysates.
Interestingly, the lysate produced via bead beating (10), despite its significantly lower protein concentration, showed a relative enzymatic activity elevated by 13–67% in all assays compared to the basic lysate (p < 0.01). This could imply a better maintenance of enzymatic activity compared to the ultrasonication protocol.
In summary, none of the tested modifications led to significant improvements in all four categories, compared to the basic lysate, although some had pronounced effects on the lysates protein concentration and individual reporter enzyme activities. Therefore, it seems appropriate to avoid the application of strongly biasing procedures (e.g. sludge pre-treatments), if the aim is to produce lysates containing a native variety of active enzymes. Due to the inconsistency of the observed trends it also became apparent that the overall enzymatic variety of a lysate and its activity towards micropollutants is difficult to assess with a small range of proxy endpoints. As a consequence, an assay for the target analytes (i.e. micropollutants) themselves was developed to further investigate the enzymatic activity of cell-free lysates and the impact of experimental parameters.
3.2. Micropollutant degradation potential of intact activated sludge (i.e. positive control)
To determine the enzymatic activity towards micropollutants, 20 compounds were spiked to the cell-free lysates under aerobic conditions and were monitored via LC-ESI-MS/MS over 72 h. The micropollutants were selected to span a wide range of different biodegradabilities observed in conventional activated sludge treatment via distinct primary degradation reactions (see 2.5.3).
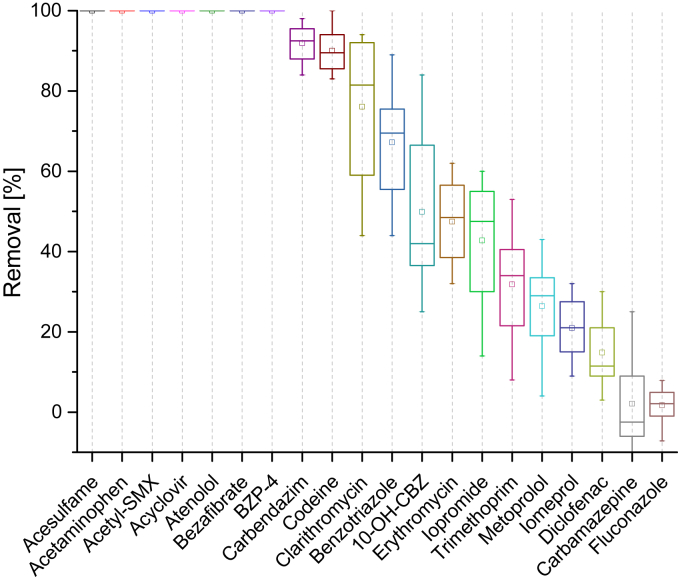
To determine the detectable enzymatic activity of the activated sludge towards micropollutants under assay conditions, washed and homogenized intact sludge flocs were used as a positive control in every assay (see section 2.3). Seven micropollutants (acesulfame, acetaminophen, acetyl-SMX, acyclovir, atenolol, bezafibrate, benzophenone-4 (BZP-4)) showed a complete primary degradation (below the limit of quantification (LOQ)) in the positive controls, while two (carbamazepine, fluconazole) were persistent and the remaining 11 compounds were removed to varying degrees (Fig. 2). These results are in accordance with the degradation potential of activated sludge from conventional WWTPs (Joss et al., 2006, Onesios et al., 2009). To confirm that the removal could be attributed to biological activity, heat-deactivated controls were run for several of these experiments (5, 6, 7, 8). These controls did not show a significant removal of any of these 20 compounds, except for clarithromycin and erythromycin. The removal of these two compounds in the positive controls and the respective negative controls therefore seems to be caused, at least partially, by abiotic effects. The absence of such effects in the cell-free lysates and their respective negative controls suggest sorption to the sludge flocs as mode of action.
Fig. 2.
Removal (%) of the 20 spiked compounds in positive controls of intact sludge flocs after 48 h of incubation. Each box represents triplicate samples from four separate positive controls (n = 12) run with sludge sampled on different days. The small square is the mean value, the solid line in the box is the median, the box edges are the 25th and 75th percentiles, and the whiskers are the fifth and 95th percentiles.
Altogether, the positive controls confirm that the activated sludge used for lysate production possesses the enzymatic machinery necessary to transform micropollutants under assay conditions.
3.3. The influence of experimental parameters on lysate activity towards micropollutants
To assess the potential of the basic lysate (without modifications) for micropollutant degradation identically produced lysates from activated sludge sampled on different days were tested in separate assays (1–3). Micropollutant degradation was confirmed by dissipation of the target compound and in some cases (10-OH-CBZ, acetyl-SMX, atenolol, erythromycin) additionally by the formation of TPs. The degradation of a compound was evaluated only, when its degradation rate constants (Table SI 3) in the active lysate significantly differed from those in the respective heat-deactivated negative control. For compounds which met these requirements, pairwise comparisons between experimental parameters were performed on the basis of degradation rate constants (significance indicated as prc, see section 2.7).
The removal percentages for seven selected compounds are summarized in Table 2. Out of 18 micropollutants which were degraded in the positive controls, five compounds (acetaminophen, acetyl-SMX, atenolol, bezafibrate, 10-OH-CBZ) showed significant degradation in the basic lysates (prc < 0.05).
Table 2.
Removal (in %) of seven selected micropollutants in cell-free lysates in dependence of experimental parameters. Only results ≥10% removal with significantly different removal rate constants from the negative control (prc < 0.05) are depicted. a substance was removed below its limit of quantification (see Table SI 1), b removal was <10%, slash (/) indicates that removal rate constants were not significantly different from negative control; n/a indicates that this substance was not spiked in the respective assay. US: ultrasonication, PF: pressure filtration, PI: peptidase inhibitors, AS: ammonium sulfate.
| ID | Category | Condition | Addition of PI | Removal [%] |
||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Acetaminophen | Acetyl-SMX | Atenolol | Bezafibrate | Codeine | 10-OH-CBZ | Erythromycin | ||||
| 1 | Basic lysate | Sludge A | >85a | 56 ± 4 | 24 ± 2 | 19 ± 4 | / | / | / | |
| 2 | Basic lysate | Sludge B | >85a | 64 ± 1 | 31 ± 3 | 24 ± 4 | / | 25 ± 4 | / | |
| 3 | Basic lysate | Sludge C | >85a | 56 ± 2 | 24 ± 5 | 18 ± 6 | / | / | / | |
| Mean (basic lysates 1–3) | >85a | 59 ± 5 | 26 ± 6 | 20 ± 8 | / | / | / | |||
| 4 | Basic lysate | Freeze/thaw | >85a | 59 ± 6 | 22 ± 1 | 21 ± 10 | / | / | / | |
| 5 | Pre-treatment | +EDTA | x | >85a | 74 ± 4 | 17 ± 8 | 10 ± 7 | / | / | / |
| 6 | Pre-treatment | −EDTA | x | >85a | 74 ± 4 | 18 ± 6 | b | / | / | / |
| 7 | Pre-treatment | +EDTA/US/PF | x | >85a | 74 ± 1 | / | / | / | / | / |
| 8 | Additives | PI | x | >85a | 34 ± 3 | / | / | / | / | / |
| 9 | Additives | AS | x | >85a | >94a | b | b | / | / | / |
| 10 | Lysis method | Bead beating | >85a | 46 ± 5 | 28 ± 4 | 34 ± 2 | b | / | / | |
| 11 | Filtration | No prefilter | >85a | 62 ± 4 | 42 ± 5 | 31 ± 6 | / | / | >96a | |
| 12 | Filtration | Unfiltered | >85a | 53 ± 8 | 54 ± 16 | 25 ± 14 | / | / | >96a | |
| 13 | Analyte mix | Reduced | >85a | n/a | 31 ± 8 | n/a | / | n/a | n/a | |
| 14 | Spike | Low | >85a | 67 ± 11 | 22 ± 6 | 23 ± 4 | / | / | / | |
| 15 | Spike | High | >85a | 58 ± 5 | 29 ± 4 | 28 ± 2 | / | / | / | |
| 16 | Cofactor | NADH | >85a | 37 ± 2 | b | b | / | / | / | |
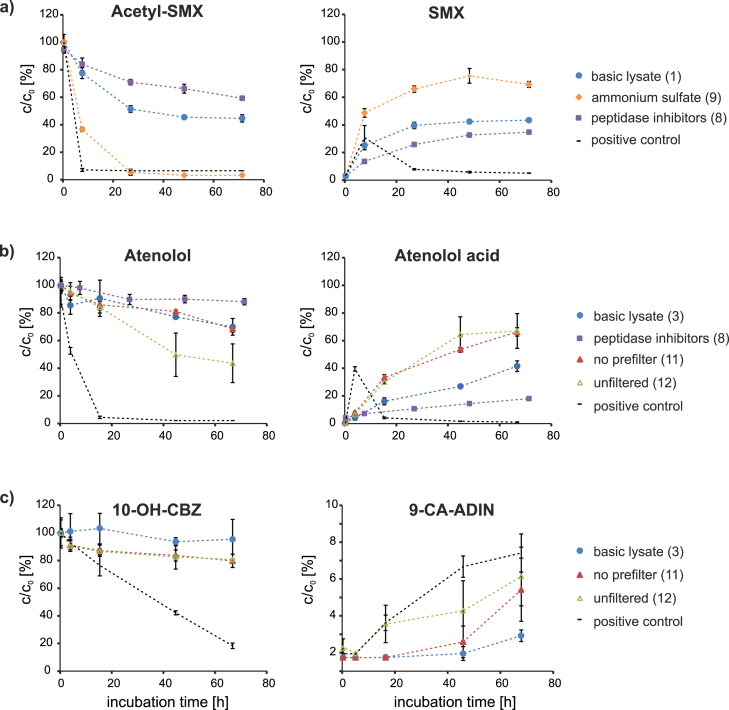
The antiphlogistic acetaminophen exhibited a complete primary degradation (<LOQ) within less than 4 h, not only in the basic lysate (1–3), but also for all tested modifications of experimental parameters (4–16). The human metabolite acetyl-SMX was also transformed in extracts of all experiments, but in contrast to acetaminophen its percentage of transformation varied for different modifications (34 – >94% removal) in comparison to the basic lysates (59 ± 5% removal). The original antibiotic sulfamethoxazole (SMX) was generated via amide hydrolysis in all experiments (as shown in Fig. 3 for selected experiments). Furthermore, the beta-blocker atenolol (26 ± 6% removal) and the lipid regulator bezafibrate (20 ± 8% removal) were degraded to a lesser but significant extent in the basic lysates (prc < 0.05). The degradation of atenolol was accompanied by formation of atenolol acid (see Fig. 3), which is in accordance with literature reporting atenolol acid as the major primary biological transformation product of atenolol (Barbieri et al., 2012, Radjenović et al., 2008). The concomitant formation of atenolol acid strongly indicated the transformation of atenolol by biologically mediated amide hydrolysis. The transformation of 10-OH-CBZ was statistically not significant for all but one basic lysate (2). However, the formation of 9-carboxylic acid acridine (9-CA-ADIN), which has been identified as one of the major TPs of the main human metabolite 10-OH-CBZ (Kaiser et al., 2014), confirmed its degradation in the other basic lysates and under the conditions shown in Fig. 3.
Fig. 3.
Effect of selected experimental conditions on the degradation of three (a–c) parent compounds (left diagrams) and the formation of their primary TPs (right diagrams) in cell-free lysates. Ratios of molar concentrations for acetyl-SMX (a), atenolol (b) and 10-OH-CBZ (c) primary transformation from experiments in which all 20 precursor compounds were spiked at a concentration of 16.7 μg L−1. The numbers in brackets refer to the experimental ID in Table 2. Each time point represents individually, incubated triplicates, which were sampled destructively.
The reduced enzymatic variety of the basic lysates, compared to the activated sludge controls, raised the question, to which extent the range of activities could be improved by simple experimental measures. Therefore the basic workflow was modified at different steps (Fig. 1) to test their individual impact on enzymatic activity and variety. The following section provides a concise overview over the results, more detailed information is provided in the SI.
While the impact of some experimental modifications was negligible (freeze/thaw cycles, number and concentration of analytes), others significantly influenced the degradation of individual micropollutants. Different sludge pre-treatments (5–7) utilizing combinations of EDTA addition, ultrasonication and/or pressure filtration increased the degradation of acetyl-SMX in lysates 5–7, but impaired the removal of atenolol and bezafibrate at the same time. In all five experiments with added peptidase inhibitors (5–9), the removal of atenolol and bezafibrate was impaired in comparison to the basic lysate. The degradation of acetaminophen (>85%), however, was not affected in the presence of peptidase inhibitors, while acetyl-SMX showed varying removal from 34 to >94%. Even though the primary degradation reaction of all four compounds is an amide hydrolysis, their degradation seems to be affected differently by the added inhibitors (see section 3.4).
Ammonium sulfate addition (9) strongly enhanced the degradation of acetyl-SMX to >94% (compared to the basic lysates with 59 ± 5%), probably due to the stabilization of specific proteins through “salting-in” effects (Richard, 2009).
Cell disruption via bead beating (10) showed promising trends regarding the maintenance of enzymatic activity. Even though the amount of protein released through bead beating was significantly lower than for sonication (see section 3.1), the enzymatic activity of both lysates was comparable.
Modifications of the lysate filtration had the biggest impact, since they increased the rate and number of degraded compounds. While the basic lysates, which were filtered through a 0.2 μm PES syringe filter with integrated glass microfiber prefilter, exhibited no significant erythromycin removal, the compound was completely removed (>96%, prc < 0.001) in unfiltered lysates (12) and lysates prepared with 0.2 μm PES syringe filters without integrated prefilter (11). Abiotic effects could be excluded since the respective negative controls showed no significant removal. These findings strongly indicate that certain proteins (e.g. enzymes degrading erythromycin) are being retained in the glass microfiber prefilters which were integrated in the routinely used syringe filters. This is further supported by the increased formation of the TPs atenolol acid and 9-CA-ADIN in experiments without prefilter (11/12), as depicted in Fig. 3. While the unfiltered lysate (12) might contain residual cells or sludge flocs, lysate 11 and all other lysates should be cell-free after filtration through 0.2 μm PES membranes. According to the manufacturer the glass microfiber prefilters have larger pore sizes (ca. 0.7–10 μm) than the subjacent PES membranes (0.2 μm), therefore proteins are probably retained by specific adsorption to the borosilicate glass surface rather than by size exclusion. These results show that different filter materials (even low protein binding ones) can have a significant effect on specific enzyme activities and therefore need to be evaluated prior to bioassays and protein analysis.
To test if the addition of cofactors could improve the degradation of compounds, the common enzyme cofactor NADH was added (16) as last experimental modification. NADH as an unbound organic cofactor (i.e. coenzyme) was selected from the multitude of existing cofactors as it is utilized by many oxidoreductases which are known to participate in micropollutant degradation such as monooxygenases (e.g. cytochrome P450 oxidases), dioxygenases and dehydrogenases (Jindrova et al., 2002, Pieper et al., 2004, Summers et al., 2011). Cofactor regenerating systems, which are frequently applied for the in situ deployment of nicotinamide cofactors (Kenneke et al., 2008, Weckbecker et al., 2010) were ruled out, as their highly active recombinant enzymes might exhibit activity towards micropollutants and cover the effects of endogenous activated sludge enzymes. However, the addition of NADH did not improve the degradation of analytes. On the contrary, the degradation of atenolol and bezafibrate were negligible in the presence of NADH. One reason for this might be a competitive inhibition of the degrading enzyme(s) through NADH, since the coenzyme also contains an amide function and therefore could serve as a substrate.
The list of the tested experimental parameters makes no claim to be complete and further parameters, e.g. the addition of mild detergents for the solubilization of membrane proteins, are being tested in ongoing experiments. Furthermore, the impact of some parameters such as lysis conditions, choice of cofactors, and the impact of various peptidase inhibitors have to be evaluated more systematically. The results illustrate, however, that single parameters can have pronounced effects on individual activities, but none or even opposing effects on others.
Furthermore, it became apparent that none of the tested conditions could restore the full enzymatic activity of intact activated sludge in a cell-free lysate. From 18 compounds which were degraded by intact activated sludge (Fig. 2), 12 compounds (e.g. acesulfame, acyclovir, codeine, BZP-4) were not degraded in cell-free lysates under any condition. Potential reasons for this reduction of enzymatic variety, such as a lack of interaction partners or membrane proteins, are discussed in section 3.5. On the other hand these results also demonstrate that cell-free lysates with significant enzymatic activity towards micropollutants can be produced and assayed reproducibly, as shown for six micropollutants (acetaminophen, acetyl-SMX, atenolol, bezafibrate, 10-OH-CBZ and erythromycin).
3.4. Observed enzymatic reactions and potentially involved enzymes
The primary degradation of the four compounds which were significantly degraded in most experiments (acetaminophen, acetyl-SMX, atenolol and bezafibrate) can be attributed to amide hydrolysis. The marked differences in their degradation efficiency in dependence of experimental parameters, especially the addition of peptidase inhibitors, indicate the involvement of different hydrolytic enzymes in the primary degradation of the four compounds. Acetaminophen and acetyl-SMX, which were degraded fastest in all experiments, are both secondary amides and more specifically belong to the group of anilides. This is in accordance with studies from Helbling et al. (2010) where it was shown, that anilides react significantly faster than other amides in sludge-seeded bioreactors. According to the authors, this indicates the presence of aryl-acylamidases (EC 3.5.1.13) with strong specificity for mono- or unsubstituted anilide substructures.
The impaired degradation of atenolol and bezafibrate in the presence of peptidase inhibitors (5–9) furthermore indicates that also enzymes acting on peptide bonds (EC 3.4.-), i.e. peptidases, are involved in amide bond hydrolysis of non-protein compounds in activated sludge. The enzymes targeted by the inhibitors belong to the class of endopeptidases which is further divided on the basis of catalytic mechanisms. A cocktail of three peptidase inhibitors was used in these studies: E-64 inhibits cysteine peptidases (EC 3.4.22) mainly of the papain family (C1), AEBSF addresses many serine peptidases (EC 3.4.21) of the chymotrypsin family (S1) and pepstatin A is an inhibitor of aspartic peptidases (EC 3.4.23), especially the pepsin family (A1). Hence, the enzyme(s) participating in the degradation of atenolol and bezafibrate might belong to one or several of these families. Altogether the Merops peptidase database (accessed on 8.5.2015 (Rawlings et al., 2014)) lists 170 known peptidases which are inhibited – mostly irreversibly, but some also reversibly – by the three used inhibitors (E-64: 70, AEBSF: 74, pepstatin: 26). This list is not specific for microorganisms from WWTPs, but contains peptidases of various taxonomic origins (including mammalian) and demonstrates the high range of peptidases which are affected by the inhibitors. This implies that many more unknown enzymes might be affected, and thus it cannot be excluded that the applied inhibitors also affect other, non-endopeptidase enzymes in their catalytic activity. Nevertheless, the impact of peptidase inhibitors on the degradation of atenolol and bezafibrate provides information on the nature of the involved enzymes and is a promising starting point for future studies.
The degradation of erythromycin indicates the presence of another catalytic activity in experiments without prefilter (11/12), since the molecule does not contain amide functions. The major transformation product was identified as TP 734, resulting from a dehydration of the parent compound. Llorca et al. (2015) found this to be the main degradation product after enzymatic treatment of erythromycin with purified EreB esterase from E. coli. The proposed degradation pathway is an initial enzymatic hydrolysis of the lactone ring ester followed by a condensation reaction. This strongly indicates the presence of esterase(s) in the respective lysates and therefore another hydrolytic activity (hydrolases acting on ester bonds (EC 3.1.-)). Finally, the formation of 9-CA-ADIN shows that the lysate also possessed the enzymatic potential for other degradation reactions than amide hydrolysis. Kaiser et al. (2014) showed that the initial reaction in the transformation pathway of 10-OH-CBZ is a biologically-mediated oxidation of a secondary alcohol to a keto group leading to the formation of oxcarbazepine, which is then further transformed by several (abiotic) reactions to 9-CA-ADIN. The exact nature of the involved enzyme(s) is still unclear, but it is assumed that the oxidation is being catalyzed by oxidoreductases acting on the CH-OH group of donors (EC 1.1.-), similar to the degradation of e.g. codeine (Wick et al., 2011).
3.5. Enzymatic variety in cell-free lysates
Taking into account all enzymatic reactions observed so far (including the conventional reporter enzymes), it is striking that eight out of nine observed enzyme activities seem to belong to the catalytic class of hydrolases (EC 3.-). Specifically: phosphatases (EC 3.1.-), galactosidases (EC 3.2.1.23) and glucuronidases (EC 3.2.1.31) as well as putative peptidases (EC 3.4.-) and/or enzymes acting on amide bonds, other than peptide bonds (EC 3.5.-) responsible for acetaminophen, acetyl-SMX, atenolol and bezafibrate degradation. Furthermore, esterases (EC 3.1.-) seem to be responsible for the removal of erythromycin. Only in the degradation of 10-OH-CBZ a non-hydrolase enzyme is responsible for the initial oxidation reaction (presumably an oxidoreductase (EC 1.-)).
Other compounds, which showed reproducible primary degradation in the positive controls, did not show visible degradation in cell-free lysates under any assay conditions. Interestingly, their primary (aerobic) degradation reactions, if known from the literature, are not hydrolytic reactions: Codeine is being oxidized (through oxidoreductases acting on the CH-OH group of donors (EC 1.1.-) (Wick et al., 2011)), trimethoprim undergoes an aryl methyl ether cleavage (i.e. demethylation, probably catalyzed by monooxygenases (EC 1.14.-)), acyclovir is being oxidized (by oxidoreductases of EC 1.1.- and 1.2.- (Prasse et al., 2011)), BZP-4 undergoes redox reactions (Beel et al., 2013) and iodinated contrast media, i.e. iomeprol and iopromide, are being acetylated after an initial oxidation reaction (carboxy-lyases (EC 4.1.1.) (Kormos et al., 2010, Schulz et al., 2008)).
These results raise the question why the observable enzymatic variety of cell-free lysates is reduced compared to the activated sludge source material and biased towards hydrolases – even if they were not specifically targeted as in other surveys.
As mentioned previously, the lysis of microbial cells for the extraction of soluble proteins naturally leads to a loss of functionality. The disruption of cellular compartmentation results in a collapse of spatial protein organization and physicochemical gradients (e.g. proton-gradients), which are essential for the interaction and function of certain enzymes.
Additionally, membrane proteins will not be extracted with a protocol focused on soluble proteins due to their hydrophobic nature. Some membrane-anchored proteins might co-extract, but without the use of detergents for solubilization, integral membrane proteins are most likely not present in the cell-free lysates.
Furthermore, different enzyme classes, e.g. cytochrome P450s, depend on cofactors such as coenzymes, prosthetic groups or metal ions (Hannemann et al., 2007). These might be deprived during the extraction process which could render enzymes inactive, even if they are present in the cell-free lysates.
The predominiant activity of hydrolases in cell-free lysates could therefore result from their insensitivity towards the abovementioned biases.
Some of the hypotheses for the reduced enzymatic variety in cell-free lysates can be substantiated through a closer look at the biochemical properties of hydrolases.
Hydrolysis is a relatively simple enzymatic reaction, which can be catalyzed through the nucleophilic and acid-base properties of the amino acids functional groups alone, while many more sophisticated enzymatic reaction types depend on interaction partners and/or non-protein cofactors to aid in catalysis (Broderick, 2001). The Swiss-Prot database query (Fig. 4) provides a general impression of the cofactor utilization of prokaryotic enzymes and shows that it varies strongly between the different catalytic groups of enzymes. While 72% of cofactor dependent oxidoreductases need one or more coenzymes only 3% of hydrolases need more than essential metal ions for catalysis. Only ligases utilize similarly few coenzymes (9% of entries) as hydrolases, but most need energy equivalents in the form of nucleoside triphosphates (e.g. ATP, GTP) for their catalytic activity, which in this function are not considered cofactors but rather cosubstrates.
Fig. 4.
Coenzyme usage of prokaryotic proteins according to catalytic activity. The local database, compiled by a Swiss-Prot database query (accessed on 28 April 2015) of reviewed bacterial and archaeal proteins featuring entries in the section “cofactor” and “EC number”, was comprised of 75,671 proteins (Boeckmann et al., 2005). Proteins without entries in these sections were not considered, recognizing that the lack of an entry only states the absence of information regarding this point, not the independence of cofactors. Coenzyme(s): proteins with entries for one or more cofactors other than metal ions (can also contain metal ions). Metal ion(s) only: proteins with entries for one or more metal ions but no other cofactors (e.g. coenzymes).
Thus, the higher resilience of hydrolases might be due to their independency of cofactors and other interaction partners, while enzyme classes which strongly rely on the properties of cofactors to complement their catalytic chemistry – like oxidoreductases, transferases or lyases – might be inactivated in lysates due to their depletion. This is in agreement with the results from this study, as the transformations which are lost in the lysates compared to the positive controls are presumably catalyzed by such enzyme classes (EC 1, 2 and 4, see above).
Besides the higher resilience of hydrolases, their predominant activity might be caused by a higher abundance in activated sludge, as suggested by their important role in organic matter degradation (e.g. Burgess and Pletschke, 2008b, Nabarlatz et al., 2012, Zhang et al., 2015). Furthermore, a better extractability due to different subcellular localization (extracellular enzymes might extract more readily) compared to other enzyme classes is possible. The enormous diversity of enzymes and organisms in environmental samples however makes it difficult to pinpoint a single reason for the reduced enzymatic variety in cell-free lysates.
Nevertheless, this cell-free approach does provide some information about the physicochemical characteristics of the enzyme(s) involved in micropollutant degradation through the experimental modifications contributing or withdrawing their activity. This information can aid the subsequent characterization and isolation of specific enzymes and provide experimental insight in the characteristics of micropollutant degrading enzymes from environmental samples.
Therefore, experiments with the systematic addition of cofactors as well as for the solubilization of membrane proteins by detergents are currently conducted. A final assessment of the mentioned reasons for the reduced enzymatic variety of the lysates (and its bias towards hydrolases) is however difficult without information on the abundance of enzymes in the lysates, independently of their activity. Metaproteomic profiling of differently produced lysates is therefore envisaged for consecutive studies. This illustrates another benefit of the cell-free approach, which allows the direct subsequent use of lysates for other protein analytical techniques, thus comprising information on the presence and activity of enzymes.
4. Conclusions
-
•
This study demonstrates the production of cell-free lysates from activated sludge which can directly transform micropollutants. The reduced complexity of these lysates compared to activated sludge facilitates the application of downstream protein analytical techniques. This proof-of-principle provides the experimental basis for further investigations of the enzymatic processes underlying wastewater treatment (e.g. micropollutant biodegradation), including the prospective identification of molecular markers for treatment efficiency.
-
•
Out of 18 compounds which were degraded in the positive control (activated sludge), six showed degradation in the cell-free extracts. While it was possible to significantly enhance specific enzymatic activities (e.g. acetyl-SMX and erythromycin) by modifying experimental parameters, the indigenous enzymatic activity towards micropollutants could not fully be restored by any of the tested modifications. For future studies it therefore seems promising to focus optimization strategies on certain catalytic groups with common requirements (e.g. cofactors, hydrophobicity, etc.), rather than to device a generic method for all types of catalytic reactions.
-
•
Different modifications in the extraction procedure, in particular pre-treatments and buffer additives significantly enhanced the enzymatic activity of some enzymes, while impairing others, suggesting that such strongly biasing procedures should be avoided if the aim is to compare the indigenous activity and variety of enzymes from activated sludge.
-
•
As the degradation assays are not restricted to designated enzymatic reactions, the assessment of micropollutants with different primary degradation reactions gives an impression of the enzymatic variety of a cell-free lysate. However, the majority of observed enzyme activities (8 out of 9, taking into account the conventional assays) belong to the catalytic class of hydrolases, while in the degradation of 10-OH-CBZ a non-hydrolase enzyme is responsible for the initial oxidation reaction (presumably an oxidoreductase). Thus, despite not being specifically targeted as in other studies, the observable enzyme activities are biased towards hydrolases.
-
•
The primary transformation rates of four micropollutants with amide hydrolysis (acetaminophen, acetyl-SMX, atenolol, bezafibrate) as primary reaction responded differently to individual experimental parameters such as peptidase inhibitors. This strongly suggests the involvement of different enzymes in their primary degradation reactions, among them possibly peptidases, and demonstrates that molecules with the same functional group can be targeted by different enzymes. The future challenge is to identify the enzymes responsible for the detected activities. Potential approaches include enrichment experiments, possibly followed by native fractionation, target analyte degradation assays and metaproteomic profiling.
Acknowledgements
We thank Sandro Castronovo for providing support and the activated sludge samples and all ATHENE project partners for the valuable discussions. Financial support from the European Research Council (ERC) through the EU-Project ATHENE (Grant agreement 267897) is gratefully acknowledged.
Footnotes
Supplementary data related to this article can be found at http://dx.doi.org/10.1016/j.watres.2016.03.037.
Appendices.
Table A.1.
Modifications (4–16) to the basic workflow. The basic workflow is described in sections 2.3–2.6 and a detailed description of the modifications is provided in the SI. Locations of the modifications in the basic workflow are visualized in Fig. 1.
| ID | Category | Condition | Description |
|---|---|---|---|
| 4 | Basic lysate | Freeze/thaw |
|
| 5 | Pre-treatment | +EDTA |
|
| 6 | Pre-treatment | −EDTA |
|
| 7 | Pre-treatment | +EDTA/US/PF |
|
| 8 | Additives | PI |
|
| 9 | Additives | AS |
|
| 10 | Lysis method | Bead beating |
|
| 11 | Filtration | No prefilter |
|
| 12 | Filtration | Unfiltered |
|
| 13 | Analyte mix | Reduced |
|
| 14 | Spike | Low |
|
| 15 | Spike | High |
|
| 16 | Cofactor | NADH |
|
Table A.2.
Chemical structures of the spiked compounds and composition of the reduced spike in experiment 13.
| Compound | Spiked in 13 | Structure | Compound | Spiked in 13 | Structure |
|---|---|---|---|---|---|
| 10,11-Dihydro-10-hydroxy-carbamazepine (10-OH-CBZ) |  |
Carbendazim |  |
||
| Acesulfame | x |  |
Clarithromycin |  |
|
| Acetaminophen | x |  |
Codeine | x |  |
| Acetyl-sulfamethoxazole (acetyl-SMX) |  |
Diclofenac |  |
||
| Acyclovir |  |
Erythromycin |  |
||
| Atenolol | x |  |
Fluconazole |  |
|
| Benzotriazole |  |
Iomeprol |  |
||
| Bezafibrate |  |
Iopromide | x |  |
|
| Benzophenone-4 (BZP-4) |  |
Metoprolol | x |  |
|
| Carbamazepine (CBZ) | x |  |
Trimethoprim |  |
Appendix A. Supplementary data
The following is the supplementary data related to this article:
References
- Abu Laban N., Selesi D., Jobelius C., Meckenstock R.U. Anaerobic benzene degradation by Gram-positive sulfate-reducing bacteria. FEMS Microbiol. Ecol. 2009;68(3):300–311. doi: 10.1111/j.1574-6941.2009.00672.x. [DOI] [PubMed] [Google Scholar]
- Agerkvist I., Enfors S.-O. Characterization of E. coli cell disintegrates from a bead mill and high pressure homogenizers. Biotechnol. Bioeng. 1990;36(11):1083–1089. doi: 10.1002/bit.260361102. [DOI] [PubMed] [Google Scholar]
- Arnosti C., Bell C., Moorhead D.L., Sinsabaugh R.L., Steen A.D. Extracellular enzymes in terrestrial, freshwater, and marine environments: perspectives on system variability and common research needs. Biogeochemistry. 2014;117(1):5–21. [Google Scholar]
- Barbieri M., Licha T., Nödler K., Carrera J., Ayora C., Sanchez-Vila X. Fate of β-blockers in aquifer material under nitrate reducing conditions: batch experiments. Chemosphere. 2012;89(11):1272–1277. doi: 10.1016/j.chemosphere.2012.05.019. [DOI] [PubMed] [Google Scholar]
- Barrett A.J., Rawlings N.D., Salvesen G., Woessner J.F. Elsevier Science Bv; Amsterdam: 2013. Handbook of Proteolytic Enzymes Introduction. [Google Scholar]
- Beel R., Eversloh C.L., Ternes T.A. Biotransformation of the UV-filter sulisobenzone: challenges for the identification of transformation products. Environ. Sci. Technol. 2013;47(13):6819–6828. doi: 10.1021/es400451w. [DOI] [PubMed] [Google Scholar]
- Bitton G. John Wiley & Sons, Inc; 2005. Wastewater Microbiology; pp. 153–171. [Google Scholar]
- Boeckmann B., Blatter M.-C., Famiglietti L., Hinz U., Lane L., Roechert B., Bairoch A. Protein variety and functional diversity: Swiss-Prot annotation in its biological context. C. R. Biol. 2005;328(10-11):882–899. doi: 10.1016/j.crvi.2005.06.001. [DOI] [PubMed] [Google Scholar]
- Broderick J.B. John Wiley & Sons, Ltd; 2001. eLS. [Google Scholar]
- Burgess J., Pletschke B. Hydrolytic enzymes in sewage sludge treatment: a mini-review. Water SA. 2008;34(3):343–349. [Google Scholar]
- Burgess J.E., Pletschke B.I. Hydrolytic enzymes in sewage sludge treatment: a mini-review. Water SA. 2008;34:343–350. [Google Scholar]
- Collado N., Buttiglieri G., Kolvenbach B.A., Comas J., Corvini P.F.X., Rodríguez-Roda I. Exploring the potential of applying proteomics for tracking bisphenol A and nonylphenol degradation in activated sludge. Chemosphere. 2013;90(8):2309–2314. doi: 10.1016/j.chemosphere.2012.10.002. [DOI] [PubMed] [Google Scholar]
- Cortés-Lorenzo C., Rodríguez-Díaz M., López-Lopez C., Sánchez-Peinado M., Rodelas B., González-López J. Effect of salinity on enzymatic activities in a submerged fixed bed biofilm reactor for municipal sewage treatment. Bioresour. Technol. 2012;121(0):312–319. doi: 10.1016/j.biortech.2012.06.083. [DOI] [PubMed] [Google Scholar]
- Daughton C.G., Ternes T.A. Pharmaceuticals and personal care products in the environment: agents of subtle change? Environ. Health Perspect. 1999;107(6):907–938. doi: 10.1289/ehp.99107s6907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ettwig K.F., van Alen T., van de Pas-Schoonen K.T., Jetten M.S.M., Strous M. Enrichment and molecular detection of denitrifying methanotrophic bacteria of the NC10 phylum. Appl. Environ. Microbiol. 2009;75(11):3656–3662. doi: 10.1128/AEM.00067-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Falås P., Wick A., Castronovo S., Habermacher J., Ternes T.A., Joss A. Tracing the limits of organic micropollutant removal in biological wastewater treatment. Water Res. 2016;95:240–249. doi: 10.1016/j.watres.2016.03.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fischer K., Majewsky M. Cometabolic degradation of organic wastewater micropollutants by activated sludge and sludge-inherent microorganisms. Appl. Microbiol. Biotechnol. 2014;98(15):6583–6597. doi: 10.1007/s00253-014-5826-0. [DOI] [PubMed] [Google Scholar]
- Flemming H.-C., Wingender J. The biofilm matrix. Nat. Rev. Microbiol. 2010;8(9):623–633. doi: 10.1038/nrmicro2415. [DOI] [PubMed] [Google Scholar]
- Garcia-Rodriguez A., Matamoros V., Fontas C., Salvado V. The ability of biologically based wastewater treatment systems to remove emerging organic contaminants-a review. Environ. Sci. Pollut. Res. 2014;21(20):11708–11728. doi: 10.1007/s11356-013-2448-5. [DOI] [PubMed] [Google Scholar]
- Gessesse A., Dueholm T., Petersen S.B., Nielsen P.H. Lipase and protease extraction from activated sludge. Water Res. 2003;37(15):3652–3657. doi: 10.1016/S0043-1354(03)00241-0. [DOI] [PubMed] [Google Scholar]
- Gómez-Silván C., Arévalo J., Pérez J., González-López J., Rodelas B. Linking hydrolytic activities to variables influencing a submerged membrane bioreactor (MBR) treating urban wastewater under real operating conditions. Water Res. 2013;47(1):66–78. doi: 10.1016/j.watres.2012.09.032. [DOI] [PubMed] [Google Scholar]
- Grekova-Vasileva M., Topalova Y. Enzyme activities and shifts in microbial populations associated with activated sludge treatment of textile effluents. Biotechnol. Biotechnol. Equip. 2009;23(1):1136–1142. [Google Scholar]
- Hannemann F., Bichet A., Ewen K.M., Bernhardt R. Cytochrome P450 systems—biological variations of electron transport chains. Biochimica et Biophysica Acta - General Subjects. 2007;1770(3):330–344. doi: 10.1016/j.bbagen.2006.07.017. [DOI] [PubMed] [Google Scholar]
- Hansen S.H., Stensballe A., Nielsen P.H., Herbst F.A. Metaproteomics: evaluation of protein extraction from activated sludge. Proteomics. 2014;14(21–22):2535–2539. doi: 10.1002/pmic.201400167. [DOI] [PubMed] [Google Scholar]
- Helbling D., Ackermann M., Fenner K., Kohler H.-P., Johnson D. The activity level of a microbial community function can be predicted from its metatranscriptome. ISME J. 2012;6(4):902–904. doi: 10.1038/ismej.2011.158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Helbling D.E., Hollender J., Kohler H.-P.E., Fenner K. Structure-based interpretation of biotransformation pathways of amide-containing compounds in sludge-seeded bioreactors. Environ. Sci. Technol. 2010;44(17):6628–6635. doi: 10.1021/es101035b. [DOI] [PubMed] [Google Scholar]
- Helbling D.E., Johnson D.R., Honti M., Fenner K. Micropollutant biotransformation kinetics associate with WWTP process parameters and microbial community characteristics. Environ. Sci. Technol. 2012;46(19):10579–10588. doi: 10.1021/es3019012. [DOI] [PubMed] [Google Scholar]
- Jansson M., Olsson H., Pettersson K. Phosphatases; origin, characteristics and function in lakes. Hydrobiologia. 1988;170:157–175. [Google Scholar]
- Jindrova E., Chocova M., Demnerova K., Brenner V. Bacterial aerobic degradation of benzene, toluene, ethylbenzene and xylene. Folia Microbiol. 2002;47(2):83–93. doi: 10.1007/BF02817664. [DOI] [PubMed] [Google Scholar]
- Johnson D.R., Helbling D.E., Men Y., Fenner K. Can meta-omics help to establish causality between contaminant biotransformations and genes or gene products? Environ. Sci. Water Res. Technol. 2015;1(3):272–278. doi: 10.1039/C5EW00016E. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joss A., Zabczynski S., Göbel A., Hoffmann B., Löffler D., McArdell C.S., Ternes T.A., Thomsen A., Siegrist H. Biological degradation of pharmaceuticals in municipal wastewater treatment: proposing a classification scheme. Water Res. 2006;40(8):1686–1696. doi: 10.1016/j.watres.2006.02.014. [DOI] [PubMed] [Google Scholar]
- Kaiser E., Prasse C., Wagner M., Broder K., Ternes T.A. Transformation of oxcarbazepine and human metabolites of carbamazepine and oxcarbazepine in wastewater treatment and sand filters. Environ. Sci. Technol. 2014;48(17):10208–10216. doi: 10.1021/es5024493. [DOI] [PubMed] [Google Scholar]
- Kenneke J.F., Mazur C.S., Ritger S.E., Sack T.J. Mechanistic investigation of the noncytochrome P450-mediated metabolism of triadimefon to triadimenol in hepatic microsomes. Chem. Res. Toxicol. 2008;21(10):1997–2004. doi: 10.1021/tx800211t. [DOI] [PubMed] [Google Scholar]
- Khunjar W.O., Mackintosh S.A., Skotnicka-Pitak J., Baik S., Aga D.S., Love N.G. Elucidating the relative roles of ammonia oxidizing and heterotrophic bacteria during the biotransformation of 17 alpha-ethinylestradiol and trimethoprim. Environ. Sci. Technol. 2011;45(8):3605–3612. doi: 10.1021/es1037035. [DOI] [PubMed] [Google Scholar]
- Kolvenbach B.A., Helbling D.E., Kohler H.-P.E., Corvini P.F.X. Emerging chemicals and the evolution of biodegradation capacities and pathways in bacteria. Curr. Opin. Biotechnol. 2014;27:8–14. doi: 10.1016/j.copbio.2013.08.017. [DOI] [PubMed] [Google Scholar]
- Kormos J.L., Schulz M., Kohler H.-P.E., Ternes T.A. Biotransformation of selected iodinated X-ray contrast media and characterization of microbial transformation pathways. Environ. Sci. Technol. 2010;44(13):4998–5007. doi: 10.1021/es1007214. [DOI] [PubMed] [Google Scholar]
- Llorca M., Rodriguez-Mozaz S., Couillerot O., Panigoni K., de Gunzburg J., Bayer S., Czaja R., Barcelo D. Identification of new transformation products during enzymatic treatment of tetracycline and erythromycin antibiotics at laboratory scale by an on-line turbulent flow liquid-chromatography coupled to a high resolution mass spectrometer LTQ-Orbitrap. Chemosphere. 2015;119:90–98. doi: 10.1016/j.chemosphere.2014.05.072. [DOI] [PubMed] [Google Scholar]
- Lolas I.B., Chen X., Bester K., Nielsen J.L. Identification of triclosan-degrading bacteria using stable isotope probing, fluorescence in situ hybridization and microautoradiography. Microbiology-Sgm. 2012;158:2796–2804. doi: 10.1099/mic.0.061077-0. [DOI] [PubMed] [Google Scholar]
- Martin H.G., Ivanova N., Kunin V., Warnecke F., Barry K.W., McHardy A.C., Yeates C., He S., Salamov A.A., Szeto E., Dalin E., Putnam N.H., Shapiro H.J., Pangilinan J.L., Rigoutsos I., Kyrpides N.C., Blackall L.L., McMahon K.D., Hugenholtz P. Metagenomic analysis of two enhanced biological phosphorus removal (EBPR) sludge communities. Nat. Biotech. 2006;24(10):1263–1269. doi: 10.1038/nbt1247. [DOI] [PubMed] [Google Scholar]
- Mills C.L., Beuning P.J., Ondrechen M.J. Biochemical functional predictions for protein structures of unknown or uncertain function. Comput. Struct. Biotechnol. J. 2015;13:182–191. doi: 10.1016/j.csbj.2015.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nabarlatz, D., Stüber, F., Font, J., Fortuny, A., Fabregat, A. and Bengoa, C. Extraction and purification of hydrolytic enzymes from activated sludge. Resour. Conserv. Recy. (0). [DOI] [PubMed]
- Nabarlatz D., Stüber F., Font J., Fortuny A., Fabregat A., Bengoa C. Extraction and purification of hydrolytic enzymes from activated sludge. Resour. Conserv. Recy. 2012;59(0):9–13. [Google Scholar]
- Onesios K., Yu J., Bouwer E. Biodegradation and removal of pharmaceuticals and personal care products in treatment systems: a review. Biodegradation. 2009;20(4):441–466. doi: 10.1007/s10532-008-9237-8. [DOI] [PubMed] [Google Scholar]
- Pieper D.H., Martins dos Santos V.t.A.P., Golyshin P.N. Genomic and mechanistic insights into the biodegradation of organic pollutants. Curr. Opin. Biotechnol. 2004;15(3):215–224. doi: 10.1016/j.copbio.2004.03.008. [DOI] [PubMed] [Google Scholar]
- Prasse C., Wagner M., Schulz R., Ternes T.A. Biotransformation of the antiviral drugs acyclovir and penciclovir in activated sludge treatment. Environ. Sci. Technol. 2011;45(7):2761–2769. doi: 10.1021/es103732y. [DOI] [PubMed] [Google Scholar]
- Prior J.E., Shokati T., Christians U., Gill R.T. Identification and characterization of a bacterial cytochrome P450 for the metabolism of diclofenac. Appl. Microbiol. Biotechnol. 2009;85(3):625–633. doi: 10.1007/s00253-009-2135-0. [DOI] [PubMed] [Google Scholar]
- Radjenović J., Pérez S., Petrović M., Barceló D. Identification and structural characterization of biodegradation products of atenolol and glibenclamide by liquid chromatography coupled to hybrid quadrupole time-of-flight and quadrupole ion trap mass spectrometry. J. Chromatogr. A. 2008;1210(2):142–153. doi: 10.1016/j.chroma.2008.09.060. [DOI] [PubMed] [Google Scholar]
- Ras M., Girbal-Neuhauser E., Paul E., Spérandio M., Lefebvre D. Protein extraction from activated sludge: an analytical approach. Water Res. 2008;42(8–9):1867–1878. doi: 10.1016/j.watres.2007.11.011. [DOI] [PubMed] [Google Scholar]
- Rawlings N.D., Waller M., Barrett A.J., Bateman A. MEROPS: the database of proteolytic enzymes, their substrates and inhibitors. Nucleic Acids Res. 2014;42(D1):D503–D509. doi: 10.1093/nar/gkt953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richard R.B. In: Methods in Enzymology. Richard R.B., Murray P.D., editors. Academic Press; 2009. pp. 331–342. [Google Scholar]
- Rühmland S., Wick A., Ternes T.A., Barjenbruch M. Fate of pharmaceuticals in a subsurface flow constructed wetland and two ponds. Ecol. Eng. 2015;80:125–139. [Google Scholar]
- Schulz M., Löffler D., Wagner M., Ternes T.A. Transformation of the X-ray contrast medium iopromide in soil and biological wastewater treatment. Environ. Sci. Technol. 2008;42(19):7207–7217. doi: 10.1021/es800789r. [DOI] [PubMed] [Google Scholar]
- Smith P.K., Krohn R.I., Hermanson G.T., Mallia A.K., Gartner F.H., Provenzano M.D., Fujimoto E.K., Goeke N.M., Olson B.J., Klenk D.C. Measurement of protein using bicinchoninic acid. Anal. Biochem. 1985;150(1):76–85. doi: 10.1016/0003-2697(85)90442-7. [DOI] [PubMed] [Google Scholar]
- Speers A.E., Wu C.C. Proteomics of integral membrane proteins theory and application. Chem. Rev. 2007;107(8):3687–3714. doi: 10.1021/cr068286z. [DOI] [PubMed] [Google Scholar]
- Summers R.M., Louie T.M., Yu C.L., Subramanian M. Characterization of a broad-specificity non-haem iron N-demethylase from Pseudomonas putida CBB5 capable of utilizing several purine alkaloids as sole carbon and nitrogen source. Microbiology. 2011;157(2):583–592. doi: 10.1099/mic.0.043612-0. [DOI] [PubMed] [Google Scholar]
- Vanwonterghem I., Jensen P.D., Ho D.P., Batstone D.J., Tyson G.W. Linking microbial community structure, interactions and function in anaerobic digesters using new molecular techniques. Curr. Opin. Biotechnol. 2014;27(0):55–64. doi: 10.1016/j.copbio.2013.11.004. [DOI] [PubMed] [Google Scholar]
- Weckbecker A., Groeger H., Hummel W., Gröger H., Wittmann C., Krull R. Regeneration of nicotinamide coenzymes: principles and applications for the synthesis of chiral compounds. Biosyst. Eng. I. 2010 doi: 10.1007/10_2009_55. [DOI] [PubMed] [Google Scholar]
- Wick A., Wagner M., Ternes T.A. Elucidation of the transformation pathway of the opium alkaloid codeine in biological wastewater treatment. Environ. Sci. Technol. 2011;45(8):3374–3385. doi: 10.1021/es103489x. [DOI] [PubMed] [Google Scholar]
- Wilmes P., Andersson A.F., Lefsrud M.G., Wexler M., Shah M., Zhang B., Hettich R.L., Bond P.L., VerBerkmoes N.C., Banfield J.F. Community proteogenomics highlights microbial strain-variant protein expression within activated sludge performing enhanced biological phosphorus removal. ISME J. 2008;2(8):853–864. doi: 10.1038/ismej.2008.38. [DOI] [PubMed] [Google Scholar]
- Xu L.H., Ikeda H., Liu L., Arakawa T., Wakagi T., Shoun H., Fushinobu S. Structural basis for the 4′-hydroxylation of diclofenac by a microbial cytochrome P450 monooxygenase. Appl. Microbiol. Biotechnol. 2015;99(7):3081–3091. doi: 10.1007/s00253-014-6148-y. [DOI] [PubMed] [Google Scholar]
- Yu G.-H., He P.-J., Shao L.-M., Lee D.-J. Enzyme activities in activated sludge flocs. Appl. Microbiol. Biotechnol. 2007;77(3):605–612. doi: 10.1007/s00253-007-1204-5. [DOI] [PubMed] [Google Scholar]
- Zhang P., Shen Y., Guo J.-S., Li C., Wang H., Chen Y.-P., Yan P., Yang J.-X., Fang F. Extracellular protein analysis of activated sludge and their functions in wastewater treatment plant by shotgun proteomics. Sci. Rep. 2015;5:12041. doi: 10.1038/srep12041. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.